Abstract
CRTC1–MAML2 and CRTC3–MAML2 fusions have been associated with favorable clinicopathological features of mucoepidermoid carcinomas. However, the significance of the MAML2 gene split has not been fully clarified. In the present study, 95 mucoepidermoid carcinomas (paraffin‐embedded materials) were analyzed for CRTC1–MAML2 and CRTC3–MAML2 fusions by RT‐PCR and for the MAML2 gene split by FISH. Quantitative RT‐PCR for the CRTC1–MAML2 transcript was performed in selected cases. MLL gene involvement, which has been reported in some leukemia cases, was examined by FISH in fusion partner‐unknown cases. CRTC1–MAML2 and CRTC3–MAML2 fusions were detected in 37 and 6 cases, respectively. The MAML2 gene split was detected in 62 cases, which included all CRTC1/3–MAML2 fusion‐positive cases. The level of CRTC1–MAML2 transcript expression was highly variable, and its clinicopathological impact was unclear. The MLL gene split was not detected. Mucoepidermoid carcinomas negative for CRTC1/3–MAML2 and positive for the MAML2 gene split (n = 19) showed favorable clinicopathological tumor features similar to those positive for CRTC1/3–MAML2 fusions. Compared with negative cases (n = 33), mucoepidermoid carcinomas positive for the MAML2 split (n = 62) were associated with lower patient age, a mild female predilection, a smaller tumor size, less frequent nodal metastasis, a lower clinical stage, a lower histological grade, and longer overall and disease‐free survival. The MAML2 gene split emerged as an independent prognostic factor for both overall and disease‐free survival in multivariate prognostic analysis. The presence of the MAML2 gene split defines a distinct mucoepidermoid carcinoma subset that is associated clinicopathologically with favorable tumor features. (Cancer Sci 2013; 104: 85–92)
Mucoepidermoid carcinomas represent 5% of all salivary gland tumors and 20% of salivary malignancies.1, 2 A subset of these carcinomas has been associated with a recurring chromosomal translocation, t(11;19)(q21;p13), which generates a fusion protein composed of the N‐terminal of cAMP response element‐binding protein (CREB)‐regulated transcription coactivator 1 (CRTC1), also called MECT1, TORC1 or WAMP1, and the C‐terminal of mastermind‐like gene 2 (MAML2).3, 4, 5, 6 Recent data suggest that CRTC1–MAML2 induced‐activation of CREB is critical for cell transformation.5, 6 CTCR1–MAML2 fusion has been detected in 40–80% of primary salivary gland mucoepidermoid carcinomas, and we and other research groups have shown that the fusion gene is associated with a distinct tumor subset with an indolent clinical course.7, 8, 9, 10 Fehr et al. (11) report a novel fusion gene, CRTC3–MAML2,11 and in Nakayama et al.12 we reveal that CRTC3–MAML2 fusion‐positive mucoepidermoid carcinoma cases have an excellent prognosis. These findings suggest that both CRTC1–MAML2 and CRTC3–MAML2 fusions may be associated with mucoepidermoid carcinoma cases with favorable clinicopathological tumor features.13
CRTC1/3–MAML2 fusions have been detected using an RT‐PCR technique, assuming that exon 1 of CRTC1/3 genes is fused to exons 2–5 of the MAML2 gene.7, 8, 12, 13 However, there would be unknown breakpoints in both CRTC1/3 and MAML2, and, therefore, the current RT‐PCR systems might not be capable of covering unknown CRTC1/3–MAML2 fusion variants. In addition, RT‐PCR may be unsuitable for detecting the gene fusions when the amount of the fusion transcripts is significantly low. Furthermore, other translocations involving MAML2, such as the MLL–MAML2 fusion that has been reported in some hematopoietic malignancies,14, 15 have not been studied in mucoepidermoid carcinomas.
In this study, we screened 95 mucoepidermoid carcinoma cases for CRTC1–MAML2 and CRTC3–MAML2 fusions by RT‐PCR and for MAML2 gene rearrangement by FISH using an MAML2 break‐apart probe. In addition, we examined MLL gene rearrangement in the CRTC1/3–MAML2‐negative and MAML2 rearrangement‐positive cases, and quantified the CRTC1–MAML2 fusion transcript in selected tumor cases. These molecular data were then correlated with clinicopathological and prognostic features of patients with mucoepidermoid carcinomas.
Materials and Methods
Case selection
Mucoepidermoid carcinomas of major and minor glands were retrieved from the pathology files of the Department of Pathology, Nagoya City University Postgraduate School of Medical Sciences. Tumors of the lung or other non‐salivary sites were not included. A total of 95 cases were included in this study, some of which were also incorporated in our previous studies.7, 12, 13 All tumor samples were fixed in formalin and embedded in paraffin. Informed consent was obtained, and the study was approved by the institutional review board of Nagoya City University and conducted in accordance with the Declaration of Helsinki. Clinicopathological data were obtained from medical records.
Clinicopathological data
The following clinicopathological factors were analyzed: age, sex, primary tumor site, tumor size, metastasis to regional lymph nodes, clinical stage, histological grade, treatment and follow up. Mucoepidermoid carcinomas were classified according to a three‐grade system that has been widely used for this carcinoma of the major and minor salivary glands.16, 17 In this system, the tumor grade is determined from the sum of the point values assigned to each of five histological factors: cystic component, neural invasion, necrosis, mitosis and anaplasia.
RT‐PCR for CRTC1/3–MAML2 fusion transcripts
CRTC1–MAML2 and CRTC3–MAML2 fusion transcripts were detected according to methods described previously.7, 12 Briefly, total RNA extracted from paraffin sections was heated to 70°C. An RT‐PCR mixture containing outer primers was then added. The thermocycler was programmed for an initial reverse transcription of 30 min at 42°C, and then for 10 min at 95°C for inactivation of reverse transcriptase as well as for activation of the polymerase. After the PCR, the products were diluted 1:50 with water, and subjected to a nested PCR with inner primers. Primers used in the present study are shown in Table 1. The PCR fragments were directly sequenced. As an internal control for RNA quality, the ubiquitously expressed ACTB mRNA fragment was amplified. Mucoepidermoid carcinomas known to possess the gene fusions and normal salivary gland tissue were used as positive and negative controls, respectively.
Table 1.
Sequences of primers
| Primer | Sequence (5′–3′) |
|---|---|
| CRTC1A (outer) | tcgcgctgcacaatcagaag |
| CRTC1Ba (inner) | gaggtcatgaaggacctgag |
| CRTC3A (outer) | tcgcgctgcacacgcagaga |
| CRTC3B (inner) | cagagacaggccgaggagac |
| MAML2A (outer) | ggtcgcttgctgttggcagg |
| MAML2Ba (inner) | ttgctgttggcaggagatag |
Primers used for quantitative RT‐PCR. CRTC 1, cAMP response element‐binding protein (CREB)‐regulated transcription coactivator 1 (CRTC1); MAML2, mastermind‐like gene 2.
Detection of CRTC1/3–MAML2 fusion variants was performed using nested RT‐PCR assays for all possible combinations of the exons. The primers were designed so that the PCR products were <300 base pairs in length (Table S1).
Quantitative real‐time RT‐PCR for the CRTC1–MAML2 fusion transcript and wild type MAML2 transcript
To examine the expression level of the fusion transcript, quantitative RT‐PCR was carried out for CRTC1–MAML2 fusion‐positive mucoepidermoid carcinoma cases. Cases were selected on the basis of whether sufficient tumor materials were available. Sections of the carcinoma and normal salivary tissue (4 μm) were deparaffinized and lightly stained with methyl green. With the sections under a dissecting microscope, the carcinoma tissues were selectively obtained by microscraping using a serial hematoxylin and eosin section as a guide. Total RNA extracted from the tissues was converted to cDNA with random hexamers. Quantitative real‐time RT‐PCR analysis of the CRTC1–MAML2 fusion transcript was carried out using SYBR Premix Ex Taq (Takara Bio, Shiga, Japan), as previously described.18 The primers used for quantitative RT‐PCR are shown in Table 1. The ACTB gene was used as an internal control. Real‐time PCR was performed in a 20 μL final reaction mixture that included diluted cDNA and 0.5 pmol/mL of each primer. Amplification conditions were 30 s at 95°C, 45 cycles of 5 s at 95°C and 30 s at 60°C. Post‐amplification melting curve analyses were performed to confirm product specificity. To ensure experimental accuracy, all reactions were performed in triplicate. To detect the level of the relative expression of the CRTC1–MAML2 fusion transcript, we used a comparative delta Ct method with which Ct values relative to ACTB mRNA were normalized. Wild type MAML2 transcript was similarly quantified in normal salivary gland and mucoepidermoid carcinoma cases. PCR primers were designed to amplify exons 1 and 2 of MAML2 mRNA, and their sequences were as follows: GGGACCAGAGGAACTCAGC and GGTCGCTTGCTGTTGG CAGG.
FISH
The tissue FISH procedure was performed as previously described.19 Formalin‐fixed, paraffin‐embedded specimens were sectioned at 4 μm. After deparaffinization, sections were digested in a protease solution at 37°C and refixed with 10% formalin. Aging was performed using 2 × saline sodium citrate/0.1% NP‐40 for 30 min at 37°C, followed by denaturation at 80°C for 5 min in 70% formamide. An MAML2 break‐apart probe (ZytoVision, Bremerhaven, Germany) was denatured for 5 min at 84°C before hybridization. Sections were incubated with the probe overnight at 37°C in a humidified chamber. After post‐hybridization washes, sections were stained with diaminophenilindole and mounted. When two signals (a green and a red) were separately observed in a tumor cell, we considered the MAML2 gene to be split. The frequencies were determined by counting split signals in more than 100 tumor cells.
Mucoepidermoid carcinoma cases known to possess CRTC1/3–MAML2 fusions were used as a positive control. For a negative control, we analyzed 10 normal parotid glands, and the signal frequency threshold was determined for the FISH probe by counting the number of split signals per nucleus in more than 100 cells. The threshold was determined as the mean + 3 SD. The cut‐off value for the MAML2 gene split thus obtained was 7%. Mucoepidermoid carcinoma cases that showed a split signal number that was greater than the cut‐off value were considered to be positive for the gene split.
For mucoepidermoid carcinoma cases positive for the MAML2 gene split but negative for CRTC1/3–MAML2 fusions by RT‐PCR, FISH analysis was further performed to detect MLL gene involvement using an MLL break‐apart probe (Kreatech Diagnostics, Amsterdam, the Netherlands). Leukemia cases positive for MLL gene splits were used as a positive control and normal parotid glands were used as a negative control. The cut‐off value for the MLL gene split was set at 6%.
Statistical analysis
Statistical evaluation of data from two groups was carried out using Fischer's exact test and Student's t‐test. To identify variables significantly associated with overall and disease‐free survival, the survival rate was evaluated using the Kaplan–Meier method. Univariate and multivariate survival analyses were performed using Cox's proportional hazard model. For the multivariate analysis, variables significant in the univariate analyses were used. All analyses were two‐tailed. A value of P < 0.05 in each test was regarded as statistically significant. All analyses were performed using the statistical package JMP (SAS Institute, Cary, NC, USA).
Results
Patient characteristics
This study included 51 males and 44 females, with ages ranging from 10 to 89 years (median, 56; mean, 53.7) (Table 2). The primary tumors were located in the major salivary glands in 37 cases and minor salivary glands in 58 cases. A total of 53 tumors were more than 2 cm in diameter, and 19 tumors showed metastasis to the regional lymph nodes. Patients with distant metastases at diagnosis were not included in this study. Forty cases were classified as clinical stage I, 34 as stage II, 13 as stage III, and 8 as stage IV. The overall and disease‐free 10‐year survival rates were 84% and 58%, respectively. All tumors were surgically resected, and some patients were additionally treated with chemotherapy and/or radiotherapy. The follow‐up period ranged from 4 to 263 mouths, with a median of 48 months. Our cases were histologically classified into low‐grade (n = 67), intermediate‐grade (n = 9) and high‐grade (n = 19) tumors.
Table 2.
Patients' characteristics
| Clinical findings | ||
| Age (years) | Mean | 53.7 |
| >60 | 35 | |
| <60 | 60 | |
| Sex | Male | 51 |
| Female | 44 | |
| Tumor site | Major | 37 |
| Minor | 58 | |
| Tumor size | >2 cm | 53 |
| <2 cm | 42 | |
| Nodal status | Positive | 19 |
| Negative | 76 | |
| Clinical stage | I, II | 74 |
| III, IV | 21 | |
| Histological findings | ||
| Histrogical grade | Low | 67 |
| Intermediate | 9 | |
| High | 19 | |
| Cystic component | >20% | 46 |
| <20% | 49 | |
| Neural invasion | Positive | 8 |
| Negative | 87 | |
| Necrosis | Positive | 18 |
| Negative | 77 | |
| Mitoses | >4/10 HPF | 19 |
| <3/10 HPF | 76 | |
| Anaplasia | Positive | 30 |
| Negative | 65 | |
RT‐PCR detection of CRTC1/3–MAML2 fusion transcripts
All tumor specimens were shown to possess RNA of satisfactory quality. RT‐PCR analysis was performed in all tumor cases, and CRTC1–MAML2 and CRTC3–MAML2 fusion transcripts were detected in 37 and 6 cases, respectively (Fig. 1, Fig. S1). The remaining 52 cases were negative for the fusion transcripts.
Figure 1.
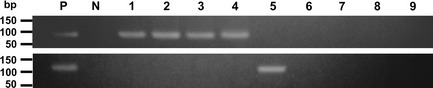
RT‐PCR detection of CRTC1–MAML2 (95 bp) and CRTC3–MAML2 (119 bp) fusions. Lanes 1–4, carcinomas positive for the CRTC1–MAML2 fusion; lane 5, a carcinoma positive for CRTC3–MAML2 fusion; lanes 6–9, carcinomas with no detectable MAML2‐associated fusion. bp, base pair; N, negative control; P, positive control.
Quantitative real‐time RT‐PCR for the CRTC1–MAML2 fusion transcript
Of 22 selected carcinoma cases, the expression of the CRTC1–MAML2 fusion transcript was successfully quantified in 12 (55%). In the remaining 10 negative cases, house‐keeping ACTB transcripts were preserved but PCR amplification failed to detect the fusion transcripts. In these negative cases, the fusion transcript levels were most likely below the detection threshold of our quantitative RT‐PCR system. Figure 2 shows the expression of the CRTC1–MAML2 fusion transcript in the quantifiable cases. The expression levels were highly variable in carcinoma cases, and the difference between the highest and lowest cases was 425‐fold. These 12 cases were divided into higher (n = 6) and lower (n = 6) expressers, and when the impact of the expression on five clinicopathological factors (age, sex, tumor site, histological grade and clinical stage) was analyzed, no factor showed a significant correlation with CRTC1/MAML2 fusion transcript expression (data not shown).
Figure 2.
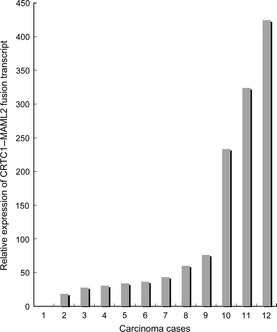
Relative expression of the CRTC1–MAML2 fusion transcript in quantifiable tumor cases (n = 12).
FISH analysis
MAML2‐associated gene abnormalities were analyzed using a dual‐color break‐apart probe for this gene, and all cases were successfully evaluated for the gene split (Fig. 3). An increased number of split signals in the MAML2 genes (ranging from 15% to 92%, median 71%) above the cut‐off value (7%) was detected in 62 cases, which were considered to be positive for the MAML2 gene split. These FISH‐positive cases included cases of CRTC1–MAML2‐positive (n = 37) and CRTC3–MAML2‐positive (n = 6) carcinomas, as detected by RT‐PCR. The remaining 19 cases were positive for the MAML2 gene split using FISH but negative for CRTC1/3–MAML2 fusions using RT‐PCR. To examine a possible MLL gene involvement in these cases, an additional FISH analysis was performed using an MLL break‐apart probe. No case showed an MLL gene split.
Figure 3.

FISH analysis for the MAML2 gene split. Arrows indicate split signals. Arrowheads indicate unsplit MAML2 genes.
Clinicopathological comparison of four groups
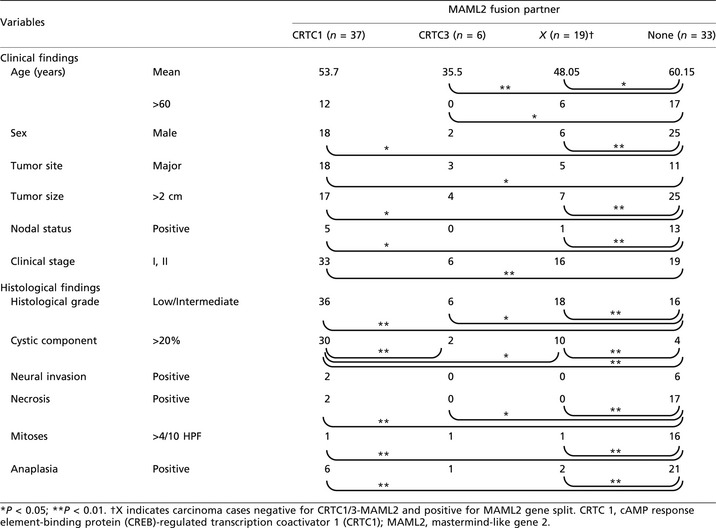
Mucoepidermoid carcinoma cases that were positive for the MAML2 gene split but negative for CRTC1/3–MAML2 fusion transcripts were designated as X‐MAML2 in this study. After RT‐PCR and FISH analyses, we divided our mucoepidermoid carcinoma cases into four groups: CRTC1–MAML2, CRTC3–MAML2, X–MAML2 and no detectable MAML2 gene abnormalities (i.e. MAML2 split‐negative), as shown in Figure 4. A clinicopathological comparison of these four groups was then performed (Table 3). The factors analyzed were as follows: six clinical factors (age, sex, tumor site, tumor size, nodal status and clinical stage) and six histopathological factors (tumor grade, cystic component, neural invasion, necrosis, mitoses and anaplasia). Compared with the group with no MAML2 split, CRTC1–MAML2, CRTC3–MAML2 and X–MAML2 showed a mild female predominance and generally favorable tumor features, including a smaller tumor size, a less advanced clinical stage, a lower histological grade, infrequent necrosis, fewer mitotic figures and less anaplasia. Some minor differences were noted among CRTC1–MAML2, CRTC3–MAML2 and X–MAML2 groups, with the CRTC3–MAML2 group associated with lower patient age and the CRTC1–MAML2 group associated histologically with a higher frequency of cystic components.
Figure 4.
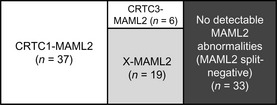
Mucoepidermoid carcinoma cases (n = 95) are divided into four groups using RT‐PCR assays for CRTC1/3–MAML2 fusions and FISH analysis for the MAML2 gene split. The X–MAML2 group may include mucoepidermoid carcinoma cases with non‐CRTC1/3 fusion partners and those with low fusion transcript expression.
Table 3.
Clinicopathological findings for MAML fusion partners in mucoepidermoid carcinoma
Detection of CRTC1/3–MAML2 fusion variants
Assuming that X–MAML2 cases include possible fusion variants of CRTC1/3–MAML2 fusions, we performed nested RT‐PCR for all exon combinations of CRTC1/3–MAML2 fusions in 19 tumor cases of this group. No variant fusion transcript was detected in our series.
Quantitative real‐time RT‐PCR for wild type MAML2 transcript
We quantified the expression of wild type MAML2 transcript in the normal salivary glands, CRTC1–MAML2 cases, X–MAML2 cases and cases with no MAML2 split (Fig. 5). Tumor cases with CRTC3–MAML2 fusion were excluded because the number of the cases was small. The expression level of wild type MAML2 transcript in the no MAML2 split group (12.3 ± 1.59, mean ± SEM) was higher than that in normal salivary glands (2.41 ± 0.48, P = 0.0006), CRTC1–MAML2 (1.56 ± 0.48, P = 0.0003) and X–MAML2 (5.89 ± 2.2, P = 0.04). No significant difference was detected in the latter three groups. Compared with the other three groups, wild type MAML2 expression levels in X–MAML2 cases were variable, but the expression levels did not correlate with age, sex, tumor site, histological grade or clinical stage (data not shown).
Figure 5.
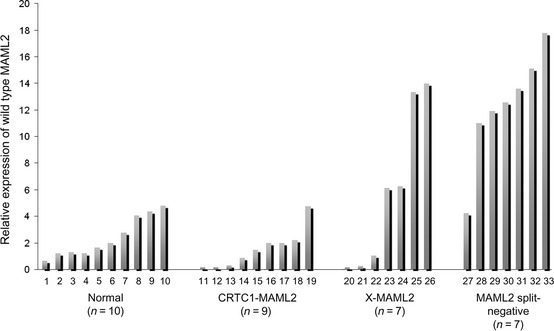
The relative levels of wild type MAML2 expression in normal salivary gland cases, CRTC1–MAML2,X–MAML2 and MAML2 split‐negative.
Comparison of prognosis of the four groups
Ten‐year overall survival rates were 100%, 100%, 93% and 59% for CRTC1–MAML2, CRTC3–MAML2, X–MAML2 and no MAML2 gene split, respectively. As shown in Figure 6, patients with MAML2 abnormalities (CRTC1–MAML2, CRTC3–MAML2 and X–MAML2) had better overall survival compared with those negative for the MAML2 gene split. No significant difference was found between overall survival rates of the three patient groups with MAML2 abnormalities. The 10‐year disease free survival rates were 60%, 100%, 71% and 37% for CRTC1–MAML2, CRTC3–MAML2, X–MAML2 and no MAML2 split, respectively. While disease‐free survival rates were not significantly different between CRTC1–MAML2, CRTC3–MAML2 and X–MAML2 groups, each of these three groups had better disease‐free survival than the group with no MAML2 split.
Figure 6.
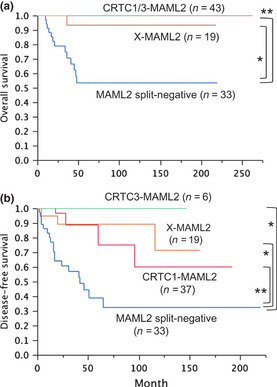
Overall (a) and disease‐free (b) survival for MAML2 fusion partners. The X–MAML2 group is negative for CRTC1/3–MAML2 fusions and positive for the MAML2 gene split. *P < 0.05, **P < 0.01.
MAML2 gene split defines a distinct subgroup
Because the CRTC1–MAML2, CRTC3–MAML2 and X–MAML2 groups had similarly favorable clinicopathological and prognostic features, we combined them as the MAML2 gene split‐positive group to compare with the MAML2 gene split‐negative group. As shown in Table 4, groups with or without the MAML2 gene split had extremely different features, and the former group was characterized by lower patient age, a mild female predilection, a smaller tumor size, less frequent nodal metastasis and a lower clinical stage. In addition, the histological grade was lower with milder features of all five histological elements (cystic component, neural invasion, necrosis, mitoses and anaplasia). The group positive for the MAML2 gene split showed much better overall and disease‐free survival than the negative group (Fig. 7). The 10‐year overall survival rates were 98% and 59%, and 10‐year disease‐free survival rates were 70% and 37% for the MAML2 gene split‐positive and negative groups, respectively. We performed univariate and multivariate prognostic analyses to identify factors useful for predicting overall and disease‐free survival using the following factors as variables: age, sex, tumor site, clinical stage, histological grade, and the presence or absence of MAML2 gene abnormalities (Table 5). For overall survival, advanced age, being male, higher histological grade and absence of MAML2 gene abnormalities emerged as significant risk factors in the univariate analysis. In a multivariate analysis using these significant factors, only the absence of MAML2 gene abnormalities was selected as an independent factor, and it was associated with unfavorable overall survival. For disease‐free survival, advanced age, higher clinical stage, higher histological grade and absence of MAML2 gene abnormalities were selected as risk factors in the univariate analysis. In the multivariate analysis, only the absence of MAML2 gene abnormalities remained as an independent risk factor.
Table 4.
Clinicopathological findings for MAML gene split in mucoepidermoid carcinoma
| Variables | MAML2 gene split | P | ||
|---|---|---|---|---|
| Positive (n = 62) | Negative (n = 33) | |||
| Clinical findings | ||||
| Age (years) | Mean | 50.21 | 60.15 | 0.0031 |
| Sex | Male | 26 | 25 | 0.0023 |
| Tumor site | Major | 26 | 11 | 0.51 |
| Tumor size | >2 cm | 28 | 25 | 0.005 |
| Nodal status | Positive | 6 | 13 | 0.001 |
| Clinical stage | III, IV | 7 | 14 | 0.0013 |
| Histological findings | ||||
| Histological grade | Low/Intermediate | 56 | 11 | <0.0001 |
| Cystic component | >20% | 42 | 4 | <0.0001 |
| Neural invasion | Positive | 2 | 6 | 0.0195 |
| Necrosis | Positive | 1 | 17 | <0.0001 |
| Mitoses | >4/10 HPF | 3 | 16 | <0.0001 |
| Anaplasia | Positive | 9 | 21 | <0.0001 |
MAML2, mastermind‐like gene 2.
Figure 7.
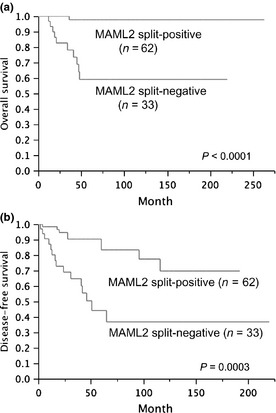
Overall (a) and disease‐free (b) survival for the MAML2 gene split.
Table 5.
Prognostic factors affecting overall and disease‐free survival
| Factor | Overall survival | Disease‐free survival | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Univariate | P | Multivariate | P | Univariate | P | Multivariate | P | ||
| Age | Years | 1.01 (0.95–1.1) | 0.013 | 1.03 (0.98–1.09) | 0.17 | 1.28 (1.00–1.06) | 0.040 | 1.01 (0.98–1.05) | 0.39 |
| Sex | Male | 3.92 (1.01–25.7) | 0.049 | 1.65 (0.41–11.1) | 0.51 | 1.14 (0.51–2.64) | 0.76 | – | – |
| Female | |||||||||
| Tumor site | Major | 1.27 (0.37–4.21) | 0.70 | – | – | 1.77 (0.76–4.59) | 0.19 | – | – |
| Minor | |||||||||
| Clinical stage | I, II | 2.91 (0.84–9.69) | 0.089 | – | – | 2.86 (1.23–6.40) | 0.016 | 1.22 (0.44–3.27) | 0.69 |
| III, IV | |||||||||
| Histrogical grade | Low/Int‐ermediate | 0.24 (0.072–0.84) | 0.027 | 0.88 (0.24–3.24) | 0.85 | 0.29 (0.13–0.69) | 0.0067 | 0.68 (0.24–1.93) | 0.46 |
| High | |||||||||
| MAML2 fusion | Positive | 0.044 (0.0024–0.23) | <.0001 | 0.070 (0.0036–0.44) | 0.0030 | 0.22 (0.092–0.50) | 0.0003 | 0.32 (0.12–0.91) | 0.033 |
| Negative | |||||||||
Discussion
In the present study of mucoepidermoid carcinoma, we analyzed MAML2‐associated gene abnormalities using RT‐PCR for the CRTC1/3–MAML2 fusion transcripts and the FISH technique for the MAML2 gene split. The advantages of being able to use routinely processed paraffin materials far outweigh any disadvantages, and both techniques are well suited to paraffin materials. Of the 95 cases examined, the CRTC1–MAML and the CRTC3–MAML2 fusion transcripts and the MAML2 gene split was detected in 37, 6 and 62 cases, respectively. All CRTC1/3–MAML2 fusion‐positive cases detected by RT‐PCR (n = 43) showed MAML2 gene splits by FISH. In the remaining 19 cases that were positive for the MAML2 gene split and negative for CRTC1/3–MAML2 fusions (designated as X–MAML2 cases in this study), we performed FISH analysis to detect the MLL gene split because the MAML2 gene has been reported to fuse with the MLL gene in some hematological malignancies.14, 15 However, the MLL gene was not involved in our carcinoma cases. Assuming that the MAML2 gene fused with unknown genes in the X–MAML2 cases, we attempted to detect possible fusion partner genes using a 5′ rapid amplification of cDNA ends.20 Unfortunately, we were unsuccessful in this analysis, partly owing to a lack of fresh tumor materials and an abundant expression of wild type MAML2 transcript in the salivary glands.
We performed quantitative real‐time RT‐PCR for CRTC1–MAML2 fusion‐positive cases using the carcinoma tissue obtained by microscraping. While all the cases examined had preserved RNA of sufficient quality, the fusion transcript was quantifiable in 45% of the cases, and no positive fusion signal was obtained in the remaining cases. In these negative cases, the amount of the fusion transcript was most likely below the detection threshold of our assay. In the cases for which quantification was successful, expression of the fusion transcript was highly variable (the highest expression was more than 400 times that of the lowest), which was supported by a previous observation that expression levels of the CRTC1–MAML2 fusion transcript varied from 8 to 10 cycles of amplification among samples.21 The CRTC1–MAML2 fusion gene is an important oncogene that underlies the development of mucoepidermoid carcinoma.5, 6 However, our finding in the present study that the amount of the CRTC1–MAML2 fusion transcript did not have any clinicopathological impact suggests that the expression level of the fusion transcript might not be crucial for maintenance or progression of the carcinoma.
The X–MAML2 group, comprised of MAML2 split‐positive and CRTC1/3–MAML2‐negative tumors, was a new mucoepidermoid carcinoma subset. Because CRTC1/3–MAML2 fusion variants were not detected in this study, this subset would include carcinomas with unknown MAML2 partners and those with low expression of CRTC1/3–MAML2 fusion transcripts. We previously reported that both CRTC1–MAML2‐positive and CRTC3–MAML2‐positive mucoepidermoid carcinomas are associated with mild histopathological features and a favorable clinical outcome compared with fusion‐negative carcinomas.7, 12, 13 Our present data show that the X–MAML2 carcinomas might also possess favorable tumor features similar to those positive for CRTC1/3–MAML2 fusions. However, it should be noted that there were minor differences between the three groups, such as differential expression of wild type MAML2 transcripts, lower patient age in the CRTC3–MAML2 fusion group and possibly poorer disease‐free survival in CRTC1–MAML2 fusion groups. We combined the three mucoepidermoid carcinoma groups harboring MAML2‐associated gene abnormalities and compared this combined series (the MAML2 gene split‐positive group) with the MAML2 gene split‐negative group. Results of this comparison showed an excellent stratification of the mucoepidermoid carcinoma cases, both clinicopathologically and prognostically. Of note, the MAML2 gene split emerged as an independent prognostic factor in analyses of both overall and disease‐free survival. Because the MAML2 gene split was detected in all three subsets of mucoepidermoid carcinoma with favorable clinicopathological features, FISH analysis of the MAML2 gene split might be more useful than RT‐PCR for CRTC1/3–MAML2 fusions for screening of a favorable subset of mucoepidermoid carcinoma cases.
In conclusion, we performed a detailed molecular study using a large number of mucoepidermoid carcinoma cases, and established that the presence of the MAML2 gene split was an important genetic marker that was associated with favorable clinicopathological features and better overall and disease‐free survival. Detection of the MAML2 gene split using the FISH technique might be the method of choice in a clinical setting because the testing procedure is easily performed using routinely processed paraffin sections. RT‐PCR assays for CRTC1/3–MAML2 fusion transcripts might also be useful to further characterize mucoepidermoid carcinoma cases. Further studies are necessary to clarify unknown MAML2 fusion partners, the clinicopathological impact of higher expression of the fusion transcript, and, finally, the molecular mechanisms of the tumorigenesis of mucoepidermoid carcinomas that are not associated with the MAML2 gene.
Disclosure Statement
The authors have no conflict of interest to declare.
Supporting information
Fig. S1. Sequence analysis of (a) CRTC1–MAML2 and (b) CRTC3–MAML2 transcripts.
Table S1. Primer sequences.
Acknowledgment
This study was supported in part by a Grant‐in‐Aid for Scientific Research from MEXT, Japan (S. M. and H. I.).
References
- 1. Goode RK, El‐Naggar AK. Mucoepidermoid carcinoma In: Barnes L, Eveson J, Reichart P, Sidransky D, eds. Pathology and Genetics of Head and Neck Tumours. Lyon: IARC Press, 2005; 219–20. [Google Scholar]
- 2. McHugh CH, Roberts DB, El‐Naggar AK et al Prognostic factors in mucoepidermoid carcinoma of the salivary glands. Cancer 2011; 118: 3928–36. [DOI] [PubMed] [Google Scholar]
- 3. Tonon G, Modi S, Wu L et al t(11;19)(q21;p13) translocation in mucoepidermoid carcinoma creates a novel fusion product that disrupts a Notch signaling pathway. Nat Genet 2003; 33: 208–13. [DOI] [PubMed] [Google Scholar]
- 4. Enlund F, Behboudi A, Andrén Y et al Altered Notch signaling resulting from expression of a WAMTP1‐MAML2 gene fusion in mucoepidermoid carcinomas and benign Warthin's tumors. Exp Cell Res 2004; 292: 21–8. [DOI] [PubMed] [Google Scholar]
- 5. Coxon A, Rozenblum E, Park YS et al Mect1‐Maml2 fusion oncogene linked to the aberrant activation of cyclic AMP/CREB regulated genes. Cancer Res 2005; 65: 7137–44. [DOI] [PubMed] [Google Scholar]
- 6. Wu L, Liu J, Gao P et al Transforming activity of MECT1–MAML2 fusion oncoprotein is mediated by constitutive CREB activation. EMBO J 2005; 24: 2391–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Okabe M, Miyabe S, Nagatsuka H et al MECT1–MAML2 fusion transcript defines a favorable subset of mucoepidermoid carcinoma. Clin Cancer Res 2006; 12: 3902–7. [DOI] [PubMed] [Google Scholar]
- 8. Behboudi A, Enlund F, Winnes M et al Molecular classification of mucoepidermoid carcinomas‐prognostic significance of the MECT1–MAML2 fusion oncogene. Genes Chromosom Cancer 2006; 45: 470–81. [DOI] [PubMed] [Google Scholar]
- 9. Tirado Y, Williams MD, Hanna EY, Kaye FJ, Batsakis JG, El‐Naggar AK. CRTC1/MAML2 fusion transcript in high grade mucoepidermoid carcinomas of salivary and thyroid glands and Warthin's tumors: implications for histogenesis and biologic behavior. Genes Chromosom Cancer 2007; 46: 708–15. [DOI] [PubMed] [Google Scholar]
- 10. Schwarz S, Stiegler C, Müller M et al Salivary gland mucoepidermoid carcinoma is a clinically, morphologically and genetically heterogeneous entity: a clinicopathological study of 40 cases with emphasis on grading, histological variants and presence of the t(11;19) translocation. Histopathology 2011; 58: 557–70. [DOI] [PubMed] [Google Scholar]
- 11. Fehr A, Röser K, Heidorn K, Hallas C, Löning T, Bullerdiek J. A new type of MAML2 fusion in mucoepidermoid carcinoma. Genes Chromosom Cancer 2008; 47: 203–6. [DOI] [PubMed] [Google Scholar]
- 12. Nakayama T, Miyabe S, Okabe M et al Clinicopathological significance of the CRTC3–MAML2 fusion transcript in mucoepidermoid carcinoma. Mod Pathol 2009; 22: 1575–81. [DOI] [PubMed] [Google Scholar]
- 13. Okumura Y, Miyabe S, Nakayama T et al Impact of CRTC1/3–MAML2 fusions on histological classification and prognosis of mucoepidermoid carcinoma. Histopathology 2011; 59: 90–7. [DOI] [PubMed] [Google Scholar]
- 14. Nemoto N, Suzukawa K, Shimizu S et al Identification of a novel fusion gene MLL–MAML2 in secondary acute myelogenous leukemia and myelodysplastic syndrome with inv(11)(q21q23). Genes Chromosom Cancer 2007; 46: 813–9. [DOI] [PubMed] [Google Scholar]
- 15. Metzler M, Staege MS, Harder L et al Inv(11)(q21q23) fuses MLL to the Notch co‐activator mastermind‐like 2 in secondary T‐cell acute lymphoblastic leukemia. Leukemia 2008; 22: 1807–11. [DOI] [PubMed] [Google Scholar]
- 16. Auclair PL, Goode RK, Ellis GL. Mucoepidermoid carcinoma of intraoral salivary glands. Evaluation and application of grading criteria in 143 cases. Cancer 1992; 69: 2021–30. [DOI] [PubMed] [Google Scholar]
- 17. Goode RK, Auclair PL, Ellis GL. Mucoepidermoid carcinoma of the major salivary glands: clinical and histopathologic analysis of 234 cases with evaluation of grading criteria. Cancer 1998; 82: 1217–24. [DOI] [PubMed] [Google Scholar]
- 18. Yamada S, Sato F, Xia H et al Forkhead box P1 overexpression and its clinicopathologic significance in peripheral T‐cell lymphoma, not otherwise specified. Hum Pathol 2012; 43: 1322–7. [DOI] [PubMed] [Google Scholar]
- 19. Xia H, Nakayama T, Sakuma H et al Analysis of API2–MALT1 fusion, trisomies, and immunoglobulin VH genes in pulmonary mucosa‐associated lymphoid tissue lymphoma. Hum Pathol 2011; 42: 1297–304. [DOI] [PubMed] [Google Scholar]
- 20. Li C, Takino H, Eimoto T et al Prognostic significance of NPM–ALK fusion transcript overexpression in ALK‐positive anaplastic large‐cell lymphoma. Mod Pathol 2007; 20: 648–55. [DOI] [PubMed] [Google Scholar]
- 21. Seethala RR, Dacic S, Cieply K, Kelly LM, Nikiforova MN. A reappraisal of the MECT1/MAML2 translocation in salivary mucoepidermoid carcinomas. Am J Surg Pathol 2010; 34: 1106–21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Sequence analysis of (a) CRTC1–MAML2 and (b) CRTC3–MAML2 transcripts.
Table S1. Primer sequences.


