Abstract
It has been more than 30 years since the discovery of human T‐lymphotropic virus type 1 (HTLV‐1), the first human retrovirus identified. Human T‐lymphotropic virus type 1 infects 15–20 million people worldwide causing two major diseases: adult T‐cell leukemia/lymphoma and HTLV‐1‐associated myelopathy/tropical spastic paraparesis. Human T‐lymphotropic virus type 1 establishes several decades of latent infection, during which viral–host interaction determines disease segregation. This review highlights non‐structural proteins that are encoded on the viral genome and manage latent infection. Latent infection is a key in HTLV pathology, so that effective inhibition of these proteins might lead to successful disease management.
Human T‐lymphotropic virus type 1 (HTLV‐1) is the first retroviridae that has been identified to infect humans1, 2 and causes adult T‐cell leukemia/lymphoma (ATL)3 and HTLV‐1‐associated myelopathy/tropical spastic paraparesis (HAM/TSP).4, 5, 6 Adult T‐cell leukemia/lymphoma in its acute/lymphoma phases is highly aggressive and resistant to current intensive chemotherapy, with median survival time of approximately 1 year after onset.7, 8 HTLV‐1‐associated myelopathy/tropical spastic paraparesis is a neuro‐degenerative disease in which patients suffer from chronic pain and loss in motility that progress irreversibly over years resulting in a low quality of life on wheelchairs. HTLV‐1 has also been found in other conditions such as infectious dermatitis, HTLV‐associated uveitis, arthritis, polymyositis and pneumonitis,9, 10 suggesting diverse and different degrees of roles in inflammation. While the diseases that are associated with HTLV‐1 infection become evident, current management is not sufficient to foresee, prevent or cure any one of those conditions.
Discovery of HTLV‐11, 11 and its association with ATL11 opened the field of human retrovirus research. It proved that the retroviridae infection to humans was not fictional; further, it could be quite common and pathogenic. It became known that HTLV‐1‐associated diseases required long latent infection, which had been making the viral etiology less apparent. While HTLV‐1 was found in conditions other than ATL, the idea of human retroviruses has led to the discovery of other retroviridae family members to date, including: HTLV‐2,12 HTLV‐313 and HTLV‐414 within the same genus deltaretroviruses, as well as human immunodeficiency viruses (HIV) that belong to the genus lentiviruses. All of these viruses can be transmitted by blood,15 breast milk16 and seminal fluid,17 and establish latent infection. Natural co‐infection of HTLV‐1/2 and HIV‐1 can occur and has been reported in the areas where both viruses are endemic, causing complications.18 Since there is no successful vaccine to any retroviruses, better guidelines in controlling HTLV‐1 are needed, especially in the developing world where uncontrolled infections remain.
As a pathogen, HTLV‐1 produces “non‐structural proteins” (NSP) that do not become parts of viral virion but are essential in causing diseases. The NSP hijack cellular machinery, which on one hand immortalizes and transforms infected cells, but on the other hand optimizes viral replication and repels immune surveillance that finds and kills infected cells in asymptomatic carriers (AC). Both of these activities are essential, enabling HTLV‐1 to establish decades of latent infection while inducing diseases.
Retroviruses other than HTLV‐1 also encode NSP that are unique to each one of them and are required for latent infection. A high homology seen between HTLV‐1 and HTLV‐2 might implicate their conserved strategies in persistence whose termination might halt disease progression.
Virus and Non‐Structural Proteins
Human T‐lymphotropic virus type 1 equips multiple genes in its limited genome size (Fig. 1).19 There are numbers of cellular and viral mechanisms that enable the proviral genome, a form of virus integrated in the host cell chromosome, to selectively express all genes and mature their end products.20 Reported mechanisms include bidirectional transcription initiated by 5′‐ and 3′‐long terminal repeats (LTR) located at both ends of the proviral genome,21, 22 alternative splicings that serve for overlapping open reading frames (ORF) with or without frame shifts,23 post‐transcriptional regulation,24 ribosomal frame‐shift during translation and post‐translational processing.25
Figure 1.
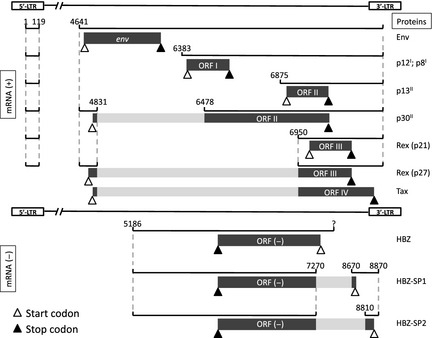
Human T‐lymphotropic virus type 1 (HTLV‐1) genome organization. The HTLV‐1 provirus uses complicated mechanisms to express mRNA and proteins. 5′‐long terminal repeat (LTR) and 3′‐LTR located on both ends operate singly or doubly spliced mRNA species with their open reading frames (ORF). The ORF relevant to non‐structural proteins are depicted. The unspliced mRNA from 5′‐LTR (not shown) encodes structural proteins other than Env and also becomes a genome in a newly formed viral virion. The origin of reported unspliced HTLV‐1 bZIP factor (HBZ) mRNA is currently unknown.
Among several transcripts and their encoding proteins, some compose viral virion enclosing its contents, which are structural components, enzymes (integrase, reverse transcriptase and protease) and linear single‐stranded RNA (+) diploid genome; while others remain in the host that produced them as non‐structural proteins (NSP). They are also referred to as “regulatory proteins” or “accessory proteins” in other reports. Importantly, cytotoxic T‐lymphocytes (CTL) directed to NSP have been detected in viral carriers.26 A list of ORF for the NSP under investigation is summarized in Table 1. In general, HTLV are conserved viruses relative to HIV.27 Human T‐lymphotropic virus type 1 and HTLV‐2 share approximately 70% homology in the nucleotide sequence,28 so that the corresponding ORF products also score a distant homology in amino acid (AA) levels.
Table 1.
Human T‐lymphotropic virus type 1 (HTLV‐1) ORF and their products (NSP) in comparison with HTLV‐2
| Transcription | ORF | NSP | Mechanisma | HTLV‐2 |
|---|---|---|---|---|
| 5′‐LTR | I | p12I; p8I | Proteolytic cleavage | p10I |
| 5′‐LTR | II | p30II; p13II | Doubly; singly spliced mRNA | p28II |
| 5′‐LTR | III | Rex (p27; p21) | Doubly; singly spliced mRNA | Rex‐2 |
| 5′‐LTR | IV | Tax | Tax‐2 | |
| 3′‐LTR | (−) | HBZ; HBZ‐SP1 or ‐SP2 | Un‐; singly spliced mRNA | APH‐2 |
Each ORF might give rise to multiple proteins by the given mechanism. HBZ, HTLV‐1 bZIP factor; LTR, long terminal repeat; NSP, non‐structural protein; ORF, open reading frame.
Both HTLV‐1 and HTLV‐2 are capable of infecting and immortalizing normal human peripheral blood cells by co‐cultivation with pre‐infected donor cells.2, 29, 30 Although there is a continuous debate,31 HTLV‐1 in vivo is more infectious to CD4+ than to CD8+ T‐cells and vice versa for HTLV‐2.32 Only HTLV‐1 causes T‐cell outgrowth and malignant transformation in vivo, indicating that viral factors unique to HTLV‐1 are essential in causing ATL. In contrast, HTLV‐2 can cause neurological disorders that might resemble HAM/TSP.33 No association of HTLV‐3/4 with diseases has been established presently, which is due to the limited number of known infections since their discoveries. Human T‐lymphotropic virus type 3 shares significant homology with Simian T‐lymphotropic virus type 3 (STLV‐3) found in non‐human primates,13 while HTLV‐4 reveals much less homology with any other known HTLV or STLV, except that antiserum developed in a carrier exhibited reactivity to HTLV‐1 antigens.14
Non‐Structural Proteins in Viral Persistence
Much like other retroviruses, HTLV‐1 integrates into host cell chromosomes after infection, that is, provirus, and persists permanently. It is true that there is no need for the virus to keep a productive replication cycle by producing infectious virions, as long as the host cell undergoes mitosis, which results in daughter cells with the same number and integration sites of the proviral copies. Given a well‐known fact that HTLV‐1 poorly self‐replicates in vivo,27 immortalization of the host appears to be a prerequisite for life‐long infection. Human T‐lymphotropic virus type 1 expresses viral proteins in the absence of adaptive immunity during primary infection. While Tax above all is believed to have the most powerful and indispensable roles in the host immortalization process through the modulation of numerous cellular transcriptions (see Disclosure Statement section), p12I was also found to potentiate activation and proliferation of T‐cells in response to different extracellular stimuli (see Open reading frame IV section). Tax, in the meantime, also activates viral gene transcription through 5′‐LTR and induces viral replication (Table 2).21
Table 2.
Non‐structural protein (NSP)‐mediated viral spread and cell growth
| NSP | Major target | Impact |
|---|---|---|
| p8I | Cell–cell contact | Increases viral transmission43 |
| p12I | Calreticulin | Induces Ca++ secretion and cell activation40 |
| p12I | IL‐2R β and γc chains | Increases sensitivity to IL‐2 of host cells37 |
| Tax; Rex | 5′‐LTR; viral mRNA | Increases viral replication21, 24 |
| Tax | NFκB; SRF | Increases IL‐2Rα, IL‐6, GM‐CSF, c‐fos, c‐egr58 |
GM‐CSF, granulocyte macrophage colony‐stimulating factor; IL, interleukin; NFκB, nuclear factor kappa B; LTR, long terminal repeat; SRF, serum response factor.
On one hand the replication benefits HTLV‐1 to spread infection de novo; however, on the other hand it increases the risk of viral antigens exposed to the immunity in AC. In fact, Tax is one of the major viral antigens that CTL target.34 Some NSP resulting from Tax‐mediated viral induction in turn self‐limit viral expression much like negative feedback mechanisms. One exception is the HTLV‐1 bZIP factor (HBZ), whose expression relies on 3′‐LTR and does not depend on Tax.35 The NSP also sabotage host immune surveillance to the virus, which ultimately promotes propagation of infected cells within AC. These adjustments would be important particularly during the primary and early infection stages, slowing infected individuals to acquire adaptive immunity against HTLV‐1. Once the infection becomes latent, the level of antisera against HTLV‐1 in general correlates with the proviral load, indicating active viral replication does induce the immune response and these two are balanced in AC.36
The NSP that have been found to limit viral replication are p8I, p30II, p13II and HBZ. P12I and p8I have also been found to interfere with cellular immune machineries (Table 3). These NSP target different phases in viral replication and immune processes and are discussed below with respect to each ORF.
Table 3.
Non‐structural protein (NSP)‐mediated viral defense against host immunity
| NSP | Major target | Impact |
|---|---|---|
| p8I | T‐cell antigen receptor | Causes anergy; inhibits viral 5′‐LTR induction25 |
| p12I | MHC‐I | Inhibits viral antigens presented to CTL38 |
| p13II | Tax | Inhibits viral replication by direct binding50 |
| p30II | Tax/Rex mRNA | Inhibits Tax/Rex production and viral replication46 |
| HBZ | CREB‐2 | Inhibits viral transcription by Tax51 |
CREB, CRE‐binding protein; CTL, cytotoxic T‐lymphocytes; HBZ, HTLV‐1 bZIP factor; LTR, long terminal repeat; MHC‐I, major histocompatibility complex class I.
Open reading frame I
It encodes hydrophobic protein of 99AA known as p12I. This protein has been shown to bind directly to interleukin‐2 receptor (IL‐2R) β and γc chains,37 major histocompatibility complex class I (MHC‐I) heavy chain,38 linker for activation of T‐cells,39 calreticulin,40 vacuolar‐ATPase41 and is present in multiple cellular compartments, most abundantly in endothelial reticulum (ER)–Golgi organelles and to a lesser extent on the plasma membrane.23, 39 Each interaction above results in sensitized IL‐2R signaling, reduction of MHC‐I on the cell surface, impaired T‐cell antigen receptor signaling and persistent leakage of Ca++ from the ER, which might confer the host aberrance in proliferation, immune evasion, anergy and activation in the absence of extracellular stimulation. P12I in the presence of phorbol myristate acetate activates nuclear factor of activated T‐cells (NFAT).42 These studies implicate significant roles of p12I during latency, but at the same time raise a question on how this protein can access different cellular compartments and exert those pleiotropic functions.
Ectopic expression of ORF I results in production of two major products by possible cleavage, whose sizes apparently differ as observed using immunoblots, that is, 12 kDa (p12I) and 8 kDa (p8I) forms.25 Given multiple functions in the different cellular compartments reported, it became necessary to reevaluate the functions that belong to each form. More importantly, analysis of proviral genomes existing in 304 HTLV‐1 carriers revealed polymorphisms in nucleotide sequences that would result in AA changes.25 Expression of those variants in vitro demonstrated a spectrum of ratios in p8I/p12I production unique to the variants. Further analyses using variants predominantly producing p12I or p8I indicated their distinct cellular distribution: p12I in the ER and Golgi organelles while p8I on the plasma membrane. It has been shown that immunological synapse (IS) recruits p8I when viral host T‐lymphocytes engage antigen‐presenting cells, causing anergic signaling. As IS induces 5′‐LTR transcription and viral replication, p8I also inhibits viral reactivation by this route.25 It was later found that p8I also increased cellular conduits and virus transmission.43 The exact cleavage sites have not been determined using terminal peptide sequencing. However, it is clear that cleavage requiring no other viral proteins co‐existed. The study of HTLV‐2 p10I protein is much behind the study for p12I; however, it has been found to bind the free chain of MHC‐I, suggesting functional overlap.44 There is no equivalent of p8I in HTLV‐2 reported. The p12I protein also shares distant homology with Nef of HIV‐1 that is important for viral infectivity. The HIV‐1 clone, in which p12I replaced Nef, retained its infectivity in vitro, demonstrating functional substitution.45
Open reading frame II
It encodes two in‐frame proteins known as p30II (241AA) and p13II (87AA). P13II is a carboxy‐terminal of p30II protein, utilizing internal methionine for its initiation and termination of p30II, operated by alternative splicing. P30II possesses nuclear localization signal (NLS) and upon expression resides in the nucleus/nucleolus. P30II does not access cytoplasm. P30II was found to specifically bind to Tax/Rex mRNA of proviral origin and retain it in the nucleus and prevent it being translated.46 By doing so, p30II suppresses viral replication, as Tax/Rex are master viral regulators. Human T‐lymphotropic virus type 2 p28II was reported to have a function similar to p30II; it has been shown to bind Tax/Rex mRNA of both HTLV‐1 and HTLV‐2, and so has p30II.47 Under genotoxic stress induced by radiation, p30II was also found to modulate activity of ataxia telangiectasia mutated, which essentially sensors DNA damage and mediates cell cycle arrest and consequently apoptosis, and increased survival. It was also found to inhibit homologous recombination favoring unfaithful DNA repair.48 P13II, which lacks amino‐terminal NLS of P30II, accumulates mostly in the inner mitochondrial membrane.49 P13II was shown to interact with Tax protein, to be rerouted in nuclear speckles where it inhibited viral replication.50 Open reading frame II products by above control Tax production and activity at the post‐transcriptional and post‐translational levels. Whether HTLV‐2 expresses HTLV‐1 p13II equivalent is currently unknown.
Open reading frame (−)
While the presence of transcription from 3′‐LTR has been known since 1989,22 the possibility of encoded proteins has not been tested or reported earlier than 2002 when HBZ of 209AA was found.51 Since then, other groups have also described its splicing variants.52, 53 Human T‐lymphotropic virus type 1 bZIP factor appears to have a bimodal effect through the mRNA and protein levels. As a protein, HBZ bound one of the activating transcription factor/CRE‐binding protein (ATF/CREB) family members, CREB‐2, and inhibited Tax‐mediated viral transcription from 5′‐LTR.51 However, at the mRNA level, HBZ promoted proliferation of a T‐cell line and, conversely, small interfering RNA against HBZ inhibited growth of ATL cells.35 As many proviral genomes in ATL cells contain defects or otherwise epigenetic changes at their 5′‐ends including LTR while 3′‐LTR remains intact with mRNA expression, these transcripts and proteins might be of relevance during late ATL development.54 Besides, the lack of 5′‐LTR might help survival of infected cells over the course of latency, among which ATL develops (Fig. 2). An mRNA encoding antisense protein of HTLV‐2 (APH‐2) with 183AA was also found in lymphocytes of HTLV‐2‐infected individuals.55 The APH‐2 localizes in the nucleus and represses Tax‐2 activity through interaction with CREB, despite the fact that it lacks the bZIP domain seen in HBZ.
Figure 2.
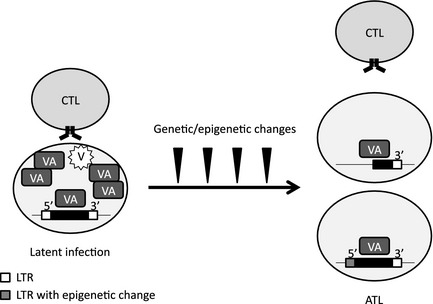
Between latent infection and adult T‐cell leukemia/lymphoma (ATL). Human T‐lymphotropic virus type 1 (HTLV‐1)‐infected cells express viral antigens (VA), that is, all viral products including non‐structural proteins, and virions (V) to survive and spread under host immunity (left). However, once immortalization of infected cells is complete the risk of having VA might exceed the benefit. In this model, the developing ATL acquires several genetic/epigenetic changes including a defect or methylation on proviral 5′‐long terminal repeat (LTR), resulting in the absence of all 5′‐LTR‐operated VA. The chance of immune detection of such cells becomes smaller, from which ATL arises (right). The exact timing and mechanism for proviral defects remain elusive.
NSP in Cell Transformation
Once it infects HTLV‐1 remains lifelong regardless of its concomitance to disease, while general longevity of any blood cell types – including viral host T‐cell subsets – would be considerably shorter.56 Thus, HTLV‐1 needs to either immortalize its host or repeat de novo infection. However, continuous replication and infection of HTLV‐1 is unlikely in vivo because of strong immune pressure even though it occurred through cell–cell transmission where no virion is produced.34 Early studies that outlined basic viral life cycle led to the findings in indispensable roles of ORF IV product Tax protein in cellular genetic changes that would substantially promote leukemia development, apart from in viral regulation in coordination with 27 kDa Rex of ORF III facilitating nuclear export of singly‐ and un‐spliced viral mRNAs.24
Open reading frame IV
It encodes 353AA protein with molecular mass of 40 kDa, transcriptional activator of pX region (Tax). Tax immortalizes T‐lymphocytes of healthy donors with high efficiency in the absence of any other viral proteins.57 It is believed that Tax, not a mutagen per se, exerts antiapoptotic effect, dysregulates cell cycle and promotes proliferation in infected cells,58 regardless of the presence of genotoxic stress and resulting mutations caused by other means. Tax‐mediated activation of nuclear factor kappa B (NFκB) may explain IL‐2Rα chain commonly overexpressed on ATL cells.59 However, Tax‐induced IL‐2 autocrine loop is currently not supported.60, 61 Tax‐2 of HTLV‐2 was also found to immortalize T‐lymphocytes associated with induction of IL‐2 and IL‐2R,62 while it activates NFAT instead of NFκB, and autocrine loop is suggested.63 Therefore, there is a significant diversity between these viral regulators even though they share 75% of homology in AA level.
Non‐Structural Proteins Tested in Viral Molecular Clones
The full length of the HTLV‐1 genome is available as molecular clones, so that ablation of specific proteins on the isogenic genome was also performed using genetic manipulation to test its effect on viral replication in vitro, ex vivo and in animal models. Loss of the ORF I message was associated with reduced infectivity in rabbit and human primary lymphocytes.64, 65 However, more recently it was reported that p12I and p30II are required for viral infectivity to human dendritic cells and in macaques, but not in rabbits.66 Lack of p13II,67 Rex68 and HBZ69 each alone was associated with significant loss in viral infectivity and persistence in rabbit. The methods used for manipulation of proviral clones as well as infectious properties of resulting viruses are summarized in Table 4. While infection studies using such clones are valuable, these mutations caused simultaneous changes in AA of other overlapping NSP and gave us an indefinite conclusion. Thus, molecular studies of individual NSP are still needed to complement what is not available in animal infection models. Of note, some mutations were reverted back to wild‐type forms in macaques, suggesting the requirement of ablated proteins during the early phase of infection.66
Table 4.
Mutant HTLV‐1 clones lacking specific non‐structural protein (NSP)
| NSPa | Manipulation | Property |
|---|---|---|
| p12I | Splice acceptor | Decreased infectivity in human PBMC and rabbits64, 65 |
| p12I | First codon | Decreased infectivity in human dendritic cells and in macaques66 |
| p13I | First codon | Loss of infectivity in rabbits67 |
| p30II | Third codon | Decreased infectivity in human dendritic cells and in macaques66 |
| Rex | Third codon | Loss of infectivity in rabbits68 |
| HBZ | Codons 11; 175 | Decreased infectivity and persistence in rabbits69 |
Knock out proteins are listed here. See references for the effects on overlapping NSP. HBZ, HTLV‐1 bZIP factor.
Conclusion
There has been no acute disease known after HTLV‐1 infection. The HTLV field in past decades saw what was thought “latent infection” was under the persistent control of NSP, many of which had previously been neglected. Relevance of NSP in HTLV pathology should therefore not be underestimated.
Disclosure Statement
The author has no conflict of interest.
(Cancer Sci 2013; 104: 983–988)
References
- 1. Poiesz BJ, Ruscetti FW, Gazdar AF, Bunn PA, Minna JD, Gallo RC. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T‐cell lymphoma. Proc Natl Acad Sci USA 1980; 77: 7415–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Miyoshi I, Kubonishi I, Yoshimoto S et al Type C virus particles in a cord T‐cell line derived by co‐cultivating normal human cord leukocytes and human leukaemic T cells. Nature 1981; 294: 770–1. [DOI] [PubMed] [Google Scholar]
- 3. Uchiyama T, Yodoi J, Sagawa K, Takatsuki K, Uchino H. Adult T‐cell leukemia: clinical and hematologic features of 16 cases. Blood 1977; 50: 481–92. [PubMed] [Google Scholar]
- 4. Rodgers PE. The clinical features and aetiology of the neuropathic syndrome in Jamaica. West Indian Med J 1965; 14: 36–47. [PubMed] [Google Scholar]
- 5. Osame M, Usuku K, Izumo S et al HTLV‐I associated myelopathy, a new clinical entity. Lancet 1986; 1: 1031–2. [DOI] [PubMed] [Google Scholar]
- 6. Gessain A, Barin F, Vernant JC et al Antibodies to human T‐lymphotropic virus type‐I in patients with tropical spastic paraparesis. Lancet 1985; 2: 407–10. [DOI] [PubMed] [Google Scholar]
- 7. Yamada Y, Tomonaga M, Fukuda H et al A new G‐CSF‐supported combination chemotherapy, LSG15, for adult T‐cell leukaemia‐lymphoma: Japan Clinical Oncology Group Study 9303. Br J Haematol 2001; 113: 375–82. [DOI] [PubMed] [Google Scholar]
- 8. Marcais A, Suarez F, Sibon D, Bazarbachi A, Hermine O. Clinical trials of adult T‐cell leukaemia/lymphoma treatment. Leuk Res Treatment 2012; 2012: 932175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goto K, Saeki K, Kurita M, Ohno S. HTLV‐I associated uveitis in central Japan. Br J Ophthalmol 1995; 79: 1018–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mahieux R, Gessain A. HTLV‐1 and associated adult T‐cell leukemia/lymphoma. Rev Clin Exp Hematol 2003; 7: 336–61. [PubMed] [Google Scholar]
- 11. Yoshida M, Miyoshi I, Hinuma Y. Isolation and characterization of retrovirus from cell lines of human adult T‐cell leukemia and its implication in the disease. Proc Natl Acad Sci USA 1982; 79: 2031–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kalyanaraman VS, Sarngadharan MG, Robert‐Guroff M, Miyoshi I, Golde D, Gallo RC. A new subtype of human T‐cell leukemia virus (HTLV‐II) associated with a T‐cell variant of hairy cell leukemia. Science 1982; 218: 571–3. [DOI] [PubMed] [Google Scholar]
- 13. Calattini S, Chevalier SA, Duprez R et al Discovery of a new human T‐cell lymphotropic virus (HTLV‐3) in Central Africa. Retrovirology 2005; 2: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wolfe ND, Heneine W, Carr JK et al Emergence of unique primate T‐lymphotropic viruses among central African bushmeat hunters. Proc Natl Acad Sci USA 2005; 102: 7994–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Inaba S, Sato H, Okochi K et al Prevention of transmission of human T‐lymphotropic virus type 1 (HTLV‐1) through transfusion, by donor screening with antibody to the virus. One‐year experience. Transfusion 1989; 29: 7–11. [DOI] [PubMed] [Google Scholar]
- 16. Hino S, Yamaguchi K, Katamine S et al Mother‐to‐child transmission of human T‐cell leukemia virus type‐I. Jpn J Cancer Res 1985; 76: 474–80. [PubMed] [Google Scholar]
- 17. Bartholomew C, Saxinger WC, Clark JW et al Transmission of HTLV‐I and HIV among homosexual men in Trinidad. J Am Med Assoc 1987; 257: 2604–8. [PubMed] [Google Scholar]
- 18. Page JB, Lai SH, Chitwood DD, Klimas NG, Smith PC, Fletcher MA. HTLV‐I/II seropositivity and death from AIDS among HIV‐1 seropositive intravenous drug users. Lancet 1990; 335: 1439–41. [DOI] [PubMed] [Google Scholar]
- 19. Seiki M, Hattori S, Hirayama Y, Yoshida M. Human adult T‐cell leukemia virus: complete nucleotide sequence of the provirus genome integrated in leukemia cell DNA. Proc Natl Acad Sci USA 1983; 80: 3618–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Journo C, Douceron E, Mahieux R. HTLV gene regulation: because size matters, transcription is not enough. Future Microbiol 2009; 4: 425–40. [DOI] [PubMed] [Google Scholar]
- 21. Sodroski JG, Rosen CA, Haseltine WA. Trans‐acting transcriptional activation of the long terminal repeat of human T lymphotropic viruses in infected cells. Science 1984; 225: 381–5. [DOI] [PubMed] [Google Scholar]
- 22. Larocca D, Chao LA, Seto MH, Brunck TK. Human T‐cell leukemia virus minus strand transcription in infected T‐cells. Biochem Biophys Res Commun 1989; 163: 1006–13. [DOI] [PubMed] [Google Scholar]
- 23. Koralnik IJ, Fullen J, Franchini G. The p12I, p13II, and p30II proteins encoded by human T‐cell leukemia/lymphotropic virus type I open reading frames I and II are localized in three different cellular compartments. J Virol 1993; 67: 2360–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Inoue J, Seiki M, Yoshida M. The second pX product p27 chi‐III of HTLV‐1 is required for gag gene expression. FEBS Lett 1986; 209: 187–90. [DOI] [PubMed] [Google Scholar]
- 25. Fukumoto R, Andresen V, Bialuk I et al In vivo genetic mutations define predominant functions of the human T‐cell leukemia/lymphoma virus p12I protein. Blood 2009; 113: 3726–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pique C, Ureta‐Vidal A, Gessain A et al Evidence for the chronic in vivo production of human T cell leukemia virus type I Rof and Tof proteins from cytotoxic T lymphocytes directed against viral peptides. J Exp Med 2000; 191: 567–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gessain A, Gallo RC, Franchini G. Low degree of human T‐cell leukemia/lymphoma virus type I genetic drift in vivo as a means of monitoring viral transmission and movement of ancient human populations. J Virol 1992; 66: 2288–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Feuer G, Green PL. Comparative biology of human T‐cell lymphotropic virus type 1 (HTLV‐1) and HTLV‐2. Oncogene 2005; 24: 5996–6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yamamoto N, Okada M, Koyanagi Y, Kannagi M, Hinuma Y. Transformation of human leukocytes by cocultivation with an adult T cell leukemia virus producer cell line. Science 1982; 217: 737–9. [DOI] [PubMed] [Google Scholar]
- 30. Chen IS, Quan SG, Golde DW. Human T‐cell leukemia virus type II transforms normal human lymphocytes. Proc Natl Acad Sci USA 1983; 80: 7006–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nagai M, Brennan MB, Sakai JA, Mora CA, Jacobson S. CD8(+) T cells are an in vivo reservoir for human T‐cell lymphotropic virus type I. Blood 2001; 98: 1858–61. [DOI] [PubMed] [Google Scholar]
- 32. Jones KS, Fugo K, Petrow‐Sadowski C et al Human T‐cell leukemia virus type 1 (HTLV‐1) and HTLV‐2 use different receptor complexes to enter T cells. J Virol 2006; 80: 8291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Murphy EL, Fridey J, Smith JW et al HTLV‐associated myelopathy in a cohort of HTLV‐I and HTLV‐II‐infected blood donors. The REDS investigators. Neurology 1997; 48: 315–20. [DOI] [PubMed] [Google Scholar]
- 34. Bangham CR. The immune control and cell‐to‐cell spread of human T‐lymphotropic virus type 1. J Gen Virol 2003; 84: 3177–89. [DOI] [PubMed] [Google Scholar]
- 35. Satou Y, Yasunaga J, Yoshida M, Matsuoka M. HTLV‐I basic leucine zipper factor gene mRNA supports proliferation of adult T cell leukemia cells. Proc Natl Acad Sci USA 2006; 103: 720–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nagai M, Usuku K, Matsumoto W et al Analysis of HTLV‐I proviral load in 202 HAM/TSP patients and 243 asymptomatic HTLV‐I carriers: high proviral load strongly predisposes to HAM/TSP. J Neurovirol 1998; 4: 586–93. [DOI] [PubMed] [Google Scholar]
- 37. Nicot C, Mulloy JC, Ferrari MG et al HTLV‐1 p12(I) protein enhances STAT5 activation and decreases the interleukin‐2 requirement for proliferation of primary human peripheral blood mononuclear cells. Blood 2001; 98: 823–9. [DOI] [PubMed] [Google Scholar]
- 38. Johnson JM, Nicot C, Fullen J et al Free major histocompatibility complex class I heavy chain is preferentially targeted for degradation by human T‐cell leukemia/lymphotropic virus type 1 p12(I) protein. J Virol 2001; 75: 6086–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fukumoto R, Dundr M, Nicot C et al Inhibition of T‐cell receptor signal transduction and viral expression by the linker for activation of T cells‐interacting p12(I) protein of human T‐cell leukemia/lymphoma virus type 1. J Virol 2007; 81: 9088–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ding W, Albrecht B, Kelley RE et al Human T‐cell lymphotropic virus type 1 p12(I) expression increases cytoplasmic calcium to enhance the activation of nuclear factor of activated T cells. J Virol 2002; 76: 10374–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Franchini G, Mulloy JC, Koralnik IJ et al The human T‐cell leukemia/lymphotropic virus type I p12I protein cooperates with the E5 oncoprotein of bovine papillomavirus in cell transformation and binds the 16‐kilodalton subunit of the vacuolar H+ ATPase. J Virol 1993; 67: 7701–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Albrecht B, D'Souza CD, Ding W, Tridandapani S, Coggeshall KM, Lairmore MD. Activation of nuclear factor of activated T cells by human T‐lymphotropic virus type 1 accessory protein p12(I). J Virol 2002; 76: 3493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Van Prooyen N, Gold H, Andresen V et al Human T‐cell leukemia virus type 1 p8 protein increases cellular conduits and virus transmission. Proc Natl Acad Sci USA 2010; 107: 20738–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Johnson JM, Mulloy JC, Ciminale V, Fullen J, Nicot C, Franchini G. The MHC class I heavy chain is a common target of the small proteins encoded by the 3′ end of HTLV type 1 and HTLV type 2. AIDS Res Hum Retroviruses 2000; 16: 1777–81. [DOI] [PubMed] [Google Scholar]
- 45. Tsukahara T, Ratner L. Substitution of HIV Type 1 Nef with HTLV‐1 p12. AIDS Res Hum Retroviruses 2004; 20: 938–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nicot C, Dundr M, Johnson JM et al HTLV‐1‐encoded p30II is a post‐transcriptional negative regulator of viral replication. Nat Med 2004; 10: 197–201. [DOI] [PubMed] [Google Scholar]
- 47. Younis I, Khair L, Dundr M, Lairmore MD, Franchini G, Green PL. Repression of human T‐cell leukemia virus type 1 and type 2 replication by a viral mRNA‐encoded posttranscriptional regulator. J Virol 2004; 78: 11077–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Baydoun HH, Pancewicz J, Nicot C. Human T‐lymphotropic type 1 virus p30 inhibits homologous recombination and favors unfaithful DNA repair. Blood 2011; 117: 5897–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. D'Agostino DM, Zotti L, Ferro T et al Expression and functional properties of proteins encoded in the x‐II ORF of HTLV‐I. Virus Res 2001; 78: 35–43. [DOI] [PubMed] [Google Scholar]
- 50. Andresen V, Pise‐Masison CA, Sinha‐Datta U et al Suppression of HTLV‐1 replication by Tax‐mediated rerouting of the p13 viral protein to nuclear speckles. Blood 2011; 118: 1549–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gaudray G, Gachon F, Basbous J, Biard‐Piechaczyk M, Devaux C, Mesnard JM. The complementary strand of the human T‐cell leukemia virus type 1 RNA genome encodes a bZIP transcription factor that down‐regulates viral transcription. J Virol 2002; 76: 12813–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cavanagh MH, Landry S, Audet B et al HTLV‐I antisense transcripts initiating in the 3′LTR are alternatively spliced and polyadenylated. Retrovirology 2006; 3: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Murata K, Hayashibara T, Sugahara K et al A novel alternative splicing isoform of human T‐cell leukemia virus type 1 bZIP factor (HBZ‐SI) targets distinct subnuclear localization. J Virol 2006; 80: 2495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Koiwa T, Hamano‐Usami A, Ishida T et al 5′‐long terminal repeat‐selective CpG methylation of latent human T‐cell leukemia virus type 1 provirus in vitro and in vivo . J Virol 2002; 76: 9389–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Halin M, Douceron E, Clerc I et al Human T‐cell leukemia virus type 2 produces a spliced antisense transcript encoding a protein that lacks a classic bZIP domain but still inhibits Tax2‐mediated transcription. Blood 2009; 114: 2427–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sprent J, Tough DF. Lymphocyte life‐span and memory. Science 1994; 265: 1395–400. [DOI] [PubMed] [Google Scholar]
- 57. Grassmann R, Berchtold S, Radant I et al Role of human T‐cell leukemia virus type 1 X region proteins in immortalization of primary human lymphocytes in culture. J Virol 1992; 6: 4570–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yoshida M. Multiple viral strategies of HTLV‐1 for dysregulation of cell growth control. Annu Rev Immunol 2001; 19: 475–96. [DOI] [PubMed] [Google Scholar]
- 59. Teshigawara K, Maeda M, Nishino K et al Adult T leukemia cells produce a lymphokine that augments interleukin 2 receptor expression. J Mol Cell Immunol 1985; 2: 17–26. [PubMed] [Google Scholar]
- 60. Inoue J, Seiki M, Taniguchi T, Tsuru S, Yoshida M. Induction of interleukin 2 receptor gene expression by p40x encoded by human T‐cell leukemia virus type 1. EMBO J 1986; 5: 2883–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hanon E, Goon P, Taylor GP et al High production of interferon gamma but not interleukin‐2 by human T‐lymphotropic virus type I‐infected peripheral blood mononuclear cells. Blood 2001; 98: 721–6. [DOI] [PubMed] [Google Scholar]
- 62. Greene WC, Leonard WJ, Wano Y et al Trans‐activator gene of HTLV‐II induces IL‐2 receptor and IL‐2 cellular gene expression. Science 1986; 232: 877–80. [DOI] [PubMed] [Google Scholar]
- 63. Niinuma A, Higuchi M, Takahashi M et al Aberrant activation of the interleukin‐2 autocrine loop through the nuclear factor of activated T cells by nonleukemogenic human T‐cell leukemia virus type 2 but not by leukemogenic type 1 virus. J Virol 2005; 79: 11925–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Albrecht B, Collins ND, Burniston MT et al Human T‐lymphotropic virus type 1 open reading frame I p12(I) is required for efficient viral infectivity in primary lymphocytes. J Virol 2000; 74: 9828–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Collins ND, Newbound GC, Albrecht B, Beard JL, Ratner L, Lairmore MD. Selective ablation of human T‐cell lymphotropic virus type 1 p12I reduces viral infectivity in vivo . Blood 1998; 91: 4701–7. [PubMed] [Google Scholar]
- 66. Valeri VW, Hryniewicz A, Andresen V et al Requirement of the human T‐cell leukemia virus p12 and p30 products for infectivity of human dendritic cells and macaques but not rabbits. Blood 2010; 116: 3809–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hiraragi H, Kim SJ, Phipps AJ et al Human T‐lymphotropic virus type 1 mitochondrion‐localizing protein p13(II) is required for viral infectivity in vivo . J Virol 2006; 80: 3469–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ye J, Silverman L, Lairmore MD, Green PL. HTLV‐1 Rex is required for viral spread and persistence in vivo but is dispensable for cellular immortalization in vitro . Blood 2003; 102: 3963–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Arnold J, Yamamoto B, Li M et al Enhancement of infectivity and persistence in vivo by HBZ, a natural antisense coded protein of HTLV‐1. Blood 2006; 107: 3976–82. [DOI] [PMC free article] [PubMed] [Google Scholar]


