ABSTRACT
Objectives:In order to cope with the rapid spread of the COVID-19 pandemic, we introduced on our in-house high-throughput molecular diagnostic platform (MDx Platform) a real-time reverse transcriptase PCR (RT-PCR) to detect the SARS-CoV-2 from any clinical specimens. The aim of this study was to compare the RT-PCR results obtain with the MDx Platform and the commercial assay cobas SARS-CoV-2 (Roche) on nasopharyngeal swab and other clinical specimens including sputum, bronchial aspirate, bronchoalveolar lavage and anal swabs. Methods: Samples received in our laboratory from patients suspected of COVID-19 (n = 262) were tested in parallel with our MDx platform SARS-CoV-2 PCR and with the cobas SARS-CoV-2 test. Results: The overall agreement between the two tests for all samples tested was 99.24% (260/262), which corresponded to agreements of 100% (178/178) on nasopharyngeal swabs, 95.45% (42/44) on lower respiratory tract specimen with discordant resultS obtained for very high cycle threshold (Ct) value and 100% (40/40) on anorectal swabs. The Ct values for nasopharyngeal swabs displayed an excellent correlation (R2 > 96%) between both tests. Conclusions: The high agreements between the cobas SARS-CoV-2 test and the MDx platform supports the use of both methods for the diagnostic of COVID-19 on various clinical samples. Very few discrepant results may occur at very low viral load.
Keywords: COVID-19; SARS-CoV-2; RT-PCR, molecular diagnostic; cobas 6800; nasopharyngeal swab; bronchial aspirate; bronchoalveolar lavage; anal swab; anorectal swab
In order to cope with the rapid spread of the COVID-19 pandemic, we compared and validated two SARS-CoV-2 RT-PCR systems.
INTRODUCTION
The rapid spread of the “Coronavirus Disease 2019” (COVID-19) pandemic caused by the “severe acute respiratory syndrome coronavirus 2” (SARS-CoV-2) illustrated the central role of diagnostic microbiology laboratories, which rapidly proposed molecular methods to detect this new coronavirus (Tadini et al. 2020). These tests were paramount for patient management and for public health strategies.
Before the report of the first COVID-19 case in Switzerland, and anticipating the spread of the disease in our country, we introduced a SARS-CoV-2 test on our high-throughput automated molecular diagnostic platform (MDx platform), which allows us to perform the RT-PCR in a 384-well format. Moreover, this open and flexible platform allows the introduction of new PCRs according to an accredited R&D process (Greub et al. 2016). So far, this platform allows to process more than 100 different PCRs to detect virus, bacteria, fungi or parasites (Greub et al. 2016). The SARS-CoV-2 diagnostic test was implemented according to the RT-PCR validated by Corman and colleagues (Corman et al. 2020).
Introduction of high-throughput SARS-CoV-2 RT-PCR and diversification of the diagnostic platforms are among the decisive interventions that contribute to an efficient and reliable response of clinical microbiology laboratory to COVID-19 pandemic; in particular in large teaching hospital (Posteraro et al. 2020). The aim of this study was to compare the cobas SARS-CoV-2 (Roche, Rotkreuz, Switzerland) test and our MDx platform: (i) to permit other laboratories to achieve the diagnostic of COVID-19, therefore increasing the overall testing capacity, (ii) to serve as backup in case of breakdown or reagent storage (iii) and to evaluate the performance of the cobas test on respiratory specimens other than nasopharyngeal swabs and on anorectal swabs. Indeed, in cases of high positive pre-test probability and negative PCR on a nasopharyngeal specimen, a lower respiratory tract specimen might be performed. The need to test various clinical samples such as LRT or anorectal swabs quickly became paramount for patients with atypical clinical presentations; for instance, anorectal swabs were tested in case of predominant gastrointestinal symptoms or in specific situations such as donor screening in the context of fecal microbiota transplantation for instance (Song et al. 2020; Xie et al. 2020; Young et al. 2020).
MATERIAL AND METHODS
Samples
Samples from patients with a suspected COVID-19 (n = 262) were tested both using our in-house high-throughput MDx platform and the cobas SARS-CoV-2 qualitative test (Roche, Rotkreuz, Switzerland). Nasopharyngeal and oropharyngeal swabs (n = 178) were collected in Copan Universal Transport Medium System (UTM-RT) or BD Universal Viral Transport System (UVT). Lower respiratory tract (LRT) specimen (n = 44) were liquefied using N-acetyl-L-cysteine prior analysis using the cobas 6800 system, or prior nucleic acid extraction using the MagNA Pure 96 instrument when samples are tested on our MDx Platform (Jaton et al. 2013; Opota et al. 2016, 2017, 2019). Anorectal swabs (n = 40) were collected as previously described (Dang et al. 2009).
RT-PCR platforms and instruments
The automated molecular diagnostic platform consists of the nucleic-acid extraction system MagNA Pure 96 instrument (Roche), associated to a liquid handling distribution system, the STARletR instrument (Hamilton, Cinnaminson, NJ), two Hamilton instruments for the assembly of the 384-well PCR plates and two QuantStudio 7 (Applied Biosystems, Waltham, MA) (Greub et al. 2016; Opota et al. 2015, 2017). For this comparison, the detection of the SARS-CoV-2 on the MDx platform was achieved using only the RT-PCR targeting the E gene using the primers and probes described by Corman and colleagues (Corman et al. 2020). The concentration of forwards and reverse primers (300 nM) and Taqman probes (100 nM) were slightly reduced as compared to the E gene PCR of Corman and colleagues. Plasmids containing the target amplicons of the RT-PCR were obtained from RD-Biotech (Besançon, France) and were used as positive controls, and to determine the analytical limit of detection of the PCR as well as the reproducibility of the PCRs as previously described (Greub et al. 2016; Opota et al. 2015, 2017). Another positive control, corresponding to SARS-CoV-2 RNA purified from cell culture supernatants, was received from the Institute of Virology of the University of Berlin, Charité (Corman et al. 2020). The analytic sensitivity was determined with the positive control plasmids diluted from 100 000 to 1 DNA copies per reaction (Table S1, Supporting Information). The intra- and inter-run reproducibility was assessed in five independent runs with dilutions of the plasmids corresponding to 100 and 10 DNA copies per reactions (Figure S1, Supporting Information). The specificity of the RT-PCR was assessed using 10 samples containing other seasonal coronavirus.
The RT-PCR targeting the RdRP gene and the N-gene were also introduced according to Corman and colleagues but showed a significantly reduced sensitivity requiring further optimization and was not used for this comparison (Pillonel et al. 2020).
The RT-PCR was established through our prerequisite real-time PCR set-up (Greub et al. 2016) with some modifications consisting on a modified mastermix to have a single step RT-PCR, i.e. by coupling retro-transcription with subsequent PCR using the obtained complementary DNA (Taqpath 1-step TRT –qPCR Master Mix, Life Technology Europe BV).
The new implemented PCR conforms to the following specifications for annealing and synthesis temperature: Hold stage: step 1: 25°C, 2 min; step 2: 50°C, 15 min; step 3: 95°C, 10 min; PCR stage 45 cycles: step 1: 95°C, 1 s; step 2: 60°C, 20 s, on a 384-wells plate Quantstudio 7 thermocycler.
The cobas SARS-CoV-2 tests were run on the cobas 6800 System (Roche) according to the manufacturer guidelines.
Statistics
Statistical analysis were performed using GraphPad Prism version 8.3.0 for Windows, GraphPad Software (San Diego, CA, www.graphpad.com). Cycle thresholds (Ct) were analysed using one-way ANOVA or using Bland–Altman analysis by ploting the difference between two measurements on the Y axis, and the average of the two measurements on the X axis. The degree of agreement was quantified by the kappa value.
Data
The data were obtained during a quality enhancement project at our institution. According to national law, the performance and publishing the results of such a project can be done without asking the permission of the competent research ethics committee.
RESULTS
Setup of the SARS-CoV-2 RT-PCR on the in-house high-throughput MDx Platform
The SARS-CoV-2 RT-PCR that we aimed to introduce in our MDx platform targeting the E, RdRP and N gene were validated by Corman and colleagues (Corman et al. 2020). We therefore decided to perform a rapid introduction and validation of these PCR to face the rapid spreading of COVID-19 in Europe. We first assessed the analytical sensitivity of the three real-time PCR using the positive control received from the Institute of Virology of the University of Berlin, Charité (Corman et al. 2020) corresponding to SARS-CoV-2 RNA purified from cell culture supernatants. The sensitivity of the test was evaluated for the three genes at 10-fold serial dilutions (103, 102 and 101, respectively). We observed a 100% positivity rate (Ct for E gene: 28.22, 31.65, 36.26; for RdRp: 29.6, 32.56, 38.58 and N gene: 29.86, 33.54 and 35.49). Because of their limited sensitivity, the complete validation of the RdRP gene and N gene PCR was postoned awaiting further optimization.
When we received the synthetic plasmid positive control for the E gene RT-PCR, the complete analytic sensitivity and reproducibility test, confirmed the reliability of the PCR. The analytic sensitivity was determined with the positive control plasmids diluted from 100 000 to 1 copies per reaction. The limit of detection (LOD) for the E gene assay was between 1 and 10 copies per reaction with respectively 36% and 100% hit rate. The intra-run variability, assessed by plotting the Ct values of the five replicates of the same run of amplification for plasmids dilutions corresponding to 100 and 10 copies per reaction, showed that the dispersion of each replicate did not exceed two standard deviation of the average (Figure S1, Supporting Information). The inter-run reproducibility was assessed in five independent runs with dilutions of the plasmids corresponding to 100 and 10 copies per reaction and also revealed that the dispersion of the average Ct value for each run did not exceed two SD of the average of all runs (Figure S1, Supporting Information). For the specificity, 10 samples containing other coronavirus (seasonal strains) were tested and showed 100% specificity without any unexpected amplification.
Agreement between the cobas SARS-CoV-2 and the MDx platform on various clinical specimens
More than 10 000 SARS-CoV-2 test have been performed in 40 days when we received the kits to test the cobas SARS-CoV-2 assay on the cobas 6800 instrument already in use in our laboratory (Fig. 1A). The comparison between the two tests included 262 samples (66 positives and 196 negatives) for a total agreement of 99.24%. The agreement between the two tests was 100% (178/178) for nasopharyngeal swabs, 95.45% (42/44) for LRT specimen and 100% (40/40) for anorectal swabs (Table 1).
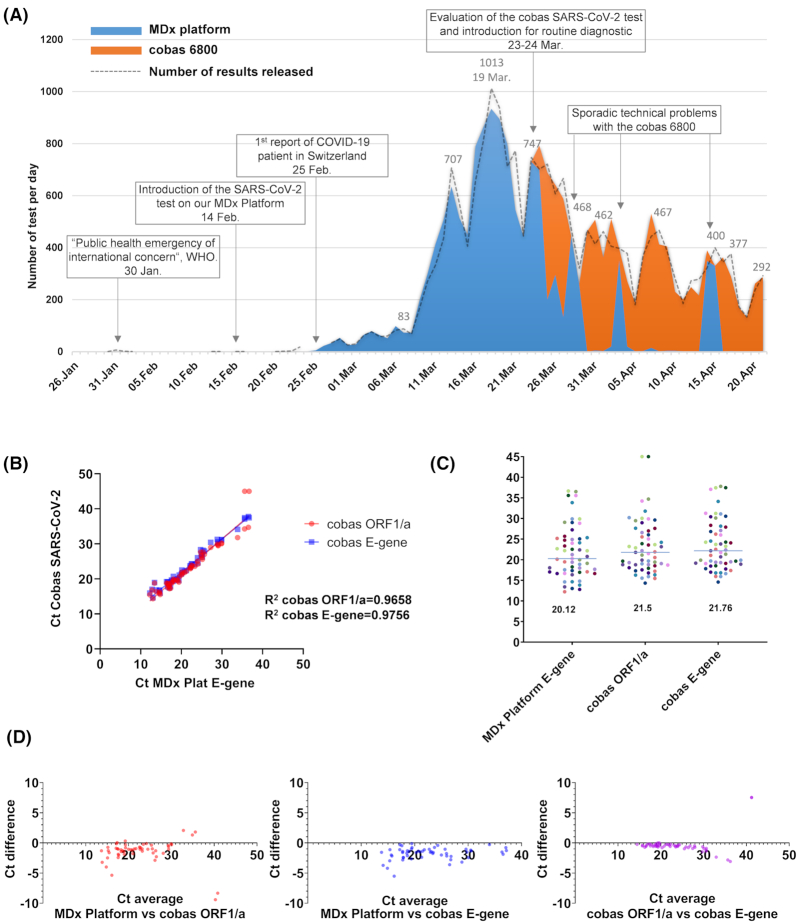
Figure 1.
SARS-CoV-2 detection using our in-house automated molecular diagnostic platform and the cobas the SARS-CoV-2 test in our laboratory. (A) Use of the in-house automated molecular diagnostic platform (blue) and the cobas the SARS-CoV-2 test (orange) during the course of the epidemic; the area represent the number of tests achieved per days while the dash line represent the results validated. Indeed, in some cases the analysis run started at J0 just before midnight and was validated at J+1. (B) Cycle threshold (Ct) values of the molecular diagnostic platform and of the cobas SARS-CoV-2 test. (C) Median of the Ct value of the molecular diagnostic platform and of the cobas SARS-CoV-2 test obtained for positive results (n = 55) (D) Bland–Altman graphic showing the (difference versus average) to compare the Ct value of the molecular diagnostic platform and of the cobas SARS-CoV-2 test.
Table 1.
Agreements between the results of the cobas SARS-COV-2 and the MDx Platform on various clinical samples.
| MDx platform | Cobas SARS-CoV-2 | Number of samples and agreement | |
|---|---|---|---|
| Nasopharyngeal secretions | |||
| Positive | Positive | 55 | |
| Positive | Negative | 0 | |
| Negative | Positive | 0 | |
| Negative | Negative | 123 | |
| Total | 178 | ||
| Agreement for nasopharyngeal secretions | 100% (178/178, Kappa = 1) | ||
| Sputum, bronchial aspirate, bronchoalveolar lavage | |||
| Positive | Positive | 7 | |
| Positive | Negative | 1 (Ct 36.49) | |
| Negative | Positive | 1 (Ct 39.91) | |
| Negative | Negative | 35 | |
| Total | 44 | ||
| Agreement for sputum, bronchial aspirates, bronchoalveolar lavages | 95.45% (42/44, Kappa = 0.847) | ||
| Anorectal swabs | |||
| Positive | Positive | 2 | |
| Positive | Negative | 0 | |
| Negative | Positive | 0 | |
| Negative | Negative | 38 | |
| Total | 40 | ||
| Agreement for anorectal swabs | 100% (40/40, Kappa = 1) | ||
| MDx Platform | Cobas SARS-CoV-2 | Number of samples tested | |
| ALL SAMPLES | |||
| Positive | Positive | 64 | |
| Positive | Negative | 1 | |
| Negative | Positive | 1 | |
| Negative | Negative | 196 | |
| Total | 262 | ||
| Agreement for all samples | 99.24% (260/262, Kappa = 0.98) | ||
Correlation between the semiquantitative results of the cobas SARS-COV-2 and the MDx platform on nasopharyngeal swabs
The semi-quantitative results of the two platform (Ct value) displayed a very good correlation (R2 = 0.96 and 0.97; Fig. 1B). The median Ct values were 20.3, 21.8 and 22.2 for the MDx platform, the cobas ORF1/a and the cobas E-gene, respectively (Fig. 1C). The Bland–Altman representation suggest a saturation of the cobas RT-PCR for Ct between 10 and 20 when compared to the MDx platform, which might explain the difference in the median Ct values (Fig. 1D). Altogether, these data suggest that the cobas and the MDx platform exhibit similar sensitivity.
Use of the MDx platform SARS-CoV-2 and the cobas SARS-CoV-2 test for the diagnosis of COVID-19
Since the introduction of the cobas SARS-CoV-2 test, 7517 tests have been performed in approximately 1 month (Fig. 1A). The cobas SARS-CoV-2 test was therefore used for the routine diagnostic of COVID-19 from upper respiratory tract (URT) mostly nasopharyngeal swabs, lower respiratory tract (LRT) and anorectal swabs. The SARS-CoV-2 test on the in-house MDx Platform was used for SARS-CoV-2 RT-PCR on other clinical specimens and as a backup. For instance, we observed 4 days of transient breakdown of the Cobas 6800 system. These days we could rely again on our MDx platform molecular diagnostic platform to achieve respectively 445, 257, 352 and 330 SARS-CoV-2 tests (Fig. 1A).
DISCUSSION
At the beginning of the COVID-19 epidemic and in the absence of test provided by industries we introduced a RT-PCR for the detection of the SARS-CoV-2 on our high-throughput open automated MDx platform according to our accredited R&D process (Greub et al. 2016). This allowed us to achieve the diagnosis of COVID-19 for our hospital as well as other hospitals of our canton (Vaud) and four other neighboring cantons, with more than 1000 analysis performed in a single day (19th March).
After 40 days of use of our MDx platform, we could test the cobas SARS-CoV-2 kit on the cobas 6800 instrument already available in our diagnostic laboratory, which revealed an excellent agreement with our MDx platform on upper respiratory tract specimen, as reported (Poljak et al. 2020). We also observed a very good correlation between the semi-quantitative results (Ct value) of both tests. We thus introduced the cobas SARS-CoV-2 test in routine and could therefore use the two methods alternatively or in parallel. This was particularly useful to manage the stock of reagent and sporadic instruments breakdowns. The correlation between the semi-quantification using both test allow us to report Ct values and corresponding viral loads to the clinicians (Jacot et al. 2020). Reporting SARS-CoV-2 viral load was very useful for both patient care and infection control strategies (Jacot et al. 2020; Moraz et al. 2020; Tom and Mina 2020; Yu et al. 2020). This was also very useful for RT-PCR results interpretation in the setting of repeated tests on the same patient or testing multiple specimen from the same patient (e.g URT versus or LRT, or URT versus anorectal swabs). Indeed, discrepant results could occur due to viral load at the limit of the detection of RT-PCR or because of the normal time course of the infection (Mueller et al. 2020). RT-PCR are very sensitive and have a detection limit of 100–1000 copies per ml of specimen (Konrad et al. 2020). However, depending on the time course of the infection with respiratory viruses, viral replication might not be (any more) detected in upper respiratory tract specimen, but can still be detected in LRT specimen as reported for influenza viruses, Middle East respiratory syndrome virus and SARS-CoV-1 (Hung et al. 2004; Oh et al. 2016; Mueller et al. 2020). SARS-CoV-2 can be detected on LRT specimen using the cobas SARS-CoV-2, as demonstrated by the high agreement with our MDx platform.
While COVID-19 is primarily associated with respiratory symptoms, atypical clinical presentations have been reported with the need to test various clinical samples, including anorectal swabs in case of diarrhoea. An excellent agreement between the cobas test and our MDx platform was also observed on anorectal swabs. Testing anorectal swabs could be useful in case of atypical clinical presentations or in specific situations such as donor screening in the context of fecal microbiota transplantation for instance (Nicco et al. 2020; Song et al. 2020; Xie et al. 2020; Young et al. 2020).
The introduction of high-throughput SARS-CoV-2 RT-PCR systems was among the decisive interventions of molecular diagnostic laboratories to face the COVID-19 pandemic (Doganay et al. 2020; Posteraro et al. 2020). However, when testing such a high number of specimen per day including different clinical specimen from the same patient, discrepant results might occur due to the normal evolution of the disease. To maintain the quality and the reliability of RT-PCR, such discrepant results can be identified and investigated using computer aided post-analytical software that are mandatory to handle such a high number of results (Mueller et al. 2020).
Other clinical specimen are routinely analysed for the diagnostic of COVID-19 on our MDx platform but not using the cobas SARS-CoV-2 test because of limited volume available (i.e. biopsy or CSF) or clotting issues (i.e. blood; Dang et al. 2009; Jaton et al. 2013).
Altogether, these data suggest that both the cobas-SARS-CoV-2 test and our MDx platform SARS-CoV-2 test are reliable to detect SARS-CoV-2 from various clinical specimen.
While this study included 262 clinical specimen, including 64 URT positive specimens, the number of positive specimen for LRT and rectal swabs received during the studied period was limited. Awaiting for future studies with a higher number of positive specimens for LRT and rectal swabs, the overall agreement between the two MDx platform and the commercial platform together with the fact that both tests contain inhibition controls support their reliability. Future studies should also include the comparison of in-house RdRP RT-PCR with commercial RT-PCR. Indeed, this comparison was not achieved as the RdRP RT-PCR needed further optimization based on recent publication that elucidated the reason of the limited sensitivity as the difference in the melting temperature of the forward and reverse primers of the initial PCR of Corman and colleagues (Corman and Drosten 2020; Muenchhoff et al. 2020; Pillonel et al.2020).
This study highlight (i) the importance of high throughput analytical platform to face the rapid spread of COVID-19, (ii) the central role of scalable molecular diagnostic platform in teaching hospital awaiting for commercial methods and (iii) the importance of having multiple analytical platforms.
The availability of all-inclusive commercial molecular diagnostic platform was important to increase the overall testing capacity. This study supports the use of RT-PCR on various clinical specimens. This will permit to address many situations for which the virus might not be detected in URT. Such data on other clinical specimen will be important for patient care and a better understanding of SARS-CoV-2 infection and pathogenesis.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all the staff of the Laboratory of Molecular Diagnostic, Mycobacteria and Biosafety of the Institute of Microbiology of the University of Lausanne.
Contributor Information
Onya Opota, Institute of Microbiology, University Hospital Center and University of Lausanne, Switzerland.
René Brouillet, Institute of Microbiology, University Hospital Center and University of Lausanne, Switzerland.
Gilbert Greub, Institute of Microbiology, University Hospital Center and University of Lausanne, Switzerland; Infectious Diseases Service, University Hospital of Lausanne, Lausanne, Switzerland.
Katia Jaton, Institute of Microbiology, University Hospital Center and University of Lausanne, Switzerland.
Conflicts of interests
None declared.
REFERENCES
- Corman VM, Drosten C. Authors' response: SARS-CoV-2 detection by real-time rt-pcr. Euro Surveill. 2020;25:2001035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman VM, Landt O, Kaiser M et al. . Detection of 2019 novel coronavirus (2019-ncov) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang T, Jaton-Ogay K, Flepp M et al. . High prevalence of anorectal chlamydial infection in HIV-infected men who have sex with men in Switzerland. Clin Infect Dis. 2009;49:1532–5. [DOI] [PubMed] [Google Scholar]
- Doganay L, Agaoglu NB, Irvem A et al. . Responding to COVID-19 in Istanbul: perspective from genomic laboratory. North Clin Istanb. 2020;7:311–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greub G, Sahli R, Brouillet R et al. . Ten years of R&D and full automation in molecular diagnosis. Fut Microbiol. 2016;11:403–25. [DOI] [PubMed] [Google Scholar]
- Hung IFN, Cheng VCC, Wu AKL et al. . Viral loads in clinical specimens and SARS manifestations. Emerg Infect Dis J. 2004;10:1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacot D, Greub G, Jaton K et al. . Viral load of SARS-CoV-2 across patients and compared to other respiratory viruses. Microbes Infect. 2020. https://www.ncbi.nlm.nih.gov/pubmed/32911086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaton K, Peter O, Raoult D et al. . Development of a high throughput PCR to detect Coxiella burnetii and its application in a diagnostic laboratory over a 7-year period. New Microbes New Infect. 2013;1:6–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad R, Eberle U, Dangel A et al. . Rapid establishment of laboratory diagnostics for the novel coronavirus SARS-CoV-2 in Bavaria, Germany, February 2020. Euro Surveill. 2020;25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraz M, Jacot D, Papadimitriou-Olivgeris M et al. . Clinical importance of reporting SARS-CoV-2 viral loads across the different stages of the COVID-19 pandemic. medRxiv. 2020, DOI: 2020.07.10.20149773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller L, Scherz V, Greub G et al. . Computer-aided medical microbiology monitoring tool: a strategy to adapt to the SARS-CoV-2 epidemic and that highlights RT-PCR consistency. medRxiv. 2020, DOI: 2020.07.27.20162123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muenchhoff M, Mairhofer H, Nitschko H et al. . Multicentre comparison of quantitative PCR-based assays to detect SARS-CoV-2, Germany, March 2020. 2020;25:2001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicco C, Paule A, Konturek P et al. . From donor to patient: collection, preparation and cryopreservation of fecal samples for fecal microbiota transplantation. Diseases. 2020;8:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh M-d, Park WB, Choe PG et al. . Viral load kinetics of MERS coronavirus infection. N Engl J Med. 2016;375:1303–5. [DOI] [PubMed] [Google Scholar]
- Opota O, Brouillet R, Greub G et al. . Methods for real-time PCR-based diagnosis of Chlamydia pneumoniae, Chlamydia psittaci, and Chlamydia abortus infections in an opened molecular diagnostic platform. Methods Mol Biol. 2017;1616:171–81. [DOI] [PubMed] [Google Scholar]
- Opota O, Jaton K, Branley J et al. . Improving the molecular diagnosis of Chlamydia psittaci and Chlamydia abortus infection with a species-specific duplex real-time PCR. J Med Microbiol. 2015;64:1174–85. [DOI] [PubMed] [Google Scholar]
- Opota O, Laurent S, Pillonel T et al. . Genomics of the new species Kingella negevensis: diagnostic issues and identification of a locus encoding a RTX toxin. Microbes Infect. 2017;19:546–52. [DOI] [PubMed] [Google Scholar]
- Opota O, Senn L, Prod'hom G et al. . Added value of molecular assay Xpert MTB/RIF compared to sputum smear microscopy to assess the risk of tuberculosis transmission in a low-prevalence country. Clin Microbiol Infect. 2016;22:613–9. [DOI] [PubMed] [Google Scholar]
- Opota O, Zakham F, Mazza-Stalder J et al. . Added value of Xpert MTB/RIF ultra for diagnosis of pulmonary tuberculosis in a low-prevalence setting. J Clin Microbiol. 2019;57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillonel T, Scherz V, Jaton K et al. . Letter to the editor: SARS-CoV-2 detection by real-time RT-PCR. Euro Surveill. 2020;25:2000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poljak M, Korva M, Knap Gasper N et al. . Clinical evaluation of the cobas SARS-CoV-2 test and a diagnostic platform switch during 48 hours in the midst of the COVID-19 pandemic. J Clin Microbiol. 2020, 58, e00599–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posteraro B, Marchetti S, Romano L et al. . Clinical microbiology laboratory adaptation to COVID-19 emergency: experience at a large teaching hospital in Rome, Italy. Clin Microbiol Infect. 2020;26:1109–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Liu P, Shi XL et al. . SARS-CoV-2 induced diarrhoea as onset symptom in patient with COVID-19. Gut. 2020;69:1143–4. [DOI] [PubMed] [Google Scholar]
- Tadini E, Papamidimitriou-Olivgeris M, Opota O et al. . SARS-CoV-2, a point in the storm. Rev Med Suisse. 2020;16:917–23. [PubMed] [Google Scholar]
- Tom MR, Mina MJ. To interpret the SARS-CoV-2 test, consider the cycle threshold value. In: Clinical Infectious Diseases: An Official Publication of the Infectious Diseases Society of America. Clin Infect Dis, 2020:ciaa619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Jiang L, Huang G et al. . Comparison of different samples for 2019 novel coronavirus detection by nucleic acid amplification tests. Int J Infect Dis. 2020;93:264–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young BE, Ong SWX, Kalimuddin S et al. . Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA. 2020;323:1488–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Yan L, Wang N et al. . Quantitative detection and viral load analysis of SARS-CoV-2 in infected patients. Clin Infect Dis. 2020;71:793–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.