Abstract
COVID-19 is a novel severe acute respiratory syndrome coronavirus. Currently, there is no effective treatment and vaccines seem to be the solution in the future. Virtual screening of potential drugs against the S protein of severe acute respiratory syndrome corona virus 2 (SARS-CoV-2) has provided small molecular compounds with a high binding affinity. Unfortunately, most of these drugs do not attach with the binding interface of the receptor-binding domain (RBD)–angiotensin-converting enzyme-2 (ACE-2) complex in host cells. Molecular modeling was carried out to evaluate the potential antiviral properties of the components of the medicinal herb Uncaria tomentosa (cat’s claw) focusing on the binding interface of the RBD–ACE-2 and the viral spike protein. The in silico approach starts with protein–ligand docking of 26 Cat’s claw key components followed by molecular dynamics simulations and re-docked calculations. Finally, we carried out drug-likeness calculations for the most qualified cat’s claw components. The structural bioinformatics approaches led to the identification of several bioactive compounds of U. tomentosa with potential therapeutic effect by dual strong interaction with interface of the RBD–ACE-2 and the ACE-2 binding site on SARS-CoV-2 RBD viral spike. In addition, in silico drug-likeness indices for these components were calculated and showed good predicted therapeutic profiles of these phytochemicals found in U. tomentosa (cat’s claw). Our findings suggest the potential effectiveness of cat’s claw as complementary and/or alternative medicine for COVID-19 treatment.
Communicated by Ramaswamy H. Sarma
Keywords: SARS-CoV-2, viral spike protein, ACE-2, cat’s claw, Uncaria tomentosa, molecular modeling, COVID-19
Introduction
The severe acute respiratory syndrome corona virus 2 (SARS-CoV-2) is a part of coronavirus family (CoV) and was initially identified in Wuhan, China. COVID-19 (coronavirus disease 2019) is highly contagious in humans, which has rapidly spread and caused an unprecedented pandemic, with a large number of deaths and economic crisis in the world (Prajapat et al., 2020). According to the latest report of the World Health Organization (WHO), over 32.7 million cases and 991 000 deaths of COVID-19 were confirmed as of September 27, 2020 (World Health Organization, 2020). In developing countries of Latin America and the Carribean, the public health has been the most affected because people do not have the opportunities to access a modern health system and medicines (Wang et al., 2020).
Phytotherapy based on natural products might be a proper alternative for treating viral diseases (Akram et al., 2018). According to WHO estimates, about 80% of the population in developing countries uses traditional medicine in primary health care, mainly medicinal plants (World Health Organization, 2018). The selection of natural products for the study of their biological properties has been addressed through three fundamental methodologies: the selection of random natural sources, the selection based on chemotaxonomy (screening of similar compounds in organisms belonging to the same family or genus) and selection based on ethnomedicine (Heinrich, 2002). Ethnomedicine has been considered the most effective therapy and consists of the study of natural products that have a long history of use in some communities for the treatment of certain diseases and are part of the phytotherapeutic arsenal of popular knowledge (Wu & Tan, 2019).
On the other hand, Uncaria tomentosa (Willd. ex Schult.) DC. named cat’s claw (‘uña de gato’ in Spanish) is a woody vine indigenous to the Peruvian Amazon and other tropical areas of South and Central America that belongs to Rubiaceae family (Sandoval et al., 2000). Currently, the raw material of U. tomentosa is dispensed in Public Hospitals of the Social Health Insurance (EsSalud-Peru) as Complementary Medicine Service (CMS) (Gonzales et al., 2010). Traditionally, extracts prepared by roots and barks decoction are used against several diseases, such as allergies, arthritis, inflammations, rheumatism infections and cancer (Araujo et al., 2018).
Bioactive constituents of U. tomentosa extracts include proanthocyanidins [proanthocyanidin B2 (the main component), proanthocyanidin B4, proanthocyanidin C1, an epicatechin trimer, epiafzelechin-4β→8-epicatechin and an epicatechin tetramer] (Batiha et al., 2020; Navarro-Hoyos et al., 2017), oxindole alkaloids (isopteropodine, pteropodine, rhynchophylline, mytraphylline, speciophylline, uncarine F and uncarine E), indole alkaloidal glucosides (cadambine, 3-dihydrocadambine and 3-isodihydrocadambine) (Batiha et al., 2020; Kuraś et al., 2009; Laus et al., 1997; Lima-Junior et al., 2013; Lock et al., 2016; Navarro et al., 2019, 2017; Snow et al., 2019), quinovic acid glycosides (Pavei et al., 2012), tannins (Ostrakhovich et al., 1997), polyphenols, catechins, beta sitosterol (Aquino et al., 1997; Navarro et al., 2019) and proteins (Lenzi et al., 2013), which individually or synergistically contribute to their therapeutic properties.
In regards to the antiviral properties of U. tomentosa, the alkaloid fraction has been demonstrated to be the most effective on human monocytes infected with dengue virus-2 (DENV) in vitro (Reis et al., 2008). Another study revealed that only the alkaloidal fraction has inhibitory activity on dengue virus, and the negative effect was observed with the nonalkaloidal fraction (Lima-Junior et al., 2013). In another study, the antiherpetic activity of U. tomentosa seems to be associated with polyphenols or with their synergistic effect with pentacyclic oxindole alkaloids or quinovic acid glycosides (Caon et al., 2014). U. tomentosa hydroethanolic extracts have demonstrated a significant in vitro inhibitory effect on the replication of herpes simplex virus type 1, and the inhibition of viral attachment in the host cells was characterized as the main mechanism of its antiviral activity (Terlizzi et al., 2016).
SARS-CoV-2 contains four structural proteins, namely the spike (S), membrane (M), envelope (E) and nucleocapsid (N) proteins. The S protein is responsible for the host attachment and fusion of the viral and host-cell membranes (Wu et al., 2020). Otherwise, the angiotensin-converting enzyme 2 receptor (ACE-2r) is the host cellular receptor with a higher affinity to SARS-CoV-2 (Jamwal et al., 2020). This process is triggered when the S1 subunit of S protein binds to a host-cell receptor (Han & Král, 2020). To engage a host-cell receptor, the receptor-binding domain (RBD) of S1 undergoes transient hinge-like conformational motions (receptor-accessible or receptor-inaccessible states). U. tomentosa’s constituents could block the virus from binding to human cell receptors and disrupt the virus cycle helping to prevent the protein maturation of SARS-CoV-2 and limit its infection spread (Li et al., 2020).
Several molecular targets have been identified as the main druggable key of SARS-Cov-2 for new antiviral discovery. Moreover, its X-ray structure has been recently released, hence allowing possible computational analysis. In fact, several computational studies have already been undertaken on this system including a long 20 μs molecular dynamics (MD) study and virtual screening of several databases (Huang et al., 2020).
With neither drugs nor vaccines approved against COVID-19 yet, finding strategies to diminish the impact of the pandemic is fundamental. Medicinal herbs and, more particularly, those with demonstrated antiviral activities as U. tomentosa could slow down the spreading of the disease. Particularly in developing countries, in which the accessibility to these plants is easier and more economically viable, adding these medicinal herbs to the general medical kit may be beneficial.
Here, our study stands on an in silico strategy reminiscent to those applied at the early stage of current state-of-the-art drug discovery pipelines and includes (1) protein–ligand docking of all bioactive compounds of U. tomentosa against focusing both on the binding interface of the RBD–ACE-2 and inside SARS-CoV-2 RBD spike protein, (2) simulations of ligand pathway of the best predicted compounds from step 1 to evaluate convenient entrance mechanism of the compounds to the binding site, (3) MD simulation to assess the stability of the best protein–ligand complexes from 1, (4) calculation of pharmacokinetics parameters for the most qualified compounds resulting from the previous parts of the docking protocol. This study demonstrates the antiviral potential of U. tomentosa-based products to be applied as a rapid phytotherapeutic option for COVID-19.
Material and methods
Protein structure and setup
Calculated binding affinity of the main constituents of the U. tomentosa (Table 1) was explored for its ability to disrupt the SARS-CoV-2/ACE-2 complex and inhibit SARS-CoV-2 spike protein of novel coronavirus findings, a facile therapeutic option for anti-coronavirus therapy. To this purpose, the crystal structures of SARS-CoV-2/ACE-2 complex and SARS-CoV-2 spike protein were downloaded from the Protein Data Bank (PDB entry code 6M17 and 6VYB, respectively) (Yan et al., 2020) and all bounded ligands, ions and solvent molecules were manually removed using the DS Visualizer 2.5 program. For docking studies, the structures of the selected proteins were parameterized using AutoDock Tools (Trott & Olson, 2009). To facilitate the formation of hydrogen bonds, polar hydrogens were added.
Table 1.
Best binding energy (kcal mol–1) based on AutoDock scoring of the main constituents of the U. tomentosa into the RBD/ACE-2 interface and SARS-CoV-2 spike protein binding domain (RBD) (PDB ID: 6VYB).
| Main constituents of cat’s claw | Best binding energy RBD/ACE-2 interface (kcal mol–1) | Best binding energy SARS-CoV-2 RBD (kcal mol–1) |
|---|---|---|
| Spiroxindole alkaloids | ||
| Uncarine F | –7.1 | –6.1 |
| Speciophylline | –6.8 | –6.8 |
| Mitraphylline | –6.8 | –6.2 |
| Pteropodine | –6.7 | –6.2 |
| Isopteropodine | –6.7 | –6.3 |
| Isomitraphylline | –6.6 | –6.1 |
| Rynchophylline | –6.0 | –5.5 |
| Isorynchophyllin | –6.0 | –5.7 |
| Indole glycosides alkaloids | ||
| 3-isodihydrocadambine | –7.6 | –6.4 |
| Cadambine | –7.6 | –6.1 |
| 3-dihydrocadambine | –7.0 | –7.1 |
| Polyhydroxylated triterpenes | ||
| Uncaric acid | –7.0 | –5.5 |
| Floridic acid | –7.0 | –5.5 |
| PHT-1 | –6.9 | –5.6 |
| Quinovic acid glycosides | ||
| QAG-2 | –8.2 | –6.8 |
| QAG-5 | –8.0 | –6.1 |
| QAG-4 | –7.8 | –6.8 |
| QAG-6 | –7.8 | –6.8 |
| QAG-1 | –7.8 | –6.1 |
| QAG-3 | –7.3 | –6.2 |
| Proanthocyanidins | ||
| Proanthocyanidin C1 | –8.6 | –7.0 |
| Epiafzelechin-4β-8 | –7.7 | –6.2 |
| Proanthocyanidin B4 | –7.6 | –7.2 |
| Proanthocyanidin B2 | –7.5 | –7.2 |
| Epicatechin | –7.1 | –6.0 |
| Chlorogenic acid | –6.1 | –6.4 |
| Reference | ||
| HepOSa | –7.1 (–7.3)b | –7.1 (–7.3)b |
HepOS: Heparin octasaccharide taken from PDB 5UE2 was used as positive control.
Estimated by Kwon et al. (2020).
Ligand dataset preparation and optimization
Ligands used in this study are major components of the U. tomentosa extracts and a sulfated heparin octasaccharide (taken from PDB 5UE2), a potent SARS-CoV-2 inhibitor in vitro reported in the literature (Kwon et al., 2020). The 2D structures of the 26 cat’s claw constituents were obtained as mol.2 files from the ZINC database (ChemAxon, 2016). The resultant compounds were submitted to MarvinSketch 8.3 (Morris et al., 1998) to correct the protonation states of the ligands at physiological pH 7.4. In addition, the geometry optimization of all ligands was carried out using the HF/6-31G* level of theory. Then, the structures were parameterized using AutodockTools to add full hydrogens to the ligands, to assign rotatable bonds and saving the resulting structure in the required format for use with AutoDock. All possible flexible torsions of the ligand molecules were defined using AUTOTORS in PDB AutoDockTools (Morris et al., 2009; Walls et al., 2020) to promote the calculated binding with the target structure.
Docking-based virtual screening
Our docking protocol was performed using AutoDock Vina and default procedures to dock a flexible ligand to a rigid protein. Docking simulation of ligands was carried out on the interface between the SARS-CoV-2 and ACE-2 (PDB code: 6M17) (Yan et al., 2020), where both proteins residues are in proximity. Next, we used the cryo-EM structure of SARS-CoV-2 spike protein (PDB code: 6VYB) in their open state (Lipinski et al., 2012) to explore the potential inhibition of components of the cat’s claw, selecting ACE-2-binding pocket to this study. Once a potential binding site was identified, 26 compounds which are the major components of the cat’s claw extracts were docked to this enzymes-site to determine the most probable and the most energetically favorable binding conformations. To accomplish rigorous docking simulations involving a grid box to the identified catalytic site, Autodock Vina 1.1.2 (Trott & Olson, 2009) was used. The exhaustiveness was 20 for each protein–ligand pair (number of internal independent runs). The active site was surrounded by a docking box of 40 × 40 × 40 Å with a grid spacing of 1 Å. Affinity scores (in kcal mol–1) given by AutoDock Vina for all compounds were obtained and ranked based on the free energy binding theory (more negative value means greater binding affinity). The resulting structures and the binding docking poses were graphically inspected to check the interactions using the DS Visualizer 2.5 (http://3dsbiovia.com/products/) or The PyMOL Molecular Graphics System 2.0 programs.
MD simulation
Molecular interaction stability of protein–ligand complexes obtained by docking simulations were verified through MD simulations by using the Gromacs program (Abraham et al., 2015) considering the SARS-CoV-2/ACE-2 interface, as well as the SARS-CoV-2 spike protein active site and the best docking pose for Proanthocyanidin C1, QAG-2, Proanthocyanidin B2 and 3-dihydrocadambine, respectively. Force field parameters for protein and ligands were derived independently. For the selected protein, the amber03 force field was selected and assigned using the pdb2gmx tool of the Gromacs program packages, meanwhile ligand force field parameters were prepared with the generalized AMBER force field (GAFF) using the molecular geometries previously optimized with the HF/6-31G* level of theory in gas phase, (Foresman et al., 1992; Glendening et al., 2012; Roothaan, 1951) with the Gamess-US program (Schmidt et al., 1993) .
In addition, each ligand was verified as a minimum through a harmonic vibrational normal mode analysis. Atomic charges were obtained with the Merz–Kollman scheme (Singh & Kollman, 1984) by fitting a restricted electrostatic potential (RESP) model by the Gamess-US program (Bayly et al., 1993), and the output file was used into the resp sub-program of the AmberTools program package (Cornell et al., 1995). Assignment of GAFF force field parameters was carried out by the Antechamber program (Wang et al., 2006) and the required input files for molecular dynamics simulations were prepared using the ACPYPE python interface. Protein and protein–ligand complexes were solvated in a rectangular box of TIP3P waters. The obtained system was neutralized adding seven-sodium counter ions to neutralize the net negative charge of the protein, and then physiological conditions (298 K, pH 7.4, 0.9% NaCl solution) were established (Hammad et al., 2020).
To remove spurious contact, molecular geometries were optimized with the steepest descent algorithm with 100,000 steps, protein backbones atoms were constrained with a force constant of 1000 kJ mol−1. Then, the MD simulations were allowed to run for 1000 ps in the NpT ensemble. In addition, 50 ns in the NpT ensemble were calculated for the production stage. All simulations were carried out under periodic boundary conditions. A cubic box with the size of 25 × 25 × 25 nm was used. A 12 Å cutoff distance was used to calculate nonbonded interactions. Electrostatic interactions were treated with the Ewald particle mesh (PME) method (Nishizawa & Nishizawa, 2010); while van der Waals interactions were introduced by using the cut-off scheme. Finally, the V-rescale thermostat at 300 K with a coupling constant of 1.0 ps was used and the pressure was kept constant at 1 atm using the Parinello–Rahman barostat (Parrinello & Rahman, 1981) with a coupling constant of 2.0 ps and a compressibility factor of 4.5 × 10−5 bar−1. All covalent bonds were constrained using the LINCS algorithm and the contact list was updated every 40 fs.
Prediction of drug-likeness indices for the most docking promissory compounds
Drug-likeness prediction along with further ADME properties presents a wide of opportunities for a rapid new antiviral drug discovery. The drug-like and ADME properties for the most active components of the U. tomentosa extract (constituents having the highest binding affinity) were screened using open-access cheminformatics platforms such as Molinspiration (for molecular weight – MW, rotatable bonds and polar surface area – PSA descriptors), ALOGPS 2.1 (for log Po/w descriptor) and the Pre-ADMET 2.0 to predict four pharmaceutical relevant properties such as intestinal permeability (App. Caco-2), albumin-binding proteins (KHSA), Madin–Darby Canine Kidney (MDCK Line) cells permeation and intestinal absorption (%HIA). These parameters establish movement, permeability, absorption and action of potential drugs (Ertl et al., 2000). The interpretation of both MDCK and Caco-2 permeability using PreADMET is as follows: (1) Permeability lower than 25: low permeability; (2) Permeability between 25 and 500: medium permeability; (3) Permeability higher 500: high permeability.
Results and discussion
Database of U. tomentosa bioactive compounds
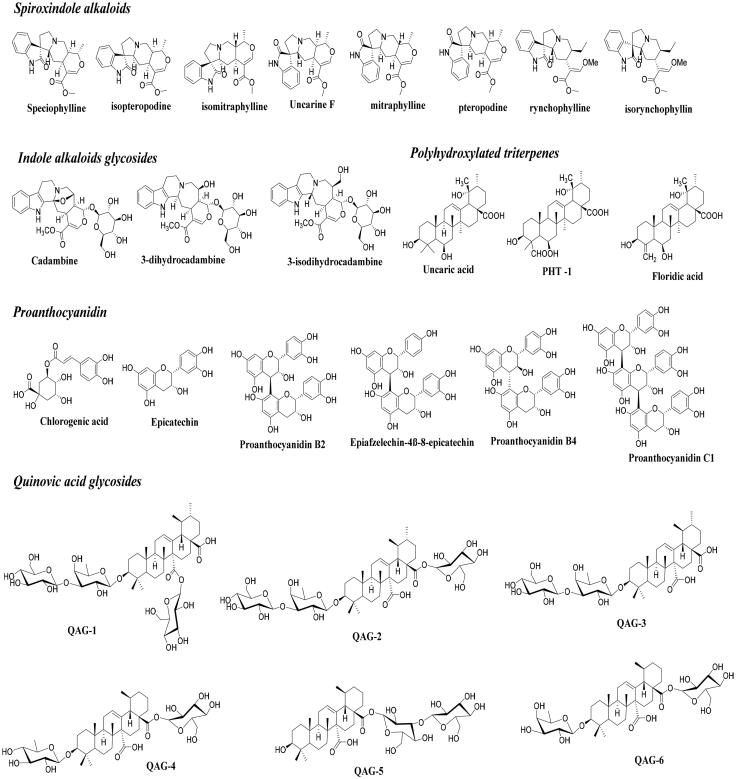
This study was performed to identify whether certain components of U. tomentosa extracts have potential therapeutic effects against COVID-19. To this purpose, a database of 26 compounds that have shown prevalence on the herbal therapeutic activity has been generated (Figure 1) (Aquino et al., 1990, 1997; Batiha et al., 2020; Keplinger et al., 1998; Kitajima et al., 2000; Lima-Junior et al., 2013; Lock et al., 2016; Montoro et al., 2004; Navarro et al., 2019, 2017; Pavei et al., 2012; Peñaloza et al., 2015; Snow et al., 2019; Vera-Reyes et al., 2015). Our initial hypothesis is that cat’s claw should contain molecules with highest therapeutic profiles against SARS-CoV-2, by disrupting SARS-CoV-2/ACE-2 association or by inhibiting SARS-CoV-2 spike protein.
Figure 1.
2D structures for the major bioactive constituents of the U. tomentosa studied as ligands against SARS-CoV-2/ACE-2 association and SARS-CoV-2 spike protein.
Structure-based virtual screening: Docking studies
During Covid-19 host infection, SARS-CoV-2 enters human epithelial cells through a first molecular recognition of RBD to the ACE-2 protein. When coronaviruses bind directly to the peptidase domain (PD) of ACE-2, it results in the loss of their primary physiological role, which includes vasoconstriction and blood pressure regulation. In consequence, binding of SARS-CoV-2 RBD to the human ACE-2 receptor is associated strongly with cardiovascular diseases, such as hypertension, heart attack and chronic nephropathies. Blocking the binding of SARS-CoV-2 to the human ACE-2 receptor may result in the most promising approach to prevent virus entry into human cells. Recently, the cryo-EM co-crystal structures of the RBD of SARS-CoV-2 with human ACE-2 have been solved (Yan et al., 2020), which open the possibility to design better and more specific inhibitors for suppression of viral infection.
Thus, to study the effectiveness of cat’s claw against SARS-CoV-2/ACE-2 complex, docking approaches were carried out in the ACE-2–RBD binding interface as the druggable site, to establish the interaction between the selected site and the main constituents of cat’s claw. We also performed molecular docking studies to find a potential association of constituents of cat’s claw to the SARS-CoV-2 spike protein. This approach also could conduce to block the SARS-CoV-2 spike protein interaction with human receptor ACE-2. Hence, in this article, the structure of spike glycoprotein (PDB ID: 6VYB) is to be considered as an additional druggable target. In addition, we have also performed the docking of a sulfated heparin octasaccharide (HepOS) as positive reference, which have been recently reported as an effective in vitro inhibitor of SARS-CoV-2 by its interaction against the spike protein RBD (Kwon et al., 2020).
Overall, docking approach revealed that the most components founded in cat’s claw could block SARS-CoV-2/ACE-2 association because they displayed significant binding affinity at interface of ACE-2–RBD complex in the range between –6.0 to –8.6 kcal mol–1 (Table 1). On the other hand, when structures were docked against the SARS-CoV-2 spike protein, good dock scores were obtained (specially, Proanthocyanidins series) ranging from –5.5 to –7.2 kcal mol–1.
Thus, we performed a rigorous exploration of the docking solutions obtained from these compounds when docking occurred against SARS-CoV-2-related enzymes. Hence, based on the analysis of these different results and visual inspection, a clear behavior appears along the molecular docking that could be summarized as follows in the following sections.
Docking profile inside RBD/ACE-2 interface
The recognition of SARS-CoV-2 by human ACE-2 can be divided into several contacts. From the ACE-2 structure: α1 helix domain comprises 12 critical aminoacids, such as GLN24, THR27, ASP30, LYS31, HIS34, GLU35, ASP38, TYR41, GLN42, GLU37, PHE390 and GLN388; α2 helix domain located around MET82 and four residues (LYS353, GLY354, ASP355 and ARG357) between β3 and β4 sheets are needed to recognize SARS-CoV-2. From SARS-CoV-2 structure, ten residues are essential for ACE-2 binding, such as LYS417, GLN493, TYR453, TYR505, TYR449, ASN501, GLN498, THR500, GLN474 and PHE486. Therefore, these key binding domains were considered in this article to explore the ability of U. tomentosa to disrupt SARS-CoV-2 RBD interaction to human ACE-2 (Yan et al., 2020).
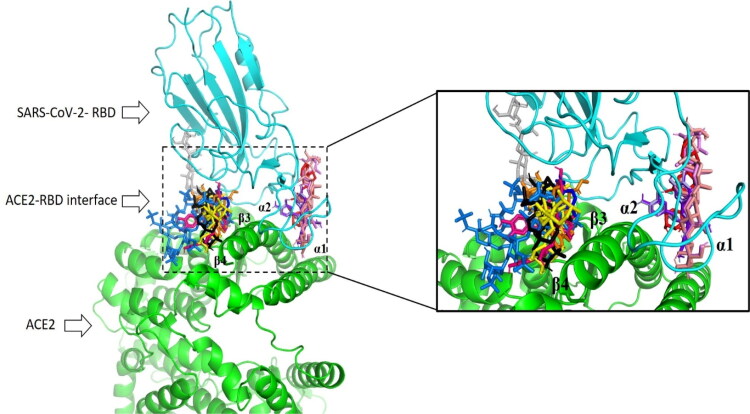
In general, all compounds as part of U. tomentosa show docked structures that fit well into the RBD/ACE-2 interface, particularly along α1, α2, β3 and β4 domains of human ACE-2 (Figure 2) and with good predicted docking scores that range from –6.0 to –8.6 kcal mol–1 (Table 1). Calculations revealed that most of ligands were located between β3 and β4 sheets, and a reduced group very close to α1 and α2 helix of ACE-2. Since during viral infection α1, α2, β3 and β4 domains are responsible for the recognition of SARS-CoV-2 by human ACE-2, our findings open the possibility to use U. tomentosa against SARS-CoV-2/ACE-2 association.
Figure 2.
Superposition of the best conformation of the most active components against ACE-2–RBD binding interface Uncarine F (in blue), Cadambine (in red), 3-isodihydrocadambine (in orange), Proanthocyanidin B2 (in light blue), Proanthocyanidin B4 (in purple blue), Proanthocyanidin C1 (in hot pink), Epiafzelechin-4β-8 (in olive), QAG-1 (in yellow), QAG-2 (in gray), QAG-4 (in violet), QAG-5 (in black), QAG-6 (in salmon) and HepOS (in bluemarine) as positive reference.
Notably, most predicted complexes have the interaction fingerprint with those of critical residues as part of ACE-2–RBD binding interface. Protein–ligand interaction analysis of the 26 constituents to the RBD/ACE-2 complex revealed that the most of constituents strongly bonded to ACE-2 through HIS34, ASP30, GLU37, PHE390, GLN388, LYS353, THR27, GLU35 residues, meanwhile with SARS-CoV-2 RBD showed critical contacts with TYR505, TYR453, LYS417, TYR449 residues. Due to these compounds as part of cat’s claw and interaction with key aminoacids inside ACE-2–RBD binding interface, we are encouraged to believe that U. tomentosa could affect the interaction of SARS-CoV-2 spike protein with ACE-2. Note that this approach may be useful as a rapid therapeutic option to prevent or treat COVID-19.
A simple view showed that all ligands were able to interact with those critical aminoacids involved in the molecular recognition of RBD to the ACE-2 protein, where at least one compound from each chemical series showed great ability to bind to the SARS-CoV-2-RBD complex along the interface. Hence, we focused on five compounds (those with highest negative energy value obtained after docking on each series) such as Proanthocyanidin C1 (–8.6 kcal mol−1), QAG-2 (–8.2 kcal mol−1), 3-isodihydrocadambine (–7.6 kcal mol−1), Uncarine F (–7.1 kcal mol–1) and Uncaric acid (–7.0 kcal mol−1), which had higher or comparable affinity than positive reference HepOS (–7.3 kcal mol−1) to the SARS-CoV-2-RBD complex (Table 1). At this point it is worth mentioning that the docking calculations involving positive reference give a value in very good agreement with experimental one (–7.3 kcal mol−1), hence providing with a certain amount of confidence regarding the Autodock scoring function of this project.
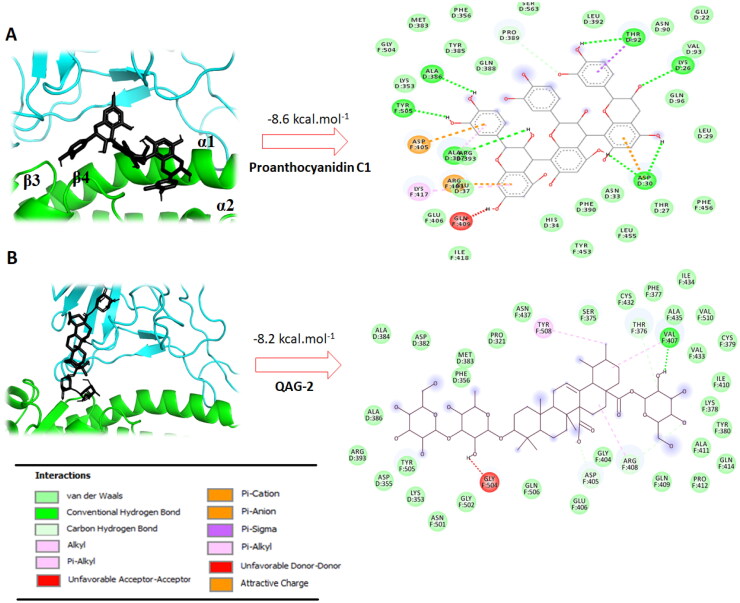
A rigorous view of 2D plots ligand interactions into ACE-2–RBD interface generated from DS visualizer program revealed which interactions are involved by the most docking active ligands and how their structures affect them. Thus, the most active docking ligand Proanthocyanidin C1 (which had affinity of −8.6 kcal mol−1) was able to interact with those critical aminoacids for binding SARS-CoV-2 spike to the human ACE-2 receptor. We found that Proanthocyanidin C1 establishes three strong hydrogen bond interactions between the hydroxyl groups in flavone moiety with TYR505 (SARS-CoV-2) and ASP30 (ACE-2 receptor) residues, one π-alkyl interaction with LYS417 residue and several hydrophobic interactions between the molecule and the GLU37, GLN388, LYS353, PHE390, THR37 and HIS34 residues. In addition to these critical aminoacids, Proanthocyanidin C1 revealed notable binding interactions with human ACE-2 receptor very close to interface through four hydrogen contact, such as ALA386, THR92, LYS26 and ALA337 that could may also affect the interaction of RBD with ACE-2 (Figure 3(A)).
Figure 3.
A,B: The best conformation of the Proanthocyanidin C1 (A) and QAG-2 (B) within ACE-2–RBD interface (PDB: 6M17). 2D interaction mode plots between the selected compounds to the ACE-2–RBD complex. Interactions between each component and amino acids residues into ACE-2–RBD binding domain are indicated by the dashed lines. D and F into circles indicated ACE-2 and Spike proteins, respectively.
Furthermore, a closer look at the best possible binding pose of QAG-2 (which display a high docking score of −8.2 kcal mol−1), reveals strong interactions at the contact interface between the two proteins. Thus, it was found that QAG-2 forms two hydrophobic interactions with critical aminoacids LYS353 and ASP355 located between β3 and β4 sheets in the ACE-2 protein. We also observed that QAG-2 displays one interaction at interface with VAL407 (from spike protein) through strong hydrogen bond. In addition, various hydrophobic contacts were observed between molecule and SER375, PHE377, VAL510, CYS379, ILE410, GLY404, ASP405, PHE356, ALA386, ARG408 and GLY502 at the SARS-CoV-2 RBD–ACE-2 interface (Figure 3(B)). Note that the deprotonated carboxylate moiety does not form any important interaction, therefore does not play any specific role in the QAG-2 binding at RBD–ACE-2 interface. This fact can be explained primarily because positively charged residues (lysine, arginine or histidine) do not surround this moiety at the interface.
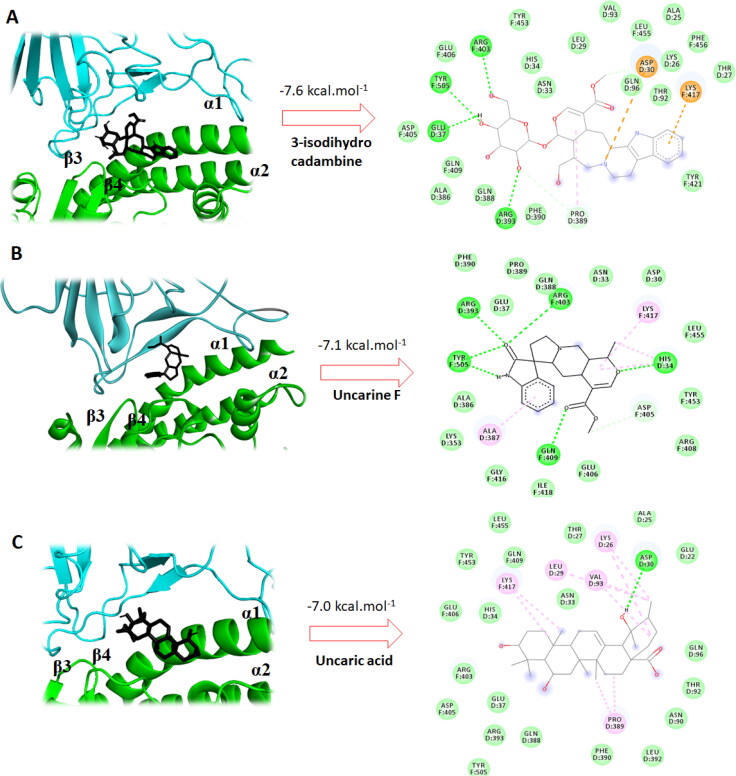
On the other side, 3-isodihydrocadambine (–7.6 kcal mol–1) has key contacts with residues in the RBD/ACE-2 binding interface (Figure 3(A)). Thus, molecule showed several interactions with critical residues of human ACE-2, which are essential for SARS-CoV-2 spike binding, among them, one hydrogen bond contact and one π-alkyl interaction with GLU37 residue. Importantly, the protonated nitrogen atom in the β-carboline moiety forms one salt bridge interaction with the carboxyl in ASP30 (from ACE-2 protein) and three hydrophobic interactions with PHE390, THR27 and GLN388. Furthermore, 3-isodihydrocadambine also had a crucial H-bond contact with TYR505 amino acid of SARS-CoV-2 spike protein, which is well-reported as an initial contact point during ACE-2 recognition. 3-Isodihydrocadambine also showed one π-cation interaction between fused aromatic ring in the β-carboline moiety and the key residue LYS417 and eight van der Waals contacts with spike protein residues, including GLU406, ASP405, GLN409, PHE456 LEU455, ASP405, TYR453 and TYR421. Further interactions were also observed that might promote the disruption of the interactions between SARS-CoV-2-RBD and ACE-2, among them two H-bonds formed per arginine residues 393 (from ACE-2 protein) and 403 (from spike protein) interacting with the sugar moiety.
Interestingly, Uncarine F binds at RBD/ACE-2 interface with binding affinity of −7.1 kcal mol−1 through several interactions with those base residues for the recognition of SARS-CoV-2 to the human ACE-2 (Figure 3(B)). Besides two strong hydrogen contacts with HIS34 of human ACE-2 and TYR505 from the SARS-CoV-2 spike protein, Uncarine F showed several hydrophobic interactions with key aminoacids at the interface, such as LYS353, ASP30, PHE390, GLU37, TYR453 and GLN388. Finally, analysis of the 2D-interaction map for Uncaric acid (which had docking score of −7.0 kcal mol−1) revealed crucial contacts with essential aminoacids residues for ACE-2–RBD binding. Thus, Uncaric acid binds at ASP30 residue of ACE-2 through a hydrogen bonding with one of the hydroxyl groups. Furthermore, hydrophobic interactions between molecule and five of those key residues at interface were displayed as follows: HIS34, TYR453, PHE390, GLN388 and TYR505. Notably, Uncaric acid had five π-alkyl interactions with other interface residues that probably could promote the ACE-2–RBD binding cleavage, such as LEU29 (D), VAL93 (D), LYS26 (D), PRO389 (D) and LYS417 (F).
In accordance to abovementioned, our docking studies shows that several components of U. tomentosa may have the ability to disrupt the association of SARS-CoV-2 spike protein with the human ACE-2 receptor. Among these components, Proanthocyanidin C1 (–8.6 kcal mol−1), QAG-2 (–8.2 kcal mol−1), 3-isodihydrocadambine (–7.6 kcal mol−1), Uncarine F (–7.1 kcal mol−1) and Uncaric acid (–7.0 kcal mol−1) had good predicted binding affinity for binding interface and they can naturally access it without noticeable energetic cost. These findings suggest that U. tomentosa may be a viable treatment option during initial stage of the COVID-19 infection.
Docking profile against SARS-CoV-2 spike RBD
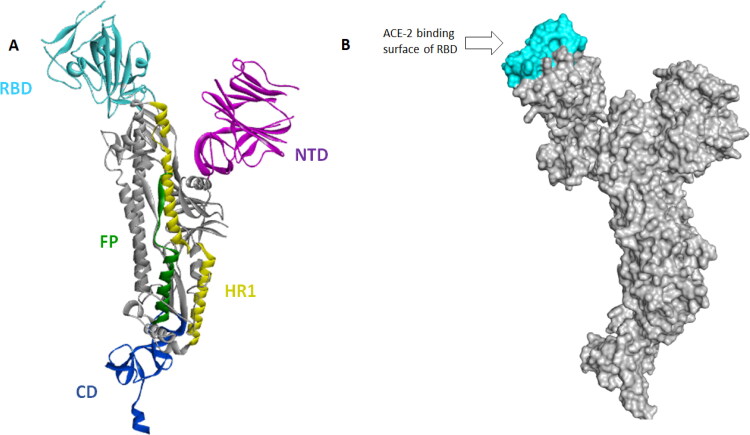
Recently, cryo-EM structure of the SARS-CoV-2 spike glycoprotein was resolved in their closed and open states (PDB ID: 6VXX and 6VYB, respectively) (Walls et al., 2020). SARS-CoV-2 spike protomer consists of five functional domains, namely as NTD, RBD, FP, HR1 and CD (Figure 4). Because RBD domain is responsible for the binding to the host cell receptor (ACE-2), we focused our docking investigations inside the ACE-2 binding surface of RBD. Hence, to carry out our docking calculations we selected as starting structure the SARS-CoV-2 spike glycoprotein in their open state (PDB ID: 6VYB), because, recently molecular dynamics simulations revealed the open state as the most active form of SARS-CoV-2 spike for binding to human ACE-2 (Gur et al., in press). Thus, docking studies on SARS-CoV-2 spike (PDB ID: 6VYB) could provide important insights into the molecular basis for coronavirus recognition in the initial step of infection.
Figure 4.
A–C: The best conformation of the 3-isodihydrocadambine (A), Uncarine F (B) and Uncaric acid (C) within ACE-2–RBD interface (PDB: 6M17). 2D interaction mode plots between the selected compounds to the ACE-2–RBD complex. Interactions between each component and amino acids residues into ACE-2–RBD binding domain are indicated by the dashed lines. The code letter ‘D’ and ‘F’ indicated ACE-2 and spike proteins, respectively.
Several residues in the SARS-CoV-2 RBD have been identified as essential in the association to the human ACE-2 during coronavirus (COVID-19) infection, including TYR449, TYR453, GLY496, THR500, TYR505, LYS417, GLN493, ASN501 and GLN498 (Lan et al., 2020). Therefore, to demonstrate the ability of constituents of U. tomentosa to block binding of the SARS-CoV-2 spike protein to human ACE-2 receptor, we performed molecular docking studies around aforementioned critical amino acids, meaning this docking runs were carried out inside ACE-2 binding surface of RBD. From a general perspective, promising docking scores were obtained when the major constituents from U. tomentosa bind to the RBD of SARS-CoV-2 (ranging from –5.5 to –7.2 kcal mol−1).
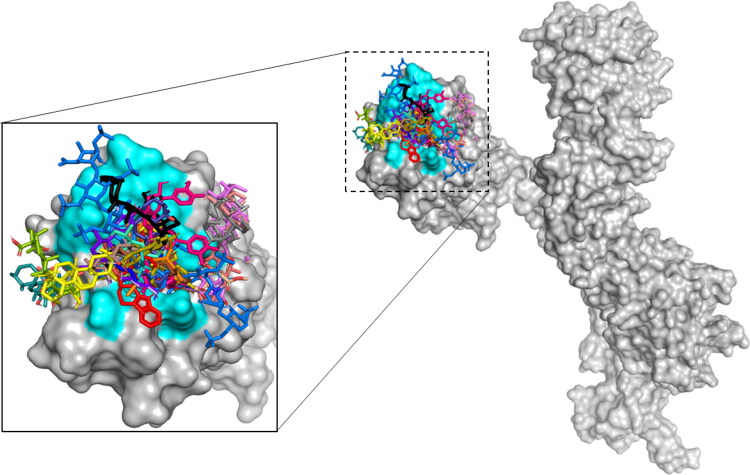
Thus, to compare the best binding pose of the most docking-active components of U. tomentosa and HepOS (positive reference) inside SARS-CoV-2 RBD, Figure 5 illustrates the most stable binding poses based on AutoDock scoring listed in Table 1 for all constituents on RBD binding domain. The superposition of the positive reference (HepOS) and the best conformation obtained theoretically for selected docked compounds showed that the major constituents in the ethanolic extract of U. tomentosa can accommodated themselves into stable conformations occupying this binding site during docking process.
Figure 5.
A: SARS-CoV-2 spike protein protomer structure. Protomer domains RBD (in cyan), NTD (in violet), HR1 (in yellow), CD (in blue) and FP (in green). B: Connolly surface of the SARS-CoV-2 spike protein showing RBD domain. The binding site appears in cyan.
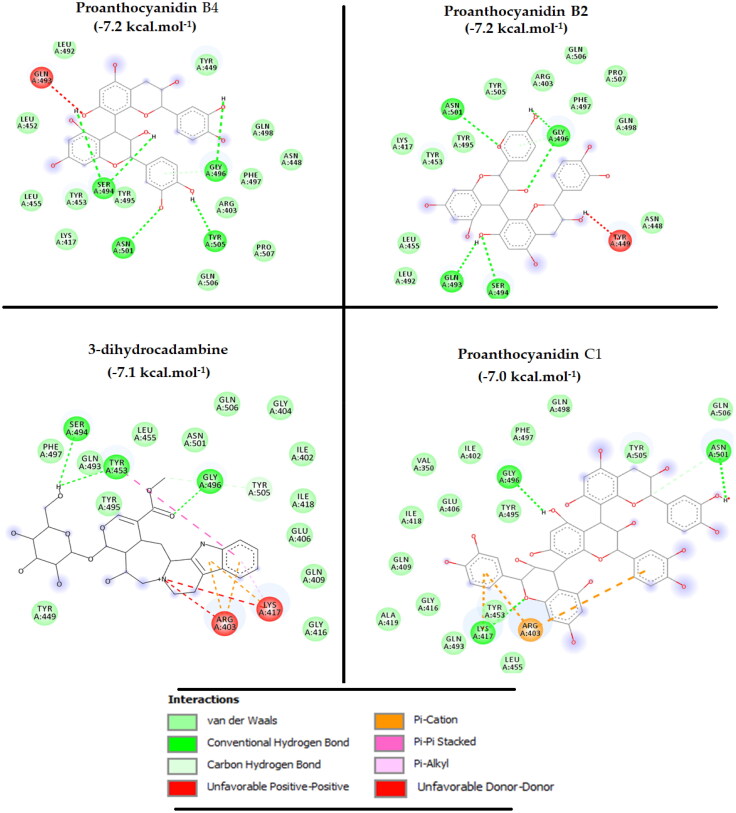
Notably, all main constituents of U. Tomentosa had at least two interactions with those key aminoacids for SARS-CoV-2 RBD binding to human ACE-2, through H-bonds, π-contacts or hydrophobic contacts, making this herb a promising treatment that may be used in the early stages of the COVID-19 infection. Among them, four exhibited high potential to bind RBD: 3-dihydrocadambine (in brown), Proanthocyanidin B4 (in purple blue), Proanthocyanidin B2 (in light blue) and Proanthocyanidin C1 (in hot pink) (Figure 5), which had the highest docking score of –7.1, –7.2, –7.2 and –7.0 kcal mol−1, respectively, that would be comparable to that reported for the potent inhibitor HepOS of –7.3 kcal mol−1. As such, in searching those critical contacts that could blocks RBD–ACE-2 interaction, an exhaustive analysis has been undertaken to the docking results for those components as mentioned in Figure 6.
Figure 6.
Superposition of the best conformation of the all constituents of U. tomentosa and positive references alongside the SARS-CoV-2 RBD binding domain (PDB: 6VYB). Critical aminoacids are represented in cyan and positive reference (HepOS) in blue marine.
As shown in Figure 7, our findings revealed that the most docking active molecules complexed with the SARS-CoV-2 RBD had an interaction fingerprint involving seven critical residues implied in the SARS-CoV-2 spike attachment to the human receptor ACE-2: TYR453, GLY496, LYS417, ASN501, TYR505, GLN498 and ASN501. Thus, 3-dihydrocadambine displays two strong H-bond through binding between sugar and acetyl moiety and key TYR453 and GLY446 residues, respectively. Notably, TYR453 also established one π–π stacking interaction with the fused aromatic ring in the β-carboline moiety. In addition, critical residues LYS417 and ARG403 from RBD was involved in the binding event by forming three π-cation contacts with 3-dihydrocadambine, while hydrophobic interactions formed with ASN501, TYR449, GLN493, TYR505 residues clearly favored its affinity for SARS-CoV-2 spike protein.
Figure 7.
2D interaction mode plots between the most active ligands inside ACE-2 active site of RBD. Interactions between each component and residues of SARS-CoV-2 RBD are indicated by the dashed lines.
However, a visual inspection to the 2D-protein–ligand interaction plot of 3-dihydrocadambine shows that the protonated nitrogen formed two unfavorable positive–positive interactions (represented in red dotted lines) that could have great significance in the stability of protein–ligand complex.
Proanthocyanidin B2 was well-fitted into the functional domain RBD and its hydroxyl groups formed four hydrogen bonding and four hydrophobic contacts with critical residues for binding to ACE-2, including GLY496, ASN501, GLN493 and TYR505, TYR453, LYS417, GLN498, respectively. One further H-bond with SER494 and one hydroxyl group were predicted by docking when Proanthocyanidin B2 binds to SARS-CoV-2 RBD. Finally, Proanthocyanidin C1 was able to bind SARS-COV-2 RBD through three strong H-bonds with those critical residues (GLY496, LYS417, ASN501) crucial to association with human ACE-2. Also, further van der Waals interactions were predicted to form between Proanthocyanidin C1 and crucial GLN498, TYR453, GLN493, TYR505, GLN498 residues that may be stabilizing the binding event to the RBD domain.
MD simulation studies
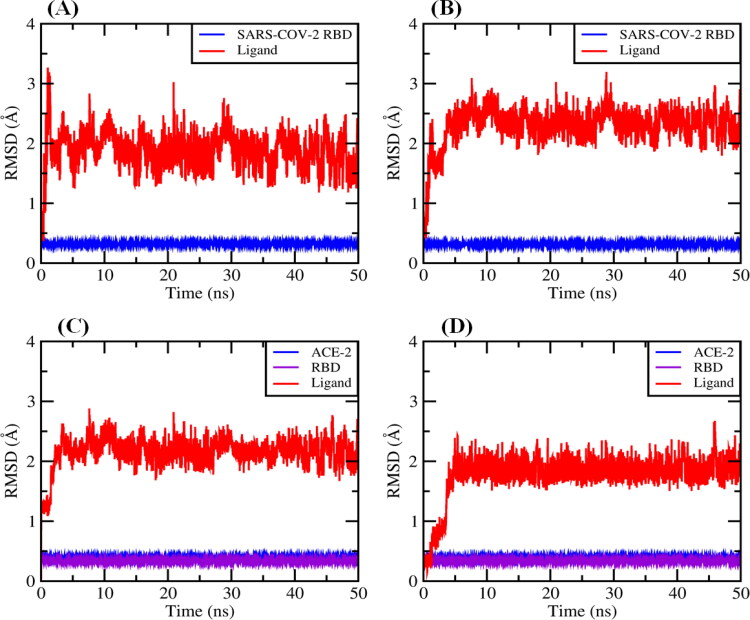
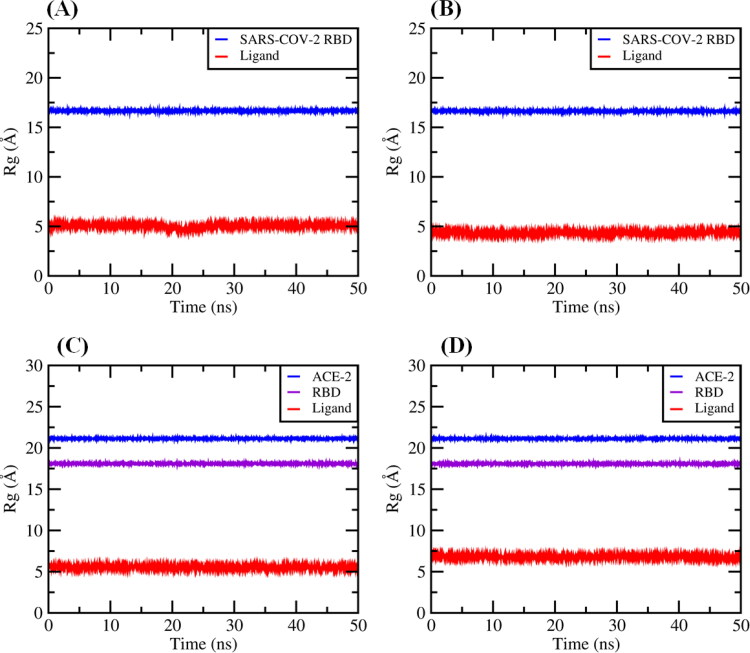
With higher confidence on the viability of our docking predictions of the best compound with highest binding affinity, such as Proanthocyanidin C1 (–8.6 kcal mol−1) and QAG-2 (–8.2 kcal mol−1) into the RBD/ACE-2 interface and 3-dihydrocadambine (–7.1 kcal mol−1) and Proanthocyanidin B2 (–7.2 kcal mol−1) within SARS-CoV-2 RBD binding domain, we further evaluated the stability of the docked complexes throughout MD simulations for 50.0 ns. To accomplish this aim, we first calculated the root mean square deviation (RMSD) for ligands for 50 ns of MD simulation at real natural conditions into the selected binding pocket. The MD simulation results (Figure 8(A–D)) showed that the RMSD of the systems reached equilibrium after around ≈5 ns of simulation time. In general, after equilibration, small fluctuations in the RMSD were observed, suggesting substantial stability for all complexes during the simulations, which fall within the ideal range around 2 Å (smaller RMSD values indicate higher stability of the simulation) (Gohlke et al., 2000; Kramer et al., 1999).
Figure 8.
Backbone RMSD values of (A) 3-dihydrocadambine within SARS-CoV-2 RBD active site (red) and protein without ligand (blue). B: Proanthocyanidin B2 within SARS-CoV-2 RBD binding site (red) and protein without ligand (blue). C: Proanthocyanidin C1 at RBD/ACE-2 interface and RBD/ACE-2 complex without ligand (blue). D: QAG-2 into RBD/ACE-2 interface (red) and RBD/ACE-2 complex without ligand (blue).
A rigorous exploration of the RMSD values for ligand–SARS-CoV-2 RBD complexes shows that structures of 3-dihydrocadambine and Proanthocyanidin B2 display good signals of stability during 50 ns of MD simulation with RMSDs values of 1.866 ± 0.300 and 2.233 ± 0.304 Å, respectively (Figure 8(A,B)). Importantly, similar behavior had the docked complex composed of Proanthocyanidin C1 and QAG-2 into the RBD/ACE-2 interface, which showed remarkable stability throughout the simulation time period at RMSD values of 2.151 ± 0.237 and 1.775 ± 0.361, respectively (Figure 8(C,D)). A closer look at RMSD plot for QAG-2 into the RBD/ACE-2 interface revealed that ligand gradually stabilized after 5 ns, which is an indication of its higher conformational flexibility within the interface between RBD and ACE-2 proteins. Taken together, these findings suggest binding stability of ligands toward the active domains of the SARS-CoV-2 RBD and RBD/ACE-2 viral targets.
The radius of gyration (Rg) represents the compactness of the protein structure and conformational stability of the whole systems (i.e. protein–ligand complexes). If the radius of gyration remained relatively constant, the complex was considered to be stably folded; otherwise, it was considered to be unfolded. In this scenario, radius of gyration values was calculated in order to observe the conformational alterations and dynamic stability of each viral protein within the 50 ns simulation time.
Figure 9(A–D) illustrates Rg values for the protein and ligand in the complexes, respectively. As shown in Figure 9(A–D), calculated Rg values for ligands into the SARS-CoV-2 RBD protein for 3-dihydrocadambine (5.065 ± 0.144 Å), Proanthocyanidin B2 (4.356 ± 0.087 Å) and inside RBD/ACE-2 binding interface for Proanthocyanidin C1 (5.542 ± 0.095 Å) and QAG-2 (6.818 ± 0.083 Å) remained relatively constant during the simulation, therefore each protein–ligand complex could be considered to be stably folded. In addition, although RMSD values would show that the ligands undergo a significant shift within the active domain, estimated Rg values suggest that overall shape of the protein was stable upon binding of the ligand during the 50-ns MD simulation.
Figure 9.
Radius of gyration (Rg) graphs: (A) for 3-dihydrocadambine into the binding cavity (red) and SARS-CoV-2 spike protein without ligand (blue). B: For Proanthocyanidin B2 onto active site (red) and SARS-CoV-2 spike protein without ligand (blue). C: For Proanthocyanidin C1 within binding cleft and RBD/ACE-2 complex without ligand (blue and violet). D: for QAG-2 into the binding site (red) and RBD/ACE-2 complex without ligand (blue and violet).
Finally, these observations are also supported by 2D-binding interactions plots and 3D representation of the selected ligands into the binding pocket, respectively (see Supporting information Figures S1–S4). In addition, to show the conformational changes of the ligands into the viral proteins active site along the first 50 ns window MD simulation (Figure 9), these plots revealed that after MD simulations the key protein–ligand interactions initially shown by the docking results were maintained and the 3-dihydrocadambine and Proanthocyanidin B2 within SARS-CoV-2 RBD, as well as Proanthocyanidin C1 at RBD/ACE-2 remained stable in the binding pocket compared to the initial docking pose. Thus, crucial binding ligand interactions with LYS417, ARG403, TYR453, GLN493, ASN501, TYR505, SER494, TYR449, TYR495 PHE497 and GLY496 were maintained after 50-ns MD simulation into the binding pocket of SARS-CoV-2 RBD. Similarly, in comparison with the docking results, those key ligand interactions with amino acid residues identified as essential for maintaining RBD-ACE-2 stability are also present after MD simulations. Besides, 3D representations of the selected ligands into each binding pocket were used with the aim to compare the best conformation poses from MD simulation and docking, hence we plotted the superposition of the docked complex 3D-structures before and after 50-ns MD simulation into the active cavity (see Supporting information Figures S5–S8). In general, there is no significant difference between the structures extracted after 50-ns MD simulation and the docking pose of ligands, only slight translational and rotational motions were observed.
The obtained MD simulation results suggest (1) the conformation of the binding pocket and ligands were stable during the MD simulations, (2) ligands do not leave the binding pocket while running MD simulation and (3) active pocket in both selected viral targets favored ligands binding, suggesting not only the rationality and validity of our docking studies, but also proposes that many of these constituents of U. tomentosa could act as a dual inhibitors of the SARS-CoV-2 spike protein and RBD/ACE-2 complex, which are mostly responsible for the attachment and internalization of the novel coronavirus in the human host.
In the final stage, selected ligands were redocked into the SARS-CoV-2 RBD (3-dihydrocadambine and Proanthocyanidin B2) or inside the RBD/ACE-2 interface (Proanthocyanidin C1 and QAG-2) starting from the mean geometries of the last 3 ns MD simulations trajectories, aiming to obtain the correct binding energies and poses. The vina re-docked results are summarized in Table 2 and the best ligand bound receptor conformations are shown in the Supporting information Figures S9–S12.
Table 2.
Scoring energies and critical interactions for the re-docking of each ligand in its active site.
| Ligand | Vina Docking score (kcal mol–1) | Vina re-docking score (kcal mol–1) | Critical ligand interactions after re-docking process |
|---|---|---|---|
| 3-Dihydrocadambinea | –7.1 | –7.5 | ASN501 (H-bond), GLY496 (three H-bond), TYR453 (H-bond), TYR505 (π-cation), and hydrophobic interactions: PHE497, TYR495, TYR449, GLN493, GLN498, GLU406. |
| Proanthocyanidin B2a | –7.2 | –6.5 | ARG403 (H-bond), SER494 (H-bond), LYS417 (H-bond), ARG403 (π-cation), GLU406 (π-anion) TYR505 (π–π stacked) and hydrophobic interactions: PHE497, TYR453, TIR495, GLN409. |
| Proanthocyanidin C1b | –8.6 | –9.0 | GLN388 (H-bond), ASP30 (two H-bond), TYR505 (H-bond), ASP405 (H-bond), ARG408 (H-bond), ARG403 (H-bond), ALA387 (amide-π stacking interaction) and hydrophobic interactions: HIS34, PHE390, GLU37, LYS353, GLY354, LYS417. |
| QAG-2b | –8.2 | –8.1 | TYR505 (H-bond), GLY354 (H-bond) and hydrophobic interactions: GLU37, GLN388, LYS353 and ASP355. |
Re-docking inside the SARS-CoV-2 RBD active site.
Re-docking at RBD/ACE-2 interface.
Notably, after re-docked process 3-dihydrocadambine displays higher binding energy (–7.5 kcal.mol−1) than the initial predicted docking score (–7.1 kcal.mol−1). 2D and 3D diagrams of protein–ligand interactions from 3D coordinates showed that 3-dihydrocadambine had a significant conformational change in their binding modes in comparison with observed binding poses initially predicted by docking. As can be seen from Table 2, interactions profile into the SARS-CoV-2 RBD binding site was substantially altered, showing contacts with those critical residues, including ASN501 (H-bond), GLY496 (three H-bond), TYR453 (H-bond), TYR505 (π-cation with cationic nitrogen). On the other hand, despite Proanthocyanidin B2 showed a re-docking score significantly lower than the initial docking. However, readers can observe that Proanthocyanidin B2 is well-accommodated inside the RBD binding pocket highlighting key contacts with TYR453, SER494, LYS417, GLN493, ARG403, TYR495, TYR505, GLU406 and PHE497 that are essential for SARS-CoV2 binding to human ACE-2 receptors. In addition, predicted binding poses were compared in Supporting information Figure S10, thus the best binding pose obtained from re-docking studies is considerably different to that retrieved from the initial docking, this finding could be strongly associated with a lower binding energy after re-docking.
Interestingly, when Proanthocyanidin C1 was re-docked using the mean geometry of the last 3 ns MD simulation trajectory, a higher binding score (–9.0 kcal mol−1) than the initial docking was obtained (–8.6 kcal mol−1). As shown in Supporting information Figure S11, Proanthocyanidin C1 fit-well into the RBD/ACE-2 interface interacting with those critical residues at the junction of SARS-CoV-2 to human ACE-2, such us GLU37, HIS34, ASP30, GLN388, TYR505, PHE390, LYS353, GLU354, LYS417. A 3D-comparison between the best initial docking pose and the best re-docking pose into the active site clearly revealed that this binding pose significantly favors Proanthocyanidin C1 binding.
Finally, re-docking calculations into RBD/ACE-2 interface for QAG-2 showed a very similar binding affinity (–8.1 kcal mol−1) to the initial docking prediction (–8.2 kcal mol−1). A close view to 2D and 3D ligand–protein diagrams plot revealed that QAG-2 fit-well inside the binding pocket, and is located closed to β3 and β4 sheets in the ACE-2 protein. Re-docked process confirms that QAG-2 is capable of binding to aminoacids residues that are critical for the recognition of the SARS-CoV-2 by full-length human ACE-2. After the re-dock analysis, we identified that QAG-2 formed two hydrogen bond interactions with key GLY354 and TYR505 residues, respectively. Similarly, ligand was able to bind through van der Waals contacts with three critical residues ASP355, GLN388 and GLU37, which could have an effect on stabilizing the binding event. As can be seen in Supporting information Figure S12, 3D-plots comparison between starting pose docking and docking pose after re-docking for QAG-2 revealed similar binding modes at RBD/ACE-2 interface, only a slight shift was observed.
Taken together, these computational results clearly evidenced that constituents of U. tomentosa are capable of binding to SARS-CoV-2 spike protein through strong interactions with those key aminoacids crucial for the viral attachment to human ACE-2 in stable complexes. Our investigations could be a new strategy for inhibiting the recognition of the SARS-COV-2 RBD by ACE-2, and therefore might interfere with the entry of coronavirus to its host cells. Computational modeling demonstrated that components of this herb may cause ACE-2 and spike protein cleavage, because they would interact with key residues within RBD/ACE-2 interface forming very stable complexes. Hence, we firmly believe that ethanolic extract of cat’s claw may be a novel herbal-based therapeutic option to treat COVID-19 infection because its components may cause disruption of SARS-CoV-2 RBD/ACE-2 complex or could block attachment of SARS-COV-2 to the entry receptor ACE-2.
Calculation of drug-likeness indices and scoring
Calculated drug-likeness profiles play a critical role in assessing the quality of novel antiviral candidates. Early predictions of pharmacokinetic behavior of the promising antiviral compounds based on its structure could help find safer and effective leads for preclinical testing. Here, we calculated and analyzed various drug-likeness indices for the most qualified components of U. tomentosa predicted by docking studies (Table 3). To this purpose, ten pharmacokinetics parameters were calculated as drug-likeness filter for Speciophylline, Uncarine F, Uncaric acid, Cadambine, 3-isodihydrocadambine, 3-dihydrocadambine, Proanthocyanidin B2, Proanthocyanidin C1, Proanthocyanidin B4, Epiafzelechin-4β-8, QAG-1, QAG-2, QAG-4, QAG-5 and QAG-6, respectively. Results obtained revealed the druggability of the selected components from ethanolic extract of cat’s claw, demonstrating their potential as likely orally active antiviral.
Table 3.
Calculated drug-likeness properties for the most qualified cat’s claw components.
| Compound | M.W.a | PSAb | n-Rot bond (0–10) | n-ON (<10)c | n-OHNHd | Log Po/we | Log KHSAf | Caco-2g (nm s–1) | App. MDCK (nm s–1)h | % HIAi | Lipinski rule of five (≤1) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Speciophylline | 368.432 | 82.804 | 1 | 8 | 1 | 1.709 | –0.044 | 307 | 153 | 81 | 0 |
| Uncarine F | 368.432 | 85.271 | 2 | 6 | 1 | 1.826 | –0.016 | 342 | 171 | 83 | 0 |
| Uncaric acid | 488.706 | 82.031 | 1 | 5 | 4 | 4.192 | 0.580 | 135 | 72 | 90 | 0 |
| Cadambine | 544.557 | 158.806 | 8 | 11 | 5 | 0.037 | –0.592 | 27 | 11 | 27 | 1 |
| 3-iDihydrcadambine | 560.600 | 156.433 | 6 | 12 | 6 | –0.707 | –0.839 | 16 | 6 | 6 | 2 |
| 3-Dihydrocadambine | 546.573 | 172.263 | 5 | 12 | 6 | –0.576 | –0.827 | 13 | 5 | 5 | 2 |
| Proanthocyanidin B2 | 578.528 | 209.177 | 3 | 12 | 10 | 0.505 | –0.300 | 1 | 1 | <25 | 2 |
| Proanthocyanidin C1 | 866.784 | 316.351 | 5 | 18 | 15 | 0.217 | –0.336 | 0 | 0 | 0 | 3 |
| Proanthocyanidin B4 | 578.528 | 209.177 | 3 | 12 | 10 | 0.505 | –0.300 | 1 | 1 | <25 | 2 |
| Epiafzelechin-4β-8 | 562.529 | 191.954 | 3 | 11 | 9 | 0.880 | –0.149 | 5 | 1 | 6 | 2 |
| QAG-1 | 957.117 | 312.05 | 10 | 19 | 11 | –0.455 | –1.266 | 1 | 0 | 0 | 3 |
| QAG-2 | 957.117 | 263.06 | 10 | 19 | 11 | –0.455 | –1.266 | 1 | 0 | 0 | 3 |
| QAG-4 | 780.948 | 204.447 | 7 | 14 | 8 | 1.491 | –0.443 | 5 | 2 | 10 | 3 |
| QAG-5 | 796.948 | 226.081 | 8 | 15 | 9 | 0.434 | –0.924 | 2 | 0 | 0 | 3 |
| QAG-6 | 780.948 | 204.507 | 7 | 14 | 8 | 1.220 | –0.677 | 6 | 2 | 9 | 3 |
| HepOSj | 2291.77 | 1373.67 | 46 | 80 | 12 | <0.0 | – | – | – | – | 5 |
Molecular weight of the hybrid (150–500).
Polar surface area (PSA) (7.0–200 Å2).
n-ON number of hydrogen bond acceptors <10.
n-OHNH number of hydrogens bonds donors ≤5.
Octanol water partition coefficient (log Po/w) (–2.0 to 5.0).
Binding-serum albumin (KHSA) (–1.5 to 1.5).
Human intestinal permeation (<25 poor, >500 great).
Madin–Darby canine kidney (MDCK) cells permeation.
Human intestinal absorption (% HIA) (>80% is high, <25% is poor).
Heparin octasaccharide used as positive reference in this work.
Despite major components had more than two violations to the rule of 5 and druggability, predicted values for Speciophylline, Uncarine F, Uncaric acid and Cadambine displayed favorable physiochemical profiles compared to positive reference HepOS, which clearly displayed important violations in ROF-5. The discussion focused on these four active compounds showed that according to Lipinski rule of five (ROF-5) (no more than one violation is acceptable) (Lipinski et al., 2012), compounds could be used as orally dosed drugs in humans. The predicted human intestinal absorption (% HIA) for Speciophylline, Uncarine F and Uncaric acid revealed greater HIA value in the range between 81% and 90%, while Cadambine having relatively low values of 27% may be acceptable within 95% of marketed drugs. Note that, greater HIA values denote that these compounds could be absorbed throughout the intestinal segments upon oral administration. On the other hand, we calculated the most important physicochemical property to correlated passive molecular transport through membranes and drug–membrane interactions, such as polar surface area (PSA) (Ertl et al., 2000). Predicted PSA showed favorable values for compounds ranging from 82 to 158 Å2, indicating they would penetrate more efficiently through the infected host cells. In addition, the partition coefficient between n-octanol and water (log Po/w) was calculated in order to explore the ability of constituents to pass through lipid bilayers (Veber et al., 2002). Notably, Speciophylline, Uncarine F and Uncaric acid presented values within ideal range for approved-drugs (ranging from 1.7 to 4.20). Binding to serum albumin (expressed as log KHSA) is the most important parameter for distribution and transport of antiviral drugs in the systemic circulation (Colmenarejo, 2003; Zhivkova, 2015). Early prediction of this parameter reduces the amount of wasted time and resources for drug development candidates in the antiviral therapy and management. We found that ligands fit well within the recommended values range (ranging from −1.5 to 1.5), showing log KHSA numbers between –0.592 and 0.58. Finally, we also predicted the passive transmembrane permeation using Caco-2 cell monolayers or MDCK cells as models. Currently, both models are used as a simplified in vitro model for intestinal absorption in drug development (Broccatelli et al., 2016; Pham-The et al., 2018; Press & Di Grandi, 2008). Our results showed that in comparison with reference drugs, Speciophylline, Uncarine F and Uncaric acid have 72–342 nm/s. From such observations, these compounds displayed great values of human intestinal absorption (% HIA) above 81%, very similar to the reference drugs values (above 96%).
Given the aforementioned results, we believe at least four components of cat’s claw here reported may provide favorable characteristics as the drug like, hence U. tomentosa may constitute itself a promissory option to fight against COVID-19 infection.
By integrating in silico approaches, this article evidenced that several components of U. tomentosa could act disrupting the association of SARS-CoV-2 spike protein with the human ACE-2 receptor or by blocking the RBD–ACE-2 interaction during COVID-19 virulence. Further, we identified that various constituents of cat’s claw bears optimal pharmacokinetics properties to be used orally as a potential antiviral response. Therefore, we believe that ethanolic extract of U. tomentosa should be taken into consideration as a rapid response to the COVID-19 during the early stages of infection.
Conclusion
COVID-19 outbreak that emerged from Wuhan, China has acquired pandemic status and severe acute respiratory syndrome requires the attention of academics to discover the possible safe and effective drug to ameliorate its effects worldwide. In the present study, 26 constituents of U. tomentosa were docked on the binding interface of the RBD–ACE-2 and inside SARS-CoV-2 RBD spike protein of novel corona virus.
It was observed that the components of U. tomentosa such as Proanthocyanidin C1, QAG-2, 3-isodihydrocadambine, Uncarine F and Uncaric acid had a good predicted binding affinity for interface of the RBD–ACE-2 as compared to the sulfated heparin octasaccharide (HepOS). Likewise, 3-dihydrocadambine, Proanthocyanidin B4, Proanthocyanidin B2 and Proanthocyanidin C1 had the highest docking score on SARS-CoV-2 spike glycoprotein in their open state, whereas MD simulations at 50 ns demonstrated both the feasibility of the binding free energy predicted by docking protocols and the stability of the docked protein–ligand complexes.
Virtual prediction ADME revealed that Speciophylline, Uncarine F and Uncaric acid presented values of druggability according to Lipinski rule, demonstrating their potential bioavailability as likely orally active antiviral. Based on our findings and its ancestral use in the traditional medicine from South American countries, U. tomentosa can be performed as an herbal supplement with the safety and efficacy parameters at both preclinical and clinical stages to evaluate its effectiveness in the treatment of novel coronavirus disease (COVID-19). Furthermore, all components found in U. tomentosa could work in synergism by different mechanisms to combat the spread of SARS-CoV-2.
Supplementary Material
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Abraham, M. J., Murtola, T., Schulz, R., Páll, S., Smith, J. C., Hess, B., & Lindah, E. (2015). Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX, 1–2, 19–25. 10.1016/j.softx.2015.06.001 [DOI] [Google Scholar]
- Akram, M., Tahir, I. M., Shah, S. M. A., Mahmood, Z., Altaf, A., Ahmad, K., Munir, N., Daniyal, M., Nasir, S., & Mehboob, H. (2018). Antiviral potential of medicinal plants against HIV, HSV, influenza, hepatitis, and coxsackievirus: A systematic review. Phytotherapy Research, 32(5), 811–822. 10.1002/ptr.6024 [DOI] [PubMed] [Google Scholar]
- Aquino, R., De Simone, F., Vincieri, F. F., Pizza, C., & Gaćs-Baitz, E. (1990). New polyhydroxylated triterpenes from Uncaria tomentosa. Journal of Natural Products, 53(3), 559–564. 10.1021/np50069a004 [DOI] [PubMed] [Google Scholar]
- Aquino, R., De Tommasi, N., De Simone, F., & Pizza, C. (1997). Triterpenes and quinovic acid glycosides from Uncaria tomentosa. Phytochemistry, 45(5), 1035–1040. 10.1016/S0031-9422(96)00716-9 [DOI] [Google Scholar]
- Araujo, L. C. C., Feitosa, K. B., Murata, G. M., Furigo, I. C., Teixeira, S. A., Lucena, C. F., Ribeiro, L. M., Muscará, M. N., Costa, S. K. P., Donato, J., Bordin, S., Curi, R., & Carvalho, C. R. O. (2018). Uncaria tomentosa improves insulin sensitivity and inflammation in experimental NAFLD. Scientific Reports, 8(1), 11013. 10.1038/s41598-018-29044-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batiha, G. E.-S., Magdy Beshbishy, A., Wasef, L., Elewa, Y. H. A., Abd El-Hack, M. E., Taha, A. E., Al-Sagheer, A. A., Devkota, H. P., & Tufarelli, V. (2020). Uncaria tomentosa (Willd. ex Schult.) DC.: A review on chemical constituents and biological activities. Applied Sciences, 10(8), 2668. 10.3390/app10082668 [DOI] [Google Scholar]
- Bayly, C. I., Cieplak, P., Cornell, W. D., & Kollman, P. A. (1993). A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: The RESP model. Journal of Physical Chemistry, 97(40), 10269-10280. 10.1021/j100142a004 [DOI] [Google Scholar]
- Cornell, W. D., Cieplak, P., Bayly, C. I., Gould, I. R., Merz, K. M., Ferguson, D. M., Spellmeyer, D. C., Fox, T., Caldwell, J. W., & Kollman, P. A. (1995). A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. Journal of the American Chemical Society, 117(19), 5179–5197. 10.1021/ja00124a002 [DOI] [Google Scholar]
- Broccatelli, F., Salphati, L., Plise, E., Cheong, J., Gobbi, A., Lee, M. L., & Aliagas, I. (2016). Predicting passive permeability of drug-like molecules from chemical structure: Where are we? Molecular Pharmaceutics, 13(12), 4199–4208. 10.1021/acs.molpharmaceut.6b00836 [DOI] [PubMed] [Google Scholar]
- Caon, T., Kaiser, S., Feltrin, C., de Carvalho, A., Sincero, T. C. M., Ortega, G. G., & Simões, C. M. O. (2014). Antimutagenic and antiherpetic activities of different preparations from Uncaria tomentosa (cat’s claw). Food and Chemical Toxicology, 66, 30–35. 10.1016/j.fct.2014.01.013 [DOI] [PubMed] [Google Scholar]
- ChemAxon. (2016). ChemAxon – Software solutions and services for chemistry and biology. MarvinSketch, Version 16.10.31. ChemAxon. [Google Scholar]
- Colmenarejo, G. (2003). In silico prediction of drug-binding strengths to human serum albumin. Medicinal Research Reviews, 23(3), 275–301. 10.1002/med.10039 [DOI] [PubMed] [Google Scholar]
- Ertl, P., Rohde, B., & Selzer, P. (2000). Fast calculation of molecular polar surface area as a sum of fragment-based contributions and its application to the prediction of drug transport properties. Journal of Medicinal Chemistry, 43(20), 3714–3717. 10.1021/jm000942e [DOI] [PubMed] [Google Scholar]
- Foresman, J. B., Head-Gordon, M., Pople, J. A., & Frisch, M. J. (1992). Toward a systematic molecular orbital theory for excited states. Journal of Physical Chemistry, 96(1), 135–149. 10.1021/j100180a030 [DOI] [Google Scholar]
- Glendening, E. D., Landis, C. R., & Weinhold, F. (2012). Natural bond orbital methods. WIREs Computational Molecular Science, 2(1), 1–42. 10.1002/wcms.51 [DOI] [Google Scholar]
- Gohlke, H., Hendlich, M., & Klebe, G. (2000). Knowledge-based scoring function to predict protein-ligand interactions. Journal of Molecular Biology, 295(2), 337–356. 10.1006/jmbi.1999.3371 [DOI] [PubMed] [Google Scholar]
- Gonzales, G. F., Aguilar, J., & Villar, M. (2010). The world summit of harmonization on traditional, alternative and complementary medicine(TACM) in Lima, Peru. Evidence-Based Complementary and Alternative Medicine, 7(2), 271–275. 10.1093/ecam/nen042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammad, S., Bouaziz-Terrachet, S., Meghnem, R., & Meziane, D. (2020). Pharmacophore development, drug-likeness analysis, molecular docking, and molecular dynamics simulations for identification of new CK2 inhibitors. Journal of Molecular Modeling, 26(6), 160. 10.1007/s00894-020-04408-2 [DOI] [PubMed] [Google Scholar]
- Han, Y., & Král, P. (2020). Computational design of ACE2-based peptide inhibitors of SARS-CoV-2. ACS Nano, 14(4), 5143–5147. 10.1021/acsnano.0c02857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich, M. (2002). Ethnomedicine and drug discovery. Journal of Ethnopharmacology, 82(2–3), 244. 10.1016/s0378-8741(02)00173-3 [DOI] [Google Scholar]
- Huang, X.,, Pearce, R., &, Zhang, Y. (2020). De novo design of protein peptides to block association of the SARS-CoV-2 spike protein with human ACE2. Aging, 12(12), 11263–11276. https://doi.org/ 10.18632/aging.103416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamwal, S.,, Gautam, A.,, Elsworth, J.,, Kumar, M.,, Chawla, R., &, Kumar, P. (2020). An updated insight into the molecular pathogenesis, secondary complications and potential therapeutics of COVID-19 pandemic. Life Sciences, 257, 118105. .10.1016/j.lfs.2020.118105 32687917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keplinger, K., Laus, G., Wurm, M., Dierich, M. P., & Teppner, H. (1998). Uncaria tomentosa (Willd.) DC. – Ethnomedicinal use and new pharmacological, toxicological and botanical results. Journal of Ethnopharmacology, 64(1), 23–34. 10.1016/S0378-8741(98)00096-8 [DOI] [PubMed] [Google Scholar]
- Kitajima, M., Hashimoto, K. I., Yokoya, M., Takayama, H., & Aimi, N. (2000). Two new 19-hydroxyursolic acid-type triterpenes from Peruvian “Una de Gato” (Uncaria tomentosa). Tetrahedron, 56(4), 547–552. 10.1016/S0040-4020(99)01048-0 [DOI] [Google Scholar]
- Kramer, B., Rarey, M., & Lengauer, T. (1999). Evaluation of the FlexX incremental construction algorithm for protein–ligand docking. Proteins: Structure, Function, and Genetics, 37(2), 228–241. [DOI] [PubMed] [Google Scholar]
- Kuraś, M., Pilarski, R., Nowakowska, J., Zobel, A., Brzost, K., Antosiewicz, J., & Gulewicz, K. (2009). Effect of alkaloid-free and alkaloid-rich preparations from Uncaria tomentosa bark on mitotic activity and chromosome morphology evaluated by Allium Test. Journal of Ethnopharmacology, 121(1), 140–147. 10.1016/j.jep.2008.10.023 [DOI] [PubMed] [Google Scholar]
- Kwon, P. S., Oh, H., Kwon, S.-J., Jin, W., Zhang, F., Fraser, K., Hong, J. J., Linhardt, R. J., & Dordick, J. S. (2020). Sulfated polysaccharides effectively inhibit SARS-CoV-2 in vitro. Cell Discovery, 6(1), 50. 10.1038/s41421-020-00192-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan, J., Ge, J., Yu, J., Shan, S., Zhou, H., Fan, S., Zhang, Q., Shi, X., Wang, Q., Zhang, L., & Wang, X. (2020). Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature, 581(7807), 215–220. 10.1038/s41586-020-2180-5 [DOI] [PubMed] [Google Scholar]
- Laus, G., Brössner, D., & Keplinger, K. (1997). Alkaloids of peruvian Uncaria tomentosa. Phytochemistry, 45(4), 855–860. 10.1016/S0031-9422(97)00061-7 [DOI] [Google Scholar]
- Lenzi, R. M., Campestrini, L. H., Okumura, L. M., Bertol, G., Kaiser, S., Ortega, G. G., Gomes, E. M., Bovo, F., Zawadzki-Baggio, S. F., Stevan-Hancke, F. R., & Maurer, J. B. B. (2013). Effects of aqueous fractions of Uncaria tomentosa (Willd.) D.C. on macrophage modulatory activities. Food Research International, 53(2), 767–779. 10.1016/j.foodres.2013.02.042 [DOI] [Google Scholar]
- Li, S.-R.,, Tang, Z.-J.,, Li, Z.-H., &, Liu, X. (2020). Searching therapeutic strategy of new coronavirus pneumonia from angiotensin-converting enzyme 2: the target of COVID-19 and SARS-CoV. European Journal of Clinical Microbiology & Infectious Diseases, 39(6), 1021–1026. https://doi.org/ 10.1007/s10096-020-03883-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima-Junior, R. S., Da Silva Mello, C., Kubelka, C. F., Siani, A. C., & Valente, L. M. M. (2013). Uncaria tomentosa alkaloidal fraction reduces aracellular permeability, il-8 and ns1 production on human microvascular endothelial cells infected with dengue virus. Natural Product Communications, 8(11), 1547–50. 10.1177/1934578X1300801112 [DOI] [PubMed] [Google Scholar]
- Lipinski, C. A., Lombardo, F., Dominy, B. W., & Feeney, P. J. (2012). Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Advanced Drug Delivery Reviews, 64, 4–17. 10.1016/j.addr.2012.09.019 [DOI] [PubMed] [Google Scholar]
- Lock, O., Perez, E., Villar, M., Flores, D., & Rojas, R. (2016). Bioactive compounds from plants used in peruvian traditional medicine. Natural Product Communications, 11(3), 315–37. [PubMed] [Google Scholar]
- Montoro, P., Carbone, V., de Dioz Zuniga Quiroz, J., De Simone, F., & Pizza, C. (2004). Identification and quantification of components in extracts of Uncaria tomentosa by HPLC-ES/MS. Phytochemical Analysis, 15(1), 55–64. 10.1002/pca.740 [DOI] [PubMed] [Google Scholar]
- Morris, G. M.,, Huey, R.,, Lindstrom, W.,, Sanner, M. F.,, Belew, R. K.,, Goodsell, D. S., &, Olson, A. J. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of Computational Chemistry, 30(16), 2785–2791. https://doi.org/ 10.1002/jcc.21256 19399780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, G. M., Goodsell, D. S., Halliday, R. S., Huey, R., Hart, W. E., Belew, R. K., & Olson, A. J. (1998). Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. Journal of Computational Chemistry, 19(14), 1639–1662. [Google Scholar]
- Navarro, M., Arnaez, E., Moreira, I., Hurtado, A., Monge, D., & Monagas, M. (2019). Polyphenolic composition and antioxidant activity of Uncaria tomentosa commercial bark products. Antioxidants, 8(9), 339. 10.3390/antiox8090339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro, M., Zamora, W., Quesada, S., Azofeifa, G., Alvarado, D., & Monagas, M. (2017). Fractioning of proanthocyanidins of Uncaria tomentosa. Composition and structure–bioactivity Relationship. Antioxidants, 6(3), 60. 10.3390/antiox6030060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Hoyos, M., Lebrón-Aguilar, R., Quintanilla-López, J., Cueva, C., Hevia, D., Quesada, S., Azofeifa, G., Moreno-Arribas, M., Monagas, M., & Bartolomé, B. (2017). Proanthocyanidin characterization and bioactivity of extracts from different parts of Uncaria tomentosa L. (cat’s claw. Antioxidants, 6(1), 12. 10.3390/antiox6010012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa, M., & Nishizawa, K. (2010). Molecular dynamics simulation analyses of viral fusion peptides in membranes prone to phase transition: Effects on membrane curvature, phase behavior and lipid–water interface destabilization. Journal of Biophysical Chemistry, 01(01), 19–32. 10.4236/jbpc.2010.11003 [DOI] [Google Scholar]
- Ostrakhovich, E. A., Mikhal'chik, E. V., Getmanskaya, N. V., & Durnev, A. D. (1997). Antioxidant activity of the extract from Uncaria Tomentosa. Pharmaceutical Chemistry Journal, 31(6), 326–329. 10.1007/BF02464127 [DOI] [Google Scholar]
- Parrinello, M., & Rahman, A. (1981). Polymorphic transitions in single crystals: A new molecular dynamics method. Journal of Applied Physics, 52(12), 7182–7190. 10.1063/1.328693 [DOI] [Google Scholar]
- Pavei, C., Kaiser, S., Verza, S. G., Borre, G. L., & Ortega, G. G. (2012). HPLC-PDA method for quinovic acid glycosides assay in cat’s claw (Uncaria tomentosa) associated with UPLC/Q-TOF-MS analysis. Journal of Pharmaceutical and Biomedical Analysis, 62, 250–257. 10.1016/j.jpba.2011.12.031 [DOI] [PubMed] [Google Scholar]
- Peñaloza, E. M. C., Kaiser, S., De Resende, P. E., Pittol, V., Carvalho, Â. R., & Ortega, G. G. (2015). Chemical composition variability in the Uncaria tomentosa (cat’s claw) wild population. Quimica Nova, 38(3), 378–386. 10.5935/0100-4042.20150007. [DOI] [Google Scholar]
- Pham-The, H., Cabrera-Pérez, M. Á., Nam, N.-H., Castillo-Garit, J. A., Rasulev, B., Le-Thi-Thu, H., & Casañola-Martin, G. M. (2018). In silico assessment of ADME properties: Advances in Caco-2 cell monolayer permeability modeling. Current Topics in Medicinal Chemistry, 18(26), 2209–2229. 10.2174/1568026619666181130140350 [DOI] [PubMed] [Google Scholar]
- Prajapat, M., Sarma, P., Shekhar, N., Avti, P., Sinha, S., Kaur, H., Kumar, S., Bhattacharyya, A., Kumar, H., Bansal, S., & Medhi, B. (2020). Drug targets for corona virus: A systematic review. Indian Journal of Pharmacology, 52(1), 56. 10.4103/ijp.IJP_115_20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Press, B., & Di Grandi, D. (2008). Permeability for intestinal absorption: Caco-2 assay and related issues. Current Drug Metabolism, 9(9), 893–900. 10.2174/138920008786485119 [DOI] [PubMed] [Google Scholar]
- Reis, S. R. I. N., Valente, L. M. M., Sampaio, A. L., Siani, A. C., Gandini, M., Azeredo, E. L., D'Avila, L. A., Mazzei, J. L., Henriques, M. d G. M., & Kubelka, C. F. (2008). Immunomodulating and antiviral activities of Uncaria tomentosa on human monocytes infected with dengue Virus-2. International Immunopharmacology, 8(3), 468–476. 10.1016/j.intimp.2007.11.010 [DOI] [PubMed] [Google Scholar]
- Roothaan, C. C. J. (1951). New developments in molecular orbital theory. Reviews of Modern Physics, 23(2), 69–89. 10.1103/RevModPhys.23.69 [DOI] [Google Scholar]
- Sandoval, M., Charbonnet, R. M., Okuhama, N. N., Roberts, J., Krenova, Z., Trentacosti, A. M., & Miller, M. J. S. (2000). Cat’s claw inhibits TNFα production and scavenges free radicals: Role in cytoprotection. Free Radical Biology and Medicine, 29(1), 71–78. 10.1016/S0891-5849(00)00327-0 [DOI] [PubMed] [Google Scholar]
- Schmidt, M. W., Baldridge, K. K., Boatz, J. A., Elbert, S. T., Gordon, M. S., Jensen, J. H., Koseki, S., Matsunaga, N., Nguyen, K. A., Su, S., Windus, T. L., Dupuis, M., & Montgomery, J. A. (1993). General atomic and molecular electronic structure system. Journal of Computational Chemistry, 14(11), 1347–1363. 10.1002/jcc.540141112 [DOI] [Google Scholar]
- Singh, U. C., & Kollman, P. A. (1984). An approach to computing electrostatic charges for molecules. Journal of Computational Chemistry, 5(2), 129–145. 10.1002/jcc.540050204 [DOI] [Google Scholar]
- Snow, A. D., Castillo, G. M., Nguyen, B. P., Choi, P. Y., Cummings, J. A., Cam, J., Hu, Q., Lake, T., Pan, W., Kastin, A. J., Kirschner, D. A., Wood, S. G., Rockenstein, E., Masliah, E., Lorimer, S., Tanzi, R. E., & Larsen, L. (2019). The Amazon rain forest plant Uncaria tomentosa (cat’s claw) and its specific proanthocyanidin constituents are potent inhibitors and reducers of both brain plaques and tangles. Scientific Reports, 9(1), 561. 10.1038/s41598-019-38645-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terlizzi, M. E., Occhipinti, A., Luganini, A., Maffei, M. E., & Gribaudo, G. (2016). Inhibition of herpes simplex type 1 and type 2 infections by Oximacro®, a cranberry extract with a high content of A-type proanthocyanidins (PACs-A). Antiviral Research, 132, 154–164. 10.1016/j.antiviral.2016.06.006 [DOI] [PubMed] [Google Scholar]
- Trott, O., & Olson, A. J. (2009). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31(2), 455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veber, D. F., Johnson, S. R., Cheng, H. Y., Smith, B. R., Ward, K. W., & Kopple, K. D. (2002). Molecular properties that influence the oral bioavailability of drug candidates. Journal of Medicinal Chemistry, 45(12), 2615–2623. 10.1021/jm020017n [DOI] [PubMed] [Google Scholar]
- Vera-Reyes, I., Huerta-Heredia, A. A., Ponce-Noyola, T., Cerda-GarcÃ‐A-Rojas, C. M., Trejo-Tapia, G., & Ramos-Valdivia, A. C. (2015). Monoterpenoid indole alkaloids and phenols are required antioxidants in glutathione depleted Uncaria tomentosa root cultures. Frontiers in Environmental Science, 3,27. 10.3389/fenvs.2015.00027 [DOI] [Google Scholar]
- Walls, A. C., Park, Y. J., Tortorici, M. A., Wall, A., McGuire, A. T., & Veesler, D. (2020). Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell, 181(2), 281–292.e6. 10.1016/j.cell.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C.,, Horby, P. W.,, Hayden, F. G., &, Gao, G. F. (2020). A novel coronavirus outbreak of global health concern. The Lancet, 395(10223), 470–473. https://doi.org/ 10.1016/S0140-6736(20)30185-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Wang, W., Kollman, P. A., & Case, D. A. (2006). Automatic atom type and bond type perception in molecular mechanical calculations. Journal of Molecular Graphics & Modelling, 25(2), 247–260. 10.1016/j.jmgm.2005.12.005 [DOI] [PubMed] [Google Scholar]
- World Health Organization. (2018). Annex 1 WHO guidelines on good herbal processing practices for herbal medicines (WHO Technical Report Series, No. 1010). WHO. [Google Scholar]
- World Health Organization. (2020). Coronavirus disease (COVID-19): weekly epidemiological update. World Health Organization. [Google Scholar]
- Wu, J.,, Yuan, X.,, Wang, B.,, Gu, R.,, Li, W.,, Xiang, X.,, Tang, L., &, Sun, H. (2020). Severe Acute Respiratory Syndrome Coronavirus 2: From Gene Structure to Pathogenic Mechanisms and Potential Therapy. Frontiers in Microbiology, 11, 1576. https://doi.org/ 10.3389/fmicb.2020.01576 32719672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X. M., & Tan, R. X. (2019). Interaction between gut microbiota and ethnomedicine constituents. Natural Product Reports, 36(5), 788–809. 10.1039/c8np00041g [DOI] [PubMed] [Google Scholar]
- Yan, R., Zhang, Y., Li, Y., Xia, L., Guo, Y., & Zhou, Q. (2020). Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science (New York, N.Y.), 367(6485), 1444–1448. 10.1126/science.abb2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhivkova, Z. (2015). Studies on drug–human serum albumin binding: The current state of the matter. Current Pharmaceutical Design, 21(14), 1817–1830. 10.2174/1381612821666150302113710 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.