ABSTRACT
One of the most rapid (less than 4 ms) transmembrane cellular mechanotransduction events involves activation of transient receptor potential vanilloid 4 (TRPV4) ion channels by mechanical forces transmitted across cell surface β1 integrin receptors on endothelial cells, and the transmembrane solute carrier family 3 member 2 (herein denoted CD98hc, also known as SLC3A2) protein has been implicated in this response. Here, we show that β1 integrin, CD98hc and TRPV4 all tightly associate and colocalize in focal adhesions where mechanochemical conversion takes place. CD98hc knockdown inhibits TRPV4-mediated calcium influx induced by mechanical forces, but not by chemical activators, thus confirming the mechanospecificity of this signaling response. Molecular analysis reveals that forces applied to β1 integrin must be transmitted from its cytoplasmic C terminus via the CD98hc cytoplasmic tail to the ankyrin repeat domain of TRPV4 in order to produce ultrarapid, force-induced channel activation within the focal adhesion.
KEY WORDS: Mechanical signaling, Mechanotransduction, Integrin, TRPV4, CD98hc
Summary: Mechanical signal transfer between cell surface β1 integrin receptors, CD98hc and TRPV4 channels within focal adhesions is shown to mediate ultrarapid transmembrane mechanochemical conversion in endothelial cells.
INTRODUCTION
Cellular mechanotransduction is crucial for maintenance of cellular growth homeostasis as well as tissue development, and deregulation of this process contributes to the etiology of numerous human diseases (Ingber, 2003). Cell surface integrin receptors that sense forces exerted on the extracellular matrix (ECM) (Wang et al., 1993) can induce mechanochemical transduction by transmitting these stresses across the cell surface, causing deformation in proteins, such as talin and vinculin, which form the cytoskeletal backbone of the focal adhesion (Choquet et al., 1997). An alternative mechanism of transmembrane mechanochemical conversion involves mechanosensitive transient receptor potential vanilloid 4 (TRPV4) ion channels on the surface membrane of endothelial cells, which are activated within 4 ms after mechanical forces are transmitted from the ECM to cell surface β1 integrin receptors (Matthews et al., 2010). This response is physiologically and clinically relevant because TRPV4 channels mediate cyclic strain-induced endothelial cell reorientation (Thodeti et al., 2009) and TRPV4 activation restores normal angiogenesis in tumors by modulating the Rho/Rho kinase pathway (Adapala et al., 2016). Mechanical activation of TRPV4 also contributes to development of pulmonary edema, and drugs that block TRPV4 channel activity prevent pulmonary vascular leakage in a human lung alveolus chip in vitro (Huh et al., 2012) as well as development of cardiogenic pulmonary edema in vivo (Thorneloe et al., 2012).
Although application of mechanical force to β1 integrins results in almost immediate activation of calcium influx through TRPV4 channels in endothelial cells, the specific path of force transfer from β1 integrin to TRPV4 remains unknown. Use of single chain integrin truncation mutants has revealed that the six terminal amino acid residues at the C terminus of the β1 integrin cytoplasmic domain are required for this rapid induction of calcium influx through TRPV4 when forces are applied directly to the extracellular domain of an engineered, single chain β1 integrin on the surface of endothelial cells using magnetic tweezers (Matthews et al., 2010). This portion of the β1 integrin C terminus is also required for adhesion strengthening and cytoskeletal tension-dependent fibronectin fibrillogenesis (Féral et al., 2007), which are mediated in part by binding of integrin to the single pass transmembrane solute carrier family 3 member 2 (CD98hc, also known as SLC3A2) glycoprotein that also associates with various other multipass transmembrane amino acid transporters through its extracellular domain (Kolesnikova et al., 2001; Zent et al., 2000). In a magnetic tweezer study, CD98hc localized to focal adhesions formed at the site of force application to integrins, and siRNA knockdown of CD98hc reduced the adhesion strength of the bound β1 integrins on the endothelial cell surface (Matthews et al., 2010). Although these studies implicate a role for CD98hc in β1 integrin-dependent stimulation of local calcium influx through TRPV4 ion channels, it remains unclear whether CD98hc is directly or indirectly involved in the transfer of mechanical forces between integrin and TRPV4 at the cell surface.
TRPV4 is a multipass transmembrane protein that forms a homo- or heterotetramer with other TRPV family proteins (White et al., 2016). The TRPV4 protein consists of a cytoplasmic N terminus, six transmembrane domains and a cytoplasmic C terminus. The cytoplasmic N terminus of TRPV4, which contains an N-terminal tail, a proline-rich domain (PRD) and six ankyrin repeat (AR) domains, has been shown to bind to cytoskeletal proteins such as actin and tubulin (Goswami et al., 2010). TRPV4 transmembrane domains also interact with cholesterol and play a role in its localization to lipid rafts (Kumari et al., 2015), whereas the cytoplasmic C terminus is involved in regulation of TRPV4 folding, maturation and trafficking (White et al., 2016).
In this study, we set out to determine whether the cytoplasmic tails of β1 integrin, CD98hc and TRPV4 bind to each other and, if so, to identify the specific domains and residues in these molecules that mediate transfer of mechanical forces from β1 integrin through CD98hc to TRPV4. Here, we report that specific amino acid (aa) residues located within the 50 aa (K160-C210) high homology (HH) domain of CD98hc bind the NPKY motif found within the β1 integrin cytoplasmic tail, whereas other residues within the cytoplasmic tail of CD98hc bind to the AR domain of TRPV4, thereby forming a physical path for mechanical signal transfer. Importantly, we also demonstrate that although this CD98hc-mediated linkage is required for activation of TRPV4 by mechanical stimulation, it is not necessary for activation of TRPV4 by chemical inducers. This new understanding of the molecular basis of mechanochemical conversion at the plasma membrane involving β1 integrin, CD98hc and TRPV4 may aid in the future development of more specific and effective mechanotherapeutics that target diseases in which mechanotransduction through TRPV4 plays an important role.
RESULTS
β1 integrin, CD98hc and TRPV4 tightly associate in focal adhesions
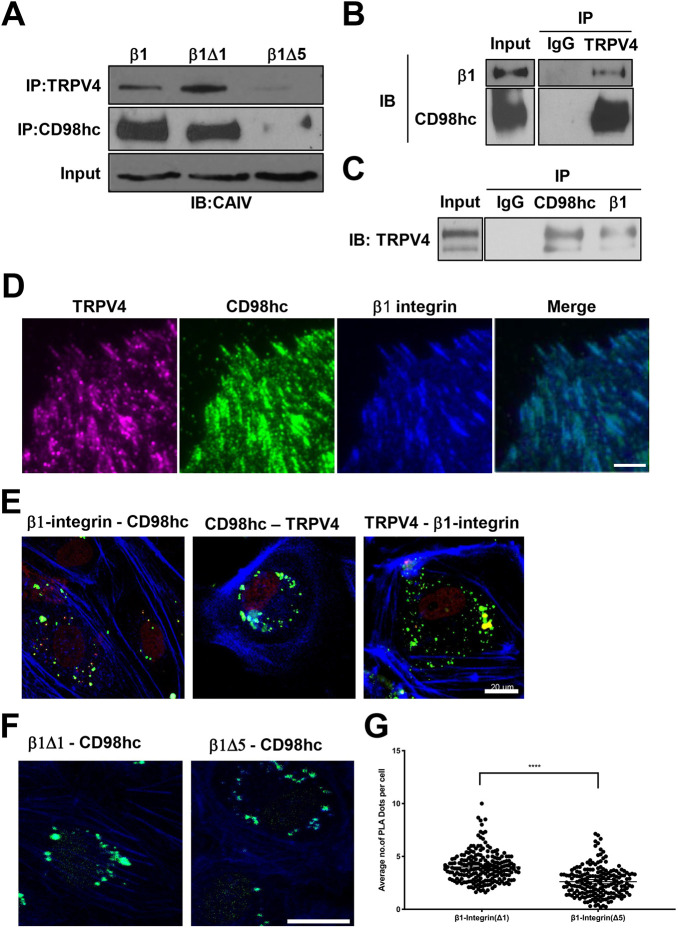
Deletion of the last six amino acids within the cytoplasmic tail of β1 integrin inhibits mechanical force-induced calcium signaling in bovine capillary endothelial cells and human dermal microvascular endothelial cells (Matthews et al., 2010). When human embryonic kidney (HEK) 293T cells were transfected with wild-type β1 integrin (β1) or single chain β1 integrin mutant constructs lacking six amino acids in the juxtamembrane region (β1Δ1) or the last six amino acids of the cytoplasmic tail (β1Δ5) (Fig. S1A), only the β1 and the β1Δ1 mutant containing the intact cytoplasmic terminus co-immunoprecipitated with both CD98hc and TRPV4 (Fig. 1A). Moreover, when similar studies were carried out in human umbilical vein endothelial (HUVE) cells using anti-TRPV4 antibody, endogenous β1 integrin and CD98hc co-precipitated (Fig. 1B). Similarly, antibodies directed against either β1 integrin or CD98hc resulted in co-precipitation of complexes containing TRPV4 (Fig. 1C).
Fig. 1.
β1 integrin, CD98hc and TRPV4 co-associate in focal adhesions in adherent cells. (A) Immunoblots showing β integrins (full length, β1Δ1, β1Δ5) immunoprecipitated with TRPV4 and CD98hc antibodies from total cell lysate of HEK293T cells transfected with CA-LDL−β1 (β1), CA-LDL−β1Δ1 (β1Δ1), and CA-LDL−β1Δ5 (β1Δ5) chimerae. (B) Immunoblots showing β1 integrin and CD98hc immunoprecipitated with TRPV4 antibody or IgG from total cell lysate of HUVE cells, with IgG used as a negative control. (C) TRPV4 immunoprecipitated with CD98hc and β1 integrin antibodies from total cell lysate of HUVE cells, with IgG used as a negative control. (D) TIRF micrographs of control HUVE cells showing localization of TRPV4 (magenta), CD98hc (green) and β1 integrin (blue) in focal adhesions, as well as a merged image of all signals. (E) Confocal micrographs of the basal regions of HUVE cells showing proximity ligation foci (green) between all endogenous molecules, including β1 integrin and CD98hc, CD98hc and TRPV4, and TRPV4 and β1 integrin. Phalloidin staining of F-actin is shown in blue. (F) Confocal micrographs of basal regions of HUVE cells showing proximity ligation foci (green) between overexpressed CA-LDL−β1Δ1 (β1Δ1) or CA-LDL−β1Δ5 (β1Δ5) and endogenous CD98hc. Phalloidin staining of F-actin is shown in blue. (G) Scatter plot showing individual number of PLA dots per slice within basal slices of images from five fields of view per biological replicate (n=6; ****P<0.0001; bars indicate the mean with 95% CI). Scale bars: 5 μm (D), 20 μm (E,F).
We then performed a proximity ligation assay (PLA), which enables detection of specific protein-protein interactions within a 40 nm radius of the target protein (Fredriksson et al., 2002). When PLA was performed in HUVE cells overexpressing β1Δ1 or β1Δ5 mutants using anti-CA and anti-CD98hc antibodies, tight interactions between CD98hc and chimeric β1 integrin were found to be reduced in cells overexpressing (β1Δ5) (Fig. 1F,G). Thus, these three surface molecules appear to form a multimolecular complex in these endothelial cells.
Consistent with the co-immunoprecipitation results, analysis of protein distribution using total internal reflection fluorescence (TIRF) microscopy confirmed that endogenous TRPV4 colocalizes with both CD98hc and β1 integrin within spontaneously formed focal adhesions along the basal membrane of adherent HUVE cells (Fig. 1D). When PLA was performed using anti-TRPV4 or anti-β1 integrin antibody in combination with anti-CD98hc antibodies, TRPV4, CD98hc and β1 integrin were found to be tightly associated, primarily at the basal cell surface (Fig. 1E) where focal adhesions are located (Fig. 1D). Taken together, these findings confirm that β1 integrin, CD98hc and TRPV4 tightly co-associate with each other in common basal focal adhesions where they colocalize within human endothelial cells.
CD98hc is required for physical coupling of β1 integrin to TRPV4 in focal adhesions
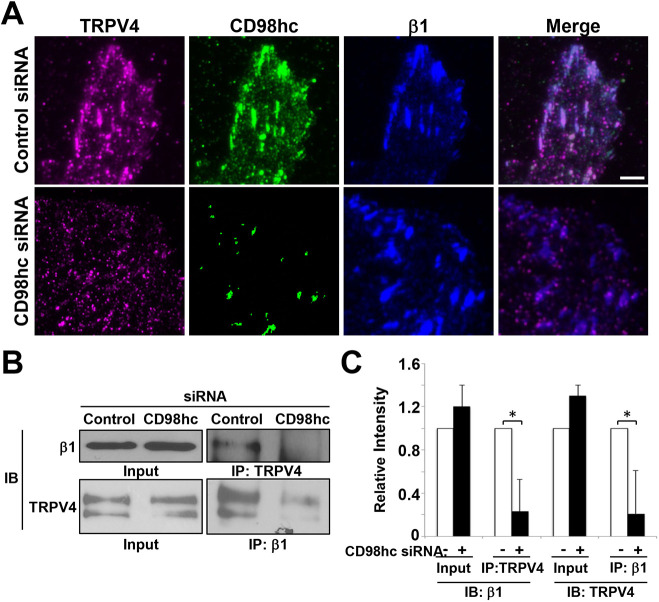
To determine whether CD98hc contributes to the mechanical linkage between β1 integrin and TRPV4, CD98hc was knocked down in HUVE cells using siRNA (Fig. S2A). This greatly reduced the amount of both CD98hc and TRPV4 within focal adhesions, although TRPV4 staining could still be detected at other regions of the cell membrane (Fig. 2A). This effect was specific, as knocking down CD98hc expression had no effect on recruitment of either integrin or vinculin to the focal adhesion (Fig. S2B). Also, CD98hc knockdown did not interfere with the formation of focal adhesions, as indicated by the absence of any suppression of cell migration in scratch wound assays (Fig. S2C). Importantly, although CD98hc was not required for focal adhesion formation, integrin recruitment to these sites or cell movement, the knockdown of CD98hc reduced the ability of β1 integrin and TRPV4 to co-associate with each other by almost 80%, as determined using a co-immunoprecipitation assay (Fig. 2B,C). These results indicate that CD98hc is required for physical coupling of β1 integrin to the subset of TRPV4 molecules that are located within focal adhesions.
Fig. 2.
CD98hc is not required for focal adhesion formation but is needed for recruitment of TRPV4. (A) TIRF micrographs of HUVE cells treated with control or CD98hc siRNA, showing localization of TRPV4 (magenta), CD98hc (green) and β1 integrin (blue) in focal adhesions, as well as a merged image of all signals. Scale bar: 5 μm. (B) Immunoblots showing β1 integrin immunoprecipitated with TRPV4 antibody or TRPV4 immunoprecipitated with β1 integrin antibody from lysates of HUVE cells treated with control or CD98hc siRNA. (C) Relative intensities of signals from the blots in B (n=3, all data shown are mean±s.e.m. *P<0.05).
Ankyrin-rich domains of TRPV4 mediate its association with CD98hc and β1 integrin
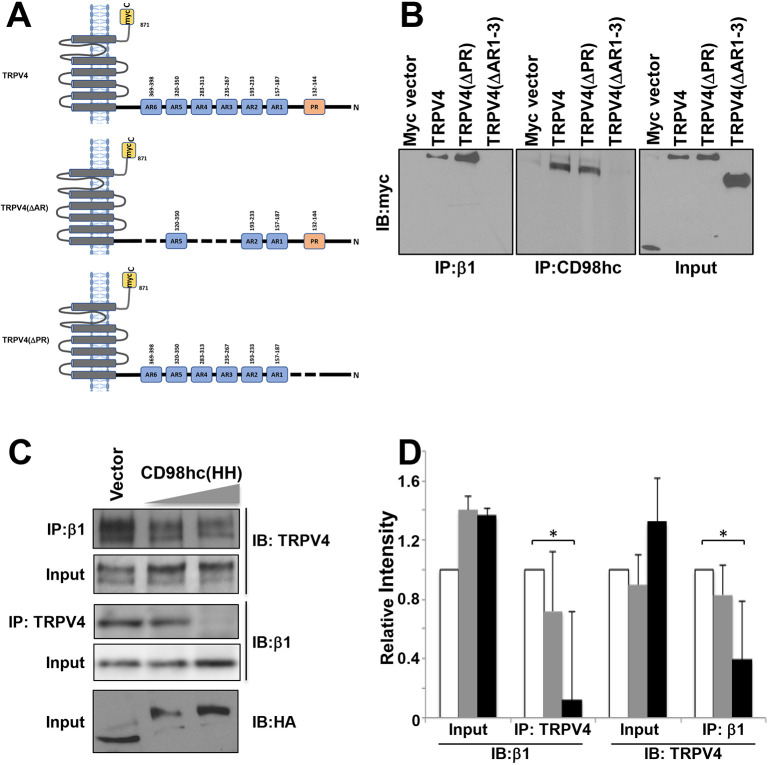
We next investigated the molecular path by which CD98hc mediates force transfer from β1 integrin to TRPV4. The highly conserved HH domain of CD98hc consists of 50 aa (160-210 aa), which span its transmembrane, extracellular juxtamembrane and intracellular juxtamembrane regions (Fig. S3A). Although the transmembrane and intracellular regions have been implicated in integrin activation (Yan et al., 2008; Fenczik et al., 2001), it is not known whether TRPV4 associates with this same domain or whether it interacts with a different region of the CD98hc protein. To better understand this mechanism, we used genetically engineered TRPV4 mutants in which the transmembrane [TRPV4(N)], C-terminal [TRPV4(ΔC)] or both transmembrane and N-terminal [TRPV4(C)] regions of the molecule were deleted to identify regions within TRPV4 that mediate its association with CD98hc (Fig. S3B). Removing the C-terminal or transmembrane domains did not alter the ability of TRPV4 to co-immunoprecipitate with either β1 integrin or CD98hc (Fig. S3C); deletion of the proline-rich domain in the TRPV4 N terminus [TRPV4(ΔPR)] also had no effect (Fig. 3B). However, deletion of three AR domains (235-267, 283-313 and 369-398 aa) in the N terminus of TRPV4 [TRPV4(ΔAR)] (Fig. 3A) was sufficient to completely inhibit association of TRPV4 with both CD98hc and β1 integrin (Fig. 3B). Furthermore, when increasing amounts of the CD98c HH domain [CD98hc (HH)] were overexpressed in HEK293T cells, it acted as a dominant negative protein and decreased the amount of β1 integrin that co-immunoprecipitated with TRPV4 in a dose-dependent manner (Fig. 3C,D). These results suggest that TRPV4 does not bind directly to the HH domain of CD98hc or β1 integrin tail, but rather to some other region of CD98hc that might be exposed via allosteric conformational changes upon its binding to β1 integrin. Alternatively, TRPV4 might bind directly to the HH domain, but such an interaction is not stable enough for co-immunoprecipitation.
Fig. 3.
CD98hc HH domain mediates the binding between β1 integrin and TRPV4. (A) Diagram of TRPV4 constructs showing transmembrane, proline-rich (PR), and ankyrin repeat (AR, blue) domains, as well as myc (yellow) tags. (B) Immunoblots showing myc-tagged TRPV4 constructs immunoprecipitated with β1 integrin (left) or CD98hc (middle) antibody in lysates from HEK293T cells transfected with the indicated DNA constructs. (C) Immunoblots showing β1 integrin and TRPV4 immunoprecipitated with β1 integrin and TRPV4 antibodies, respectively, in lysates from HEK293T cells transfected with increasing amounts (2.5, 5 µg) of the CD98hc(HH) domain construct. (D) Histogram showing the densitometric quantification of immunoblot signals from experiments described in C. White bars, vector control; grey bars, CD98hc(HH) low; black bars, CD98hc(HH) high (n=3, all data shown are mean±s.e.m. *P<0.05).
Molecular modeling of β1 integrin-CD98hc-TRPV4 interactions
Molecular dynamics simulation (MDS) was then used to identify the sites within CD98hc where TRPV4 binds. Because the crystal structure of the CD98hc cytoplasmic tail containing the HH domain is not available, PEP-FOLD3.0 MDS software was used to predict the structure of 46 aas adjacent to the HH domain [CD98hc(P121-S166)] de novo. In silico rigid docking was performed using the known crystal structure of the TRPV4 AR domain and the best fit model of CD98hc(P121-S166) using ClusPro2.0. Calculation of the frequency at which each residue in the human TRPV4 AR domain and CD98hc(P121-S166) forms H-bonds in the models analyzed during computational docking experiments (Table S1) predicted that the amino acid residues most crucial for CD98hc-TRPV4 complex formation are R224, E218, R219, K310, R316 and Y236 in the AR domain of TRVP4 and D152, E153, E155, K148, K141, K146 and K140 in CD98hc(P121-S166) (Fig. 4A; Table S1). A similar approach was used to determine the amino acid residues involved in the binding of the β1 integrin cytoplasmic tail to the HH domain of CD98hc (Fig. 4B; Table S2), which correspond to K794, Y795 and E796 of β1 integrin and E168, W179 and R181 in CD98hc.
Fig. 4.
Molecular modeling of β1-CD98hc-TRPV4 interactions. (A) Ribbon representation of the predicted complex between human TRPV4 (blue) and the fragment of CD98hc (P121-S166; green). Hydrogen bonds are shown as red broken lines. The crystal structure of human TRPV4 ankyrin was taken from PDB database (PDB ID: 4DX2). Simulation for the cytosolic domain of CD98hc (P121-S166) was performed by PEP-FOLD 3.0. CD98hc (P121-S166) was docked into the 3D structure of human TRPV4 using ClusPro 2.0. The highest scoring model with good topologies is shown. Enlarged stick representation to the right highlights the interacting residues between human TRPV4 and CD98hc (P121-S166). (B) Ribbon representation shows molecular modeling of complex between the C terminus of β1 integrin (N792-K798; red) and the fragment of CD98hc (K161-R210; green). Hydrogen bonds are shown as pink broken lines. Simulations for C terminus of β1 integrin (N792-K798) and the fragment of CD98hc (K161-R210) were performed using PEP-FOLD 3.0. Integrin (N792-K798) was docked into CD98hc (K161-R210) using ClusPro 2.0. All images were generated using PyMol. (C) Immunoblots showing FLAG-tagged CD98hc constructs immunoprecipitated with β1 integrin (bottom) or TRPV4 (top) antibody in lysates from untransfected (left) or HEK293T cells transfected with wild-type (middle) or mutant CD98hc constructs (right) (n=3 Co-IP experiments).
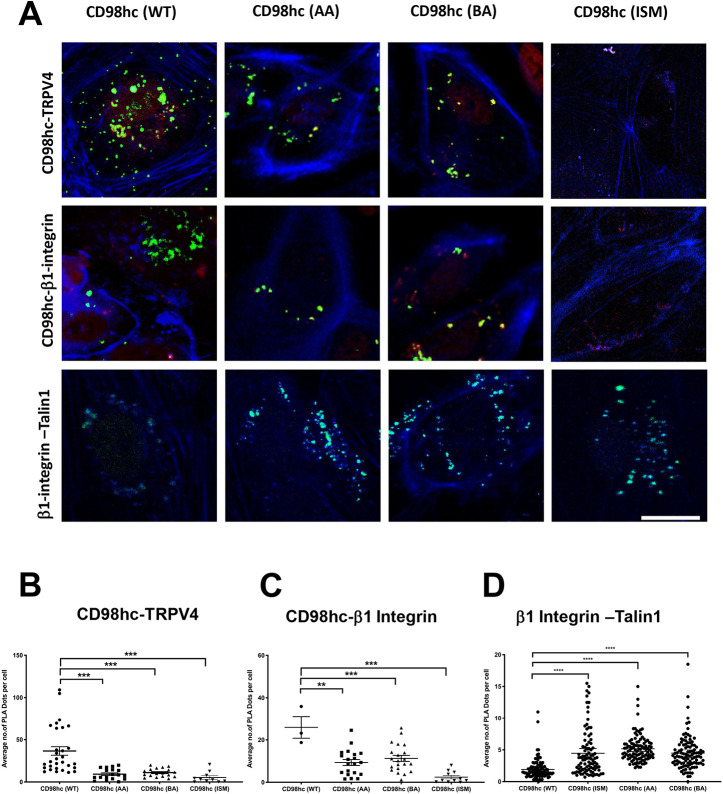
CD98hc constructs with specific point mutations were then engineered to validate these predictions experimentally (Fig. S1B). FLAG-tagged wild-type CD98c [CD98hc(WT)] or CD98hc containing mutated TRPV4-interacting acidic [CD98hc(AA)] or basic [CD98hc(BA)] residues, or altered β1 integrin-interacting residues [CD98hc(ISM)], were overexpressed in HEK293T cells, along with full-length β1 integrin and TRPV4. Immunoprecipitation using anti-FLAG antibody and immunoblotting with antibodies against TRPV4 or β1 integrin showed that CD98hc(ISM) failed to pull down TRPV4 or β1 integrin (Fig. 4C). Use of the proximity ligation assay demonstrated that expression of CD98hc containing either mutated TRPV4-interacting acidic or basic residues significantly decreased (P<0.001) the appearance of tight CD98hc-TRPV4 associations that normally appear evenly distributed across the apical and basal surface of HUVE cells (Fig. 5A,B). Thus, these residues appear to be required for physical associations between these molecules in situ. Similarly, expressing CD98hc containing mutated β1 integrin-interacting residues significantly decreased (P<0.001) the β1 integrin-CD98hc associations along the basal surface of HUVE cells (Fig. 5A,B). This decrease in β1 integrin-CD98hc binding interactions was accompanied by significantly increased (P<0.0001) association between β1 integrin and talin-1 along the basal cell surface, and similar results were obtained by overexpressing CD98hc containing mutated residues responsible for its interactions with TRPV4 (Fig. 5A,B).
Fig. 5.
Proximity profiling of β1-CD98hc-TRPV4 interactions. (A) Confocal micrographs of basal slices of HUVE cells showing proximity ligation foci (green) between FLAG-CD98hc wild type (WT), mutated acidic residues (AA) or mutated basic residues (BA) and endogenous TRPV4 (top row), endogenous β1 integrin (middle row) or endogenous β1 integrin and endogenous Talin1 (bottom row). Phalloidin staining of F-actin is shown in blue. Scale bar: 20 μm. (B) Scatter plots showing individual number of PLA dots per cell within basal sections of images from five fields of view per biological replicate, corresponding to experiments shown in A (n=3; **P<0.01, ***P<0.001, ****P<0.0001; bars indicate the mean with 95% CI).
CD98hc is required for mechanical activation of TRPV4
To explore the functional relevance of these findings, we knocked down CD98hc using specific siRNA in HUVE cells grown on flexible ECM-coated substrates (Flexcell dishes) to which static mechanical strain (15%) was applied briefly (4 s). This level of strain is physiologically relevant as it mimics levels experienced by lung endothelium during ventilator-induced lung injury. Inhibition of CD98hc expression resulted in reduced activation of calcium influx, whereas transfection with a control scrambled siRNA had no effect (Fig. 6A,B), as previously described (Matthews et al., 2010). Importantly, inhibiting this mechanically induced, integrin-dependent TRPV4 activation mechanism by knocking down CD98hc did not interfere with chemical stimulation of the channel by the known TRPV4 inducer, 4α-phorbol 12,13-didecanoate (4α-PDD) (Fig. 6A,B). Similar specific inhibition of mechanical, but not chemical, signaling through TRPV4 was demonstrated using an RGD peptide that inhibits integrin binding (Fig. S4). Moreover, overexpression of the dominant negative CD98hc HH domain (Fig. 6C), knockdown of CD98hc (Fig. 6C,D) or overexpression of wild-type CD98hc, but not point mutant CD98hc (ISM) (Fig. 6E,F), also altered cell physiology in a similar manner. The normal ability of the endothelial cells to reorient their shape and internal actin cytoskeleton in a perpendicular direction when exposed to uniaxial cyclic mechanical strain was inhibited, which was previously shown to be mediated by mechanical activation of TRPV4 through β1 integrin (Thodeti et al., 2009). Hence, the role of CD98hc in mechanically induced activation of calcium signaling through TRPV4 and regulation of endothelial cell physiology correlates directly with its ability to physically associate with TRPV4 through its cytoplasmic tail, and through neighboring sites within this same cytoplasmic region of the CD98hc molecule, which also mediates its recruitment into β1 integrin-containing focal adhesions (Fig. 1B).
Fig. 6.
CD98hc is required for mechanical activation of TRPV4. (A) Top graph shows relative changes in cytosolic calcium (Ca2+ F/F0) in Fluo-4-loaded HUVE cells treated with control or CD98hc siRNA in response to static 15% strain for 4 s (arrow indicates time of force application). Lower graph shows the effect of addition of the TRPV4 activator 4α-PDD (10 mM; arrow indicates time of addition) using the same assay. (B) Average maximum relative changes in cytosolic calcium (Max Ca2+ F/F0) in cells described in A (n=3, *P<0.05). (C) Histogram showing quantification of aligned cells (oriented 90°±30° relative to direction of stretch) under control conditions or after transfection with CD98hc(HH) or CD98hc siRNA (*P<0.05). (D) Phase contrast and fluorescence micrographs of cells stained for F-actin with AlexaFluor 488-phalloidin showing cell reorientation in HUVE cells treated with control or CD98hc siRNA in response to cyclic strain (15%, 1 Hz, 2 h). Arrows indicate the direction of applied strain. (E) Fluorescence micrographs of cells stained for F-actin showing cell orientation in response to application in uniaxial cyclic strain (11%, 1 Hz, 4 h) in HUVE cells transfected with CD98hc (WT) or CD98hc (ISM); arrows indicate direction of applied strain. (F) Scatter plot showing percentage of cells aligned (oriented 90°±30° relative to direction of stretch) for each field of view, from five fields of view per biological replicate (n=4; **P<0.01, bars indicate s.e.m.). Scale bars: 50 µm (D, top; E), 5 µm (D, bottom).
This form of transmembrane mechanotransduction through local mechanical force transfer across these three closely associated focal adhesion molecules helps to explain the observed rapidity (<4 ms) of mechanical activation of TRPV4 by forces applied to integrins and why calcium influx localizes to the focal adhesion where force is applied (Matthews et al., 2010). This mechanism is supported by our finding that integrin, CD98hc and TRPV4 bind directly together to form heteroprotein complexes within focal adhesions, but TRPV4 channel activation is also known to be modulated (i.e. enhanced), but not activated, by its phosphorylation state (Fan et al., 2009). Indeed, we found that mechanical deformation of the ECM substrate on which HUVE cells are cultured leads to increased phosphorylation of TRPV4 and that this can be inhibited by treatment with siRNA targeting CD98hc (Fig. S5), again confirming that mechanical force transfer within the focal adhesion is upstream of this signaling response.
DISCUSSION
Cells contain a tensionally integrated cytoskeleton that interacts physically with the ECM through specific mechanoreceptors, including β1 integrins (Wang et al., 1993; Ingber, 1997). Mechanical stresses can therefore determine cell and nuclear shape (Maniotis et al., 1997) and control numerous cellular functions, including growth, differentiation, migration and apoptosis (Chen et al., 1997), as well as stem cell lineage commitment (Moore et al., 2002; Ingber, 2005; Sanchez-Esteban et al., 2006; Mammoto and Ingber, 2009; Mammoto et al., 2011). Cellular mechanotransduction involves integrin-mediated force transmission between cells and the ECM, which results in force-induced structural changes in proteins, resulting in mechanochemical conversion and triggering subsequent mechanical signaling pathways (Geiger et al., 2009; Hoffman et al., 2011). Past studies have demonstrated that mechanical forces transmitted across the cell surface induce deformations in proteins, such as talin and vinculin, within focal adhesions (Choquet et al., 1997). However, little is known about the molecular mechanism of ion channel activation on the cell surface by mechanical forces applied to ECM that trigger mechano-electrical conversion processes, which are important for control of cell, tissue and organ physiology. The major advance of this study is the mapping of mechanical signal transfer across cell surface β1 integrins to TRVP4 ion channels, which are activated almost immediately (<4 ms) after force application to ECM adhesions (Matthews et al., 2010). This has important clinical relevance as this form of mechanical signaling through TRPV4 has been shown to be involved in diseases ranging from pulmonary edema (Willette et al., 2008) to cancer (Adapala et al., 2016).
Our results show that β1 integrin, CD98hc and TRPV4 colocalize and tightly associate within spontaneously formed mature focal adhesions at the base of the cell, which is consistent with past work showing similar co-association in nascent focal adhesions formed at the interface of ligand-coated beads that bind to cell surface β1 integrin receptors (Wang and Ingber, 1995; Matthews et al., 2006, 2010). However, although intact CD98hc was required for TRPV4 colocalization to focal adhesions, it was not necessary for the formation of these adhesions. This contrasts with a previous study that reported defective integrin-dependent cell spreading, defective cell migration and increased apoptosis in CD98hc null mouse embryonic stem cells and fibroblasts (Féral et al., 2005). This suggests that CD98hc might be essential for integrin-dependent survival signaling in actively migrating cells, but less so in stably anchored cells that formed mature focal adhesions. Moreover, our proximity ligation assay results similarly showed that close interactions between β1 integrin and CD98hc are restricted to focal adhesions, whereas CD98hc and TRPV4 associate tightly at sites distributed across the entire cell membrane. These findings suggest that β1 integrin availability is the limiting factor that controls recruitment of CD98hc-TRPV4 complexes into focal adhesions and formation of a heterocomplex containing these three transmembrane proteins.
Consistent with a past study, we found that the transmembrane and cytoplasmic juxtamembrane domains of CD98hc mediate CD98hc binding to β1 integrin (Cai et al., 2005). However, we extended these findings by demonstrating that the 50 aa HH domain within CD98hc is responsible for the ability of CD98hc to bind to the β1 integrin tail, and by identifying W179 and R181 as the key residues that mediate this interaction. In addition, we discovered that the TRPV4 AR domain is required for CD98hc-TRPV4 binding. This was based on our MDS studies, which predicted that residues E153, K148, D152, K141, K146, E155 and K161 in CD98hc interact with residues R224, E218, R219, K310, R316 and Y236 located in a pocket within the second AR domain of TRPV4. This pocket is close to the site where phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] binds and induces rearrangements in the cytosolic tail of TRPV4, thereby facilitating its activation by heat and osmotic stimuli (Garcia-Elias et al., 2013), whereas it is distinct from the site in the AR domain where binding of PI(4,5)P2 binding has been shown to reduce channel activity (Takahashi et al., 2014). It is important to note, however, that although our MDS models predict that β1 integrin, CD98hc and TRPV4 bind directly to one another, our experimental methods cannot rule out the possibility that other molecules are also involved in the formation of this mechanical signaling complex.
Talin-1 is also a key component of the integrin adhesome and binds to the tail of β integrin at two sites – the membrane proximal NPXY motif and an adjacent membrane proximal region (Wegener et al., 2007); another adhesome protein, kindlin, also binds to the membrane distal NxxY motif (Ma et al., 2008). MDS studies have revealed that kindlin interactions with the membrane distal NxxY motif of the integrin tail indirectly pushes talin-1 towards the membrane proximal region, helping it bind more strongly to integrin (Haydari et al., 2020; Moser et al., 2009). Our model suggests that CD98hc binds the integrin tail using the membrane distal NxxY motif as well. The proximity ligation data showing increased engagement of talin-1 by CD98hc mutants compared to wild-type CD98hc therefore suggest that CD98hc and kindlin compete for binding to the membrane distal NxxY motif. Further studies will be needed to clarify the role of CD98hc in kindlin-mediated regulation of talin-1 engagement of β integrin tails.
Various stimuli promote TRPV4 channel opening and they utilize multiple signaling pathways and second messenger systems (Vriens et al., 2016), and many chemical ligands that modulate TRPV4 activity target its transmembrane domain in areas close to the pore-forming region (White et al., 2016) (Fig. 7). In contrast, our results suggest that mechanical stresses transmitted across integrins alter TRPV4 activity by inducing conformational changes in the cytoplasmic AR domains of the molecules that associate with CD98hc, which are also sites where other physical stimuli, such as heat and osmotic changes, promote TRPV4 activation (Garcia-Elias et al., 2013). This hypothesis is further strengthened by our recent study in which we used AAV vectors to deliver the dominant negative HH domain of CD98hc, which blocked mechanical stimulus-induced calcium signaling in primary human microvascular endothelial cells (Li et al., 2019). The ultrarapid (<4 ms) activation of TRPV4 by forces applied to integrins appears to rule out the involvement of soluble second messenger systems that appear to be upstream mediators of TRVP4 activation by heat and osmotic stress (Gao et al., 2003; Garcia-Elias et al., 2013). This is also consistent with our finding that chemical activation of TRPV4 by 4α-PDD does not require CD98hc, further emphasizing the mechanospecificity of this β1 integrin-CD98hc-TRPV4 signaling axis.
Fig. 7.
Mapping the force transfer from β1 integrin through CD98hc to TRPV4. Under basal conditions, integrin binding to extracellular matrix initiates outside-in signaling within focal adhesions. CD98hc along with TRPV4 form part of the β1 integrin adhesome within a subset of focal adhesions.TRPV4 channels in these sites exhibit baseline activity due to numerous environmental stimuli, including temperature and resting tension on the channel through the CD98hc-β1 integrin axis. When the cells are treated with TRPV4 chemical agonists (e.g. 4α-PDD) that bind to the transmembrane domain of TRPV4 and activate the channel directly, calcium enters into the cell through the channel. When cells are subject to mechanical deformation, force is transferred from outside to inside through the β1 integrin-CD98hc-TRPV4 axis, resulting in further opening of TRPV4 channel and increased entry of calcium into the cell.
Alterations in mechanosensitive ion channel activity commonly result from changes in the 3D conformation of the channel that alter its opening and closing kinetics (Ranade et al., 2015), and this appears to be sensitized (further enhanced) by kinase-induced phosphorylation of TRPV4 (Fan et al., 2009). In this study, we found that mechanical forces are transferred between integrin, CD98hc and TRPV4 and produce conformational changes that may directly influence TRPV4 channel activity, which is consistent with the rapidity of this response (<4 ms). We also show that knocking down CD98hc inhibits TRPV4 phosphorylation and, thus, either force transfer to TRPV4 or its recruitment to focal adhesions is required for this kinase-mediated enhancement response. Taken together, these results suggest that mechanical forces transferred across the integrin-CD98hc-TRPV4 axis modulate TRPV4 channel activity, in part through transfer of mechanical forces that induce a conformational change in TRPV4 and make it susceptible to modification by an associated kinase. Alternatively, TRPV4 may need to be present within focal adhesions for this modulating phosphorylation to take place. If this kinase always tightly associates with TRPV4, then mechanical force transfer to the kinase via the tightly associated multimolecular complex containing TRPV4, CD98hc and β1 integrin could also result in conformation changes in the kinase that increase its activity and, hence, its ability to phosphorylate TRPV4. One of the many kinases that associate with the cytoplasmic portion of β1 integrin (e.g. FAK, Src, ILK, etc.) in focal adhesions could phosphorylate TRPV4 in response to mechanical strain application, but the kinetics of FAK and Src activation are much slower in response to mechanical force application to integrins than activation of TRPV4, and TRPV4 was shown to be upstream of FAK activation (Thodeti et al., 2009). If ILK or another integrin-associated kinase mediates this activation, then it must be triggered directly by mechanical deformation of integrins or one of its associated molecules given the incredible rapidity of the response. Serine/threonine kinases also could be involved because protein kinase C and protein kinase A have been similarly shown to enhance activation of the TRPV4 ion channel by inducing its phosphorylation, and this depends on assembly of these two kinases into a signaling complex with TRPV4 (Fan et al., 2009). However, it is important to reemphasize that this phosphorylation mechanism is not involved in direct mechanical activation of TRPV4, but rather only in amplification of this response once triggered mechanically.
A direct mechanism of force transfer between surface receptor (β1 integrin), bound linker protein (CD98hc) and mechanochemical transducer molecule (TRPV4) within the focal adhesion can explain why this integrin-specific mechanotransduction mechanism is so fast, occurring within a few milliseconds rather than seconds or minutes observed for most other integrin-associated mechanochemical conversion mechanisms in nonspecialized (non-mechanosensor) cells. Because force-dependent activation of TRPV4 depends on mechanical strain and not isometric tension, this arrangement also explains why partially decreasing the pre-stress (isometric tension) in the cytoskeleton can enhance force-dependent distortion of this multiprotein complex (Matthews et al., 2010) and, hence, how cytoskeletal tension can ‘tune’ the ultimate mechanochemical conversion response (Wang et al., 2009; Mammoto and Ingber, 2009). TRPV4 and PIEZO1 play important roles during normal mechanoelectrical transduction in response to substrate deflections, but PIEZO1 alone is involved in membrane stretch-mediated channel gating (Servin-Vences et al., 2017). Together with our findings, this suggests that mechanical force transferred from tethered integrins through CD98hc may gate TRPV4 channels within focal adhesions, whereas forces transferred through membrane lipids gate PIEZO1 channels. Finally, our findings that CD98hc siRNA and integrin antagonists inhibit TRPV4 activation induced by mechanical strain application through cell-ECM adhesions, but not by the chemical activator 4α-PDD that stimulates TRPV4 independently of protein kinase C (Vriens et al., 2016), suggest that these two TRPV4 activation pathways are distinct. This raises the possibility of developing mechanotherapeutics that specifically interfere with mechanical induction of TRPV4 without inhibiting its activation by physiological chemical cues, thereby producing more effective and specific therapeutics for treatment of various diseases, including pulmonary edema.
MATERIALS AND METHODS
Reagents
Key reagents are listed in Table S3.
Experimental model and subject details
Pooled human umbilical vein endothelial (HUVE) cells were obtained from Lonza and HEK293T cells were obtained from the American Type Culture Collection (ATCC). HUVE cells were cultured in endothelial growth medium supplemented with 5% fetal bovine serum (FBS) and growth factors (EGM-2; Lonza). HEK293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS and 100 units/ml penicillin.
Method details
Plasmid construction
CD98hc-HA, CD98hc(ΔHH)-HA and CD98hc(HH) were constructed using RT-PCR with cDNA from HUVE cells, and subcloned into pCMV-HA vector (Clontech) at the SalI/XhoI sites. TRPV4-myc, TRPV4(ΔPR)-myc and TRPV4(ΔAR1-3)-myc were constructed using PCR with template plasmids from Open Biosystems subcloned into pCMV-myc vector (Clontech) at the SalI/XhoI sites. FLAG-tagged CD98hc(WT) was constructed by cloning a synthesized gene block containing the sequence corresponding to amino acids 1-210 of CD98hc and dsredexpress2 connected by a T2A linker into the NotI-EcoRI sites of pcDNA3.1(-)zeo. FLAG-tagged CD98hc(ISM), CD98hc(AA) and CD98hc(BA) mutant constructs were constructed by site-directed mutagenesis of CD98hc(WT) at the specific residues listed in Fig. S1B.
Protein colocalization experiments
Colocalization experiments were performed on HUVE cells seeded on fibronectin-coated (5 µg/ml; R&D Systems) No. 1 glass bottom dishes (Mattek) and maintained in culture for 24 h. Cells were fixed with 4% PFA for 30 min, permeabilized with 0.1% PBS-T for 5 min and blocked with blocking buffer (0.03% PBS-T containing 10% donor donkey serum) for 1 h at room temperature. Cells were incubated with the above-listed combinations of anti-β1 integrin (RRID:AB_2128060 at 1:200), anti-CD98hc (RRID:AB_638284 at 1:50) and anti-TRPV4 (RRID:AB_2040264 at 1:100) overnight at 4°C. Cells were washed and stained with appropriate secondary antibodies and imaged using an inverted laser scanning confocal microscope (TCS SP5 X; Leica) and Leica Application Software (LAS; Leica) or using a Zeiss TIRF/LSM 710 confocal microscope equipped with a Hamamatsu ImagEM-1K Back Thinned EMCCD camera controlled by ZEN imaging software (Zeiss).
Gene overexpression and knockdown
HUVE cells were transfected using silentFect Lipid Reagent (BioRad) for siRNA studies or Targefect-HUVEC (Targeting Systems) for wild-type and mutant construct overexpression studies, according to the manufacturer's instructions. HEK293T cells were transfected using Lipofectamine 2000 according to the manufacturer's instructions. Our studies involving siRNA-mediated knockdown were performed in HUVE cells transfected with 50 nM of siRNA against CD98hc or scrambled siRNA duplex (Qiagen). Wild-type and mutant construct overexpression studies were carried out in HUVE cells seeded on glass-bottomed eight-well chamber slides, transfected with 0.5 µg of corresponding plasmid DNA per well and fixed 36 h after transfection for immunofluorescent staining and imaging. For gene overexpression studies, HEK293T cells were seeded in 10 cm cell culture dishes, transfected with 10 µg of corresponding plasmid DNA per dish and lysed 48 h after transfection.
Co-immunoprecipitation studies
HUVE cells were extracted in ice-cold Triton buffer (50 mM Tris-HCl, pH 7.4, containing 150 mM NaCl, 1% Triton X-100 and 5 mM EDTA). Cell extracts were centrifuged at 10,000×g for 15 min at 4°C and incubated with magnetic beads (Dynabeads, Invitrogen) conjugated with specific antibodies at 4°C for 2 h. After washing the beads with Triton buffer, immunoprecipitated protein was detected by immunoblotting. HEK293T cells were extracted using the above-mentioned ice-cold Triton buffer, sonicated (30% amplitude, two pulses of 10 s each), centrifuged, precleared with control IgG beads for 15 min and incubated with anti-FLAG agarose beads (RRID: AB_10063035) at 4°C for 1 h. Beads were washed three times with ice-cold Triton buffer and immunoprecipitated protein detected by immunoblotting.
Molecular dynamics simulation
The three-dimensional (3D) structures of the C-terminal tail of β1 integrin (N792-K798) and the cytoplasmic fragments of CD98hc (P121-S166) and CD98hc (K161-R210) were predicted by PEP-FOLD 3.0 (http://bioserv.rpbs.univ-paris-diderot.fr/services/PEP-FOLD) (Thevenet et al., 2012). In this method, models were clustered by the sOPEP energy value (the coarse-grained energy) and then ranked based on their cluster scores. Among the top five ranks, the models with best scores were selected for docking experiments.
Molecular docking procedure
Docking experiments were performed using the ClusPro 2.0 program (Comeau et al., 2004a,b). The 3D crystal structure of human TRPV4 ankyrin (PDB ID: 4DX2) was obtained from the PDB Data Bank. The simulated model of CD98hc (P121-S166) was then docked into the human TRPV4 to generate predicted binding models of CD98hc (P121-S166)–TRPV4. The simulated model of the C-terminal tail of β1 integrin (N792-K798) was docked into the cytoplasmic fragment of CD98hc (K161-R210) to generate predicted binding models of β1 integrin (N792-K798)-CD98hc (K161-R210). Models with the highest scores and good topologies were selected for the proposed models of the interaction between TRPV4 and CD98hc, β1 integrin and CD98hc.
Proximity ligation assay
HUVE cells seeded on IBIDI µ-slide were transfected with wild-type and mutant CD98hc constructs using Targefect-HUVEC reagent as per manufacturer's recommendation. Cells were fixed 36 h after transfection with 4% PFA for 30 min at room temperature. The proximity ligation assay was performed using mouse or rabbit anti-FLAG, mouse anti-β1 integrin and rabbit anti-TRPV4 antibodies as per manufacturer's protocol. Wells were mounted with ProLong Glass Antifade and confocal microscopy performed using Zeiss LSM 880 airyscan or inverted laser scanning confocal microscope (TCS SP5 X, Leica). Images were analyzed using Cell Profiler software (Carpenter et al., 2006).
Wound scratch assay
For the scratch assay, HUVE cells were seeded on fibronectin-coated (5 µg/ml; R&D Systems) No. 1 glass-bottomed dishes (Mattek) and maintained (2-3 days) in culture and until confluent for 24 h. Wounds were created using plastic pipette tips (200 µl; Denville) and images of the scratched monolayers were captured at the indicated time points using a Nikon Eclipse TE200 inverted microscope equipped with an RT Monochrome SPOT camera controlled using Spot imaging software (v4.0.9, Diagnostics Instruments).
Calcium imaging
HUVE cells were seeded on stageFlexer-collagen type IV-membranes (Cat# SFM-C-IV-Pack, Flexcellint) and, 24 h later, incubated with Fluo-4 dye for 45 min at 37°C as per manufacturer's recommendations. Cells were washed with live cell imaging solution (Cat#A14291DJ, Thermofisher), mounted onto a Flexcell StageFlexer device (Flexcellint, Cat# SF-3000), mechanical strain was applied using Flexcell FX-4000 Tension System (Cat# FX-4000T, Flexcellint) and calcium imaging performed using a Nikon eclipse upright microscope fitted with a Hamamatsu EMCCD camera. For mechanical strain-induced phosphorylation studies, cells were grown on UniFlex culture plates coated with collagen type I (Cat# UF-4001C, Flexcellint) and mechanical strain was applied using the Flexcell FX-4000 Tension System (Cat# FX-4000T, Flexcellint). Cells were lysed, immunoprecipitated and immunoblotted for phosphorylated proteins using pan anti-phosphoserine antibody (RRID:AB_11210897).
Cell reorientation experiments
HUVE cells were seeded on collagen type I-coated uniflex plates (Flexcellint) and 24 h later transfected with various plasmids or siRNA. siRNA-transfected cells were subject to cyclic strain (15%, 1 Hz, 2 h) at 24 h post transfection. Plasmid transfected cells were subject to cyclic strain (11%, 1 Hz, 4 h) at 36 h post transfection. The percentage of perpendicularly oriented cells was measured using Cell Profiler software and defined by an orientation of 90°±30° to the x-axis (the direction of strain).
Quantification and statistical analysis
All values are expressed as the mean±standard error of the mean (s.e.m.) or mean±95% confidence interval mean (95% CI). All experiments were repeated at least three times. Statistical comparisons were performed using Welch's t-test for experiments with two conditions or ANOVA for experiments with more than two conditions, using GraphPad Prism.
Supplementary Material
Acknowledgements
We thank Thomas Ferrante and Sasan Jalili-Firoozinezhad for assistance with TIRF microscopy, Aram Ghalili for assistance with proximity ligation assay and Oren levy for valuable guidance and critic in manuscript preparation.
Footnotes
Competing interests
D.E.I. holds equity in Emulate, Inc. and chairs its Advisory Board; he also holds multiple patents that are licensed to the company.
Author contributions
Conceptualization: R.P., M.H.-K., A.M., B.D.M., D.E.I.; Methodology: R.P., M.H.-K., H.W., H.C., B.D.M.; Software: R.P., H.W., B.D.M.; Validation: R.P., M.H.-K., H.W., B.D.M.; Formal analysis: R.P., M.H.-K.; Investigation: M.H.-K., A.M., B.D.M., D.E.I.; Resources: A.M., B.D.M., D.E.I.; Data curation: R.P., M.H.-K.; Writing - original draft: R.P.; Writing - review & editing: R.P., A.M., D.E.I.; Visualization: R.P.; Supervision: H.C., A.M., D.E.I.; Project administration: A.M.; Funding acquisition: M.H.-K., B.D.M., D.E.I.
Funding
This study was supported by research grants from the National Institutes of Health (R01-EB020004 to D.E.I and R01-HL146134, R01-HL141853, R01-HL130845, R01-HL093242 to H.C.) and fellowships from the UEHARA Memorial Foundation and the Japan Society for the Promotion of Science (JSPS) to M.H.-K. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at https://jcs.biologists.org/lookup/doi/10.1242/jcs.248823.supplemental
Peer review history
The peer review history is available online at https://jcs.biologists.org/lookup/doi/10.1242/jcs.248823.reviewer-comments.pdf
References
- Adapala R. K., Thoppil R. J., Ghosh K., Cappelli H. C., Dudley A. C., Paruchuri S., Keshamouni V., Klagsbrun M., Meszaros J. G., Chilian W. M. et al. (2016). Activation of mechanosensitive ion channel TRPV4 normalizes tumor vasculature and improves cancer therapy. Oncogene 35, 314-322. 10.1038/onc.2015.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S., Bulus N., Fonseca-Siesser P. M., Chen D., Hanks S. K., Pozzi A. and Zent R. (2005). CD98 modulates integrin beta1 function in polarized epithelial cells. J. Cell Sci. 118, 889-899. 10.1242/jcs.01674 [DOI] [PubMed] [Google Scholar]
- Carpenter A. E., Jones T. R., Lamprecht M. R., Clarke C., Kang I. H., Friman O., Guertin D. A., Chang J. H., Lindquist R. A., Moffat J. et al. (2006). CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100 10.1186/gb-2006-7-10-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. S., Mrksich M., Huang S., Whitesides G. M. and Ingber D. E. (1997). Geometric control of cell life and death. Science (New York N.Y.) 276, 1425-1428. 10.1126/science.276.5317.1425 [DOI] [PubMed] [Google Scholar]
- Choquet D., Felsenfeld D. P. and Sheetz M. P. (1997). Extracellular matrix rigidity causes strengthening of integrin-cytoskeleton linkages. Cell 88, 39-48. 10.1016/S0092-8674(00)81856-5 [DOI] [PubMed] [Google Scholar]
- Comeau S. R., Gatchell D. W., Vajda S. and Camacho C. J. (2004a). ClusPro: a fully automated algorithm for protein-protein docking. Nucleic Acids Res. 32, W96-W99. 10.1093/nar/gkh354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeau S. R., Gatchell D. W., Vajda S. and Camacho C. J. (2004b). ClusPro: an automated docking and discrimination method for the prediction of protein complexes. Bioinformatics 20, 45-50. 10.1093/bioinformatics/btg371 [DOI] [PubMed] [Google Scholar]
- Fan H.-C., Zhang X. and Mcnaughton P. A. (2009). Activation of the TRPV4 ion channel is enhanced by phosphorylation. J. Biol. Chem. 284, 27884-27891. 10.1074/jbc.M109.028803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenczik C. A., Zent R., Dellos M., Calderwood D. A., Satriano J., Kelly C. and Ginsberg M. H. (2001). Distinct domains of CD98hc regulate integrins and amino acid transport. J. Biol. Chem. 276, 8746-8752. 10.1074/jbc.M011239200 [DOI] [PubMed] [Google Scholar]
- Féral C. C., Nishiya N., Fenczik C. A., Stuhlmann H., Slepak M. and Ginsberg M. H. (2005). CD98hc (SLC3A2) mediates integrin signaling. Proc. Natl. Acad. Sci. USA 102, 355-360. 10.1073/pnas.0404852102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Féral C. C., Zijlstra A., Tkachenko E., Prager G., Gardel M. L., Slepak M. and Ginsberg M. H. (2007). CD98hc (SLC3A2) participates in fibronectin matrix assembly by mediating integrin signaling. J. Cell Biol. 178, 701-711. 10.1083/jcb.200705090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksson S., Gullberg M., Jarvius J., Olsson C., Pietras K., Gústafsdóttir S. M., Östman A. and Landegren U. (2002). Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 20, 473-477. 10.1038/nbt0502-473 [DOI] [PubMed] [Google Scholar]
- Gao X., Wu L. and O'neil R. G. (2003). Temperature-modulated diversity of TRPV4 channel gating: activation by physical stresses and phorbol ester derivatives through protein kinase C-dependent and -independent pathways. J. Biol. Chem. 278, 27129-27137. 10.1074/jbc.M302517200 [DOI] [PubMed] [Google Scholar]
- Garcia-Elias A., Mrkonjić S., Pardo-Pastor C., Inada H., Hellmich U. A., Rubio-Moscardó F., Plata C., Gaudet R., Vicente R. and Valverde M. A. (2013). Phosphatidylinositol-4,5-biphosphate-dependent rearrangement of TRPV4 cytosolic tails enables channel activation by physiological stimuli. Proc. Natl. Acad. Sci. USA 110, 9553-9558. 10.1073/pnas.1220231110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger B., Spatz J. P. and Bershadsky A. D. (2009). Environmental sensing through focal adhesions. Nat. Rev. Mol. Cell Biol. 10, 21-33. 10.1038/nrm2593 [DOI] [PubMed] [Google Scholar]
- Goswami C., Kuhn J., Heppenstall P. A. and Hucho T. (2010). Importance of non-selective cation channel TRPV4 interaction with cytoskeleton and their reciprocal regulations in cultured cells. PLoS ONE 5, e11654 10.1371/journal.pone.0011654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydari Z., Shams H., Jahed Z. and Mofrad M. R. K. (2020). Kindlin assists talin to promote integrin activation. Biophys. J. 118, 1977-1991. 10.1016/j.bpj.2020.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman B. D., Grashoff C. and Schwartz M. A. (2011). Dynamic molecular processes mediate cellular mechanotransduction. Nature 475, 316-323. 10.1038/nature10316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh D., Leslie D. C., Matthews B. D., Fraser J. P., Jurek S., Hamilton G. A., Thorneloe K. S., Mcalexander M. A. and Ingber D. E. (2012). A human disease model of drug toxicity–induced pulmonary edema in a lung-on-a-chip microdevice. Sci. Transl. Med. 4, 159ra147 10.1126/scitranslmed.3004249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingber D. E. (1997). Tensegrity: the architectural basis of cellular mechanotransduction. Annu. Rev. Physiol. 59, 575-599. 10.1146/annurev.physiol.59.1.575 [DOI] [PubMed] [Google Scholar]
- Ingber D. E. (2003). Mechanobiology and diseases of mechanotransduction. Ann. Med. 35, 564-577. 10.1080/07853890310016333 [DOI] [PubMed] [Google Scholar]
- Ingber D. E. (2005). Mechanical control of tissue growth: function follows form. Proc. Natl. Acad. Sci. USA 102, 11571-11572. 10.1073/pnas.0505939102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnikova T. V., Mannion B. A., Berditchevski F. and Hemler M. E. (2001). Beta1 integrins show specific association with CD98 protein in low density membranes. BMC Biochem. 2, 10 10.1186/1471-2091-2-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari S., Kumar A., Sardar P., Yadav M., Majhi R. K., Kumar A. and Goswami C. (2015). Influence of membrane cholesterol in the molecular evolution and functional regulation of TRPV4. Biochem. Biophys. Res. Commun. 456, 312-319. 10.1016/j.bbrc.2014.11.077 [DOI] [PubMed] [Google Scholar]
- Li J., Wen A. M., Potla R., Benshirim E., Seebarran A., Benz M. A., Henry O. Y. F., Matthews B. D., Prantil-Baun R., Gilpin S. E. et al. (2019). AAV-mediated gene therapy targeting TRPV4 mechanotransduction for inhibition of pulmonary vascular leakage. APL Bioeng 3, 046103 10.1063/1.5122967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y.-Q., Qin J., Wu C. and Plow E. F. (2008). Kindlin-2 (Mig-2): a co-activator of beta3 integrins. J. Cell Biol. 181, 439-446. 10.1083/jcb.200710196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammoto A. and Ingber D. E. (2009). Cytoskeletal control of growth and cell fate switching. Curr. Opin. Cell Biol. 21, 864-870. 10.1016/j.ceb.2009.08.001 [DOI] [PubMed] [Google Scholar]
- Mammoto T., Mammoto A., Torisawa Y.-S., Tat T., Gibbs A., Derda R., Mannix R., De Bruijn M., Yung C. W., Huh D. et al. (2011). Mechanochemical control of mesenchymal condensation and embryonic tooth organ formation. Dev. Cell 21, 758-769. 10.1016/j.devcel.2011.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniotis A. J., Chen C. S. and Ingber D. E. (1997). Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc. Natl. Acad. Sci. USA 94, 849-854. 10.1073/pnas.94.3.849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews B. D., Overby D. R., Mannix R. and Ingber D. E. (2006). Cellular adaptation to mechanical stress: role of integrins, Rho, cytoskeletal tension and mechanosensitive ion channels. J. Cell Sci. 119, 508-518. 10.1242/jcs.02760 [DOI] [PubMed] [Google Scholar]
- Matthews B. D., Thodeti C. K., Tytell J. D., Mammoto A., Overby D. R. and Ingber D. E. (2010). Ultra-rapid activation of TRPV4 ion channels by mechanical forces applied to cell surface β1 integrins. Integr. Biol. 2, 435-442. 10.1039/c0ib00034e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore K. A., Huang S., Kong Y., Sunday M. E. and Ingber D. E. (2002). Control of embryonic lung branching morphogenesis by the Rho activator, cytotoxic necrotizing factor 1. J. Surg. Res. 104, 95-100. 10.1006/jsre.2002.6418 [DOI] [PubMed] [Google Scholar]
- Moser M., Legate K. R., Zent R. and Fässler R. (2009). The tail of integrins, talin, and kindlins. Science 324, 895-899. 10.1126/science.1163865 [DOI] [PubMed] [Google Scholar]
- Ranade S. S., Syeda R. and Patapoutian A. (2015). Mechanically activated ion channels. Neuron 87, 1162-1179. 10.1016/j.neuron.2015.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Esteban J., Wang Y., Filardo E. J., Rubin L. P. and Ingber D. E. (2006). Integrins beta1, alpha6, and alpha3 contribute to mechanical strain-induced differentiation of fetal lung type II epithelial cells via distinct mechanisms. Am. J. Physiol. Lung Cell. Mol. Physiol. 290, L343-L350. 10.1152/ajplung.00189.2005 [DOI] [PubMed] [Google Scholar]
- Servin-Vences M. R., Moroni M., Lewin G. R. and Poole K. (2017). Direct measurement of TRPV4 and PIEZO1 activity reveals multiple mechanotransduction pathways in chondrocytes. Elife 6, e21074 10.7554/eLife.21074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N., Hamada-Nakahara S., Itoh Y., Takemura K., Shimada A., Ueda Y., Kitamata M., Matsuoka R., Hanawa-Suetsugu K., Senju Y. et al. (2014). TRPV4 channel activity is modulated by direct interaction of the ankyrin domain to PI(4,5)P₂. Nat. Commun. 5, 4994 10.1038/ncomms5994 [DOI] [PubMed] [Google Scholar]
- Thevenet P., Shen Y., Maupetit J., Guyon F., Derreumaux P. and Tuffery P. (2012). PEP-FOLD: an updated de novo structure prediction server for both linear and disulfide bonded cyclic peptides. Nucleic Acids Res. 40, W288-W293. 10.1093/nar/gks419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thodeti C. K., Matthews B., Ravi A., Mammoto A., Ghosh K., Bracha A. L. and Ingber D. E. (2009). TRPV4 channels mediate cyclic strain-induced endothelial cell reorientation through integrin-to-integrin signaling. Circ. Res. 104, 1123-1130. 10.1161/CIRCRESAHA.108.192930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorneloe K. S., Cheung M., Bao W., Alsaid H., Lenhard S., Jian M.-Y., Costell M., Maniscalco-Hauk K., Krawiec J. A., Olzinski A. et al. (2012). An orally active TRPV4 channel blocker prevents and resolves pulmonary edema induced by heart failure. Sci. Transl. Med. 4, 159ra148 10.1126/scitranslmed.3004276 [DOI] [PubMed] [Google Scholar]
- Vriens J., Watanadapala R. K., Thoppil R. J., Ghosh K., Cappelli H. C., Dudley A. C., Paruchuri S., Keshamouni V., Klagsbrun M., Meszaros J. G., et al. (2016). Activation of mechanosensitive ion channel TRPV4 normalizes tumor vasculature and improves cancer therapy. Oncogene 35, 314-322. 10.1038/onc.2015.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegener K. L., Partridge A. W., Han J., Pickford A. R., Liddington R. C., Ginsberg M. H. and Campbell I. D. (2007). Structural basis of integrin activation by talin. Cell 128, 171-182. 10.1016/j.cell.2006.10.048 [DOI] [PubMed] [Google Scholar]
- Willette R. N., Bao W., Nerurkar S., Yue T.-L., Doe C. P., Stankus G., Turner G. H., Ju H., Thomas H., Fishman C. E. et al. (2008). Systemic activation of the transient receptor potential vanilloid subtype 4 channel causes endothelial failure and circulatory collapse: Part 2. J. Pharmacol. Exp. Ther. 326, 443-452. 10.1124/jpet.107.134551 [DOI] [PubMed] [Google Scholar]
- Wang N. and Ingber D. E. (1995). Probing transmembrane mechanical coupling and cytomechanics using magnetic twisting cytometry. Biochem. Cell Biol. 73, 327-335. 10.1139/o95-041 [DOI] [PubMed] [Google Scholar]
- Wang N., Butler J. P. and Ingber D. E. (1993). Mechanotransduction across the cell surface and through the cytoskeleton. Science (New York N.Y.) 260, 1124-1127. 10.1126/science.7684161 [DOI] [PubMed] [Google Scholar]
- Wang N., Tytell J. D. and Ingber D. E. (2009). Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat. Rev. Mol. Cell Biol. 10, 75-82. 10.1038/nrm2594 [DOI] [PubMed] [Google Scholar]
- White J. P., Cibelli M., Urban L., Nilius B., Mcgeown J. G. and Nagy I. (2016). TRPV4: molecular conductor of a diverse orchestra. Physiol. Rev. 96, 911-973. 10.1152/physrev.00016.2015 [DOI] [PubMed] [Google Scholar]
- Yan Y., Vasudevan S., Nguyen H. T. and Merlin D. (2008). Intestinal epithelial CD98: an oligomeric and multifunctional protein. Biochim. Biophys. Acta 1780, 1087-1092. 10.1016/j.bbagen.2008.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zent R., Fenczik C. A., Calderwood D. A., Liu S., Dellos M. and Ginsberg M. H. (2000). Class- and splice variant-specific association of CD98 with integrin β cytoplasmic domains. J. Biol. Chem. 275, 5059-5064. 10.1074/jbc.275.7.5059 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.