Abstract
Mutations in the epidermal growth factor receptor (EGFR) gene confer it with cancer driver gene functions in non‐small cell lung cancer (NSCLC). Epidermal growth factor receptor ‐tyrosine kinase inhibitors are effective agents against NSCLC with a mutated EGFR gene. Accordingly, many guidelines recommend the use of an EGFR mutation test in NSCLC. However, not all patients are tested in most countries where tissue samples are mainly used for the test. As of 2011, most of the patients with advanced NSCLC are tested in Japan, and the use of cytological samples has significantly contributed to this success. A portion of samples used to determine a definite diagnosis of NSCLC, either tissue samples or cytological samples, is ensured to contain cancer cells, and is then investigated by an EGFR mutation test that is applicable to both tissue samples and cytological samples. Cytological samples now account for one‐third of all the samples investigated. EGFR mutation is detected in cytological samples at a similar rate with tissue samples. The criterion ensuring an EGFR mutation test to have satisfactory sensitivity and specificity for use in both tissue and cytological samples is presented. Cytological samples are valuable clinical sources being collected less invasively than tissue samples, and should therefore be extensively used in EGFR mutation testing.
Cancer driver genes are mutated genes that confer a significant growth advantages on cells and play key roles in the cancer development.1, 2 Therapies targeting cancer driver genes have presented dramatic responses in many malignancies, including lung cancer,3, 4, 5, 6 leukemia,7 and melanoma.8 Information on cancer driver gene is indispensable for selecting an appropriate treatment for particular cancers.
Somatic mutations in the epidermal growth factor receptor (EGFR) gene are frequently observed in non‐small cell lung cancer (NSCLC).9, 10, 11 The mutated EGFR gene is a cancer driver gene and NSCLCs harboring it responds well to treatment with EGFR‐tyrosine kinase inhibitors (EGFR‐TKIs) such as gefitinib and erlotinib.3, 4, 5, 6 Many therapeutic guidelines recommend the use of EGFR‐TKIs for the treatment of NSCLC with mutated EGFR.12, 13, 14 Accordingly, an increasing number of patients with NSCLC have been tested for EGFR mutations. The procedures for testing have been discussed.15, 16, 17 However, a significant proportion of patients are still untested in many counties, simply because tissue samples are not available. In contrast, almost all patients have been tested in Japan, where either tissue samples or cytological samples are used for the mutation test. Cytological samples have advantages over tissue samples: the former is collected using less‐invasive procedures than the latter, while the former is suited to EGFR mutation test similarly to the latter. Here, we summarize the sampling and testing scheme enabling EGFR mutation test in cytological samples. The scheme may be useful worldwide and applicable to many solid tumors other than NSCLC.
Importance of cytological samples for EGFR mutation test in NSCLC
Figure 1 shows the sequence of events in NSCLC diagnosis and treatment in clinical practice. First, lung cancer is provisionally diagnosed by the imaging studies. Next, samples are collected from the legion suspicious of cancer. Pathologists examine the sample and determine a definite diagnosis. Treatment is started thereafter.
Figure 1.
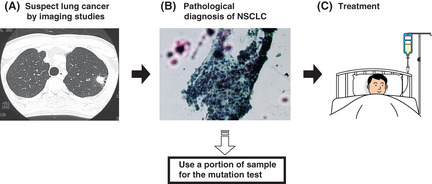
Flow chart showing the routine clinical practice for non‐small cell lung cancer (NSCLC) diagnosis and treatment. (A) Lung cancer is suspected by imaging studies. (B) A definite diagnosis of NSCLC is determined by pathological examination. Because a definite diagnosis is mandatory before initiating cancer treatment, all patients provide either tissue samples or cytological samples containing cancer cells. At this point, access to the cancer cells is available and we are able to perform the mutation test. (C) Treatment is initiated after a definite diagnosis is determined.
By dividing the samples submitted for pathological examination into aliquots [Fig. 1(B)], the mutation test can be performed for all patients without the need to collect additional samples. Moreover, information on the mutation status is readily applicable to the determination of the treatment regimens. Determination of EGFR mutation status at this timing is the most practical and useful.
Either a tissue sample or a cytological sample is submitted to the pathologists. Tissue samples include surgically resected samples and biopsy samples. Cytological samples include sputum, bronchoscopy samples (obtained by brushing or washing), pleural effusion, and samples obtained by fine needle aspiration. Tissue samples are collected from only a portion of patients, while cytological samples are collected from almost all patients. For example, a cytological sample (i.e. pleural effusion) is easily aspirated from patients with malignant pleural effusion, while a tissue sample is very difficult to obtain from such patients. Moreover, the invasive procedures required to collect tissue samples are often contraindicated in patients with a poor performance status.18 EGFR mutation tests that are applicable only to tissue samples exclude the patients described above and thus are unacceptable.
Contamination of the normal cells
Collecting samples that solely contain cancer cells is almost impossible. Stromal cells and blood cells are normal cells that inevitably contaminate cancer samples. Normal EGFR gene sequence in the genomic DNA derived from normal cells obscures the somatic mutations carried in cancer cells. Figure 2 shows the percentage of the cancer cells to the total number of cells in clinical samples.19 Tissue samples contained many normal cells, and cytological samples contain more of these cells. Empirically, the lowest percentage of cancer cells in pathologically cancer‐positive samples is 1%. Samples with a percentage of <1% may also exist. However, Figure 2 suggests that pathologists hesitate to determine a definite diagnosis using such samples and thus request re‐sampling. Therefore, 1% is a good estimate of the detection limit of pathological examination, and thus is a detection limit obligatory for an EGFR mutation test to be applicable to all pathologically cancer‐positive samples. This is the theoretical consensus in our country and constitutes qualification criterion for EGFR mutation tests.19, 20
Figure 2.
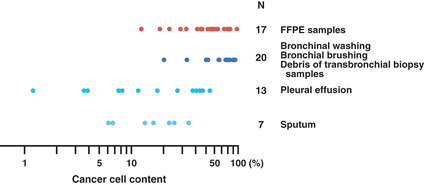
Ratios of cancer cells to normal cells in pathologically cancer‐positive samples. The ratios of the number of cancer cells to the total number of cells in a variety of samples are shown (modified from Tanaka et al.19) Archival slides that had enabled a definite cancer diagnosis were randomly chosen, and the numbers of cancer cells and normal cells were counted. Tissue samples (i.e. formalin‐fixed, paraffin‐embedded [FFPE] samples) are indicated by a warm color and cytological samples (i.e. the others) are indicated by cold colors.
Procedure ensuring the presence of cancer cells
Figure 3 illustrates sample submission procedures. The presence of cancer cells should be confirmed before performing EGFR mutation test, otherwise false‐negative results are obtained. For tissue samples (Fig. 3A), serial thin sections are made: the presence of cancer cells is confirmed in one section, and the test is performed with the other sections. For cytological samples (Fig. 3B), the cells are suspended and mixed well in a saline buffer. The suspension is then divided into two aliquots. The presence of cancer cells is confirmed in one aliquot, and the other is kept frozen or stored in a DNA‐isolation solution (e.g. AL buffer; Qiagen, Hilden, Germany) until the pathological examination is complete. Tissue samples may be treated in the same manner as cytological samples (Fig. 3C). In the last procedure, formalin fixation, which fragments DNA into small pieces, is avoided, as well as quick penetration of the DNA‐isolation solution into the cells is enabled. Tissues processed as shown in Figure 3C thus yields more definitive results in the test than those treated as shown in Figure 3A.
Figure 3.
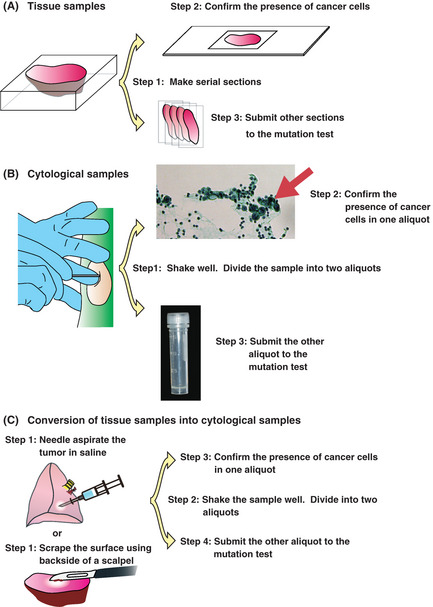
Sample preparation procedures. (A) Tissue samples. Step 1: Serial sectioning. Step 2: The presence of cancer cells is confirmed in 1 section. Step 3: The EGFR mutation is investigated using other sections. Macro‐dissection may be required to remove normal tissue before step 1. (B) Cytological samples. Step 1: Suspend the cells in saline. Divide the samples into two aliquots. Step 2: Confirm the presence of cancer cells in one aliquot. Step 3: Investigate the EGFR mutation using the other aliquot. (C) Preparation of cytological samples from tissue. Step 1: Scrape the surface of the tissue. Suspend the cells in saline. Step 2: Divide the samples into two aliquots. Step 3: Confirm the presence of cancer cells in one aliquot. Step 4: Investigate the EGFR mutation using the other aliquot.
The procedure shown in Figure 3B,C also applies when cytological samples are subjected to clinical tests based on reverse transcriptase‐PCR (RT‐PCR) reaction, for example, detection of the fusion genes such as EML4‐ALK.21 In such case, the following modifications should be made: cells in the aliquot for the mutation test should be collected by centrifugation (1300g, 5 min) at the earliest convenience after the sample collection (e.g. 20 min) and stored in a RNA protect reagent (e.g. RNAprotect Cell Reagent; Qiagen). Many RNA protect reagents allow us to isolate both DNA and RNA, and thus to perform both PCR‐ and RT‐PCR‐based investigations.
Recently, liquid‐based cytology is often used for the diagnosis of NSCLC. It has been reported that EGFR mutation test is reliably performed for liquid‐based cytology samples when combined with high sensitivity detection methods.22 The procedure shown in Figure 3B,C is applicable to liquid‐based cytology samples, and should be strictly observed.
EGFR mutation test statistics in Japan
Figure 4A shows the cumulative number of EGFR mutation tests performed in the three major, commercial Japanese laboratories. An additional 2000 or more samples are tested in university or hospital laboratories. The cost of the test was reimbursed by the National Health Insurance on an once‐in‐a‐lifetime basis until March 2012. Currently, it is reimbursed on an every‐exacerbation basis. Therefore, the numbers before March 2012 are almost equal to the numbers of the patients tested. The number of patients newly diagnosed with advanced NSCLC is estimated to be 50 000/year.23, 24 Altogether, most of the patients with an advanced disease, and thus are the targets of EGFR‐TKIs, were tested in 2011.
Figure 4.
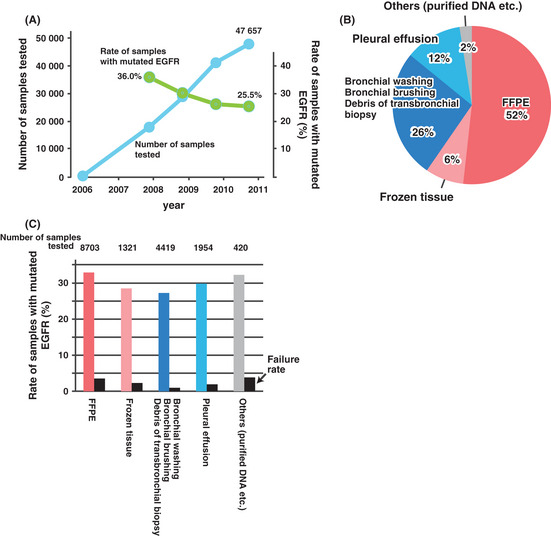
The EGFR mutation test in Japan. (A) The number of the EGFR mutation tests performed in three major commercial laboratories in Japan. The rate of EGFR mutation‐positive samples, which was curated from the database of one of the laboratories, is also shown. (B) The sample categories, which were summarized from approximately 17 000 samples submitted to one of the laboratories in 2009.36 Tissue samples (i.e. formalin‐fixed, paraffin‐embedded [FFPE] and frozen tissue) are indicated in warm colors, while cytological samples (i.e. bronchoscopy specimens and pleural effusion) are indicated in cold colors. (C) The rate of EGFR mutations according to sample category summarized from the data for approximately 17 000 samples.36 The failure rate represents the proportion of samples for which polymerase chain reaction (PCR) fails to amplify the target DNA. Tissue samples are indicated in warm colors, and cytological samples are indicated in cold colors.
The percentage of the samples with a mutated EGFR gene decreased as the number of the tests approached the number of patients with advanced NSCLC. This is likely because of an increase in the number of samples with fewer mutations; that is, samples with non‐adenocarcinoma histology, or samples collected from aged male patients.
Figure 4B shows the fraction of samples submitted to the test according to category. Almost 40% were cytological samples. This demonstrates that the use of cytological samples is indispensable for testing all advanced NSCLC patients. Accordingly, almost all EGFR mutation tests have been performed by one of three highly sensitive, PCR‐based methods that include the PNA–LNA PCR clamp,4, 18, 19, 25, 26, 27 the Cycleave method,5 and the PCR invader,20 all of which detect EGFR mutations in samples with a ratio of cancer cells of 1%.20
Figure 4C shows the rates of EGFR mutations according to sample categories. The mutation rates for tissue samples and cytological samples were similar. A direct comparison between each category may be inappropriate because an inherent difference should exist in the mutation rate between the categories. For example, the mutation rate for pleural effusion is likely to be high because malignant pleural effusion is mostly caused by adenocarcinoma. The rate of samples in which DNA failed to be amplified by PCR is high in formalin‐fixed, paraffin‐embedded (FFPE) samples, probably because of the fragmentation of DNA by formalin.28, 29
Clinical studies and cytological samples
The use of cytological samples enables a rapid accrual of patients for a variety of clinical trials. Clinical studies in which mutation in the EGFR gene have mostly been tested in cytological samples include a phase II study,30 a randomized phase III study,4 and a phase II study for patients with poor performance status.18 The last study is particularly important because cytological samples were the only samples available for many of the patients.
Criterion required for the kits testing EGFR mutation
After years of clinical investigations and discussions, Japanese clinicians treating NSCLC have reached a consensus that comprises the following elements: (i) cytological samples are valuable clinical specimens for testing EGFR mutations; (ii) a complete review of all patients with advanced NSCLC for EGFR mutations is very difficult to achieve without employing cytological samples; and (iii) in order to test both tissue and cytological samples, the EGFR mutation test should be able to detect mutations in samples with a ratio of cancer cells of 1%.
To attain the consensus above, we describe our provisional criterion that the kit used for EGFR mutation test is required to satisfy (Table 1).
Table 1.
Specifications for the EGFR mutation tests that can be used in the clinical practice
| Criterion |
| Kits used for EGFR mutation test are required to detect the type of mutations described in the Mutations section (see below) from the samples with a ratio of the cancer cells of 1%. To attain this, the kits are required to pass the assay described in the Assay section. |
| Mutations |
| Mandatory† |
| E746‐A750del (2235–2249delGGAATTAAGAGAAGC) |
| E746‐A750del (2236–2250delGAATTAAGAGAAGCA) |
| L858R |
| G719S |
| T790M |
| Recommended† |
| L747‐S752del P753S (2240–2257delTAAGAGAAGCAACATCTC) |
| L747‐E749del A750P (2239–2247delTTAAGAGAA, 2248G > C) |
| G719A |
| G719C |
| L861Q |
| Assay |
| Mutations that occur at the same position are usually detected at similar sensitivity. Therefore, only a single exon 19 deletion is included in the assay. The assay uses plasmid constructs each containing Del E746–A750 (2235–2249delGGAATTAAGAGAAGC), L858R, G719S, or T790M. Each plasmid DNA is mixed with normal human genomic DNA (10 ng/μL) to make the Assay Samples by achieving a copy number ratio of 1–200 of mutant EGFR sequence to normal EGFR sequence (Fig. S1). This simulates the test conditions in which the ratio of cancer cells to normal cells is 1–100 (Fig. 2). For the assay, 100 Assay Samples comprising 20 samples for each of the four mutants, and 20 Assay Samples containing only the normal human genomic DNA (10 ng/μL), are set up. There are randomized, and then investigated. This test is expected to correctly identify both presence and type of mutations in 95% of the samples (Fig. S2). Use of more than 5 ng of DNA from each Assay Sample is mandatory. Because the copy number of the mutant EGFR gene sequence conforms to a binomial distribution, use of <5 ng DNA causes significant sampling errors (see Fig. 5). |
Issues associated with the DNA‐based mutation test
We discuss some of the issues frequently raised in relation to EGFR mutation test. Detection of somatic mutations in organs other than the lung may share common issues.
DNA amount
When cells are sampled from a mixture of cancer cells and normal cells, the number of cancer cells conforms to a binomial distribution. When 100 cells (~650 pg DNA) are sampled from a cell mixture in which the ratio of cancer cells is 1%, there is a 37% chance that no cancer cells are sampled. When 800 cells (5 ng DNA) are sampled, there is more than a 96% chance that the ratio of cancer cells in the sample is more than 0.4%, and there is more than a 90% chance that the ratio is more than 0.6% (Fig. 5). Considering sampling errors, the mutation test should be performed using more than 5 ng DNA.
Figure 5.
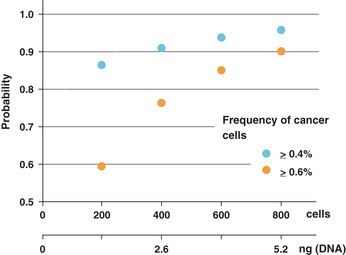
Sampling errors. The number of cancer cells sampled from a mixture of cancer cells and normal cells conforms to a binomial distribution. It is assumed that the ratio of cancer cells to normal cells in the cell suspension is 1:100. When 800 cells are collected from the suspension, there is a 96% chance that the ratio of cancer cells in the collection is more than 0.4% (i.e. 32 cells) and a 90% chance that the ratio is more than 0.6% (i.e. 48 cells).
Use of serum samples for mutation detection
Several studies have reported the detection of mutated genes in serum.31, 32, 33 The use of serum is attractive because serum collection is less invasive than many other sampling procedures. However, a serious concern arises when mutated genes are not detected in serum, because the reason for this is difficult to ascertain. Possible explanations include (i) the serum does not contain sufficient cancer‐derived DNA; and (ii) the cancer cells do not contain the mutated gene. The rate at which serum is shown to contain an insufficient amount of cancer‐derived DNA is significant,34 which inflates the false‐negative rate. The mutation test for detecting mutated gene in serum is currently unacceptable for clinical practice.
Use of circulating tumor cells for the detection of mutations
Circulating tumor cells (CTCs) are the cells detached from the tumor, enter the blood stream, and circulate throughout the body. Circulating tumor cells are a very attractive target for the mutation testing because they may be readily collected from peripheral blood.35 However, a simple calculation casts doubt on their clinical utility. The pulmonary capillaries have a diameter of 5 μm and trap particles with a size of 10–60 μm, which is the size of the 99mTc‐macro‐aggregated albumin that is used to embolize and image the pulmonary capillaries in pulmonary perfusion scintigraphy. The diameter of NSCLC cells is usually much larger than 5 μm, and they are thus considered unable to pass through the pulmonary capillaries. Rather, they are likely to be trapped at the entrance of the capillaries and subsequently eliminated. It is thus assumed that CTCs are eliminated during a single passage through the pulmonary circulation. Therefore, for 10 CTCs to be detected in 1 mL of blood, 10 (CTCs)/mL × 5000 (mL/min: cardiac output) × 1440 (min/day) = 7.2 × 107 CTCs/day (i.e. almost a gram of cells) are required to enter into circulation. Considering that cancer cells have a doubling time of more than 24 h, this formula indicates that a gram of cancer tissue should be present in the patients that doubles in 24 h and release half of the descendant cells into the circulation. This suggests that the patient has a large tumor burden, and thus is in a very advanced stage of the disease. Circulating tumor cells are considered difficult to isolate from patients in the early stages of NSCLC and thus may have limited clinical utility.
A detection system with a higher sensitivity
A mutation test may detect mutations in a sample in which the ratio of cancer cell is 0.1%. However, because the copy number of genomic DNA conforms to a binomial distribution, more than 50 ng of genomic DNA (DNA from 8000 cells) should be used for a successful test. The requirement for a large amount of DNA may increase the stress associated with sample collection on patients. An increase in the sensitivity of the test may not parallel an increase in its clinical utility.
Clinical samples in which the ratio of cancer cells is <1%
While ascertaining the presence of cancer cells in one aliquot of the sample (Fig. 3), pathologists may notice that the ratio of cancer cells in the sample may be <1%. On such occasions, the pathologists should notify clinicians that the sample may not be suitable for mutation testing and that re‐sampling may be required. Cooperation of clinicians and pathologists is highly recommended for reducing the false‐negative rate that stems from samples of unsatisfactory quality.
Future perspectives
Our ever‐expanding understanding of cancer driver genes lengthens the list of the gene mutations to be tested. In contrast, patients desire clinical procedures to be less stressful by the collection of smaller or fewer samples. Mutation testing aims to select patients suitable for specific treatments. At the same time, it excludes patients not suitable for certain treatments. If negative for mutations, excluded patients may be disappointed recalling their undergoing stressful sampling procedures only to obtain negative results. A long list of genes, therefore, does not justify the collection procedures much more stressful than those currently used.
The sample quality should be determined at the time of sampling. If inappropriately handled, DNA or RNA may be degraded immediately after sampling. Clinician training is very important such that they are prepared to handle the samples for the mutation test. Currently, most clinicians are aware of sample‐processing methods for pathological examinations. However, most of these procedures are inappropriate for DNA and RNA. For example, formalin fixation fragments DNA, while paraffin embedding makes DNA purification difficult. Following suitable procedures for DNA or RNA examination significantly reduces the amount of sample required for the test.
Next‐generation sequencing is being introduced for mutation testing. We anticipate an increase in the number of genes tested and a reduction in the cost of testing. However, whatever method is used, sensitivity of the test is limited by the amount of DNA available (Fig. 5), and the amount of DNA is limited by the size and type of cancer lesion and sampling procedures. As a result, sensitivity of the mutation tests stays, at least for the time being, around the current level. Development of sampling procedures that is far less invasive to the patient than those currently used and thus, able to collect more cancer‐derived DNA, is wanted to overcome the limitation of sensitivity, and will contribute greatly to future mutation testing.
Supporting information
Fig. S1. Preparation of assay samples.
Fig. S2. Assay.
Disclosure statement
Koichi Hagiwara holds a patent on the PNA‐LNA PCR clamp method and received royalties from the Mitsubishi Chemical Medience. Koichi Hagiwara and Kunihiko Kobayashi received lecture fees from AstraZeneca.
(Cancer Sci, doi: 10.1111/cas.12081, 2013)
References
- 1. Futreal PA, Coin L, Marshall M et al A census of human cancer genes. Nat Rev Cancer 2004; 4: 177–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pao W, Girard N. New driver mutations in non‐small‐cell lung cancer. Lancet Oncol 2011; 12: 175–80. [DOI] [PubMed] [Google Scholar]
- 3. Mok TS, Wu YL, Thongprasert S et al Gefitinib or carboplatin‐paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009; 361: 947–57. [DOI] [PubMed] [Google Scholar]
- 4. Maemondo M, Inoue A, Kobayashi K et al Gefitinib or chemotherapy for non‐small‐cell lung cancer with mutated EGFR. N Engl J Med 2010; 362: 2380–8. [DOI] [PubMed] [Google Scholar]
- 5. Mitsudomi T, Morita S, Yatabe Y et al Gefitinib versus cisplatin plus docetaxel in patients with non‐small‐cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol 2010; 11: 121–8. [DOI] [PubMed] [Google Scholar]
- 6. Rosell R, Carcereny E, Gervais R et al Erlotinib versus standard chemotherapy as first‐line treatment for European patients with advanced EGFR mutation‐positive non‐small‐cell lung cancer (EURTAC): a multicentre, open‐label, randomised phase 3 trial. Lancet Oncol 2012; 13: 239–46. [DOI] [PubMed] [Google Scholar]
- 7. O'Brien SG, Guilhot F, Larson RA et al Imatinib compared with interferon and low‐dose cytarabine for newly diagnosed chronic‐phase chronic myeloid leukemia. N Engl J Med 2003; 348: 994–1004. [DOI] [PubMed] [Google Scholar]
- 8. Flaherty KT, Puzanov I, Kim KB et al Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med 2010; 363: 809–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lynch TJ, Bell DW, Sordella R et al Activating mutations in the epidermal growth factor receptor underlying responsiveness of non‐small‐cell lung cancer to gefitinib. N Engl J Med 2004; 350: 2129–39. [DOI] [PubMed] [Google Scholar]
- 10. Paez JG, Janne PA, Lee JC et al EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004; 304: 1497–500. [DOI] [PubMed] [Google Scholar]
- 11. Kosaka T, Yatabe Y, Endoh H, Kuwano H, Takahashi T, Mitsudomi T. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res 2004; 64: 8919–23. [DOI] [PubMed] [Google Scholar]
- 12. D'Addario G, Fruh M, Reck M, Baumann P, Klepetko W, Felip E. Metastatic non‐small‐cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow‐up. Ann Oncol 2010; 21 (Suppl 5): v116–9. [DOI] [PubMed] [Google Scholar]
- 13. Clinical practice guideline for lung cancer. The Japan Lung Cancer Society, 2010. [Cited 9 Sep 2012.] Available from URL: http://www.haigan.gr.jp. [Google Scholar]
- 14. NCCN Clinical Practice Guidlines in Oncology, Non‐Small Cell Lung Cancer. [Cited 9 Sep 2012.] Available from URL: 2010. http://www.nccn.org.
- 15. Eberhard DA, Giaccone G, Johnson BE. Biomarkers of response to epidermal growth factor receptor inhibitors in Non‐Small‐Cell Lung Cancer Working Group: standardization for use in the clinical trial setting. J Clin Oncol 2008; 26: 983–94. [DOI] [PubMed] [Google Scholar]
- 16. Marchetti A, Normanno N, Pinto C et al Recommendations for mutational analysis of EGFR in lung carcinoma. Pathologica 2010; 102: 119–26. [PubMed] [Google Scholar]
- 17. Pirker R, Herth FJ, Kerr KM et al Consensus for EGFR mutation testing in non‐small cell lung cancer: results from a European workshop. J Thorac Oncol 2010; 5: 1706–13. [DOI] [PubMed] [Google Scholar]
- 18. Inoue A, Kobayashi K, Usui K et al First‐line gefitinib for patients with advanced non‐small‐cell lung cancer harboring epidermal growth factor receptor mutations without indication for chemotherapy. J Clin Oncol 2009; 27: 1394–400. [DOI] [PubMed] [Google Scholar]
- 19. Tanaka T, Nagai Y, Miyazawa H et al Reliability of the peptide nucleic acid‐locked nucleic acid polymerase chain reaction clamp‐based test for epidermal growth factor receptor mutations integrated into the clinical practice for non‐small cell lung cancers. Cancer Sci 2007; 98: 246–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goto K, Satouchi M, Ishii G et al An evaluation study of EGFR mutation tests utilized for non‐small‐cell lung cancer in the diagnostic setting. Ann Oncol 2012; 23: 2914–9. [DOI] [PubMed] [Google Scholar]
- 21. Takeuchi K, Choi YL, Soda M et al Multiplex reverse transcription‐PCR screening for EML4‐ALK fusion transcripts. Clin Cancer Res 2008; 14: 6618–24. [DOI] [PubMed] [Google Scholar]
- 22. Malapelle U, de Rosa N, Bellevicine C et al EGFR mutations detection on liquid‐based cytology: is microscopy still necessary? J Clin Pathol 2012; 65: 561–4. [DOI] [PubMed] [Google Scholar]
- 23. Journal of Health and Welfare Statistics, Health and Welfare Statistics Association. Tokyo: Koseido, 2009. (in Japanese). [Google Scholar]
- 24. Ministry of Health, Labour and Welfare, Japan, 2011. [Cited 9 Sep 2012.] Available from URL: http://www.mhlw.go.jp/toukei/saikin/hw/jinkou/kakutei10/dl/11_h7.pdf (in Japanese).
- 25. Nagai Y, Miyazawa H, Huqun H et al Genetic heterogeneity of the epidermal growth factor receptor in non‐small cell lung cancer cell lines revealed by a rapid and sensitive detection system, the peptide nucleic acid‐locked nucleic acid PCR clamp. Cancer Res 2005; 65: 7276–82. [DOI] [PubMed] [Google Scholar]
- 26. Miyazawa H, Tanaka T, Nagai Y et al Peptide nucleic acid‐locked nucleic acid polymerase chain reaction clamp‐based detection test for gefitinib‐refractory T790M epidermal growth factor receptor mutation. Cancer Sci 2008; 99: 595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tanaka T, Matsuoka M, Sutani A et al Frequency of and variables associated with the EGFR mutation and its subtypes. Int J Cancer 2010; 126: 651–5. [DOI] [PubMed] [Google Scholar]
- 28. Greer CE, Wheeler CM, Manos MM. Sample preparation and PCR amplification from paraffin‐embedded tissues. PCR Methods Appl 1994; 3: S113–22. [DOI] [PubMed] [Google Scholar]
- 29. Liu D, Nakano J, Ueno M et al A useful protocol for analyses of mutations of the epidermal growth factor receptor gene. Oncol Rep 2006; 15: 1503–5. [PubMed] [Google Scholar]
- 30. Sutani A, Nagai Y, Udagawa K et al Gefitinib for non‐small‐cell lung cancer patients with epidermal growth factor receptor gene mutations screened by peptide nucleic acid‐locked nucleic acid PCR clamp. Br J Cancer 2006; 95: 1483–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kimura H, Kasahara K, Kawaishi M et al Detection of epidermal growth factor receptor mutations in serum as a predictor of the response to gefitinib in patients with non‐small‐cell lung cancer. Clin Cancer Res 2006; 12: 3915–21. [DOI] [PubMed] [Google Scholar]
- 32. Gormally E, Caboux E, Vineis P, Hainaut P. Circulating free DNA in plasma or serum as biomarker of carcinogenesis: practical aspects and biological significance. Mutat Res 2007; 635: 105–17. [DOI] [PubMed] [Google Scholar]
- 33. Bai H, Mao L, Wang HS et al Epidermal growth factor receptor mutations in plasma DNA samples predict tumor response in Chinese patients with stages IIIB to IV non‐small‐cell lung cancer. J Clin Oncol 2009; 27: 2653–9. [DOI] [PubMed] [Google Scholar]
- 34. Goto K, Ichinose Y, Ohe Y et al Epidermal growth factor receptor mutation status in circulating free DNA in serum: from IPASS, a phase III study of gefitinib or carboplatin/paclitaxel in non‐small cell lung cancer. J Thorac Oncol 2012; 7: 115–21. [DOI] [PubMed] [Google Scholar]
- 35. Maheswaran S, Sequist LV, Nagrath S et al Detection of mutations in EGFR in circulating lung‐cancer cells. N Engl J Med 2008; 359: 366–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fujimoto H, Furumoto N, Takeda M et al EGFR mutation test using the PNA‐LNA PCR clamp performed in our company. Byori Gijutsu 2010; 73: 67–9. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Preparation of assay samples.
Fig. S2. Assay.


