While V. parahaemolyticus and V. vulnificus cause shellfish-associated morbidity and mortality among shellfish consumers, current regulatory assays for vibrios are complex, time-consuming, labor-intensive, and relatively expensive. In this study, the rapid, simple, and inexpensive COPP assay was identified as a possible alternative to MPN-PCR for shellfish monitoring. This paper shows differences in total Vibrionaceae and pathogenic vibrios found in seawater and oysters from the commercially important Delaware and Chesapeake Bays. Vibrio parahaemolyticus isolates from the Delaware Bay were more likely to contain commonly recognized pathogenicity genes than those from the Chesapeake Bay.
KEYWORDS: Vibrio, oysters, Chesapeake Bay, Delaware Bay, Mid-Atlantic, COPP assay, MPN-PCR, direct plating, Vibrio parahaemolyticus, Vibrio vulnificus
ABSTRACT
Oyster and seawater samples were collected from five sites in the Chesapeake Bay, MD, and three sites in the Delaware Bay, DE, from May to October 2016 and 2017. Abundances and detection frequencies for total and pathogenic Vibrio parahaemolyticus and Vibrio vulnificus were compared using the standard most-probable-number–PCR (MPN-PCR) assay and a direct-plating (DP) method on CHROMagar Vibrio for total (tlh+) and pathogenic (tdh+ and trh+) V. parahaemolyticus genes and total (vvhA) and pathogenic (vcgC) V. vulnificus genes. The colony overlay procedure for peptidases (COPP) assay was evaluated for total Vibrionaceae. DP had high false-negative rates (14 to 77%) for most PCR targets and was deemed unsatisfactory. Logistic regression models of the COPP assay showed high concordances with MPN-PCR for tdh+ and trh+ V. parahaemolyticus and vvhA+ V. vulnificus in oysters (85.7 to 90.9%) and seawater (81.1 to 92.7%) when seawater temperature and salinity were factored into the model, suggesting that the COPP assay could potentially serve as a more rapid method to detect vibrios in oysters and seawater. Differences in total Vibrionaceae and pathogenic Vibrio abundances between state sampling sites over different collection years were contrasted for oysters and seawater by MPN-PCR. Abundances of tdh+ and trh+ V. parahaemolyticus were ∼8-fold higher in Delaware oysters than in Maryland oysters, whereas abundances of vcgC+ V. vulnificus were nearly identical. For Delaware oysters, 93.5% were both tdh+ and trh+, compared to only 19.2% in Maryland. These results indicate that pathogenic V. parahaemolyticus was more prevalent in the Delaware Bay than in the Chesapeake Bay.
IMPORTANCE While V. parahaemolyticus and V. vulnificus cause shellfish-associated morbidity and mortality among shellfish consumers, current regulatory assays for vibrios are complex, time-consuming, labor-intensive, and relatively expensive. In this study, the rapid, simple, and inexpensive COPP assay was identified as a possible alternative to MPN-PCR for shellfish monitoring. This paper shows differences in total Vibrionaceae and pathogenic vibrios found in seawater and oysters from the commercially important Delaware and Chesapeake Bays. Vibrio parahaemolyticus isolates from the Delaware Bay were more likely to contain commonly recognized pathogenicity genes than those from the Chesapeake Bay.
INTRODUCTION
Members of the family Vibrionaceae are naturally occurring marine bacteria containing some pathogenic species that negatively impact aquatic systems and human health (1). Vibrio parahaemolyticus and Vibrio vulnificus are important human pathogens responsible for morbidity and mortality, most often associated with the consumption of molluscan shellfish. Most illnesses from V. vulnificus and V. parahaemolyticus occur during warmer months, which follow seasonal peaks in their densities in seawater and shellfish. Vibrio densities in oysters are influenced by environmental factors, principally temperature and, to a lesser extent, salinity (2–5).
Shellfish safety is regulated in the United States according to the requirements set forth by the Interstate Shellfish Sanitation Conference (ISSC) under the objective to “Adopt sound, uniform methods into a National Shellfish Sanitation Program (NSSP) that is accepted by participating shellfish control authorities” (6). Under the NSSP guidelines, total levels of V. parahaemolyticus and V. vulnificus can be identified from positive most-probable-number–PCR (MPN-PCR) tubes based on the presence of the species-specific thermolabile hemolysin gene (tlh) and V. vulnificus hemolysin gene A (vvhA), respectively (7–9). The detection of pathogenic strains of V. parahaemolyticus relies on the molecular detection of the thermostable direct hemolysin (tdh) and tdh-related hemolysin (trh) genes, which encode two major virulence factors (7). For V. vulnificus, the virulence-correlated gene (vcgC) serves as a reliable biomarker to screen for potentially virulent strains (8, 9). MPN followed by PCR testing requires several days and makes the identification and enumeration of total and pathogenic vibrios not only slow but also labor-intensive and costly. From 1998 to 2019, a number of significant V. vulnificus and V. parahaemolyticus outbreaks, oyster recalls, and closures of shellfish harvesting areas have occurred in the United States, resulting in an increased interest in the development of more-rapid, simpler, and less costly methods to monitor these pathogens (10–14).
Our research group developed a simple, rapid, and inexpensive enzyme-based fluorogenic procedure, known as the colony overlay procedure for peptidases (COPP) assay, to identify and quantify total Vibrionaceae (TV) in seawater and shellfish in less than 24 h (15, 16). Currently, this is the only agar plate, culture-based method using nonselective and nondifferential media for specific detection of TV abundances. While the COPP procedure detects and enumerates TV, the predictive value of using TV as an indicator of pathogenic Vibrio species has not been determined, but it would be analogous to MPN monitoring for total or fecal coliforms as indicators for the possible presence of enteric pathogens, e.g., Escherichia coli or Salmonella, under NSSP guidelines (17). Unfortunately, quantification of pathogenic V. vulnificus and V. parahaemolyticus using conventional MPN-PCR analyses is labor-intensive and costly and requires several days (18). A 24-h direct-plating (DP) method is available, using selective and differential CHROMagar Vibrio (CHROMagar, Paris, France) to differentiate potentially pathogenic Vibrio species (19, 20).
The objectives of this study were as follows: (i) to simultaneously evaluate the distribution of TV and pathogenic V. parahaemolyticus and V. vulnificus in oysters and seawater collected from the Delaware and Chesapeake Bays using the COPP assay, the CHROMagar Vibrio direct-plating method, and MPN-PCR; (ii) to model the use of the COPP assay or direct-plating method as potential substitutes for MPN-PCR assays; (iii) to compare and contrast abundances, detection frequencies, and dynamics of TV and pathogenic vibrios in the Delaware and Chesapeake Bays; (iv) to evaluate seasonal and geographical differences in TV and pathogenic vibrios in the bays; and (v) to evaluate the influence of physicochemical parameters of seawater on TV and pathogenic Vibrio levels in the bays.
RESULTS
Correlation of COPP assay, MPN-PCR, and DP methods.
The first step in our analysis was to examine simple correlation between the methods. The COPP assay for TV correlated moderately well with total V. vulnificus and V. parahaemolyticus as determined by both direct plating (DP) and MPN-PCR in water (r values = 0.50 to 0.69) (Table 1). In oysters, similar relationships were observed for total V. vulnificus (r = 0.56 to 0.62), but poor correlation was noted for total V. parahaemolyticus (r = 0.23 to 0.34). Correlations with virulence markers tdh, trh, and vcgC were all <0.45. Notably, the relationships between MPN-PCR and the COPP assay for tdh and trh in water were 0.32 and 0.38, respectively. Overall, there is a general trend of slightly stronger correlation of the COPP assay with direct plating than with MPN-PCR (Table 1).
TABLE 1.
Pearson’s correlation coefficients comparing the use of the colony overlay procedure for peptidases assay with direct plating and most-probable-number–PCR assays for total (vvhA) and virulent (vcgC) V. vulnificus and total (tlh) and virulent (tdh and trh) V. parahaemolyticus
| Pathogen and gene | Comparison | Correlation coefficient (r value, significance level)a
|
|
|---|---|---|---|
| Seawater | Oysters | ||
| Total | |||
| V. vulnificus vvhA | COPP vs DP | 0.69, <0.0001 | 0.62, <0.0001 |
| COPP vs MPN-PCR | 0.50, <0.0001 | 0.56, <0.0001 | |
| DP vs MPN-PCR | 0.42, <0.0001 | 0.59, <0.0001 | |
| V. parahaemolyticus tlh | COPP vs DP | 0.63, <0.0001 | 0.34, 0.0003 |
| COPP vs MPN-PCR | 0.65, <0.0001 | 0.23, 0.177 | |
| DP vs MPN-PCR | 0.66, <0.0001 | 0.67, <0.0001 | |
| Pathogenic | |||
| V. vulnificus vcgC | COPP vs DP | 0.42, <0.0001 | 0.40, <0.0001 |
| COPP vs MPN-PCR | 0.38, <0.0001 | 0.33, <0.0003 | |
| DP vs MPN-PCR | 0.40, <0.0001 | 0.40, <0.0001 | |
| V. parahaemolyticus | |||
| tdh | COPP vs DP | 0.31, 0.0009 | 0.32, 0.0006 |
| COPP vs MPN-PCR | 0.32, 0.0006 | −0.04, 0.6214 | |
| DP vs MPN-PCR | 0.54, <0.0001 | 0.31, 0.0008 | |
| trh | COPP vs DP | 0.44, <0.0001 | 0.39, <0.0001 |
| COPP vs MPN-PCR | 0.38, <0.0001 | −0.02, 0.7791 | |
| DP vs MPN-PCR | 0.49, <0.0001 | 0.34, 0.0003 | |
Numbers in bold are correlation coefficients of 0.40 or higher.
DP and MPN-PCR showed variable relationships depending on the Vibrio species and the gene marker evaluated. Correlation was stronger for total V. parahaemolyticus (r = 0.66 and 0.67 in water and oysters, respectively) than for total V. vulnificus (r = 0.42 and 0.59, respectively) (Table 1). For the virulence markers, correlation was similar among all gene targets and in both matrices (oysters and seawater), ranging from 0.31 to 0.54.
Sensitivity, specificity, and accuracy of DP in comparison to MPN-PCR.
The second step in our evaluation of the two methods was to further examine the performance of DP versus MPN-PCR to offer an explanation of the moderate to poor correlations observed. Sensitivity, or the measure of the proportion of instances when both methods provided positive results, was highest in oysters for total (tlh+) V. parahaemolyticus (87%) and total (vvhA+) V. vulnificus (85%) and was consistently higher in oysters (58%) than in water (37%) across all gene targets (Table 2). Generally poor results were obtained for all virulence markers in both matrices (19 to 51%). Again, with the exception of total V. parahaemolyticus and total V. vulnificus in oysters (13 and 15%, respectively), DP failed to detect all other Vibrio targets 42 to 81% of the time. Thus, for the remainder of this study, only MPN-PCR results were used for COPP assay model development, Vibrio species and strain enumeration, and environmental comparisons.
TABLE 2.
Performance of CHROMagar (direct plating [DP]) method for the detection of total and pathogenic Vibrio parahaemolyticus and Vibrio vulnificusa
| Sample type/gene | No. of samples |
% (lower value, upper value) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total | True positive | False positive | False negative | True negative | Sensitivity | Specificity | False-positive rate | False-negative rate | Accuracy | |
| Oyster/tlh | 110 | 90 | 3 | 14 | 3 | 87 (78, 92) | 50 (19, 81) | 50 (19, 81) | 13 (7, 20) | 86 (78, 91) |
| Water/tlh | 110 | 58 | 2 | 46 | 5 | 56 (46, 65) | 71 (35, 92) | 29 (2, 55) | 44 (35, 54) | 57 (45, 66) |
| Oyster/tdh | 110 | 35 | 1 | 34 | 40 | 51 (39, 62) | 98 (86, 100) | 2 (0, 7) | 49 (38, 61) | 68 (60, 77) |
| Water/tdh | 110 | 12 | 5 | 30 | 64 | 29 (17, 44) | 93 (84, 97) | 7 (1, 13) | 71 (58, 84) | 69 (60, 77) |
| Oyster/trh | 110 | 18 | 2 | 30 | 60 | 38 (25, 52) | 97 (88, 100) | 3 (0, 7) | 63 (49, 76) | 71 (62, 79) |
| Water/trh | 110 | 7 | 3 | 30 | 71 | 19 (9, 35) | 96 (88, 99) | 4 (0, 8) | 81 (69, 93) | 70 (62, 79) |
| Oyster/vvhA | 110 | 93 | 0 | 17 | 0 | 85 (76, 90) | 0 (IDb ) | 0 (ID) | 15 (9, 22) | 85 (78, 91) |
| Water/vvhA | 110 | 63 | 1 | 46 | 0 | 58 (48, 67) | 0 (0, 71) | 100 (ID) | 42 (33, 51) | 57 (48, 66) |
| Oyster/vcgC | 110 | 31 | 1 | 71 | 7 | 30 (22, 40) | 88 (51, 99) | 13 (0, 31) | 70 (61, 78) | 35 (26, 43) |
| Water/vcgC | 110 | 21 | 1 | 77 | 11 | 21 (14, 31) | 92 (62, 100) | 8 (0, 22) | 79 (71, 87) | 29 (21, 38) |
True positive, positive by both methods; false positive, positive only by CHROMagar method; false negative, positive only by MPN-PCR method; true negative, negative by both methods. CHROMagar-negative and MPN-PCR-positive results were interpreted as CHROMagar false negative. MPN-PCR-negative and CHROMagar-positive results were considered CHROMagar false positive. Samples positive and negative for V. parahaemolyticus and V. vulnificus by both methods were considered true positive and true negative, respectively.
ID, indeterminate.
COPP assay as an indicator of Vibrio species and virulence marker abundance.
The final step for method development was the examination of the potential utility of the COPP assay to serve as an “indicator” of elevated abundances of the two Vibrio species and virulence markers that were evaluated. For the purpose of this initial evaluation, “elevated” was defined as the top 25%, or quantile, of the data for each species/marker, and COPP was examined alone and in combination with measurements of temperature and salinity at the depth of sample collection (bottom for oyster and surface for water). The results are presented in Table 3 and are ordered by the Akaike information criterion corrected for small sample size (AICc) (21), as described in Materials and Methods, with AICc values shown from closest to the “truth” for this candidate set of predictors to furthest from the truth. AICc weights, as shown in Table 3, provide the likelihood that a given model is the best, and concordance represents the classification error or, in this case, the proportion of times the model correctly classifies the dependent variable as elevated.
TABLE 3.
Summary statistics of logistic regression model fits for prediction of the top quartile of Vibrio abundance in oysters and watera
| Sample and dependent variable | Predictor variables | K | −2log(L) | AICc | ΔAICc | AICc wt | C (%) | HL (P) |
|---|---|---|---|---|---|---|---|---|
| Oysters | ||||||||
| Elevated V. parahaemolyticus (>2.96 log MPN/g) | COPP + BWT × Bsal | 3 | 109.47 | 115.71 | 0.0 | 0.65114 | 73.7 | 0.52 |
| COPP + BWT + Bsal | 4 | 109.50 | 117.89 | 2.2 | 0.218924 | 73.4 | 0.29 | |
| COPP + BWT | 3 | 113.62 | 119.84 | 4.1 | 0.082576 | 70.7 | 0.12 | |
| COPP | 2 | 117.58 | 121.70 | 6.0 | 0.032581 | 68.6 | 0.86 | |
| BWT + Bsal | 3 | 117.37 | 123.37 | 7.7 | 0.014136 | 66.3 | 0.34 | |
| Intercept only | 1 | 127.51 | 129.55 | 13.8 | 0.000643 | |||
| Elevated V. vulnificus (>3.96 log MPN/g) | COPP + BWT + Bsal | 4 | 87.12 | 95.51 | 0.0 | 0.708921 | 86.5 | 0.44 |
| COPP + BWT | 3 | 91.28 | 97.51 | 2.0 | 0.260797 | 84.2 | 0.38 | |
| COPP + BWT × Bsal | 3 | 96.22 | 102.45 | 6.9 | 0.02206 | 82.6 | 0.37 | |
| COPP | 2 | 100.58 | 104.70 | 9.2 | 0.007162 | 81.5 | 0.65 | |
| BWT + Bsal | 3 | 102.29 | 108.52 | 13.0 | 0.001061 | 80.5 | 0.78 | |
| Intercept only | 1 | 131.48 | 133.52 | 38.0 | 3.95E−09 | |||
| Elevated tdh (>0.52 log MPN/g) | BWT + Bsal | 3 | 79.01 | 85.24 | 0 | 0.577265 | 85.2 | 0.16 |
| COPP + BWT + Bsal | 4 | 78.16 | 86.56 | 1.32 | 0.29836 | 85.7 | 0.11 | |
| COPP + BWT × Bsal | 3 | 82.08 | 88.31 | 3.07 | 0.124375 | 85.9 | 0.43 | |
| COPP + BWT | 3 | 123.20 | 129.43 | 44.19 | 1.46E−10 | 58.0 | 0.86 | |
| Intercept only | 1 | 127.51 | 129.55 | 44.31 | 1.38E−10 | |||
| COPP | 2 | 125.69 | 129.80 | 44.56 | 1.22E−10 | 56.9 | 0.72 | |
| Elevated trh (>0.40 log MPN/g) | BWT + Bsal | 3 | 63.39 | 69.62 | 0.00 | 0.514814 | 90.1 | 0.45 |
| COPP + BWT + Bsal | 4 | 62.70 | 71.10 | 1.48 | 0.245625 | 90.9 | 0.22 | |
| COPP + BWT × Bsal | 3 | 64.92 | 71.15 | 1.53 | 0.239561 | 89.0 | 0.02 | |
| COPP | 2 | 120.44 | 124.55 | 54.93 | 6.08E−13 | 61.0 | 0.32 | |
| COPP + BWT | 3 | 118.33 | 124.56 | 54.94 | 6.05E−13 | 59.9 | 0.35 | |
| Intercept only | 1 | 123.16 | 125.20 | 55.58 | 4.39E−13 | |||
| Elevated vcgC (>1.64 log MPN/g) | COPP + BWT × Bsal | 3 | 106.18 | 112.42 | 0 | 0.775304 | 74.8 | 0.67 |
| COPP + BWT + Bsal | 4 | 106.92 | 115.31 | 2.89 | 0.182775 | 74.3 | 0.76 | |
| COPP + BWT | 3 | 113.07 | 119.30 | 6.88 | 0.02486 | 69.5 | 0.45 | |
| COPP | 2 | 116.31 | 120.42 | 8.00 | 0.0142 | 69.1 | 0.38 | |
| BWT + Bsal | 3 | 117.50 | 123.73 | 11.31 | 0.002714 | 66.2 | 0.52 | |
| Intercept only | 1 | 127.51 | 129.55 | 17.13 | 0.000148 | |||
| Water | ||||||||
| Elevated V. parahaemolyticus (>1.397 log MPN/ml) | COPP + SWT + Ssal | 4 | 46.29 | 54.68 | 0.0 | 0.981729 | 96.1 | 0.63 |
| COPPTV + SWT | 3 | 56.68 | 62.91 | 8.2 | 0.016028 | 94.6 | 0.20 | |
| COPP + SWT × Ssal | 3 | 60.69 | 66.92 | 12.2 | 0.002155 | 92.2 | 0.34 | |
| COPP | 2 | 69.53 | 73.64 | 19.0 | 7.5E−05 | 91.3 | 0.12 | |
| SWT + Ssal | 3 | 70.90 | 77.12 | 22.4 | 1.32E−05 | 90.1 | 0.36 | |
| Intercept only | 1 | 125.39 | 127.42 | 72.7 | 1.57E−16 | |||
| Elevated V. vulnificus (>1.968 log MPN/ml) | COPP + SWT + Ssal | 4 | 94.71 | 103.10 | 0.0 | 0.727875 | 81.1 | 0.49 |
| COPP + SWT × Ssal | 3 | 99.67 | 105.91 | 2.8 | 0.178597 | 77.9 | 0.60 | |
| COPP + SWT | 3 | 101.38 | 107.61 | 4.5 | 0.076335 | 78.7 | 0.67 | |
| COPP | 2 | 105.52 | 110.63 | 7.5 | 0.016863 | 75.8 | 0.48 | |
| SWT + Ssal | 3 | 112.29 | 118.52 | 15.4 | 0.000326 | 68.1 | 0.45 | |
| Intercept only | 1 | 125.39 | 127.42 | 24.3 | 3.81E−06 | |||
| Elevated tdh (>0.00689 log MPN/ml) | COPP + SWT + Ssal | 4 | 72.65 | 80.96 | 0 | 0.951142 | 87.7 | 0.42 |
| SWT + Ssal | 3 | 80.77 | 87.00 | 6.05 | 0.046301 | 84.6 | 0.14 | |
| COPP + SWT | 3 | 86.67 | 92.91 | 11.96 | 0.002411 | 85.0 | 0.02 | |
| COPP + SWT × Ssal | 3 | 83.53 | 125.39 | 44.43 | 2.14E−10 | 83.4 | 0.18 | |
| COPP | 2 | 94.41 | 98.52 | 17.57 | 0.000146 | 81.6 | 0.11 | |
| Intercept only | 1 | 125.39 | 127.42 | 46.47 | 7.74E−11 | |||
| Elevated trh (>0.00389 log MPN/ml) | COPP + SWT + Ssal | 4 | 58.87 | 67.27 | 0.00 | 0.985699 | 92.7 | 0.10 |
| COPP + SWT | 3 | 70.75 | 76.98 | 9.71 | 0.007678 | 90.8 | 0.14 | |
| COPP + SWT × Ssal | 3 | 71.37 | 77.60 | 10.33 | 0.005631 | 89.5 | 0.37 | |
| SWT + Ssal | 3 | 75.20 | 81.43 | 14.16 | 0.00083 | 87.3 | <0.01 | |
| COPP | 2 | 80.59 | 84.70 | 17.43 | 0.000162 | 88.0 | 0.51 | |
| Intercept only | 1 | 125.39 | 127.42 | 60.15 | 8.55E−14 | |||
| Elevated vcgC (>0.154 log MPN/ml) | COPP + SWT × Ssal | 3 | 97.17 | 103.41 | 0 | 0.697358 | 82.0 | 0.69 |
| COPP + SWT + Ssal | 4 | 97.79 | 106.18 | 2.77 | 0.174565 | 80.9 | 0.65 | |
| SWT + Ssal | 3 | 101.05 | 107.28 | 3.87 | 0.100715 | 78.6 | 0.73 | |
| COPP | 2 | 106.61 | 110.73 | 7.32 | 0.017945 | 76.0 | 0.63 | |
| COPP + SWT | 3 | 105.79 | 112.02 | 8.61 | 0.009415 | 76.3 | 0.78 | |
| Intercept only | 1 | 127.51 | 129.55 | 26.14 | 1.47E−06 |
Model selection based on Akaike’s information criterion corrected for small sample size (AICc), change in AICc, and AICc weights for logistic regression models for prediction of the top quartile Vibrio abundance in oysters and water. −2log(L), two log likelihood or deviance; K, number of estimated parameters. COPP, colony overlay procedure for peptidase assay; BWT and SWT, water temperature at bottom or surface, respectively; Bsal and Ssal, salinity at bottom or surface, respectively. The concordance of logistic regression models coefficient (C) and Hosmer-Leemshow (HL) goodness-of-fit statistics are also presented.
For oysters, the COPP assay alone never provided the best model (concordance = 56.9 to 81.5%), and with the noted exception of V. vulnificus (81.5% concordance), it generally provided moderate to low predictive power on its own (Table 3). However, in combination with bottom water temperature and salinity, all final models exceeded 73% concordance and were generally more than twice as likely to be the best model as competing models. The noted exceptions are models for tdh and trh, for which the addition of the COPP assay afforded little improvement over temperature and salinity alone (<1% concordance, ΔAICc < 2). However, there is little downside to incorporating COPP assay results, as these models demonstrated high concordances of 85.7 and 90.9%, respectively, for oysters.
Water models generally showed higher concordance than oysters, with all over 81% (Table 3). COPP alone was never the best-performing model, but for V. parahaemolyticus (tlh, tdh, and trh) it demonstrated potentially useful concordances of 91.3, 81.6, and 88.0%, respectively. In all cases, the top-performing model was at least four times more likely to be the best model over the other candidates. In two instances, individual models failed the Hosmer-Lemeshow (HL) goodness-of-fit test (Table 3), where HL (P) was ≤0.05; however, neither model was selected as the top performer, and these results are included only for reference.
Environmental characterization.
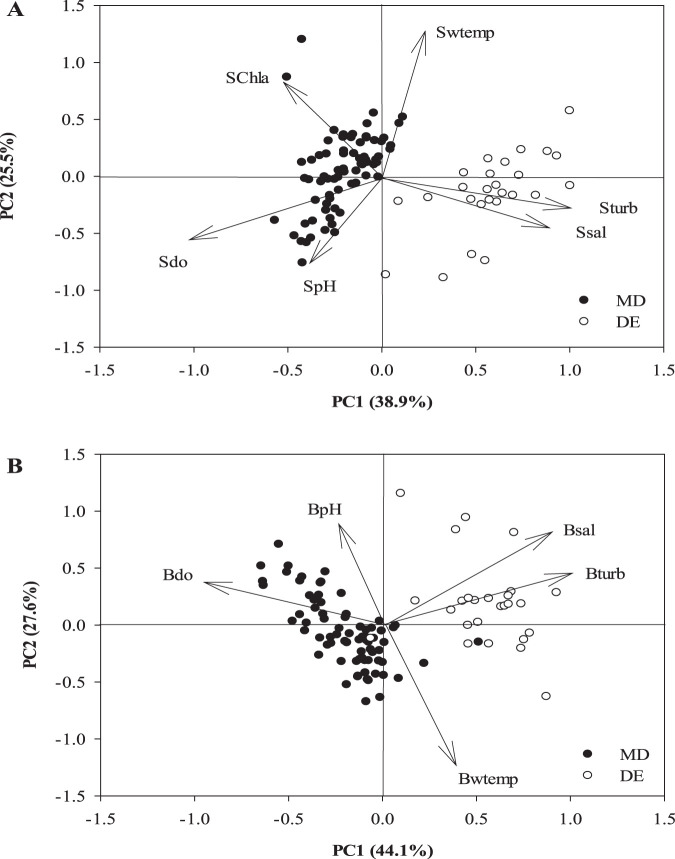
The second goal of this study was to compare Vibrio dynamics between the estuaries in both water and oysters. To begin, we first characterized the physicochemical environment to determine the major gradients that exist between the two systems. Overall, 64.4% of the environmental variation in surface water characteristics could be explained by the first two principal components (PCs) (Fig. 1A), with PC1 (38.9%) representing a salinity/turbidity gradient and PC2 (25.5%) a temperature/oxygen/pH gradient. PC1 clearly separates the states, with Delaware sites being more turbid and having a greater salt content than those sampled in Maryland. Nearly identical results were obtained for bottom waters, with PC1 (salinity/turbidity) explaining 44.1% of the overall variation and PC2 (temperature/oxygen/pH) contributing 27.6% (71.7% overall) (Fig. 1B).
FIG 1.
Principal components (PCs) representing the environmental gradients encountered in Maryland (closed circles) and Delaware (open circles) in this study in surface waters (A) and bottom waters (B). Abbreviations: S and Sw, surface water; B and Bw, bottom water; Chla, chlorophyll a; do, dissolved oxygen; temp, temperature; sal, salinity; tur, turbidity; MD, Maryland; DE, Delaware.
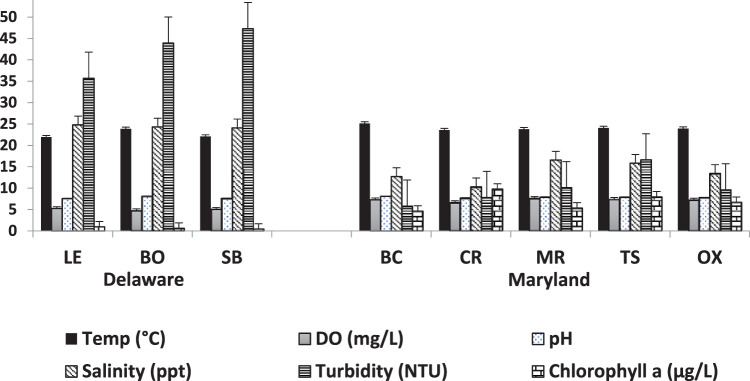
Surface and bottom water temperature and pH did not differ significantly between the states; however, all other water quality parameters measured did (P < 0.05) (Fig. 2). Dissolved oxygen was low during the summer and relatively high in the winter, with values ranging from 2.8 mg/liter (July) to 10.1 mg/liter (November). The annual mean dissolved oxygen values were 7.23 and 5.02 mg/liter, respectively, in Maryland and Delaware surface waters and 7.20 and 4.90 mg/liter, respectively, in bottom waters. The chlorophyll a values for water at both Maryland and Delaware sites ranged from 0.1 to 41.1 μg/liter; they were significantly higher in Maryland as noted from the principal-component analysis (PCA). The most striking differences separating the bays are in salinity and turbidity. Delaware sites were over 10 ppt saltier than Maryland sites on average and over four times more turbid.
FIG 2.
Average surface water physicochemical parameters for eight sampling sites. Abbreviations: LE, Lewes; BO, Bowers; SB, Slaughter Beach; BC, Broad Creek; CR, Chester River; MR, Manokin River; TS, Tangier Sound; OX, Oxford; Temp, temperature; DO, dissolved oxygen.
Comparative abundances of total Vibrionaceae in the Chesapeake Bay, MD, and in the Delaware Bay, DE.
In our initial analysis of environmental conditions in the two bays, a clear separation of Maryland and Delaware was noted along a salinity/turbidity gradient (PC1) (Fig. 1A and B). This difference in environmental conditions is confounded with the designation of “state” as a predictor variable, precluding determination of the variability in abundance associated with both. Indeed, evaluation of models including both PC1 and state as independent variables for multicollinearity demonstrated variance inflation in exceedance of an acceptable range. Therefore, the analysis presented below uses only PC2 as a covariate, and differences highlighted between the states may be due to either differences in turbidity/salinity or other factors unique to the water bodies—factors that were not measured.
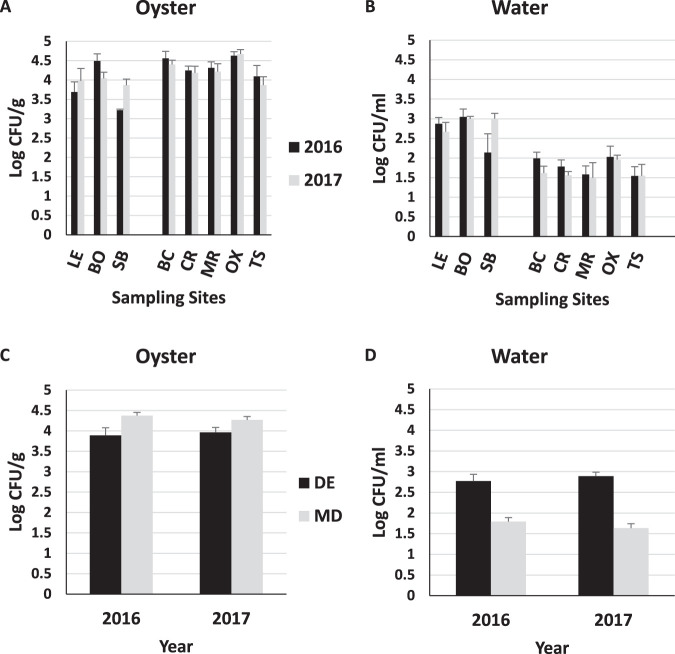
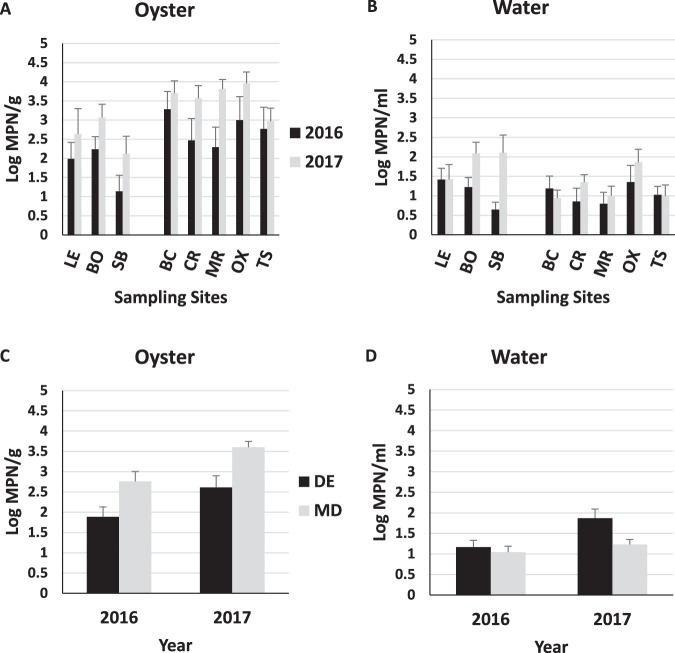
All oyster samples were positive for total Vibrionaceae (TV) in both states, with concentrations typically ranging from 2.78 to 5.18 log CFU/g (Table 4; Fig. 3). State, site, and PC2 were significant (P < 0.05), with the effect size of PC2 (ω2 = −0.14) and site (ω2 = −0.13) greater than that of state (ω2 = −0.02). Overall, Maryland oysters were marginally higher in TV than Delaware oysters (4.33 CFU/g versus 3.95 CFU/g, respectively; P = 0.04) (Fig. 3C), with the Slaughter Beach and Lewes sites generally having lower concentrations of TV in oysters than the other sites (Fig. 3). In comparison to physicochemical variables, TV in oysters correlated somewhat with temperature (r = 0.49) but not with any other factors (Table 5).
TABLE 4.
Abundance and frequency of detection of total and pathogenic Vibrio parahaemolyticus and Vibrio vulnificus in oysters and seawater based on most-probable-number–PCR and total Vibrionaceae
| Sampling location | Sample | Genea | Log MPN/g or log MPN/ml |
Detection frequency (%) | |
|---|---|---|---|---|---|
| Range | Avg | ||||
| Delaware | Oyster | tlh | 1.38–4.97 | 2.59 | 100 |
| tdh | NDb –2.97 | 0.94 | 96.8 | ||
| trh | ND–2.38 | 0.95 | 93.5 | ||
| vvhA | 0.52–5.34 | 2.31 | 100 | ||
| vcgC | ND–3.16 | 1.07 | 93.5 | ||
| TV | 2.84–4.91 | 3.95 | 100 | ||
| Water | tlh | 0.53–3.63 | 1.86 | 100 | |
| tdh | ND–1.64 | 0.20 | 87.1 | ||
| trh | ND–1.40 | 0.29 | 83.9 | ||
| vvhA | 0.09–3.38 | 1.57 | 100 | ||
| vcgC | ND–1.97 | 0.39 | 90.3 | ||
| TV | 1.54–3.50 | 2.90 | 100 | ||
| Maryland | Oyster | tlh | ND–5.03 | 1.97 | 92.4 |
| tdh | ND–0.66 | 0.15 | 48.1 | ||
| trh | ND–0.66 | 0.07 | 22.8 | ||
| vvhA | 0.13–5.26 | 3.19 | 100 | ||
| vcgC | ND–3.59 | 1.06 | 92.4 | ||
| TV | 2.78–5.18 | 4.33 | 100 | ||
| Water | tlh | ND–1.8 | 0.29 | 88.6 | |
| tdh | ND–0.02 | <0.01 | 13.9 | ||
| trh | ND–0.3 | <0.01 | 3.8 | ||
| vvhA | ND–3.63 | 1.14 | 98.7 | ||
| vcgC | ND–0.94 | 0.09 | 79.7 | ||
| TV | ND–3.43 | 1.71 | 100 | ||
tlh, thermolabile hemolysin gene of V. parahaemolyticus (indicator of total V. parahaemolyticus); tdh, thermostable direct hemolysin gene of pathogenic strains of V. parahaemolyticus; trh, tdh-related hemolysin gene of pathogenic strains of V. parahaemolyticus; vvhA, cytolytic hemolysin gene of V. vulnificus (indicator of total V. vulnificus); vcgC, virulence-correlated gene of V. vulnificus (indicator of pathogenic V. vulnificus); TV, total Vibrionaceae as determined by the COPP assay.
ND, nondetectable.
FIG 3.
Average abundances of total Vibrionaceae in oysters (A and C) and seawater (B and D) determined using the COPP assay. Abbreviations: DE, Delaware; MD, Maryland; LE, Lewes; BO, Bowers; SB, Slaughter Beach; BC, Broad Creek; CR, Chester River; MR, Manokin River; OX, Oxford; TS, Tangier Sound.
TABLE 5.
Pearson correlation coefficients comparing the physicochemical parameters of the harvest water with the colony overlay procedure for peptidases assay or the most-probable-number–PCR method
| Physicochemical parameter | Comparison | Correlation coefficient (r value, significance level)a
|
|
|---|---|---|---|
| Water | Oysters | ||
| Temp | COPP vs temp | 0.36, <0.0001 | 0.49, <0.0001 |
| Salinity | COPP vs salinity | 0.46, <0.0001 | −0.27, 0.3254 |
| Dissolved oxygen | COPP vs dissolved oxygen | −0.70, <0.0001 | −0.10, 0.3254 |
| Turbidity | COPP vs turbidity | 0.63, <0.0001 | −0.30, 0.0024 |
| Chlorophyll a | COPP vs chlorophyll a | −0.18, 0.0705 | 0.13, 0.2005 |
| pH | COPP vs pH | −0.22, 0.0231 | −0.06, 0.5175 |
| Temp | V. vulnificus (MPN-PCR) vs temp | 0.41, <0.0001 | 0.48, <0.0001 |
| Salinity | V. vulnificus (MPN-PCR) vs salinity | 0.07, 0.4445 | −0.24, 0.0112 |
| Dissolved oxygen | V. vulnificus (MPN-PCR) vs dissolved oxygen | −0.33, 0.0005 | 0.02, 0.8564 |
| Turbidity | V. vulnificus (MPN-PCR) vs turbidity | 0.25, 0.0105 | −0.24, 0.0112 |
| Chlorophyll a | V. vulnificus (MPN-PCR) vs chlorophyll a | 0.04, 0.7065 | −0.02, 0.0179 |
| pH | V. vulnificus (MPN-PCR) vs pH | −0.19, 0.0606 | −0.02, 0.8395 |
| Temp | V. parahaemolyticus (MPN-PCR) vs temp | −0.02, <0.0001 | 0.19, 0.0536 |
| Salinity | V. parahaemolyticus (MPN-PCR) vs salinity | 0.68, <0.0001 | 0.34, 0.0003 |
| Dissolved oxygen | V. parahaemolyticus (MPN-PCR) vs dissolved oxygen | −0.59, <0.0001 | −0.16, 0.1119 |
| Turbidity | V. parahaemolyticus (MPN-PCR) vs turbidity | 0.69, <0.0001 | −0.16, 0.1058 |
| Chlorophyll a | V. parahaemolyticus (MPN-PCR) vs chlorophyll a | −0.28, 0.0038 | −0.13, 0.1766 |
| pH | V. parahaemolyticus (MPN-PCR) vs pH | −0.14, 0.1715 | 0.01, 0.8938 |
Correlations reflect surface water comparisons for water and bottom water comparisons for oysters, with the exception of those with chlorophyll a, which are surface water for both. Numbers in bold are r values of >0.40.
The vast majority of water samples were also positive for TV, although generally at concentrations more than a log lower than found in oysters (Table 4). The largest effect size was attributed to state (ω2 = 0.56), with PC2 (ω2 = 0.17) and site (ω2 = 0.04) also contributing significantly (P < 0.05). In contrast to the case for oysters, TV in Delaware waters averaged over a log greater than in Maryland (2.90 and 1.71 log CFU/ml, respectively) (Fig. 3D). Correlation of TV with PC1 was very strong (r = 0.72, P < 0.0001), suggesting that state level effects may largely be associated with higher salinity and turbidity in Delaware. Further, TV correlated to an extent with salinity of the seawater (r = 0.46) and moderately with turbidity (r = 0.63), while a strong negative correlation with dissolved oxygen was observed (r = −0.70) (Table 5).
Vibrio parahaemolyticus and V. vulnificus: detection frequencies, abundance, and association with environmental parameters.
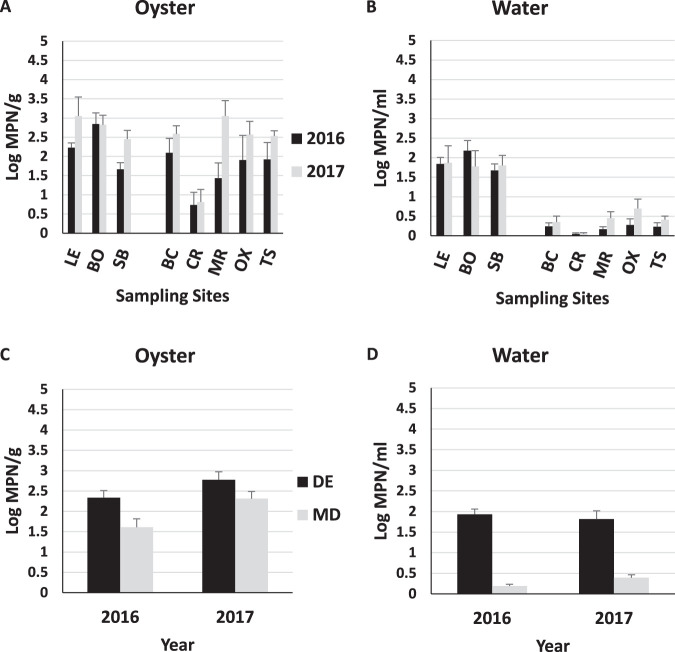
Frequencies of detection of V. parahaemolyticus in oysters as determined by tlh-positive samples were similar between the states, occurring in all Delaware samples and 92.4% of Maryland oysters (Table 4). Vibrio parahaemolyticus concentrations in oysters were marginally higher in Delaware than in Maryland (2.59 and 1.97 log MPN/g, respectively) (Fig. 3). The largest effect size was attributed to site (ω2 = 0.20) and year (ω2 = 0.09), with state (ω2 = 0.05) and PC2 (ω2=0.03) also contributing to a lesser extent (P < 0.05). Site differences were largely attributable to consistently low levels in oysters from the Chester River, MD, compared to the other sites (Fig. 4). None of the water’s physicochemical factors correlated particularly well with V. parahaemolyticus in oysters (Table 5).
FIG 4.
Average abundances of total Vibrio parahaemolyticus in oysters (A and C) and seawater (B and D) determined using MPN-PCR. Abbreviations: DE, Delaware; MD, Maryland; LE, Lewes; BO, Bowers; SB, Slaughter Beach; BC, Broad Creek; CR, Chester River; MR, Manokin River; OX, Oxford; TS, Tangier Sound.
In water, V. parahaemolyticus was detected 100% of the time in Delaware and slightly less frequently (88.6%) in Maryland (P = 0.11, Fisher’s exact test). Difference among the states (1.86 versus 0.29 log MPN/g in Delaware and Maryland, respectively) was the only component of those examined that explained a significant proportion of variance (ω2 = 0.63) (Fig. 4). Similar to the case for TV in water, correlation with PC1 was very strong (r = 0.75, P < 0.0001), suggesting that state differences may, in fact, be due to the salinity/turbidity gradient. Vibrio parahaemolyticus in the water correlated moderately with salinity and turbidity (r = 0.68 and 0.69, respectively) and correlated negatively with dissolved oxygen (r = −0.59) (Table 5).
A high detection frequency was also observed for total V. vulnificus in oysters (vvhA+), with 100% occurrence in both states (Table 4). Year, state, and PC2 were all significant (P < 0.05), with the largest effect size attributed to year (ω2 = 0.16) and PC2 (ω2 = 0.16). The Vibrio vulnificus concentration was higher in 2017 (3.39 log MPN/g) than in 2016 (2.49 log MPN/g) in both states. The average V. vulnificus concentration was nearly 1 log higher in Maryland samples than in Delaware samples (3.19 versus 2.31 log MPN/g, respectively) (Fig. 5).
FIG 5.
Average abundances of total Vibrio vulnificus in oysters (A and C) and seawater (B and D) determined using MPN-PCR. Abbreviations: DE, Delaware; MD, Maryland; LE, Lewes; BO, Bowers; SB, Slaughter Beach; BC, Broad Creek; CR, Chester River; MR, Manokin River; OX, Oxford; TS, Tangier Sound.
Vibrio vulnificus in water responded similarly in terms of annual differences (2017 > 2016, P < 0.05) but was marginally higher in Delaware than Maryland (P < 0.05) (Fig. 5D). Overall, mean concentrations were 1 to 2 logs lower than those observed in oysters (1.57 and 1.14 log MPN/g in Delaware and Maryland, respectively). PC2 (ω2 = 0.18) demonstrated the largest effect size, reflecting increasing abundance with temperature. The frequency of detection was 100% in Delaware and 98.7% in Maryland (Table 4). Vibrio vulnificus counts in seawater and oysters correlated somewhat with water temperature (r = 0.41 and 0.48, respectively) but poorly with all other variables (Table 5).
Virulence genes: detection frequencies, abundance, and association with environmental parameters.
While overall concentrations of V. vulnificus were higher in Maryland oysters than in Delaware oysters (3.19 versus 2.31 log MPN/g), the prevalence (92.4 versus 93.5%) and concentration (1.06 versus 1.07 log MPN/g) of vcgC+ strains were nearly identical (Table 4). Of the candidate predictors, PC2 and site explained most of the variance, but the overall model was not significant (P > 0.05).
Potentially pathogenic (tdh+) V. parahaemolyticus strains were found in 96.8% of Delaware oyster samples, compared to 48.1% of Maryland oysters (P < 0.0001, Fisher’s exact test) (Table 4). State demonstrated the largest effect size (ω2 = 0.39), with site also contributing (ω2 = 0.06) (P < 0.05). Concentrations were low in oysters from both states, averaging 0.94 and 0.15 log MPN/g in Delaware and Maryland, respectively (Table 4). High correlation was noted for PC1 (r = 0.62, P < 0.0001) and specifically salinity (r = 0.69, P < 0.0001), suggesting that state differences may be attributable to environmental differences.
Potentially pathogenic V. parahaemolyticus (trh+ strains) was detected in 93.5% of Delaware oyster samples, compared to only 22.8% of Maryland oysters (P < 0.0001, Fisher’s exact test) (Table 4). In Delaware oysters, 93.5% had both pathogenicity markers (tdh and trh), compared to just 19.2% in Maryland (P < 0.0001, Fisher’s exact test). The same trend as tdh is apparent for trh in oysters, with low concentrations of the strains overall but significantly higher in Delaware than in Maryland (0.95 ± 0.07 and 0.07 ± 0.04 log MPN/g, respectively; P < 0.0001, ω2 = 0.49) (Table 4). No other factor explained a significant portion of variance. As with tdh, trh correlated well with PC1 (r = 0.63, P < 0.0001) and salinity (r = 0.69, P < 0.0001) (Table 6), suggesting that higher salinity in Delaware may account for the observed differences.
TABLE 6.
Pearson correlation coefficients comparing the physicochemical parameters of the harvest water with abundance of vibrios carrying virulence markers in water and oysters
| Physicochemical parameter | Comparisona | Correlation coefficients (r value and significance level)b
|
|
|---|---|---|---|
| Water | Oysters | ||
| Temp | tdh vs temp | −0.02, 0.84 | 0.06, 0.54 |
| Salinity | tdh vs salinity | 0.45, <0.0001 | 0.69, <0.0001 |
| Dissolved oxygen | tdh vs dissolved oxygen | −0.35, 0.0003 | −0.61, <0.0001 |
| Turbidity | tdh vs turbidity | 0.23, 0.0173 | 0.42, <0.0001 |
| Chlorophyll a | tdh vs chlorophyll a | −0.17, 0.0934 | −0.29, 0.0032 |
| pH | tdh vs pH | 0.02, 0.8474 | −0.02, 0.8797 |
| Temp | trh vs temp | 0.01, 0.9163 | 0.04, 0.7024 |
| Salinity | trh vs salinity | 0.42, <0.0001 | 0.69, <0.0001 |
| Dissolved oxygen | trh vs dissolved oxygen | −0.48, <0.0001 | −0.60, <0.0001 |
| Turbidity | trh vs turbidity | 0.32, 0.0008 | 0.47, <0.0001 |
| Chlorophyll a | trh vs chlorophyll a | −0.21, 0.0317 | −0.30, 0.0019 |
| pH | trh vs pH | −0.18, 0.0637 | −0.10, 0.3017 |
| Temp | vcgC vs temp | 0.14, 0.1461 | 0.37, <0.0001 |
| Salinity | vcgC vs salinity | 0.39, <0.0001 | 0.15, 0.1360 |
| Dissolved oxygen | vcgC vs dissolved oxygen | −0.40, <0.0001 | −0.15, 0.1158 |
| Turbidity | vcgC vs turbidity | 0.45, <0.0001 | 0.07, 0.4834 |
| Chlorophyll a | vcgC vs chlorophyll a | −0.08, 0.4128 | 0.11, 0.2554 |
| pH | vcgC vs pH | −0.20, 0.0438 | 0.003, 0.9740 |
tlh, thermolabile hemolysin gene of V. parahaemolyticus (indicator of total V. parahaemolyticus); tdh, thermostable direct hemolysin gene of pathogenic strains of V. parahaemolyticus; trh, tdh-related hemolysin of pathogenic strains of V. parahaemolyticus; vvhA, cytolytic hemolysin gene of V. vulnificus (indicator of total V. vulnificus); vcgC, virulence-correlated gene of V. vulnificus (indicator of pathogenic V. vulnificus).
Correlations reflect surface water comparisons for water and bottom water comparisons for oysters, with the exception of those with chlorophyll a, which are surface waters for both. Numbers in bold are r values of >0.40.
Frequencies of detection of virulence genes in water samples followed a trend similar to that in oysters, with 90.3 and 79.7% vcgC positive in Delaware and Maryland, respectively (P < 0 0.05, Fisher’s exact test) (Table 4). State (ω2 = 0.25), year (ω2 = 0.04), and PC2 (ω2 = 0.06) were all significant (P < 0.05), with Delaware having higher average concentrations than Maryland (0.39 versus 0.09 log MPN/g) (Table 4) and overall concentrations being slightly higher in 2017. A moderate correlation with turbidity was observed (r = 0.45, P < 0.05) (Table 6).
For V. parahaemolyticus in water, tdh+ and trh+ strains were present in a higher proportion of samples in Delaware (87.1% tdh+, 83.9% trh+) than in Maryland (13.9% tdh+, 3.8% trh+) (P < 0.0001, Fisher’s exact test) (Table 4). Both tdh+ and trh+ strains were more prevalent in 2017 than in 2016 (P < 0.05, Fisher’s exact test). As with oysters, state carried the largest effect size for both markers (ω = 0.20 and 0.31 for tdh and trh, respectively), with moderate correlations with PC1 and salinity (r = 0.40 to 0.51, P < 0.05). A weak year effect was also notable with tdh (ω2 = 0.03), being greater in 2017 than in 2016 (P = 0.04).
DISCUSSION
This is the most comprehensive study comparing abundances and frequencies of detection of total vibrios and total and pathogenic V. parahaemolyticus and V. vulnificus in oysters and water in the Chesapeake Bay and Delaware Bay. Multiple detection methods were compared from May to October, when the highest abundances of Vibrio typically occur in the United States. Five Maryland and three Delaware oyster-harvesting sites that provided high and low salinity ranges were selected in order to maximize the range of collection conditions encountered during the study.
In this study, DP on CHROMagar Vibrio was much less sensitive in identifying vcgC-positive isolates than MPN-PCR. A similar trend was also observed for the tdh- and trh-positive isolates of V. parahaemolyticus. This might be due to the testing of 10 to 20% of the isolates using real-time PCR instead of all colonies recovered from a sample. One limitation in using CHROMagar Vibrio is that the agar contains 6% NaCl, which may be inhibitory to many vibrios, including V. parahaemolyticus and V. vulnificus, especially if the vibrios are already stressed. MPN-PCR is more sensitive than direct plating of samples on CHROMagar Vibrio for detecting/enumerating these pathogens (22, 23), due in large part to the initial enrichment step which resuscitates weakened vibrios under more normal salt levels (18).
In our DP tests, we picked mauve colonies (presumptive V. parahaemolyticus) and green colonies (presumptive V. vulnificus) from plates for species confirmation by real-time PCR. The high percentage of false-negative results (ranging from 49% to 81%) for pathogenic strains of V. parahaemolyticus and V. vulnificus (Table 2) limits the utility of DP to serve as a viable method for monitoring pathogenic vibrios in oysters and seawater. False-negative results were somewhat less for total V. parahaemolyticus and V. vulnificus in oysters (13 and 15%, respectively). Sensitivity and specificity rates higher than 95% are usually considered acceptable measures of a validated method (24). None of the sensitivity results for the DP method in oysters or seawater met this threshold (Table 2).
The COPP assay for total Vibrionaceae detection was developed not to discriminate between pathogenic and nonpathogenic vibrios but rather as a rapid screening technique to assess the general levels of Vibrionaceae present in oysters, seawater, and environmental samples, including well water (15, 16, 25). The Vibrionaceae family of bacteria contains, among others, members of the genera Vibrio, Photobacterium, and Listonella. Because of genetic similarities between Shewanella spp. and Vibrionaceae, Shewanella was also recommended for inclusion in the Vibrionaceae family (26). To date, all of these genera have been shown to test positive by the rapid, simple, and inexpensive COPP assay. In contrast, the standard MPN-PCR for vibrios is designed to detect total V. parahaemolyticus (tlh+) and total V. vulnificus (vvhA+). Consequently, this study evaluated whether total Vibrionaceae as determined by the COPP assay demonstrated predictive potential for total or pathogenic V. parahaemolyticus and V. vulnificus in either seawater or oysters.
Direct correlation of total Vibrionaceae (COPP) abundance with total or pathogenic V. parahaemolyticus and V. vulnificus in either seawater or oysters was variable with both MPN-PCR and DP (Table 1). Given the multitude of differences in media, enrichment selectivity, and endpoint sensitivity previously discussed, this is not surprising. However, when combined with the simple-to-measure environmental parameters of temperature and salinity, predictive models demonstrate potentially useful concordance (Table 3). Notably, these models do not predict absolute abundance but rather predict the probability of exceeding a predetermined threshold. For initial model development, we used the data to define thresholds (top quartile), which may or may not have relevance to human health. Currently, there are no criteria for Vibrio in water or for pathogenic strains. While criteria have been proposed for V. parahaemolyticus in oysters, most oyster samples encountered in this study exceeded the recommended pathogenic limit of 100 MPN/g (27), and a few reached the total V. parahaemolyticus threshold of 10,000 MPN/g (17). While there is ongoing debate as to dose response or the relationship between total V. parahaemolyticus and illness, thresholds for virulence genes (trh and tdh) may offer a more direct means of assessing risk. The COPP assay in combination with temperature and salinity was extremely reliable in predicting elevated levels of these genes and offers promise for future development of a rapid “indicator” of oysters more likely to cause illness.
The second goal of this study was to compare Vibrio levels in Maryland and Delaware. In order to reduce the number of variables in the analysis and visualize major environmental differences among the systems, we used principal-component analysis. The exercise highlighted major differences between the systems along a salinity-turbidity gradient, with Delaware samples coming from significantly more turbid and saline environments. Inclusion of this principal component (PC1) in subsequent statistical models resulted in excessive variance inflation, and thus it was removed. Therefore, we are left only to speculate through correlation on whether differences in Vibrio concentration between the states are due to this gradient or to factors not measured.
Although sites and study years had no overall effect on the prevalence of TV, they had effects on the prevalence of V. parahaemolyticus and V. vulnificus. The Chester River had the lowest level of V. parahaemolyticus compared to all other sites (P < 0.05). This might be due to the low salinity (9.2 to 13.9 ppt) at this site, as the optimum growth of V. parahaemolyticus occurs within the salinity range of 10 to 23 ppt (3, 5, 28). Samples recovered from Maryland had higher V. vulnificus levels, and those from Delaware had higher V. parahaemolyticus levels. The salinity of the Delaware sites on average was over 10 ppt higher than that at the Maryland sites, which may explain the differences. It has been also reported that V. vulnificus is more susceptible to high salinity than V. parahaemolyticus (29–31). In 2017, differences in abundance were about one-half log for both V. parahaemolyticus and V. vulnificus between the two geographical regions. Several studies (4, 29–32) also reported that the prevalence of these bacteria may vary from site to site and that physicochemical parameters may be responsible for this variation. However, we did not find any clear explanation based on the physicochemical parameters of the water. This suggests that there are yet-unidentified factors that contribute to the frequency and distribution of these bacteria in oysters and water.
Compared to seawater, oyster samples contained more TV, V. parahaemolyticus, and V. vulnificus at all sites in Delaware and Maryland. This is commonly observed due to the ability of oysters to bioaccumulate large numbers of microorganisms as they filter the water. Our results are consistent with the finding of previous studies that reported higher levels of V. parahaemolyticus and V. vulnificus in oysters than in seawater (4, 29, 30).
In this study, tdh- and trh-positive samples were more prevalent in Delaware than in Maryland. In addition, more than 90% of Delaware samples were positive for both tdh and trh, compared to 19% in Maryland. There was a significant (P < 0.05) difference between the prevalences of tdh-positive V. parahaemolyticus in 2016 and 2017. In contrast, there was no significant difference (P > 0.05) in the prevalence of vcgC-positive samples in Delaware or Maryland. These results indicate that V. parahaemolyticus isolates recovered from the Delaware Bay may be significantly (P < 0.05) more virulent than those from the Chesapeake Bay, based on the detection of commonly recognized pathogenicity genes. Previous studies reported that levels of pathogenic vibrios may vary from region to region and year to year, which is consistent with results of our study (4, 29, 33). The majority of the pathogenic V. parahaemolyticus and V. vulnificus samples were recovered from June through August. On the other hand, a previous study in the Chesapeake Bay reported that most of the pathogenic V. parahaemolyticus isolates were observed in both warmer and colder months (32) suggesting that environmental factors may affect the temporal changes in the prevalence of these strains of V. parahaemolyticus. However, we did not observe any correlation between the prevalence of the pathogenic strains in oysters and environmental parameters, except for a weak correlation between seawater turbidity and vcgC+ V. vulnificus levels. Correlations were moderate for pathogenic V. parahaemolyticus and salinity and turbidity and were negative for dissolved oxygen (Table 6). Additional research is needed to confirm whether these strains are consistently more abundant in Delaware and, if so, why. The high incidence of pathogenic V. parahaemolyticus in oyster and water samples is a huge concern from food safety and public health standpoints, and the results are inconsistent with previous studies that reported that 2 to 40% of oyster and water samples were positive for these pathogens. The prevalence of these strains may vary by sample type as well as the sensitivity of the detection methodology (4, 34).
Previous studies have indicated that there was a positive correlation between vibrios in oysters and seawater temperature (4, 29, 30, 32). In our current study, we did not observe any strong positive correlation between vibrios in the samples and temperature, which is likely due to our collection of samples only during the warmer months (May to October). Salinity is one of the variables most often correlated with levels of vibrios in water. Recently, Froelich et al. (30) reported that salinity did correlate with the levels of V. parahaemolyticus and V. vulnificus in oysters and water in a North Carolina estuary. In our study, TV and V. parahaemolyticus correlated with salinity, but V. vulnificus, which is less salt tolerant than V. parahaemolyticus, did not correlate with salinity. Another study conducted in the Chesapeake Bay reported no correlation between salinity and V. parahaemolyticus levels in oysters or water (4). Johnson et al. (29) conducted a multiyear study in three different regions of the United States and observed that the correlation between salinity and the Vibrio level in oysters and water depended on the range of the salinity in water and the sample size in the studies.
Like in previous studies (4, 31), TV and V. parahaemolyticus showed negative and positive relationships with dissolved oxygen (32) and turbidity, respectively. Chlorophyll a and pH values did not correlate with TV and V. parahaemolyticus or V. vulnificus in Delaware or Maryland, which is also consistent with previous studies conducted in Maryland (4, 32).
In summary, two methods were compared with standard MPN-PCR assays as potentially simpler, less costly, and more rapid alternatives to the MPN-PCR for V. parahaemolyticus and V. vulnificus detection. The DP method showed high false-negative rates, which precluded its further evaluation. In contrast, logistic regression models of the COPP assay, factoring in seawater temperature and salinity, showed strong potential as an indicator for tdh+ and trh+ V. parahaemolyticus with very high concordances. Good concordances were also obtained for total and pathogenic strains of V. vulnificus in oysters. Interestingly, concordances for all total and pathogenic vibrios were consistently higher in seawater than in oysters, again factoring into account seawater temperature and salinity. These findings suggest that the COPP assay has good potential to serve as a predictive index of total and pathogenic Vibrio levels in oysters and seawater. Further studies to evaluate this possibility are warranted. This study also compared the abundances and detection frequencies of vibrios in oysters and seawater in the Delaware and Chesapeake Bays. Although abundances were low for pathogenic V. parahaemolyticus in both regions, the detection frequency was high and significantly greater in the Delaware Bay than in the Chesapeake Bay. Pathogenic V. vulnificus strains were not significantly different in abundance or prevalence between the two regions and were also generally low. Additional studies are needed to assess the influence of seawater salinity, turbidity, temperature, and other environmental factors on the development of a prediction model for pathogenic vibrios in oysters.
MATERIALS AND METHODS
Sampling sites and collection of samples.
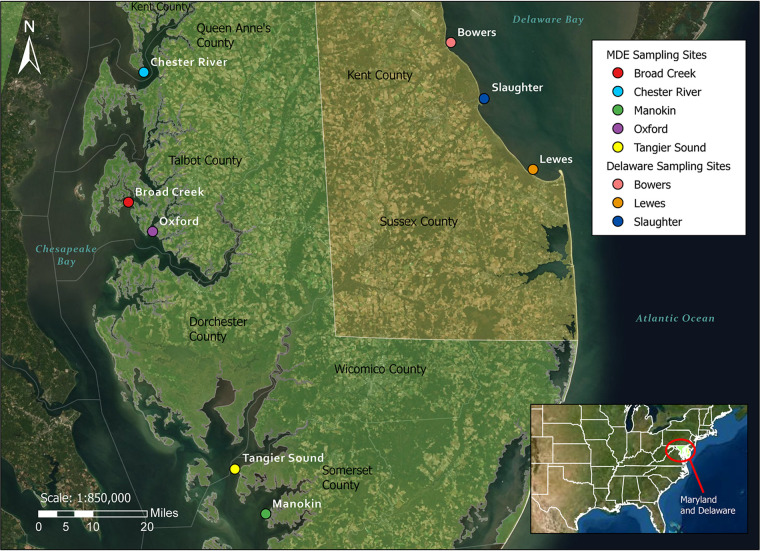
Study sites and locations were selected to represent the range of salinities where Vibrio species may be present. The sites in Delaware represent the higher salinity range (generally 21 to 30 ppt), and the sites in Maryland represent the lower salinity range (10 to 18 ppt). For this project, three Delaware Bay study sites (Lewes, Bowers, and Slaughter Beach) in Delaware and five Chesapeake Bay sites (Broad Creek, Chester River, Manokin River, Oxford, and Tangier Sound) in Maryland were selected (Fig. 6) based on salinities, accessibility, and availability of oysters. Salinities at selected sites were representative of oyster-growing areas nationally.
FIG 6.
The five sample collection sites in Maryland and three sample collection sites in Delaware.
The Delaware Bay is located on the east coast of the United States and is bordered by the states of Delaware and New Jersey. It is 84 km (52 miles) long and covers an area of approximately 2,025 km2 (782 miles2). The shoreline is composed mainly of salt marsh lowlands. Salinity is highest at the mouth of the bay where it meets the Atlantic Ocean. The middle of the bay is brackish, and the northern part of the bay is freshwater with salinity of less than 1 ppt (35).
The Chesapeake Bay is located on the east coast of the United States within the states of Maryland and Virginia. The Chesapeake Bay is the largest estuary in North America. It is 314 km (195 miles) long and between 6 and 48 km (4 to 30 miles) wide, covering an area of 8,384 km2 (3,237 miles2) with 4,470 km2 (1,726 miles2) in Maryland. Salinities increase from north to south as one moves downstream from Maryland’s portion of the bay into Virginia, where the Chesapeake Bay meets the Atlantic Ocean.
Oyster and water samples were collected from three study sites in Delaware (monthly) and five sites in Maryland (every 3 weeks) from May through October for 2 years (2016 and 2017). The five sampling sites in Maryland are classified as approved by the National Shellfish Sanitation Program (NSSP) for commercial harvesting of oysters. Twelve market-size oysters and one container (1 liter) of seawater were collected from each study site, yielding a total of 110 oyster and 110 water samples. These samples were then divided into three subsamples (a total of 330 oysters and 330 water samples) for analysis of TV by the COPP assay and of TV and pathogenic Vibrio species by microbiological and molecular methods. During the collection of samples, temperature, salinity, turbidity, dissolved oxygen, chlorophyll a, and pH in the surface and bottom water were measured with a YSI 6600 multiparameter meter (Yellow Springs Instrument Co., Yellow Springs, OH). The depth of the oyster-harvesting areas in Maryland and Delaware ranged from 2.4 to 5.5 m and 0.25 to 1.65 m, respectively. Water samples were collected following the method of the American Public Health Association (36) in sterile 1-liter wide-mouth containers (Thermo Fisher Scientific, Waltham, MA). Immediately after harvest, oysters were bagged and chilled in an insulated chest with ice, and a sheet of bubble wrap was used to prevent direct contact of samples with the ice. The shipping temperature was monitored using a data logger (Dickson, Addison, IL) to verify that the temperature was less than 10°C (minimum temperature for Vibrio growth) during transport. The oyster and water samples were transported to the laboratories at the University of Maryland Eastern Shore and Delaware State University and analyzed within 24 h of collection (4, 37).
Analysis of samples for total Vibrionaceae using the COPP assay for oyster and water samples.
Total Vibrionaceae counts were determined according to the procedures described by Richards et al. (15). The COPP assay detects a lysyl-aminopeptidase produced by Vibrionaceae family members. In brief, the 12 oysters from each sample were subdivided into 3 sets of 4 oysters. Each set was blended, and 25 g was removed and homogenized with 225 ml of 0.1% peptone buffer (Thermo Fisher Scientific, Waltham, MA). The remaining homogenate was used for the MPN. Serial dilutions of each homogenate (to a final dilution of 10−6 to 10−9 depending upon the expected Vibrio counts at the time of sampling) and water sample (to a final dilution of 10−4) were prepared in phosphate-buffered saline (PBS) or 0.1% peptone buffer. One hundred microliters of each dilution, as well as 100 μl of undiluted water samples, was spread plated in duplicate onto tryptic soy agar (Becton, Dickinson, Sparks, MD) containing 1% NaCl (TSA-N) and incubated overnight at 37°C. After incubation, one countable plate (preferably with 20 to 200 colonies) from each subsample was subjected to the COPP assay by overlaying the colonies with a cellulose acetate membrane (Sartorius, Edgewood, NY) containing the fluorogenic substrate l-lysyl-7-amino-4-trifluoromethylcoumarin (MP Biomedicals, Solon, OH) (15). After precisely 10 min, the membrane was carefully removed from the plate and observed under long-wave UV light for fluorescence. The number of fluorescent foci, representing colonies of Vibrionaceae, was enumerated. Total Vibrionaceae organisms per gram of oyster and per milliliter of seawater were calculated.
Analysis of samples of total and pathogenic V. parahaemolyticus and V. vulnificus using DP.
For direct plating (DP), the same serial dilutions that were prepared for the enumeration of total Vibrionaceae (up to 10−6 to 10−9), as described in the previous section, were plated onto CHROMagar Vibrio (CHROMagar, Paris, France). One hundred microliters of each dilution was spread plated, and the plates were incubated at 37°C for 24 h. After incubation, presumptive colonies of V. parahaemolyticus (mauve) and V. vulnificus (green) were counted, and all or 10% of colonies (depending on the number of colonies on each plate) were confirmed using real-time PCR (38, 39). In brief, isolates were grown overnight in tryptic soy broth (Thermo Fisher Scientific, Waltham, MA) with 1% NaCl and then were boiled for 10 min. The aqueous DNA from cellular materials was separated by centrifugation at 10,000 × g for 2 min. Supernatants were used as PCR templates and were stored at –80°C until they were tested. All presumptive V. parahaemolyticus isolates were tested for tlh (total) and tdh and trh (pathogenic) hemolysin genes, while all presumptive V. vulnificus isolates were tested for vvhA (total) and vcgC (pathogenic) genes. Probes, primers, and cycling conditions for all genes are described in the section below. The presumptive colonies were then multiplied by the percentage of isolates that were confirmed by real-time PCR to be either V. parahaemolyticus or V. vulnificus, and the data were converted to CFU/g or CFU/ml of sample.
Analysis of samples of total and pathogenic V. parahaemolyticus and V. vulnificus using MPN–real-time-PCR.
Three-tube most-probable-number (MPN) procedures were performed in alkaline peptone water (APW), and multiplex real-time PCR assays were used for enumeration of total (tlh) and pathogenic (tdh and trh) V. parahaemolyticus and total (vvhA) and pathogenic (vcgC) V. vulnificus in oyster and water samples (38, 39). In brief, oyster homogenates from the same sample, consisting of three subsets of four oysters each, prepared for the COPP assay and direct plating as mentioned in the previous sections were combined and reblended for 30 s. This homogenate (12 oysters) was serially diluted 10-fold to 10−6 to 10−9 (depending upon the expected Vibrio counts at the time of sampling) in 0.1% peptone. One gram of homogenized tissue and 1 ml of each dilution were inoculated into triplicate MPN tubes containing sterile APW consisting of 1% peptone and 1% NaCl, pH 8.5. Incubation was at 37°C for 18 to 24 h. PCR was performed on MPN tubes showing signs of microbial growth (increased turbidity) for tlh+, tdh+, and trh+ V. parahaemolyticus using PCR primers and probe sequences and PCR amplification conditions described in a previous study (39). An internal amplification control (IAC) was used to ensure PCR integrity to detect any false-negative samples (39). The same MPN tubes were screened for the vvhA and vcgC genes of V. vulnificus by PCR. Probes, primers, and cycling conditions for these genes were described by Baker-Austin et al. (38), except that the initial denaturation time was changed from 10 to 3 min. All primers and probes were manufactured by Thermo Fisher Scientific, except the IAC probe, which was synthesized by Integrated DNA Technologies (Skokie, IL). PCR was performed using iTaq Universal Probes Supermix (Bio-Rad Laboratories, Hercules, CA) containing iTaq DNA polymerase, deoxynucleoside triphosphates (dNTPs), and MgCl2 according to the manufacturer’s instructions in 25-μl reaction volumes. All PCR was performed on an Applied Biosystems (ABI) cycler (7500 RT PCR system; Thermo Fisher Scientific). Positive and negative controls were used for each PCR cycle. After completion of the MPN–real-time PCR, the number of MPN-positive tubes that showed amplification of virulence genes was recorded, and an MPN index was generated using standard U.S. Food and Drug Administration (FDA) procedures and tables (https://www.fda.gov/food/foodscienceresearch/laboratorymethods/ucm109656.htm).
Statistical analyses.
Pearson's correlation (Proc Corr, SAS v 9.4; SAS Institute, Cary, NC) was employed to examine the potential association of the COPP total Vibrio (TV) assay with the more specific methods for V. parahaemolyticus and V. vulnificus. Correlations were considered weak when the r values were between 0.40 and 0.49, moderate when r values were between 0.50 and 0.69, and high when r values were ≥0.70. All data were transformed using log(CFU + 1) for statistical analysis, and an alpha level of 0.05 was considered the minimum level for statistical significance.
To compare DP on CHROMagar Vibrio with MPN-PCR, the sensitivity (true positive rate), specificity (true negative rate), and accuracy (method agreement) of DP were calculated with standard formulae, and MPN-PCR was used as the standard method (24, 40). DP-negative and MPN-PCR-positive results were interpreted as DP false negative, whereas MPN-PCR-negative and DP-positive results were considered DP false positive. Samples that were positive and negative for total and pathogenic V. parahaemolyticus and V. vulnificus by both methods were considered true positive and true negative, respectively (24).
For the statistical methods component of this project, we used an information-theoretic approach (21) for model selection (i.e., Akaike information criterion corrected for small sample size [AICc]) to determine the potential for using the COPP TV assay and known environmental determinants of Vibrio growth to predict elevated occurrence of the species and genetic markers evaluated in this study. Models were constructed for 10 outcome variables (dependent variables): total V. parahaemolyticus (tlh), total V. vulnificus (vvhA), and pathogenicity markers vcgC, trh, and tdh in both water and oyster tissue as determined by the MPN-PCR method. The predictor variables (independent variables) for the candidate models were selected a priori and are combinations of the COPP assay and surface (water) and bottom (oyster) water temperature and salinity. Two categories were created to represent the top quantile of the data distribution (>75th quantile) and the remainder (<75th quantile). Logistic regression was used in all cases to model the categorical data using both Proc Logistic and Proc Genmod (logit link) to obtain the range of diagnostic statistics reported (SAS Institute, Cary, NC).
Under the information-theoretic approach, a candidate set of models is fit and then compared using a model selection criterion. The best model is determined by examining their relative distance to the “truth.” We used AICc weight as a measure of relative plausibility of the models within the candidate set. We interpret the AICc weight (wi) of evidence that model i is the best approximating model, given the data and set of candidate models (21). While model selection was done based on AICc weight, tests of concordance (how often the model correctly categorized the dependent variable) and the Hosmer-Lemeshow goodness-of-fit test were also evaluated (α > 0.05 was considered an adequate fit).
For state comparisons of Vibrio abundance, principal-component Analysis (PCA) was first employed to characterize the physicochemical gradients that exist within and between Delaware and Maryland. This was done to illuminate major differences in water quality parameters that could explain variability in Vibrio levels between the states and to consolidate the number of independent variables. Principal component (PC1) and PC2 were evaluated in subsequent multivariate analysis of variance (ANOVA) (Proc GLM; SAS Institute, Cary, NC). Models were developed for each species and gene target with the explanatory variables state, year, site nested within state, and PC1 and PC2 as covariates, for the COPP TV assay and MPN-PCR method only. All values were log transformed prior to analysis, with model residuals examined to ensure no violations of normality and homogeneity of variance assumptions. Variance inflation was also examined for each model to assess multicollinearity. Tukey’s test was used for within-class comparisons in all cases. While P values are provided, we focused primarily on effect size of significant components as a means of understanding the relative contribution of each explanatory variable and in recognition of their increased use in other fields of science (41). Partial omega squared (ω2) was employed in all cases, following the criteria of Field (42), with 0.01 considered a small effect, 0.06 a medium effect, and ≥0.14 a large effect. Partial omega squared is preferred for multifactorial analyses due to the calculation of effect size relative to error variance rather than total variance. The latter diminishes the effect size of all factors as each new factor is entered (43). Percent data explored for comparison of detection were examined using frequency tables and Fisher’s exact test to account for small sample size (Proc Freq; SAS Institute, Cary, NC).
ACKNOWLEDGMENTS
We are grateful to the Maryland Department of the Environment (MDE) for the collection of samples and to Debtra Rosales for technical assistance. We thank John Bowers, U.S. FDA, for statistical advice, for helpful suggestions, and for reviewing the manuscript, and we thank Jessica Jones, U.S. FDA, for helpful suggestions for MPN–real-time PCR.
We thank the U.S. Department of Agriculture Capacity Building Grants (CBG) Program for funding (award number 2014-38821-22430).
The scientific results and conclusions, as well as any views or opinions expressed herein, are those of the authors and do not necessarily reflect the views of NOAA or the Department of Commerce. The use of trade names or names of commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture (USDA). The USDA is an equal opportunity provider and employer.
REFERENCES
- 1.Daniels NA, MacKinnon L, Bishop R, Altekruse S, Ray B, Hammond RM, Thompson S, Wilson S, Bean NH, Griffin PM, Slutsker L. 2000. Vibrio parahaemolyticus infections in the United States, 1973–1998. J Infect Dis 181:1661–1666. doi: 10.1086/315459. [DOI] [PubMed] [Google Scholar]
- 2.Motes ML, DePaola A, Cook DW, Veazey JE, Hunsucker JC, Garthright WE, Blodgett RJ, Chirtel SJ. 1998. Influence of water temperature and salinity on V. vulnificus in Northern Gulf and Atlantic Coast oysters (Crassostrea virginica). Appl Environ Microbiol 64:1459–1465. doi: 10.1128/AEM.64.4.1459-1465.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DePaola A, Nordstrom JL, Bowers JC, Wells JG, Cook DW. 2003. Seasonal abundance of total and pathogenic Vibrio parahaemolyticus in Alabama oysters. Appl Environ Microbiol 69:1521–1526. doi: 10.1128/AEM.69.3.1521-1526.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parveen S, Hettiarachchi KA, Bowers JC, Jones JL, Tamplin ML, McKay R, Beatty W, Brohawn K, Dasilva LV, Depaola A. 2008. Seasonal distribution of total and pathogenic Vibrio parahaemolyticus in Chesapeake Bay oysters and waters. Int J Food Microbiol 128:354–361. doi: 10.1016/j.ijfoodmicro.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 5.Johnson CN, Flowers AR, Noriea NF, Zimmerman AM, Bowers JC, DePaola A, Grimes DJ. 2010. Relationships between environmental factors and pathogenic vibrios in the northern Gulf of Mexico. Appl Environ Microbiol 76:7076–7084. doi: 10.1128/AEM.00697-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Interstate Shellfish Sanitation Conference. 2018. Constitution, bylaws and procedures. http://www.issc.org/Data/Sites/1/media/2018-conference-forms/issc-constitution-bylaws-procedures-8-2018.pdf. Accessed 31 July 2020.
- 7.Shirai H, Ito H, Hirayama T, Nakamoto Y, Nakabayashi N, Kumagai K, Takeda Y, Nishibuchi M. 1990. Molecular epidemiologic evidence for association of thermostable direct hemolysin (TDH) and TDH-related hemolysin of Vibrio parahaemolyticus with gastroenteritis. Infect Immun 58:3568–3573. doi: 10.1128/IAI.58.11.3568-3573.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosche TM, Yano Y, Oliver JD. 2005. A rapid and simple PCR analysis indicates there are two subgroups of Vibrio vulnificus which correlate with clinical or environmental isolation. Microbiol Immunol 49:381–389. doi: 10.1111/j.1348-0421.2005.tb03731.x. [DOI] [PubMed] [Google Scholar]
- 9.Warner E, Oliver JD. 2008. Population structure of two genotypes of Vibrio vulnificus in oysters (Crassostrea virginica) and seawater. Appl Environ Microbiol 74:80–85. doi: 10.1128/AEM.01434-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.CDC. 2007. Vibrio foodborne illnesses increased in 2006. Morb Mortal Wkly Rep 56:336–339. [Google Scholar]
- 11.CDC. 2007. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—10 states, 2006. Morb Mortal Wkly Rep 55:392–395. [PubMed] [Google Scholar]
- 12.CDC. 1999. Outbreaks of Vibrio parahaemolyticus infections associated with eating raw oysters and clams harvested from Long Island Sound—Connecticut, New Jersey, and New York, 1998. Morb Mortal Wkly Rep 48:48–51. [PubMed] [Google Scholar]
- 13.Centers for Science in the Public Interest. 2020. Outbreaks and recalls. Vibrio: raw oysters from Hood Canal (WA). Food Safety Outbreak Alert. http://www.cspinet.org/foodsafety/outbreak_report.html. Accessed 11 April 2020.
- 14.U.S. Food and Drug Administration. 13 November 2009. FDA statement on Vibrio vulnificus in raw oysters. FDA news release.
- 15.Richards GP, Watson MA, Parveen S. 2005. Development of a simple and rapid fluorogenic procedure for identification of Vibrionaceae family members. Appl Environ Microbiol 71:3524–3527. doi: 10.1128/AEM.71.7.3524-3527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richards GP, Watson MA. 2006. A simple fluorogenic method to detect Vibrio cholerae and Aeromonas hydrophila in well water impacted by catastrophic disasters. Am J Trop Med Hyg 75:516–521. doi: 10.4269/ajtmh.2006.75.516. [DOI] [PubMed] [Google Scholar]
- 17.U.S. Food and Drug Administration. 2015. National Shellfish Sanitation Program (NSSP) guide for the control of molluscan shellfish (revision). U.S. Department of Health and Human Services, Washington, DC. [Google Scholar]
- 18.Smith CJ, Osborn AM. 2009. Advantages and limitations of quantitative PCR (Q-PCR) based approaches in microbial ecology. FEMS Microbiol Ecol 67:6–20. doi: 10.1111/j.1574-6941.2008.00629.x. [DOI] [PubMed] [Google Scholar]
- 19.Griffitt KJ, Grimes DJ. 2013. A novel agar formulation for isolation and direct enumeration of Vibrio vulnificus from oyster tissue. J Microbiol Methods 94:98–102. doi: 10.1016/j.mimet.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 20.Froelich BA, Weiss MJ, Noble RT. 2014. The evaluation of four recent culture-based methods for the isolation and enumeration of Vibrio vulnificus bacteria from oyster meat. J Microbiol Methods 97:1–5. doi: 10.1016/j.mimet.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 21.Burnham KP, Anderson DR, Huyvaert KP. 2011. AIC model selection and multimodel inference in behavioral ecology: some background, observations, and comparisons. Behav Ecol Sociobiol 65:23–35. doi: 10.1007/s00265-010-1029-6. [DOI] [Google Scholar]
- 22.Blanco-Abad V, Ansede-Bermejo J, Rodriguez-Castro A, Martinez-Urtaza J. 2009. Evaluation of different procedures for the optimized detection of Vibrio parahaemolyticus in mussels and environmental samples. Int J Food Microbiol 129:229–236. doi: 10.1016/j.ijfoodmicro.2008.11.028. [DOI] [PubMed] [Google Scholar]
- 23.Staley C, Chase E, Harwood VJ. 2013. Detection and differentiation of Vibrio vulnificus and V. sinaloensis in water and oysters of a Gulf of Mexico estuary. Environ Microbiol 15:623–633. doi: 10.1111/1462-2920.12045. [DOI] [PubMed] [Google Scholar]
- 24.National Institute of Standards and Technology. 1993. Standard reference materials no. 2201: sodium chloride (ion-selective). NIST, Gaithersburg, MD. [Google Scholar]
- 25.Richards GP, Watson MA. 2010. Fluorogenic membrane overlays to enumerate total and fecal Escherichia coli and total Vibrionaceae in shellfish and seawater. Int J Microbiol 2010:910486–910489. doi: 10.1155/2010/910486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacDonell MT, Colwell RR. 1985. Phylogeny of the family Vibrionaceae and recommendations for two new genera: Listonella and Shewanella. Syst Appl Microbiol 6:171–182. doi: 10.1016/S0723-2020(85)80051-5. [DOI] [Google Scholar]
- 27.Food and Agriculture Organization of the United Nations World Health Organization). 2020. Risk assessment tools for Vibrio parahaemolyticus and Vibrio vulnificus associated with seafood. Microbiological risk assessment series no. 20. https://apps.who.int/iris/bitstream/handle/10665/330867/9789240000186-eng.pdf?sequence=1&isAllowed=y. Accessed 11 April 2020.
- 28.Elmahdi S, Parveen S, Ossai S, DaSilva LV, Jahncke M, Bowers J, Jacobs J. 2018. Vibrio parahaemolyticus and Vibrio vulnificus recovered from oysters during an oyster relay study. Appl Environ Microbiol 84:e01790-17. doi: 10.1128/AEM.01790-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson NC, Bowers CJ, Griffitt JK, Molina CV, Clostio WR, Pei S, Laws E, Paranjpye NR, Strom SM, Chen A, Hasan AN, Huq A, Noriea FN, Grimes CJ, Colwell RR. 2012. Ecology of Vibrio parahaemolyticus and Vibrio vulnificus in the coastal and estuarine waters of Louisiana, Maryland, Mississippi, and Washington (United States). Appl Environ Microbiol 78:7249–7257. doi: 10.1128/AEM.01296-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Froelich BA, Phippen B, Fowler P, Noble RT, Oliver JD. 2017. Differences in abundances of total Vibrio spp., V. vulnificus, and V. parahaemolyticus in clams and oysters in North Carolina. Appl Environ Microbiol 83:e02265-16. doi: 10.1128/AEM.02265-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davis BJK, Jacobs JM, Davis MF, Schwab KJ, DePaola A, Curriero FC. 2017. Environmental determinants of Vibrio parahaemolyticus in the Chesapeake Bay. Appl Environ Microbiol 83:e01147-17. doi: 10.1128/AEM.01147-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hartwick MA, Urquhart EA, Whistler CA, Cooper VS, Naumova EN, Jones SH. 2019. Forecasting seasonal Vibrio parahaemolyticus concentrations in New England shellfish. Int J Environ Res Public Health 16:4341. doi: 10.3390/ijerph16224341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nilsson WB, Paranjpye RN, Hamel OS, Hard C, Strom MS, Nilsson WB, Paranjpye RN. 2019. Vibrio parahaemolyticus risk assessment in the Pacific Northwest: it's not what's in the water. FEMS Microbiol Ecol 95:fiz027. doi: 10.1093/femsec/fiz027. [DOI] [PubMed] [Google Scholar]
- 34.Chen AJ, Hasan NA, Haley BJ, Taviani E, Tarnowski M, Browan K, Johnson CN, Colwell RR, Huq A. 2017. Characterization of pathogenic Vibrio parahaemolyticus from the Chesapeake Bay, Maryland. Front Microbiol 8:2460. doi: 10.3389/fmicb.2017.02460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neilan B. 2015. Studying the Delaware Bay, 2015 report. Division of Fish and Wildlife, NJDEP, Trenton, NJ: http://www.state.nj.us/dep/fgw/artdelbaystudy15.htm. Accessed 20 April, 2020. [Google Scholar]
- 36.Greenberg AE, Clesceri LS, Eaton AD (ed). 1992. Standard methods for the examination of water and wastewater, 18th ed American Public Health Association, Washington, DC. [Google Scholar]
- 37.DaSilva L, Parveen S, DePaola A, Bowers J, Brohawn K, Tamplin ML. 2012. Development and validation of a predictive model for the growth of Vibrio vulnificus in post-harvest shell stock oysters. Appl Environ Microbiol 78:1675–1681. doi: 10.1128/AEM.07304-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baker-Austin C, Gore A, Oliver JD, Rangdale R, McArthur JV, Lees DN. 2009. Rapid in situ detection of virulent Vibrio vulnificus strains in raw oyster matrices using real-time PCR. Environ Microbiol Rep 2:76–80. doi: 10.1111/j.1758-2229.2009.00092.x. [DOI] [PubMed] [Google Scholar]
- 39.Nordstrom JL, Vickery MCL, Blackstone GM, Murray SL, DePaola A. 2007. Development of a multiplex real-time PCR assay with an internal amplification control for the detection of total and pathogenic Vibrio parahaemolyticus bacteria in oysters. Appl Environ Microbiol 73:5840–5847. doi: 10.1128/AEM.00460-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dawson-Saunders B, Trapp RG. 1994. Basic and clinical biostatistics, 2nd ed Appleton and Lange, Norwalk, CT. [Google Scholar]
- 41.Hentshke H, Stuttgen MC. 2011. Computation of measures of effect size for neuroscience data sets. Euro J Neuosci 34:1887–1894. doi: 10.1111/j.1460-9568.2011.07902.x. [DOI] [PubMed] [Google Scholar]
- 42.Field A. 2013. Discovering statistics using IBM SPSS Statistics, 4th ed Sage, London, UK. [Google Scholar]
- 43.Maxwell SE, Camp CJ, Arvey RD. 1981. Measures of strength of association: a comparative examination. J Appl Psychol 66:525–534. doi: 10.1037/0021-9010.66.5.525. [DOI] [Google Scholar]