Abstract
Gastric cancer (GC) is one of the most common malignant tumors with a high rate of recurrence, which results in surgery being unsuccessful. Therefore, it is important to find the reason for the surgery failing. The purpose of the present study was to investigate a factor related to recurrence. Expression of KIAA0101 was assessed in 61 paired human primary GC and non‐cancerous gastric tissue using immunohistochemistry. After surgery, all 61 patients were followed regularly for more than 24 months or until death to analyze the 2‐year survival rate and recurrence. After suppressing KIAA0101 by RNA interference in human GC cell lines, the cell viability was detected using MTT. We are first to find that KIAA0101 was elevated in GC tissues compared with paired non‐cancerous gastric tissues. Immunohistochemical staining also revealed the predominant nuclear localization of KIAA0101 protein. Despite these findings, GC patients with elevated KIAA0101 expression levels exhibited a high recurrence and subsequently poor prognosis in the survival study. Also, cell viability was significantly inhibited after suppressing KIAA0101 in GC cells, suggesting that KIAA0101 might promote cancer cell proliferation. KIAA0101 is increased in human GC and is a marker of recurrence.
Gastric cancer (GC) is one of the most common malignant tumors in the world, yet the prognosis for patients with advanced disease remains very poor.1, 2, 3 Even after surgery, approximately 40–65% of GC patients will experience a recurrence and die from the disease.4 A variety of clinical and biological variables have been proposed as risk prognostic factors for GC, but the precise molecular mechanisms underlying the development and progression of GC remain unclear. The identification of novel molecular markers for the prediction of recurrence risk in tumor prognosis will contribute to the development of better strategies for patient management.
KIAA0101 was first identified as p15PAF (proliferating cell nuclear antigen [PCNA] associated factor) based on its ability to bind PCNA in a yeast two‐hybrid screen in 2001.5 It is alternatively referred to as L5,6 OEATC‐17 or NS5ATP9.8 In our earlier study, we used suppression subtractive hybridization (SSH) to show that KIAA0101 was upregulated by hepatitis C virus non‐structural protein 5A.8 We then identified the promoter region of this gene, screened KIAA0101 gene promoter‐binding proteins and verified that rhNF‐kB could bind to the KIAA0101 promoter and participate in the regulation of KIAA0101 expression.9 Moreover, recent reports have shown that KIAA0101 can also interact with BRCA1 and regulate centrosome number.10 Another report demonstrated that KIAA0101 is an APC/C‐regulated protein involved in both cell cycle progression and the DNA damage response.11 Overexpression of KIAA0101 in mammalian cells was also found to protect cells from UV‐induced cell death.12, 13, 14 Interestingly, Hosokawa et al.15 found that KIAA0101 had oncogenic activity, as exogenous overexpression of KIAA0101 promoted cancer cell growth and transformed mouse fibroblast NIH3T3 cells into tumor cells in vivo. These findings suggest that KIAA0101 might play an important role in tumor proliferation, which contributes to carcinogenesis and tumor recurrence.
In recent years, several reports have demonstrated that changes in KIAA0101 expression occurred in several cancers.7, 10, 16, 17, 18 Elevated KIAA0101 expression was associated with more advanced tumor stages and contributes to poor prognosis.17, 18 Despite these observations, the expression pattern of KIAA0101 and its prognostic significance in human gastric cancer remain unclear. In the present study, we investigated the expression of KIAA0101 at the mRNA and protein levels and the localization of KIAA0101 protein in GC tissues to elucidate the prognostic significance of this gene, and then clarified its role in cell viability in vitro. Also, we explored the 2‐year survival rate of GC patients.
Materials and Methods
Tissue samples
A total of 61 surgically resected, unifocal, primary human gastric cancers that were diagnosed by pathology were collected in the Oncology Surgery Department of the First Affiliated Hospital of Xi'an Jiaotong University between 1 January 2008 and 1 January 2009. In addition, 17 chronic gastritis tissue samples were collected from gastroscopies at the same time. All 139 tissues (61 GC tissues, 61 matched adjacent non‐cancerous gastric tissues and 17 gastritis tissues) were snap frozen at the time of surgery and stored at −80°C for further experiments. The 61 GC patients were treated with the same therapeutic strategy: complete resection with negative margins (R0 resection) and extensive lymphadenectomy. In addition, patients with an advanced tumor stage were treated with the same adjunctive treatment (5‐fluorouracil, oxaliplatin and folinic acid; four times; once per month) after surgery. All patients were followed regularly for more than 24 months or until death. The follow up for all cases was terminated in September 2011. Tumor stages and histological grades were recorded using the classification guidelines of the American Joint Committee on Cancer (AJCC)6. All samples were collected after written informed consent was obtained from the patients, and the experiments were approved by the Conduct of Human Ethics Committee of the First Affiliated Hospital of Xi'an Jiaotong University.
Immunohistochemistry
All tissue specimens were embedded in paraffin and serial 4‐μm sections were cut, deparaffinized and rehydrated. Sections were incubated with primary antibody specific for KIAA0101 (H0009768‐M01, 1:500 dilution; Abnova, Taipei, Taiwan) and PCNA (ab29, 1:1000 dilution; Abcam, Cambridge, UK) at 4°C overnight. They were then incubated with secondary antibody (Goat Anti‐Mouse IgG, 1:200 dilution; Pierce Biotechnology, Inc., Rockford, IL, USA) and 3,3′‐diaminobenzidine was used as the chromogen. The slides were counterstained with hematoxylin solution and then dehydrated and coverslipped. Images of stained sections were obtained using an optical microscope (BX51; Olympus, Tokyo, Japan) equipped with a digital camera (PD71; Olympus). KIAA0101 was scored by multiplication of the percentage of positive tumor cells and staining intensity reported in previous studies.17 The percentage of positive cells was scored as: 0, 0%; 1+, 1–10%; 2+, 11–50%; and 3+, 51–100%. Intensity of staining was graded as follows: grade 0, negative; 1+, weak positive; 2+, moderate positive; and 3+, strong positive. According to these immunohistochemical scores, KIAA0101 expression was then divided into two groups: negative or weak KIAA0101 expression (score ≤3); and positive or strong KIAA0101 expression (score ≥4). All slides were independently evaluated by three investigators (Dr Gang Cui, Dr Shemin Lu and Dr Leilei Pei) and agreement was reached after careful discussion.
RNA extraction and q‐PCR
Total RNA was extracted using the Trizol procedure (Invitrogen, Carlsbad, CA, USA) and reverse transcribed using a First‐strand cDNA Synthesis Kit (Fermentas, Burlington, ON, Canada). SYBR Premix Ex Taq (TaKaRa, Ohtsu, Shiga, Japan) was used for q‐PCR analysis. All PCR reactions were performed in a final volume of 20 μL containing 1 μL of cDNA template on an iQ5 system (Bio‐Rad, Hercules, CA, USA). Each sample was analyzed in triplicate and analyses were repeated three times. As an internal standard, a fragment of human endogenous β‐actin was amplified simultaneously in each PCR. The primer sequences were as follows: KIAA0101, F, 5′‐TCC TGA AGA GGC AGG AAG CAG T‐3′, R, 5′‐ TTG TGT GAT CAG GTT GCA AAG GA‐3′; and β‐actin, F, 5′‐TGG CAC CCA GCA CAA TGA A‐3′, R, 5′‐CTA AGT CAT AGT CCG CCT AGA AGC A‐3′. The cycle parameters were as follows: denaturing for 15 s at 95°C, annealing for 20 s at a specific temperature 62°C, and extending for 20 s at 72°C for a total of 40 cycles. To quantitatively analyze the differential expression of KIAA0101, a panel of tumor tissues and paired non‐cancerous tissues were used. Values were calculated using the comparative threshold cycle (C t) method after normalization to the control housekeeping gene β‐actin and are reported as T : N ratios. KIAA0101 was defined as overexpressed in GC tissues when the T : N ratio was >2 or in paired non‐cancerous tissues when the T : N ratio was <0.5.
Protein extraction and western blotting
The tissues were lysed in buffer (50 mM Tris‐HCl, pH 8.0, 150 mM NaCl, 100 μg/mL PMSF, 1% Triton X‐100). After the removal of tissue debris by centrifugation, the protein concentration of lysates was quantified using a BCA kit (Pierce Biotechnology, Inc.). Protein samples (50 μg) were separated using 15% SDS‐PAGE and transferred onto polyvinylidene difluoride membranes (Sigma, St Louis, MO, USA). The membranes were incubated with primary antibodies specific for KIAA0101 (H0009768‐M01, 1:2000 dilution; Abnova) or β‐actin (1:1000; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) at 4°C overnight, and then incubated with secondary antibodies (Goat Anti‐Mouse/Rabbit IgG; 1:2000 dilution; Pierce Biotechnology, Inc.) for 1 h. Reactive proteins were detected using ECL reagents (Pierce Biotechnology, Inc.) and the G:BOX Bio Imaging System (Syngene, Cambridge, UK). The KIAA0101 expression level was determined by normalizing the signal intensity of KIAA0101 to that of β‐actin using Gel‐Pro Analyzer Software (Media Cybernetics, Washington, DC, USA). Western blotting for each sample was performed at least three times. For each sample, the KIAA0101 expression level was scored as high (ratio > 1.0), intermediate (ratio 0.5–1.0) or low (0.2 < ratio < 0.5). Samples with ratios >0.5 were defined as overexpressing KIAA0101.
Cell culture and transfection
Human GC cell lines (MGC80‐3 cells and AGS cells) conserved by our laboratory were cultured in DMEM (Hyclone, Logan, UT, USA) containing 10% fetal calf serum (Hyclone). Transfection of plasmid to cells was performed with X‐tremeGENE HP (Roche, Basel, Switzerland) according to the manufacturer's instructions.
Plasmid constructs
To knockdown endogenous KIAA0101 expression in GC cells, we used pSIREN‐Shuttle vector for expression of short hairpin RNA against a target gene. The sequence of the synthetic oligonucleotides against KIAA0101 was 5′‐ GCA ACC TGA TCA CAC AAA TGA‐3′, and scramble oligonucleotides was 5′‐ GAC GGA CAA CAT AAC TCT CAA‐3′ as a negative control.
Cell viability assay
Cell viability after transfection of shKIAA0101 was determined by the colorimetric 3‐(4,5‐diamethylthiazol‐2‐yl)‐2,5‐dipphenytetrazolium bromide (MTT) assay. One day before transfection the cells were seeded into 96‐well plates (5 × 103 cells/well). The cells were then transfected with shKIAA0101 or negative control plasmid. At 6, 24, 48 and 72 h after transfection, the culture medium was removed and 20 μL MTT solution (5 mg/mL in PBS) was dissolved in 200 μL DMEM, then it was added into each well and incubated at 37°C for 4 h. The supernatant was then carefully removed and 150 μL DMSO (Sigma) was added to each well. Spectrophotometric absorbance was measured at a wavelength of 490 nm using a microtitre reader (Thermo Electron Corporation, Vantaa, Finland). Each experiment was performed three times.
Transwell assay
The filter membrane with 8 μm pores between two chambers was coated with Matrigel BD Biosciences (San Jose, CA, USA). AGS cells transfected with shKIAA0101 or negative control plasmid were starved for 6 h and suspended in DMEM at 1 × 105/mL. Next, 200 μL supernatant of cells were put into the upper chambers, with 600 μL FBS‐containing medium in the lower chamber and incubated for 24 h. The cells in the upper chamber were then carefully removed from the upper membrane. Cells on the lower surface of the membrane were stained with 0.1% crystal violet in methanol for 10 min and observed with a light microscope (IX71; Olympus) and photographed at ×200 magnification.
Scratch test
After transfection with shKIAA0101 or negative control plasmid, AGS cell substrates were wounded with 200 μL pipette tips. After washing, the cells were cultured with 2 mL DMEM for 24 h. Cell migration was observed with a light microscope (IX71; Olympus) and photographed at ×200 magnification.
Statistical analysis
The data analyses were performed using spss 13.0 Software (SPSS, Chicago, IL, USA). Two‐tailed χ2 test and Student's t‐test were used to assess the statistical significance of differences between groups. Multivariate stepwise logistic regression analysis was performed to identify independent variables that were correlated with KIAA0101 overexpression. The Kaplan–Meier method was used to estimate the distribution of survival curves and log‐rank tests were used to compare the distributions between groups. Cox proportional hazards regression analysis was performed to identify independent variables that were correlated with patient survival. P < 0.05 was considered statistically significant.
Results
KIAA0101 was elevated in GC tissue at mRNA and protein levels
To determinate the role of KIA0101, we first detected the KIAA0101 protein level in human GC tissue and human normal gastric tissue. The results of the immunohistochemistry showed that KIA0101 was elevated in human GC tissue, including both early and advanced‐stage gastric cancer tissue, compared with the paired non‐cancerous tissue (Fig. 1). Moreover, we found that a positive expression of KIAA0101 was detected in 35 of 61 (57.4%) GC tissues, whereas only 10 of 61 (16.4%) matched adjacent non‐cancer gastric tissues and four of 17 (23.5%) chronic gastritis tissues displayed KIAA0101 staining. KAA0101 was expressed at higher levels in GC tissue samples than in matched adjacent non‐cancerous gastric tissues (P < 0.001) and chronic gastritis tissues (P = 0.014). In addition, KIAA0101 positive expression was detected in 21 of 32 (65.6%) stage I and II tumor tissues and in 14 of 29 (48.3%) stage III and IV tumor tissues. A comparison of stage III and IV tumors with stage I and II tumors did not reveal any differences in the expression of KIAA0101 (P = 0.171).
Figure 1.
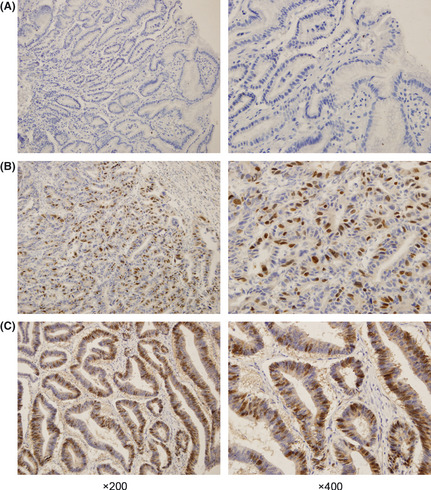
Immunohistochemistry analysis of KIAA0101 protein expression in tissue samples. (A) Expression of KIAA0101 protein is indicated by blue nuclear staining counterstaining in paired non‐cancerous tissue. (B) Elevated expression of KIAA0101 protein is indicated by dark brown nuclear staining in an early stage gastric cancer tissue sample (from a pathological tumor/node/metastasis [pTNM] stage I cancer patient). (C) Strong expression of KIAA0101 protein is indicated by dark brown nuclear staining in advanced‐stage gastric cancer tissue (from a pTNM stage III cancer patient). The images on the right show higher magnification views of representative regions of the images on the left.
To further explore the KIAA010 level in human GC tissue, we obtained 33 paired GC samples and matched adjacent non‐cancerous samples from 61 GC patients who underwent gastrectomy. q‐PCR was used to quantitatively compare the differential expression of KIAA0101 in the paired tissue samples. The data presented in Figure 2A show the ratios of KIAA0101 mRNA expression levels in cancerous tissues to those in non‐cancerous tissues relative to the levels of β‐actin in each tissue. Through the q‐PCR, we found KIAA0101 mRNA was relatively overexpressed in 18 of the 33 (54.5%) GC tissues, while in five of the 33 (15.2%) paired non‐cancerous gastric tissues, only 10 of the 33 (30.3%) pairs of tissues showed no differences in KIAA0101 expression levels between the cancerous and non‐cancerous tissues. Thus, KIAA0101 mRNA expression levels were higher in the majority of human GC tissues than in the paired non‐cancerous tissues (P = 0.001). Because these changes in KIAA0101 mRNA expression might not reflect changes in protein content, we then assessed the expression of KIAA0101 at the protein level in the same 33 paired human GC tissues and matched non‐cancerous gastric tissues. Using western blot analysis, we found that KIAA0101 protein expression was higher in 17 of the 33 (51.5%) GC tissues while in five of the 33 (15.2%) matched adjacent non‐cancerous gastric tissues the expression of KIAA0101 protein was higher in human GC tissues compared with the matched adjacent non‐cancerous gastric tissues (P = 0.002). These data show that KIAA0101 was remarkably overexpressed in GC tissues at both the mRNA and protein levels. Western blot results for four of 33 paired samples are shown in Figure 2B.
Figure 2.
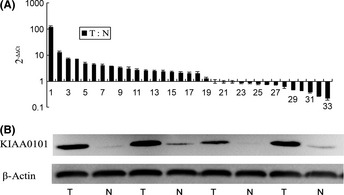
KIAA0101 mRNA and protein expression in gastric tissue samples. (A) q‐PCR analysis of KIAA0101 expression in gastric tumor samples relative to matched non‐cancerous samples. KIAA0101 mRNA was overexpressed in 18 of 33 gastric cancer tissues as the T : N ratio was >2, while KIAA0101 mRNA was overexpressed in five of 33 paired non‐cancerous gastric cancer tissues as the T : N ratio was <0.5. (B) Western blot analysis of KIAA0101 expression in paired tissue samples. KIAA0101 was overexpressed in tumor tissues compared with paired non‐cancerous samples; β‐actin was used as an internal control. N, non‐cancerous tissue; T, tumor tissue.
Correlations of KIAA0101 expression with associated factors
The expression of KIAA0101 was detected more frequently in GC tissues than in non‐cancerous tissues. To better understand the significance of KIAA0101 overexpression, we next explored whether KIAA0101 expression was correlated with GC patient‐associated factors such as age, gender, histological grade, tumor invasion, lymph node invasion, TNM tumor stages and tumor recurrence. Twenty‐six of 61 (42.6%) patients found tumor recurrence during the 2‐year follow up. Correlations of KIAA0101 expression with associated factors was performed using logistic regression analysis in 61 paired tissue samples and the results are shown in Table 1. The results demonstrated that elevated KIAA0101 expression correlated with tumor recurrence (P = 0.008). Unfortunately, we did not find any other significant factors association with KIAA0101 expression in the 61 paired GC tissues. Multivariate stepwise logistic regression analysis did not identify any other independent variables except for tumor recurrence that were significantly correlated with KIAA0101 overexpression (P = 0.008) (Table 1).
Table 1.
Associations between KIAA0101 expression and clinicopathological features in patients with gastric cancer (n = 61)
| Variables | No. cases | T† (%) | P (Uni/Multi)** | N ‡ (%) | P (Uni/Multi)** |
|---|---|---|---|---|---|
| Age (years) | 0.852/0.949 | 0.865/0.708 | |||
| <60 | 29 | 17 (58.6 | 5 (17.2) | ||
| ≥60 | 32 | 18 (56.3) | 5 (15.6) | ||
| Gender | 0.211/0.14 | 0.429/0.462 | |||
| Male | 47 | 29 (61.7) | 9 (19.1) | ||
| Female | 14 | 6 (42.9) | 1 (7.1) | ||
| pTNM stage | 0.171/0.999 | 0.496/0.524 | |||
| I–II | 32 | 21 (65.6) | 4 (12.5) | ||
| III–IV | 29 | 14 (48.3) | 6 (20.6) | ||
| Tumor invasion | 0.956/0.179 | 0.710/0.124 | |||
| T1–T2 | 19 | 11 (57.9) | 4 (21.1) | ||
| T3–T4 | 42 | 24 (57.1) | 6 (14.3) | ||
| Lymph node | 0.481/0.999 | 0.307/0.631 | |||
| Negative | 29 | 18 (62.1) | 3 (10.3) | ||
| Positive | 32 | 17 (53.1) | 7 (21.9) | ||
| Histological grade | 0.315/0.228 | 0.082/0.069 | |||
| Well differentiated | 35 | 22 (62.9) | 3 (8.6) | ||
| Poorly differentiated | 26 | 13 (50.0) | 7 (26.9) | ||
| Tumor recurrence | 0.008/0.008* | 0.854 | |||
| Negative | 35 | 15 (42.9) | 6 (17.1) | ||
| Positive | 26 | 20 (76.9) | 4 (15.4) | ||
*Correlation significant at the 0.01 level (two tailed). **P‐value analyzed using univariate (Uni) and multivariate (Multi) logistic regression analysis. †Number of KIAA0101‐positive expression cases and the percentage in gastric cancer tissues. ‡Number of KIAA0101‐positive expression cases and the percentage in matched adjacent non‐cancerous gastric tissues. pTNM, pathological tumor/node/metastasis.
Increased KIAA0101 expression decreased the survival rate
During follow up, 23 of 61 (37.7%) patients died. We analyzed the effect of elevated expression of KIAA0101 on survival by comparing KIAA0101‐high patients with patients who were not. A significant difference in survival was observed; patients with elevated KIAA0101 expression had poor outcomes and experienced a recurrence (Table 1), and the survival time was short based on the log‐rank test (P = 0.008) (Fig. 3). In addition, survival time was correlated with tumor invasion (P = 0.004), lymph node invasion (P < 0.001) and TNM stages (P < 0.001) based on the log‐rank survival analysis. However, in GC patients, survival time was not influenced by the patient's age (P = 0.914), gender (P = 0.450) or histological grade (P = 0.378). Also, survival time was not influenced by KIAA0101 expression in adjacent non‐cancerous gastric tissues (P = 0.842). However, based on the Cox regression analysis, survival was influenced by the TNM stage (P < 0.001) and KIAA0101 expression (P < 0.001). The results are shown in Table 2. Based on the Cox regression analysis of patients with elevated KIAA0101 expression, the risk ratio was very high, as shown in Table 2.
Figure 3.
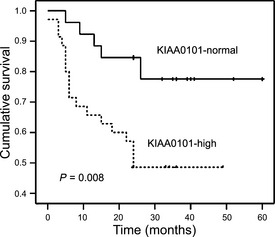
Kaplan–Meier analysis of 2‐year survival in 61 gastric cancer patients. The survival rate was lower in patients with high KIAA0101 expression than patients with normal KIAA0101 expression (P = 0.008).
Table 2.
Cox proportional hazards regression model of prognostic variables for overall survival
| Prognostic variable | P‐value | Risk ratio (95% CI) |
|---|---|---|
| KIAA0101 | <0.001a | 7.68 (2.52–23.36) |
| TNM stage | <0.001a | 18.88 (5.37–66.37) |
| Histological grade | 0.13 | 0.50 (0.21–1.23) |
| Gender | 0.38 | 1.77 (0.50–6.30) |
| Age | 0.66 | 0.82 (0.35–1.93) |
Correlation significant at the 0.01 level (two tailed). CI, confidence interval.
KIAA0101 promoted the viability of GC cells
We detected the KIAA0101 expression levels in two GC cell lines (MGC80‐3 cells and AGS cells), all this two GC cell lines could express KIAA0101. To investigate the biological significance of KIAA0101 overexpression in GC cells, we constructed shKIAA0101 and transfected it into GC cells. After 48 h, a significant knockdown effect was observed using western blot; KIAA0101 protein was significantly inhibited compared with the scramble oligonucleotides group (Fig. 4A). In addition, MTT assays revealed a significant reduction in the number of cells transfected with shKIAA0101 compared with the scramble oligonucleotides group, which revealed no knockdown effect. MTT assays performed at 6, 24, 48 and 72 h after transfection (Fig. 4B,C) suggested that KIAA0101 could serve as a proliferation marker. Also, there was a linear relationship between KIAA0101 and PCNA (Fig. 4D). All of these results suggest that KIAA0101 could serve as a marker of cell proliferation.
Figure 4.
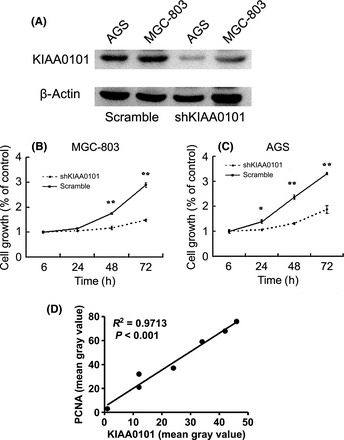
Effect of KIAA0101 knockdown by shRNA on viability in gastric cancer cells. (A) Knockdown effect on KIAA0101 transcript was validated using western blot with β‐actin expression as a quantitative control. Transfection with shKIAA0101 showed significant knockdown effect, whereas scramble oligonucleotide plasmid showed no effect on the level of KIAA0101 expression. (B,C) Cell viability was detected using a MTT assay with a set of six wells for each experiment. Transfection with shKIAA0101 resulted in significant reduction of the cell viability in gastric cancer cells. Levels of significance were calculated using the Student's t‐test. *Correlation significant at the 0.05 level (two tailed). **Correlation significant at the 0.01 level (two tailed). (D) Immunohistochemistry results of KIAA0101 and proliferating cell nuclear antigen (PCNA) were analyzed using software Image 5.1 (Media Cybernetics). The relationship between KIAA0101 and PCNA mean gray value was analyzed.
KIAA0101 promoted the invasion and migration of GC cells
To further explore the role of KIAA0101 in GC cells, we detected the invasion and migration of AGS transfected with shKIAA0101 or scramble oligonucleotide plasmid using the transwell test and scratch test. The result of the transwell test showed that in the shKIAA0101 group, AGS cells on the low surface of the membrane decreased (Fig. 5A), suggesting that KIAA0101 could promote cell invasion. The scratch test results also showed that KIA0101 could promote the direct migration of AGS cells (Fig. 5B).
Figure 5.
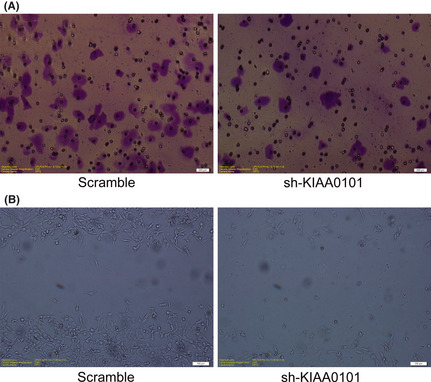
Effect of KIAA0101 knockdown by shRNA on invasion and migration in gastric cancer cells. (A) The invasiveness of AGS was evaluated using matrigel‐coated transwell assay after sh‐KIAA0101 or scramble oligonucleotide plasmid transfection for 24 h. Cells invading into the lower surface of the membrane were stained with 0.1% crystal violet. (B) The migration of AGS was evaluated using the scratch test after sh‐KIAA0101 or scramble oligonucleotide plasmid transfection for 24 h. Original magnification, ×200.
Discussion
In the present study, we found that KIAA0101 was significantly overexpressed in GC samples. What is more important, patients with tumors with elevated KIAA0101 expression showed a higher recurrence rate and significantly decreased survival. After suppressing KIAA0101 by shRNA in GC cells, the cell viability, cell invasion and migration were significantly inhibited. Although there was enough data to show that GC cells are dependent on overexpression of KIAA0101, these two independent findings in vivo and in vitro suggest that KIAA0101 overexpression is an important factor related to the enhanced proliferative potential of GC cells and a predictive marker of poor prognosis. Although similar results have been demonstrated in other cancers, to our knowledge we are the first to demonstrate the connection between KIAA0101 overexpression and GC.
KIAA0101 encodes a 15 kDa protein that contains a conserved PCNA‐binding motif.5 The PCNA interacts with several DNA replication proteins and is an essential scaffold molecule for DNA replication and repair.20, 21 KIAA0101 shares the PCNA‐binding motif with many important cell cycle regulatory PCNA‐binding proteins, such as p21, p57 and p33ING1b.5, 12, 15, 22 Taken together, these findings suggest that KIAA0101 is likely a member of a DNA replication complex in replication foci and positive related to PCNA. During tumorigenesis, PCNA and Ki‐67 serve as wellness proliferation markers and are standard analysis with many tumors, these indicate that KIAA0101 also serves as a proliferation marker. In the present study, we demonstrated that KIAA0101 was remarkably overexpressed in GC tissues at both the mRNA and protein levels, and by suppressing KIAA0101 using shRNA, the GC cell viability was significantly inhibited. These findings imply that KIAA0101 has growth‐promoting properties. However, KIAA0101 expression was still detected in some non‐cancerous tissues or chronic gastritis tissue samples. In a previous study, Simpson et al.12 demonstrated localization of KIAA0101 in the basal cell layer of the skin and the proliferation zone in the crypts of the colon; a similar result has been reported by Jain et al.23 in adrenocortical cells. This pattern was particularly apparent in areas of high cell proliferation, suggesting that KIAA0101 overexpression has the potential to facilitate cell proliferation both under physiological conditions and in human cancers.12, 23 In the present study, elevated KIAA0101 expression in non‐cancerous gastric tissues and chronic gastritis tissues were expected to reflect a growth advantage for cells.
In the present study, GC patients with elevated KIAA0101 expression had poor outcomes, the survival time was short and the risk ratio was very high. These findings show that KIAA0101 overexpression is an important factor related to poor prognosis. We analyzed the relationship between KIAA0101 expression and patients’ associated factors such as age, gender, tumor histological grade, tumor invasion, lymph node invasion and tumor stage. Unfortunately, we did not find any significant factors associated with KIAA0101 expression in the 61 paired GC tissues. However, patients with tumors with elevated KIAA0101 expression showed a higher recurrence rate after surgery. Taken together with previous findings, KIAA0101 is likely an important factor associated with cell proliferation and contributes to surgery failure in GC patients, might not be a function in tumor invasion and varied in tumor stages. In other studies, epigenetic silencing of tumor‐associated genes by promoter hypermethylation is increasingly recognised to play an instrumental role in cancer development.24 Kolesnikova et al. and Leung et al.25, 26 observed KIAA0101 promoter methylation in the blood of GC patients; methylation of this gene was found to be higher in patients with advanced tumor stages. To fully understand the different expression of KIAA0101 in GC, a large‐scale study of patients is needed to rule out statistical differences.
In a previous study, suppression of KIAA0101 expression in pancreatic cancer cells or inhibition of the KIAA0101‐PCNA interaction using cell‐permeable dominant‐negative peptides resulted in growth suppression of cancer cells, suggesting that KIAA0101 might be a promising target for the development of novel anticancer therapies.15 Moreover, cells expressing functional KIAA0101 might be susceptible to programmed cell proliferation, leading to tumor progression and recurrence. Thus, KIAA0101 expression levels might be considered when deciding whether further treatment should be administered to patients with local disease, because chemotherapy mainly affects dividing cells and therefore should be most successful in rapidly proliferating tumors.27 However, in other studies, KIAA0101 was downregulated and found to be a growth inhibitory gene.28 Based on these results and the present study, KIAA0101 appears to be involved in disease progression of various cancers; however, its contribution might differ in different cancers.
Conclusion
The present study indicates that KIAA0101 expression is upregulated in the majority of GC. In addition, a high level of KIAA0101 expression is correlated with a poor prognosis. Assessment of KIAA0101 expression might provide prognostic information that might be helpful in determining the most appropriate duration and intensity of treatment. More importantly, this factor might play a role in the recurrence of GC.
Disclosure Statement
The authors have no conflict of interest.
Acknowledgments
The authors express their gratitude to Dr Gang Cui, Dr Shemin Lu and Dr Leilei Pei for their expertise and assistance with the experiments. The authors are grateful to Dr Ni Hou and Dr Peixiang Lan for making valuable suggestions for the experiments. The present study was supported by grants from the National Natural Scientific Foundation of China (no. 81001089 [to K.L.], no. 30973489 [to C.D.] and no. 81000728 [to L.S.]) and Shaanxi research project (no.2010K14‐03 to [KZ]).
(Cancer Sci, doi: 10.1111/cas.12083, 2013)
References
- 1. Crew KD, Neugut AI. Epidemiology of upper gastrointestinal malignancies. Semin Oncol 2004; 31: 450–64. [DOI] [PubMed] [Google Scholar]
- 2. Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin 2009; 59: 225–49. [DOI] [PubMed] [Google Scholar]
- 3. Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol 2006; 24: 2137–50. [DOI] [PubMed] [Google Scholar]
- 4. Macdonald JS, Smalley SR, Benedetti J et al Chemoradiotherapy after surgery compared with surgery alone for adenocarcinoma of the stomach or gastroesophageal junction. N Engl J Med 2001; 345: 725–30. [DOI] [PubMed] [Google Scholar]
- 5. Yu P, Huang B, Shen M et al p15(PAF), a novel PCNA associated factor with increased expression in tumor tissues. Oncogene 2001; 20: 484–9. [DOI] [PubMed] [Google Scholar]
- 6. Petroziello J, Yamane A, Westendorf L et al Suppression subtractive hybridization and expression profiling identifies a unique set of genes overexpressed in non‐small‐cell lung cancer. Oncogene 2004; 23: 7734–45. [DOI] [PubMed] [Google Scholar]
- 7. Mizutani K, Onda M, Asaka S et al Overexpressed in anaplastic thyroid carcinoma‐1 (OEATC‐1) as a novel gene responsible for anaplastic thyroid carcinoma. Cancer 2005; 103: 1785–90. [DOI] [PubMed] [Google Scholar]
- 8. Shi L, Zhang SL, Li K et al NS5ATP9, a gene up‐regulated by HCV NS5A protein. Cancer Lett 2008; 259: 192–7. [DOI] [PubMed] [Google Scholar]
- 9. Li K, Ma Q, Shi L et al NS5ATP9 gene regulated by NF‐kappaB signal pathway. Arch Biochem Biophys 2008; 479: 15–9. [DOI] [PubMed] [Google Scholar]
- 10. Kais Z, Barsky SH, Mathsyaraja H et al KIAA0101 interacts with BRCA1 and regulates centrosome number. Mol Cancer Res 2011; 9: 1091–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Emanuele MJ, Ciccia A, Elia AE, Elledge SJ. Proliferating cell nuclear antigen (PCNA)‐associated KIAA0101/PAF15 protein is a cell cycle‐regulated anaphase‐promoting complex/cyclosome substrate. Proc Natl Acad Sci U S A 2011; 108: 9845–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Simpson F, Lammerts van Bueren K, Butterfield N et al The PCNA‐associated factor KIAA0101/p15(PAF) binds the potential tumor suppressor product p33ING1b. Exp Cell Res 2006; 312: 73–85. [DOI] [PubMed] [Google Scholar]
- 13. Turchi L, Fareh M, Aberdam E et al ATF3 and p15PAF are novel gatekeepers of genomic integrity upon UV stress. Cell Death Differ 2009; 16: 728–37. [DOI] [PubMed] [Google Scholar]
- 14. van Bueren KL, Bennetts JS, Fowles LF, Berkman JL, Simpson F, Wicking C. Murine embryonic expression of the gene for the UV‐responsive protein p15(PAF). Gene Expr Patterns 2007; 7: 47–50. [DOI] [PubMed] [Google Scholar]
- 15. Hosokawa M, Takehara A, Matsuda K et al Oncogenic role of KIAA0101 interacting with proliferating cell nuclear antigen in pancreatic cancer. Cancer Res 2007; 67: 2568–76. [DOI] [PubMed] [Google Scholar]
- 16. Notterman DA, Alon U, Sierk AJ, Levine AJ. Transcriptional gene expression profiles of colorectal adenoma, adenocarcinoma, and normal tissue examined by oligonucleotide arrays. Cancer Res 2001; 61: 3124–30. [PubMed] [Google Scholar]
- 17. Kato T, Daigo Y, Aragaki M, Ishikawa K, Sato M, Kaji M. Overexpression of KIAA0101 predicts poor prognosis in primary lung cancer patients. Lung Cancer 2012; 75: 110–8. [DOI] [PubMed] [Google Scholar]
- 18. Yuan RH, Jeng YM, Pan HW et al Overexpression of KIAA0101 predicts high stage, early tumor recurrence, and poor prognosis of hepatocellular carcinoma. Clin Cancer Res 2007; 13: 5368–76. [DOI] [PubMed] [Google Scholar]
- 19. American Joint Committee on Cancer (AJCC) , 7th edn. Berlin, Germany: Springer Science and Business Media LLC, 2010. [Google Scholar]
- 20. Bravo R, Frank R, Blundell PA, Macdonald‐Bravo H. Cyclin/PCNA is the auxiliary protein of DNA polymerase‐delta. Nature 1987; 326: 515–7. [DOI] [PubMed] [Google Scholar]
- 21. Zhang Z, Shibahara K, Stillman B. PCNA connects DNA replication to epigenetic inheritance in yeast. Nature 2000; 408: 221–5. [DOI] [PubMed] [Google Scholar]
- 22. Li R, Waga S, Hannon GJ, Beach D, Stillman B. Differential effects by the p21 CDK inhibitor on PCNA‐dependent DNA replication and repair. Nature 1994; 371: 534–7. [DOI] [PubMed] [Google Scholar]
- 23. Jain M, Zhang L, Patterson EE, Kebebew E. KIAA0101 is overexpressed, and promotes growth and invasion in adrenal cancer. PLoS ONE 2011; 6: e26866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature 2004; 429: 457–63. [DOI] [PubMed] [Google Scholar]
- 25. Kolesnikova EV, Tamkovich SN, Bryzgunova OE et al Circulating DNA in the blood of gastric cancer patients. Ann N Y Acad Sci 2008; 1137: 226–31. [DOI] [PubMed] [Google Scholar]
- 26. Leung WK, To KF, Chu ES et al Potential diagnostic and prognostic values of detecting promoter hypermethylation in the serum of patients with gastric cancer. Br J Cancer 2005; 92: 2190–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Miller WR. Clinical, pathological, proliferative and molecular responses associated with neoadjuvant aromatase inhibitor treatment in breast cancer. J Steroid Biochem Mol Biol 2010; 118: 273–6. [DOI] [PubMed] [Google Scholar]
- 28. Guo M, Li J, Wan D, Gu J. KIAA0101 (OEACT‐1), an expressionally down‐regulated and growth‐inhibitory gene in human hepatocellular carcinoma. BMC Cancer 2006; 6: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]


