Abstract
Background
Management and control of coronavirus disease 2019 (COVID-19) relies on reliable diagnostic testing.
Objectives
To evaluate the diagnostic test accuracy (DTA) of nucleic acid amplification tests (NAATs) for the diagnosis of coronavirus infections.
Data sources
PubMed, Web of Science, the Cochrane Library, Embase, Open Grey and conference proceeding until May 2019. PubMed and medRxiv were updated for COVID-19 on 31st August 2020.
Study eligibility
Studies were eligible if they reported on agreement rates between different NAATs using clinical samples.
Participants
Symptomatic patients with suspected upper or lower respiratory tract coronavirus infection.
Methods
The new NAAT was defined as the index test and the existing NAAT as reference standard. Data were extracted independently in duplicate. Risk of bias was assessed using the Quality Assessment of Diagnostic Accuracy Studies 2 tool. Confidence regions (CRs) surrounding summary sensitivity/specificity pooled by bivariate meta-analysis are reported. Heterogeneity was assessed using meta-regression.
Results
Fifty-one studies were included, 22 of which included 10 181 persons before COVID-19 and 29 including 8742 persons diagnosed with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The overall summary sensitivity was 89.1% (95%CR 84.0–92.7%) and specificity 98.9% (95%CR 98.0–99.4%). Nearly all the studies evaluated different PCRs as both index and reference standards. Real-time RT PCR assays resulted in significantly higher sensitivity than other tests. Reference standards at high risk of bias possibly exaggerated specificity. The pooled sensitivity and specificity of studies evaluating SARS-COV-2 were 90.4% (95%CR 83.7–94.5%) and 98.1% (95%CR 95.9–99.2), respectively. SARS-COV-2 studies using samples from the lower respiratory tract, real-time RT-PCR, and tests targeting the N or S gene or more than one gene showed higher sensitivity, and assays based on reverse transcriptase loop-mediated isothermal amplification (RT-LAMP), especially when targeting only the RNA-dependent RNA polymerase (RdRp) gene, showed significantly lower sensitivity compared to other studies.
Conclusions
Pooling all studies to date shows that on average 10% of patients with coronavirus infections might be missed with PCR tests. Variables affecting sensitivity and specificity can be used for test selection and development.
Keywords: Acute respiratory tract infection, Coronavirus, COVID-19, Nucleic acid amplification tests, SARS-CoV-2
Introduction
Six coronaviruses (CoVs) have been identified as infectious to humans. The α-CoVs HCoV-229E and HCoV-NL63 and the β-CoVs HCoV-HKU1 and HCoV-OC43 have low pathogenicity and cause mild respiratory symptoms similar to those of the common cold. The other two β-CoVs—severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV)—and the current severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can lead to severe and potentially fatal respiratory tract infections.
The accuracy of tests to diagnose coronavirus infections is crucial for patient management and to control the pandemic. The SARS-CoV-2 real-time reverse-transcriptase (RT)-PCR tests were developed under emergency conditions, and were based on analytic performance in the laboratory and not in real-life conditions. Several tests are currently available, most targeting the nucleocapsid protein (N) or spike protein (S) genes, combining them with the envelope protein gene (E) or the RNA-dependent RNA polymerase gene (RdRP). The N gene provided lower analytical sensitivity (technical limit of detection of 8.3 copies) than the RdRP and E genes (3.6 and 3.9 copies, respectively) [1]. The Food and Drugs Administration (FDA) approved the CDC test targeting the N gene under emergency conditions [2]. Since then, several nucleic acid amplification tests (NAATs) have received FDA emergency use authorization (EUA).
We aimed to summarize studies evaluating the diagnostic test accuracy (DTA) of NAATs performed on respiratory samples for the diagnosis of upper or lower acute respiratory tract infections (ARTIs) caused by coronaviruses, with special emphasis on the type of specimen.
Methods
This was a DTA systematic review with meta-analysis, performed as part the Value Dx Innovative Medicines Initiative (IMI) project examining the overall value of diagnostics to combat antimicrobial resistance. The protocol was registered on the International Prospective Register of Systematic Reviews (https://www.crd.york.ac.uk/PROSPERO/CRD42019145282).
Data sources and searches
We searched PubMed, Web of Science, the Cochrane Library, Embase and Open Grey until May 2019. A search string was developed for PubMed (Supplement 1) and adapted for the other databases as appropriate. This search targeted NAATs or antigen-based tests for any community-acquired respiratory tract infection for a large review performed within the Value-Dx IMI project; we selected studies evaluating NAATs for coronavirus infections from the database of all studies. Given the coronavirus disease 2019 (COVID-19) pandemic, a PubMed and medRxiv update was performed to include studies examining NAATs for COVID-19 until 31st August 2020, using the following search string: "(coronavirus OR covid OR covid-19 OR sars-cov OR mers-cov) AND (sensitivity[ti] OR specificity[ti] OR diagnostic[ti]) AND (pcr OR polymerase OR sequencing OR naat OR nucleic-acid)". Preprint (not peer-reviewed) studies were included. The references of all included studies were searched for additional studies.
Study selection
We included clinical studies evaluating NAATs among symptomatic patients with and without coronavirus infection, reporting quantitatively on both sensitivity and specificity. We included both cohort and case–control studies published until 31st August 2020, with no language restriction. We excluded animal or in vitro studies, case series including fewer than 20 patients, and case reports. We included studies where the index test was not performed in real time and did not affect decision-making, but we excluded studies where the index test was not relevant for real-time decision making.
Participants
These included patients of any age in the outpatient or inpatient setting with upper or lower acute respiratory tract infections or symptoms of COVID-19. The target condition was ARTI caused by any species of coronavirus. The index test was any coronavirus NAAT performed on respiratory-tract specimens. In studies assessing several respiratory viruses or bacteria, we extracted only the data on coronaviruses. If multiple species of coronavirus were evaluated in the same study, we used the data for the most prevalent species to avoid population duplication. However, we conducted a separate sensitivity analysis where all species were compared. In studies assessing more than one index test (comparison between tests), we used the data for the newer or better reported test. Since there is no reference standard for the diagnosis of coronavirus infection, we accepted any NAAT as reference standard. In studies that examined agreement or concordance rates between different NAATs without defining the index test and reference standard, we used the newer test as index and the test in clinical use as the reference standard. We defined that reference standards based on an algorithm using more than one NAAT test or whole-genome sequencing, with clinical/radiological features, were likely to correctly classify the target condition. In studies evaluating SARS-CoV-2 RT-PCR tests targeting two or more genes, a result of one positive gene was addressed as evaluated in the study (according to confirmatory testing or excluded from the analysis), but was not considered as a negative test in our review.
Data extraction
One reviewer performed the search and identified potentially eligible studies. Two reviewers independently applied inclusion/exclusion criteria to the eligible studies and extracted descriptive and diagnostic test accuracy data. Discrepancies were resolved by discussion. The crude number of patients with true-positive (TP), false-positive, true-negative and false-negative (FN) test results were extracted. Other data collected included study design, years (<2011, 2011–2019, 2020), location (US/Canada, East Asia and others), setting (limited to emergency department/hospitalized patients and other populations), participants' age (children and adults), and target condition. Respiratory tract infections were classified as upper (e.g. influenza-like illness), lower (e.g. pneumonia), or combined. We also collected data on the type of specimen tested (nasopharyngeal swab, aspirate or lower respiratory sample). The commercial name, types and methodology of NAATs were extracted and PCR tests were classified as real-time or not and multiplex tests or not.
Quality assessment
We evaluated the study design, including whether prospective or retrospective. We assessed risk of bias and concerns regarding applicability using the Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2) tool adapted for our review (Supplementary Material Supplement 2) [3].
Data synthesis and analysis
DTA meta-analysis was performed using the bivariate model, a hierarchical meta-regression method incorporating both sensitivity and specificity while taking into account the correlation [4]. The model estimates the parameters for the logit sensitivity, logit specificity, their variance and correlation. The summary sensitivity and specificity are reported with 95% confidence regions (CRs). Possible sources of heterogeneity were included as covariates in the meta-regression model to explain variation in accuracy, threshold or shape of the curve. We evaluated the following factors: study design, age, study year, location and settings, type of PCR, type of infection and specimen, all subgrouped as defined under the data extraction section. Following the onset of the SARS-CoV-2 pandemic, we repeated the analyses for SARS-CoV-2 alone and analysed the index test used and gene targeted as additional covariates. Analyses including three studies or more are reported. The statistical analysis was conducted using R v3.5.1 (R Core Team, 2018) and the two packages for meta-analysis meta [5] and mada [6] for DTA meta-analysis.
Results
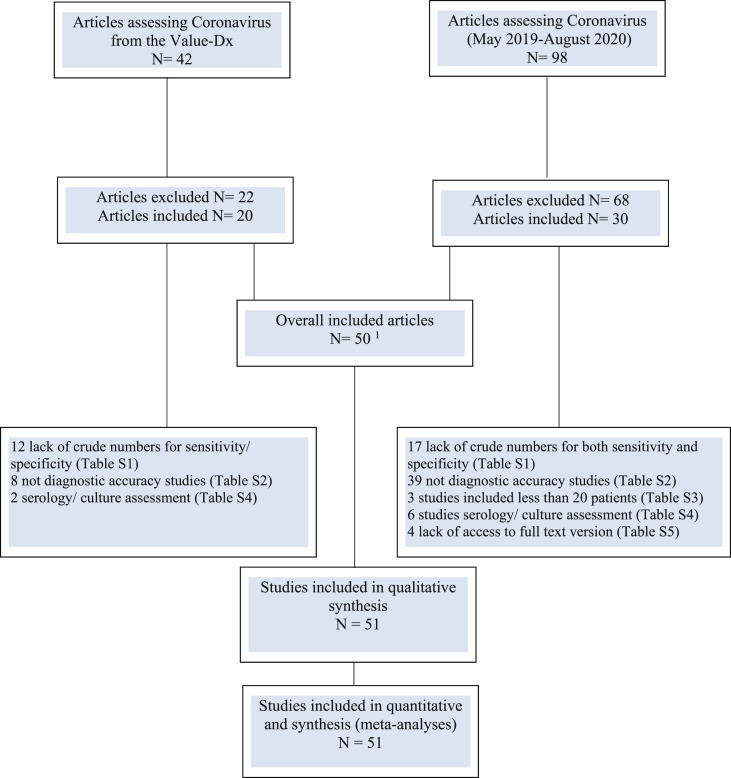
Altogether, 138 full-text articles assessing NAATs for the diagnosis of acute respiratory tract infections caused by different coronaviruses were evaluated (Fig. 1 ). Excluded studies are described in the Supplementary Material (Supplement 3). Fifty articles, published between 2004 and 2020, were included (Table 1 ) [[7], [8], [9], [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42], [43], [44], [45], [46], [47], [48], [49], [50], [51], [52], [53], [54], [55], [56]]. One article included two different studies [43]. The 51 studies analysed 18 923 persons, of these 10 181 persons (22 studies) before COVID-19 and 8742 persons for SARS-CoV-2 (29 studies) [7,9,13,14,[17], [18], [19],22,23,[25], [26], [27],[34], [35], [36],38,39,41,43,[45], [46], [47],50,51,[53], [54], [55], [56]]. The studies evaluated mostly patients with non-specific influenza-like illness or suspected COVID-19, the latter including both upper ARTI and pneumonia. Five studies included only children <2 years [16,21,31,32,48], 25 studies included a mixed age range, and 21 did not address patients' age. Eighteen studies reported a hospital setting, usually the emergency department, while others did not report the setting in which the samples were taken.
Fig. 1.
PRISMA (Preferred Reporting Items for Systematic Reviews and Meta- Analyses) flow chart. 1 One article included two studies.
Table 1.
Characteristics of included studiesa
| First author, year | Coronavirus b | Study design | Infection | n patients/samples | Index test type | Index test name | Type of specimen | Reference standard |
|---|---|---|---|---|---|---|---|---|
| Nolte FS, 2007 [40] | OC43, NL63 | Retrospective | ILI | 27 | Multiplex RT-PCR | EraGen Bioscience2 | Nasopharyngeal, nose, throat, and lung tissue, BAL, sputum | Real-time RT-PCR |
| Gadsby NJ, 2010 [20] | OC43, 229E, NL63, HKU1 | Retrospective | ILI | 286 | Multiplex real-time RT-PCR | Luminex Molecular Diagnostics | NPS, BAL | Real-time RT-PCR |
| Gharabaghi F, 2011 [21] | OC43/HKU1, 229E/NL63 | Retrospective | ILI | 750 | Multiplex RT-PCR | Seegene | NPS | Multiplex RT-PCR |
| Pabbaraju K, 2011 [42] | 229E, HKU1, NL63, OC43 | Prospective | NR | 334 | Multiplex RT-PCR | Luminex NxTAG Respiratory | NPS, NPA, nasal swab, throat swab, BAL, sputum, unkown respiratory origin | Multiplex RT-PCR |
| Bierbaum S, 2012 [12] | HKU1, NL63, OC43, 229E | Prospective | Upper and lower tract infection symptoms | 300 | Multiplex RT-PCR | Qiagen | Pharyngeal swabs/nasopharyngeal spirates | Monoplex real-time RT-PCR |
| Li J, 2012 [31] | OC43, NL63, 229E, HKU1 | Prospective | Pneumonitis, broncho-pneumonia | 126 | Multiplex RT-PCR | GeXP multiplex RT-PCR assay | Nasopharyngeal aspirate | Multiplex RT-PCR |
| Puppe W, 2012 [44] | OC43, 229E | Retrospective | NR | 178 | Multiplex RT-PCR | BioRad iCycler, Perkin-Elmer GeneAmp | Nasopharyngeal aspirate, NPS, BAL | Culture, RT-PCR |
| Sakthivel SK, 2012 [48] | OC43, 229E, NL63 | Prospective | ILI | 308 | Multiplex real-time RT-PCR | Applied Biosystems | Nasopharyngeal aspirates | Real-time RT-PCR |
| Choudhary ML, 2013 [16] | OC43 | Retrospective | ILI/severe acute respiratory illness | 843 | Multiplex RT-PCR | GeneAmp PCR System 9700 | Nasal, nasopharyngeal, throat swab | Real-time RT-PCR |
| Kim HK, 2013 [28] | OC43/HKU1 | Mixed | ILI | 482 | Multiplex real-time RT-PCR | Seegene | Nasopharyngeal aspirate, NPS, BAL | Multiplex RT-PCR |
| Li J, 2013 [32] | OC43, NL63, 229E, HKU1 | Prospective | ILI, Pneumonia | 247 | Multiplex RT-PCR | Qiagen | Nasopharyngeal aspirate | Multiplex RT-PCR |
| Bierbaum S, 2014 [11] | OC43, NL63, 229E | Prospective | ILI | 369 | Multiplex real-time RT-PCR | Inhouse | 214 pharyngeal swab, 152 nasopharyngeal aspirates, 3 BAL | Monoplex real-time RT-PCR |
| Salez N, 2015 [49] | Any COV, 229E, NL63, OC43, HKU1 | Prospective | ILI | 166 | Multiplex RT-PCR | RespiFinder SMART 22 | NPS | Duplex PCR or RT-PCR |
| Beckmann C, 2016 [10] | OC43, 229E, HKU1, NL63 | Mixed | ILI | 282 | Multiplex RT-PCR | Luminex NxTAG Respiratory | NPS, BAL, throat swabs, tracheal secretion, sputum | Multiplex RT-PCR (MLPA) |
| Chen H, 2017 [15] | OC43, 229E, HKU1, NL63 | Prospective | CAP | 74 | Multiplex RT-PCR | BioFire FilmArray Respiratory | Nasal swab | Multiplex real-time RT-PCR |
| Ko DH, 2017 [29] | OC43/HKU1, NL63, 229E | Retrospective | NR | 254 | Multiplex RT-PCR | Luminex NxTAG Respiratory | Sputum, NPS | Multiplex real-time RT-PCR |
| Mohamed DH, 2017 [37] | MERS-CoV | Retrospective | ILI | 234 | Real-time RT-PCR | NR | NPS/oropharyngeal swab | Real-time RT-PCR |
| Babady NE, 2018 [8] | NR | Mixed | ILI | 2908 | Multiplex RT-PCR | GenMark ePlex Respiratory | NPS | Multiplex RT-PCR |
| Leber AL, 2018 [30] | HKU1, 229E, NL63, OC43 | Prospective | ILI | 1612 | Multiplex RT-PCR | BioFire FilmArray RP2 | NPS | Multiplex RT-PCR |
| Vos LM, 2018 [52] | NR | Prospective | ILI | 62 | Multiplex RT-PCR | BioFire FilmArray Respiratory | NPS | Real-time RT-PCR |
| Hecht LS, 2019 [24] | MERS-CoV | Retrospective | ILI | 29 | Real-time RT-PCR | RealStar MERS-CoV | Nasal swab, nasopharyngeal aspirates | Real-time RT-PCR |
| Li X, 2019 [33] | NR | Prospective | CAP | 289 | Multiplex RT-PCR | Ningbo HEALTH Gene | Sputum, BAL, pharyngeal swab | Multiplex RT-PCR |
| Assennato SM, preprint [7] | SARS-CoV-2 | Retrospective | Symptoms of COVID-19 | 172 | RT-LAMP | SAMBA-II | NPS | Real-time RT-PCR |
| Basu A, 2020 [9] | SARS-CoV-2 | Prospective | Symptoms of COVID-19 | 101 | Isothermal amplification | Abbott ID NOW | Nasal swab | Real-time RT-PCR |
| Bisoffi Z, preprint [13] | SARS-CoV-2 | Prospective | Symptoms of COVID-19 | 345 | Real-time RT-PCR | CDC 2019-Novel Coronavirus | NPS | Real-time RT-PCR, serology + clinical |
| Brandsma E, preprint [14] | SARS-CoV-2 | Retrospective | Symptoms of COVID-19 | 378 | RT-LAMP + Cas12 | DETECTR | NPS, BAL, sputum | qRT-PCR |
| Collier D, preprint [17] | SARS-CoV-2 | Prospective | Symptoms of COVID-19 | 149 | RT-LAMP | SAMBA-II | NPS | RT-PCR |
| Cradic K, 2020 [18] | SARS-CoV-2 | Prospective | Symptoms of COVID-19 | 184 | Isothermal amplification | Abbott ID NOW | NPS | Real-time RT-PCR |
| Dao Thi VL, preprint [19] | SARS-CoV-2 | Retrospective | NR | 775 | RT-LAMP | Inhouse | Pharyngeal swabs | RT-PCR |
| Ghofrani M, preprint [22] | SARS-CoV-2 | Prospective | Symptoms of COVID-19, proven COVID-19 | 113 | Isothermal amplification | Abbott ID NOW | NPS, nasal, other clinical | PCR |
| Harrington A, 2020 [23] | SARS-CoV-2 | Prospective | Symptoms of COVID-19 | 524 | Isothermal amplification | Abbott ID NOW | Nasal swab | Real-time RT-PCR |
| Hogan CA, 2020 [25] | SARS-CoV-2 | Retrospective | NR | 100 | RT-PCR + lateral flow | Accula SARS-CoV-2 POCT | NPS | Real-time RT-PCR |
| Hou T, 2020 [26] | SARS-CoV-2 | Retrospective | NR | 114 | CRIPSR | CRIPSR-COVID | NPS, BAL | Metagenomic NGS |
| Jiang M, 2020 [27] | SARS-CoV-2 | Prospective | Symptoms of COVID-19 | 260 | RT-LAMP | Inhouse | NPS, sputum, tears | qRT-PCR |
| Loeffelholz MJ, 2020 [34] | SARS-CoV-2 | Prospective | Symptoms of COVID-19 | 481 | Real-time RT-PCR | Cepheid Xpert/GeneXpert | NPS, pharyngeal swab, tracheal aspirate | Real-time RT-PCR |
| Matzkies, LM 2020 [35] | SARS-CoV-2 | Retrospective | Symptoms of COVID-19, asymptomatic | 95 | RT-PCR | VIASURE SARSCoV-2 | NPS/oropharyngeal swab | qRT-PCR |
| Mitchell SL, 2020 [36] | SARS-CoV-2 | Retrospective | NR | 61 | Isothermal amplification | Abbott ID NOW | NPS | Real-time RT-PCR |
| Moore NM, preprint [38] | SARS-CoV-2 | Retrospective | Symptoms of COVID-19 | 198 | Isothermal amplification | Abbott ID NOW | NPS | Real-time RT-PCR + clinical |
| Moran A, 2020 [39] | SARS-CoV-2 | Retrospective | NR | 103 | Real-time RT-PCR | Cepheid Xpert/GeneXpert | NPS and nasal swa | qRT-PCR |
| Österdahl MF, preprint [41] | SARS-CoV-2 | Prospective | COVID-19 contacts in nursing home | 21 | RT-LAMP with magnetic bead capture | RT-LAMP with magnetic bead capture | NPS | RT-PCR |
| Poljak M, 2020 [43] | SARS-CoV-2 | Prospective | Symptoms of COVID-19 | 501 | qRT-PCR | Cobas 6800, Roche | Nasopharyngeal/oropharyngeal swab | Real-time RT-PCR |
| Poljak M, 2020 [43] | SARS-CoV-2 | Retrospective | Symptoms of COVID-19 | 215 | qRT-PCR | Cobas 6800, Roche | Nasopharyngeal/oropharyngeal swab | Real-time RT-PCR |
| Ridgday JP, 2020 [45] | SARS-CoV-2 | Prospective | Symptoms of COVID-19 | 2442 | Real-time RT-PCR | Cepheid Xpert/GeneXpert and Roche cobas SARS-CoV-2 | NPS | Real-time RT-PCR |
| Rodriguez-Manzano J, preprint [46] | SARS-CoV-2 | Retrospective | Symptoms of COVID-19 | 181 | RT-qLAMP | Inhouse | NPS, pharyngeal, nasal swabs | Real-time RT-PCR |
| Rohaim MA, preprint [47] | SARS-CoV-2 | Retrospective | NR | 199 | RT-LAMP | RT-LAMP with automatic AI based color interpretation | NPS | Real-time RT-PCR |
| Smithgall MC, 2020 [50] | SARS-CoV-2 | Retrospective | NR | 113 | Real-time RT-PCR | Cepheid Xpert/GeneXpert | NPS | RT-PCR |
| Suo T, preprint [51] | SARS-CoV-2 | Prospective | Symptoms of COVID-19 | 58 | Droplet Digital PCR | Inhouse | Pharyngeal swabs | RT-PCR + clinical |
| Wei S, preprint [53] | SARS-CoV-2 | Retrospective | Symptoms of COVID-19, close contact | 20 | RT-LAMP | Inhouse | NPS | qRT-PCR |
| Williams E, preprint [54] | SARS-CoV-2 | Retrospective | Symptoms of COVID-19, close contact | 675 | Heminested, multiplex, tandem real-time RT-PCR | Inhouse | NPS 98% | RT-PCR |
| Wolters F, 2020 [55] | SARS-CoV-2 | Retrospective | NR | 60 | Real-time RT-PCR | Cepheid Xpert/GeneXpert | NPS | RT-PCR |
| Zhen W, 2020 [56] | SARS-CoV-2 | Mixed | Symptoms of COVID-19 | 104 | Real-time RT-PCR | Applied Biosystems ThermoFisher Scientific | NPS | RT-PCR |
NR, not reported; ILI, influenza-like illness; CAP, community-acquired pneumonia; NPS, nasopharyngeal swab; BAL, bronchoalveolar lavage; MLPA, multiplex ligation-dependent probe amplification technology; NGS, next-generation sequencing; RT-LAMP, reverse transcriptase loop-mediated isothermal amplification.
Studies are sorted by year of publication and author.
In bold: the species selected for the main analysis.
The studies evaluated different PCRs as index tests; a single study described the development and testing of a CRISPR-based rapid assay based on Cas13a for SARS-CoV-2 detection [26]. Real-time RT-PCR tests were used in 18/51 studies (Table 1). Assays based on RT loop-mediated isothermal amplification (RT-LAMP) or other isothermal amplification for the detection of SARS-CoV-2 were assessed in 15 studies. All studies used a different PCR as reference standard, typically an approved commercial test that was in use in the laboratory performing the study or the reference laboratory. The reference standard was deemed optimal for coronavirus detection in seven studies using more than one PCR assay, serial testing, or next-generation sequencing alongside clinical presentation [7,13,26,38,41,45,51]. The specific species of the coronaviruses were reported in all but three of the studies before COVID-19 (Table 1). The target gene(s) were described in only 5/22 studies before COVID-19 [16,24,31,32,44] and in all of the COVID-19 studies (Supplementary Material Supplement 4). Different specimens were taken, with nasopharyngeal swabs being the most common. None of the studies reported who took the sample or how it was taken.
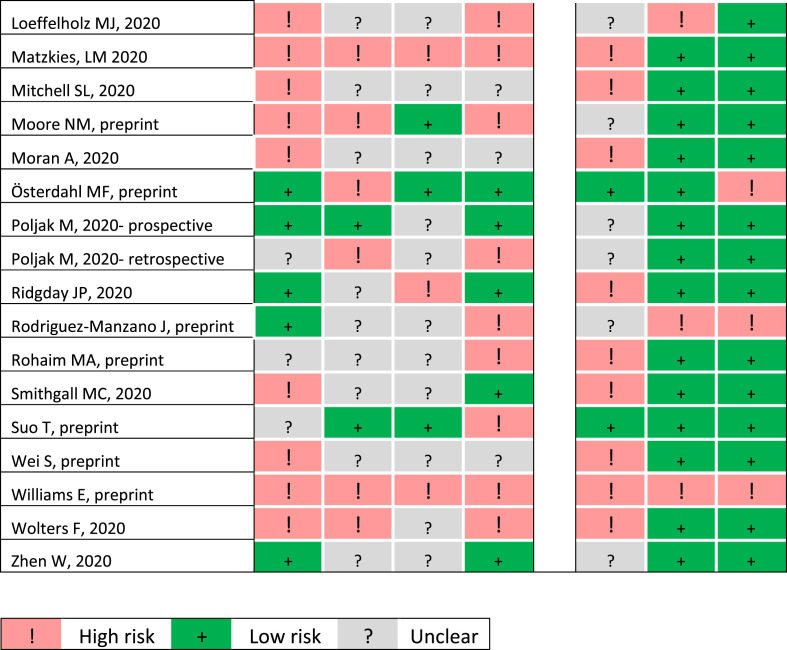
Twenty-three studies were prospective (12/29 COVID-19 studies) and the remainder were retrospective or mixed, typically using stored samples for analysis (Table 1). A case–control design was not avoided in 14/51 studies, among them 13 assessing SARS-CoV-2. The QUADAS-2 grading is presented in Fig. 2 and Supplementary Material Fig. S1. In general, studies were at higher risk of bias than at risk of poor applicability. Patient selection procedures were mostly at high or unclear risk of bias, considering that most studies did not describe a consecutive cohort, and some studies were enriched for positive samples. The index tests were at high risk of bias, since it was usually unclear whether the index tests were interpreted without knowledge of the results of the reference standard, and results were reported for different combined samples. The reference standard was deemed at low risk of bias in only four studies, complying with our definitions (see Methods), and was interpreted mostly without knowledge of the index test results. The flow and timing were downgraded, due mainly to unclear intervals between the index test and the reference standard (typically performed on the same sample, but with the index test performed after the reference standard) and patient exclusion in case of undetermined index test or reference standard results.
Fig. 2.
Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2) summary items for risk of bias and applicability for all studies.
The summary sensitivity was 89.1% (95%CR 84.0–92.7%) and the specificity was 98.9% (95%CR 98.0–99.4%). Sensitivity was more heterogeneous than specificity, as seen in Supplementary Material Figs S2 and S3. The sensitivity of studies evaluating SARS-CoV-2 was not significantly higher (90.4%, 95%CR 83.7–94.5%) than that of studies evaluating other coronavirus species (86.2%, 95%CR 77.1–92.1%), with statistical but not clinically significant lower specificity (Fig. 3 and Table 2 ). The covariate best explaining heterogeneity of the overall analysis was the NAAT test type real-time RT-PCT, resulting in higher sensitivity compared to other NAATs (Table 2). No other clinical or laboratory covariate explained significantly heterogeneity, including setting, type of sample taken, year or location. Study design and all risk of bias domains were not associated with test performance, apart from a high-risk reference standard, which was associated with significantly higher (probably exaggerated) specificity than an unclear or adequate reference standard (blinded to the index test and deemed likely to appropriately classify the target condition (see methods)).
Fig. 3.
Summary receiver operating characteristic (ROC) plot of studies evaluating PCR on respiratory samples for diagnosis of coronavirus infections, by species type. (A) All species. Studies reporting separately on different coronaviruses (all pre-COVID-19) included more than once, but each species-specific analysis includes each study only once. SARS-CoV-2 in red. (B) SARS-CoV-2 versus all other coronaviruses (each study included only once).
Table 2.
Factors underlying heterogeneity of the diagnostic accuracy of nucleic acid amplification tests (NAATs) for the diagnosis of coronavirus infection
| Variable | Sensitivity (%) with 95%CR | Specificity (%) with 95%CR | Significance a |
|---|---|---|---|
| All coronavirus species | |||
| SARS-CoV-2 versus others | 90.4 (83.7–94.5) versus 86.2 (77.1–92.1) | 98.1 (95.9–99.2) versus 99.4 (99.1–99.6) | SP p 0.002 |
| Real-time RT-PCR versus other PCR | 95.2 (90.5–97.6) versus 82.8 (75.8–88.1) | 98.9 (97.3–99.6) versus 98.8 (97.7–99.4) | SE p < 0.001 |
| Reference standard risk of bias (high versus low versus unclear) | 86.9 (78.5–92.3) versus 89.6 (61.4–97.9) versus 91.6 (83.6–95.9) | 99.3 (98.8–99.6) versus 94.3 (50.9–99.6) versus 98.2 (95.4–99.3) | SP p 0.009 |
| SARS-CoV-2 | |||
| Nasopharyngeal sample versus others b | 88.0 (79.5–93.3) versus 95.8 (88.1–98.6) | 98.0 (94.9–99.3) versus 98.3 94.1–99.5) | SE p 0.04 |
| Index test type (GeneXpert versus RT-LAMP/ isothermal versus others) | 98.9 (96.2–99.7) versus 84.2 (75.0–90.5) versus 93.8 (78.1–98.5) | 95.5 (91.8–97.5) versus 97.7 (92.8–99.3) versus 98.6 (94.4–99.7) | SE p 0.017 |
| Real-time RT-PCR versus other PCR | 96.2 (91.0–98.4) versus 82.7 (73.1–89.4) | 98.5 (95.2–99.6) versus 97.8 (93.7–99.3) | SE p < 0.001 |
| Single gene target versus more than one gene | 82.3 (72.4–89.2) versus 95.6 (89.6–98.2) | 97.6 (91.9–99.3) versus 98.5 (96.4–99.4) | SE p 0.001 |
| E gene included in test versus not included | 97.8 (95.6–98.9) versus 85.3 (77.3–90.9) | 98.6 (93.9–99.7) versus 98.0 (94.8–99.2) | SE p < 0.001 |
| N gene included in test versus not included | 93.9 (86.5–97.3) versus 84.6 (72.6–91.9) | 98.2 (95.8–99.3) versus 98.0 (92.4–99.5) | SE p 0.045 |
| RdRp gene alone versus other one or more genesc | 77.0 (65.7–85.4) versus 93.2 (86.8–96.6) | 97.5 (84.6–99.6) versus 98.3 (96.1–99.3) | SE 0.014 |
P values for sensitivity (SE) or specificity (SP). Only statistically significant differences are shown.
Studies in which samples taken from the upper respiratory tract (nasal, pharyngeal or nasopharyngeal) compared to studies reporting a mix of upper and lower respiratory tract samples.
All the studies using the RNA-dependent RNA polymerase (RdRp) gene targeted it as a single gene and all assessed different reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) or isothermal tests as index test.
More factors explained heterogeneity in the analysis limited to SARS-CoV-2 (Table 2). Studies evaluating nasopharyngeal swabs showed lower sensitivity than studies using lower respiratory tract or combined samples. RT-LAMP or isothermal assays, evaluated in 15 studies, resulted in lower sensitivity (84.2%, 75.0–90.5%) than GeneXpert 98.9% (96.2–99.7%) or other NAATs (93.8%, 78.1–98.5%), all with high specificity (Fig. 4 ). As for the overall analysis, real-time RT-PCR assays provided better sensitivity than other RT-PCRs (Fig. 4). Tests targeting the N gene or E gene had higher sensitivity than other tests, while the RdRp gene, always targeted by RT-LAMP or isothermal assays, had significantly lower sensitivity. Tests targeting more than one gene had better sensitivity than tests targeting a single gene (Supplementary Material Fig. S4). Three studies showed a specificity >90% [41,47,51], and no covariate explained the heterogeneity. Preprint publication (13 studies) was not associated with significantly different results than peer-reviewed published studies (16 studies).
Fig. 4.
Summary receiver operating characteristic (ROC) plot for SARS-CoV-2 nucleic acid amplification tests (NAATs) by type of PCR test. (A) Tests classified to real-time RT-PCR (12 studies, blue) versus non-quantitative assays (17 studies, red). (B) Types of test classified to Cepheid Xpert/GeneXpert (four studies, blue), different reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) or isothermal assays (15 studies, red) and others (eight NAATs studies, green).
Discussion
In this systematic review of studies assessing NAAT of respiratory samples for the diagnosis of coronavirus ARTIs, we identified 51 studies examining mostly agreement rates between different PCR tests in clinical samples. Typically, a newly developed or introduced test was compared with the commonly used reference standard. The search was completed on 31st August 2020 and identified 29 studies examining NAATs for COVID-19 diagnosis, beyond the analytical phase. The studies included patients with suspected coronavirus infection, examined mostly at the onset of the disease for the initial diagnosis. The pooled sensitivity of the new test in bivariate analysis was 89.1 (95%CI 84.0–92.7%), with large heterogeneity. Real-time RT-PCRs were significantly more sensitive (95.2%, 95%CR 90.5–97.6%) than other PCRs. The specificity was >98% in 45/51 studies (pooled specificity 98.9, 95%CR 98.0–99.4%), with SARS-CoV-2 PCRs and reference standards with low risk of bias associated with slightly lower specificity than other studies within this very narrow range of excellent specificity in the context of the initial diagnosis of coronavirus infection.
Analysing the agreement rates between different NAATs to diagnose COVID-19, heterogeneity could be explained by several factors related to the sample taken and the type and methods of the PCR test. Notably, RT-LAMP-based PCRs (especially when targeting the RdRp gene only) resulted in lower sensitivity (86.3%, 95%CR 74.0–93.3%) than other PCRs, while real-time PCRs had higher sensitivity (96.2%, 95%CR 91.0–98.4%). Tests targeting more than one gene, specifically the N or S genes, showed higher sensitivity. Studies evaluating upper respiratory samples alone (nasopharyngeal swabs) had slightly lower sensitivity than studies evaluating mixed upper/lower respiratory samples. All the differences in sensitivity did not affect the typically excellent specificity shown in these studies (pooled specificity 98.1%, 95%CR 95.9–99.2%). Two of the three studies with <90% specificity concluded that the new tests (Droplet Digital PCR [51] and Artificial Intelligence-Assisted Loop Mediated Isothermal Amplification [47]) were more sensitive than the reference standard commercial PCR, resulting in the low negative agreement rate.
Currently there is interest in the utility of PCR tests to screen populations for COVID-19 as a containment strategy [57,58]. In this context, near perfect specificity is required rather than optimal sensitivity. However, our review addressed symptomatic patients suspected of coronavirus infection and tested for this indication, where excellent sensitivity is required. Rapid testing is crucial in this setting, thus multiple studies have examined the Abbott ID NOW assay or in-house RT-LAMP-based assays, which can provide results with 30–60 minutes. Although resulting in imperfect sensitivity, missing about 15% of truly positive patients, their specificity was similar to that of other PCRs (pooled false-positive rate of about 2%). In the clinical workflow, such a test can be used in emergency departments to rapidly detect and isolate most positive patients, with confirmatory testing of the negative patients using real-time PCR to detect those missed by the rapid test. Although correct sampling probably affects the yield of diagnostics on respiratory samples, the sampling techniques were not reported in the included studies. Nevertheless, considering that the index test and the reference standard were always performed on the same sample, this should not have affected the reported diagnostic test accuracy. Results were not available by time from symptom onset and by disease severity, all potentially related to viral load and thus potentially affecting test performance. Although some of the studies reported the performance of real-time RT-PCR test by threshold cycle (Ct) value as a correlate of viral load, we do not present an analysis on this level but report the overall results of all patients/samples included in the study.
We have included studies examining test agreement/concordance, and present the data as sensitivity/specificity, maintaining a direction of new test versus reference standard. However, the latter corresponds to positive and negative agreements, and should be interpreted as such, considering that in most studies the reference standard could not perfectly determine whether patients had COVID-19. In the main analysis we include each study once to avoid population duplication, selecting the numbers reported for one of the coronavirus species (in studies reporting on non-SARS coronaviruses) or a pair of tests (for studies comparing agreement of several tests). The heterogeneity assessment was limited to a single covariate at a time; obviously these are not independent. Thus, the analysis by species is obviously linked with year, NAAT method, and improved reporting methods. In the analysis of SARS-CoV-2, the gene targeted by the assay was linked with the test type. Furthermore, heterogeneity assessment of the index test is limited by the fact that studies used different reference standards. We included only studies reporting on both sensitivity and specificity; therefore, we excluded studies such as that by Dong et al., claiming a sensitivity advantage of a newly developed digital RT-PCR over commercial tests among sick patients, all diagnosed with COVID-19 [59]. Finally, intensive research is ongoing in the COVID-19 pandemic and new studies appear daily. The evidence will need to be updated.
In summary, the pooled evidence shows imperfect sensitivity of respiratory PCR tests for the diagnosis of coronavirus acute respiratory tract infections, including COVID-19. The best performing tests will miss about 4% of positive patients and, overall, all assessed tests missed about 10%. In the context of a suspected disease, nearly all PCRs showed excellent specificity. The factors identified as underlying heterogeneity in the COVID-19 analyses can used to select the optimal test for clinical use and for further test development. To examine sensitivity and specificity, rather than test agreement, an optimized reference standard should be defined that can be used consistently in future studies.
Author contributions
MMH, FM, EC, EG, PDN, IP, MP: search, data extraction, validation. AG, MML: data analysis. ET, MML, MP: supervision. ET: project administration and funding acquisition. MMH, MP: writing, original draft. All authors contributed to the conception and design of the study and to review and editing of the manuscript.
Transparency declaration
All authors have no conflicts of interest to declare. Mariska Leeflang is co-convenor of Cochrane's Screening and Diagnostic Test Methods Group. Funding source: Innovative Medicines Initiative-2 Joint Undertaking, grant agreement No 820755 (Value-Dx). This Joint Undertaking receives support from the European Union's Horizon 2020 research and innovation programme and EFPIA and bioMérieux SA, Janssen Pharmaceutica NV, Accelerate Diagnostics SL, Abbott, Bio-Rad Laboratories, BD Switzerland Sàrl, and The Wellcome Trust Limited. The commercial companies had no part in the design, analysis, writing or decision to publish the results.
Editor: L. Leibovici
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2020.11.002.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y.C. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020 doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention . Centers for Disease Control and Prevention; 2020. CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel 2020.https://www.fda.gov/media/134922/download Available from: [Google Scholar]
- 3.Whiting P.F., Rutjes A.W., Westwood M.E., Mallett S., Deeks J.J., Reitsma J.B. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 4.Reitsma J.B., Glas A.S., Rutjes A.W., Scholten R.J., Bossuyt P.M., Zwinderman A.H. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. J Clin Epidemiol. 2005;58:982–990. doi: 10.1016/j.jclinepi.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 5.Schwarzer G., Carpenter J., Rücker G. Springer International Publishing AG; 2015. Meta-analysis with R. [Google Scholar]
- 6.Doebler P., Holling H. 2012. Meta-analysis of diagnostic accuracy with mada.https://cran.r-project.org/web/packages/mada/vignettes/mada.pdf Available from: [DOI] [PubMed] [Google Scholar]
- 7.Assennato S.M., Ritchie A.V., Nadala C., Goel N., Zhang H., Datir R. Performance evaluation of the point-of-care SAMBA II SARS-CoV-2 Test for detection of SARS-CoV-2. medRxiv. 2020;2020 doi: 10.1128/JCM.01262-20. 5.24.20100990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Babady N.E., England M.R., Jurcic Smith K.L., He T., Wijetunge D.S., Tang Y.W. Multicenter evaluation of the ePlex Respiratory Pathogen Panel for the detection of viral and bacterial respiratory tract pathogens in nasopharyngeal swabs. J Clin Microbiol. 2018;56 doi: 10.1128/JCM.01658-17. e01658-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Basu A., Zinger T., Inglima K., Woo K.M., Atie O., Yurasits L. Performance of Abbott ID Now COVID-19 Rapid Nucleic Acid Amplification Test using nasopharyngeal swabs transported in viral transport media and dry nasal swabs in a New York City academic institution. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.01136-20. e01136-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beckmann C., Hirsch H.H. Comparing luminex NxTAG-respiratory pathogen panel and RespiFinder-22 for multiplex detection of respiratory pathogens. J Med Virol. 2016;88:1319–1324. doi: 10.1002/jmv.24492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bierbaum S., Forster J., Berner R., Rucker G., Rohde G., Neumann-Haefelin D. Detection of respiratory viruses using a multiplex real-time PCR assay in Germany, 2009/10. Arch Virol. 2014;159:669–676. doi: 10.1007/s00705-013-1876-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bierbaum S., Konigsfeld N., Besazza N., Blessing K., Rucker G., Kontny U. Performance of a novel microarray multiplex PCR for the detection of 23 respiratory pathogens (SYMP-ARI study) Eur J Clin Microbiol Infect Dis. 2012;31:2851–2861. doi: 10.1007/s10096-012-1639-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bisoffi Z., Pomari E., Deiana M., Piubelli C., Ronzoni N., Beltrame A. Sensitivity, specificity and predictive values of molecular and serological tests for COVID-19. A longitudinal study in emergency room. medRxiv. 2020:2020. doi: 10.3390/diagnostics10090669. 08.09.20171355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brandsma E., Verhagen H.J., van de Laar Tjw, Claas E.C.J., Cornelissen M., van den Akker E. Rapid, sensitive and specific SARS coronavirus-2 detection: a multi-center comparison between standard qRT-PCR and CRISPR based DETECTR. medRxiv. 2020:20147249. doi: 10.1093/infdis/jiaa641. 2020.07.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen H., Weng H., Lin M., He P., Li Y., Xie Q. The clinical significance of FilmArray respiratory panel in diagnosing community-acquired pneumonia. Biomed Res Int. 2017;2017:7320859. doi: 10.1155/2017/7320859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choudhary M.L., Anand S.P., Heydari M., Rane G., Potdar V.A., Chadha M.S. Development of a multiplex one step RT-PCR that detects eighteen respiratory viruses in clinical specimens and comparison with real time RT-PCR. J Virol Methods. 2013;189:15–19. doi: 10.1016/j.jviromet.2012.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collier D.A., Assennato S.M., Sithole N., Sharrocks K., Ritchie A., Ravji P. Rapid point of care nucleic acid testing for SARS-CoV-2 in hospitalised patients: a clinical trial and implementation study. medRxiv. 2020:2020. doi: 10.1016/j.xcrm.2020.100062. 05.31.20114520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cradic K., Lockhart M., Ozbolt P., Fatica L., Landon L., Lieber M. Clinical evaluation and utilization of multiple molecular in vitro diagnostic assays for the detection of SARS-CoV-2. Am J Clin Pathol. 2020;154:201–207. doi: 10.1093/ajcp/aqaa097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dao Thi V.L., Herbst K., Boerner K., Meurer M., Kremer L.P.M., Kirrmaier D. Screening for SARS-CoV-2 infections with colorimetric RT-LAMP and LAMP sequencing. medRxiv. 2020:2020. 05.05.20092288. [Google Scholar]
- 20.Gadsby N.J., Hardie A., Claas E.C., Templeton K.E. Comparison of the Luminex Respiratory Virus Panel fast assay with in-house real-time PCR for respiratory viral infection diagnosis. J Clin Microbiol. 2010;48:2213–2216. doi: 10.1128/JCM.02446-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gharabaghi F., Hawan A., Drews S.J., Richardson S.E. Evaluation of multiple commercial molecular and conventional diagnostic assays for the detection of respiratory viruses in children. Clin Microbiol Infect. 2011;17:1900–1906. doi: 10.1111/j.1469-0691.2011.03529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghofrani M., Casas M.T., Pelz R.K., Kroll C., Blum N., Foster S.D. Performance characteristics of the ID NOW COVID-19 assay: a regional health care system experience. medRxiv. 2020:2020. 06.03.20116327. [Google Scholar]
- 23.Harrington A., Cox B., Snowdon J., Bakst J., Ley E., Grajales P. Comparison of Abbott ID Now and Abbott m2000 methods for the detection of SARS-CoV-2 from nasopharyngeal and nasal swabs from symptomatic patients. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.00798-20. e00798-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hecht L.S., Jurado-Jimenez A., Hess M., Halas H.E., Bochenek G., Mohammed H. Verification and diagnostic evaluation of the RealStar((R)) Middle East respiratory syndrome coronavirus (N gene) reverse transcription-PCR kit 1.0. Future Microbiol. 2019;14:941–948. doi: 10.2217/fmb-2019-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hogan C.A., Garamani N., Lee A.S., Tung J.K., Sahoo M.K., Huang C. Comparison of the Accula SARS-CoV-2 Test with a laboratory-developed assay for detection of SARS-CoV-2 RNA in clinical nasopharyngeal specimens. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.01072-20. e01072-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hou T., Zeng W., Yang M., Chen W., Ren L., Ai J. Development and evaluation of a rapid CRISPR-based diagnostic for COVID-19. PLOS Pathogens. 2020;16 doi: 10.1371/journal.ppat.1008705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang M., Pan W., Arastehfar A., Fang W., Ling L., Fang H. Development and validation of a rapid single-step reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) system potentially to be used for reliable and high-throughput screening of COVID-19. medRxiv. 2020:2020. doi: 10.3389/fcimb.2020.00331. 03.15.20036376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim H.K., Oh S.H., Yun K.A., Sung H., Kim M.N. Comparison of Anyplex II RV16 with the xTAG respiratory viral panel and Seeplex RV15 for detection of respiratory viruses. J Clin Microbiol. 2013;51:1137–1141. doi: 10.1128/JCM.02958-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ko D.H., Kim H.S., Hyun J., Kim H.S., Kim J.S., Park K.U. Comparison of the luminex xTAG respiratory viral panel fast v2 assay with anyplex II RV16 detection kit and AdvanSure RV real-time RT-PCR assay for the detection of respiratory viruses. Ann Lab Med. 2017;37:408–414. doi: 10.3343/alm.2017.37.5.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leber A.L., Everhart K., Daly J.A., Hopper A., Harrington A., Schreckenberger P. Multicenter evaluation of BioFire FilmArray Respiratory Panel 2 for detection of viruses and bacteria in nasopharyngeal swab samples. J Clin Microbiol. 2018;56 doi: 10.1128/JCM.01945-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J., Mao N.Y., Zhang C., Yang M.J., Wang M., Xu W.B. The development of a GeXP-based multiplex reverse transcription-PCR assay for simultaneous detection of sixteen human respiratory virus types/subtypes. BMC Infect Dis. 2012;12:189. doi: 10.1186/1471-2334-12-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J., Qi S., Zhang C., Hu X., Shen H., Yang M. A two-tube multiplex reverse transcription PCR assay for simultaneous detection of sixteen human respiratory virus types/subtypes. Biomed Res Int. 2013;2013:327620. doi: 10.1155/2013/327620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X., Chen B., Zhang S., Li X., Chang J., Tang Y. Rapid detection of respiratory pathogens for community-acquired pneumonia by capillary electrophoresis-based multiplex PCR. SLAS Technol. 2019;24:105–116. doi: 10.1177/2472630318787452. [DOI] [PubMed] [Google Scholar]
- 34.Loeffelholz M.J., Alland D., Butler-Wu S.M., Pandey U., Perno C.F., Nava A. Multicenter evaluation of the cepheid xpert xpress SARS-CoV-2 test. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.00926-20. e00926-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matzkies L.M., Leitner E., Stelzl E., Assig K., Bozic M., Siebenhofer D. Lack of sensitivity of an IVD/CE-labelled kit targeting the S gene for detection of SARS-CoV-2. Clin Microbiol Infect. 2020;26:1417.e1–1417.e4. doi: 10.1016/j.cmi.2020.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitchell S.L., George K.S. Evaluation of the COVID19 ID NOW EUA assay. J Clin Virol. 2020;128:104429. doi: 10.1016/j.jcv.2020.104429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mohamed D.H., AlHetheel A.F., Mohamud H.S., Aldosari K., Alzamil F.A., Somily A.M. Clinical validation of 3 commercial real-time reverse transcriptase polymerase chain reaction assays for the detection of Middle East respiratory syndrome coronavirus from upper respiratory tract specimens. Diagn Microbiol Infect Dis. 2017;87:320–324. doi: 10.1016/j.diagmicrobio.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moore N.M., Li H., Schejbal D., Lindsley J., Hayden M. Comparison of two commercial molecular tests and a laboratory-developed modification of the CDC 2019-nCOV RT-PCR assay for the qualitative detection of SARS-CoV-2 from upper respiratory tract specimens. medRxiv. 2020:2020. doi: 10.1128/JCM.00938-20. 05.02.20088740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moran A., Beavis K.G., Matushek S.M., Ciaglia C., Francois N., Tesic V. Detection of SARS-CoV-2 by use of the cepheid xpert xpress SARS-CoV-2 and roche cobas SARS-CoV-2 assays. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.00772-20. e00772-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nolte F.S., Marshall D.J., Rasberry C., Schievelbein S.S., Banks G.G., Storch G.A. MultiCode-PLx system for multiplexed detection of seventeen respiratory viruses. J Clin Microbiol. 2007;45:2779–2786. doi: 10.1128/JCM.00669-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Osterdahl M.F., Lee K.A., Ni Lochlainn M., Wilson S., Douthwaite S., Horsfall R. Detecting SARS-CoV-2 at point of care: preliminary data comparing Loop-mediated isothermal amplification (LAMP) to PCR. medRxiv. 2020:2020. doi: 10.1186/s12879-020-05484-8. 04.01.20047357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pabbaraju K., Wong S., Tokaryk K.L., Fonseca K., Drews S.J. Comparison of the Luminex xTAG respiratory viral panel with xTAG respiratory viral panel fast for diagnosis of respiratory virus infections. J Clin Microbiol. 2011;49:1738–1744. doi: 10.1128/JCM.02090-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poljak M., Korva M., Knap Gasper N., Fujs Komlos K., Sagadin M., Ursic T. Clinical evaluation of the cobas SARS-CoV-2 test and a diagnostic platform switch during 48 hours in the midst of the COVID-19 pandemic. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.00599-20. e00599-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Puppe W., Weigl J., Grondahl B., Knuf M., Rockahr S., von Bismarck P. Validation of a multiplex reverse transcriptase PCR ELISA for the detection of 19 respiratory tract pathogens. Infection. 2013;41:77–91. doi: 10.1007/s15010-012-0298-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ridgway J.P., Pisano J., Landon E., Beavis K.G., Robicsek A. Clinical sensitivity of severe acute respiratory syndrome coronavirus 2 nucleic acid amplification tests for diagnosing coronavirus disease 2019. Open Forum Infect Dis. 2020;7 doi: 10.1093/ofid/ofaa315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodriguez-Manzano J., Malpartida-Cardenas K., Moser N., Pennisi I., Cavuto M., Miglietta L. A handheld point-of-care system for rapid detection of SARS-CoV-2 in under 20 minutes. medRxiv. 2020:20142349. doi: 10.1021/acscentsci.0c01288. 2020.06.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rohaim M.A., Clayton E., Sahin I., Vilela J., Khalifa M.E., Al-Natour M.Q. Artificial intelligence-assisted loop mediated isothermal amplification (ai-LAMP) for rapid and reliable detection of SARS-CoV-2. medRxiv. 2020:2020. doi: 10.3390/v12090972. 07.08.20148999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sakthivel S.K., Whitaker B., Lu X., Oliveira D.B., Stockman L.J., Kamili S. Comparison of fast-track diagnostics respiratory pathogens multiplex real-time RT-PCR assay with in-house singleplex assays for comprehensive detection of human respiratory viruses. J Virol Methods. 2012;185:259–266. doi: 10.1016/j.jviromet.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salez N., Vabret A., Leruez-Ville M., Andreoletti L., Carrat F., Renois F. Evaluation of four commercial multiplex molecular tests for the diagnosis of acute respiratory infections. PLoS One. 2015;10 doi: 10.1371/journal.pone.0130378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smithgall M.C., Scherberkova I., Whittier S., Green D.A. Comparison of cepheid xpert xpress and Abbott ID now to roche cobas for the rapid detection of SARS-CoV-2. J Clin Virol. 2020;128:104428. doi: 10.1016/j.jcv.2020.104428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suo T., Liu X., Feng J., Guo M., Hu W., Guo D. ddPCR: a more sensitive and accurate tool for SARS-CoV-2 detection in low viral load specimens. medRxiv. 2020:2020. doi: 10.1080/22221751.2020.1772678. 02.29.20029439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vos L.M., Riezebos-Brilman A., Schuurman R., Hoepelman A.I.M., Oosterheert J.J. Syndromic sample-to-result PCR testing for respiratory infections in adult patients. Neth J Med. 2018;76:286–293. [PubMed] [Google Scholar]
- 53.Wei S., Kohl E., Djandji A., Morgan S., Whittier S., Mansukhani M. Direct diagnostic testing of SARS-CoV-2 without the need for prior RNA extraction. medRxiv. 2020:2020. doi: 10.1038/s41598-021-81487-y. 05.28.20115220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams E., Bond K., Chong B., Giltrap D., Eaton M., Kyriakou P. Implementation and evaluation of a novel real-time multiplex assay for SARS-CoV-2: in-field learnings from a clinical microbiology laboratory. medRxiv. 2020:2020. doi: 10.1016/j.pathol.2020.08.004. 06.03.20117267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wolters F., van de Bovenkamp J., van den Bosch B., van den Brink S., Broeders M., Chung N.H. Multi-center evaluation of cepheid xpert® xpress SARS-CoV-2 point-of-care test during the SARS-CoV-2 pandemic. J Clin Virol. 2020;128:104426. doi: 10.1016/j.jcv.2020.104426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhen W., Manji R., Smith E., Berry G.J. Comparison of four molecular in vitro diagnostic assays for the detection of SARS-CoV-2 in nasopharyngeal specimens. J Clin Microbiol. 2020 doi: 10.1128/JCM.00743-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mina M.J., Parker R., Larremore D.B. Rethinking Covid-19 test sensitivity - a strategy for containment. New Engl J Med. 2020;383:e120. doi: 10.1056/NEJMp2025631. [DOI] [PubMed] [Google Scholar]
- 58.Kimball A., Hatfield K.M., Arons M., James A., Taylor J., Spicer K. Asymptomatic and presymptomatic SARS-CoV-2 infections in residents of a long-term care skilled nursing facility—king County, Washington, March 2020. MMWR Morb Mortal Wkly Rep. 2020;69:377–381. doi: 10.15585/mmwr.mm6913e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dong L., Zhou J., Niu C., Wang Q., Pan Y., Sheng S. Highly accurate and sensitive diagnostic detection of SARS-CoV-2 by digital PCR. medRxiv. 2020:2020. doi: 10.1016/j.talanta.2020.121726. 03.14.20036129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.