Abstract
The epithelial–mesenchymal transition (EMT) contributes to the malignant progression of cancer cells including acquisition of the ability to undergo metastasis. However, whereas EMT‐related transcription factors (EMT‐TF) are known to play an important role in the malignant progression of epithelial tumors, their role in mesenchymal tumors remains largely unknown. We show that expression of the gene for Twist2 is downregulated in human osteosarcoma and correlates inversely with tumorigenic potential in mouse osteosarcoma. Forced expression of Twist2 in highly tumorigenic murine osteosarcoma cells induced a slight inhibition of cell growth in vitro but markedly suppressed tumor formation in vivo. Conversely, knockdown of Twist2 in osteosarcoma cells with a low tumorigenic potential promoted tumor formation in vivo, suggesting that Twist2 functions as a tumor suppressor in osteosarcoma cells. Furthermore, Twist2 induced expression of fibulin‐5, which has been reported as a tumor suppressor. Medium conditioned by mouse osteosarcoma cells overexpressing Twist2 inhibited expression of the MMP9 gene as well as invasion in mouse embryonic fibroblasts, and forced expression of Twist2 in osteosarcoma cells suppressed MMP9 gene expression in tumor tissue. Data from the present study suggest that Twist2 inhibits formation of a microenvironment conducive to tumor growth and thereby attenuates tumorigenesis in osteosarcoma.
The epithelial–mesenchymal transition (EMT) plays a central role in morphogenesis during embryonic development as well as in the malignant progression of cancer including acquisition of the metastatic phenotype.1, 2 Although the underlying molecular mechanisms are complex, the EMT is regulated by three major groups of transcription factors belonging to the Zeb, Snail and Twist families.3 Upregulation of the expression of these EMT‐inducing transcription factors (EMT‐TF) promotes tumor invasiveness and correlates with disease relapse and poor prognosis in individuals with various types of epithelial tumor.4 In addition, EMT‐TF are associated with resistance to chemotherapy and stem‐like characteristics in cancer cells.3 The EMT‐TF are implicated in the malignant progression of epithelial tumors, but their role in mesenchymal tumors remains unclear.
Osteosarcoma is the most frequent type of malignant bone tumor and primarily affects children and adolescents. Although the prognosis of individuals with osteosarcoma has improved with chemotherapy, a significant proportion of patients remains incurable as a result of the development of metastatic lesions.5, 6 While EMT‐TF are thought to contribute to the maintenance of mesenchymal features of osteosarcoma, it remains largely unknown whether they might promote metastasis and malignant behavior in this tumor type as they do in epithelial tumors. Amplification or deletion of the TWIST1 locus has been detected in pediatric osteosarcoma, with such deletion being associated with a poor clinical outcome.7, 8 Twist1 and Twist2 are highly conserved basic helix–loop–helix transcription factors that play essential roles during embryogenesis.9 Twist proteins inhibit skeletal development through direct and indirect inhibition of Runx2,10, 11 a key regulator of osteogenic differentiation. Expression of Twist1 and Twist2 in mesenchymal cells has been suggested to promote adipogenesis.12 Together, these various observations suggest that Twist proteins might play an important role in normal bone marrow stromal cells (BMSC) as well as in malignant mesenchymal tumors.
We recently established osteosarcoma‐initiating cells by introducing the c‐Myc gene into BMSC of Ink4a/Arf knockout (Ink4a KO) mice.13, 14 Mice injected with these cells develop osteosarcoma with pathological features similar to those of human osteosarcoma, including the production of osteoid and highly metastatic behavior. In addition, we established two distinct types of cell lines from the c‐Myc‐overexpressing Ink4a KO BMSC: low‐tumorigenic cells, which can differentiate into osteogenic, chondrogenic and adipogenic lineages; and high‐tumorigenic cells, which can differentiate into osteogenic and chondrogenic lineages. Moreover, we found that the tumorigenic capacity was suppressed by forced expression of peroxisome proliferator‐activated receptor γ (PPARγ), a master regulator of adipogenesis. These findings suggest that the loss of adipogenic potential is a key factor for osteosarcoma development in vivo.
We have now analyzed the expression of EMT‐TF in clinical specimens of human osteosarcoma and found that TWIST2 expression is frequently downregulated in this cancer. We further found that Twist2 inhibits tumor formation in our mouse osteosarcoma model, most likely by inducing expression of fibulin‐5, a tumor suppressor. Our results therefore implicate Twist2 as a tumor suppressor in osteosarcoma.
Materials and Methods
Cell culture
The BMSC from wild‐type (WT) or Ink4a KO mice as well as AO, AOT1, AOT2, AX, AXPP and AXPPT cells were established as previously described.13 Further details are provided in Supporting Information Data S1.
Human specimens
For the experimental use of osteosarcoma surgical samples, informed consent was obtained from patients in accordance with the Hospital Ethical Guidelines of Keio University Hospital and the National Cancer Center Hospital. Normal human BMSC were purchased from Lonza (Basel, Switzerland).
RT and real‐time PCR analysis
Total RNA preparation and real‐time PCR analysis was performed as previously described.13 The sequences of the PCR primers are shown in Table S1. Data were normalized by the corresponding amount of Gapdh mRNA. All data are means ± SD from three independent experiments.
Tumor xenograft model
All animal care and procedures were performed in accordance with the guidelines of Keio University. For xenograft experiments, cells in culture were isolated by exposure to trypsin and centrifugation, suspended in PBS and injected subcutaneously (3 × 106 cells/100 μL) into 8‐week‐old female syngeneic C57BL/6 or nude mice.
Gene expression profiling
Analysis of gene expression in human osteosarcoma specimens was performed with a GeneChip Human Genome U133 plus 2.0 array (Affymetrix, Santa Clara, CA, USA). Gene expression profiling of AX‐Mock and AX‐Twist2 cells was performed with a 3D‐Gene Chip (Toray, Tokyo, Japan) as previously described.13
Plasmid and retroviral gene transfer, knockdown of Twist2, differentiation assays, cell growth assay, invasion assay, immunohistochemistry, flow cytometry and immunoblot analysis
Details are described in Data S1.
Statistical analysis
Data were compared between groups with the Student's t test. A P‐value of <0.05 was considered statistically significant.
Results
Downregulation of TWIST2 expression in human osteosarcoma
We initially characterized the gene expression profiles for six EMT‐TF (Snail [also known as SNAI1], Slug [also known as SNAI2], Twist1, Twist2, Zeb1 and Zeb2) in 23 human osteosarcoma specimens and nine normal human BMSC with the use of microarray analysis. The expression level of ZEB2 was upregulated, whereas those of ZEB1 and TWIST2 were significantly downregulated in osteosarcoma compared with BMSC (Fig. 1). In particular, the expression level of TWIST2 was consistently and markedly attenuated in all osteosarcoma specimens compared with BMSC. Therefore we further evaluated the role of Twist2 in osteosarcoma.
Figure 1.
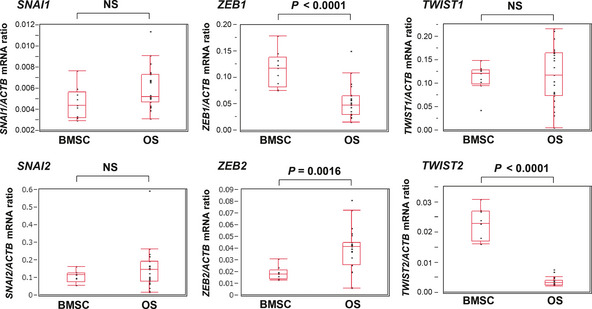
Expression of EMT‐TF genes in human osteosarcoma. Expression of EMT‐TF in 23 human osteosarcoma (OS) specimens and nine human normal bone marrow stromal cells (BMSC) was evaluated using microarray analysis. The ratio of the abundance of each mRNA to that of β‐actin (ACTB) mRNA is presented for each specimen as well as in box‐and‐whisker plots. EMT, epithelial–mesenchymal transition; NS, not significant; TF, transcription factor.
Downregulation of TWIST2 correlates with increased tumorigenic ability and loss of adipogenic differentiation potential in a mouse osteosarcoma model
We recently established two osteosarcoma cell lines by introducing the c‐Myc gene into BMSC of Ink4a KO mice.13 AO cells possess a low tumorigenic potential as well as a trilineage (adipogenic, osteogenic and chondrogenic) differentiation potential similar to that of mesenchymal stem cells (MSC). In contrast, AX cells exhibit a high tumorigenic potential as well as a bilineage (osteogenic and chondrogenic) differentiation potential similar to that of osteochondrocyte‐committed progenitor cells. Therefore we then examined the expression of Twist1 and Twist2 using real‐time PCR analysis in BMSC from WT and Ink4a KO mice as well as in AO and AX cells (Fig. 2a). Similar to the results obtained with human osteosarcoma, the expression of Twist2 was significantly downregulated in AX cells compared with that in WT and Ink4a KO BMSC or AO cells, whereas the expression of Twist1 did not differ among the four cell types. Immunohistochemical staining of AX‐derived subcutaneous tumors revealed that nuclei of GFP‐positive tumor cells were negative for Twist2, while those of the surrounding stromal cells were positive (Fig. 2b).
Figure 2.
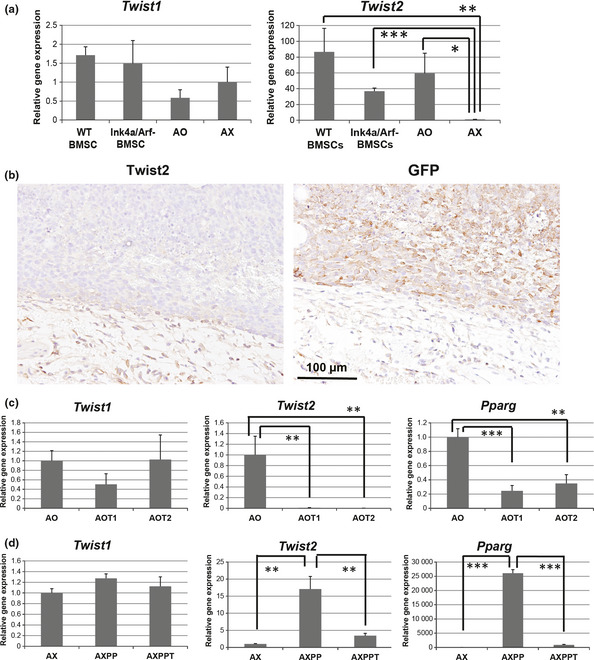
Expression of Twist2 correlates negatively with tumorigenic potential and positively with adipogenic differentiation ability in mouse osteosarcoma cells. (a) Twist1 and Twist2 expression in bone marrow stromal cells (BMSC) from WT or Ink4a/Arf knockout (Ink4a KO) mice as well as AO and AX cells. (b) Twist2 and GFP staining of serial sections of AX‐derived subcutaneous tumor. (c) Twist1,Twist2 and Pparg expression in AO, AOT1 and AOT2 cells. (d) Twist1,Twist2 and Pparg expression in AX, AXPP and AXPPT cells. *P < 0.05. **P < 0.01. ***P < 0.001.
Twist1 and Twist2 are key regulators of osteoblast differentiation.10, 11, 12, 15 Expression of Twist2 was previously shown to decrease markedly during osteoblastic differentiation of MC3T3 cells.15 We observed a marked reduction in the level of Twist2 expression during osteogenic differentiation of AO cells (Fig. S1), suggesting that Twist2 expression is associated with a differentiation state of the osteoblast lineage. We confirmed that expression of Twist2 correlates positively with adipogenic differentiation potential and negatively with tumorigenic ability with the use of two additional cell lines. We established AOT1 and AOT2 cells from independent late‐onset osteosarcomas derived from AO cells.13 AOT1 and AOT2 cells have a higher tumorigenic ability than AO cells and have lost adipogenic differentiation potential while retaining osteogenic differentiation potential. The expression level of Twist2 as well as that of the gene for PPARγ (Pparg) was greatly reduced in AOT1 and AOT2 cells compared with AO cells (Fig. 2c). In addition, the expression level of Twist2 in AXPP cells (AX cells with restored adipogenic differentiation potential due to overexpression of PPARγ) was higher than that in parental AX cells (Fig. 2d). In contrast, the expression level of Twist2 in AXPPT cells, which were established from an AXPP cell‐derived osteosarcoma13 and are highly tumorigenic, was again attenuated together with that of Pparg compared with that in AXPP cells (Fig. 2d). These results suggest that the expression of Twist2 is associated with a trilineage differentiation potential and that the downregulation of Twist2 expression is a key event in osteosarcoma tumorigenesis.
Overexpression of Twist2 attenuates tumorigenic potential in mouse osteosarcoma
To examine further the effect of Twist2 on adipogenic differentiation potential and tumorigenic ability of osteosarcoma cells, we introduced Twist2 into AX cells by retroviral gene transfer. Overexpression of Twist2 in AX cells resulted in upregulation of the mRNA for C/EBPα (Cebpa), a key regulator of adipogenesis,16, 17 whereas the expression of Pparg and adipogenic differentiation potential were not affected (Fig. 3a,c). In contrast, expression of the gene for the osteogenic marker osteocalcin (Bglap) as well as the osteogenic differentiation potential were attenuated by overexpression of Twist2 (Fig. 3b,c).
Figure 3.
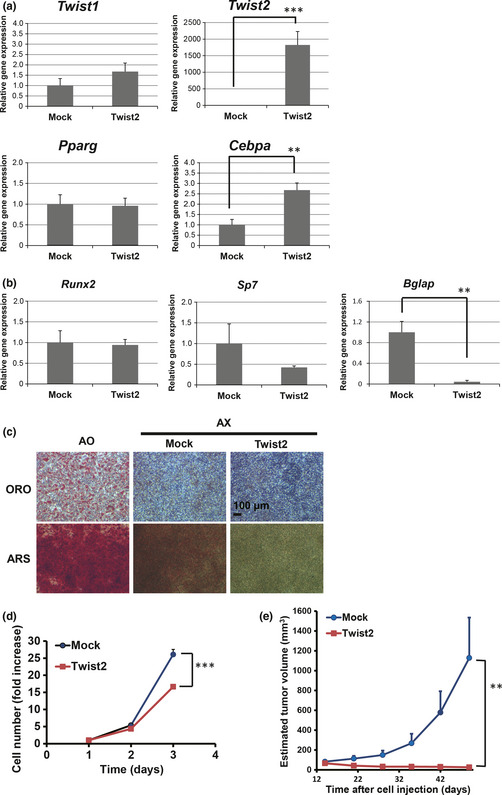
Effects of Twist2 overexpression in AX cells on differentiation and tumorigenic potential. (a) Twist1,Twist2,Pparg and Cebpa expression in AX‐Twist2 and AX‐Mock. (b) The expression of the osteogenic differentiation marker genes Runx2,Sp7 and Bglap in AX‐Mock and AX‐Twist2 cells. (c) Adipogenic and osteogenic differentiation potential in AO, AX‐Mock and AX‐Twist2 cells was evaluated with oil red O (ORO) and alizarin red S (ARS) staining, respectively. (d) Cell growth assay for AX‐Mock and AX‐Twist2 cells. (e) Growth of tumors derived from subcutaneously injected AX‐Mock or AX‐Twist2 cells in syngeneic mice. Data are means ± SEM for four tumors. **P < 0.01. ***P < 0.001.
We then determined the effect of Twist2 on the growth of osteosarcoma cells in vitro and in vivo. The cell growth of AX cells was slightly but significantly inhibited by overexpression of Twist2 (Fig. 3d), whereas overexpression of Twist2 did not affect anchorage‐independent growth and invasion in the in vitro assays (Fig. S2). In contrast, overexpression of Twist2 resulted in marked inhibition of tumorigenesis by AX cells implanted subcutaneously into syngeneic mice (Fig. 3e). Collectively, these data suggest that the expression of Twist2 did not affect the adipogenic differentiation potential but directly inhibited tumorigenic activity in mouse osteosarcoma cells.
Knockdown of Twist2 enhances the tumorigenic potential of AO cells
To further examine the effect of Twist2 on tumor formation, we introduced a shRNA specific for Twist2 mRNA into the AO cells and established three sublines by performing single‐cell cloning. The expression levels of Pparg and Cebpa as well as the adipogenic differentiation capacity were attenuated in all sublines compared with the control AO cells (Fig. 4a,b). In contrast, osteogenic marker gene expression and osteogenic differentiation potential remained intact after depletion of Twist2. These results therefore suggest that AO cells acquired a phenotype similar to that of osteoblast‐committed progenitors after knockdown of Twist2.
Figure 4.
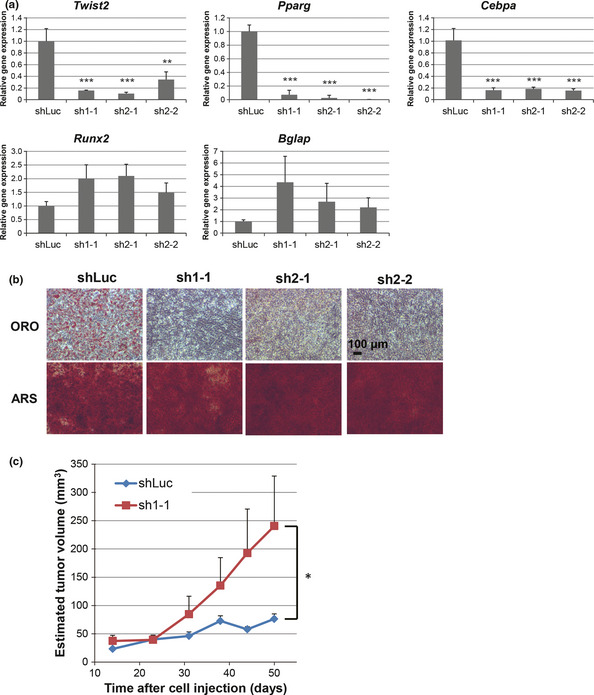
Effects of Twist2 knockdown in AO cells on differentiation and tumorigenic potential. (a) Twist2,Pparg,Cebpa,Runx2 and Bglap expression in AO cells expressing luciferase (shLuc) or Twist2 (sh1‐1, sh2‐1 and sh2‐2) shRNA. (b) Adipogenic (ORO) and osteogenic (ARS) differentiation potential of AO‐shLuc, AO‐sh1‐1, AO‐sh2‐1 and AO‐sh2‐2 cells. (c) Growth of tumors derived from subcutaneously injected AO‐shLuc or AO‐sh1‐1 cells in nude mice. Data are means ± SEM for five tumors. *P < 0.05. **P < 0.01. ***P < 0.001.
The AO cells expressing luciferase (control) or Twist2 shRNA failed to form tumors after subcutaneous implantation in syngeneic mice (data not shown). The Twist2‐depleted cells formed tumors in nude mice, but the control cells did not (Fig. 4c). These findings suggest that knockdown of Twist2 promotes tumorigenic activity in AO cells and that Twist2 plays an important inhibitory role in the growth of osteosarcoma cells in vivo.
Upregulation of Fbln5 induced by Twist2 overexpression in mouse osteosarcoma
To gain insight into the mechanism by which Twist2 suppresses tumor growth, we examined gene expression in AX cells overexpressing Twist2 and the corresponding control (AX‐Mock) cells. To investigate functional annotations enriched in upregulated genes induced in AX‐Twist2 cells, we conducted a gene‐enrichment analysis with Gene Ontology terms (gene‐GO term enrichment analysis) (Fig. 5a; the process is described in detail in Data S1 and Table S2). Consequently, we further analyzed a gene, Fbln5, which was annotated with both significant gene annotations and showed the highest fold change. Fibulin‐5 was previously shown to suppress tumor growth and metastasis through inhibition of tumor angiogenesis18, 19, 20 and the production of matrix metalloproteinase 9 (MMP9)21, respectively. We confirmed that the expression of Fbln5 in AX‐Twist2 cells was upregulated at both the mRNA and protein levels (Fig. 5b,c). Similarly, the amounts of fibulin‐5 mRNA and protein were found to be reduced in AX cells compared with BMSC or AO cells (Fig. 5b,c). Moreover, siRNA‐mediated knockdown of Twist2 in AO cells resulted in downregulation of the abundance of fibulin‐5 mRNA and protein (Fig. 5d), again suggesting that Twist2 positively regulates expression of fibulin‐5.
Figure 5.
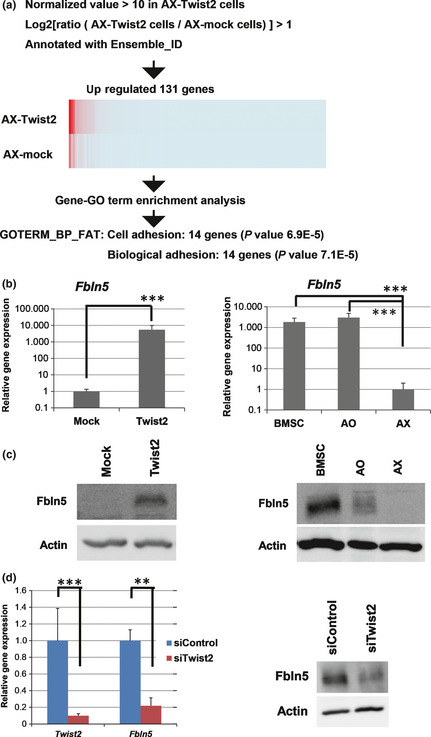
Induction of Fbln5 expression by Twist2. (a) Selection flow to identify the significant gene annotations upregulated by overexpression of Twist2. (b) Expression of Fbln5 in AX‐Mock and AX‐Twist2 cells as well as in mouse bone marrow stromal cells (BMSC), AO and AX cells. (c) Immunoblot analysis of fibulin‐5 expression in AX‐Mock and AX‐Twist2 cells as well as in mouse BMSC, AO and AX cells. β‐Actin was examined as a loading control. (d) Twist2 and Fbln5 expression as well as immunoblot analysis of fibulin‐5 in AO cells transfected with control or Twist2 siRNA. **P < 0.01. ***P < 0.001.
Overexpression of Twist2 downregulates Mmp9 expression in osteosarcoma
Overexpression of fibulin‐5 in tumor cells was previously shown to suppress metastatic organ colonization by tumor cells as well as the invasive activity of fibroblasts by blocking the induction of MMP9.21 We therefore tested whether the forced expression of Twist2 in osteosarcoma affects the expression of Mmp9 as well as the invasive ability in fibroblasts. Exposure of mouse embryonic fibroblasts (MEF) to the conditioned medium from AX‐Twist2 cells resulted in significant suppression of the expression of Mmp9 upregulated by TNF‐α treatment (Fig. 6a), as well as MEF invasion (Fig. 6b), suggesting that overexpression of Twist2 in osteosarcoma cells affects their tumor environment.
Figure 6.
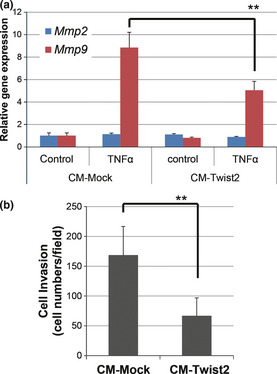
Conditioned medium of AX‐Twist2 cells inhibits Mmp9 expression and invasion in MEF. (a) The expression of Mmp2 and Mmp9 in MEF exposed for 24 h to medium conditioned by AX‐Mock or AX‐Twist2 cells with or without mouse tumor necrosis factor‐alpha (TNF‐α) was determined using real‐time PCR analysis. (b) The invasive ability of MEF treated conditioned medium (CM) of AX‐Mock or AX‐Twist2 cells with mouse TNF‐α was analyzed using a Matrigel invasion assay. Data are means ± SD from six independent experiments. **P < 0.01.
We then analyzed the expression of Mmp9 in subcutaneous tumors formed by AX‐Mock or AX‐Twist2 cells in syngeneic mice at 7 days after cell injection. The expression level of Fbln5 in cells expressing GFP encoded by the retroviral vector used to establish AX cells was greater for tumors derived from AX‐Twist2 cells than for those derived from AX‐Mock cells (Fig. 7a). Furthermore, expression of Mmp9 was downregulated in total tumor tissue derived from AX‐Twist2 cells compared with that derived from AX‐Mock cells (Fig. 7b). These data suggest that overexpression of Twist2 in tumor cells influences tumor‐associated stromal cells and thereby inhibits tumor formation in mouse osteosarcoma.
Figure 7.
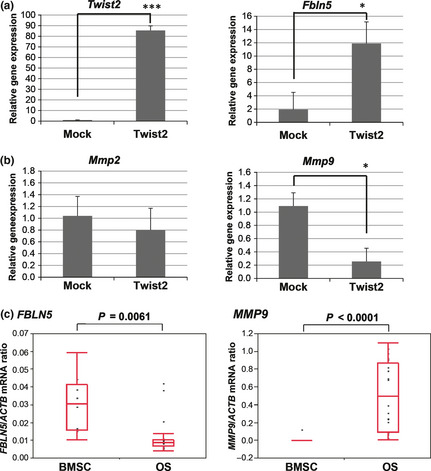
Reduced Mmp9 expression in tumors derived from AX cells overexpressing Twist2. (a) Expression of Twist2 and Fbln5 in GFP‐positive cells isolated from tumors formed in syngeneic mice at 7 days after subcutaneous injection of AX‐Mock or AX‐Twist2 cells. (b) Mmp9 expression in total tumor tissue formed as in (a). (c) The expression of FBLN5 and MMP9 in human osteosarcoma (OS) specimens and bone marrow stromal cells (BMSC) was evaluated using microarray analysis. The ratio of the abundance of each mRNA to that of β‐actin (ACTB) mRNA is presented for each specimen as well as in box‐and‐whisker plots. *P < 0.05. ***P < 0.001.
Finally, we investigated the expression of FBLN5 and MMP9 in human osteosarcoma samples. The comparison of microarray data revealed that the expression level of FBLN5 was downregulated, whereas that of MMP9 was significantly upregulated in human osteosarcoma specimens compared with normal BMSC (Fig. 7c). These findings support our hypothesis that Twist2 functions as a tumor suppressor mediated by the induction of FBLN5.
Discussion
The EMT promotes malignant progression in epithelial tumors by increasing invasive capacity and conferring stem‐like features, whereas the expression or function of EMT‐TF in mesenchymal tumors has remained largely unexplored. We have now shown that loss of Twist2 expression is associated with progression of osteosarcoma, possibly as a result of downregulation of the tumor suppressor fibulin‐5.
Signaling by transforming growth factor‐β (TGF‐β) plays a key role in the EMT and serves to either suppress or promote tumor growth depending on the cellular context.22 It is possible that EMT‐TF also function as tumor suppressors in specific types or at certain stages of cancer. In acute lymphoblastic leukemia, expression of TWIST2 was found to be downregulated as a result of epigenetic inactivation, whereas overexpression of the gene resulted in inhibition of cell growth, induction of apoptosis and increased sensitivity to chemotherapeutic agents.23 In prostate and bladder cancer cell lines, epithelial‐like subpopulations possess high metastatic tumor‐initiating activity, whereas mesenchymal‐like subpopulations have a reduced tumor‐initiating capacity, with Snail being a key EMT‐TF and functioning as a tumor suppressor in these cells.24 Thus, accumulating evidence suggests that EMT‐TF play a suppressive role in tumorigenesis as well as contribute to the regulation of mesenchymal features.
The EMT‐TF have recently been proposed to function as master regulators of adult stem cells. For example, Slug was found to regulate the mammary stem cell state.25 In the present study, we focused on the role of Twist2 in osteosarcoma and BMSC. Twist proteins play an essential role in osteoblast differentiation and maintenance of stem cell properties, with Twist2 being a candidate marker for MSC.26 We found that Twist2 inhibited osteogenic differentiation and that expression of Twist2 was correlated with the MSC state and in our model.
The results of the present study suggest that loss of Twist2 expression is a key event in osteosarcoma formation, but it is unclear how expression of Twist2 is regulated in the process of tumorigenesis. In general, epigenetic changes are known to control stem cell differentiation. DNA methylation and histone modifications are critical events for MSC differentiation toward osteogenic and adipogenic lineages.27 It was also reported that Twist2 was repressed through DNA methylation in several cancer cell lines.28, 29 Therefore, we assumed that repression of Twist2 by epigenetic modification might be essential for osteosarcoma formation from MSC.
Given that our mouse osteosarcoma model has a Ink4a KO background, we investigated the relationship between the expression of TWIST2 and INK4A/ARF in human osteosacoma specimens. However, we could not find any significant relationship (R = −0.189, P = 0.388) between them.
It remains unclear whether inhibition of osteogenic differentiation by Twist2 is directly related to its tumor‐suppressive action. However, we found that Twist2 modulates not only osteogenic and adipogenic differentiation but also expression of fibulin‐5 in osteosarcoma cells. Fibulin‐5 is an ECM protein that contributes to the formation and stabilization of basement membrane, elastic fibers and connective tissue.18 It suppresses tumor formation through control of tumor cell proliferation and motility as well as angiogenic sprouting.18, 19, 20, 21, 30 Thus, Twist2 might be expected to inhibit tumor angiogenesis or formation of an appropriate tumor microenvironment by various mechanisms. In contrast, fibulin‐5 also stimulates cell growth and motility in fibroblasts and fibrosarcoma cells, suggesting that it might exhibit opposite effects on tumorigenesis in a manner dependent on cell context.31 The fibulin‐5 gene is also a target of TGF‐β signaling.32, 33 Together with our data showing that Twist2 regulates the expression of fibulin‐5, these various observations suggest that fibulin‐5 expression might be modulated during the EMT in various cell contexts.
Twist1 and Twist2 are highly conserved and have redundant functions and Twist1 also has an important role in osteogenic differentiation.10, 11, 12 We confirmed that overexpression of Twist1 in AX cells could also induce fibulin‐5 and suppress in vivo tumor growth (Fig. S3). Deletion of the TWIST1 locus has been reported in osteosarcoma patients7 and some of our clinical samples showed a low expression of TWIST1 (Fig. 1), suggesting that Twist1 has a similar tumor suppressive role to Twist2 in osteosarcoma formation.
The outcome of the EMT is thought to be regulated in a complex manner dependent on cell context and the microenvironment. Further investigation into the roles of EMT‐TF in various tumor types on different genetic backgrounds or in different microenvironments should provide insight into tumor formation and malignant progression as well as provide a basis for selection of patients for treatment with anticancer drugs including those that target the EMT.
Disclosure Statement
T.I. is an employee of Daiichi Sankyo Co. Ltd. H.S. received research grants from Diichi Sankyo Inc., Kyowa Hakko‐Kirin Inc., LinkGenomics Inc. and Shimadzu Corp. The other authors declare no conflict of interest.
Supporting information
Fig. S1. Downregulation of Twist2 expression during osteogenic differentiation of AO cells.
Fig. S2. Effects of Twist2 overexpression in AX cells on anchorage‐independent growth and invasion potential.
Fig. S3. Effects of Twist1 overexpression in AX cells on tumorigenic potential.
Table S1. Sequences of PCR primers and sense oligonucleotides for shRNA.
Table S2. Selected genes that are more highly expressed in AX‐Twist2 cells than in AX‐Mock cells.
Data S1. Supplementary materials and methods.
Acknowledgments
The authors thank I. Ishimatsu for technical assistance as well as K. Arai for secretarial assistance. This work was supported in part by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to T.S. and H.S.) and by a grant from the National Institute of Biomedical Innovation of Japan (to H.S.).
(Cancer Sci 2013; 104: 880–888)
References
- 1. Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial–mesenchymal transitions in development and disease. Cell 2009; 139: 871–90. [DOI] [PubMed] [Google Scholar]
- 2. Kalluri R, Weinberg RA. The basics of epithelial–mesenchymal transition. J Clin Invest 2009; 119: 1420–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sanchez‐Tillo E, Liu Y, de Barrios O et al EMT‐activating transcription factors in cancer: beyond EMT and tumor invasiveness. Cell Mol Life Sci 2012; 69: 3429–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Peinado H, Olmenda D, Cano A. Snail, ZEB and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer 2007; 7: 415–28. [DOI] [PubMed] [Google Scholar]
- 5. Kansara M, Thomas DM. Molecular pathogenesis of osteosarcoma. DNA Cell Biol 2007; 26: 1–18. [DOI] [PubMed] [Google Scholar]
- 6. Clark JCM, Dass CR, Choong PFM. A review of clinical and molecular prognostic factors in osteosarcoma. J Cancer Res Clin Oncol 2008; 134: 281–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Entz‐Werle N, Stoetzel C, Berard‐Marec P et al Frequent genomic abnormalities at TWIST in human pediatric osteosarcomas. Int J Cancer 2005; 117: 349–55. [DOI] [PubMed] [Google Scholar]
- 8. Entz‐Werle N, Lavaux T, Metzger N et al Involvement of MET/TWIST/APC combination or the potential role of ossification factors in pediatric high‐grade osteosarcoma oncogenesis. Neoplasia 2007; 9: 678–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Castanon I, Baylies MK. A Twist in fate: evolutionary comparison of Twist structure and function. Gene 2002; 287: 11–22. [DOI] [PubMed] [Google Scholar]
- 10. Isenmann S, Arthur A, Zannettio ACW et al TWIST family of basic helix‐loop‐helix transcription factors mediate human mesenchymal stromal/stem cell growth and commitment. Stem Cells 2009; 27: 2457–68. [DOI] [PubMed] [Google Scholar]
- 11. Bialek P, Kern B, Yang X et al A twist code determines the onset of osteoblast differentiation. Dev Cell 2004; 6: 423–35. [DOI] [PubMed] [Google Scholar]
- 12. Miraoui H, Marie PJ. Pivotal role of Twist in skeletal biology and pathology. Gene 2010; 468: 1–7. [DOI] [PubMed] [Google Scholar]
- 13. Shimizu T, Ishikawa T, Saya H et al c‐Myc overexpression with loss of Ink4a/Arf transforms bone marrow stromal cells into osteosarcoma accompanied by loss of adipogenesis. Oncogene 2010; 29: 5687–99. [DOI] [PubMed] [Google Scholar]
- 14. Shimizu T, Ishikawa T, Iwai S et al Fibroblast growth factor‐2 is an important factor that maintains cellular immaturity and contributes to aggressiveness of osteosarcoma. Mol Cancer Res 2012; 10: 454–68. [DOI] [PubMed] [Google Scholar]
- 15. Zhang Y, Hassan MQ, Li ZY et al Intricate gene regulatory networks of helix‐loop‐helix (HLH) proteins support regulation of bone‐tissue related genes during osteoblast differentiation. J Cell Biochem 2008; 105: 487–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rosen ED, Walkey CJ, Puigserver P, Spiegelman BM. Transcriptional regulation of adipogenesis. Genes Dev 2000; 14: 1293–307. [PubMed] [Google Scholar]
- 17. Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol 2006; 7: 885–96. [DOI] [PubMed] [Google Scholar]
- 18. Albig AR, Neil JR, Schiemann WP. Fibulin 3 and 5 antagonize tumor angiogenesis in vivo . Cancer Res 2006; 66: 2621–9. [DOI] [PubMed] [Google Scholar]
- 19. Sullivan KM, Bissonnette R, Yanagisawa H, Hussain SN, Davis EC. Fibulin‐5 functions as an endogenous angiogenesis inhibitor. Lab Invest 2007; 87: 818–27. [DOI] [PubMed] [Google Scholar]
- 20. Xie L, Palmsten K, MacDonald B et al Basement membrane derived fibulin‐1 and fibulin‐5 function as angiogenesis inhibitors and suppress tumor growth. Exp Biol Med 2008; 233: 155–62. [DOI] [PubMed] [Google Scholar]
- 21. Møller HD, Ralfkjær U, Cremers N et al Role of fibulin‐5 in metastatic organ colonization. Mol Cancer Res 2011; 9: 553–63. [DOI] [PubMed] [Google Scholar]
- 22. Ikushima H, Miyazono K. TGFβ signaling: a complex web in cancer progression. Nat Rev Cancer 2010; 10: 415–24. [DOI] [PubMed] [Google Scholar]
- 23. Thathia SH, Ferguson S, Gautrey HE et al Epigenetic inactivation of TWIST2 in acute lymphoblastic leukemia modulates proliferation, cell survival and chemosensitivity. Haematologia 2011; 97: 371–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Celia‐Terrassa T, Meca‐Cortes O, Thomson TM et al Epithelial–mesenchymal transition can suppress major attributes of human epithelial tumor‐initiating cells. J Clin Invest 2012; 122: 1849–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guo W, Keckesova Z, Donaher JL et al Slug and Sox9 cooperatively determine the mammary stem cell state. Cell 2012; 148: 1015–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu Y, Wang L, Fatahi R et al Isolation of murine bone marrow derived mesenchymal stem cells using Twist2 Cre transgenic mice. Bone 2010; 47: 916–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Teven CM, Liu X, Hu N et al Epigenetic regulation of mesenchymal stem cells: a focus on osteogenic and adipogenic differentiation. Stem Cells Int 2011; 2011: 201371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mao Y, Toh HB, Ding Z et al Differential methylation of CpG islands within the dermo1 gene promoter in several cancer cell lines. Oncol Rep 2011; 25: 107–11. [PubMed] [Google Scholar]
- 29. Raval A, Lucas DM, Matkovic JJ et al TWIST2 demonstrates differential methylation in immunoglobulin variable heavy chain mutated and unmutated chronic lymphocytic leukemia. J Clin Oncol 2005; 23: 3877–85. [DOI] [PubMed] [Google Scholar]
- 30. Timpl R, Sasaki T, Kostka G, Chu ML. Fibulins: a versatile family of extracellular matrix proteins. Nat Rev Mol Cell Biol 2003; 4: 479–89. [DOI] [PubMed] [Google Scholar]
- 31. Albig AR, Schiemann WP. Fibulin‐5 function during tumorigenesis. Future Oncol 2005; 1: 23–35. [DOI] [PubMed] [Google Scholar]
- 32. Schiemann WP, Blobe GC, Kalume DE, Pandey A, Lodish HF. Context‐specific effects of fibulin‐5 (DANCE/EVEC) on cell proliferation, motility, and invasion: fibulin‐5 is induced by TGF‐β and affects protein kinase cascades. J Biol Chem 2002; 277: 27367–77. [DOI] [PubMed] [Google Scholar]
- 33. Lee YH, Albig AR, Regner MA, Schiemann BJ, Schiemann WP. Fibulin‐5 initiates epithelial–mesenchymal transition (EMT) and enhances EMT induced by TGF‐β in mammary epithelial cells via a MMP‐dependent mechanism. Carcinogenesis 2008; 29: 2243–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Downregulation of Twist2 expression during osteogenic differentiation of AO cells.
Fig. S2. Effects of Twist2 overexpression in AX cells on anchorage‐independent growth and invasion potential.
Fig. S3. Effects of Twist1 overexpression in AX cells on tumorigenic potential.
Table S1. Sequences of PCR primers and sense oligonucleotides for shRNA.
Table S2. Selected genes that are more highly expressed in AX‐Twist2 cells than in AX‐Mock cells.
Data S1. Supplementary materials and methods.


