Abstract
Molecular abnormalities involved in the multistep leukemogenesis of adult T‐cell leukemia (ATL) remain to be clarified. Based on our integrated database, we focused on the expression patterns and levels of Ikaros family genes, Ikaros,Helios, and Aiolos, in ATL patients and HTLV‐1 carriers. The results revealed profound deregulation of Helios expression, a pivotal regulator in the control of T‐cell differentiation and activation. The majority of ATL samples (32/37 cases) showed abnormal splicing of Helios expression, and four cases did not express Helios. In addition, novel genomic loss in Helios locus was observed in 17/168 cases. We identified four ATL‐specific short Helios isoforms and revealed their dominant‐negative function. Ectopic expression of ATL‐type Helios isoform as well as knockdown of normal Helios or Ikaros promoted T‐cell growth. Global mRNA profiling and pathway analysis showed activation of several signaling pathways important for lymphocyte proliferation and survival. These data provide new insights into the molecular involvement of Helios function in the leukemogenesis and phenotype of ATL cells, indicating that Helios deregulation is one of the novel molecular hallmarks of ATL.
Adult T‐cell leukemia (ATL) is a highly aggressive malignancy of mature CD4+ T cells and is caused by HTLV‐1. After HTLV‐1 infection, ATL is thought to develop following a multitude of events, including both genetic and epigenetic changes in the cells. Although many aspects of HTLV‐1 biology have been elucidated, the detailed molecular mechanism of ATL leukemogenesis remains largely unknown.1, 2 Therefore, to precisely define the comprehensive abnormalities associated with ATL leukemogenesis, we previously carried out global mRNA and miRNA profiling of ATL cells derived from a large number of patients.3, 4 In this study, we focused on Ikaros family genes, especially Helios, on the basis of our integrated profiling of expression and gene copy number in ATL cells, which revealed the deregulated expression of this family of genes and genomic loss of Helios locus.
Ikaros family genes are specifically expressed in the hematopoietic system and play a vital role in regulation of lymphoid development and differentiation.5, 6, 7, 8, 9, 10, 11 In addition, they are known to function as tumor suppressors during leukemog‐enesis according to several genetic studies carried out in mouse models.12, 13, 14, 15 Recently, many studies reported the deregulated splicing of Ikaros and the deletion of Ikaros locus in several human leukemias.16, 17, 18, 19, 20, 21, 22, 23 These abnormalities are associated with poor prognoses.24, 25, 26, 27 Helios is mainly expressed in the T‐cell lineage.10, 11 Genomic changes and abnormal expression of Helios are also observed in some patients with T‐cell malignancies.18, 28, 29, 30, 31 However, in contrast to Ikaros, the substantial impact of aberrant Helios expression remains to be elucidated because of the absence of functional information, including the target genes of Helios.
In this study, we carried out a detailed expression analysis of Ikaros family genes in a large panel of clinical samples from ATL patients and HTLV‐1 carriers and consequently identified a novel molecular characteristic, that is, abnormal splicing of Helios and loss of expression, which seems to be a significant key factor in leukemogenesis affecting the regulation of T‐cell proliferation.
Materials and Methods
Cell lines and clinical samples
HeLa and 293T cells were cultivated in DMEM supplemented with 10% FCS. Human leukemic T cells, Jurkat, Molt‐4, and CEM, ATL‐derived, MT‐1 and TL‐Om1, and HTLV‐1‐infected MT‐2 and Hut‐102 cell lines were all maintained in RPMI‐1640 with 10% FCS. The PBMCs from ATL patients of four clinical subtypes32 and healthy volunteers were a part of those collected with informed consent as a collaborative project of the Joint Study on Prognostic Factors of ATL Development. The project was approved by the Institute of Medical Sciences, University of Tokyo Human Genome Research Ethics Committee (Tokyo, Japan). Clinical information of ATL individuals is provided in Table S1.
RNA isolation and RT‐PCR analysis
The preparation of total RNA and synthesis of the first strand of cDNA were described previously.3 The mRNAs of Ikaros family genes were examined by PCR with Platinum Taq DNA Polymerase High Fidelity (Invitrogen, Carlsbad, CA, USA). The PCR products were sequenced by automated DNA sequencer. Nested PCR amplification was carried out with diluted full‐length PCR products by Accuprime Taq DNA polymerase High Fidelity (Invitrogen). Quantitative PCR was carried out as previously described.3 The specific primer sets for each PCR are described in Table S2.
Immunoblot analysis
Cells were collected, washed with PBS, and lysed with RIPA buffer. For immunoprecipitation, cells were lysed with TNE buffer and incubated with specific antibody. Proteins samples were then analyzed by immunoblots with specific antibodies: anti‐tubulin, anti‐Ikaros, and anti‐Helios antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Mouse anti‐FLAG antibody (M2) was from Sigma‐Aldrich (St. Louis, MO, USA). Rabbit polyclonal anti‐HA antibody was from MBL (Nagoya, Japan). Anti‐mouse, rabbit, and goat secondary antibodies were from Promega (Fitchburg, WI, USA).
Immunostaining
HeLa cells were cultured on coverslip slides and transfected with the indicated expression vectors by Lipofectamine LTX (Invitrogen). At 24 h post transfection, cells were washed three times with PBS, fixed in 4% paraformaldehyde, and permeabilized with 0.1% Triton X‐100. Then, cells were stained with primary antibodies (diluted 1:500 to 1:2000). Alexa‐488 or 546‐conjugated secondary antibodies (Molecular Probes, Life Technologies, Carlsbad, CA, USA) were used for detection of specific targets, and DAPI was used for nuclear staining. Images were acquired by using a Nikon A1 confocal microscope (Nikon, Tokyo, Japan).
Electrophoretic mobility‐shift assay
Experimental conditions and detail methods were previously reported.3 For evaluation of DNA binding activity, 3–5 μg nuclear extracts from each transfectant were used per each lane of electrophoresis. The oligonucleotide sequences used as a probe are provided in Table S2.
Luciferase assay
The pGL4.10‐firefly vector (Promega) containing Hes1 promoter was used as a reporter vector and RSV‐renilla vector was used as a control vector. HeLa cells were transiently transfected with these reporters and each Ikaros or/and Helios expression vector by Lipofectamine 2000 reagent (Invitrogen). The luciferase activities were quantified by the Dual‐Luciferase Reporter Assay System (Promega) at 24 h post‐transfection.
Retroviral construction and transduction
The FLAG‐Hel‐5 cDNA sequence was subcloned into retrovirus vector pRx‐puro. Stable cell populations expressing Hel‐5 were selected by puromycin. The shRNA‐expressing retroviral vectors and virus production procedures have been established.3 The shRNA sequences are listed in Table S2. Stable cell populations were obtained by puromycin or G418 selection.
Proliferation assays
Cells (0.5 or 1.0 × 104) were plated in 96‐well plates with media supplemented with 10% or 0.2% FCS. The cell numbers were evaluated for 4 days by Cell Counting Kit‐8 (Dojindo, Kumamoto, Japan). The averages of at least three independent experiments are shown.
Gene expression microarray analyses
Gene expression microarray used the 4 × 44K Whole Human Genome Oligo Microarray (Agilent Technologies, Santa Clara, CA, USA); detailed methods were previously reported.3 Coordinates have been deposited in the Gene Expression Omnibus database with accession numbers GSE33615 (gene expression microarray), GSE33602 (copy number analyses), and GSE41796 (Jurkat models).
Results
Abnormal expression of short Helios transcripts in primary ATL cells
To characterize the gene expression signature in primary ATL cells, we previously carried out mRNA microarray analyses on a large number of samples. The comprehensive survey unveiled deregulated expression of Ikaros family genes; transcription levels of Ikaros and Aiolos were downregulated in ATL samples, whereas Helios was upregulated (Fig. S1). Thus, we examined the detailed expression patterns and levels of Ikaros family members in PBMCs derived from a panel of ATL patients and HTLV‐1 carriers (Fig. 1a). Compared with control PBMCs from normal volunteers (Fig. 1b), the expression levels of Ikaros and Aiolos seemed to be downregulated in ATL samples, consistent with our microarray results. However, there were obvious abnormalities in the expression patterns of Helios. The main isoform of Helios was changed from full‐length Hel‐1 to Hel‐2, which lacks exon 3 that contains the first N‐terminal zinc finger in the DNA‐binding domain. In addition, four ATL‐specific Helios short transcripts were identified (Fig. 1c). Among them, Hel‐5 and Hel‐6 have been reported to be expressed in ATL.29 We also identified two novel variants, Hel‐v1 that lacks exons 3 and 4 and Hel‐v2 that lacks exons 2, 3, and 6. These abnormal Helios variants were also expressed in the samples of high‐risk HTLV‐1 carriers, who subsequently developed ATL in the next few years. Furthermore, nested PCR revealed that Hel‐5 or Hel‐6 were expressed in a majority of ATL samples (17/22 acute cases, 10/10 chronic cases, and 5/5 smoldering cases; total, 32/37 cases) (Fig. 1d, upper panels), whereas Hel‐v1 was expressed only in limited cases of ATL (Fig. 1d, lower panels). In four cases, Helios was not expressed. Collectively, our mRNA analysis showed that Helios expression was generally deregulated in ATL cells.
Figure 1.

(On the next page) Abnormal expression of Helios mRNA in primary adult T‐cell leukemia (ATL) cells. (a) Expression analysis of Ikaros family genes in PBMCs by full‐length RT‐PCR (Acute, n = 22; Chronic, n = 10; Smoldering, n = 5; HTLV‐1 carriers, n = 5; High‐risk carriers, n = 4). To detect and distinguish alternative splicing variants, PCR analyses were carried out with the sense and antisense primer sets designed in the first and final exons of each full‐length transcript of Ikaros family genes. Obtained cDNAs were cloned and their sequences were analyzed. The samples acute #4, 4', and 4'' were derived from the same patient, but were studied independently. (b) Expression of Ikaros family genes in PBMCs from normal volunteers (n = 10). (c) Schematic representation of Hel‐1, Hel‐2, and ATL‐type Helios isoforms identified in this study. Hel‐variant 1 (Hel‐v1) and Hel‐variant 2 (Hel‐v2) are novel isoforms in ATL. Arrows indicate primer locations of full‐length PCR for Helios. Ex, exon; F1–F6, functional zinc‐finger domains. (d) Nested PCR with specific primer sets, which were designed at exon junction of exon 1–5 or exon 2–5 for detection of Hel‐5 and Hel‐6 (upper panel), or detection of Hel‐v1 (lower panel), respectively. Arrows indicate primer locations.
Genomic abnormalities at the Helios locus in primary ATL cells
To investigate the Helios locus in ATL, we retrieved data from our gene copy number analysis3 and found that specific genomic deletion was accumulated at the Helios locus in ATL samples (17/168 cases, Fig. 2). All 17 cases were aggressive‐type ATL (12/17 lymphoma types and 5/17 acute types). Furthermore, we found that two acute ATL cases in Figure 1(a) (#9 and #14), which showed severely deregulated or lost Helios expression, had a genomic deletion of the Helios locus.
Figure 2.
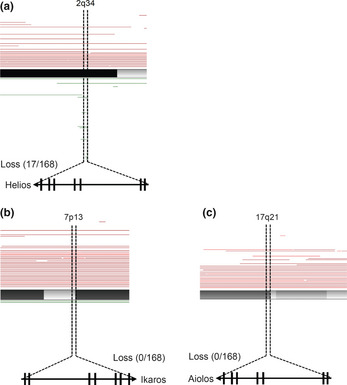
Genetic abnormalities in Helios locus in primary adult T‐cell leukemia cells. The results of our copy number analyses3 (total number, n = 168; acute type, n = 35; chronic type, n = 41; lymphoma type, n = 44; smoldering type, n = 10; intermediate, n = 1; unknown diagnosis, n = 37). Tumor‐associated deletion of Helios region (17/168) was detected (a). No specific genomic losses were observed in Ikaros (b) or Aiolos loci (c). Recurrent genetic changes are depicted by horizontal lines based on Copy Number Analyser for GeneChip output of the single nucleotide polymorphism array analysis.
Dimerization ability of ATL‐type Helios isoforms with wild‐type Helios or Ikaros
Consistent with a previously published report,33 co‐immunoprecipitation analyses confirmed that wild‐type Hel‐1 formed homodimers with themselves and heterodimers with wild‐type Ikaros (Ik‐1) protein (Fig. 3a, top panel, lane 1 and lane 4). In contrast, the dimerization activity of another artificial Helios mutant (Hel‐ΔC), which lacks the dimerization domain at the C‐terminal region, was dramatically declined (Fig. 3b, top panel, lane 1 and lane 4). We confirmed that all ATL‐type Helios proteins could interact with Hel‐1 and Ik‐1, despite the fact that all of them lack various sets of the N‐terminal exons (Fig. 3c–f).
Figure 3.

Dimerization ability of adult T‐cell leukemia (ATL)‐type Helios isoforms. In vitro dimerization assays by co‐immunoprecipitation between ATL‐type Helios and wild‐type Helios or Ikaros proteins. 293T cells were transfected with the indicated combination of expression vectors and subjected to co‐immunoprecipitation analyses (top panels). Arrowheads indicate the complex of FLAG and HA‐tagged proteins. Middle and bottom panels show the input samples. Hel‐1 (a) and Hel‐ΔC (b) included as positive and negative controls, respectively. ATL‐specific isoforms, Hel‐5 (c), Hel‐6 (d), Hel‐v1 (e), and Hel‐v2 (f) were tested. IB, immunoblot; IP, immunoprecipitant.
Cytoplasmic localization of ATL‐type Helios isoforms lacking exon 6
Ectopically expressed Hel‐1 and Ik‐1 were localized in the nucleus (Fig. 4a, top two panels). Regarding the ATL‐type Helios isoforms, we found that Hel‐5 and Hel‐v1 were localized in the nucleus, whereas Hel‐6 and Hel‐v2, both of which lack exon 6, were substantially localized in the cytoplasm (Fig. 4a, middle four panels). We also confirmed the cytoplasmic localization of Hel‐Δexon 6, which is an artificial Helios mutant lacking only exon 6 (Fig. 4a, bottom panel). Thus, exon 6 appears to be critical for nuclear localization of Helios proteins. Furthermore, defect of exon 6 led to disruption of the cellular localization of binding partners. When Hel‐6 or Hel‐v2 were co‐expressed with Hel‐1 or Ik‐1, they were co‐localized in the cytoplasm (Fig. 4b, Fig. S2).
Figure 4.
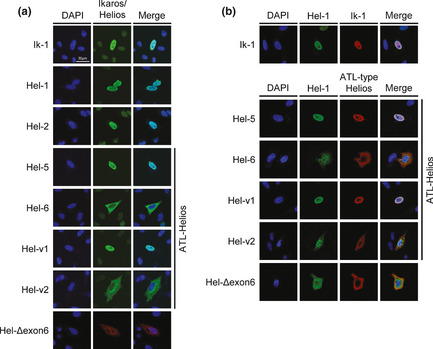
Subcellular localization of adult T‐cell leukemia (ATL)‐type Helios isoforms. Immunostaining analyses of Helios and Ikaros proteins. HeLa cells were transfected with each individual expression vector (a) or the indicated combination of expression vectors (b). Each protein was visualized with anti‐FLAG (green) or anti‐HA antibodies (red). Nuclei were detected by DAPI staining (blue). Colocalization between Ik‐1 and ATL‐type Helios was shown in Fig. S2. Hel‐v1, Hel‐variant 1; Hel‐v2, Hel‐variant 2.
Dominant‐negative function of ATL‐type Helios isoforms against wild‐type Helios and Ikaros
We next examined the functional aspects of these ATL‐type Helios isoforms by evaluating their DNA‐binding capacities. For EMSA, we used an oligonucleotide probe derived from the promoter region of human Hes1, which was a direct target of Ikaros.34, 35 Ectopically expressed Hel‐1 or Ik‐1 could bind human Hes1 promoter DNA (Fig. 5a). Supershift assays confirmed the binding specificity (Fig. 5b). In contrast, all ATL‐type Helios isoforms did not show any specific binding to the Hes1 promoter (Fig. 5a). This impossibility of specific DNA binding of ATL‐type Helios was confirmed with another independent DNA probe, IkBS433, 36 (data not shown). In addition, it was found in co‐expression experiments that Hel‐5 had antagonistic effects on the DNA binding capacity of Ik‐1 in a dose‐dependent manner (Fig. 5c). Reporter assays showed that Hel‐1 and Ik‐1 suppressed Hes1 promoter activity. However, ATL‐type Helios isoforms did not show any suppressive activity, and actually slightly activated the promoter (Fig. 5d). Furthermore, they also inhibited the suppressive function of Hel‐1 and Ik‐1 in a dose‐dependent manner (Fig. 5e, Fig. S3). These data clearly indicate that ATL‐type Helios isoforms are functionally defective because of a DNA binding deficiency and act dominant‐negatively in transcriptional suppression induced by Hel‐1 or Ik‐1. We also confirmed that Hel‐2, which lacks only exon 3 and is a major isoform in ATL cells, did not possess suppressive activity against Hes1 promoter in spite of having binding activity (Fig. 5a,d).
Figure 5.

Dominant‐negative function of adult T‐cell leukemia (ATL)‐type Helios isoforms. (a) DNA‐binding activities of wild‐type Helios or Ikaros and ATL‐type Helios proteins. Each FLAG‐tagged Helios or Ikaros isoforms were ectopically expressed in 293T cells and their nuclear extracts were subjected to EMSA with a [γ‐32P]‐labeled Hes1 promoter probe. Oct‐1 probe was used as an internal control. Arrowheads indicate Helios or Ikaros complexes. *Non‐specific bands. Hel‐v1, Hel‐variant 1; Hel‐v2, Hel‐variant 2. (b) Results of supershift assays. Anti‐FLAG (0, 0.5, 1 μg) or control IgG (1 μg) antibodies were added to each nuclear extract prior to electrophoresis. The black and white arrowheads indicate the supershifted bands of Ik‐1 and Hel‐1, respectively. (c) Antagonistic effects of Hel‐5 on DNA‐binding of Ik‐1 tested by EMSA. The molar ratios of Ik‐1 to Hel‐5 plasmids are 1:1, 1:4, and 1:8. Expression levels of FLAG‐Ik‐1 and HA‐Hel‐5 were assessed by immunoblotting. The arrowheads indicate the Ik‐1 specific band. AP‐1 probe was used as an internal control. WB, western blot. (d) Transcriptional suppression activities of various Helios or Ikaros isoforms tested by Hes1 promoter‐luciferase reporter systems (n = 3, mean ± SD). Basal Hes1 promoter activity was defined as firefly/renilla ratio, and suppression activities of Helios or Ikaros are relatively presented. Statistical significance was evaluated by unpaired Student's t‐test (*P < 0.05; **P < 0.01). (e) Inhibitory function of Hel‐5 against Ik‐1 and Hel‐1 tested by Hes1 promoter assay (n = 3, mean ± SD). The molar ratios of Ik‐1 or Hel‐1 to Hel‐5 plasmids are 1:1, 1:2, and 1:3. Relative luciferase activities were defined as firefly/renilla ratio.
Major ATL‐type Helios variant, Hel‐5, promotes T cell growth
Given the tumor‐suppressive roles of Ikaros family members,12, 13, 14, 15 it was expected that abnormal splicing of Helios could contribute to T cell leukemogenesis. The mRNA level of Helios was significantly downregulated in ATL‐related cell lines compared with that in T‐cell lines without HTLV‐1 (Fig. 6a, Fig. S4). Moreover, Helios protein was not detected in any ATL‐derived or HTLV‐1‐infected cell lines used in this study (Fig. 6b). In contrast, the expression levels of Ikaros mRNA did not show major differences between HTLV‐1‐infected and uninfected T‐cell lines. Those of Aiolos were low in most cell lines irrespective of HTLV‐1 infection (Fig. 6a, Fig. S4). Ikaros protein was detected in all T‐cell lines used in this study (Fig. 6b). To elucidate the cellular effects of the expression of dominant‐negative ATL‐type Helios isoforms in T cells, we established stable Jurkat cells expressing Hel‐5 (Fig. 6c). A cell proliferation assay confirmed that Hel‐5 expression significantly promoted Jurkat cell proliferation (Fig. 6d). To examine whether the cellular effect of Hel‐5 was due to its dominant‐negative function against Hel‐1 and Ik‐1, we carried out further knockdown analyses with specific shRNAs (Fig. 6e). The results showed that knockdown of wild‐type Helios or Ikaros led to enhanced cell growth (Fig. 6f), which was consistent with the results of enforced Hel‐5 expression. These results collectively suggested that counteraction of Ikaros or Helios by dominant‐negative isoforms contributed to T cell growth.
Figure 6.

Hel‐5 functions in T cell growth and survival. (a) Expression patterns and levels of Ikaros family genes in various cell lines examined by RT‐PCR. ATL, adult T‐cell leukemia; T‐ALL, acute T lymphoblastic leukemia. (b) Results of immunoblotting analyses of the immunoprecipitants (top panel) and cell lysates (lower panels). Positive control (p.c.), Hel‐1 transfectant. IB, immunoblot; IP, immunoprecipitant. (c) Establishment of Jurkat cells stably expressing Hel‐5. The Hel‐5 level was quantified by quantitative RT‐PCR (top, n = 3, mean ± SD) and immunoblotting (bottom). (d) Cell proliferation analysis of control cells (▲) and Hel‐5‐expressing Jurkat cells (■) under two FCS conditions (n = 3, mean ± SD). Statistical significance was observed (*P <0.01, Student's t‐test). (e) Knockdown analyses of Helios or Ikaros in Jurkat cells. The Helios and Ikaros levels were evaluated by quantitative RT‐PCR (top, n = 3, mean ± SD) and immunoblotting (bottom), respectively. (f) Cell proliferation curves of scrambled shRNA (shSc) cells (▲), shIkaros (shIk) cells (●), and shHelios (shHel) cells (■) were examined in two FBS conditions (n = 3, mean ± SD; *P < 0.01).
Helios deficiency causes expression of various genes in T cells
We globally searched mRNA expression changes using microarray analysis of Jurkat cells expressing Hel‐5 and those of knocked‐down Helios or Ikaros (Fig. 7a,b). The results clearly showed differentially expressed gene sets between the trans‐formants and control cells (Fig. 7c). Furthermore, pathway analysis37 of each upregulated gene set identified activation of several signaling cascades. In particular, we focused on six common pathways identified in both Hel‐5 transduced and Helios or Ikaros knocked‐down Jurkat cells (Fig. 7d). These pathways are important for various T cell regulations, for example, cell growth, apoptosis resistance, and migration activity. Among these pathways, it has not been reported that the shingosine‐1‐phosphate (S1P) pathway is regulated by the Ikaros family. We confirmed overexpressed S1PR1 and S1PR3, which are critical receptors for the activation of the S1P pathway, in manipulated Jurkat samples (Fig. 7e).
Figure 7.

Comprehensive search for Helios target genes by microarray analysis. (a,b) Gene expression analysis of Jurkat stable cells. The gene expression patterns of Jurkat cells expressing Hel‐5 (n = 3), shIkaros (n = 3), and shHelios (n = 3) were comprehensively analyzed by microarray technique. The obtained 2D hierarchical clusters and Pearson's correlation between the cells expressing Hel‐5 or not (a) and the cells introducing shHel, shIk, or shSc (b). (c) Venn diagram of differential gene expression pattern in the Jurkat sublines. The each differential expression gene set (5‐fold changes, P < 1 × 10−5) was compared. (d) Venn diagram depicting the overlap between the outputs of pathway analysis in Jurkat sublines. The analysis was based on the NCI‐Nature Pathway Interaction Database.37 Each differential pathway set (t‐test, P < 0.01) was compared and the common pathways listed. (e) Results of quantitative RT‐PCR of shingosine‐1‐phosphate receptor 1 (S1PR1) and receptor 3 (S1PR3) in Jurkat sublines (n = 3, mean ± SD). HDAC, histone deacetylase; VEGFR, vascular endothelial growth factor receptor.
Discussion
In the present study, on the basis of the integrated analysis of ATL cells using our biomaterial bank in Japan, we revealed a novel molecular characteristic of ATL cells, which is a profound abnormality in the expression of Helios. The abnormal alternative splicing and, in some cases, loss of Helios expression appear to be a part of the basis for advantageous cell growth and survival in ATL cells. We also showed the tumor‐suppressive function and target genes, as well as pathways of Helios, in mature human T cells.
Characterization of Ikaros family members revealed profound abnormalities in Helios expression in ATL cells: (i) biased and increased expression of alternatively spliced variants; (ii) suppression of Hel‐1 expression; (iii) lack of Helios expression in some cases; and (iv) frequent genomic defects of the Helios locus. Our results also revealed that alternatively spliced Helios variants are expressed in PBMCs of HTLV‐1 carriers, suggesting that the abnormal splicing of Helios may occur in HTLV‐1‐infected cells at the carrier state until progression to leukemia development. However, the genomic deletions appear to be one of the important genetic events during the latter stages of leukemia development, as they were observed only in aggressive subtypes of ATL.
The structural characteristics of the ATL‐type Helios variants involve a selective lack of one or more zinc fingers in the N‐terminal domain. The results of this study indicated that these variant proteins lost DNA binding activity, whereas the capacity of dimerization was preserved. Therefore, these variant proteins hindered transcriptional activities of Ikaros family proteins, showing dominant‐negative effects. In addition, a part of ATL‐type Helios isoform, which lacks exon 6, is linked to abnormal localization of wild‐type Helios and Ikaros. We confirmed that Helios isoforms lacking exon 6 were overexpressed in primary ATL cells (Fig. S5). Interestingly, Hel‐2 has reduced transcriptional suppressive activity compared with Hel‐1, although it can bind to the target sequence as well as Hel‐1. This is similar to a previous report,36 which noted that the activity of mouse Ik‐2 protein for the reporter gene was remarkably lower than that of Ik‐1, whereas the binding affinities of Ik‐1 and Ik‐2 were similar. The exon 3 skip occurred more frequently in ATL cells, compared to PBMCs from normal volunteers (Fig. S6). These results collectively indicate that all abnormalities of Helios expression, including loss of or decreased Hel‐1 expression and upregulated Hel‐2 and ATL‐type Helios, result in abrogation of Ikaros family functions in ATL cells.
We also confirmed that Hes1, a target gene of the Notch pathway, is one of the targets of Helios as well as Ikaros.34, 35 A recent study reported that activated Notch signaling may be important to ATL pathogenesis and that Hes1 is upregulated in ATL cells.38 Thus, we examined expression levels of Hes1 mRNA by quantitative RT‐PCR and confirmed the upregulation in our ATL samples (Fig. S7). Hes1 has been reported to directly promote cell proliferation through the transcriptional repression of p27kip1.39 Taken together, our results suggest a possibility that abnormalities in Helios expression are one of the causes of Hes1 activation, which may be one of the genetic events involved in ATL leukemogenesis.
Our results show that the Hel‐5 variant may have an oncogenic role, whereas the wild‐type Helios, Hel‐1, shows tumor suppressor‐like activity. These findings are consistent with previous findings in mice.15 Furthermore, our description of expression profiles of stable cells followed by pathway analyses showed activation of several important pathways in lymphocytes for the regulation of proliferation, survival, and others. In particular, we discovered novel molecular cross‐talk between the Ikaros family and the S1P pathway. The S1P–S1PR1 axis is known to play important roles in regulation of the immune system, apoptosis, cell cycle, and migration of lymphocytes.40, 41, 42 Recently, activation of the S1P pathway in various diseases, including leukemia, has been reported, and the therapeutic potential of S1PR1 inhibitors was suggested.42 Studies of functional roles of S1P pathway activation in ATL cells are now underway in our laboratory.
In conclusion, our present study revealed a novel aspect of molecular abnormalities in ATL cells: a profound deregulation in Helios expression, which appears to play an important role in T‐cell proliferation. Our experimental approaches also imply that, in addition to genetic and epigenetic abnormalities, ATL shows abnormal splicing, which has been observed in various human diseases including cancers.43, 44, 45
Disclosure Statement
The authors have no conflict of interest.
Supporting information
Fig. S1. Deregulated expression of Ikaros family genes in primary adult T‐cell leukemia cells.
Fig. S2. Colocalization of wild‐type Ikaros and adult T‐cell leukemia‐type Helios.
Fig. S3. Dominant‐negative inhibition of Hel‐6, Hel‐v1, and Hel‐v2 in the suppressive activities of wild‐type Helios and Ikaros.
Fig. S4. Downregulation of the expression of Helios mRNA in HTLV‐1‐positive T cell lines.
Fig. S5. Overexpression of abnormal Helios isoforms lacking exon 6 in adult T‐cell leukemia samples.
Fig. S6. Relative value of Helios transcripts skipping exon 3 to all is upregulated in primary adult T‐cell leukemia cells.
Fig. S7. Upregulated expression of Hes1 in primary adult T‐cell leukemia cells.
Table S1. Clinical characteristics of adult T‐cell leukemia patients and HTLV‐1 carriers.
Table S2. Primer list and probe sequences.
Acknowledgments
We thank Mr. M. Nakashima and Ms. T. Akashi for support and maintenance of the Joint Study on Prognostic Factors of ATL Development. This work is supported by JSPS KAKENHI Grant Numbers 24790436 (M.Y.), 23390250 (T.W.), 23659484 (T.W.), 23・6291 (S.A.), NEXT KAKENHI Grant Number 221S0001 (T.W.), and a Grant‐in‐Aid from the Ministry of Health, Labor and Welfare of Japan H24‐G‐004 (M.Y. and T.W.).
(Cancer Sci 2013; 104: 1097–1106)
References
- 1. Yamaguchi K, Watanabe T. Human T lymphotropic virus type‐I and adult T‐cell leukemia in Japan. Int J Hematol 2002; 76: 240–45. [DOI] [PubMed] [Google Scholar]
- 2. Iwanaga M, Watanabe T, Utsunomiya A et al Human T‐cell leukemia virus type I (HTLV‐1) proviral load and disease progression in asymptomatic HTLV‐1 carriers: a nationwide prospective study in Japan. Blood 2010; 116: 1211–19. [DOI] [PubMed] [Google Scholar]
- 3. Yamagishi M, Nakano K, Miyake A et al Polycomb‐mediated loss of miR‐31 activates NIK‐dependent NF‐κB pathway in adult T cell leukemia and other cancers. Cancer Cell 2012; 21: 121–35. [DOI] [PubMed] [Google Scholar]
- 4. Yamagishi M, Watanabe T. Molecular hallmarks of adult T cell leukemia. Front Microbiol 2012; 3: 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lo K, Landau NR, Smale ST. LyF‐1, a transcriptional regulator that interacts with a novel class of promoters for lymphocyte‐specific genes. Mol Cell Biol 1991; 11: 5229–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Georgopoulos K, Moore DD, Derfler B. Ikaros, an early lymphoid‐specifictranscription factor and a putative mediator for T cell commitment. Science 1992; 258: 808–12. [DOI] [PubMed] [Google Scholar]
- 7. Hahm K, Ernst P, Lo K, Kim GS, Turck C, Smale ST. The lymphoid transcription factor LyF‐1 is encoded by specific, alternatively spliced mRNAs derived from the Ikaros gene. Mol Cell Biol 1994; 14: 7111–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sun L, Liu A. Zinc finger‐mediated protein interactions modulate Ikaros activity, a molecular control of lymphocyte development. EMBO J 1996; 15: 5358–69. [PMC free article] [PubMed] [Google Scholar]
- 9. Morgan B, Sun L, Avitahl N et al Aiolos, a lymphoid restricted transcription factor that interacts with Ikaros to regulate lymphocyte differentiation. EMBO J 1997; 16: 2004–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kelley CM, Ikeda T, Koipally J et al Helios, a novel dimerization partner of Ikaros expressed in the earliest hematopoietic progenitors. Curr Biol 1998; 8: 508–15. [DOI] [PubMed] [Google Scholar]
- 11. Cobb BS, McCarty AS, Brown KE et al Helios, a T cell‐restricted Ikaros family member that quantitatively associates with Ikaros at centromeric heterochromatin. Genes Dev 1998; 12: 782–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Winandy S, Wu P, Georgopoulos K. A dominant mutation in the Ikaros gene leads to rapid development of leukemia and lymphoma. Cell 1995; 83: 289–99. [DOI] [PubMed] [Google Scholar]
- 13. Wang JH, Nichogiannopoulou A, Wu L et al Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity 1996; 5: 537–49. [DOI] [PubMed] [Google Scholar]
- 14. Wang JH, Avitahl N, Cariappa A et al Aiolos regulates B cell activation and maturation to effector state. Immunity 1998; 9: 543–53. [DOI] [PubMed] [Google Scholar]
- 15. Zhang Z, Swindle CS, Bates JT, Ko R, Cotta CV, Klug CA. Expression of a non‐DNA‐binding isoform of Helios induces T‐cell lymphoma in mice. Blood 2007; 109: 2190–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sun L, Crotty ML, Sensel M et al Expression of dominant‐negative Ikaros isoforms in T‐cell acute lymphoblastic leukemia. Clin Cancer Res 1999; 5: 2112–20. [PubMed] [Google Scholar]
- 17. Nakase K, Ishimaru F, Avitahl N et al Dominant negative isoform of the Ikaros gene in patients with adult B‐cell acute lymphoblastic leukemia. Cancer Res 2000; 60: 062–4065. [PubMed] [Google Scholar]
- 18. Takanashi M, Yagi T, Imamura T et al Expression of the Ikaros gene family in childhood acute lymphoblastic leukaemia. Br J Haematol 2002; 117: 525–30. [DOI] [PubMed] [Google Scholar]
- 19. Nishii K, Katayama N, Miwa H. Non‐DNA‐binding Ikaros isoform gene expressed in adult B‐precursor acute lymphoblastic leukemia. Leukemia 2002; 16: 1285–92. [DOI] [PubMed] [Google Scholar]
- 20. Tonnelle C, Imbert M‐C, Sainty D, Granjeaud S, N'Guyen C, Chabannon C. Overexpression of dominant‐negative Ikaros 6 protein is restricted to a subset of B common adult acute lymphoblastic leukemias that express high levels of the CD34 antigen. Hematol J 2003; 4: 104–9. [DOI] [PubMed] [Google Scholar]
- 21. Klein F, Feldhahn N, Herzog S et al BCR‐ABL1 induces aberrant splicing of IKAROS and lineage infidelity in pre‐B lymphoblastic leukemia cells. Oncogene 2006; 25: 1118–24. [DOI] [PubMed] [Google Scholar]
- 22. Zhou F, Mei H, Jin R, Li X, Chen X. Expression of ikaros isoform 6 in chinese children with acute lymphoblastic leukemia. J Pediatr Hematol Oncol 2011; 33: 429–32. [DOI] [PubMed] [Google Scholar]
- 23. Mullighan CG, Miller CB, Radtke I et al BCR‐ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature 2008; 453: 110–14. [DOI] [PubMed] [Google Scholar]
- 24. Kano G, Morimoto A, Takanashi M et al Ikaros dominant negative isoform (Ik6) induces IL‐3‐independent survival of murine pro‐B lymphocytes by activating JAK‐STAT and up‐regulating Bcl‐xl levels. Leuk Lymphoma 2008; 49: 965–73. [DOI] [PubMed] [Google Scholar]
- 25. Iacobucci I, Lonetti A, Messa F et al Expression of spliced oncogenic Ikaros isoforms in Philadelphia‐positive acute lymphoblastic leukemia patients treated with tyrosine kinase inhibitors: implications for a new mechanism of resistance. Blood 2008; 112: 3847–55. [DOI] [PubMed] [Google Scholar]
- 26. Mullighan CG, Su X, Zhang J et al Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med 2009; 360: 470–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kuiper RP, Waanders E, van der Velden VHJ et al IKZF1 deletions predict relapse in uniformly treated pediatric precursor B‐ALL. Leukemia 2010; 24: 1258–64. [DOI] [PubMed] [Google Scholar]
- 28. Nakase K, Ishimaru F, Fujii K et al Overexpression of novel short isoforms of Helios in a patient with T‐cell acute lymphoblastic leukemia. Exp Hematol 2002; 30: 313–17. [DOI] [PubMed] [Google Scholar]
- 29. Fujii K, Ishimaru F, Tabayashi T et al Over‐expression of short isoforms of Helios in patients with adult T‐cell leukaemia/lymphoma. Br J Haematol 2003; 120: 986–9. [DOI] [PubMed] [Google Scholar]
- 30. Fujiwara SI, Yamashita Y, Nakamura N et al High‐resolution analysis of chromosome copy number alterations in angioimmunoblastic T‐cell lymphoma and peripheral T‐cell lymphoma, unspecified, with single nucleotide polymorphism‐typing microarrays. Leukemia 2008; 22: 1891–8. [DOI] [PubMed] [Google Scholar]
- 31. Fujimoto R, Ozawa T, Itoyama T, Sadamori N, Kurosawa N, Isobe M. HELIOS‐BCL11B fusion gene involvement in a t(2;14)(q34;q32) in an adult T‐cell leukemia patient. Cancer Genet 2012; 205: 356–64. [DOI] [PubMed] [Google Scholar]
- 32. Shimoyama M. Diagnostic criteria and classification of clinical subtypes of adult T‐cell leukaemia‐lymphoma. A report from the Lymphoma Study Group (1984–87). Br J Haematol 1991; 79: 428–37. [DOI] [PubMed] [Google Scholar]
- 33. Tabayashi T, Ishimaru F, Takata M et al Characterization of the short isoform of Helios overexpressed in patients with T‐cell malignancies. Cancer Sci 2007; 98: 182–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kathrein KL, Chari S, Winandy S. Ikaros directly represses the notch target gene Hes1 in a leukemia T cell line: implications for CD4 regulation. J Biol Chem 2008; 283: 10476–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kleinmann E, Geimer Le Lay AS, Sellars M, Kastner P, Chan S. Ikaros represses the transcriptional response to Notch signaling in T‐cell development. Mol Cell Biol 2008; 28: 7465–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Molnár A, Georgopoulos K. The Ikaros gene encodes a family of functionally diverse zinc finger DNA‐binding proteins. Mol Cell Biol 1994; 14: 8292–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schaefer CF, Anthony K, Krupa S et al PID: the pathway interaction database. Nucleic Acids Res 2009; 37: D674–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pancewicz J, Taylor JM, Datta A. Notch signaling contributes to proliferation and tumor formation of human T‐cell leukemia virus type 1‐associated adult T‐cell leukemia. Proc Natl Acad Sci USA 2010; 107: 16619–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Murata K, Hattori M, Hirai N et al Hes1 directly controls cell proliferation through the transcriptional repression of p27Kip1. Mol Cell Biol 2005; 25: 4262–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Maeda Y, Seki N, Sato N, Sugahara K, Chiba K. Sphingosine 1‐phosphate receptor type 1 regulates egress of mature T cells from mouse bone marrow. Int Immunol 2010; 22: 515–25. [DOI] [PubMed] [Google Scholar]
- 41. Spiegel S, Milstien S. The outs and the ins of sphingosine‐1‐phosphate in immunity. Nat Rev Immunol 2011; 11: 403–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Maceyka M, Harikumar KB, Milstien S, Spiegel S. Sphingosine‐1‐phosphate signaling and its role in disease. Trends Cell Biol 2012; 22: 50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ghigna C, Valacca C, Biamonti G. Alternative splicing and tumor progression. Curr Genomics 2008; 9: 556–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. David CJ, Manley JL. Alternative pre‐mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev 2010; 24: 2343–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Blair CA, Zi X. Potential molecular targeting of splice variants for cancer treatment. Indian J Exp Biol 2011; 49: 836–9. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Deregulated expression of Ikaros family genes in primary adult T‐cell leukemia cells.
Fig. S2. Colocalization of wild‐type Ikaros and adult T‐cell leukemia‐type Helios.
Fig. S3. Dominant‐negative inhibition of Hel‐6, Hel‐v1, and Hel‐v2 in the suppressive activities of wild‐type Helios and Ikaros.
Fig. S4. Downregulation of the expression of Helios mRNA in HTLV‐1‐positive T cell lines.
Fig. S5. Overexpression of abnormal Helios isoforms lacking exon 6 in adult T‐cell leukemia samples.
Fig. S6. Relative value of Helios transcripts skipping exon 3 to all is upregulated in primary adult T‐cell leukemia cells.
Fig. S7. Upregulated expression of Hes1 in primary adult T‐cell leukemia cells.
Table S1. Clinical characteristics of adult T‐cell leukemia patients and HTLV‐1 carriers.
Table S2. Primer list and probe sequences.


