Abstract
Smoking cessation is the most effective measure for reducing the risk of smoking-related diseases. However, switching to less harmful products (modified-risk tobacco products [MRTP]) can be an alternative to help reduce the risk for adult smokers who would otherwise continue to smoke. In an 18-month chronic carcinogenicity/toxicity study in A/J mice (OECD Test Guideline 453), we assessed the aerosol of Tobacco Heating System 2.2 (THS 2.2), a candidate MRTP based on the heat-not-burn principle, compared with 3R4F cigarette smoke (CS). To capture toxicity- and disease-relevant mechanisms, we complemented standard toxicology endpoints with in-depth systems toxicology analyses. In this part of our publication series, we report on integrative assessment of the apical and molecular exposure effects on the respiratory tract (nose, larynx, and lungs). Across the respiratory tract, we found changes in inflammatory response following 3R4F CS exposure (eg, antimicrobial peptide response in the nose), with both shared and distinct oxidative and xenobiotic responses. Compared with 3R4F CS, THS 2.2 aerosol exerted far fewer effects on respiratory tract histology, including adaptive tissue changes in nasal and laryngeal epithelium and inflammation and emphysematous changes in the lungs. Integrative analysis of molecular changes confirmed the substantially lower impact of THS 2.2 aerosol than 3R4F CS on toxicologically and disease-relevant molecular processes such as inflammation, oxidative stress responses, and xenobiotic metabolism. In summary, this work exemplifies how apical and molecular endpoints can be combined effectively for toxicology assessment and further supports findings on the reduced respiratory health risks of THS 2.2 aerosol.
Keywords: cigarette smoke, heated tobacco product, mouse, respiratory, histology, gene expression, protein expression
Smoking causes several diseases, including those of the cardiovascular and respiratory systems. Chronic obstructive pulmonary disease (COPD) is a global health problem, for which tobacco smoke exposure is a major risk factor (Postma et al. 2015). Smoking cessation is the most effective measure for reducing the risk of smoking-related diseases (Godtfredsen et al. 2008). However, switching to less harmful products can be an alternative to help address the harm caused by smoking.
Modified-risk tobacco products (MRTP) are defined as “any tobacco product that is sold or distributed for use to reduce harm or the risk of tobacco-related disease associated with commercially marketed tobacco products” (US Family Smoking Prevention and Tobacco Control [Family Smoking Prevention and Tobacco Control Act [FSPTCA] 2009]). Tobacco Heating System (THS) 2.2 is a candidate MRTP developed by Philip Morris International. It leverages the heat-not-burn principle (Smith et al. 2016): tobacco is electronically heated in a controlled fashion to release nicotine and volatiles that contribute to tobacco flavors, while preventing combustion. This produces an aerosol with a lower number and lower levels of harmful and potentially harmful constituents than cigarette smoke (CS) (Schaller et al. 2016).
Most diseases caused by CS develop after an extended period of exposure. Thus, chronic toxicity studies are a relevant component of the comparative assessment of MRTP aerosols and CS. OECD Test Guideline 453 provides recommendations for the conduct of combined chronic toxicity/carcinogenicity studies (OECD 2018); the objective of such studies is to identify possible health hazards that arise from repeated exposure for a period lasting up to the lifespan of the investigated species. However, studies that measure only standard toxicological endpoints can miss low-level effects and gain only limited insights into toxicological mechanisms (Ellinger-Ziegelbauer et al. 2011; Titz et al. 2018). Thus, systems toxicology studies complement these standard endpoints by extensive molecular measurements using omics technologies, such as transcriptomics and proteomics (Hartung et al. 2017; Titz et al. 2014, 2016).
The lungs are the main target organs in smoking-related diseases in the respiratory tract. However, CS exposure has been demonstrated to affect the whole respiratory tract. In rodent studies, CS exposure commonly affects tissue architecture and molecular responses in the nasal epithelia (Oviedo et al. 2016; Phillips et al. 2019b; Wong et al. 2016a), including both adaptive (eg, squamous differentiation) and adverse (eg, tissue atrophy) changes. The larynx of rodents is especially sensitive to irritating exposures and, for example, responds with squamous differentiation and basal cell hyperplasia (Osimitz et al. 2007; Oviedo et al. 2016; Phillips et al. 2019b). In smokers, CS has been found to cause a “field of molecular injury throughout the airway epithelium” (Gower et al. 2011; Sridhar et al. 2008), which includes buccal and nasal epithelium. Although CS-triggered molecular responses differ across the respiratory tract, the upper airway has been proposed as a potential convenient sampling source for biomarkers of lung diseases (Gower et al. 2011; Talikka et al. 2017).
Because of its high susceptibility to lung tumors, the A/J mouse model is widely used as a screening system in carcinogenicity testing, including for CS exposure (Stinn et al. 2010, 2013; Witschi et al. 2002). Furthermore, A/J mice are well suited for studying smoking-induced COPD by induction of pronounced lung inflammation and emphysematous changes (Cabanski et al. 2015; Stinn et al. 2013).
Here, we report on the respiratory effects of chronic exposure to THS 2.2 aerosol compared with those of 3R4F CS exposure in A/J mice. By using systems toxicology approaches, the current work focuses on the histological and molecular effects of exposure across the respiratory tract, with a special emphasis on the relationship between these apical and molecular endpoints. For more details on the overall study execution and data on OECD endpoints for chronic toxicity and carcinogenicity, the reader is referred to our accompanying publication (Wong et al. 2020).
MATERIALS AND METHODS
Inhalation study
This study was conducted to evaluate lung tumor incidence and multiplicity, extent of lung inflammation and emphysematous changes, and toxicity from lifetime chronic inhalation of mainstream aerosol from the candidate MRTP THS 2.2 in A/J mice relative to those from mainstream smoke from the reference cigarette 3R4F. The study design was based on OECD Test Guideline 453 (OECD 2018), adapted for exposure of substances by inhalation. The study design and details of generation and administration of the test atmospheres are provided in an accompanying publication (Wong et al. 2020) and the Supplementary Material.
Briefly, female A/J mice were exposed to fresh air (sham), 3 concentrations of THS 2.2 aerosol corresponding to nicotine concentrations of 6.7 (low; L), 13.4 (medium; M), and 26.8 (high; H) µg/l test atmosphere, or 1 concentration of 3R4F CS (13.4 µg nicotine/l test atmosphere) in whole-body exposure chambers for 6 h per day, 5 days per week, and up to 18 months. Interim dissections were scheduled for months 1, 5, and 10. Male mice were exposed to either fresh air (sham) or THS 2.2 (H) aerosol for 15 months. Chronic toxicity and carcinogenicity endpoints outlined in the OECD protocol were assessed, and the detailed findings are included in Wong et al. (2020). Several tissues were collected for further analysis for evaluation of non-OECD endpoints by following a systems toxicology approach (proteomics, transcriptomics, and genomics).
Housing and all procedures involving animals were performed in accordance with the approved Institutional Animal Care and Use Committee protocol in a facility licensed by the Agri-Food & Veterinary Authority of Singapore and accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International, where the protocols for care and use of animals for scientific purposes were in accordance with the National Advisory Committee for Laboratory Animal Research (NACLAR) Guideline (NACLAR, 2004).
Animals allocated to omics endpoints (N = 8–20) were dissected within 16–24 h of the last exposure and following randomization of all planned necropsies for the dissection time point in question. All efforts were made to minimize potential nucleic acid and protein degradation, and samples were frozen as rapidly as possible once ex vivo. Respiratory nasal epithelia (RNE), larynx, and lung tissues were collected for investigating the effects of chronic THS 2.2 aerosol exposure on the respiratory tract.
Because the most proximal part of the trachea was dissected together with the larynx, and further trimming and separation of the tissues was not desirable in favor of preserving nucleic acid integrity, laryngeal and tracheal epithelia were combined (termed “larynx” herein) for transcriptomics analysis.
Lungs collected after month 1 were snap frozen, and the entire left lung was dedicated to transcriptomics analysis, whereas the right caudal lobe was used for proteomics analysis. From month 5 onwards, the lungs were instilled with and embedded in Tissue-Tek optimum cutting temperature compound (InLab Supplies Pte Ltd, Singapore), and lung parenchymal or tumor cells were specifically collected by subsequent laser-capture microdissection. The analysis reported here employed microarray data from lung parenchyma samples.
Histopathological evaluation
Histoprocessing details are summarized in the Supplementary Material. Histological sections of respiratory tract organs were prepared at defined levels: nose transverse sections at posterior to the upper incisor, posterior to incisive papilla, and at the first molar teeth; larynx transverse sections at the base of the epiglottis and arytenoid projections; trachea, 1 transverse and 1 longitudinal section at bifurcation; lung, 4-µm serial sections taken at intervals of 300 µm, starting from the first visible tissue to the last visible tissue in the paraffin block. All sections were stained with hematoxylin and eosin (MilliporeSigma, Burlington, Massachusetts). Sections from nose level 1, tracheal bifurcation, and lungs at the main bronchus level were additionally stained with Alcian blue-periodic acid-Schiff reagent (all Merck, Kenilworth, New Jersey, except periodic acid [MilliporeSigma]) for goblet cells. Lung sections at the main bronchus level were also stained with resorcin-fuchsin (Electron Microscopy Sciences, Hatfield, Pennsylvania) for elastic fibers and collagen.
Histopathological evaluation of the upper respiratory organ sections was performed by using scanned digital slides (Aperio, Leica Biosystems, Wetzlar, Germany), and the study pathologist was blinded to the exposure groups. No coding was assigned with respect to dissection time point, sex, or date of necropsy. Decoding was performed from decoding lists provided by the test facility upon completion of the histopathological evaluation. Incidences of histopathological findings were recorded, and the severity of lesions was scored in accordance with a defined severity scale from 0 to 5, with 0 indicating findings within normal limits; 1, minimal changes; 2, minimal to moderate changes; 3, moderate changes; 4, moderate to severe changes; and 5, severe changes.
The histopathological findings were peer-reviewed by Comparative Biosciences, Inc. (Sunnyvale, California) to confirm the interpretation.
Gene and protein expression analyses
Details on gene and protein expression analyses are provided in the Supplementary Material. Briefly, Affymetrix microarrays (Thermo Fisher Scientific, Waltham, Massachusetts) were used to generate mRNA and miRNA expression profiles for the RNE, larynx, and lungs. An iTRAQ-based quantitative proteomics approach (AB Sciex, Framingham, Massachusetts) was used to generate protein expression profiles for the RNE and lungs. Multiplexed iTRAQ labeling sets were separated on a 50-cm Acclaim PepMap 100 C18 LC column (2-μm particle size; Thermo Fisher Scientific) and analyzed on a Q Exactive mass analyzer (Thermo Fisher Scientific).
Statistical and computational analyses
For pairwise statistical analysis, a linear model was fitted for each exposure condition and the respective sham group. p values were calculated from moderated t statistics with the empirical Bayes approach (Gentleman et al. 2004), and genes/proteins with a Benjamini-Hochberg false discovery rate (FDR)-adjusted p value < .05 were considered differentially expressed.
The ordinal principal component (PC) analysis for the histopathology score matrix was generated by using the princals function of the Gifi R package (de Leeuw 2005). To associate individual histopathology scores with PC scores, we fitted ordinal logistic regression models and visualized the predicted scores dependent on the PC score.
Transcriptomic data were also analyzed in the context of hierarchically structured network models describing the molecular mechanisms underlying essential biological processes in the respiratory tract (Boué et al. 2015; Hoeng et al. 2012). By leveraging the “cause-and-effect” network models together with network perturbation amplitude (NPA) algorithms, gene expression fold changes were translated into differential values for each network node (Martin et al. 2012, 2014). These were, in turn, summarized into a quantitative NPA measure, and NPA values were aggregated into a biological impact factor; details have been described elsewhere (Kogel et al. 2014; Phillips et al. 2015).
To study the structure of the transcriptomic response to exposure in respiratory tract tissues, we first calculated differential expression matrices, which are obtained from expression matrices by subtracting from each sample the average expression in the time-matched sham group. We then used the t-distributed stochastic neighbor embedding (t-SNE) algorithm to perform a dimensional reduction of the differential expression matrices from 17 473 genes to 2 dimensions. Unlike principal component analysis, this nonlinear unsupervised approach provides information about the data structure at multiple scales (van der Maaten and Hinton 2008). We used this essentially qualitative information to define sample “super-groups” and a selection of their relative contrasts to be used in the same statistical analysis framework as that for the sample group-based pairwise comparisons described above. The t-SNE calculations were performed by using the R package Rtsne available on CRAN (https://cran.r-project.org/web/packages/Rtsne). We used the default options and considered 19 perplexity values from 5 to 95 to select the most representative one. To further investigate these tissue and time dependencies, we used 2 linear models, one for comparing mean tissue responses over time and another for assessing the time dependencies within each tissue. Gene-set enrichment analysis was performed by using the fgsea algorithm (Sergushichev 2016) and piano package for R (Väremo et al. 2013). Enrichment of gene sets from the Reactome database was evaluated (Fabregat et al. 2018).
Ordinal logistic regression models were used to associate changes in histopathological endpoints and gene expression across the 3 respiratory tract organs. For this, the glmnetcr function in the glmnetcr package was used with a ridge penalty, and the lambda constraint was selected by using the Bayesian information criterion (Archer and Williams 2012). To leverage data across respiratory tract locations, we concatenated the gene expression data matrices for the RNE, larynx, and lungs and selected representative histopathological endpoints for each category for each location. For “Cornification,” we selected “Epithelium, cornification” at the “Larynx—base of epiglottis” and “Respiratory epithelium, cornification” at “Nose level 1” (absent in the lungs). For “Squamous metaplasia”, we selected “Epithelium, squamous epithelial metaplasia” at the “Larynx—base of epiglottis” and “Respiratory epithelium, squamous epithelial metaplasia” at “Nose level 1” (absent in the lungs). For “Yellow macrophages”, we selected “Alveolar lumen, yellow pigmented macrophages” in “left lung” (absent in other organs). For “Emphysema”, we selected “Emphysema” in “left lung” (absent in other organs). For “Hyperplasia”, we selected “Epithelium, hyperplasia” at “Larynx—base of epiglottis”, “Respiratory epithelium, hyperplasia” at “Nose level 1”, and “Bronchi, reserve cell hyperplasia” in “left lung”. Because histopathological and molecular data were only available from separate dissection groups, we matched histopathology scores to molecular samples at the group level by employing random procedure sampling from the empirical score distribution of the respective group. For predictor estimation, we repeated this sampling 10 times. In addition, we performed 5-fold cross-validation to assess the overall performance of the models on the basis of a single, randomly selected score sample.
Multi-omics factor analysis (MOFA) was performed with the corresponding package (version 0.99.8) in the R statistical environment (version 3.5.1) by using the default model and train options (Argelaguet et al. 2018). For each evaluated tissue type, a separate MOFA model was established, including the available data modalities for that tissue type. Because lung miRNA and protein profiling was only conducted for month 1, only data from this time point were considered for the lung MOFA model. Gene set enrichment analysis was performed by using the enrichment algorithm implemented in the MOFA package (local statistics, loading; transformation, absolute value; global statistics, mean difference; statistical test, parametric; FDR threshold, 0.01). Enrichment of gene sets from the Reactome database was evaluated (Fabregat et al. 2018).
RESULTS
Study Design and Exposure Characterization
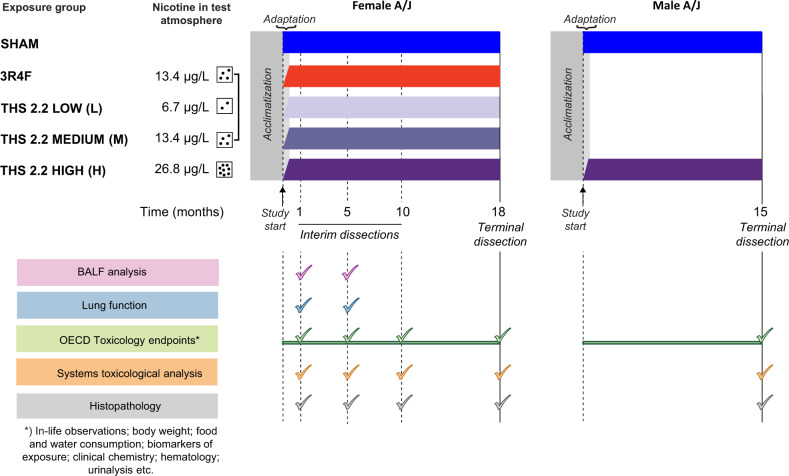
This 18-month assessment study included 7 study arms (Figure 1): fresh air (sham) exposure and high THS 2.2 aerosol exposure (THS H) for female and male mice and 3R4F CS, low THS 2.2 (THS L), and medium THS 2.2 (THS M) exposure for female mice. Female mice were evaluated for effects on a battery of molecular and apical endpoints after 1, 5, 10, and 18 months of exposure; male mice were evaluated after 15 months of exposure (Table 1).
Figure 1.
Study design. Female A/J mice were exposed to filtered air (sham), 3 concentrations of Tobacco Heating System (THS) 2.2 aerosol (6.7, 13.4, and 26.8 µg/l nicotine), or 3R4F cigarette smoke (13.4 µg/l nicotine). Additionally, male mice were exposed to filtered air or THS 2.2 aerosol (26.8 µg/l nicotine). The experimental period was preceded by 25 days of acclimatization. Interim dissections of subgroups of female mice were performed after 1, 5, and 10 months of exposure. Terminal dissections of the male and female mice were performed in months 15 and 18, respectively. At selected time points, the animals were allocated for analysis of the following endpoints: OECD toxicology endpoint analyses (mortality, hematological analysis, clinical chemistry analysis, and urinalysis), bronchoalveolar lavage fluid (BALF) analysis by flow cytometry and multi-analyte (cytokine/chemokine, growth factor) profiling, histopathological evaluation of respiratory and nonrespiratory tract organs, lung function tests, lung morphometry, lung tumor analysis, and systems toxicology analysis (transcriptomics, proteomics, and DNA sequencing). THS, tobacco heating system; CS, cigarette smoke; OECD, Organisation for Economic Co-operation and Development; BALF, bronchoalveolar lavage fluid.
Table 1.
Tissues and Main Endpoints
| Tissue | Endpoint | Time Pointsa (months) |
|---|---|---|
| Respiratory nasal epithelium | Histopathology | 1 m (N = 11), 5 m (N = 12), 10 m (N = 11–12), 18 m (N = 54–99) |
| mRNA expression | 1 m (N = 8), 5 m (N = 8), 10 m (N = 10–12), 18 m (N = 16–20) | |
| miRNA expression | 1 m (N = 8), 5 m (N = 8), 10 m (N = 10–12), 18 m (N = 16–20) | |
| Protein expression | 1 m (N = 8), 5 m (N = 8), 10 m (N = 10–12), 18 m (N = 16–20) | |
| Larynx | Histopathology | 1 m (N = 10–11), 5 m (N = 11–12), 10 m (N = 11–12), 18 m (N = 54–95) |
| mRNA expression | 1 m (N = 8), 5 m (N = 8), 10 m (N = 10–12), 18 m (N = 16–20) | |
| miRNA expression | 1 m (N = 8), 5 m (N = 8), 10 m (N = 10–12), 18 m (N = 16–20) | |
| Lung | Histopathology | 1 m (N = 10–11), 5 m (N = 12), 10 m (N = 11–12), 18 m (N = 53–98) |
| mRNA expression | 1 m (N = 8), 5 m (N = 8), 10 m (N = 10–12), 18 m (N = 16–20) | |
| miRNA expression | 1 m (N = 8) | |
| Protein expression | 1 m (N = 8) |
Number of replicates (N) refers to the number of animals.
For THS (M) and 3R4F, nicotine exposure concentrations in aerosol/smoke were matched at 13.4 µg nicotine/l. This nicotine dose—assuming a 0.03-l/min minute volume, 25-g body weight, and complete uptake of nicotine and considering the exposure regimen and a body surface conversion factor of 12.3 (CDER 2005)—equates to approximately 140 µg nicotine per day, corresponding to a human equivalent nicotine dose of 77 mg or about 3 packs of cigarettes per day. Measurement of nicotine metabolites in urine confirmed aerosol/smoke uptake and a dose-dependent increase in nicotine metabolites with increasing THS 2.2 aerosol concentrations (Table 2). In line with the reduced levels of harmful and potentially harmful constituents in THS 2.2 aerosol relative to those in 3R4F CS (Schaller et al. 2016), the levels of blood carboxyhemoglobin and urinary markers of acrolein (hydroxypropyl mercapturic acid), benzene (S-phenylmercapturic acid), acrylonitrile (2-cyanoethylmercapturic acid), and nicotine-derived nitrosamine ketone [4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol] exposure were substantially lower in the THS 2.2 groups than in the 3R4F group, and these levels generally remained close to those in sham-exposed animals. The high basal levels of hydroxypropyl mercapturic acid, including those in the sham group, can be explained by endogenous acrolein production (Stevens and Maier 2008).
Table 2.
Exposure characterization.
| Matrix | Endpoint | Time (days) | Sham/F | 3R4F/F | THS (L)/F | THS (M)/F | THS (H)/F | Sham/M | THS (H)/M |
|---|---|---|---|---|---|---|---|---|---|
| Urine | Total nicotine metabolites (nmol) | 37–54 | 2.81 ± 0.46 [12] | 492.37 ± 46.28 [12]* | 235.38 ± 20.23 [12]*# | 435.08 ± 57.52 [12]* | 695.32 ± 61.14 [10]*# | 2.18 ± 0.22 [12] | 1023.31 ± 83.49 [12]* |
| 156–180 | 15.6 ± 14.3 [11] | 388.93 ± 27.38 [11]* | 250.6 ± 17.32 [11]*# | 638.76 ± 86.1 [10]*# | 723.65 ± 66.67 [12]*# | 1.88 ± 0.16 [12] | 756.53 ± 94.43 [9]* | ||
| 268–292 | 1.28 ± 0.21 [12] | 247.47 ± 33.51 [11]* | 308.28 ± 23.66 [13]* | 476.36 ± 40.18 [12]*# | 636.49 ± 73.17 [10]*# | 1.57 ± 0.4 [14] | 682.86 ± 101.73 [8]* | ||
| 387–411 | 1.54 ± 0.15 [11] | 279.04 ± 29.39 [11]* | 243.2 ± 28.6 [11]* | 480.45 ± 57.7 [12]*# | 843.99 ± 75.55 [14]*# | 2.03 ± 0.23 [16] | 1053.14 ± 134.74 [11]* | ||
| Hydroxypropyl mercapturic acid (ng) | 37–61 | 3,505.83 ± 535.84 [8] | 8,071.93 ± 827.84 [8]* | 3,629.32 ± 335.2 [8]# | 3,744.14 ± 522.57 [8]# | 4,686.37 ± 420.74 [8]# | |||
| 156–180 | 2,780.82 ± 267.2 [8] | 7,230.68 ± 514.64 [8]* | 2,904.94 ± 341.46 [8]# | 3,069.31 ± 226.25 [8]# | 3,875.35 ± 476.11 [8]# | ||||
| 268–292 | 3,072.79 ± 378.82 [8] | 7,078.7 ± 1167.21 [8]* | 2,917.82 ± 448.25 [8]# | 2,345.8 ± 321.16 [8]# | 3,177.94 ± 397.37 [8]# | ||||
| 387–411 | 1,562.3 ± 253.04 [8] | 5,143.63 ± 1232.02 [8]* | 1,814.98 ± 316.1 [8]# | 2,574.59 ± 403.44 [8] | 3,376.89 ± 565.27 [8]* | 5,219.38 ± 1,024.69 [7] | 9,166.31 ± 921.25 [8]* | ||
| S-Phenylmercapturic acid (ng) | 37–61 | 1.95 ± 0.28 [8] | 293.96 ± 21.05 [8]* | 3.72 ± 0.43 [8]*# | 6.37 ± 0.74 [8]*# | 9.2 ± 1.44 [8]*# | |||
| 156–180 | 1.32 ± 0.2 [8] | 202.16 ± 17.43 [8]* | 2.71 ± 0.36 [8]*# | 3.53 ± 0.35 [8]*# | 5.73 ± 0.6 [8]*# | ||||
| 268–292 | 3.79 ± 0.84 [8] | 207.08 ± 32.12 [8]* | 4.23 ± 0.43 [8]# | 5.24 ± 0.35 [8]# | 7.39 ± 1.07 [8]*# | ||||
| 387–411 | 1.43 ± 0.14 [8] | 164.42 ± 24.77 [8]* | 2.51 ± 0.38 [8]*# | 4.55 ± 0.32 [8]*# | 6.54 ± 0.63 [8]*# | 1.75 ± 0.25 [7] | 6.9 ± 1.05 [8]* | ||
| 2-Cyanoethylmercapturic acid (ng) | 37–61 | 3.92 ± 0.51 [8] | 753.83 ± 66.87 [8]* | 7.97 ± 0.84 [8]*# | 12.38 ± 1.34 [8]*# | 18.79 ± 2.92 [8]*# | |||
| 156–180 | 5.45 ± 0.7 [8] | 704.4 ± 57.77 [8]* | 7.95 ± 0.75 [8]*# | 9.95 ± 0.87 [8]*# | 14.32 ± 1.17 [8]*# | ||||
| 268–292 | 4.88 ± 0.81 [8] | 821.47 ± 153.13 [8]* | 6.83 ± 0.46 [8]# | 8.89 ± 0.47 [8]*# | 16.05 ± 1.76 [8]*# | ||||
| 387–411 | 3.76 ± 0.62 [8] | 684.73 ± 126.09 [8]* | 6.96 ± 0.81 [8]*# | 9.69 ± 1.42 [8]*# | 14.06 ± 1.67 [8]*# | 4.83 ± 0.52 [7] | 16.95 ± 2.22 [8]* | ||
| Total 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (ng) | 37–61 | 10.01 ± 3.17 [8] | 2052.67 ± 184.1 [8]* | 46.52 ± 28.56 [8]# | 71.72 ± 7.74 [8]*# | 177.54 ± 39.97 [8]*# | |||
| 156–180 | 16.4 ± 7.89 [8] | 1800.95 ± 189.05 [8]* | 59.4 ± 11.88 [8]*# | 94.16 ± 32.04 [8]# | 184.57 ± 42.78 [8]*# | ||||
| 268–292 | 6.82 ± 0.44 [8] | 1303.65 ± 248.43 [8]* | 27.04 ± 12.41 [8]*# | 51.98 ± 13.64 [8]*# | 69.37 ± 19.47 [8]*# | ||||
| 387–411 | 7.86 ± 0.4 [8] | 940.02 ± 161.6 [8]* | 26.42 ± 6.67 [8]*# | 79.43 ± 13.86 [8]*# | 91.11 ± 10.36 [8]*# | 6.41 ± 0.46 [7] | 71.44 ± 9.01 [8]* | ||
| Blood | Carboxyhemoglobin (%) | 92–100 | 3.38 ± 0.04 [8] | 32.31 ± 0.91 [9]* | 3.85 ± 0.05 [10]*# | 4.16 ± 0.07 [9]*# | 4.6 ± 0.07 [10]*# | 3.52 ± 0.02 [10] | 4.76 ± 0.12 [10]* |
| 218–227 | 3.46 ± 0.03 [9] | 28.99 ± 1.32 [10]* | 3.56 ± 0.03 [10]# | 3.91 ± 0.05 [10]*# | 4.36 ± 0.11 [10]*# | 3.49 ± 0.02 [10] | 4.56 ± 0.06 [8]* | ||
| 337–346 | 3.36 ± 0.03 [10] | 30.64 ± 1.06 [10]* | 3.59 ± 0.04 [10]*# | 4.02 ± 0.07 [9]*# | 4.03 ± 0.06 [10]*# | 3.69 ± 0.04 [9] | 4.38 ± 0.1 [8]* | ||
| 470–477 | 3.55 ± 0.04 [10] | 28.76 ± 1.43 [9]* | 3.93 ± 0.04 [10]*# | 4.07 ± 0.05 [10]*# | 4.36 ± 0.09 [10]*# |
Data are presented as mean ± SEM. The number of replicates (N) is given in square brackets. 3R4F, reference cigarette smoke; F, female mice; M, male mice; THS, tobacco heating System.
p < .05 versus Sham.
p < .05 versus 3R4F cigarette smoke exposure.
Overall, these measurements confirmed a well-controlled exposure in this 18-month study in A/J mice. For more details on the exposure characterization and full range of endpoints recommended by OECD Test Guideline 453, the reader is referred to Wong et al. (2020).
THS 2.2 Aerosol Exposure Induces Fewer Tissue Changes Across the Respiratory Tract Than CS Exposure
CS exposure induces adaptive changes in the upper respiratory tract of rodents, which prominently include squamous cell metaplasia and reserve cell hyperplasia (Phillips et al. 2019a; Phillips et al. 2018; Titz et al. 2018). Previous studies have reported differences in sensitivity to CS exposure among various locations in the respiratory tract, with the larynx identified as a particularly sensitive tissue (Mowat et al. 2017).
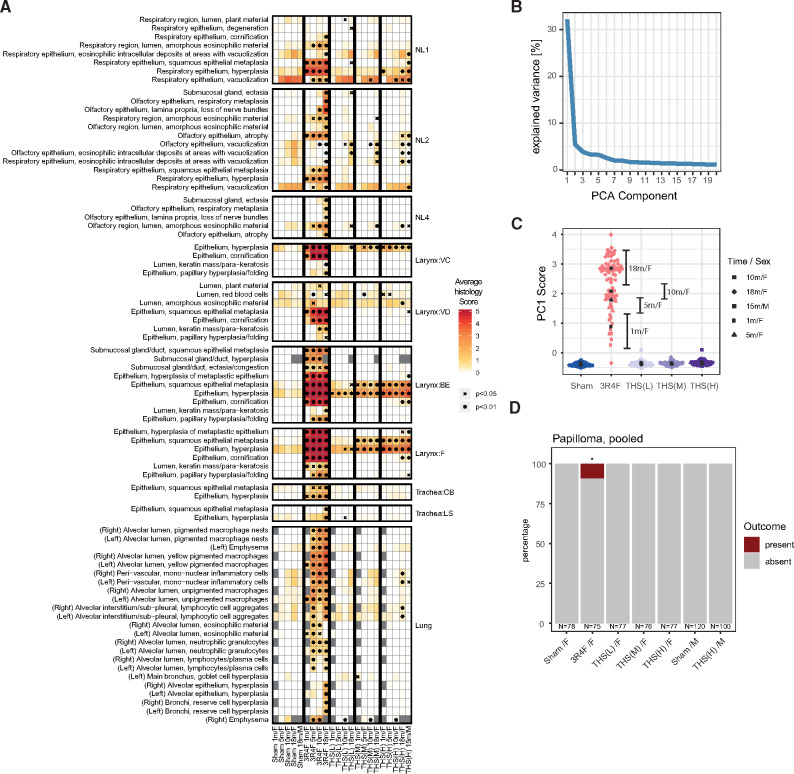
To assess how the exposures affected the respiratory tract tissues, we performed comprehensive histopathological evaluation using a semi-quantitative severity grading scheme (Figure 2A). 3R4F CS exposure induced substantial histopathological changes across the entire respiratory tract, including squamous epithelial metaplasia, epithelium hyperplasia, and epithelium cornification. The effects of THS 2.2 aerosol exposure were less severe and commonly did not reach significance, even at the highest concentration (Figure 3 and Supplementary Figure 1).
Figure 2.
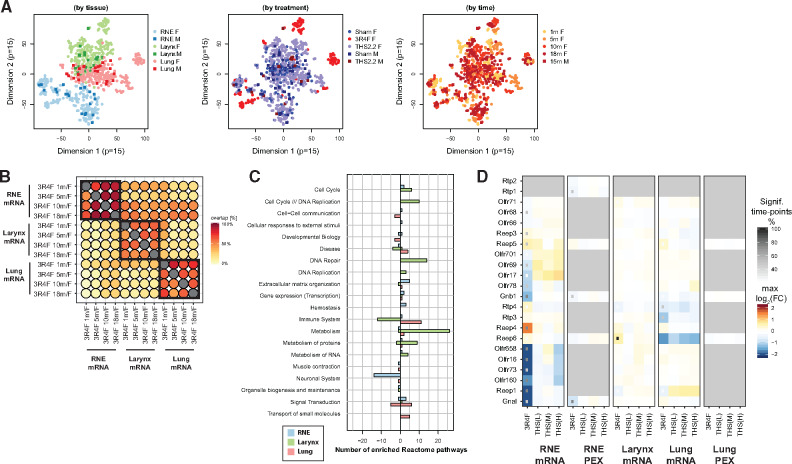
Tobacco Heating System (THS) 2.2 aerosol exposure induces fewer tissue alterations in the respiratory tract than cigarette smoke exposure. A, Histology score profiles. Average histology scores are color coded for each endpoint (rows) and exposure group (columns) (see color key). Histology scores reflect semi-quantitative severity grading: 0, within normal limits; 1–5, minimal to severe changes. Statistical significance versus sham is indicated: xp < .05; *p < .01. Histological endpoints are grouped by region of the respiratory tract: nose levels 1–4 (NL1–NL4), laryngeal regions (VC, vocal cords; VD, ventral depression; BE, base of the epiglottis; F, floor), tracheal regions (CB, carina of bifurcation; LS, longitudinal section), and the lungs. Endpoint with significant changes versus sham for any group is shown. B, Variance explained by the components of a (ordinal) principal component (PC) model for the histology scores. C, Scores for each sample/animal on the first PC. Shape of the points indicates time point and sex (see key). See Supplementary Figure 1 for PCs 2–5. D, Pooled papilloma incidence across various regions of the larynx.*p < .05.
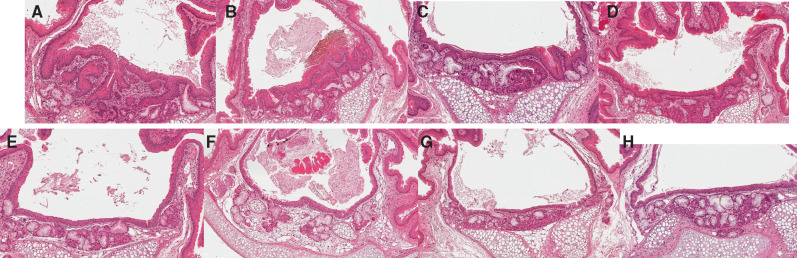
Figure 3.
THS 2.2 aerosol exposure induces fewer tissue changes across the respiratory tract than CS exposure. Representative images of laryngeal epithelial changes over time. Shown is the base of the epiglottis from a female sham animal at 18 months, a female 3R4F group animal (upper panel), and a female THS 2.2 Medium group animal (lower panel) at 1, 5, 10, and 18 months. Note the replacement of stratified with squamous metaplastic epithelium, the markedly increased epithelial thickness, and folding in the 3R4F CS-exposed animal as early as month 1. Magnification: 20×. Hematoxylin & eosin.
The histology score profile (Figure 2A) showed clear severity gradients across exposure types, exposure durations, and respiratory tract locations. We investigated whether this complex response pattern could be more concisely summarized as an overall net measure of exposure strength. The findings of principal component analysis demonstrated that the majority of the variance was captured by a single (the first) PC (Figure 2B and Supplementary Figure 2). The sample scores of the first PC suggested an increasing effect of 3R4F CS exposure over time and emphasized the much smaller effects of exposure in the THS 2.2 groups (Figure 2C).
As mentioned before, the larynx is very sensitive to irritation. After only 1 month of 3R4F CS exposure, the severity scores for several histological findings in the larynx reached the maximum, including those for squamous epithelial metaplasia, epithelium hyperplasia, and epithelium cornification (Figure 2A). Altthough these endpoints were also observed in the THS 2.2 groups, a further response gradation was apparent: Hyperplasia appeared to be the most responsive endpoint (already detected in response to low THS 2.2 exposure at early time points); squamous metaplasia appeared to be slightly less responsive; and cornification only occurred at the last time point in the high THS 2.2 exposure (Figure 3). To more formally evaluate and rank the responsiveness of the various endpoints to exposure, we predicted the expected histology score for each endpoint across PC1 (as the derived measure for net irritation strength) (Supplementary Figure 3). As expected, this analysis highlighted the selected endpoints in the larynx as the most responsive, followed by inflammation-related (eg, pigmented macrophages) and emphysema endpoints in the lungs. The latter were strongly affected by 3R4F CS exposure and to a much lesser degree by THS 2.2 aerosol exposure: Only exposure to the high THS 2.2 dose for 18 months resulted in low-severity findings of perivascular mononuclear cells and lymphocytic cell aggregates.
Because of the sensitivity of the larynx, we observed effects that went beyond the common adaptive respiratory tissue changes to aerosol/smoke exposure. In particular, 3R4F CS exposure for 10 and 18 months resulted in papillary hyperplasia/folding of the epithelium (Figure 2A). Likely pushing the tissue further along this trajectory, at month 18, only 3R4F CS exposure was associated with a significant incidence of papilloma in the larynx (Figure 2D).
In summary, analysis of the global histology profile of the respiratory tract allowed a fine-graded assessment of the exposure impact. Compared with 3R4F CS exposure, THS 2.2 aerosol exposure was associated with substantially smaller effects. Although several endpoints likely reflected adaptive responses to the exposures, a subset of endpoints triggered by 3R4F CS exposure was more severe, with implications for disease, and included emphysema and laryngeal papilloma.
THS 2.2 Aerosol Exposure Induces Fewer Molecular Changes Across the Respiratory Tract Than CS Exposure
Systems toxicology complements apical endpoint measurements with in-depth molecular profiling to elucidate toxicologically relevant mechanisms (Hartung et al. 2017; Talikka et al. 2016; Titz et al. 2018). Here, we profiled mRNA and miRNA changes across the RNE, larynx, and lungs. In addition, we measured protein changes in RNE and lung tissues. miRNA and protein changes in the lungs were only measured after 1 month of exposure, which, for proteomics, was attributable to the incompatibility of the method with the embedding medium (optimum cutting temperature compound) used for subsequent sample collection.
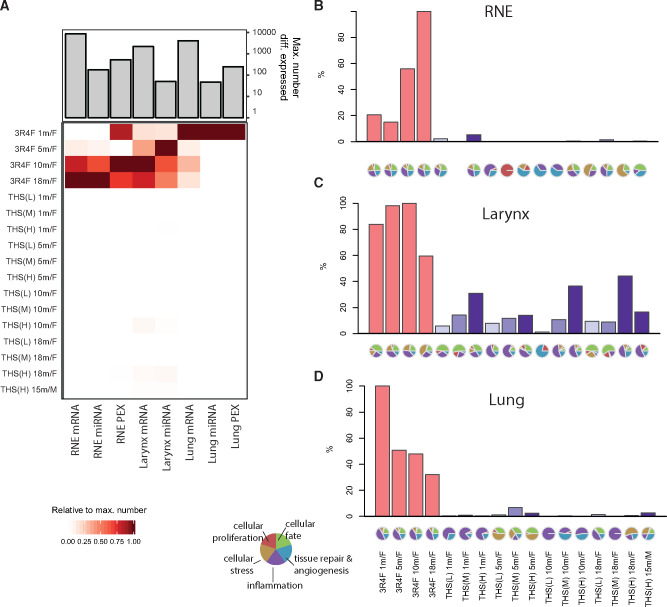
First, we calculated differential expression profiles comparing each exposure group with its corresponding sham group (FDR < 0.05). 3R4F CS exposure resulted in a clear differential expression response across all tissues and the 3 data modalities (Figure 4A). In contrast, THS 2.2 aerosol exposure induced a much more limited differential expression response, with a maximum of 212 differentially expressed genes in the larynx of female mice after 10 months of THS (H) aerosol exposure, in contrast to 2168 differentially expressed genes after 10 months of 3R4F CS exposure.
Figure 4.
Tobacco Heating System (THS) 2.2 aerosol induces fewer molecular changes in the respiratory tract than cigarette smoke. A, Number of differentially expressed molecules in the analyzed tissues. Heatmap shows the number of differentially expressed molecules per exposure group (rows) and data type (columns) relative to the maximum number of significantly affected molecules for the data type (bar chart). Note that miRNA and protein (PEX) expression in the lungs were only measured after 1 month (missing values in grey). B–D, Relative biological impact factors in the respiratory nasal epithelium (RNE), larynx, and lung calculated from transcriptomic data and causal biological network models. Relative biological impact factors are represented for each group versus sham. Note that none of the evaluated causal network models was significantly affected in the THS(M) 1 m/F versus sham 1 m/F group. See Supplementary Figure 3 for individual causal network responses.
We complemented this analysis using a previously published causal network enrichment approach (Iskandar et al. 2017; Martin et al. 2014). This approach does not depend on gene-level FDR thresholds and instead evaluates gene expression changes directly in the context of curated, biologically relevant causal network models (Boué et al. 2015). The NPA is calculated for each causal network, and the NPA values are aggregated to derive overall relative biological impact factors (Hoeng et al. 2014) (Figs. 4B–D; see Supplementary Figure 4 for the individual network responses). Across the 3 tissues, the results of the network enrichment approach confirmed the trends observed in the differential expression profiles: 3R4F CS exposure strongly perturbed the evaluated biological mechanisms, whereas the effects of THS 2.2 aerosol exposure were much milder, with the clearest effects observed with the highest THS 2.2 concentration in the larynx.
3R4F CS Exposure Induces Shared and Distinct Molecular Responses Across the Respiratory Tract
To obtain an overview of exposure responses across conditions and tissues, we visualized the sample relationships with t-SNE (van der Maaten and Hinton 2008) (Figure 5A). t-SNE, a machine learning algorithm, is a nonlinear dimension-reduction technique that embeds high-dimensional data (here, the gene expression data across all samples) in a low-dimensional space (here, in 2-dimensional plots) for visualization and investigation of sample relationships (eg, similarity clusters). t-SNE analysis prominently highlighted the tissue differences (Figure 5A, left), with 3R4F CS exposure causing distinct changes in each tissue (Figure 5A, middle), and only a minor modulation of these effects over time (Figure 5A, right). Correlation analysis of 3R4F CS treatment profiles across tissues and time points confirmed this finding of the highest similarity of 3R4F CS treatment responses within each tissue and partially shared responses across tissues (Figure 5B).
Figure 5.
Global exposure response profiles. A, Visualization of sample relationships by using t-distributed stochastic neighbor embedding (t-SNE) plots, with colors highlighting tissue (left), treatment (middle), and time (right). B, Overlap between sets of differentially expressed genes across the 3 tissues. For each pair of indicated exposure groups, the percent overlap is calculated with respect to the exposure group in the columns (see color key). C. Number of significantly up- or down-regulated gene sets for the mean effects of 3R4F cigarette smoke over time per tissue (global contrasts). Results are grouped by high-level pathways of the Reactome pathway collection (see Supplementary Figure 3 for details). (D) Tissue expression heatmap for the olfactory signaling pathway. For each gene/protein, the differential expression results are summarized across tissues and treatments. For each exposure arm, the changes detected are summarized as the maximum log2 fold change across time points and the percent of time points with significant changes (see key). PEX, protein expression; RNE, respiratory nasal epithelium; THS, tobacco heating system.
We performed gene-set enrichment analysis with the Reactome pathway collection to further investigate biological response categories that might be overemphasized in one tissue versus another (Figure 5C). For example, the analysis highlighted the downregulation of several neuronal gene sets following 3R4F CS exposure in RNE, with increasing downregulation over the course of the study (Supplementary Figure 5). This also included downregulation of the olfactory signaling pathway, possibly reflecting the loss of olfactory epithelium over time (Figure 5D). Gene sets related to formation of cornified cell envelope were among those upregulated in the RNE, whereass immune system-related gene sets (T-cell and B-cell related) were downregulated in the larynx; both gene sets were most affected after 18 months of exposure. In addition, several gene sets likely reflective of direct responses to exposure were upregulated in the larynx, including genes related to xenobiotic metabolism, DNA repair, and the cell cycle (Supplementary Figure 5). The effects of 3R4F CS exposure on the lungs, for example, included upregulation of innate immune response gene sets and downregulation of cell-cell communication gene sets, possibly reflecting the adverse impact of 3R4F CS exposure on lung tissue integrity. Having established the general tissue response patterns, we next investigated these responses in more detail.
THS 2.2 Aerosol Exposure Induces Less Tissue Adaptation Across the Respiratory Tract Than CS Exposure
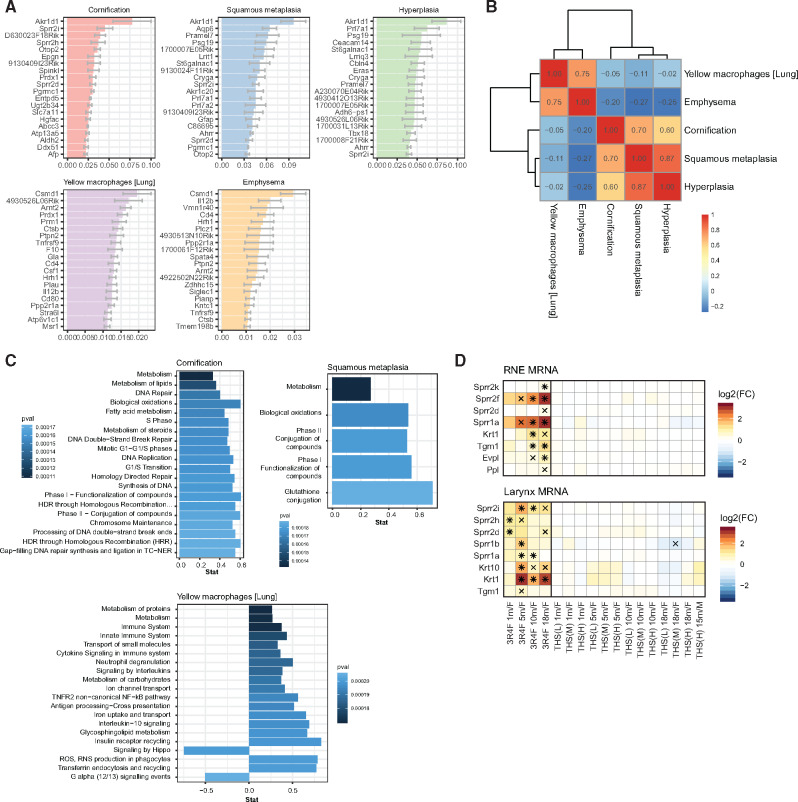
As outlined in the previous sections, histopathological and molecular endpoints exhibited similar global response profiles, demonstrating substantially smaller effects following THS 2.2 aerosol than 3R4F CS exposure as well as the higher sensitivity of the larynx. We studied whether more direct associations between molecular and histopathological changes could be established beyond these global trends. To this end, we fitted regularized ordinal logistic regression models for 5 key endpoint categories: cornification, squamous metaplasia, hyperplasia, emphysema, and yellow macrophages (Figure 6A and Supplementary Figure 6). For example, among the top 20 predictor molecules for cornification were the Sprr genes Sprr2i, Sprr2h, and Sprr2d, whose protein products are cross-linked to and stabilize the cornified membrane (Cabral et al. 2001; Dakir et al. 2008; Finkelman et al. 2005; Zheng et al. 2009). However, reflecting the correlation (and functional association) among the histology scores, the gene associations we discovered were also highly correlated for lung (yellow macrophages and emphysema) and upper respiratory tract (cornification, squamous metaplasia, and hyperplasia) endpoints (Figure 6B). These associations included several common molecules already present among the top 20 predictors, such as Ark1d1 for the upper respiratory tract and Csmd1 for lung endpoints (Figure 6A). Gene-set enrichment analysis showed that the predictor molecules discovered for yellow macrophages and emphysema were associated with immune-related functions (Figure 6C and Supplementary Figure 7), whereas those for upper respiratory tract endpoints were associated with several gene sets reflecting a direct response to the xenobiotic challenge, including phase I and II xenobiotic responses. Thus, although the exposure responses do not necessarily support unique associations between molecule sets and specific endpoints, these molecular associations reflect relevant biological responses to the exposures.
Figure 6.
Tobacco Heating System (THS) 2.2 aerosol induces fewer adaptive tissue adaptations in the respiratory tract than cigarette smoke. (A) Association between gene expression profiles and histological changes in the respiratory tract. Mean ± standard error of the mean of coefficients for the regularized ordinal logistic regression model linking the indicated histology endpoints to gene expression across the respiratory tissues (top 20). B, Clustered Pearson correlation heatmap for the mean coefficients of the ordinal logistic regression model. C, Top enriched gene sets (by p-value) for mean coefficients of the ordinal logistic regression models (filtered for false discovery rate [FDR] < 0.05; maximum 20 gene sets shown). See Supplementary Figure 4 for emphysema and hyperplasia. D, Gene expression profiles for cornification-related genes in respiratory nasal epithelium (RNE) and the trachea. Log2 fold changes (FCs) versus sham are color coded, and statistical significance is indicated: *FDR-adjusted p-value < .01; xFDR adjusted p-value < .05. Cornified cell envelope-related genes were obtained from a previous publication (Kalinin et al. 2001).
Finally, to provide a concrete example for the association between molecular changes and a histopathological endpoint, we further evaluated the association between cornification and Sprr gene expression by assessing the expression profiles of genes (including Sprr genes) that have a known role in the formation of cornified cell envelope (Figure 6D) (Kalinin et al. 2001). For example, in RNE, 3R4F CS exposure resulted in increased expression of several Sprr genes, the cross-linking transglutaminase 1 gene (Tgm1), and a cornified envelope precursor protein-encoding gene (Evpl). Consistent with the histopathological findings for cornification, THS 2.2 aerosol exposure did not cause significant upregulation of these genes.
Taken together, although not necessarily providing a specific set of marker genes for each histological endpoint, this analysis suggests functionally relevant associations between the molecular and apical layers.
THS 2.2 Aerosol Exposure Induces a Milder Immune Response Across the Respiratory Tract Than CS Exposure
CS induces a strong immune response across the respiratory tract. In both humans and rodents (Oviedo et al. 2016; Phillips et al. 2016; Rovina et al. 2013; Zuo et al. 2014), the immune response elicited by CS in the lungs is extensive and has been linked to the pathogenesis of COPD (Zuo et al. 2014). At the same time, CS also induces inflammatory features in the nasal mucosa (Vachier et al. 2004), which, however, differ and can even behave contrarily among different respiratory tract tissues (Talikka et al. 2017).
The molecular profiles generated in the present study enabled us to assess immune-related tissue responses across the respiratory tract. In particular, MOFA effectively highlighted the important and distinct contributions of immune-related processes in the nose (Figure 7A), larynx (Figure 7C), and lungs (Figure 7E and Supplementary Figure 8). MOFA represents a multivariate framework for unsupervised integration of multi-omics datasets and was used to decompose the data matrices per tissue into one latent factor matrix and weight matrices for each data modality (Argelaguet et al. 2018). Similar to principal component analysis for single-omics data, MOFA identifies interpretable low-dimensional representations of the data, with latent factors capturing the major sources of variation across data modalities.
Figure 7.
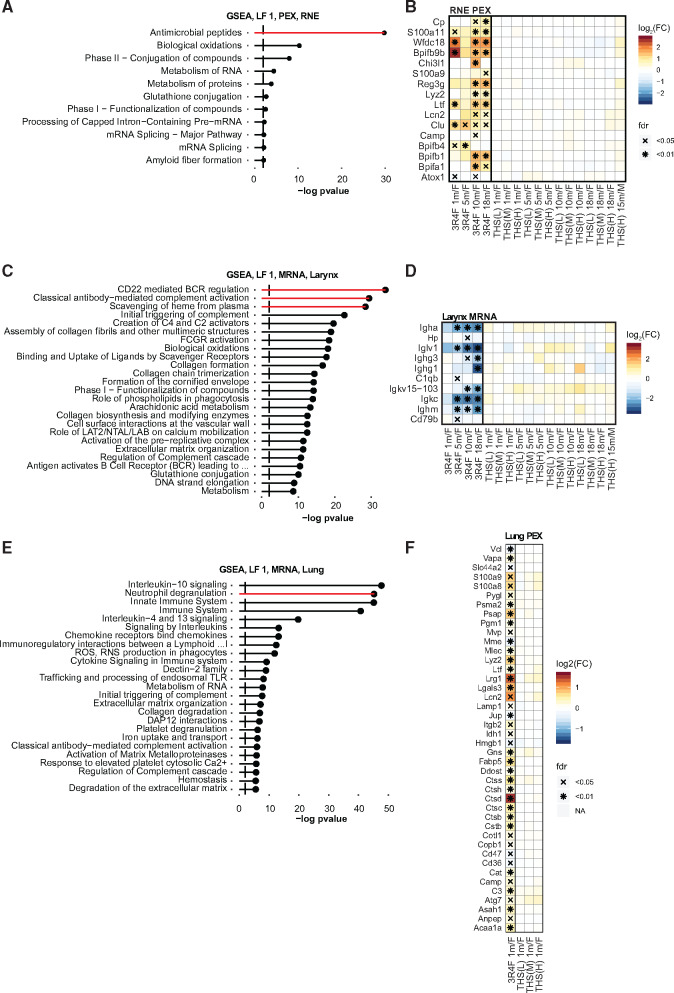
Tobacco Heating System (THS) 2.2 aerosol induces a smaller immune response in the respiratory tract than cigarette smoke. A, Gene sets associated with latent factor (LF) 1 of the multi-omics factor analysis (MOFA) model for protein expression (PEX) in the nose (respiratory nasal epithelium, RNE). Gene-set enrichment analysis (GSEA) was performed for the Reactome gene-set collection. Significant gene sets (false discovery rate [FDR] < 0.05) are ranked by −log10 FDR-adjusted p-values. See Supplementary Figure 5 for more details on MOFA analysis. B, Protein expression profile of antimicrobial peptides/proteins in RNE. Log2 fold changes (FCs) versus sham are color coded, and statistical significance is indicated: *FDR-adjusted p-value < .01; xFDR-adjusted p-value < .05. Gene set from the Reactome (antimicrobial peptides) and Uniprot (antimicrobial [KW-0929]) databases (including Bpifb9b, Wfdc18, S100a11, and Cp from MOFA analysis). C, As in panel A, but for the larynx. D, Gene expression profile in the larynx. E, As in panel A, but for the lungs. F, Protein expression profile of the neutrophil degranulation pathway in the lungs.
In RNE tissues, MOFA highlighted the induction of antimicrobial peptides following exposure to 3R4F CS but not THS 2.2 aerosol (Figs. 7A–7B and Supplementary Figure 9). The tissue profile indicated that this response was the strongest in RNE, although several antimicrobial peptides were also induced in the larynx and lungs (Supplementary Figure 9). In particular, 3R4F CS exposure induced several genes of the BPI fold-containing family (Bpifa1/b1/b4/b9b)—a protein superfamily with immune response functions in the respiratory tract (Britto and Cohn 2015)—of which 2 members, Bpifa1 and Bpifb1, have been associated with COPD (De Smet et al. 2017; Titz et al. 2015). Other antimicrobial peptides that were upregulated following 3R4F CS exposure included regenerating islet-derived 3 γ (Reg3g), lysozyme C2 (Lyz2), lactotransferrin (Ltf), and lipocalin 2 (Lcn2).
MOFA also highlighted an association between 3R4F CS exposure and several B-cell- and antibody-related gene sets in the larynx (Figure 7C), further supporting the observed downregulation of T-cell- and B-cell-related gene sets (Figure 5C and Supplementary Figs. 5 and 10). 3R4F CS exposure resulted in downregulation of several antibody-related genes, specifically in the larynx (Figure 7D).
In the lungs, 3R4F CS-induced inflammatory response was already apparent in the histopathological and association analyses. MOFA demonstrated that 3R4F CS exposure affected innate immune responses, including neutrophil degranulation and interleukin 3, 10, and 13 signaling (Figs. 7E–7F and Supplementary Figs. 7F–H).
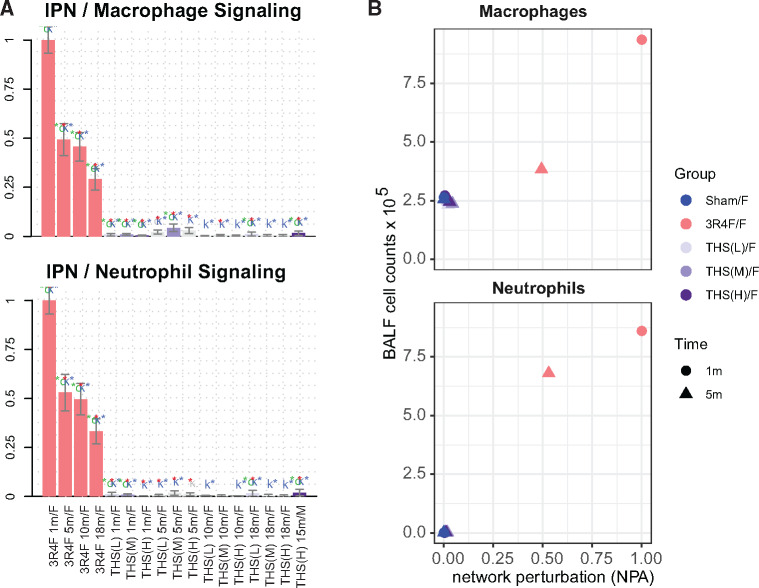
This observation was also supported by the causal network enrichment approach, which demonstrated activation of several inflammatory networks, such as the neutrophil and macrophage signaling networks, in the lungs of 3R4F CS-exposed mice (Figure 8A and Supplementary Figure 4). In addition, neutrophil and macrophage cell counts in bronchoalveolar lavage fluid, indicative of increased inflammatory lung responses, were substantially higher in the 3R4F CS versus sham group (Figure 8B). In contrast, THS 2.2 aerosol exposure had only very limited effects on these molecular and cellular inflammation parameters.
Figure 8.
Tobacco Heating System (THS) 2.2 aerosol is associated with lower macrophage and neutrophil numbers and activation in the lungs than cigarette smoke. A, Network enrichment for the neutrophil and macrophage signaling networks in the lungs. The bars show the overall network perturbation amplitude calculated from transcriptomic data; error bars denote 95% confidence intervals. Three statistical measures are shown: The red, green (o statistic), and blue (k statistic) stars indicate statistical significance with respect to biological replicates, permutation of genes downstream of the network nodes, and permutation of the network topology, respectively (p-value < .05). B, Macrophage and neutrophil counts in bronchoalveolar lavage fluid as markers of inflammation plotted against the corresponding network perturbation amplitude scores from panel I. IPN, inflammatory process network.
Overall, 3R4F CS exposure induced a clear immune response in the 3 respiratory tract tissues we investigated, and this response had both shared and distinct features. In contrast, THS 2.2 aerosol exposure was associated with much smaller to no effects on the lungs of exposed mice, a response further corroborated by the lower immune cell counts in bronchoalveolar lavage fluid.
THS 2.2 Aerosol Exposure Induces Lower Cell Stress Responses Across the Respiratory Tract Than CS Exposure
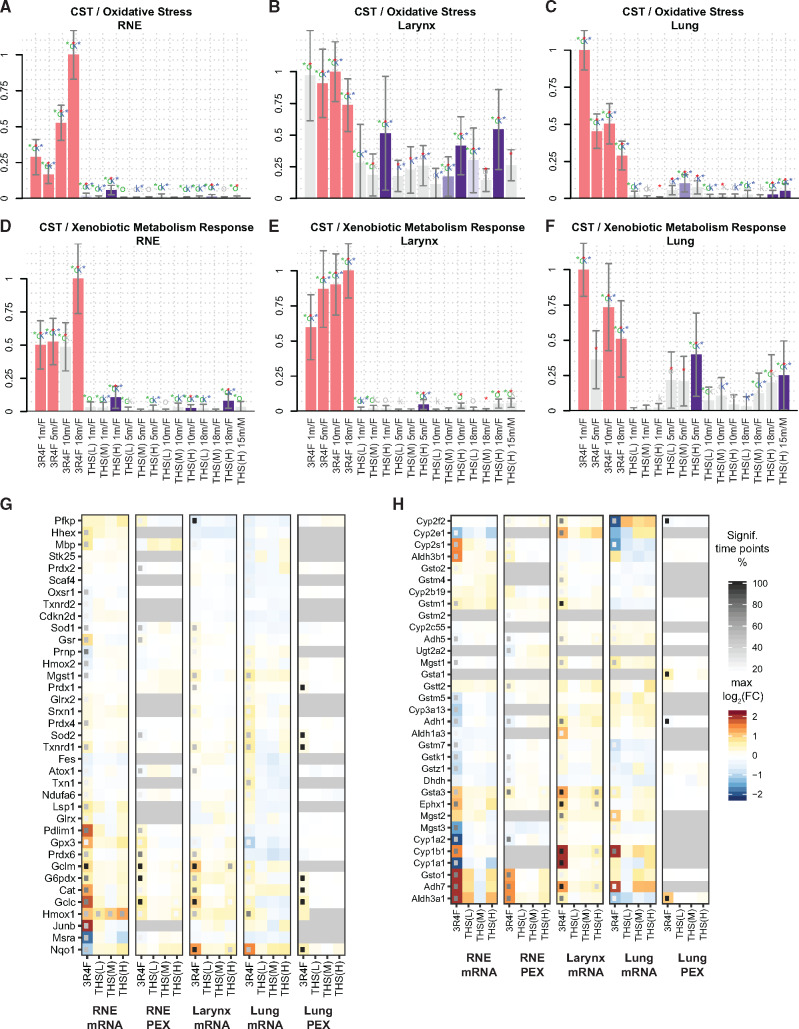
Substantially contributing to its adverse effects, CS exposure represents an oxidative challenge for tissue and induces an extensive oxidative stress response (Fischer et al. 2011; Kirkham and Barnes 2013; Rahman et al. 2006; Titz et al. 2015, 2016). 3R4F CS induced substantial cellular stress responses in the 3 respiratory tract tissues assessed in this study (Supplementary Figure 4). These included perturbation of oxidative stress (Figs. 9A–C) and xenobiotic metabolism (Figs. 9D–F) response networks. In contrast, THS 2.2 aerosol exposure was generally associated with a smaller impact on these cellular stress networks. However, in the most sensitive respiratory tissue, the larynx, the highest concentration of THS 2.2 aerosol also triggered a clear activation of the oxidative stress network, albeit less than the effect of 3R4F CS, at twice the nicotine concentration.
Figure 9.
Tobacco Heating System (THS) 2.2 aerosol induces fewer cell stress responses in the respiratory tract than cigarette smoke. Perturbation of the oxidative stress (A–C) and xenobiotic metabolism response (D–F) networks in the respiratory nasal epithelium (RNE) (A, D), trachea (B, E), and lungs (C, F). The bars show the overall network perturbation amplitude calculated from transcriptomic data. See caption of Figure 7A for details. (G) Tissue expression profile for oxidative stress response genes/proteins. See caption of Figure 4D for details. Genes are part of the reactive oxygen species pathway of the mSigDB hallmark collection (Liberzon et al. 2015). See Supplementary Figure 8 for detailed expression responses. (H) Tissue expression profile for xenobiotic metabolism genes/proteins. Genes are part of the metabolism of xenobiotics by cytochrome P450 gene set from the KEGG database (Kanehisa et al. 2019). See Supplementary Figure 9 for detailed expression responses. CST, cell stress; FC, fold change; PEX, protein expression.
Specifically, the oxidative stress response triggered by 3R4F CS exposure involved the glutathione system (eg, glutamate-cysteine ligase [Gclc/Gclm] and glutathione reductase [Gsr]), the thioredoxin system (thioredoxin [Txn1] and thioredoxin reductase 1 [Txnrd1]), catalase (Cat), heme oxygenase (Hmox1/2), quinone NAD(P)H dehydrogenase 1 (Nqo1), and metabolic adaptations supporting NAPDH generation (glucose-6-phosphate dehydrogenase [G6pdx]) (Figure 9G and Supplementary Figure 11). A decreasing gradient of oxidative stress response activation, supported by causal network analysis, was apparent from RNE to the larynx to the lungs (Figure 9G). For example, whereas glutathione and thioredoxin systems were induced in all tissues following 3R4F CS exposure, other responses, such as that of Nqo1, appeared to be tissue-specific (downregulated in RNE but upregulated in the larynx and lungs). Compared with 3R4F CS, THS 2.2 aerosol exposure had more limited to no effects on the oxidative stress response. Of note, among all oxidative stress response genes, Hmox1 was the most sensitive in RNE response, responding even to the low concentration of THS 2.2 aerosol.
The xenobiotic metabolism response to 3R4F CS exposure included upregulation of cytochrome P450 1B1 (Cyp1b1), glutathione S-transferase a3 (Gsta3), and alcohol dehydrogenase 7 (Adh7) in the 3 tissues of the respiratory tract (Figure 9D and Supplementary Figure 12). However, for many xenobiotic metabolism enzymes, the expression responses to 3R4F CS exposure differed among the 3 tissues. For example, aldehyde dehydrogenase 3b1 (Aldh3b1) and Cyp2s1 were only upregulated in RNE; epoxide hydrolase 1 (Ephx1) was upregulated in RNE and the larynx, but not in the lungs, possibly reflecting the different requirements for coping with xenobiotic challenges along the respiratory tract. In contrast to 3R4F CS, THS 2.2 aerosol exerted much weaker effects, only inducing significant expression changes in a few xenobiotic enzymes at the highest applied dose (corresponding to twice the nicotine concentration in 3R4F CS).
In summary, exposure to 3R4F CS elicited substantial cellular stress responses in the 3 respiratory tract tissues, causing differential expression of both oxidative stress response and xenobiotic metabolism enzymes. In contrast, THS 2.2 aerosol exposure elicited much smaller to no effects.
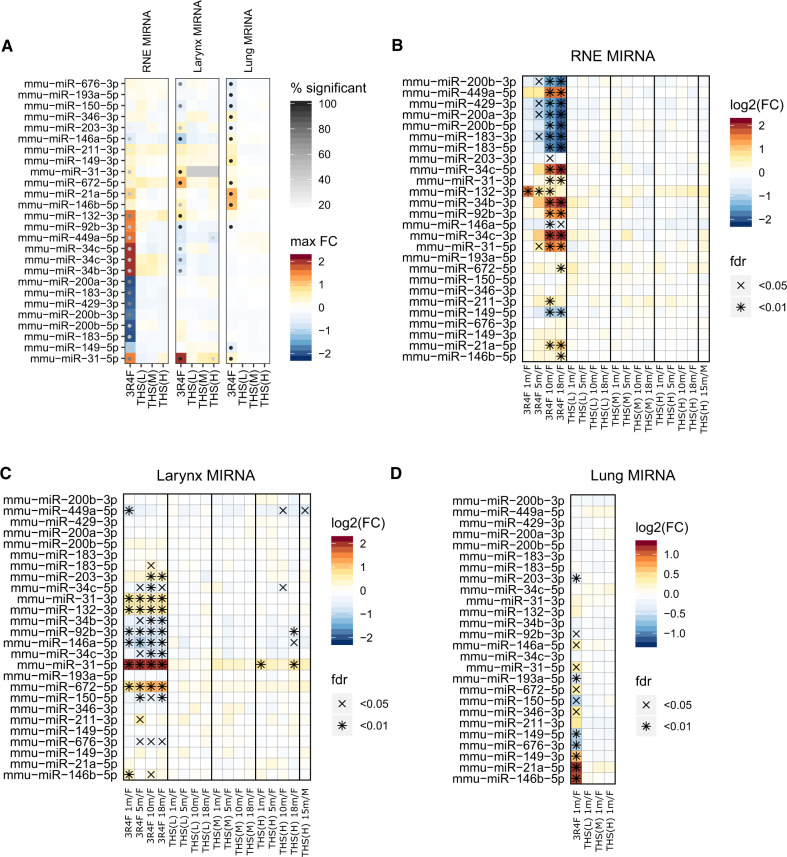
THS 2.2 Aerosol Exposure Induces Lower miRNA Changes Across the Respiratory Tract Than CS Exposure
Micro-RNAs (miRNAs) contribute to the post-transcriptional regulation of various biological processes (Pritchard et al. 2012). Changes in miRNA levels upon CS exposure have been reported in vitro, in vivo, and in clinical studies (Pottelberge et al. 2011; Titz et al. 2020; Zanetti et al. 2018).
Across the 3 tissues investigated in the current study, we observed distinct miRNA response profiles (Figure 10A). For example, a cluster of related miRNAs –miR-34b and miR-34c – showed increased levels upon 3R4F CS versus sham exposure, whereas the levels of these miRNAs were suppressed upon 3R4F CS exposure in the larynx. Another cluster of miRNAs, including miR-200a and miR-200b, was only downregulated in the 3R4F CS groups in RNE. MicroRNA-31 demonstrated the strongest (by fold change) and most robust (by significance) increase upon 3R4F CS exposure in larynx. Finally, the miRNAs with the strongest upregulation upon 3R4F CS exposure in lung included miR-21a, miR-146b, and miR-149. Of note, although miRNA changes for lung were only measured for month 1, distinct time courses of the miRNA response were apparent for RNE and the larynx (Figs. 10B and 10C). For example, in RNE several miRNAs, including miR-34, were strongly induced by 3R4F CS exposure only after 10 and 18 months of exposure, whereas miR-132 was already induced after 1 month of 3R4F CS exposure.
Figure 10.
Tobacco Heating System (THS) 2.2 aerosol induces fewer miRNA changes in the respiratory tract than cigarette smoke. miRNA expression profiles across tissues are shown. Top 10 high-confidence miRNAs correlated with 3R4F CS response in MOFA models in each tissue selected for visualization. (A) miRNA expression across tissues. Max fold change and % significant time points visualized. (B) Differential miRNA expression for the RNE, (C) larynx, and (D) lungs.
In contrast to 3R4F CS exposure, the miRNA responses to THS 2.2 aerosol exposure were more limited. Indeed, neither in lung nor in RNE, THS 2.2 aerosol induced any significant miRNA changes compared with sham. However, consistent with the generally higher sensitivity of the larynx (see above), for example, 5 miRNAs demonstrated significant changes upon THS 2.2 exposure in females at the highest concentration at the last time point (compared with 25 for 3R4F CS exposure). This included miR-31, which also showed the strongest fold-change response upon 3R4F CS exposure, and the responses to high THS 2.2 remained below those observed for 3R4F CS.
DISCUSSION
We comparatively assessed the effects of THS 2.2 aerosol and CS exposure across the respiratory tract of A/J mice. Details of the overall study execution, standard OECD endpoints, and lung carcinogenicity evaluated in this study are reported in our accompanying publication (Wong et al. 2020).
Because standard toxicological endpoints can lack sensitivity and only yield limited insights into toxicologically relevant mechanisms (Ellinger-Ziegelbauer et al. 2011; Titz et al. 2018), we performed an integrated systems toxicology assessment (Hartung et al. 2017; Talikka et al. 2016), in which we complemented classical toxicological endpoints with in-depth molecular profiling by mRNA/miRNA transcriptomics and proteomics analyses. Robust computational analysis approaches are key to deriving relevant insights from these data. For transcriptomic data, we leveraged a previously developed causal biological network enrichment approach, which quantitatively and statistically evaluates the perturbation of context-relevant causal network models (Hoeng et al. 2012; Iskandar et al. 2017; Martin et al. 2012, 2014). To achieve a comprehensive, integrative view of the molecular exposure effects, we complemented the network enrichment approach with other dedicated computational approaches such as single and multi-omics latent factor identification (Argelaguet et al. 2018b), multivariate data mapping (van der Maaten and Hinton 2008), multivariate regression, and gene-set analyses.
We began our investigation by generating an integrative multivariate view of the semi-quantitative histopathology scores across the respiratory tract tissues, which revealed clear qualitative and quantitative trends in the exposure responses. As expected from previous studies (Oviedo et al. 2016; Phillips et al. 2019b), squamous metaplasia and basal cell hyperplasia were the most responsive endpoints in nasal and laryngeal epithelia, whereas inflammation-related endpoints and emphysema were more prominent in the lungs (parenchyma). Differentiation into multilayered squamous epithelium, possibly with cornification, is a common adaptive strategy of respiratory epithelia for protection against toxicants (Burger et al. 1989; Harkema et al. 2006). In contrast, an immune response is a common reaction of lung parenchyma to toxicological challenges, intended to not only clear toxicants but also promote lung diseases such as COPD when these responses are chronic (Wong et al. 2016b).
Evaluation of histopathology scores across all conditions showed a gradation of exposure sensitivities across the respiratory tract. The most sensitive response was observed in the larynx, particularly for epithelium hyperplasia and squamous epithelial metaplasia at the base of the epiglottis and larynx floor. This finding is expected in rodent inhalation studies (Mowat et al. 2017), with squamous epithelial metaplasia being commonly recognized as the most responsive endpoint in repeated-dose inhalation studies (Osimitz et al. 2007). The toxicological relevance of squamous epithelial metaplasia in the larynx has been discussed previously. Osmitz et al. described it as an adaptive response, which is not indicative of human risk (Osimitz et al. 2007). A European Society of Toxicologic Pathology workshop concluded that moderate to severe laryngeal squamous metaplasia can indeed exert an adverse impact on the function of the larynx in rodents; but, for nongenotoxic compounds, laryngeal squamous metaplasia by itself should not be regarded as a precancerous lesion (Kaufmann et al. 2009). In addition, the direct translatability of these findings to humans has been questioned because the rodent larynx is more susceptible to toxicants (Mowat et al. 2017).
Regardless of the direct relevance of these laryngeal endpoints to humans, we argue here that these endpoints can help define the overall strength or tissue irritability to an exposure. In this context, it is critical to jointly assess exposure responses across all endpoints and respiratory tract tissues. Here, we demonstrated that principal component analysis can effectively capture the histopathological effects in the dominant PC as a summary of the overall exposure strength. 3R4F CS exposure resulted in strong effects along this axis that were amplified over time, culminating, for example, in laryngeal papillomas. In contrast, in this integrative evaluation of histopathological findings, the effects of THS 2.2 aerosol exposure remained close to those observed in the sham group at all nicotine concentrations assessed.
The molecular profiles observed in the 3 respiratory tract tissues—the nose, larynx, and lungs—further suggested the much lower effects of THS 2.2 aerosol than 3R4F CS. Causal network analysis of mRNA transcriptomic data confirmed that THS 2.2 aerosol exposure affected toxicologically and disease-relevant processes—including inflammation, cellular stress, and tissue repair—to a lesser degree than 3R4F CS exposure. These findings are consistent with those of our previous (shorter) assessment studies of THS 2.2 aerosol in the ApoE−/− mouse model (Supplementary Figure 13) (Phillips et al. 2016, 2019b; Titz et al. 2016).
Global comparison of the tissues and exposure responses analyzed in this study revealed both shared and distinct responses to 3R4F CS across the tissues. We observed increased cornification in RNE, downregulation of T-cell and B-cell responses in the larynx, and upregulation of innate immune-related processes in the lungs.
As a potential avenue for future investigations, the similarity of responses across time points suggests that it is feasible to predict apical endpoints from molecular data at earlier time points. However, more diverse datasets (eg, a panel of toxicants triggering diverse toxicological mechanisms) are required to stringently test such approaches in a robust fashion. For example, a previous proof-of-concept study predicted histopathology scores from gene expression data in the Open TG-GATEs database (Eastman and Pande 2019). In the current study, we established ordinal logistic regression models to identify associations between core histopathology endpoints and the underlying gene expression changes. Among the caveats here are the facts that these 2 types of endpoint measurements were not obtained from the same animals but from separate dissection groups and that the datasets mostly reflected a single dominant force driving the effects, namely 3R4F CS exposure. With this, the associations we identified were not necessarily fully specific to a given histopathology endpoint; rather, the identified genes associated with squamous metaplasia and hyperplasia were strongly correlated. Nevertheless, using this approach, we were able to identify a clear association between the yellow macrophages observed by histopathological analysis with immune-related gene sets in the lungs as well as between cornification and upregulation of Sprr genes, which are essential for the formation of cornified cell envelope (Kalinin et al. 2001).
Beyond the genes involved in adaptive processes of the tissue structure, molecular profiling also yielded insights into the immune and cellular stress responses in the 3 respiratory tract tissues. Guided by MOFA, we identified different aspects of the immune response triggered in the nose, larynx, and lungs following 3R4F CS exposure. The nasal epithelium reacted to 3R4F CS exposure with the strongest upregulation of antimicrobial peptides, which was observed on both protein and mRNA levels. Interestingly, among the upregulated members of the BPI fold-containing family (Bpifa1/b1/b4/b9b), 2 proteins, Bpifa1 and Bpifb1, have been associated with COPD (De Smet et al. 2017; Titz et al. 2015). Another example is LCN2, which is upregulated in mucosal tissues during inflammation and has direct antimicrobial activity (Chang et al. 2019; Goetz et al. 2002). Reg3g is an antimicrobial peptide with a critical role in the defense of the lungs against Staphylococcus aureus infection (Choi et al. 2013). Of note, a previous proteomics study identified several of these antimicrobial peptides among proteins most expressed in the nasal cavity of mice, including Bpifb9b, Camp, Lyz2, and Lcn2 (Kuntová et al. 2018). With this, our results indicate a strong induction by 3R4F CS of an antimicrobial peptide-mediated innate immune response in the nose, whereas these effects are limited or even absent following exposure to THS 2.2 aerosol.
In the larynx, we identified an association between 3R4F CS exposure and downregulation of T-cell-, B-cell-, and antibody-related genes and gene sets. Reports on the role of B cells in the larynx are scarce; however, the effects of smoking on CD4+ T-cell numbers, either positive (Rees et al. 2006) or negative (Jetté et al. 2017), have been reported in clinical studies. This points to the previously noted immunological role of the larynx, even if the functional role of the observed changes in A/J mice are currently unclear (Jetté et al. 2017; Thibeault et al. 2009).
In the lungs, consistent with previous findings in ApoE−/− mice (Phillips et al. 2016, 2019b; Titz et al. 2016), 3R4F CS exposure induced extensive inflammation-related changes, which were not observed in the THS 2.2 aerosol-exposed groups. Multi-omics analysis highlighted effects on innate immune responses, including neutrophil degranulation, which were confirmed by increased neutrophil and macrophage numbers in bronchoalveolar lavage fluid. These findings were expected for the 3R4F group—in line with previous findings on CS-induced lung inflammation in both human and rodents (Oviedo et al. 2016; Phillips et al. 2016; Rovina et al. 2013; Zuo et al. 2014)—and likely constitute a mechanistic link between CS exposure and COPD pathogenesis (Zuo et al. 2014). Thus, these results demonstrate that THS 2.2 aerosol triggers a much milder inflammatory response in the respiratory tract and, therefore, suggest a much reduced contribution of THS 2.2 aerosol to disease-relevant tissue changes.
THS 2.2 aerosol exposure triggered substantially lower cellular stress responses (oxidative stress and xenobiotic metabolism) than 3R4F CS exposure in the respiratory tract tissues we investigated. Oxidative stress, which is induced directly or indirectly by 3R4F CS exposure, represents a toxicologically relevant challenge for exposed tissues (Fischer et al. 2011; Kirkham and Barnes 2013; Rahman et al. 2006; Titz et al. 2015, 2016) and has also been implicated in COPD pathogenesis (McGuinness and Sapey 2017). The lower to no oxidative stress response to THS 2.2 aerosol exposure is consistent with the lower levels of oxidative stress-inducing chemicals and free radicals in THS 2.2 aerosol (Schaller et al. 2016; Shein and Jeschke 2019) and also—representing an indirect mechanism for oxidative stress (Zuo et al. 2014)—with the much milder inflammatory responses associated with THS 2.2 aerosol exposure. Although 3R4F CS activated these cellular stress responses in all 3 respiratory tract tissues, we noted several differences in the molecular responses. For example, Nqo1 was downregulated in RNE but upregulated in the larynx and lungs; Aldh3b1 and Cyp2s1 were only upregulated in RNE; and Ephx1 was upregulated in RNE and the larynx but not in the lungs. Overall, these differences are likely explained by the varying quantitative and qualitative exposures in these tissues as well as by different adaptive response strategies (such as the ability to shield the tissue by squamous metaplastic differentiation and cornification in nasal and laryngeal epithelia but not in lung parenchyma).
Finally, miRNAs showed consistent exposure response profiles. 3R4F CS exposure was associated with a distinct miRNA response across the 3 investigated tissues (RNE, larynx, and lung). MicroRNA-21a, miR-146b, and miR-149 were highlighted as those miRNAs with the most prominent increase upon 3R4F CS versus sham in lung compared with the other tissues. For all 3 miRNAs, an association with immune-responses have been reported: miR-21a is a negative immune regulator in mouse liver regeneration (via nuclear factor kappa B [NF-κB] inhibition) (Marquez et al. 2010) and of the macrophage response in peritonitis (Barnett et al. 2016), miR-146b is an innate immune cell-associated miRNA (Hou et al. 2009), and miR-149 has been implicated in immune-regulatory feedback loops (Shi et al. 2017; Xu et al. 2014). Overall, this is consistent with the prominent immune response observed in the lung upon 3R4F CS exposure. In the larynx, 3R4F CS exposure elicited a preferential upregulation of miR-31, which was also significantly upregulated upon high THS 2.2 aerosol exposure. MicroRNA-31 has been associated with epithelial proliferation via regulation of signaling axes such as Wnt signaling (Lv et al. 2017; Tian et al. 2019). Thus, miR-31 upregulation might contribute to restorative epithelial proliferation and, possibly, the observed tissue architecture changes upon exposure. Finally, in RNE, 3R4F CS preferentially induced a miRNA cluster including miR-34b and miR-34c. miR-34 family members have been identified as p53 target genes, with an implicated role in the apoptotic response and cell cycle arrest (Hermeking 2010). With this, the miR-34 family could be involved in the 3R4F CS—DNA damage—p53 response axis in RNE.
Taken together, miRNA profiling revealed distinct responses of the 3 tissues to 3R4F CS exposure—and further supported the overall lower molecular response to THS 2.2 aerosol compared with 3R4F CS exposure.
Limitations of the Study
We investigated the chronic effects of exposure to THS 2.2 aerosol and 3R4F CS in A/J mice. This mouse strain is susceptible to both lung tumor development (Stinn et al. 2010, 2013; Witschi et al. 2002) and induction of pronounced lung inflammation and emphysematous changes following CS exposure (Cabanski et al. 2015; Stinn et al. 2013). Importantly, although the current study substantially expands the time frame of exposure to up to 18 months, the findings are overall consistent with those of our previous studies in ApoE−/− mice (Phillips et al. 2019a,b; Titz et al. 2016) (Supplementary Figure 13). As discussed before, the lack of animal-level matched data from histopathological and molecular analyses made the association across data modalities challenging. However, observing related effect reductions such as cornification and cornification-related gene expression in 2 separate dissection groups further supports the robustness of our conclusions on the reduced effects of THS 2.2 aerosol exposure. By design, we did not collect a full dataset for each data modality across all time points. For example, proteome changes in the lungs were only assessed after 1 month, because the embedding medium for laser capture microdissection prevented mass spectrometry-based proteome analysis during the later time points. Inclusion of additional omics data modalities, such as lipidomics and metabolomics, might have yielded further molecular insights into the exposure effects. Finally, the molecular analyses were conducted for bulk tissues, which were intended to comprehensively cover the relevant exposure effects; but, the lack of cell type (or single-cell) data makes the elucidation of molecular mechanisms challenging.
CONCLUSIONS
Using a systems toxicology approach, we assessed how exposure to THS 2.2 aerosol, in comparison with 3R4F CS, for up to 18 months affects the respiratory tract in A/J mice. This report complements our accompanying publication on OECD chronic toxicity and carcinogenicity endpoints (Wong et al. 2020). Our study demonstrated that integrative systems toxicology approaches can yield deep insights into toxicologically relevant mechanisms, such as the relevance of differing inflammatory responses across the respiratory tract and the expression of a diverse set of antimicrobial peptides in the nose. Moreover, our analysis revealed both commonalities and differences in oxidative and xenobiotic responses across the respiratory tract.
Unlike 3R4F CS, THS 2.2 aerosol had only limited effects on the histological characteristics of the respiratory tract, including adaptive tissue changes in nasal and laryngeal epithelia and inflammation and emphysematous changes in the lungs. Integrative analysis of the molecular changes in the nose, larynx, and lungs confirmed the substantially lower impact of THS 2.2 aerosol on toxicologically and disease-relevant molecular processes, including inflammation, oxidative stress responses, and activation of xenobiotic metabolism. Overall, the current findings support a reduced impact of THS 2.2 aerosol relative to that of CS on the respiratory tract.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
DATA AVAILABILITY
Datasets from the main inhalation study, further details on the protocols, and additional data visualizations are available on the INTERVALS platform at https://doi.org/10.26126/intervals.3pcrrx.1.
Raw, processed, and contrast data for the omics datasets analyzed are also available on INTERVALS (https://www.intervals.science/) (Boué et al. 2017).
Mass spectrometry proteomic data are available from the ProteomeXchange Consortium through the PRIDE partner repository (http://www.ebi.ac.uk/pride/archive/) (Vizcaíno et al. 2013), with the identifiers PXD016466 (RNE data) and PXD016460 (lung data).
Transcriptomic data are available from ArrayExpress (www.ebi.ac.uk/arrayexpress) (Kolesnikov et al. 2015), with accession numbers E-MTAB-8540 (mRNA) and E-MTAB-8541 (miRNA).
Supplementary Material
ACKNOWLEDGMENTS
The authors thank the study team and especially acknowledge the technical assistance and support of the Bioresearch and Aerosol Teams at Philip Morris International Research Laboratories Pte. Ltd., Singapore and PMI R&D, Philip Morris Products S.A., Neuchâtel, Switzerland. The authors thank Sam Ansari for managing the biobanking and acknowledge the support of Nick Karoglou and Sindhoora Bhargavi Gopala Reddy for editing a draft of the manuscript.
FUNDING
Philip Morris International is the sole source of funding and sponsor of this research.
DECLARATION OF CONFLICTING INTERESTS
All authors except A.B. are employees of Philip Morris International. A.B. is an employee of Histovia, which was contracted and paid by Philip Morris International to perform the histopathology analysis.
Contributor Information
Bjoern Titz, PMI R&D, Philip Morris Products S.A, CH-2000 Neuchâtel, Switzerland.
Alain Sewer, PMI R&D, Philip Morris Products S.A, CH-2000 Neuchâtel, Switzerland.
Karsta Luettich, PMI R&D, Philip Morris Products S.A, CH-2000 Neuchâtel, Switzerland.
Ee Tsin Wong, Philip Morris International Research Laboratories Pte. Ltd, Singapore 117406.
Emmanuel Guedj, PMI R&D, Philip Morris Products S.A, CH-2000 Neuchâtel, Switzerland.
Catherine Nury, PMI R&D, Philip Morris Products S.A, CH-2000 Neuchâtel, Switzerland.
Thomas Schneider, Biognosys AG, 8952 Schlieren, Switzerland.
Yang Xiang, PMI R&D, Philip Morris Products S.A, CH-2000 Neuchâtel, Switzerland.
Keyur Trivedi, PMI R&D, Philip Morris Products S.A, CH-2000 Neuchâtel, Switzerland.
Grégory Vuillaume, PMI R&D, Philip Morris Products S.A, CH-2000 Neuchâtel, Switzerland.
Patrice Leroy, PMI R&D, Philip Morris Products S.A, CH-2000 Neuchâtel, Switzerland.
Ansgar Büttner, Histovia GmbH, 51491 Overath, Germany.
Florian Martin, PMI R&D, Philip Morris Products S.A, CH-2000 Neuchâtel, Switzerland.
Nikolai V Ivanov, PMI R&D, Philip Morris Products S.A, CH-2000 Neuchâtel, Switzerland.
Patrick Vanscheeuwijck, PMI R&D, Philip Morris Products S.A, CH-2000 Neuchâtel, Switzerland.
Julia Hoeng, PMI R&D, Philip Morris Products S.A, CH-2000 Neuchâtel, Switzerland.
Manuel C Peitsch, PMI R&D, Philip Morris Products S.A, CH-2000 Neuchâtel, Switzerland.
REFERENCES
- Archer K. J., Williams A. A. (2012). L 1 penalized continuation ratio models for ordinal response prediction using high‐dimensional datasets. Stat. Med. 31, 1464–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argelaguet R., Velten B., Arnol D., Dietrich S., Zenz T., Marioni J. C., Buettner F., Huber W., Stegle O. (2018). Multi-omics factor analysis: A framework for unsupervised integration of multi-omics data sets. Mol. Syst. Biol. 14, e8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett R. E., Conklin D. J., Ryan L., Keskey R. C., Ramjee V., Sepulveda E. A., Srivastava S., Bhatnagar A., Cheadle W. G. (2016). Anti‐inflammatory effects of mir‐21 in the macrophage response to peritonitis. J. Leuk. Biol. 99, 361–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boué S., Exner T., Ghosh S., Belcastro V., Dokler J., Page D., Boda A., Bonjour F., Hardy B., Vanscheeuwijck P., et al. (2017). Supporting evidence-based analysis for modified risk tobacco products through a toxicology data-sharing infrastructure. F1000Res 6, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boué S., Talikka M., Westra J. W., Hayes W., Di Fabio A., Park J., Schlage W. K., Sewer A., Fields B., Ansari S., et al. (2015). Causal biological network database: A comprehensive platform of causal biological network models focused on the pulmonary and vascular systems. Database J. Biol. Databases Curation 2015, bav030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britto C. J., Cohn L. (2015). Bactericidal/permeability-increasing protein fold-containing family member a1 in airway host protection and respiratory disease. Am. J. Respir. Cell Mol. Biol. 52, 525–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G. T., Renne R. A., Sagartz J. W., Ayres P. H., Coggins C. R., Mosberg A. T., Hayes A. W. (1989). Histologic changes in the respiratory tract induced by inhalation of xenobiotics: Physiologic adaptation or toxicity? Toxicol. Appl. Pharmacol. 101, 521–542. [DOI] [PubMed] [Google Scholar]
- Cabanski M., Fields B., Boue S., Boukharov N., DeLeon H., Dror N., Geertz M., Guedj E., Iskandar A., Kogel U., et al. (2015). Transcriptional profiling and targeted proteomics reveals common molecular changes associated with cigarette smoke-induced lung emphysema development in five susceptible mouse strains. Inflam. Res. 64, 471–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral A., Voskamp P., Cleton-Jansen A. M., South A., Nizetic D., Backendorf C. (2001). Structural organization and regulation of the small proline-rich family of cornified envelope precursors suggest a role in adaptive barrier function. J. Biol. Chem. 276, 19231–19237. [DOI] [PubMed] [Google Scholar]
- CDER. (2005). Guidance for industry: Estimating the maximum safe starting dose in initial clinical trials for therapeutics in adult healthy volunteers. Food and Drug Administration Center for Drug Evaluation and Research, Rockville, MD, pp. 1–27.
- Chang Y.-L. Wang Z. Igawa S. Choi J. E. Werbel T. Di Nardo A. (2019). Lipocalin 2: A new antimicrobial in mast. Cells Int. J. Mol. Sci. 20, 2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S.-M., McAleer J. P., Zheng M., Pociask D. A., Kaplan M. H., Qin S., Reinhart T. A., Kolls J. K. (2013). Innate stat3-mediated induction of the antimicrobial protein reg3γ is required for host defense against MRSA pneumonia. J. Exp. Med. 210, 551–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dakir E. H., Feigenbaum L., Linnoila R. I. (2008). Constitutive expression of human keratin 14 gene in mouse lung induces premalignant lesions and squamous differentiation. Carcinogenesis 29, 2377–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw J.(2011). Multivariate Analysis with Optimal Scaling. UCLA: Department of Statistics, UCLA. Available at: https://escholarship.org/uc/item/95c2w5wd. [Google Scholar]
- De Smet E. G., Seys L. J., Verhamme F. M., Vanaudenaerde B. M., Brusselle G. G., Bingle C. D., Bracke K. R. (2017). Association of innate defense proteins bpifa1 and bpifb1 with disease severity in COPD. Int. J. Chronic Obstruct. Pulmonary Dis. 13, 11–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman P., Pande V. S. (2019). Predicting toxicity from gene expression with neural networks. arXiv preprint: 190200060.
- Ellinger-Ziegelbauer H., Adler M., Amberg A., Brandenburg A., Callanan J. J., Connor S., Fountoulakis M., Gmuender H., Gruhler A., Hewitt P., et al. (2011). The enhanced value of combining conventional and “omics” analyses in early assessment of drug-induced hepatobiliary injury. Toxicol. Appl. Pharmacol. 252, 97–111. [DOI] [PubMed] [Google Scholar]
- Fabregat A., Jupe S., Matthews L., Sidiropoulos K., Gillespie M., Garapati P., Haw R., Jassal B., Korninger F., May B., et al. (2018). The reactome pathway knowledgebase. Nucleic Acids Res. 46, D649–D655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Family Smoking Prevention and Tobacco Control Act (FSPTCA). (2009). Public Law No 111–31.
- Finkelman F. D., Yang M., Perkins C., Schleifer K., Sproles A., Santeliz J., Bernstein J. A., Rothenberg M. E., Morris S. C., Wills-Karp M. (2005). Suppressive effect of il-4 on il-13-induced genes in mouse lung. J. Immunol. 174, 4630–4638. [DOI] [PubMed] [Google Scholar]
- Fischer B. M., Pavlisko E., Voynow J. A. (2011). Pathogenic triad in COPD: Oxidative stress, protease-antiprotease imbalance, and inflammation. Int. J. Chron. Obstruct. Pulmon. Dis. 6, 413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., et al. (2004). Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 5, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godtfredsen N., Lam T., Hansel T., Leon M., Gray N., Dresler C., Burns D., Prescott E., Vestbo J. (2008). COPD-related morbidity and mortality after smoking cessation: Status of the evidence. Eur. Respir. J. 32, 844–853. [DOI] [PubMed] [Google Scholar]
- Goetz D. H., Holmes M. A., Borregaard N., Bluhm M. E., Raymond K. N., Strong R. K. (2002). The neutrophil lipocalin ngal is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol. Cell 10, 1033–1043. [DOI] [PubMed] [Google Scholar]
- Gower A. C., Steiling K., Brothers J. F., Lenburg M. E., Spira A. (2011). Transcriptomic studies of the airway field of injury associated with smoking-related lung disease. Proc. Am. Thoracic Soc. 8, 173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkema J. R., Carey S. A., Wagner J. G. (2006). The nose revisited: A brief review of the comparative structure, function, and toxicologic pathology of the nasal epithelium. Toxicol. Pathol. 34, 252–269. [DOI] [PubMed] [Google Scholar]
- Hartung T., FitzGerald R. E., Jennings P., Mirams G. R., Peitsch M. C., Rostami-Hodjegan A., Shah I., Wilks M. F., Sturla S. J. (2017). Systems toxicology: Real world applications and opportunities. Chem. Res. Toxicol. 30, 870–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermeking H. (2010). The mir-34 family in cancer and apoptosis. Cell Death Differ. 17, 193–199. [DOI] [PubMed] [Google Scholar]
- Hoeng J., Deehan R., Pratt D., Martin F., Sewer A., Thomson T. M., Drubin D. A., Waters C. A., de Graaf D., Peitsch M. C. (2012). A network-based approach to quantifying the impact of biologically active substances. Drug Discov. Today 17, 413–418. [DOI] [PubMed] [Google Scholar]
- Hoeng J., Talikka M., Martin F., Sewer A., Yang X., Iskandar A., Schlage W. K., Peitsch M. C. (2014). Case study: The role of mechanistic network models in systems toxicology. Drug Discov. Today 19, 183–192. [DOI] [PubMed] [Google Scholar]
- Hou J., Wang P., Lin L., Liu X., Ma F., An H., Wang Z., Cao X. (2009). Microrna-146a feedback inhibits rig-i-dependent type i ifn production in macrophages by targeting traf6, irak1, and irak2. J. Immunol. 183, 2150–2158. [DOI] [PubMed] [Google Scholar]
- Iskandar A., Titz B., Sewer A., Leroy P., Schneider T., Zanetti F., Mathis C., Elamin A., Frentzel S., Schlage W., et al. (2017). Systems toxicology meta-analysis of in vitro assessment studies: Biological impact of a candidate modified-risk tobacco product aerosol compared with cigarette smoke on human organotypic cultures of the aerodigestive tract. Toxicol. Res. 6, 631–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetté M. E., Seroogy C. M., Thibeault S. L. (2017). Laryngeal t regulatory cells in the setting of smoking and reflux. Laryngoscope 127, 882–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinin A., Marekov L. N., Steinert P. M. (2001). Assembly of the epidermal cornified cell envelope. J. Cell Sci. 114, 3069–3070. [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Sato Y., Furumichi M., Morishima K., Tanabe M. (2019). New approach for understanding genome variations in KEGG. Nucleic Acids Res. 47, D590–D595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann W., Bader R., Ernst H., Harada T., Hardisty J., Kittel B., Kolling A., Pino M., Renne R., Rittinghausen S., et al. (2009). 1st international estp expert workshop: “Larynx squamous metaplasia”. A re-consideration of morphology and diagnostic approaches in rodent studies and its relevance for human risk assessment. Exp. Toxicol. Pathol. 61, 591–603. [DOI] [PubMed] [Google Scholar]
- Kirkham P. A., Barnes P. J. (2013). Oxidative stress in COPD. Chest 144, 266–273. [DOI] [PubMed] [Google Scholar]
- Kogel U., Schlage W. K., Martin F., Xiang Y., Ansari S., Leroy P., Vanscheeuwijck P., Gebel S., Buettner A., Wyss C., et al. (2014). A 28-day rat inhalation study with an integrated molecular toxicology endpoint demonstrates reduced exposure effects for a prototypic modified risk tobacco product compared with conventional cigarettes. Food Chem. Toxicol. 68, 204–217. [DOI] [PubMed] [Google Scholar]
- Kolesnikov N., Hastings E., Keays M., Melnichuk O., Tang Y. A., Williams E., Dylag M., Kurbatova N., Brandizi M., Burdett T., et al. (2015). ArrayExpress update–simplifying data submissions. Nucleic Acids Res. 43, D1113–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntová B. Stopková R. Stopka P. (2018). Transcriptomic and proteomic profiling revealed high proportions of odorant binding and antimicrobial defense proteins in olfactory tissues of the house mouse. Front. Genet. 9, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J. P., Tamayo P. (2015). The molecular signatures database hallmark gene set collection. Cell Syst. 1, 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv C., Li F., Li X., Tian Y., Zhang Y., Sheng X., Song Y., Meng Q., Yuan S., Luan L., et al. (2017). Mir-31 promotes mammary stem cell expansion and breast tumorigenesis by suppressing wnt signaling antagonists. Nat. Commun. 8, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maaten L., Hinton G. (2008). Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605. [Google Scholar]
- Marquez R. T., Wendlandt E., Galle C. S., Keck K., McCaffrey A. P. (2010). Microrna-21 is upregulated during the proliferative phase of liver regeneration, targets pellino-1, and inhibits nf-κb signaling. Am. J. Physiol. Gastroint. Liver Physiol. 298, G535–G541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F., Sewer A., Talikka M., Xiang Y., Hoeng J., Peitsch M. C. (2014). Quantification of biological network perturbations for mechanistic insight and diagnostics using two-layer causal models. BMC Bioinformatics 15, 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F., Thomson T. M., Sewer A., Drubin D. A., Mathis C., Weisensee D., Pratt D., Hoeng J., Peitsch M. C. (2012). Assessment of network perturbation amplitude by applying high-throughput data to causal biological networks. BMC Syst. Biol. 6, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuinness A. J. A., Sapey E. (2017). Oxidative stress in COPD: Sources, markers, and potential mechanisms. J. Clin. Med. 6, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowat V., Alexander D. J., Pilling A. M. (2017). A comparison of rodent and nonrodent laryngeal and tracheal bifurcation sensitivities in inhalation toxicity studies and their relevance for human exposure. Toxicol. Pathol. 45, 216–222. [DOI] [PubMed] [Google Scholar]
- OECD. (2018). 453: Combined Chronic Toxicity/Carcinogenicity Studies. OECD Guidelines for the Testing of Chemicals, Section, 4.
- Osimitz T. G., Droege W., Finch J. M. (2007). Toxicologic significance of histologic change in the larynx of the rat following inhalation exposure: A critical review. Toxicol. Appl. Pharmacol. 225, 229–237. [DOI] [PubMed] [Google Scholar]
- Oviedo A., Lebrun S., Kogel U., Ho J., Tan W. T., Titz B., Leroy P., Vuillaume G., Bera M., Martin F., et al. (2016). Evaluation of the tobacco heating system 2.2. Part 6: 90-day OECD 413 rat inhalation study with systems toxicology endpoints demonstrates reduced exposure effects of a mentholated version compared with mentholated and non-mentholated cigarette smoke. Regul. Toxicol. Pharmacol. 81, S93–S122. [DOI] [PubMed] [Google Scholar]
- Phillips B., Szostak J., Titz B., Schlage W. K., Guedj E., Leroy P., Vuillaume G., Martin F., Buettner A., Elamin A., et al. (2019. a). A six-month systems toxicology inhalation/cessation study in ApoE−/− mice to investigate cardiovascular and respiratory exposure effects of modified risk tobacco products, CHTP 1.2 and THS 2.2, compared with conventional cigarettes. Food Chem. Toxicol. 126, 113–141. [DOI] [PubMed] [Google Scholar]
- Phillips B., Szostak J., Titz B., Schlage W. K., Guedj E., Leroy P., Vuillaume G., Martin F., Buettner A., Elamin A., et al. (2019. b). 6-month systems toxicology inhalation/cessation study with CHTP 1.2 and THS 2.2 in ApoE-/- mice. Available at: https://www.intervals.science/studies/#/apoe-chtp12-ths22. Accessed August 15, 2020.
- Phillips B., Veljkovic E., Boue S., Schlage W. K., Vuillaume G., Martin F., Titz B., Leroy P., Buettner A., Elamin A., et al. (2016). An 8-month systems toxicology inhalation/cessation study in ApoE-/- mice to investigate cardiovascular and respiratory exposure effects of a candidate modified risk tobacco product, THS 2.2, compared with conventional cigarettes. Toxicol. Sci. 149, 411–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips B., Veljkovic E., Peck M. J., Buettner A., Elamin A., Guedj E., Vuillaume G., Ivanov N. V., Martin F., Boue S., et al. (2015). A 7-month cigarette smoke inhalation study in c57bl/6 mice demonstrates reduced lung inflammation and emphysema following smoking cessation or aerosol exposure from a prototypic modified risk tobacco product. Food Chem. Toxicol. 80, 328–345. [DOI] [PubMed] [Google Scholar]
- Phillips B. W., Schlage W. K., Titz B., Kogel U., Sciuscio D., Martin F., Leroy P., Vuillaume G., Krishnan S., Lee T., et al. (2018). A 90-day OECD TG 413 rat inhalation study with systems toxicology endpoints demonstrates reduced exposure effects of the aerosol from the carbon heated tobacco product version 1.2 (CHTP1. 2) compared with cigarette smoke. I. Inhalation exposure, clinical pathology and histopathology. Food Chem. Toxicol. 116, 388–413. [DOI] [PubMed] [Google Scholar]
- Postma D. S., Bush A., van den Berge M. (2015). Risk factors and early origins of chronic obstructive pulmonary disease. Lancet 385, 899–909. [DOI] [PubMed] [Google Scholar]
- Pottelberge G. R. V., Mestdagh P., Bracke K. R., Thas O., Durme YMv., Joos G. F., Vandesompele J., Brusselle G. G. (2011). MicroRNA expression in induced sputum of smokers and patients with chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 183, 898–906. [DOI] [PubMed] [Google Scholar]
- Pritchard C. C., Cheng H. H., Tewari M. (2012). MicroRNA profiling: Approaches and considerations. Nat. Rev. Genet. 13, 358–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman I., Biswas S. K., Kode A. (2006). Oxidant and antioxidant balance in the airways and airway diseases. Eur. J. Pharmacol. 533, 222–239. [DOI] [PubMed] [Google Scholar]
- Rees L. E., Jones P. H., Ayoub O., Gunasekaran S., Rajkumar K., Stokes C. R., Haverson K., Bailey M., Birchall M. A. (2006). Smoking influences the immunological architecture of the human larynx. Clin. Immunol. 118, 342–347. [DOI] [PubMed] [Google Scholar]
- Rovina N., Koutsoukou A., Koulouris N. G. (2013). Inflammation and immune response in COPD: Where do we stand? Mediat. Inflam. 2013, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller J.-P., Keller D., Poget L., Pratte P., Kaelin E., McHugh D., Cudazzo G., Smart D., Tricker A. R., Gautier L., et al. (2016). Evaluation of the tobacco heating system 2.2. Part 2: Chemical composition, genotoxicity, cytotoxicity, and physical properties of the aerosol. Regul. Toxicol. Pharmacol. 81, S27–S47. [DOI] [PubMed] [Google Scholar]
- Sergushichev A. (2016). An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. BioRxiv.060012.
- Shein M., Jeschke G. (2019). Comparison of free radical levels in the aerosol from conventional cigarettes, electronic cigarettes, and heat-not-burn tobacco products. Chem. Res. Toxicol. 32, 1289–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Shan S., Li H., Song G., Li Z. (2017). Anti-inflammatory effects of millet bran derived-bound polyphenols in lps-induced ht-29 cell via ros/mir-149/akt/nf-κb signaling pathway. Oncotarget 8, 74582–74594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. R., Clark B., Lüdicke F., Schaller J.-P., Vanscheeuwijck P., Hoeng J., Peitsch M. C. (2016). Evaluation of the tobacco heating system 2.2. Part 1: Description of the system and the scientific assessment program. Regul. Toxicol. Pharmacol. 81, S17–S26. [DOI] [PubMed] [Google Scholar]
- Sridhar S., Schembri F., Zeskind J., Shah V., Gustafson A. M., Steiling K., Liu G., Dumas Y.-M., Zhang X., Brody J. S., et al. (2008). Smoking-induced gene expression changes in the bronchial airway are reflected in nasal and buccal epithelium. BMC Genomics 9, 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J. F., Maier C. S. (2008). Acrolein: Sources, metabolism, and biomolecular interactions relevant to human health and disease. Mol. Nutr. Food Res. 52, 7–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinn W., Arts J. H., Buettner A., Duistermaat E., Janssens K., Kuper C. F., Haussmann H.-J. (2010). Murine lung tumor response after exposure to cigarette mainstream smoke or its particulate and gas/vapor phase fractions. Toxicology 275, 10–20. [DOI] [PubMed] [Google Scholar]
- Stinn W., Berges A., Meurrens K., Buettner A., Gebel S., Lichtner R. B., Janssens K., Veljkovic E., Xiang Y., Roemer E., et al. (2013). Towards the validation of a lung tumorigenesis model with mainstream cigarette smoke inhalation using the A/J mouse. Toxicology 305, 49–64. [DOI] [PubMed] [Google Scholar]
- Talikka M., Boué S., Frentzel S., Kogel U., Sierro N., Titz B., Veljkovic E., Hoeng J., Peitsch M. C. (2016). Systems toxicology. In Encyclopedia of Drug Metabolism and Interactions (A. Lyubimov, Ed.). John Wiley & Sons, Inc, New Jersey.
- Talikka M., Martin F., Sewer A., Vuillaume G., Leroy P., Luettich K., Chaudhary N., Peck M. J., Peitsch M. C., Hoeng J. (2017). Mechanistic evaluation of the impact of smoking and chronic obstructive pulmonary disease on the nasal epithelium. Clin. Med. Insights Circ. Respir. Pulmonary Med. 11, 1179548417710928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibeault S. L., Rees L., Pazmany L., Birchall M. A. (2009). At the crossroads: Mucosal immunology of the larynx. Mucosal Immunol. 2, 122–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y., Xu J., Li Y., Zhao R., Du S., Lv C., Wu W., Liu R., Sheng X., Song Y., et al. (2019). MicroRNA-31 reduces inflammatory signaling and promotes regeneration in colon epithelium, and delivery of mimics in microspheres reduces colitis in mice. Gastroenterology 156, 2281–2296.e6. [DOI] [PubMed] [Google Scholar]
- Titz B., Boue S., Phillips B., Talikka M., Vihervaara T., Schneider T., Nury C., Elamin A., Guedj E., Peck M. J., et al. (2016). Effects of cigarette smoke, cessation and switching to two heat-not-burn tobacco products on lung lipid metabolism in c57bl/6 and ApoE-/- mice—An integrative systems toxicology analysis. Toxicol. Sci. 149, 441–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titz B., Elamin A., Martin F., Schneider T., Dijon S., Ivanov N. V., Hoeng J., Peitsch M. C. (2014). Proteomics for systems toxicology. Comput. Struct. Biotechnol. J. 11, 73–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titz B., Kogel U., Martin F., Schlage W. K., Xiang Y., Nury C., Dijon S., Baumer K., Peric D., Bornand D., et al. (2018). A 90-day OECD TG 413 rat inhalation study with systems toxicology endpoints demonstrates reduced exposure effects of the aerosol from the carbon heated tobacco product version 1.2 (CHTP1. 2) compared with cigarette smoke. Ii. Systems toxicology assessment. Food Chem. Toxicol. 115, 284–301. [DOI] [PubMed] [Google Scholar]
- Titz B., Sewer A., Schneider T., Elamin A., Martin F., Dijon S., Luettich K., Guedj E., Vuillaume G., Ivanov N. V., et al. (2015). Alterations in the sputum proteome and transcriptome in smokers and early-stage COPD subjects. J. Proteomics 128, 306–320. [DOI] [PubMed] [Google Scholar]
- Titz B., Szostak J., Sewer A., Phillips B., Nury C., Schneider T., Dijon S., Lavrynenko O., Elamin A., Guedj E., et al. (2020). Multi-omics systems toxicology study of mouse lung assessing the effects of aerosols from two heat-not-burn tobacco products and cigarette smoke. Comput. Struct. Biotechnol. J. 18, 1056–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vachier I., Vignola A. M., Chiappara G., Bruno A., Meziane H., Godard P., Bousquet J., Chanez P. (2004). Inflammatory features of nasal mucosa in smokers with and without COPD. Thorax 59, 303–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Väremo L., Nielsen J., Nookaew I. (2013). Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res. 41, 4378–4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno J. A., Côté R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., et al. (2013). The proteomics identifications (pride) database and associated tools: Status in 2013. Nucleic Acids Res. 41, D1063–D1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witschi H., Espiritu I., Dance S. T., Miller M. S. (2002). A mouse lung tumor model of tobacco smoke carcinogenesis. Toxicol. Sci. 68, 322–330. [DOI] [PubMed] [Google Scholar]
- Wong E. T., Kogel U., Veljkovic E., Martin F., Xiang Y., Boue S., Vuillaume G., Leroy P., Guedj E., Rodrigo G., et al. (2016. a). Evaluation of the tobacco heating system 2.2. Part 4: 90-day OECD 413 rat inhalation study with systems toxicology endpoints demonstrates reduced exposure effects compared with cigarette smoke. Regul. Toxicol. Pharmacol. 81, S59–S81. [DOI] [PubMed] [Google Scholar]
- Wong E. T., Luettich K., Krishnan S., Wong S. K., Lim W. T., Yeo D., Büttner A., Leroy P., Vuillaume G., Boué S., et al. (2020). Reduced chronic toxicity and carcinogenicity in A/J mice in response to life-time exposure to aerosol from a heated tobacco product compared with cigarette smoke. Toxicol. Sci. doi: 10.1093/toxsci/kfaa131. [DOI] [PMC free article] [PubMed]
- Wong J., Magun B. E., Wood L. J. (2016. b). Lung inflammation caused by inhaled toxicants: A review. Int. J. Chronic Obstruct. Pulmonary Dis. 11, 1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G., Zhang Z., Xing Y., Wei J., Ge Z., Liu X., Zhang Y., Huang X. (2014). Microrna‐149 negatively regulates tlr‐triggered inflammatory response in macrophages by targeting myd88. J. Cell. Biochem. 115, 919–927. [DOI] [PubMed] [Google Scholar]
- Zanetti F., Sewer A., Scotti E., Titz B., Schlage W. K., Leroy P., Kondylis A., Vuillaume G., Iskandar A. R., Guedj E., et al. (2018). Assessment of the impact of aerosol from a potential modified risk tobacco product compared with cigarette smoke on human organotypic oral epithelial cultures under different exposure regimens. Food Chem. Toxicol. 115, 148–169. [DOI] [PubMed] [Google Scholar]
- Zheng L., Zhou Z., Lin L., Alber S., Watkins S., Kaminski N., Choi A. M., Morse D. (2009). Carbon monoxide modulates alpha-smooth muscle actin and small proline rich-1a expression in fibrosis. Am. J. Respir. Cell Mol. Biol. 41, 85–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L., He F., Sergakis G. G., Koozehchian M. S., Stimpfl J. N., Rong Y., Diaz P. T., Best T. M. (2014). Interrelated role of cigarette smoking, oxidative stress, and immune response in COPD and corresponding treatments. Am. J. Physiol. Lung Cell. Mol. Physiol. 307, L205–L218. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Datasets from the main inhalation study, further details on the protocols, and additional data visualizations are available on the INTERVALS platform at https://doi.org/10.26126/intervals.3pcrrx.1.
Raw, processed, and contrast data for the omics datasets analyzed are also available on INTERVALS (https://www.intervals.science/) (Boué et al. 2017).
Mass spectrometry proteomic data are available from the ProteomeXchange Consortium through the PRIDE partner repository (http://www.ebi.ac.uk/pride/archive/) (Vizcaíno et al. 2013), with the identifiers PXD016466 (RNE data) and PXD016460 (lung data).
Transcriptomic data are available from ArrayExpress (www.ebi.ac.uk/arrayexpress) (Kolesnikov et al. 2015), with accession numbers E-MTAB-8540 (mRNA) and E-MTAB-8541 (miRNA).