Abstract
Given the highly contagious nature of SARS-CoV-2, it has resulted in an unprecedented number of COVID-19 infected and dead people worldwide. Since there is currently no vaccine available in the market, the identification of potential drugs is urgently needed to control the pandemic. In this study, 92 phytochemicals from medicinal plants growing in the Andean region were screened against SARS-CoV-2 3 C-like protease (3CLpro) and RNA-dependent RNA polymerase (RdRp) in their active sites through molecular docking. The cutoff values were set from the lowest docking scores of the FDA-approved drugs that are being used to treat COVID-19 patients (remdesivir, lopinavir, and ritonavir). Compounds with docking scores that were lower than cutoff values were validated by molecular dynamics simulation with GROMACS, using root mean square deviation (RMSD), root mean square fluctuation (RMSF), radius of gyration (Rg), and intermolecular hydrogen bonds (H-bonds). Furthermore, binding free energies were estimated using the MM-PBSA method, and ADMET profiles of potential inhibitors were assessed. Computational analyses revealed that the interaction with hesperidin (theoretical binding energies, ΔGbind = −15.18 kcal/mol to 3CLpro and ΔGbind = −9.46 kcal/mol to RdRp) remained stable in both enzymes, unveiling its remarkable potential as a possible multitarget antiviral agent to treat COVID-19. Importantly, lupinifolin with an estimated binding affinity to 3CLpro higher than hesperidin (ΔGbind = −20.93 kcal/mol) is also a potential inhibitor of the 3CLpro. These two compounds displayed suitable pharmacological and structural properties to be drug candidates, demonstrating to be worthy of further research.
Communicated by Ramaswamy H. Sarma
Keywords: SARS-CoV-2, molecular docking, molecular dynamics, MM-PBSA, ADMET
1. Introduction
Coronaviruses (CoVs) are a group of enveloped, large non-segmented positive-sense RNA viruses able to cause enteric, respiratory, and central nervous diseases in animals, including humans (McIntosh, 1974; Weiss & Navas-Martin, 2005). Whereas most of human CoVs cause mild respiratory infections, the outbreaks of severe acute respiratory syndrome (SARS), Middle East respiratory syndrome (MERS), and coronavirus disease 2019 (COVID-19) which emerged in 2003, 2012, and 2019, respectively, have demonstrated how devastating and life-threatening they can be.
COVID-19 is caused by a new type of transmissible pathogenic SARS-CoV (SARS-CoV-2) - formerly 2019-nCoV - which belongs to the zoonotic CoVs of the genus Betacoronavirus in the family Coronaviridae. Symptoms include fever, dry cough, fatigue, shortness of breath, and in some cases gastrointestinal distress (Guo et al., 2020).
The SARS-CoV-2 genome size of about 30 kb encodes multiple structural and non-structural proteins (Ahmed et al., 2020). The spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins are considered essential to produce structurally complete viral particles (Ahmed et al., 2020; Mangar et al., 2020). The 3 C-like protease (3CLpro), also known as main protease, is one of the two proteases that SARS-CoVs utilize for replication and infection processes (Pillaiyar et al., 2016). Unlike the catalytic tryad found in serine proteases and other cysteine proteases, a catalytic dyad is present in the active site of SARS-CoVs main protease (Anand et al., 2003; Zhang et al., 2020). The RNA-dependent RNA polymerase (RdRp), also named nsp12, is the most highly conserved protein among RNA viruses and its role is to catalyze the viral genome replication and transcription (Jácome et al., 2020). RdRp activity is dependent on magnesium ions and requires the non-structural proteins nsp7 and nsp8 for complete activity (Kirchdoerfer & Ward, 2019). The active site of the SARS-CoV-2 RdRp is formed by conserved polymerase motifs (A-G), where the motif A and motif C contain the divalent-cation-binding amino acid D618, and the catalytic residues 759SDD761, respectively (Gao et al., 2020). Taken together, these enzymes are attractive drug targets to treat these human CoVs.
Potential COVID-19 treatments include ivermectin, a broad-spectrum anti-parasitic agent (Caly et al., 2020); type I interferons, a group of cytokines with a broad spectrum activity against RNA viruses (Mantlo et al., 2020); favipiravir, a viral RdRp inhibitor (Cai et al., 2020); and convalescent plasma. The protease inhibitors lopinavir and ritonavir, used to treat HIV infections, and recommended by the National Health Commission (NHC) of China to treat COVID-19, did not significantly accelerate clinical improvement, reduce mortality, or decrease throat viral RNA detection in a randomized, controlled, open-label, not blinded trial involving hospitalized patients with severe COVID-19 (Cao et al., 2020). However, the work had several limitations and larger studies with greater variety of patients are needed to evaluate the efficacy of this treatment. Remdesivir, a nucleoside analog used to block viral RdRps including filoviruses, paramyxoviruses, pneumoviruses, as well as animal and human CoVs, was used to treat severely ill patients with COVID-19 in a randomized, double-blind, placebo-controlled, multicentre trial, showing a numerically faster time to clinical improvement but not statistically significant (Wang et al., 2020). Using insect cells, it was found to be a direct-acting antiviral inhibiting with the same potency and mechanism of action the RdRp of SARS-CoV-2 and its ortholog in SARS-CoV (Gordon et al., 2020). Remdesivir was approved on May 1, 2020, by the United States Food and Drug Administration (FDA) to treat hospitalized COVID-19 patients. More recently, in a large trial including hospitalized patients critically ill with COVID-19, the steroid dexamethasone reduced deaths by one-third in patients receiving invasive mechanical ventilation and by one-fifth in patients receiving oxygen but were not on ventilators. Importantly, this steroid had not effect on patients with less severe cases those not receiving respiratory support (Horby et al., 2020).
The high diversity and complex molecular structures of natural compounds make them an abundant biological source for drug discovery, even more, considering the limited number of plant species that have been explored for pharmaceutical purposes (Saklani & Kutty, 2014). Flavonoids are naturally occurring compounds that have been proposed as attractive antiviral agents given their broad mechanisms of action including transcription and translation blocking, as well as their general safety and non-cytotoxicity to human cells (Lalani & Poh, 2020; Paduch & Kandefer-Szerszen, 2014). Herbacetin, rhoifolin and pectolinarin were demonstrated to inhibit the protease activity of the SARS-CoV 3CLpro in vitro (Jo et al., 2020). We focused our research to the Andean region considering that it is one of the most biodiverse regions in the world due to the different environmental niches resulting from its great elevational and latitudinal diversity gradient (Anthelme et al., 2014; Mutke et al., 2014; Pennington et al., 2010). The present work was aimed to perform molecular docking, molecular dynamics simulations, binding free energy estimations, and to predict ADMET profiles of natural compounds identified in medicinal plants growing in the Andean region as potential inhibitors of the 3CLpro and RdRp of SARS-CoV-2.
2. Methodology
2.1. Preparation of SARS-CoV-2 protein targets
Two SARS-CoV-2 proteins were selected as targets based on the key role they played in the replication/transcription machinery. The 3 D structures of the 3CLpro (PDB ID: 6LU7) and RdRp (PDB ID: 6M71) were downloaded from the Protein Data Bank. Water molecules and inhibitor were removed from 6LU7 using Discovery Studio Visualizer (2016). Similarly, co-factors nsp7 and nsp8 were removed from 6M71. Missing atoms for residues F70, K73, R74, E83, K98, F101, F102, I114, R365, and D824, in the RdRp file, were modeled using the Swiss-Pdb Viewer v4.1.0 (Guex & Peitsch, 1997). AutoDock Tools v1.5.6 (Sanner, 1999) was used to add polar hydrogens to the structures and convert them in pdbqt format.
2.2. Library of ligands
Ligands used for molecular docking included 92 phytochemicals from 20 medicinal plants growing in the Andean region in South America, as well as some FDA-approved drugs with potential anti-SARS-CoV-2 (darunavir, nelfinavir, paritaprevir, saquinavir, setrobuvir, simeprevir, and sofosbuvir), the aminoquinolines chloroquine and hydroxychloroquine, and cinnamaldehyde as negative controls due to its poor affinity to these SARS-CoV-2 viral targets (Elfiky, 2020). Natural compounds and synthetic drugs were downloaded from PubChem and DrugBank, respectively. Ligands were converted to PDB format using Open Babel (O’Boyle et al., 2011) and later prepared to pdbqt format using AutoDock Tools v1.5.6 (Sanner, 1999).
2.3. Determination of active sites
The active pockets on the target proteins were identified using the CASTp (Computed Atlas of Surface Topography of proteins) server (Tian et al., 2018).
2.4. Molecular docking
Docking analysis was performed using the AutoDock Vina software (Trott & Olson, 2010). Grid boxes of 24 Å x 26 Å x 30 Å centered at x, y, z = −10, 12, 69 and 36 Å x 34 Å x 19 Å centered at x, y, z = 113, 113, 130 were set for 3CLpro and RdRp, respectively. A grid spacing of 1 Å and default exhaustiveness were used. After docking, the structures were examined in Discovery Studio Visualizer (2016).
2.5. Molecular dynamics
The resulting protein-ligand complex structures from molecular docking were used to perform molecular dynamics (MD) simulations in GROMACS 2018 (Van Der Spoel et al., 2005) with MPI support at the Oklahoma State University HPC system. Ligand and protein parameters were generated using CHARMM general force-field (Vanommeslaeghe et al., 2010) and CHARMM36 force-field (Huang & MacKerell, 2013), respectively; the topologies for both 3CLpro and RdRp were generated using the pdb2gmx utility included in GROMACS, while the CGenFF online program was used for the ligands (https://cgenff.umaryland.edu) (Vanommeslaeghe et al., 2012; Vanommeslaeghe & MacKerell, 2012). In order to prepare the system for MD simulations, all protein-ligand and protein with no ligand systems were centered in a rhombic dodecahedron box with a box-system distance of 1.0 nm and solvated with three-point transferable intermolecular potential (TIP3P) water (Bjelkmar et al., 2010).
Charge neutralization was carried out for the 3CLpro system by adding four sodium ions, while six sodium ions were added for the RdRp system. Then, the systems were relaxed through 50000 steps of the steepest descent algorithm for energy minimization calculations at a tolerance value of 1000 kJ/(mol.nm). This was followed by the equilibration with position restraint on the protein and ligand molecules for 0.1 ns using NVT and NPT ensembles, wherein the systems were heated to 300 K using Berendsen thermostat (Berendsen et al., 1984) with a coupling time of 0.1 ps, and the pressure was maintained with a coupling to a reference pressure of 1 bar. For energy minimization, NVT, and NPT relaxation simulation, a smooth force-switch 1.2 nm cutoff was used in short-range interactions, and long-range electrostatics were evaluated using the PME (Particle-Mesh-Ewald) (Darden et al., 1993); additionally, hydrogen-bonds were restrained with the LINCS algorithm(Hess et al., 1997).
Final MD simulations of 200 ns were performed without restraints using an integration time-step of 2 fs, and trajectory snapshots were captured at every 1 ps. Root mean square deviation (RMSD), root mean square fluctuation (RMSF), radius of gyration (Rg), and number of intermolecular hydrogen bonds (H-bonds) were chosen as parameters to analyze MD trajectories using GROMACS utilities and plotted with Xmgrace. To further confirm the stability of protein-ligand complexes, changes in secondary structures during MD simulations were assessed using the Dictionary of Secondary Structure of Proteins (DSSP) algorithm.
2.6. Binding free energy estimation
The Molecular Mechanics Poisson–Boltzmann surface area (MM-PBSA) method (Kollman et al., 2000), implemented in the g_mmpbsa tool (Kumari et al., 2014) of GROMACS, was used to estimate the relative binding free energy of each protein-ligand complex. Briefly, the binding free energy (ΔGbind) of a protein-ligand complex in solvent is calculated as the free energy difference between the complex (Gcomplex) and the summation of the free energy of the protein (Gprotein) and ligand (Gligand):
| (1) |
The analyses were performed for 500 snapshots collected consecutively at an interval of 40 ps from the last 20 ns of MD simulations. The solute dielectric constant () was assigned based on the protein-ligand interfaces by using the scheme: 1 for non-polar residues, 2 for polar not charged residues, and 4 for polar charged (positively and negatively) residues (Ravindranathan et al., 2011; Wang et al., 2019).
2.7. Admet prediction
A total of 19 molecular descriptors were calculated to predict the absorption, distribution, metabolism, elimination, and toxicity (ADMET) profile of the selected potential inhibitors of the 3Clpro and RdRp of SARS-CoV-2, using the Web servers SwissADME (Daina et al., 2017) and pkCSM (Pires et al., 2015) which have been extensively validated with experimental data. The Lipinski's rule of five considering key physicochemical parameters (molecular weight, lipophilicity, polar surface area, hydrogen bonding, and charge) was used to evaluate the drug likeness of the potential inhibitors.
3. Results and discussion
3.1. Molecular docking
Docking scores of FDA-approved protease and RdRp inhibitors which are used to treat COVID-19 patients were calculated in order to set cutoff values. Remdesivir docking scores (-8.2 kcal/mol to 3CLpro and −7.5 kcal/mol to RdRp) were lower than or equal to those found for lopinavir (-7.9 kcal/mol to 3CLpro and −6.9 kcal/mol to RdRp) and ritonavir (-8.2 kcal/mol to 3CLpro and − 7.2 kcal/mol to RdRp). Then, −8.2 kcal/mol to 3CLpro and −7.5 kcal/mol to RdRp were set as cutoff values.
The docking scores to RdRp of all tested FDA-approved drugs which are being proposed as candidates against SARS-CoV-2, excepting sofosbuvir, were lower than remdesivir. Moreover, paritaprevir, setrobuvir, simeprevir, and nelfinavir also showed higher affinity to 3CLpro than ritonavir and remdesivir (Supplementary Table 1). The low affinities of chloroquine and hydroxychloroquine to both enzymes (Supplementary Table 1) indicate that they do not interact effectively with these viral targets. In fact, the mode of action of these aminoquinolines is not related to RdRp or 3CLpro inhibition. CQ impairs the autophagosome fusion with lysosomes (Mauthe et al., 2018), and interferes with the glycosylation of viral or host proteins. More specifically, CQ inhibits the glycosylation of HIV viral particles or SARS-CoV human receptor angiotensin-converting enzyme 2 (ACE2) (Savarino et al., 2004; Vincent et al., 2005)
Table 1.
Theoretical MM-PBSA free energies (kcal/mol) of the stable complexes.
| Complex | Van der Waal energy | Electrostatic energy | Polar solvation energy | SASA energy | Binding energy |
|---|---|---|---|---|---|
| 3CLpro-hesperidin | −38.79 (2.99) | −14.16 (3.23) | 42.23 (5.99) | −4.46 (0.31) | −15.18 (4.82) |
| 3CLpro-lupinifolin | −43.44 (3.26) | −2.78 (2.76) | 30.31 (4.40) | −5.02 (0.28) | −20.93 (3.45) |
| RdRp-hesperidin | −31.96 (3.94) | −19.92 (-8.40) | 47.20 (11.58) | −4.77 (0.54) | −9.46 (7.23) |
Standard deviations are in parentheses.
From the analysis of the 92 phytochemicals, 10 exhibited docking scores lower than the cutoff for 3CLpro (corosolic acid, friedelin, hesperidin, guaijaverin, hyperin, lupinifolin, mangiferin, pinocembrin-7-O-rutinoside, quercitrin, and rutin) while 12 had docking scores lower than the cutoff for RdRp (Supplementary Table 1). These compounds showing such affinities were selected to perform MD simulations in order to evaluate the stability of protein-ligand complexes.
Interestingly, unlike their aglycones, hesperidin (hesperetin-7-O-rutinoside), rutin (quercetin-3-O-rutinoside), and quercitrin (quercetin-3-O-α-rhamnoside) showed significant affinities to both viral targets (Supplementary Table 1). In addition, the quercetin glycosides guaijaverin (quercetin-3-O-α-arabinoside) and hyperin (quercetin-3-O-β-galactoside) revealed significant affinities to 3CLpro. By comparing the protein-ligand interactions, the glycoside moiety was involved in molecular interactions.
3.2. Molecular dynamics
Molecular Dynamics (MD) is a computational approach used to analyze the dynamic behaviour of complex systems where atoms and molecules interact as a function of time. In general, computational methods improve the efficiency in drug discovery, Importantly, unlike general molecular docking methods, MD simulations consider the flexibility of the targets and combined with binding energy calculations a more accurate prediction of potential inhibitors can be developed (Liu et al., 2018). Structural parameters including RMSD, RMSF, Rg, and number of intermolecular H-bonds were used to evaluate the stability, dynamic behaviour, and compactness of protein-drug complexes.
3.2.1. Root mean square deviation
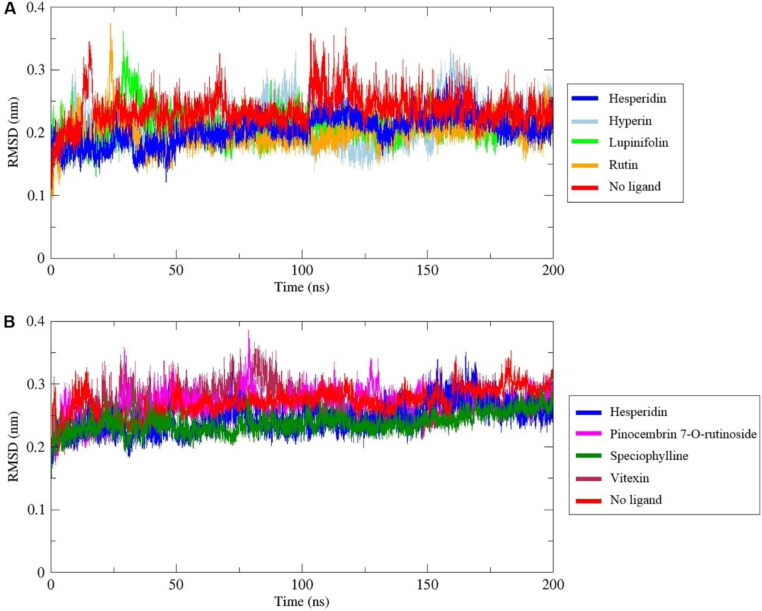
Root mean square deviation (RMSD) of the protein backbone was used to analyze the stability of 3CLpro and RdRp in complex with the selected phytochemicals, where a value lower than that calculated to the free protein indicates a stable complex (Liu et al., 2017). The mean RMSD values for free 3CLpro and 3CLpro in complex with hesperidin, hyperin, lupinifolin, and rutin were 2.37 Å, 2.13 Å, 2.11 Å, 2.04 Å, and 2.00 Å, respectively. Similarly, the mean RMSD for free RdRp and RdRp in complex with hesperidin, pinocembrin-7O-rutinoside, speciophylline, and vitexin were 2.72 Å, 2.44 Å, 2.69 Å, 2.38 Å, and 2.67 Å, respectively. Then, these phytochemicals did not disturb the structural stability of these enzymes (Figure 1). Backbones with mean RMSD higher than free protein are plotted in Figure S1.
Figure 1.
Backbone root mean square deviation (RMSD) of the 3CLpro (A) and RdRp (B) of SARS-CoV-2 in complex with phytochemicals, during 200 ns of the molecular dynamics simulation period. Backbones with mean RMSD lower than free protein are plotted.
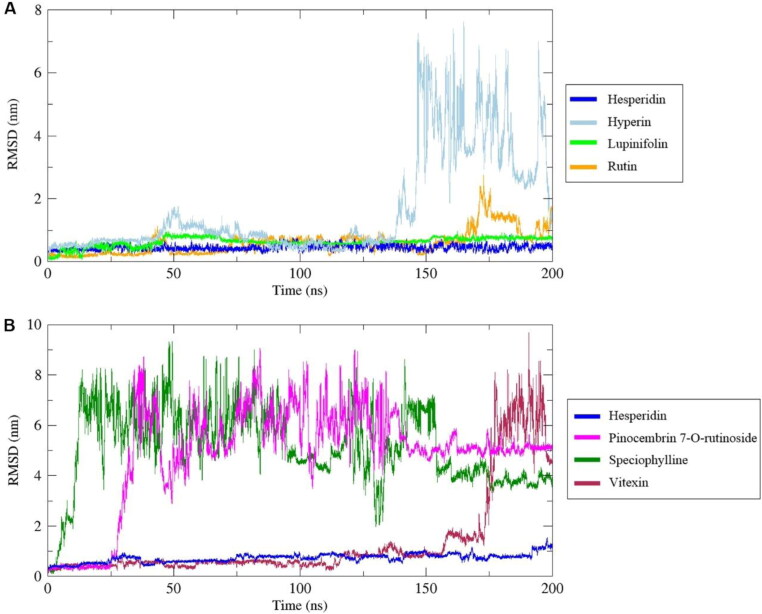
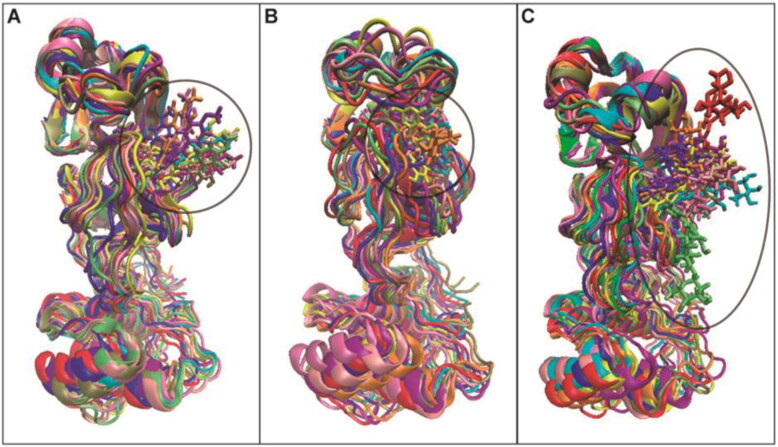
To ensure the binding stability of these compounds in the active site, ligand positional RMSD was calculated. In complex with 3CLpro, high fluctuations were found for hyperin, corosolic acid, friedelin, guaijaverin, mangiferin, pinocembrin-7-O-rutinoside, and quercitrin (Figure 2(A), Figure S2). In contrast, hesperidin and lupinifolin were stable throughout the simulation (Figure 2(A)). Since rutin fluctuated from 0.2 nm to 2.3 nm at the end of the simulation, ten snapshots were downloaded in the interval of 20 ns during the entire simulation wherein it was observed that rutin had moved considerably from the active site, while hesperidin and lupinifolin remained firmly bound (Figure 3). It suggests the inability of rutin to inhibit efficiently the 3CLpro of SARS-CoV-2. Among the ligands that formed stable complexes with RdRp, only hesperidin showed stable binding in the active site (Figure 2(B), Figure 4). Following analyses were done for hesperidin and lupinifolin which did not disturb the protein stability and remained firmly bound to the active site.
Figure 2.
Backbone root mean square deviation (RMSD) of phytochemicals from complexes with 3CLpro (A) and RdRp (B), during 200 ns of the molecular dynamics simulation period.
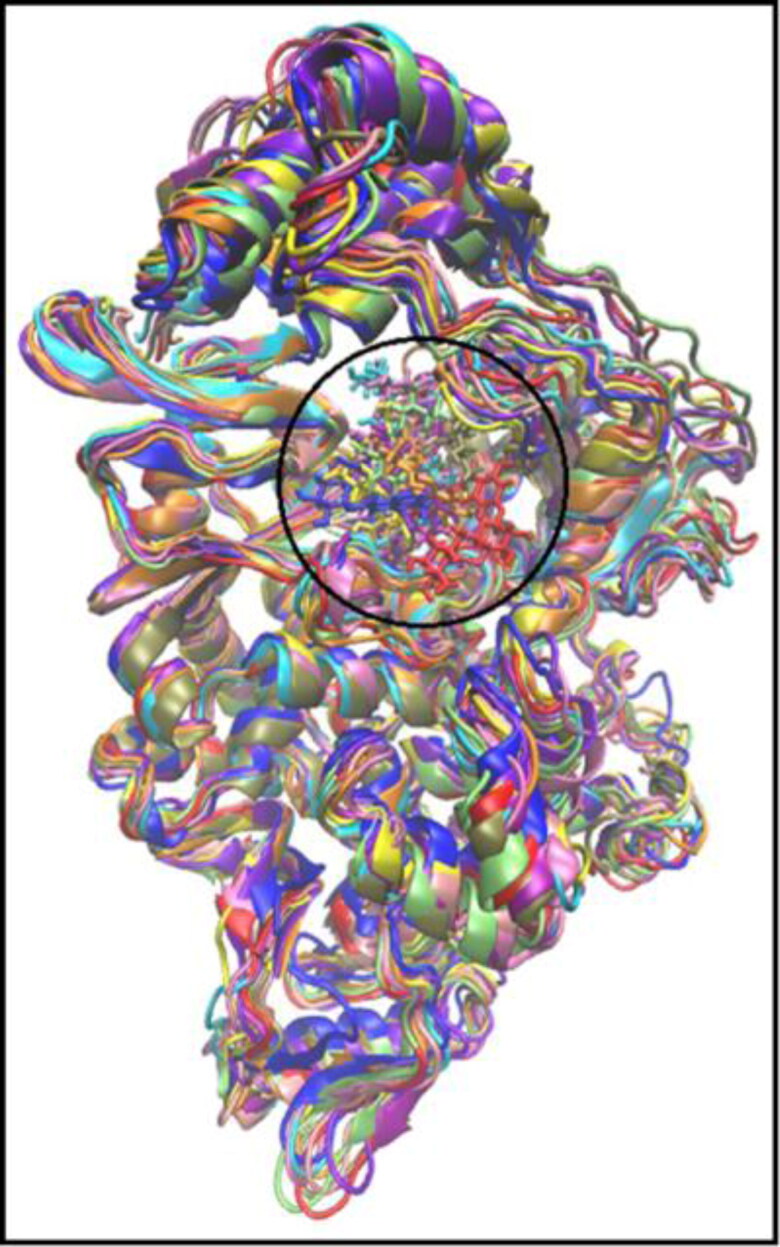
Figure 3.
Snapshot of the superimposed structures of 3CLpro in complex with hesperidin (A), lupinifolin (B), and rutin (C). Structures were obtained from the trajectory file in the interval of 20 ns.
Figure 4.
Snapshot of the superimposed structures of RdRp-hesperidin complex. Structures were obtained from the trajectory file in the interval of 20 ns.
3.2.2. Root mean square fluctuation and DSSP
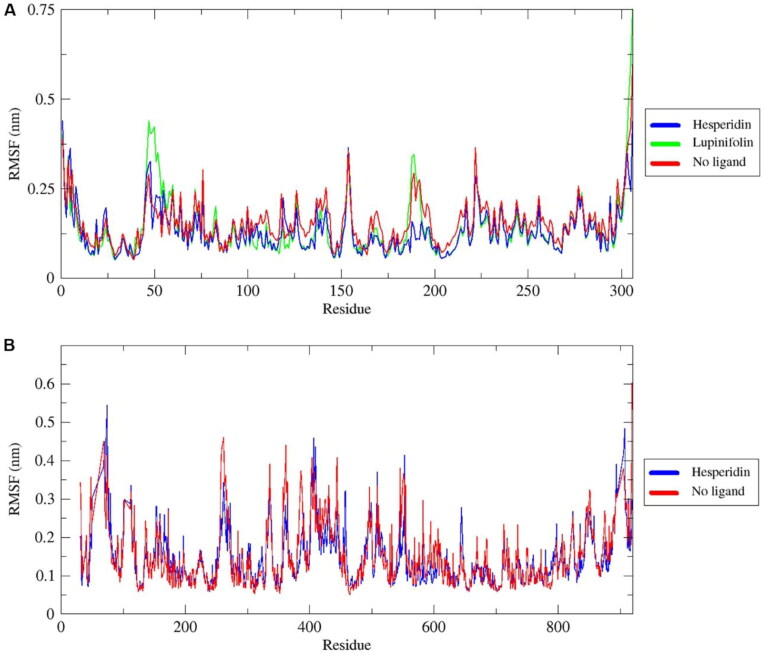
In order to calculate the residual mobility in the absence and presence of the phytochemicals, RMSF analysis was calculated and plotted against the residue number and was supported with the DSSP analysis. Part of the noticeable fluctuations occurring to backbones of 3CLpro complexes around position 50 corresponds to residues belonging to a loop surrounding the active site (Figure 5(A)). It has been demonstrated that loops can be involved in ligand binding and conformational changes can often take place in these sites (Lee et al., 2012; Teague, 2003). In contrast, residues interacting with stable ligands including the catalytic dyad H41 and C145 for 3CLpro, as well as the catalytic residues 759SDD761 and the divalent-cation-binding residue D618 for RdRp were among the most stable residues. Missing residues between K50-Y69, F102-P112, and L895-M906 in the crystal structure of RdRp were evidenced by ambiguous fluctuations in those positions (Figure 5(B)). In all complexes, there were no significant structural changes during the throughout simulation. The helical and β-sheet content remained stable (Figure S3, Figure S4).
Figure 5.
Root mean square fluctuation (RMSF) of backbone atoms of 3CLpro (A) and RdRp (B) as free proteins and forming complexes with phytochemicals, during the simulation period.
3.2.3. Radius of gyration
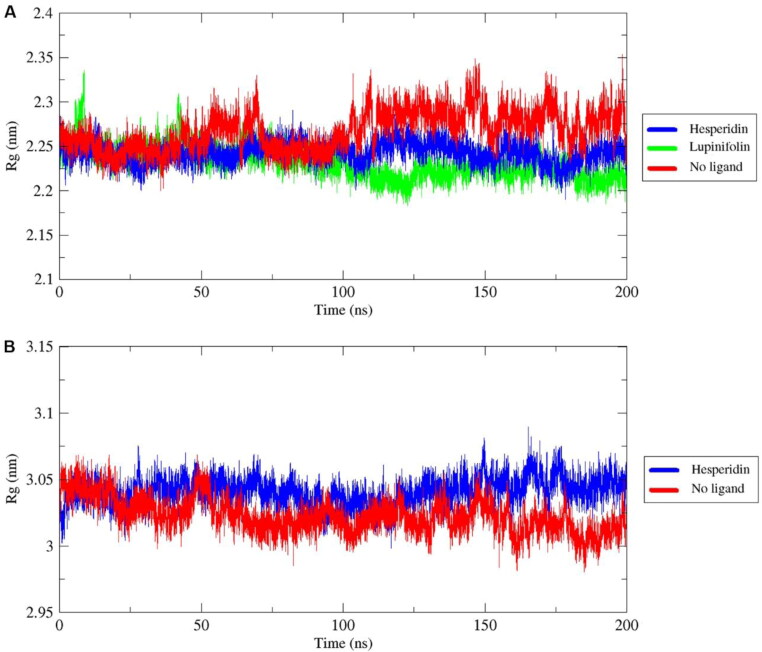
Radius of gyration (Rg) is a measure of the compactness of a protein which allows to understand its folding properties (Lobanov et al., 2008). Small Rg values indicate a tight packing whereas high Rg values show a floppy packing. A relative constant Rg value through time indicates that the ligands hold the folding behavior of the protein whereas abrupt fluctuations of the Rg values denote protein folding instability (Khan et al., 2020).
The 3Clpro in complex with hesperidin (mean Rg = 2.24 nm) and lupinifolin (mean Rg = 2.23 nm) presented more compactness compared with the free protein (mean Rg = 2.27 nm). On the other hand, the RdRp in complex with hesperidin (Rg = 3.04 nm) was slightly less compact than the free RdRp (Rg = 3.02 nm). In all these complexes, the Rg kept constant with no abrupt fluctuations through the time, indicating that hesperidin, rutin, and lupinifolin maintain the folding behavior of 3Cl pro while hesperidin maintains RdRp folding (Figure 6).
Figure 6.
Backbone radius of gyration of 3CLpro (A) and RdRp (B) as free proteins and forming complexes with phytochemicals, during 200 ns of the molecular dynamics simulation period.
3.2.4. Hydrogen bonds
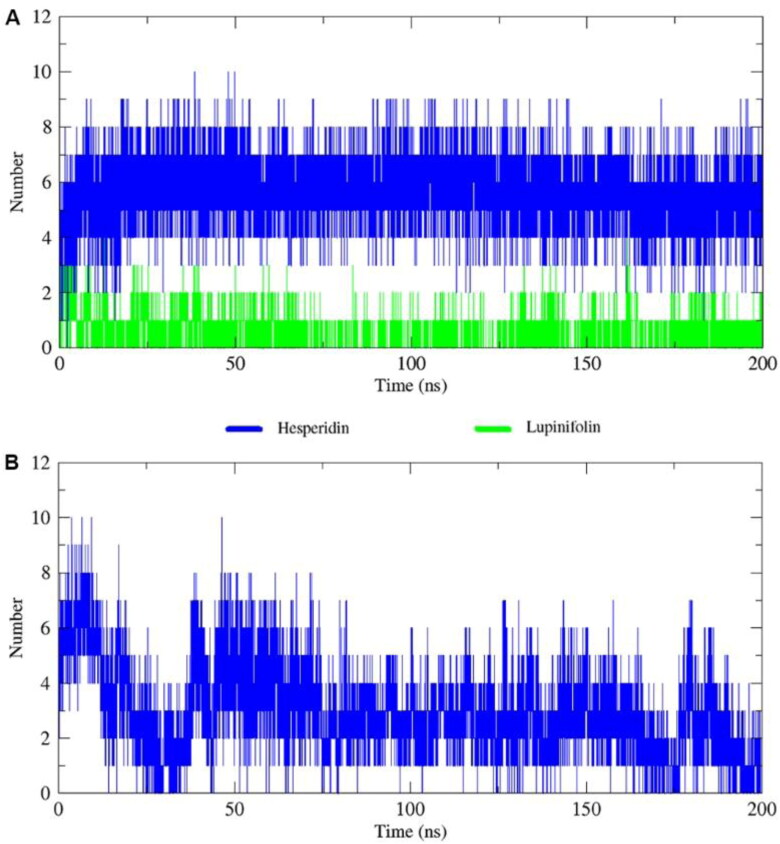
It is well known that H-bonds are responsible for the secondary and tertiary structural protein motifs. Formation of H-bonds between a ligand and a protein motif explains the binding affinity of a drug towards a protein target in molecular dynamics simulations; so, the more number of H-bonds the stronger interactions (Menendez et al., 2016).
For 3CLpro, a mean number of hydrogen bonds of 5-6, and 1, were found for hesperidin and lupinifolin, respectively (Figure 7(A)). The stability of lupinifolin can be explained by the six hydrophobic interactions revealed from the docking analysis. In addition, the contribution of van der Waals interactions to the binding energy of lupinifolin was the lowest among the complexes (Table 1). Even though pinocembrin 7-O-rutinoside formed 4 hydrogen bonds, it was discarded because of the backbone instability obtained in the RMSD analysis. On the other hand, for RdRp in complex with hesperidin, an average of 2-3 hydrogen bonds was found (Figure 7(B)). The formation of these hydrogen bonds explains the high affinities of the ligands to 3Clpro and RdRp, as well as the stability of these complexes over time. In general, small non-covalent or reversible covalent inhibitors, such as phytochemicals, display several advantages regarding side effects and toxicity compared with covalent inhibitors (Pillaiyar et al., 2016).
Figure 7.
Total number of hydrogen bonds interactions between phytochemicals (hesperidin and lupinifolin) and proteins 3CLpro (A) and RdRp (B).
3.3. Binding energy estimation
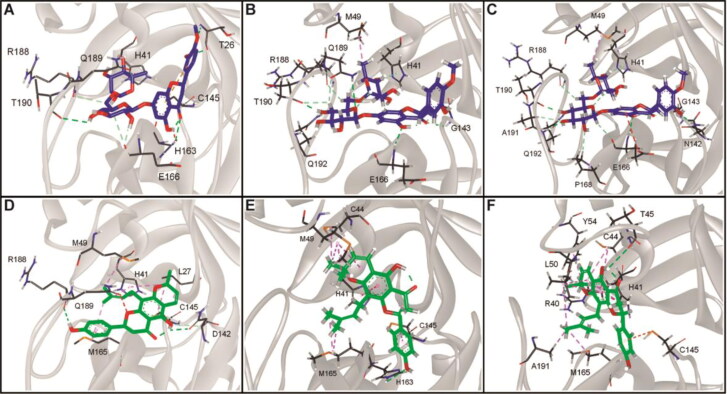
By rescoring docked complexes, the MM-PBSA method has demonstrated to be a valuable tool in drug discovery to remove possible false positive compounds obtained by docking analysis performed with standard methods (Kumari et al., 2014; Rastelli et al., 2009). Binding energies (ΔGbind) of the stable complexes with ligands firmly bound to the active site of 3CLpro and RdRp were estimated according to the MM-PBSA method (Table 1). Molecular interactions of hesperidin and lupinifolin in complex with 3CLpro taken from molecular docking and during the simulation at 100 ns and 200 ns included mostly polar not charged and non-polar residues (Figure 8), then a value of = 2 was set for the MM-PBSA calculations of these complexes. On the other hand, a value of = 4 was used for the RdRp-hesperidin complex because the highly charged protein-ligand interface including aspartic acid, glutamic acid, arginine, and lysine residues (Figure 9).
Figure 8.
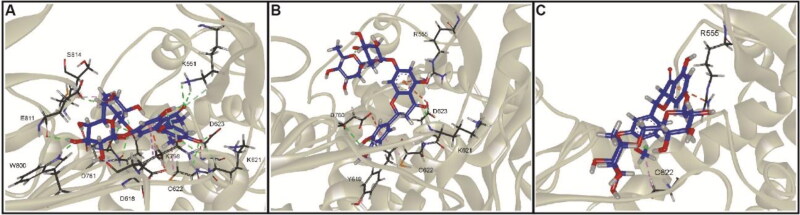
Molecular interactions of hesperidin (blue) and lupinifolin (green) in complex with the 3CLpro of SARS-CoV-2 taken from the molecular docking (A,D) and during the simulation at 100 ns (B,E) and 200 ns (C,F).
Figure 9.
Molecular interactions of hesperidin in complex with the RdRp of SARS-CoV-2 taken from molecular docking (A) and during the simulation at 100 ns (B) and 200 ns (C).
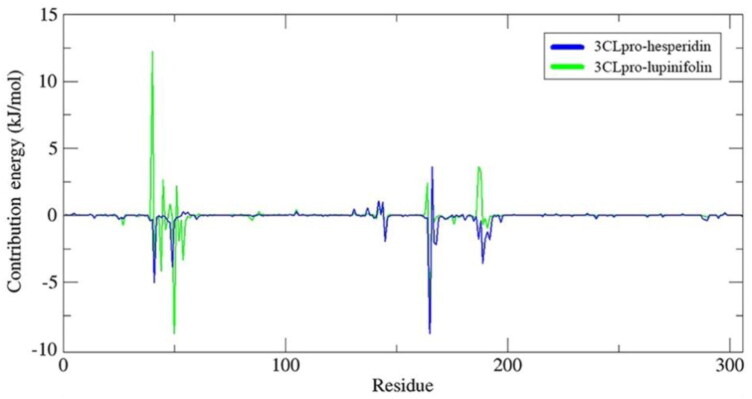
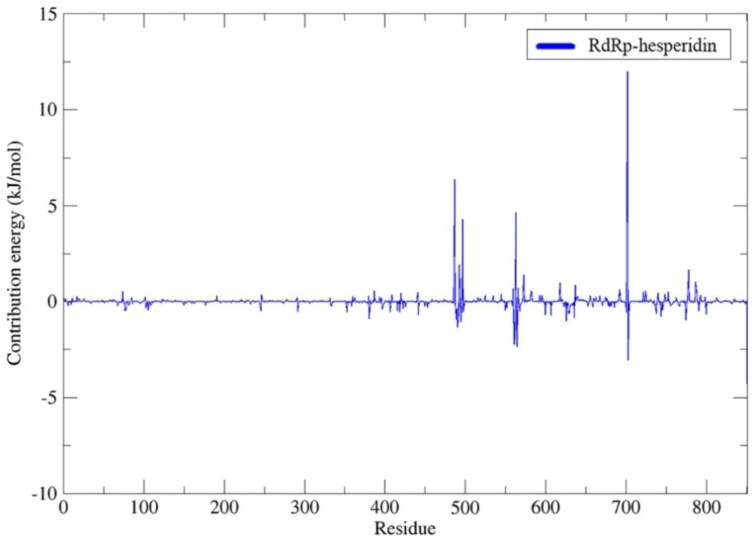
Results indicated that lupinifolin showed more affinity to 3CLpro compared to hesperidin (Table 1). However, hesperidin also formed a stable complex with RdRp showing a ΔGbind = −9.46 kcal/mol. In these complexes, the contribution of van der Waals interactions to the binding energy was lower than electrostatic and non-polar solvation energies. To understand protein-ligand associations, the total binding energies were decomposed into the contribution made by each residue. For 3CLpro complexes, H41, C145, and M165 were found to strongly interact with both hesperidin and lupinifolin, (Figure 10). For RdRp-hesperidin complex, residues S561 and T565 surrounding the active site as well as D703 were part of those contributing negatively to the binding energy (Figure 11).
Figure 10.
MM-PBSA binding free energy contribution per residue of 3CLpro in complex with hesperidin and lupinifolin.
Figure 11.
MM-PBSA binding free energy contribution per residue of RdRp in complex with hesperidin.
3.4. Admet prediction
In silico ADMET analysis is used to predict the pharmacokinetic profile of compounds, before experimental procedures (Jayaraj et al., 2020). The bioavailability of any compound is highly influenced by its physicochemical properties of the compound. According to Lipinski's rule, a poor absorption and a low permeation are more likely when the molecule meets the following properties: the molecular weight of the compound is greater than 500 g/mol, there are more than 10 H-bond donors and more than 5 H-bond acceptors, and the logarithm of octanol-water partition coefficient (Log P) is greater than 5 (Benet et al., 2016). Whereas the values for lupinifolin lie between the ranges, hesperidin violated molecular weight and number of H-bond acceptors (Table 2). The low lipophilicity of hesperidin (Log P = −0.72) and poor water solubility of lupinifolin (Log p = 4.38) implying a low oral bioavailability can be overcome by using oral drug delivery systems or intravenous administration.
Table 2.
Predicted ADMET profile of hesperidin and lupinifolin.
| Descriptor | Hesperidin | Lupinifolin |
|---|---|---|
| Molecular weight (g/mol) | 610.56 | 406.47 |
| Number of rotatable bonds | 7 | 3 |
| Number of H-bond donors | 8 | 2 |
| Number of H-bond acceptors | 15 | 5 |
| Log P | −0.72 | 4.38 |
| Gastrointestinal absorption | Low | High |
| BBB permeability | No | No |
| Substrate of P-gp | Yes | No |
| CYP1A2 inhibitor | No | No |
| CYP2C19 inhibitor | No | Yes |
| CYP2C9 inhibitor | No | Yes |
| CYP2D6 inhibitor | No | No |
| CYP3A4 inhibitor | No | Yes |
| Bioavailability score | 0.17 | 0.55 |
| Log CLtot (mL/min/kg) | 0.211 | 0.31 |
| Hepatotoxicity | No | No |
| Ames toxicity | No | No |
| hERG I channel inhibition | No | No |
| hERG II channel inhibition | Yes | No |
The analysis of distribution took into consideration both the permeability across the blood-brain barrier (BBB) and whether the compounds could be substrates of the P-glycoprotein (P-gp) (Prachayasittikul & Prachayasittikul, 2016). Since the inability of hesperidin and lupinifolin to permeate across the BBB (Table 2), the central nervous system is protected from their action. Importantly, the distribution of hesperidin could be limited by the P-gp-mediated efflux.
Metabolism of drugs is carried out by the cytochrome P450 (CYP) superfamily. Among the analyzed CYPs (Table 2), whereas hesperidin might not inhibit any of these enzymes, lupinifolin could inhibit CYP2C19, CYP2C9, and CYP3A4, but not neither CYP1A2 nor CYP2D6. Because of these inhibitions, lupinifolin may affect the metabolism and clearance of the potential drugs resulting into bioaccumulation (Terao & Mukai, 2014), so optimal drug doses must be defined.
Excretion analysis considered the total clearance (CLtot) because it influences the half-life and bioavailability of drugs. Moreover, CLtot provides a framework for the initial dose for first in human studies (Berellini et al., 2012). The CLtot of lupinifolin was higher than hesperidin (Table 2). Neither hesperidin nor lupinifolin was found to be carcinogenic or to present hepatotoxicity. However, hesperidin revealed weak inhibition of the human ether-a-go-go-related gene II (hERG II) potassium channel.
ADMET: Absorption, distribution, metabolism, excretion, toxicity; Log P: logarithm of octanol-water partition coefficient; BBB: Blood-brain barrier; P-gp: P-glycoprotein; Log CLtot: logarithm of total clearance; hERG: human ether-a-go-go-related gene
3.5. Potential inhibitors of the 3CLpro and RdRp of SARS-CoV-2
From the combination of molecular docking to score binding poses, molecular dynamics to evaluate the interaction and stability of protein-ligand complexes, and MM-PBSA to estimate binding energies, among 92 phytochemicals, hesperidin was found to be a promising multitarget antiviral against SAS-CoV-2. More specifically, this flavonoid rutinoside showed high affinities and stability in complex with the 3CLpro and RdRp in their active sites. Structurally, hesperidin is constituted of the flavanone hesperitin (aglycone) and the disaccharide rutinoside (rhamnose linked to glucose), and it is mainly found in citrus fruits. Pharmacological effects of hesperidin include antimicrobial, antiviral, antihyperlipidemic, cardioprotective, antihypertensive, antidiabetic, and anti-inflammatory activities (Man et al., 2019; Zanwar et al., 2014). By performing cell-free and cell-based assays, hesperitin was found to significantly inhibit the cleavage processing of SARS-CoV 3CLpro while being less toxic to Vero cells when compared to other phenolic compounds (Lin et al., 2005). Considering that SARS-CoV-2 3CLpro is 96% identical with its SARS-CoV ortholog (Gao et al., 2020; Zhang et al., 2020), the estimated binding energies, and the stability with both 3CLpro and RdRp revealed by molecular dynamics simulation, hesperidin becomes a remarkable candidate to act as a multitarget antiviral, inhibiting two essential enzymes of the causative agent of COVID-19.
Lupinifolin was found to be a potential inhibitor of the main protease of SARS-CoV-2 because of its affinity and stability in complex with 3CLpro. This prenylated flavonoid has been identified in several medicinal plants belonging mostly to the family Fabaceae. Other sources include plants in the family Rutaceae and Apocynaceae (Ganapaty et al., 2006; Joycharat et al., 2016; Lin et al., 1991; Mahidol et al., 1997; Smalberger et al., 1974; Soonthornchareonnon et al., 2004). The wide range of pharmacological effects of lupinifolin includes antimicrobial activities against multidrug-resistant enterococci and Mycobacterium tuberculosis (Sianglum et al., 2019; Soonthornchareonnon et al., 2004), cytotoxicity against breast cancer, human small-cell lung (NCI-H187) and oral human epidermoid carcinoma (KB), with limited or not determined cytotoxicity against Vero cells (Soonthornchareonnon et al., 2004; Sutthivaiyakit et al., 2009), and antiviral activity against herpes simplex virus type 1 (HSV-1) (Soonthornchareonnon et al., 2004). Lupinifolin was found to inhibit bacterial growth via cell membrane disruption, increasing its permeability while decreasing salt tolerance (Sianglum et al., 2019; Yusook et al., 2017). On the other hand, its antiviral mechanism has not been studied yet.
Protein-interacting residues taken from molecular docking and during the simulation at 100 ns and 200 ns were annotated in order to obtain deeper insight into the interaction pattern. In the 3CLpro complexes, residues forming the catalytic dyad, C145 and H41, were found to interact with hesperidin and lupinifolin (Figure 8). In the stable RdRp complex, residues of the catalytic site (759SDD761) as well as the divalent-cation-binding residue D618 interacted with hesperidin (Figure 9).
It is worth mentioning that the oral bioavailability of flavonoids is restricted by the extensive first-pass metabolism in the intestine and liver (Gonzales, 2017). For instance, flavonoid rutinosides such as hesperidin are metabolized via methylation, demethylation, glucuronidation, sulfation, and sulfoglucuronidation in the intestine and liver, and hydrolyzed by microbial enzymes in the large intestine (Boyle et al., 2000; Nectoux et al., 2019). Moreover, the transport of prenylated flavonoids to blood circulation is lowered by prenyl groups and a long-term dietary intake can derive in tissue accumulation (Terao & Mukai, 2014). Hence, the route of administration and delivery are crucial to consider. Moreover, in vitro and in vivo assays are required to evaluate the efficacy of these natural compounds.
4. Conclusions
Using a virtual screening approach, the present study aimed to elucidate natural compounds as potential inhibitors of the SARS-CoV-2 3CLpro and RdRp which are enzymes that play a key role in the virus replication cycle. After performing molecular docking to estimate the binding affinities and molecular dynamics simulation to evaluate the stability of the protein-ligand complexes, hesperidin (estimated binding affinities, ΔGbind = −15.18 kcal/mol to 3CLpro and ΔGbind = −9.46 kcal/mol to RdRp) was identified as a lead candidate because of its potential to efficiently inhibit both 3CLpro and RdRp. Furthermore, lupinifolin exhibited potential to inhibit the main protease showing a higher affinity than hesperidin (ΔGbind = −20.93 kcal/mol) while displaying suitable pharmacokinetics and physicochemical properties. Based on the safety and low toxicity to normal cells reported in the literature, the predicted ADMET profile, and its affinity and stability when forming complexes with these SARS-CoV-2 enzymes, hesperidin becomes an attractive multitarget drug candidate to treat COVID-19. However, further in vivo/in vitro assays are required to evaluate the proposed therapeutic use.
Supplementary Material
Acknowledgements
The authors thank Dr. Francisco Javier Flores for revising and improving our manuscript. Portions of the computing for this project was performed at the OSU High-Performance Computing Center at Oklahoma State University.
Disclosure statement
The authors declare no conflict of interest.
References
- Ahmed, S. F., Quadeer, A. A., & McKay, M. R. (2020). Preliminary identification of potential vaccine targets for the COVID-19 Coronavirus (SARS-CoV-2) based on SARS-CoV Immunological studies. Viruses, 12(3), 254. 10.3390/v12030254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand, K., Ziebuhr, J., Wadhwani, P., Mesters, J. R., & Hilgenfeld, R. (2003). Coronavirus main proteinase (3CLpro) structure: Basis for design of anti-SARS drugs. Science (New York, NY), 300(5626), 1763–1767. 10.1126/science.1085658 [DOI] [PubMed] [Google Scholar]
- Anthelme, F., Jacobsen, D., Macek, P., Meneses, R. I., Moret, P., Beck, S., & Dangles, O. (2014). Biodiversity patterns and continental insularity in the tropical high andes. Arctic, Antarctic, and Alpine Research, 46(4), 811–828. 10.1657/1938-4246-46.4.811 [DOI] [Google Scholar]
- Benet, L. Z., Hosey, C. M., Ursu, O., & Oprea, T. I. (2016). BDDCS, the rule of 5 and drugability. Advanced Drug Delivery Reviews, 101(1), 89–98. 10.1016/j.addr.2016.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berellini, G., Waters, N. J., & Lombardo, F. (2012). In silico prediction of total human plasma clearance. Journal of Chemical Information and Modeling, 52(8), 2069–2078. 10.1021/ci300155y [DOI] [PubMed] [Google Scholar]
- Berendsen, H. J. C., Postma, J. P. M., van Gunsteren, W. F., DiNola, A., & Haak, J. R. (1984). Molecular dynamics with coupling to an external bath. The Journal of Chemical Physics, 81(8), 3684–3690. 10.1063/1.448118 [DOI] [Google Scholar]
- Bjelkmar, P., Larsson, P., Cuendet, M. A., Hess, B., & Lindahl, E. (2010). Implementation of the CHARMM force field in GROMACS: Analysis of protein stability effects from correction maps, virtual interaction sites, and water models. Journal of Chemical Theory and Computation, 6(2), 459–466. 10.1021/ct900549r [DOI] [PubMed] [Google Scholar]
- Boyle, S. P., Dobson, V. L., Duthie, S. J., Hinselwood, D. C., Kyle, J. A., & Collins, A. R. (2000). Bioavailability and efficiency of rutin as an antioxidant: A human supplementation study. European Journal of Clinical Nutrition, 54(10), 774–782. 10.1038/sj.ejcn.1601090 [DOI] [PubMed] [Google Scholar]
- Cai, Q., Yang, M., Liu, D., Chen, J., Shu, D., Xia, J., Liao, X., Gu, Y., Cai, Q., Yang, Y., Shen, C., Li, X., Peng, L., Huang, D., Zhang, J., Zhang, S., Wang, F., Liu, J., Chen, L., … Liu, L. (2020). Experimental treatment with favipiravir for COVID-19: An open-label control study. Engineering. 10.1016/j.eng.2020.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caly, L., Druce, J. D., Catton, M. G., Jan, D. A., & Wagstaff, K. M. (2020). The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Research, 178, 104787. 10.1016/j.antiviral.2020.104787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, B., Wang, Y., Wen, D., Liu, W., Wang, J., Fan, G., Ruan, L., Song, B., Cai, Y., Wei, M., Li, X., Xia, J., Chen, N., Xiang, J., Yu, T., Bai, T., Xie, X., Zhang, L., Li, C., … Wang, C. (2020). A trial of lopinavir–ritonavir in adults hospitalized with severe Covid-19. The New England Journal of Medicine, 382(19), 1787–1799. 10.1056/NEJMoa2001282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daina, A., Michielin, O., & Zoete, V. (2017). SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Scientific Reports, 7, 42717. 10.1038/srep42717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darden, T., York, D., & Pedersen, L. (1993). Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. The Journal of Chemical Physics, 98(12), 10089–10092. 10.1063/1.464397 [DOI] [Google Scholar]
- Discovery Studio Visualizer . (2016). 20.1.0.19295 Edition; Dassault Systèmes Biovia Corp: Dassault Systèmes.
- Elfiky, A. A. (2020). Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life Sciences, 253, 117592. 10.1016/j.lfs.2020.117592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganapaty, S., Josaphine, J. S., & Thomas, P. S. (2006). Anti-inflammatory activity of Derris scandens. Journal of Natural Remedies, 6(1), 73–76. 10.18311/jnr/2006/282 [DOI] [Google Scholar]
- Gao, Y., Yan, L., Huang, Y., Liu, F., Zhao, Y., Cao, L., Wang, T., Sun, Q., Ming, Z., Zhang, L., Ge, J., Zheng, L., Zhang, Y., Wang, H., Zhu, Y., Zhu, C., Hu, T., Hua, T., Zhang, B., … Rao, Z. (2020). Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science(New York, NY), 368(6492), 779–782. 10.1126/science.abb7498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales, G. B. (2017). In Vitro bioavailability and cellular bioactivity studies of flavonoids and flavonoid-rich plant extracts: Questions, considerations and future perspectives. The Proceedings of the Nutrition Society, 76(3), 175–181. 10.1017/S0029665116002858 [DOI] [PubMed] [Google Scholar]
- Gordon, C. J., Tchesnokov, E. P., Woolner, E., Perry, J. K., Feng, J. Y., Porter, D. P., & Götte, M. (2020). Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. Journal of Biological Chemistry, 295(20), 6785–6797. 10.1074/jbc.RA120.013679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex, N., & Peitsch, M. C. (1997). SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling . Electrophoresis, 18(15), 2714–2723. 10.1002/elps.1150181505 [DOI] [PubMed] [Google Scholar]
- Guo, Y.-R., Cao, Q.-D., Hong, Z.-S., Tan, Y.-Y., Chen, S.-D., Jin, H.-J., Tan, K.-S., Wang, D.-Y., & Yan, Y. (2020). The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Military Medical Research, 7(1), 11. 10.1186/s40779-020-00240-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess, B., Bekker, H., Berendsen, H. J. C., & Fraaije, J. (1997). LINCS: A linear constraint solver for molecular simulations. Journal of Computational Chemistry, 18(12), 1463–1472 [Google Scholar]
- Horby, P., Lim, W. S., Emberson, J., Mafham, M., Bell, J., Linsell, L., Staplin, N., Brightling, C., Ustianowski, A., Elmahi, E., Prudon, B., Green, C., Felton, T., Chadwick, D., Rege, K., Fegan, C., Chappell, L. C., Faust, S. N., Jaki, T., … Landray, M. J. (2020). Effect of dexamethasone in hospitalized patients with COVID-19: Preliminary report. medRxiv. 10.1101/2020.06.22.20137273 [DOI] [Google Scholar]
- Huang, J., & MacKerell, A. D. (2013). CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. Journal of Computational Chemistry, 34(25), 2135–2145. 10.1002/jcc.23354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jácome, R., Campillo-Balderas, J. A., Ponce-De León, S., Becerra, A., & Lazcano, A. (2020). Sofosbuvir as a potential alternative to treat the SARS-CoV-2 epidemic. Scientific Reports, 10, 9294. 10.21203/rs.3.rs-21002/v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraj, J. M., Reteti, E., Kesavan, C., & Muthusamy, K. (2020). Structural insights on Vitamin D receptor and screening of new potent agonist molecules: Structure and ligand-based approach. Journal of Biomolecular Structure and Dynamics, 1–12. 10.1080/07391102.2020.1775122 [DOI] [PubMed] [Google Scholar]
- Jo, S., Kim, S., Shin, D. H., & Kim, M. S. (2020). Inhibition of SARS-CoV 3CL protease by flavonoids. Journal of Enzyme Inhibition and Medicinal Chemistry, 35(1), 145–151. 10.1080/14756366.2019.1690480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joycharat, N., Boonma, C., Thammavong, S., Yingyongnarongkul, B., Limsuwan, S., & Voravuthikunchai, S. P. (2016). Chemical constituents and biological activities of Albizia myriophylla wood. Pharmaceutical Biology, 54(1), 62–73. 10.3109/13880209.2015.1014920 [DOI] [PubMed] [Google Scholar]
- Khan, R., Jha, R. K., Amera, G. M., Jain, M., Singh, E., Pathak, A., Singh, R. P., Muthukumaran, J., & Singh, A. K. (2020). Targeting SARS-CoV-2: A systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2′-O-ribose methyltransferase. Journal of Biomolecular Structure & Dynamics, 1–14. 10.1080/07391102.2020.1753577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer, R. N., & Ward, A. B. (2019). Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nature Communications, 10(1), 2343. 10.1038/s41467-019-10280-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman, P. A., Massova, I., Reyes, C., Kuhn, B., Huo, S., Chong, L., Lee, M., Lee, T., Duan, Y., Wang, W., Donini, O., Cieplak, P., Srinivasan, J., Case, D. A., & Cheatham, T. E. (2000). Calculating structures and free energies of complex molecules: Combining molecular mechanics and continuum models. Accounts of Chemical Research, 33(12), 889–897. 10.1021/ar000033j [DOI] [PubMed] [Google Scholar]
- Kumari, R., Kumar, R., & Lynn, A., Open Source Drug Discovery Consortium . (2014). g_mmpbsa-a GROMACS tool for high-throughput MM-PBSA calculations . Journal of Chemical Information and Modeling, 54(7), 1951–1962. 10.1021/ci500020m [DOI] [PubMed] [Google Scholar]
- Lalani, S., & Poh, C. L. (2020). Flavonoids as antiviral agents for Enterovirus A71 (EV-A71). Viruses, 12(2), 184. 10.3390/v12020184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, G. R., Shin, W. H., Park, H.-B., Shin, S., & Seok, C. (2012). Conformational sampling of flexible ligand-binding protein loops. Bulletin of the Korean Chemical Society, 33(3), 770–774. 10.5012/bkcs.2012.33.3.770 [DOI] [Google Scholar]
- Lin, C. W., Tsai, F. J., Tsai, C. H., Lai, C. C., Wan, L., Ho, T. Y., Hsieh, C. C., & Chao, P. D. (2005). Anti-SARS coronavirus 3C-like protease effects of Isatis indigotica root and plant-derived phenolic compounds. Antiviral Research, 68(1), 36–42. 10.1016/j.antiviral.2005.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y.-L., Chen, Y.-L., & Kuo, Y.-H. (1991). Three new flavonoids, 3′-methoxylupinifolin, laxifolin, and isolaxifolin from the roots of Derris laxiflora Benth. Chemical & Pharmaceutical Bulletin, 39(12), 3132–3135. 10.1248/cpb.39.3132 [DOI] [Google Scholar]
- Liu, K., Watanabe, E., & Kokubo, H. (2017). Exploring the stability of ligand binding modes to proteins by molecular dynamics simulations. Journal of Computer-Aided Molecular Design, 31(2), 201–211. 10.1007/s10822-016-0005-2 [DOI] [PubMed] [Google Scholar]
- Liu, X., Shi, D., Zhou, S., Liu, H., Liu, H., & Yao, X. (2018). Molecular dynamics simulations and novel drug discovery. Expert Opinion on Drug Discovery, 13(1), 23–37. 10.1080/174604411403419 [DOI] [PubMed] [Google Scholar]
- Lobanov, M. Y., Bogatyreva, N. S., & Galzitskaya, O. V. (2008). Radius of gyration as an indicator of protein structure compactness. Molecular Biology, 42(4), 623–628. 10.1134/S0026893308040195 [DOI] [PubMed] [Google Scholar]
- Mahidol, C., Prawat, H., Ruchirawat, S., Lihkitwitayawuid, K., Lin, L.-Z., & Cordell, G. A. (1997). Prenylated flavanones from Derris reticulata. Phytochemistry, 45(4), 825–829. 10.1016/S0031-9422(97)00001-0 [DOI] [Google Scholar]
- Man, M. Q., Yang, B., & Elias, P. M. (2019). Benefits of hesperidin for cutaneous functions. Evidence-Based Complementary and Alternative Medicine: ECAM, 2019, 2676307. 10.1155/2019/2676307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangar, P., Pradhan, S., Rai, S., Lepcha, K., & Ranjan, V. K. (2020). Comparative analysis based on the spike glycoproteins of SARS-CoV2 isolated from COVID 19 patients of different countries. Preprints. 10.20944/preprints202004.0154.v1 [DOI] [Google Scholar]
- Mantlo, E., Bukreyeva, N., Maruyama, J., Paessler, S., & Huang, C. (2020). Antiviral activities of Type I interferons to SARS-CoV-2 infection. Antiviral Research, 179, 104811. 10.1101/2020.04.02.022764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauthe, M., Orhon, I., Rocchi, C., Zhou, X., Luhr, M., Hijlkema, K., Coppes, R. P., Engedal, N., Mari, M., & Reggiori, F. (2018). Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy, 14(8), 1435–1455. 10.1080/15548627.2018.1474314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh, K. (1974). Coronaviruses: A comparative review. Current Topics in Microbiology and Inmunology, 3, 85–129. 10.1007/978-3-642-65775-7_3 [DOI] [Google Scholar]
- Menendez, C. A., Accordino, S. R., Gerbino, D. C., & Appignanesi, G. A. (2016). Hydrogen bond dynamic propensity studies for protein binding and drug design. PloS One, 11(10), e0165767 10.1371/journal.pone.0165767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutke, J., Jacobs, R., Meyers, K., Henning, T., & Weigend, M. (2014). Diversity patterns of selected Andean plant groups correspond to topography and habitat dynamics, not orogeny. Frontiers in Genetics, 5, 351 10.3389/fgene.2014.00351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nectoux, A. M., Abe, C., Huang, S. W., Ohno, N., Tabata, J., Miyata, Y., Tanaka, K., Tanaka, T., Yamamura, H., & Matsui, T. (2019). Absorption and metabolic behavior of hesperidin (Rutinosylated Hesperetin) after single oral administration to sprague-dawley rats. Journal of Agricultural and Food Chemistry, 67(35), 9812–9819. 10.1021/acs.jafc.9b03594 [DOI] [PubMed] [Google Scholar]
- O’Boyle, N. M., Banck, M., James, C. A., Morley, C., Vandermeersch, T., & Hutchison, G. R. (2011). Open Babel: An open chemical toolbox. Journal of Cheminformatics, 3, 33. 10.1186/1758-2946-3-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paduch, R., & Kandefer-Szerszen, M. (2014). Antitumor and antiviral activity of pentacyclic triterpenes. Mini-Reviews in Organic Chemistry, 11(3), 262–268. 10.2174/1570193X1103140915105240 [DOI] [Google Scholar]
- Pennington, R. T., Lavin, M., Särkinen, T., Lewis, G. P., Klitgaard, B. B., & Hughes, C. E. (2010). Contrasting plant diversification histories within the Andean biodiversity hotspot. Proceedings of the National Academy of Sciences of the United States of America, 107(31), 13783–13787. 10.1073/pnas.1001317107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillaiyar, T., Manickam, M., Namasivayam, V., Hayashi, Y., & Jung, S.-H. (2016). An overview of severe acute respiratory syndrome-coronavirus (SARS-CoV) 3CL protease inhibitors: Peptidomimetics and small molecule Chemotherapy. Journal of Medicinal Chemistry, 59(14), 6595–6628. 10.1021/acs.jmedchem.5b01461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires, D. E., Blundell, T. L., & Ascher, D. B. (2015). pkCSM: Predicting small-molecule pharmacokinetic and toxicity properties using graph-based Signatures. Journal of Medicinal Chemistry, 58(9), 4066–4072. 10.1021/acs.jmedchem.5b00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prachayasittikul, V., & Prachayasittikul, V. (2016). P-glycoprotein transporter in drug development. EXCLI Journal, 15, 113–118. 10.17179/excli2015-768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastelli, G., Degliesposti, G., Del Rio, A., & Sgobba, M. (2009). Binding estimation after refinement, a new automated procedure for the refinement and rescoring of docked ligands in virtual screening. Chemical Biology & Drug Design, 73(3), 283–286. 10.1111/j.1747-0285.2009.00780.x [DOI] [PubMed] [Google Scholar]
- Ravindranathan, K., Tirado-Rives, J., Jorgensen, W. L., & Guimarães, C. R. (2011). Improving MM-GB/SA Scoring through the Application of the Variable Dielectric Model. Journal of Chemical Theory and Computation, 7(12), 3859–3865. 10.1021/ct200565u [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saklani, A., & Kutty, S. K. (2014). Herbal medicine as inducers of apoptosis in cancer treatment. Advanced Pharmaceutical Bulletin, 4(Suppl 1), 421–427. 10.5681/apb.2014.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanner, M. F. (1999). Python: A programming language for software integration and development. Journal of Molecular Graphics, 17, 57–61. [PubMed] [Google Scholar]
- Savarino, A., Lucia, M. B., Rastrelli, E., Rutella, S., Golotta, C., Morra, E., Tamburrini, E., Perno, C. F., Boelaert, J. R., Sperber, K., & Cauda, R. (2004). Anti-HIV effects of chloroquine: Inhibition of viral particle glycosylation and synergism with protease inhibitors. Journal of Acquired Immune Deficiency Syndromes, 35(3), 223–232. 10.1097/00126334-200403010-00002 [DOI] [PubMed] [Google Scholar]
- Sianglum, W., Muangngam, K., Joycharat, N., & Voravuthikunchai, S. P. (2019). Mechanism of action and biofilm inhibitory activity of lupinifolin against multidrug-resistant enterococcal clinical isolates. Microbial Drug Resistance, 25(10), 1391–1400. 10.1089/mdr.2018.0391 [DOI] [PubMed] [Google Scholar]
- Smalberger, T. M., Vleggaar, R., & Weber, J. C. (1974). Flavonoids from Tephrosia—VII: The constitution and absolute configuration of lupinifolin and lupinifolinol, two flavanones from Tephrosia lupinifolia Burch (DC). Tetrahedron, 30(21), 3927–3931. 10.1016/S0040-4020(01)97085-1 [DOI] [Google Scholar]
- Soonthornchareonnon, N., Ubonopas, L., Kaewsuwan, S., & Wuttiudomlert, M. (2004). Lupinifolin, a bioactive flavanone from Myriopteron extensum (Wight) K. Schum. stem. Thai Journal of Phytopharmacy, 11(2), 19–28. [Google Scholar]
- Sutthivaiyakit, S., Thongnak, O., Lhinhatrakool, T., Yodchun, O., Srimark, R., Dowtaisong, P., & Chuankamnerdkarn, M. (2009). Cytotoxic and antimycobacterial prenylated flavonoids from the roots of Eriosema chinense. Journal of Natural Products, 72(6), 1092–1096. 10.1021/np900021h [DOI] [PubMed] [Google Scholar]
- Teague, S. J. (2003). Implications of protein flexibility for drug discovery. Nature Reviews. Drug Discovery, 2(7), 527–541. 10.1038/nrd1129 [DOI] [PubMed] [Google Scholar]
- Terao, J., & Mukai, R. (2014). Prenylation modulates the bioavailability and bioaccumulation of dietary flavonoids. Archives of Biochemistry and Biophysics, 559, 12–16. 10.1016/j.abb.2014.04.002 [DOI] [PubMed] [Google Scholar]
- Tian, W., Chen, C., Lei, X., Zhao, J., & Liang, J. (2018). CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Research, 46(W1), W363–W367. 10.1093/nar/gky473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott, O., &, Olson, A. J. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31(2), 455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Spoel, D., Lindahl, E., Hess, B., Groenhof, G., Mark, A. E., & Berendsen, H. J. (2005). GROMACS: Fast, flexible, and free. Journal of Computational Chemistry, 26(16), 1701–1718. 10.1002/jcc.20291 [DOI] [PubMed] [Google Scholar]
- Vanommeslaeghe, K., & MacKerell, A. D. (2012). Automation of the CHARMM General Force Field (CGenFF) I: Bond perception and atom typing. Journal of Chemical Information and Modeling, 52(12), 3144–3154. 10.1021/ci300363c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanommeslaeghe, K., Hatcher, E., Acharya, C., Kundu, S., Zhong, S., Shim, J., Darian, E., Guvench, O., Lopes, P., Vorobyov, I., & Mackerell, A. D. (2010). CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields . Journal of Computational Chemistry, 31(4), 671–690. 10.1002/jcc.21367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanommeslaeghe, K., Raman, E. P., & MacKerell, A. D. (2012). Automation of the CHARMM General Force Field (CGenFF) II: Assignment of bonded parameters and partial atomic charges. Journal of Chemical Information and Modeling, 52(12), 3155–3168. 10.1021/ci3003649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent, M. J., Bergeron, E., Benjannet, S., Erickson, B. R., Rollin, P. E., Ksiazek, T. G., Seidah, N. G., & Nichol, S. T. (2005). Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virology Journal, 2, 69. 10.1186/1743-422X-2-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, E., Weng, G., Sun, H., Du, H., Zhu, F., Chen, F., Wang, Z., & Hou, T. (2019). Assessing the performance of the MM/PBSA and MM/GBSA methods. 10. Impacts of enhanced sampling and variable dielectric model on protein-protein Interactions. Physical Chemistry Chemical Physics: PCCP, 21(35), 18958–18969. 10.1039/C9CP04096J [DOI] [PubMed] [Google Scholar]
- Wang, Y., Zhang, D., Du, G., Du, R., Zhao, J., Jing, Y., Fu, S., Gao, L., Cheng, Z., Lu, Q., Hu, Y., Luo, G., Wang, K., Lu, Y., Li, H., Wang, S., Ruan, S., Yang, C., Mei, C., ... Wang, C. (2020). Remdesivir in adults with severe COVID-19: A randomised, double-blind, placebo-controlled, multicentre trial. TheLancet, 395, 1569-1578. 10.1016/S0140-6736(20)31022-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, S. R., & Navas-Martin, S. (2005). Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiology and Molecular Biology Reviews: MMBR, 69(4), 635–664. 10.1128/MMBR.69.4.635-664.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusook, K., Weeranantanapan, O., Hua, Y., Kumkrai, P., & Chudapongse, N. (2017). Lupinifolin from Derris reticulata possesses bactericidal activity on Staphylococcus aureus by disrupting bacterial cell membrane. Journal of Natural Medicines, 71(2), 357–366. 10.1007/s11418-016-1065-2 [DOI] [PubMed] [Google Scholar]
- Zanwar, A. A., Badole, S. L., Shende, P. S., Hegde, M. V., & Bodhankar, S. L. (2014). Chapter 76 - cardiovascular effects of hesperidin: A Flavanone glycoside. In Watson R. R., Preedy V. R., & Zibadi S. (Eds.), Polyphenols in human health and disease (pp. 989–992). Academic Press. [Google Scholar]
- Zhang, L., Lin, D., Sun, X., Curth, U., Drosten, C., Sauerhering, L., Becker, S., Rox, K., & Hilgenfeld, R. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science (New York, NY), 368(6489), 409–412. 10.1126/science.abb3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.