Bacterial communities encased by self-produced extracellular polymeric substances (EPSs), known as biofilms, have a wide influence on human health and environmental problems. The importance of biofilm research has increased, as biofilms are the preferred bacterial lifestyle in nature. Furthermore, in recent years it has been noted that the contribution of phenotypic heterogeneity within biofilms requires analysis at the single-cell or subpopulation level to understand bacterial life strategies. In Streptococcus mutans, a cariogenic bacterium, extracellular DNA (eDNA) contributes to biofilm formation. However, it remains unclear how and where the cells produce eDNA within the biofilm. We focused on LytF, an autolysin that is induced by extracellular peptide signals. We used single-cell level imaging techniques to analyze lytF expression in the biofilm population. Here, we show that S. mutans generates eDNA by inducing lytF expression near the bottom of the biofilm, thereby enhancing biofilm adhesion and structural stability.
KEYWORDS: Streptococcus mutans, biofilms, cell death, cell-to-cell communication, extracellular DNA
ABSTRACT
Extracellular DNA (eDNA) is a biofilm component that contributes to the formation and structural stability of biofilms. Streptococcus mutans, a major cariogenic bacterium, induces eDNA-dependent biofilm formation under specific conditions. Since cell death can result in the release and accumulation of DNA, the dead cells in biofilms are a source of eDNA. However, it remains unknown how eDNA is released from dead cells and is localized within S. mutans biofilms. We focused on cell death induced by the extracellular signaling peptide called competence-stimulating peptide (CSP). We demonstrate that nucleic acid release into the extracellular environment occurs in a subpopulation of dead cells. eDNA production induced by CSP was highly dependent on the lytF gene, which encodes an autolysin. Although lytF expression was induced bimodally by CSP, lytF-expressing cells further divided into surviving cells and eDNA-producing dead cells. Moreover, we found that lytF-expressing cells were abundant near the bottom of the biofilm, even when all cells in the biofilm received the CSP signal. Dead cells and eDNA were also abundantly present near the bottom of the biofilm. The number of lytF-expressing cells in biofilms was significantly higher than that in planktonic cultures, which suggests that adhesion to the substratum surface is important for the induction of lytF expression. The deletion of lytF resulted in reduced adherence to a polystyrene surface. These results suggest that lytF expression and eDNA production induced near the bottom of the biofilm contribute to a firmly attached and structurally stable biofilm.
IMPORTANCE Bacterial communities encased by self-produced extracellular polymeric substances (EPSs), known as biofilms, have a wide influence on human health and environmental problems. The importance of biofilm research has increased, as biofilms are the preferred bacterial lifestyle in nature. Furthermore, in recent years it has been noted that the contribution of phenotypic heterogeneity within biofilms requires analysis at the single-cell or subpopulation level to understand bacterial life strategies. In Streptococcus mutans, a cariogenic bacterium, extracellular DNA (eDNA) contributes to biofilm formation. However, it remains unclear how and where the cells produce eDNA within the biofilm. We focused on LytF, an autolysin that is induced by extracellular peptide signals. We used single-cell level imaging techniques to analyze lytF expression in the biofilm population. Here, we show that S. mutans generates eDNA by inducing lytF expression near the bottom of the biofilm, thereby enhancing biofilm adhesion and structural stability.
INTRODUCTION
Extracellular DNA (eDNA), one of the major components of biofilms, contributes to biofilm formation and the mechanical stability of biofilms (1). eDNA is predicted to be generated primarily by lysis of a subpopulation of cells and the subsequent release of genomic DNA from the cytoplasm into the extracellular environment. In Gram-positive bacteria, eDNA production is mediated by quorum sensing (2, 3), autolysis (3, 4), H2O2 production (5–7), and bacteriophages (8, 9). However, how bacterial cells produce eDNA is poorly documented.
In Streptococcus mutans, an etiological agent of dental caries and infective endocarditis, eDNA is one of the major biofilm components (2, 10). eDNA is thought to be used for initial attachment to substratum surfaces in S. mutans biofilm formation (11). DNase I treatment shows an inhibitory effect on the biofilm formation of S. mutans (2). Thus, eDNA production contributes to biofilm formation and the pathogenesis of S. mutans. The main production system of eDNA is considered to be an autolysis-dependent mechanism (12, 13). eDNA is also produced via a membrane vesicle-mediated pathway that is independent of autolysis (14). lytF, which is a conserved gene in the genus Streptococcus, encodes an autolysin in S. mutans (15). LytF has a group B streptococcal secreted protein (Bsp) domain and a cysteine, histidine-dependent amidohydrolase/peptidase (CHAP) domain (15). The expression of lytF is regulated by two types of secretory peptide signals called competence-stimulating peptide (CSP) and sigX-inducing peptide (XIP) (16–21). CSP signaling is necessary for eDNA production (22), however it is still unclear how CSP-regulated lytF contributes to eDNA production in biofilms.
The precursor of CSP is encoded by the comC gene. It is secreted into the extracellular environment via ComAB (also known as NlmTE) and then processed by SepM, an extracellular protease (23). The processed CSP is an active form and is received by a two-component regulatory system ComDE, which induces the expression of the ComE regulon. The cipB gene in the ComE regulon encodes bacteriocin, and CipI is reported as its immune protein (12). cipB is essential for the induction of the downstream comRS-sigX system, which directly upregulates lytF expression when exogenous CSP is supplemented in the medium (24). comS encodes the precursor of XIP. Extracellular XIP is taken up by cells through oligopeptide permease (Opp) and binds to ComR. The ComR-XIP complex induces the expression of sigX, an alternative sigma factor, resulting in the expression of the SigX regulon, including lytF. Synthetic CSP (sCSP) induces the expression of the sigX regulon but not XIP in complex media (17, 25). In addition, in planktonic cells, sCSP induces the expression of the ComE regulon in a unimodal manner but induces the expression of the sigX regulon in a bimodal manner (26). The SigX regulon contains late competence genes and several genes of unknown function. The lytF gene in the SigX regulon encodes autolysin, which is involved in cell death in planktonic cells (15). Cell death is expected to result in eDNA production, which contributes to the structural stability of biofilms. Thus, the distribution of the bimodal expression of the sigX regulon within the biofilm may be strongly related to the physiological function and the pathogenesis, though little is currently known regarding the localization of the expression of the sigX regulon within biofilms.
S. mutans biofilms are composed of numerous microcolonies, the formation of which requires insoluble glucan synthesis (22). The gtfB and gtfC genes encode the enzymes responsible for insoluble glucan synthesis, which is a major characteristic of S. mutans. Glucans synthesized by GtfB (Gtf-I) are responsible for microcolony formation, while glucans synthesized by GtfC (Gtf-SI) strongly attach cells and microcolonies to the substratum surfaces (27). There is an acidic microenvironment inside the microcolony due to the accumulation of lactic acid produced by S. mutans. The central portion and the bottom layer of the microcolony (close to the substratum surface) are hardly neutralized even when treated with a neutral buffer (28). The acidic environment is reported to result in increased eDNA production in planktonic cells (11). However, the localization of eDNA within biofilms has not been clarified. In addition, the comDE gene is highly expressed near the bottom of the biofilm (29). Given the heterogeneity within the biofilm, the induction of cell death and eDNA production would be spatiotemporally regulated. However, the direct link between lytF expression, cell death, and eDNA production in the biofilm remains unclear.
In this study, we focused on lytF-mediated cell death induced by CSP to elucidate eDNA production within the biofilm, which is the dominant state of S. mutans in the human oral cavity. Using time-lapse imaging, we analyzed lytF expression in the biofilm and the location where cells release eDNA. Here, we succeeded in visualizing eDNA production at the single-cell level. We revealed the heterogeneous localization of lytF-expressing cells, dead cells, and eDNA in the biofilm. These results provide insights into how S. mutans forms structurally stable biofilms.
RESULTS
CSP signaling induces cell death and the production of eDNA in a subpopulation of cells.
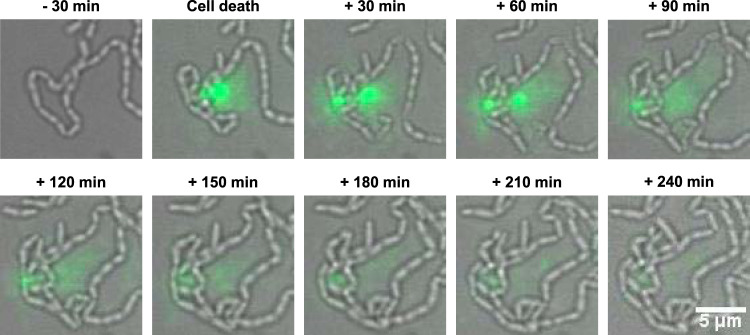
The primary production mechanism of eDNA is the release of genomic DNA from dead cells (22, 30, 31). However, no direct observation of eDNA production in S. mutans has been documented. Therefore, we observed the process of eDNA production at the single-cell level using a high-aspect ratio quasi-two-dimensional microfluidic chamber. Dead cells and extracellular nucleic acids were visualized using SYTOX Green, a fluorescent marker of cell death. When cells were cultured in brain heart infusion (BHI) medium, we observed cell death (green fluorescent cells) but not the release of nucleic acids (Movie S1A in the supplemental material). Then, we focused on CSP-mediated cell death (32). Bright SYTOX Green fluorescence was detected around the dead cells and diffused gradually in the presence of sCSP (Fig. 1, Movie S1B). This result indicates that the production of extracellular nucleic acids occurs in a dead-cell subpopulation in the presence of sCSP. We quantitatively confirmed that sCSP significantly induced eDNA as well as cell death in wild-type (WT) cells (Fig. 2). Thus, CSP induces cell death in a cell subpopulation and the release of DNA into the extracellular environment.
FIG 1.
A subpopulation of cells releases nucleic acids into the extracellular environment. We inoculated WT cells at the exponential phase into a microfluidic device, which allowed the cells to grow two-dimensionally at 37°C in BHI with sCSP under flow conditions. The medium flowed from left to right in these images at a constant rate (150 μl/h). SYTOX Green dye (1.25 μM) was added to the medium to visualize dead cells and extracellular nucleic acids. Images before and after cell death are displayed. Green indicates dead cells and extracellular nucleic acids.
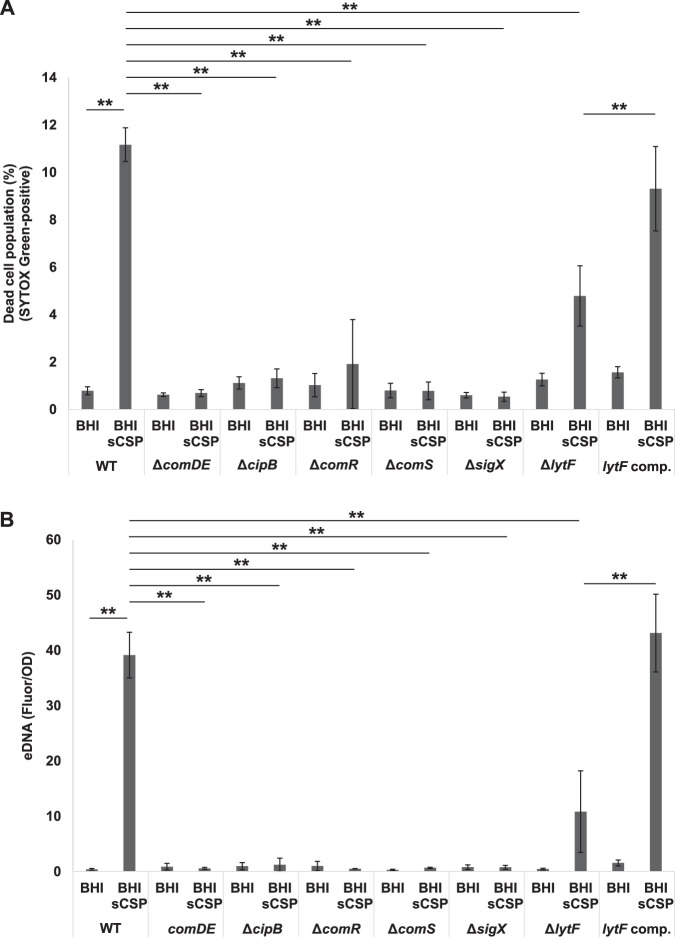
FIG 2.
Cell death and eDNA production induced by sCSP depend on lytF expression via the Com system. Cells were grown in BHI with or without sCSP in an aerobic atmosphere containing 5% CO2 at 37°C for 6 h. (A) Dead cells were stained with 500 nM SYTOX Green for 15 min and quantified by flow cytometry. (B) eDNA was extracted from the 6-h culture supernatants and quantified using a Quant-iT PicoGreen dsDNA assay kit. The results were standardized by the OD600 value at 6 h of culture, since sCSP affects growth. These data indicate the mean ± standard deviations of the results from three independent experiments. The asterisks indicate a significant difference; **, adjusted P < 0.01 (post hoc Bonferroni test).
Induction of cell death and eDNA production by CSP mainly depends on lytF.
CSP induces the expression of the ComE regulon in a unimodal manner and the subsequent SigX regulon in a bimodal manner (26). In the strains carrying mutated versions of comDE, cipB, comR, comS, and sigX, which are required for expression of the sigX regulon, sCSP did not induce cell death and eDNA production (Fig. 2A and B). The SigX regulon contains an autolysin-encoding gene, lytF (15). In the presence of sCSP, cell death and eDNA production in ΔlytF mutant cells were significantly reduced compared with those in WT cells (Fig. 2A and B). In addition, sCSP-induced cell death and eDNA production were restored in the lytF-complemented strain (Fig. 2A and B). Given the hierarchy of ComDE-CipB-ComRS-SigX, cell death and eDNA production in response to sCSP are dependent on sigX and, among the genes of the SigX regulon, autolysin-encoding lytF remarkably contributes to cell death and eDNA production.
Cell death occurs in a subpopulation of the lytF-expressing population.
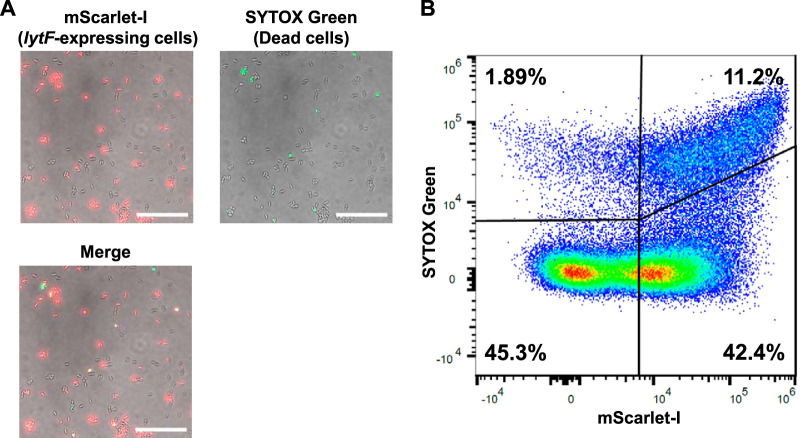
lytF expression is induced in a subpopulation of cells when S. mutans is cultured in complex medium containing sCSP (17). In addition, LytF of S. mutans is likely a self-acting autolysin (15). Thus, we examined whether cell death occurs in lytF-expressing cells. The results of microscopic observation of the PlytF reporter strain showed that lytF expression was induced in a subpopulation of cells (Fig. 3A), and these results are consistent with the results of a previous report (17). Notably, microscopic observation and flow cytometry analysis showed that most cell death occurred in lytF-expressing cells (Fig. 3A and B). In addition, a subpopulation of the lytF-expressing population (mScarlet-I-positive) were dead cells (i.e., SYTOX Green-positive), and more than half of the lytF-expressing cells were alive (i.e., SYTOX Green-negative) (Fig. 3B). The comRSX system, which regulates lytF expression, induces genetic competence (19). lytF is coexpressed with comS in cells (17). Therefore, we speculated that lytF-expressing cells might be divided into surviving competent cells and dead cells that contribute to eDNA production.
FIG 3.
Cell death occurs in a subpopulation of lytF-expressing cells. The PlytF reporter strain was grown in BHI with sCSP in an aerobic atmosphere containing 5% CO2 at 37°C for 6 h. Dead cells were stained with 500 nM SYTOX Green for 15 min. (A) Cells observed with differential interference contrast (DIC) and fluorescence microscopy. mScarlet-I (lytF-expressing cells) and SYTOX Green (dead cells) are shown in red and green, respectively. The scale bars indicate 20 μm. (B) lytF-expressing cells and dead cells were quantified by flow cytometry. The x axis and y axis show the fluorescence intensities of mScarlet-I and SYTOX Green, respectively. Gating was determined based on unstained and single fluorescent samples. Representative data from independent experiments are presented.
LytF-expressing cells, dead cells, and eDNA are localized near the bottom of the biofilm.
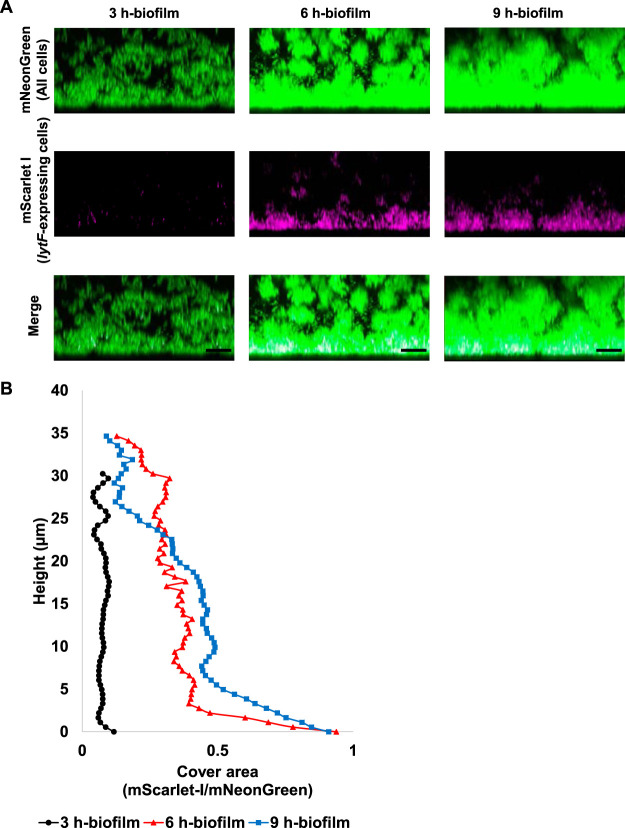
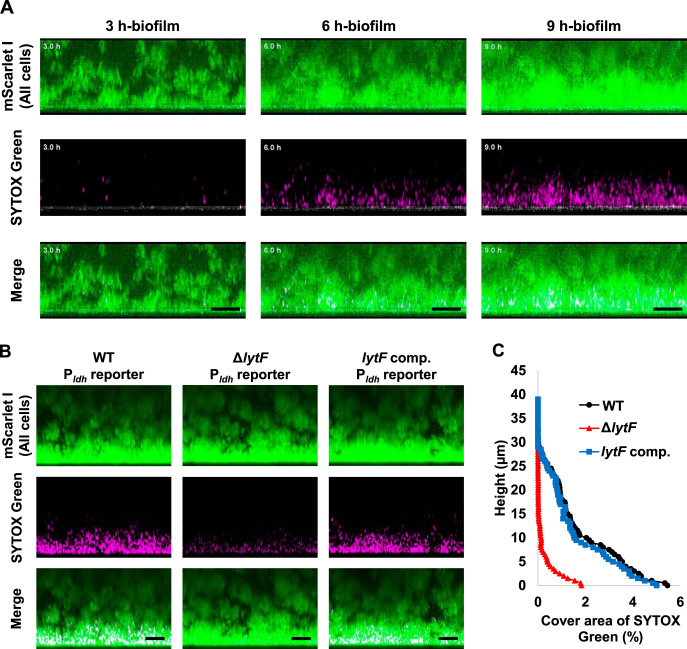
Within biofilms, spatially heterogeneous gene expression enables the collective function of a bacterial population, which could contribute to the tolerance and structural stability of the biofilm. S. mutans survives in the form of biofilms in the actual environment, such as the oral cavity; thus, we analyzed the localization of lytF-expressing cells in biofilms with three-dimensional imaging by confocal laser scanning microscopy (CLSM). We cultured S. mutans in BHIs (BHI containing sucrose) to induce biofilm formation. In accordance with the analysis of planktonic cells, lytF expression was induced in a subpopulation of biofilm cells in the presence of sCSP (Fig. S2A). In contrast, we observed few lytF-expressing cells in the absence of sCSP (Fig. S2B). To determine the localization of lytF-expressing cells in the biofilm, we calculated the ratio of the lytF-expressing cells to the whole cells in the xy-section at each height of the CLSM images (Fig. S2C and D). The image analysis showed the lytF-expressing cells were particularly abundant near the bottom of the biofilm in the presence of sCSP. We then focused on the bottom of the biofilm at the early growth phase. The lytF-expressing cells began to appear in the 3-h biofilm and gradually increased (Fig. 4A and B). Notably, the appearance of the lytF-expressing cells was limited to near the bottom, even at 9 h (Fig. 4A and B). Observation of the PcipB reporter strain, an indicator of sCSP signals, confirmed that most cells in the biofilm received the sCSP signal (Fig. S3A and B). Even in the absence of sCSP, we detected a few lytF-expressing cells in the 6-h biofilm, and these cells were also localized near the bottom of the biofilm (Fig. S4). Therefore, endogenous CSP signaling within biofilms also induces localized lytF expression at the bottom.
FIG 4.
The distribution of lytF-expressing cells in the biofilm is maintained near the bottom even when the culture time is extended. The Pldh and PlytF dual-reporter strain was grown in an aerobic atmosphere containing 5% CO2 at 37°C in BHIs with sCSP for 3, 6, or 9 h in glass-bottom dishes. Nonadherent cells were removed by washing twice with PBS. Areas of approximately 40 μm from the surface were observed using an inverted CLSM. Z-stacks were acquired at 0.5 μm intervals. (A) Images showing side views of the biofilms. Since the samples for each culture time were individually prepared, these images are not in the same field of view. mNeonGreen (all cells) and mScarlet-I (lytF-expressing cells) are shown in green and magenta, respectively. The scale bars indicate 10 μm. Representative images from independent experiments are presented. (B) Image analysis of the biofilms in panel A. In each two-dimensional image, the cover areas showing the fluorescence of mScarlet-I (lytF-expressing cells) and mNeonGreen (all cells) were calculated by ImageJ and Fiji.
Next, we investigated whether dead cells and eDNA were also localized there. We performed time-lapse imaging analysis of dead cells and extracellular nucleic acids during biofilm formation with SYTOX Green staining. The Pldh (a constitutive promoter) reporter strain was used for CLSM observation. We acquired images up to a height of approximately 40 μm from the glass surface to analyze the area near the bottom. Dead cells and extracellular nucleic acids increased near the bottom as the biofilm grew in the presence of sCSP (Fig. 5A, Movie S2A). To investigate whether the cell death and production of extracellular nucleic acid are dependent on lytF, we observed the 6-h biofilm of a lytF deletion mutant and its complemented strain. The localized dead cells and extracellular nucleic acid observed near the bottom of the biofilm were reduced in ΔlytF and restored by complementation of lytF (Fig. 5B and C). Therefore, these results suggest that cell death and eDNA production near the bottom of the biofilm depend on lytF expression. In addition, even in the absence of sCSP, we detected similar localization of cell death and extracellular nucleic acids in the biofilm, albeit at a lower frequency than that in the presence of sCSP (Fig. S5, Movie S2B).
FIG 5.
Dead cells and eDNA are also abundant near the bottom of the biofilm. (A) The Pldh reporter was grown in a glass-bottom dish in BHIs with sCSP at 37°C. The medium was supplemented with 1.25 μM SYTOX Green to stain dead cells and extracellular nucleic acids. The same field of view within a range of approximately 40 μm from the glass surface was observed while culturing at 37°C. Z-stacks were acquired at 0.55 μm intervals. These images show side views of the biofilms, and the white line below each image shows the glass surface obtained by the reflection. (B) The WT, lytF deficient mutant (ΔlytF), and lytF-complemented (lytF comp) strains with a Pldh reporter background were grown in an aerobic atmosphere containing 5% CO2 at 37°C in BHIs with sCSP for 6 h in glass-bottom dishes. Planktonic cells were removed by washing twice with PBS. These images show side views of the biofilms. Z-stacks were acquired at 0.5 μm intervals. mScarlet-I (all cells) and SYTOX Green (dead cells and extracellular nucleic acids) are shown in green and magenta, respectively. The scale bars indicate 10 μm. Representative images from independent experiments are presented. (C) Image analysis of the 3D images shown in panel B. In each two-dimensional image, the cover areas showing the fluorescence of SYTOX Green were calculated by ImageJ and Fiji.
Adhesion to the substratum surface may contribute to the heterogeneous expression of lytF within the biofilm.
To investigate whether lytF expression is induced by biofilm formation, we quantified the percentage of lytF-expressing cells in planktonic and biofilm cells in the same culture medium by flow cytometry. The percentage of lytF-expressing cells among biofilm cells was significantly increased compared with that in planktonic cells (planktonic cells, 26.65 ± 2.13%; biofilm cells, 58.08 ± 3.92%; P < 0.01), indicating that biofilm formation causes an increase in the lytF-expressing population.
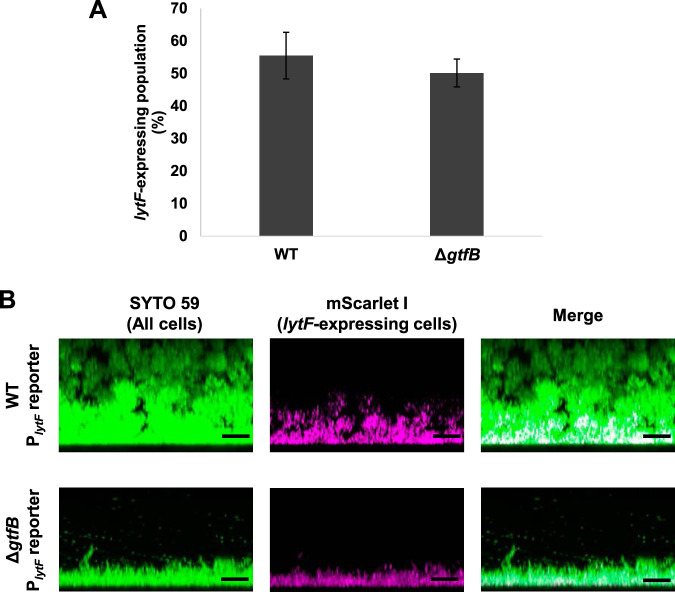
Physiological heterogeneity is generated by the three-dimensional structure of the biofilm (33). At the bottom of biofilms, metabolites accumulate and/or nutrients are depleted. We hypothesized that lytF expression was affected by the chemical gradient within the biofilm. Accordingly, if our hypothesis is correct, disrupting the three-dimensional structure of the biofilm should affect the distribution of lytF-expressing cells. In S. mutans, the gtfB-null mutant loses microcolony formation and forms a thin biofilm (27). We constructed a PlytF reporter strain with a gtfB-deficient background (ΔgtfB PlytF reporter) to investigate the localization of lytF-expressing cells in the biofilm of ΔgtfB cells. First, we found that gtfB deficiency did not affect the population of lytF-expressing cells in the planktonic culture (Fig. 6A). In addition, we observed thin biofilm formation of the ΔgtfB PlytF reporter strain (Fig. 6B). In this biofilm, lytF-expressing cells were still abundant near the bottom (Fig. 6B). The frequency of lytF-expressing cells in ΔgtfB cells at the bottom port within 5 μm of the surface was comparable to that in the WT cells (WT, 80.62 ± 8.35%; ΔgtfB, 79.2 ± 1.28%). These results suggest the 3D biofilm structure is not required for the induction of lytF gene expression. Thus, the 3D biofilm formation-mediated chemical gradients, such as a pH gradient, are unlikely to trigger lytF expression.
FIG 6.
lytF-expressing cells are abundant near the bottom of the biofilm even in a thin biofilm formed by ΔgtfB cells. (A) PlytF reporter and ΔgtfB PlytF reporter strains were grown in an aerobic atmosphere containing 5% CO2 at 37°C in BHIs with sCSP for 6 h in a 6-well plate. Sonication was performed to suspend aggregated cells. lytF-expressing cells were quantified by flow cytometry. These data indicate the mean ± standard deviations of the results from three independent experiments. (B) Images showing side views of the biofilms for strains grown in an aerobic atmosphere containing 5% CO2 at 37°C in BHIs with sCSP for 6 h in a glass-bottom dish. Nonadherent cells were removed by washing twice with PBS. The biofilm cells were stained with 5 μM SYTO 59 and observed with inverted CLSM. Z-stacks were acquired at 0.5 μm intervals. SYTO 59 (all cells) and mScarlet-I (lytF-expressing cells) are shown in green and magenta, respectively. The scale bars indicate 10 μm. Representative data from independent experiments are presented.
Oral bacteria, including S. mutans, adhere to saliva-coated surfaces in the oral cavity. We observed the biofilm formed on the saliva-coated glass surface to examine whether the properties of the attachment surface affect lytF expression in biofilms. The CLSM observation and image analysis results showed there were many lytF-expressing cells near the bottom of the biofilm, even on the saliva-coated glass surface (Fig. S6 and S7). Thus, saliva is unlikely to affect lytF expression.
Recently, it has been reported that adhesion to the substratum surface resulted in altered gene expression in S. mutans (29). The altered gene expression occurs not only in cells directly contacting surfaces, but also in cells within a certain proximity to the attached cells. The signaling between surface-attached cells and adjacent cells is mediated by quorum sensing by AI-2. We investigated the distribution of lytF-expressing cells in ΔluxS cells, which do not produce AI-2. Our imaging analysis showed that deletion of the luxS gene did not change the distribution of lytF-expressing cells in the biofilm (Fig. S7 and S8). Thus, quorum sensing via AI-2 does not appear to affect the distribution of lytF-expressing cells within the biofilm. From these data, physical contact of the cells with the surfaces was expected to result in a heterogeneous distribution of lytF expression within the biofilm.
lytF contributes to cell adhesion to the substratum surface.
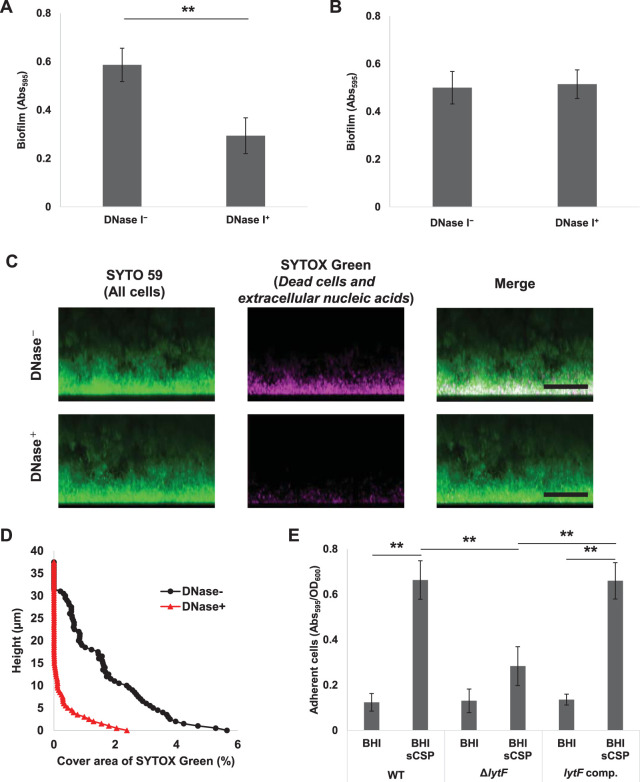
eDNA contributes to the attachment of bacterial cells to the substratum (34, 35). DNase I has an inhibitory effect on the biofilm formation of S. mutans (2). We show that the addition of DNase I prior to culture significantly inhibits biofilm formation in the presence of sCSP (Fig. 7A). We confirmed that the addition of DNase I to the medium reduced the detection area of dead cells and extracellular nucleic acids in the biofilm by CLSM observation and image analysis (Fig. S9A and B). Thus, we suggest that DNase I inhibits biofilm attachment to the surface by degrading the eDNA localized at the bottom of the biofilm. However, compared to no treatment, DNase I treatment of preformed biofilms did not significantly decrease the amount of biofilm (Fig. 7B). In mature biofilms, the EPS matrix, mainly consisting of insoluble glucans, may protect eDNA at the bottom from DNase I enzymatic attack. To test this possibility, we performed DNase I treatment on the preformed biofilm and observed the dead cells and extracellular nucleic acids within the biofilm. The SYTOX Green signals in the biofilm were reduced by DNase I treatment (Fig. 7C and D). Therefore, we consider that eDNA is not necessarily required for the structural integrity in the S. mutans mature biofilm, and the biofilm structure can be maintained even when subjected to external DNase I attack.
FIG 7.
CSP-induced lytF expression and eDNA production contribute to biofilm adhesion to surfaces. (A) DNase I inhibits biofilm formation. The overnight culture was diluted to an OD600 of 0.05 with BHIs with 1 μM CSP and cultured in an aerobic atmosphere containing 5% CO2 at 37°C for 6 h. DNase I was added to the medium at a concentration of 50 U/ml (DNase+) before the incubation. The biofilms were stained with CV. The absorbance at 595 nm (Abs595) was measured on a plate reader. (B to D) eDNA within the preformed biofilm was degraded by DNase I, but the biofilm structure was maintained. Biofilms were formed in BHIs with sCSP medium in an aerobic atmosphere containing 5% CO2 at 37°C for 6 h. The culture solutions were removed and washed twice with PBS. (B) The biofilms were treated with incubation buffer with or without DNase I (50 U/ml) at 37°C for 6 h. They were then stained with CV and the absorbance at 595 nm (Abs595) was measured on a plate reader. (C) Side views of biofilm cells stained with 5 μM SYTO 59 and 500 μM SYTOX Green for 30 min and observed with inverted CLSM. Z-stacks were acquired at 0.5 μm intervals. SYTO 59 (all cells) and SYTOX Green (eDNA and extracellular nucleic acids) are shown in green and magenta, respectively. The scale bars indicate 20 μm. (D) In each two-dimensional image in panel C, the cover areas showing the fluorescence of SYTOX Green were calculated by ImageJ and Fiji. (E) Quantification of adhered cells to the surface in the WT, ΔlytF, and lytF-complemented (lytF comp.) strains. The cells grown in BHI with sCSP were allowed to adhere to the surface by standing for 6 h at 4°C. Adherent cells were stained with CV for 15 min. The absorbance at 595 nm (Abs595) was measured with a plate reader. The results were standardized by the OD600 value at 6 h of culture. These data indicate the mean ± standard deviations of the results from three independent experiments. The asterisks indicate a significant difference; **, adjusted P < 0.01 (post hoc Bonferroni test).
We then considered that localized lytF expression and consequent eDNA release at the bottom of the S. mutans biofilm might confer adhesive properties to the substratum surface. To investigate the possibility that lytF expression and eDNA production contribute to the initial attachment to the substratum surface, we compared the attachment property of the WT, ΔlytF, and lytF-complemented cells to a polystyrene 96-well plate surface. We observed that the adherent cells were significantly increased in the WT and lytF-complemented strains in response to sCSP, whereas they were not increased in the lytF mutant (Fig. 7E). This result indicates that lytF contributes to the enhancement of cell adhesion to the substratum surface during the early stage of biofilm formation.
DISCUSSION
We succeeded in observing the eDNA production process at the single-cell level and experimentally showed that eDNA is released to the extracellular milieu from dead cells. In S. mutans, since random amplified polymorphic DNA indicates that eDNA is derived from genomic DNA, eDNA has been suggested to be mainly genomic DNA released from dead cells (10). Our data show that cell death that occurs in a subpopulation is a source of eDNA in S. mutans. Additionally, we found that not all cells stained with SYTOX Green release nucleic acids into the extracellular environment (Movie S1B). In addition, in the absence of sCSP we observed a few dead cells but did not see eDNA released from these cells. This indicates that not all dead cells are employed to release eDNA. In pyroptosis of mammalian cells, a model has been proposed in which cell death and cell lysis are not necessarily coupled (36). Similarly, bacterial cell death may not always lead to eDNA production. S. mutans possesses a thick cell wall, which provides the cell with a fairly rigid structure and could interfere with genomic DNA release from dead cells unless the cell walls are lysed. Therefore, we suggest that both cell death and autolysin expression, such as lytF, are necessary for the release of eDNA.
lytF deletion resulted in a significant reduction in cell death induction and eDNA production in the presence of sCSP, but did not completely abolish them. Besides lytF, cipB is a gene that is induced by CSP and is predicted to be involved in its own cell death in S. mutans (12). The deficiency of cipB completely abolished cell death induction and eDNA production in response to sCSP. cipB is essential for the CSP-induced expression of the ComR regulon and SigX regulon in complex media, such as the BHI medium that was used in this study (24). This could be why the cipB mutant displays decreased cell death and eDNA production. In contrast, in chemically defined medium (CDM), synthetic XIP can induce lytF without cipB (17). CDM could be useful to assess the impact of CipB and LytF on cell death and eDNA production. However, SigX activates the cipB expression in the presence of XIP in CDM (17). Thus, it is difficult to completely distinguish lytF-dependent cell death from cipB-dependent cell death in both complex medium and CDM. In the present study, we also observed that the deletion of comS, comR, or sigX completely abolished cell death and eDNA production in response to sCSP. Given that the comRS-sigX system is not necessary for cipB expression in complex media (19), the SigX regulon is mainly responsible for cell death and eDNA production under our experimental conditions. Therefore, we suggest that lytF-dependent cell death, rather than cipB, is dominant in response to CSP. In addition, our data imply the presence of cell death-related factors other than LytF in the SigX regulon.
We showed that cell death occurs in a subpopulation of lytF-expressing cells. This result strongly supports the idea that the lytF of S. mutans encodes a self-acting autolysin (15). The communication system via CSP is conserved in streptococci, and several competence-associated murein hydrolases have been reported (37–41). In Streptococcus pneumoniae, competent cells secrete the muralytic enzyme CbpD and kill noncompetent cells (42, 43). This suggests there is a strategy to make it easier for competent cells to acquire new genes, which are released from noncompetent siblings and related species (44). Most streptococcal species that do not have CbpD, such as S. mutans, have the lytF gene, which is predicted to be a functional analog of cbpD (45). However, S. mutans LytF acts within the produced cells rather than intercellularly, as does CbpD. Streptococcus sanguinis also has LytF, which is composed of the Bsp and CHAP domains, although the number of Bsp domains in LytF of S. sanguinis is different from the number in LytF of S. mutans. Since LytF of S. sanguinis can digest not only its own peptidoglycans but also the peptidoglycans of Streptococcus gordonii and S. mutans, it has been hypothesized that this LytF may kill other bacteria to induce eDNA production (40). For these reasons, there is a possibility that the physiological role of murein hydrolase associated with genetic competence may vary depending on the species. In addition, the finding that not all lytF-expressing cells cause cell death to produce eDNA implies the presence of other factor(s) that determine the survival or death of lytF-expressing cells. Therefore, this suggests that cell death mediated by LytF autolysin activity is controlled at posttranscriptional and/or posttranslational levels by unknown factor(s) that are necessary for the activity of LytF or to counteract the function of LytF.
lytF-expressing cells were observed near the bottom of the 2-h biofilm and gradually increased in the presence of sCSP. In addition, the deficiency of lytF reduced the adhesion of cells to the surface. In S. sanguinis, a strain overexpressing lytF by plasmid complementation was shown to decrease the biofilm formation (40). Thus, proper regulation of lytF expression within the biofilm may contribute to biofilm stability. The promoter activity of lytF is increased by heat shock, low pH, and oxidative stress, suggesting that CSP is an alarmone induced by stress (15). Cells located in the deep part of biofilms are exposed to various stresses, such as low pH, metabolic substrate limitations, and metabolite accumulation. It has been reported that the degradation of exopolysaccharides by dextranase treatment results in the disappearance of the acidic environment in microcolonies (28). Thus, the chemical gradient formation within the biofilms requires the exopolysaccharides produced by S. mutans. We show that the population of lytF-expressing cells in ΔgtfB cells, which were unable to produce the insoluble polysaccharides necessary for microcolony formation, was comparable to that in WT cells. Furthermore, our data show that even if lytF-expressing cells increase in the biofilm, these cells stay near the bottom of the biofilm. Recently, AI-2-mediated quorum sensing was found to control the expression of several genes in cells close to a substratum surface in S. mutans (29). However, the heterogeneous distribution of lytF-expressing cells in the biofilm was maintained with ΔluxS, which was unable to produce AI-2. Therefore, it is thought that other environmental factor(s) surrounding the cells near the bottom of the biofilm contribute to the induction of the localized lytF expression.
In addition, similar distributions of lytF-expressing cells were observed on surfaces with different properties, such as polystyrene, glass, and saliva-coated glass. These points suggest the heterogeneous expression of lytF in the biofilm is not dependent on the nature of substratum surface. Bacterium-substratum physicochemical interactions and cell wall deformation have been shown to affect gene expression (46). In Pseudomonas aeruginosa, the expression of algC, a gene involved in alginate biosynthesis, is induced by surface attachment (47). In Staphylococcus aureus, it is suggested that adhesion force regulates biofilm matrix production (48). Although the surface recognition mechanism is unclear in S. mutans, SpaP and GbpC on the cell surface attach cells to the tooth surface by binding to salivary agglutinin glycoprotein (49, 50). Since our data indicate that the presence or absence of the saliva coat does not affect the localization of lytF-expressing cells and eDNA production within the biofilm, SpaP- and GbpC-mediated adherence to the substratum surface is unlikely to cause these localizations. In addition, Wang et al. reported that adherence to the substratum surface and AI-2-mediated quorum sensing triggered the increased expression of comDE and brpA near the bottom of the biofilm (29). However, our data show that the localization of lytF-expressing cells was not altered even in the biofilm formed by ΔluxS cells. BrpA is involved in the regulation of the response to cell surface stress (51). Comparison of gene expression by DNA microarrays of S. mutans UA159 and the brpA mutant shows no change in the expression of most SigX regulon genes, including lytF (51). Therefore, surface recognition appears to play a role in the localization of lytF-expressing cells in the biofilm, though the detailed mechanism needs to be elucidated in future studies.
In addition to surface recognition, the bottom of the biofilm has particularly high cell density and cell proximity and these may also be involved in lytF expression. In Streptococcus pyogenes, peptide signaling-mediated cell density-dependent gene expression alters the expression of many genes, including pathogenic factors (52). In addition, CSP signaling in S. pneumoniae occurs through cell contact (53). Thus, differences in cell density and cell proximity within S. mutans biofilms may also be involved in inducing lytF expression.
Consistent with the localization of lytF-expressing cells, eDNA was also localized near the bottom of the biofilm. It has been reported that eDNA is mainly involved in initial attachment to the substratum surface in S. mutans biofilm formation (11). Our data show that eDNA contributes to the early stages of biofilm formation, while the mature biofilm can retain the structure when eDNA is removed. After the initial attachment by eDNA, insoluble glucan is mainly responsible for the development of the biofilm and the maintenance of the structure of the mature biofilm. Thus, we hypothesize that S. mutans effectively utilizes the eDNA produced at the expense of a subpopulation of cells by locally producing the eDNA required for initial attachment near the bottom of the biofilm.
Even without the addition of sCSP, lytF-expressing cells were observed in the 6-h biofilm. In previous studies, the expression of the SigX regulon, including lytF, was analyzed with the external addition of the sCSP signal. Our data are important and show that the CSP-mediated communication system works in actual biofilms without the external addition of CSP.
In conclusion, we have shown that CSP induces lytF-dependent cell death and eDNA production. lytF expression in biofilms is induced at the bottom, which contributes to cell adhesion to the substratum surface. Our data suggest that the heterogeneous lytF expression in the biofilm was due to physical contact with the surfaces rather than environmental heterogeneity, based on the three-dimensional structure of the biofilm. However, we could not exclude the possibility that other environmental factors at the bottom of the biofilm, such as high cell density and cell proximity, affect lytF expression, which should be addressed in future work. Moreover, we also show that a subpopulation of cells expressing lytF undergoes cell death and produces eDNA, while lytF-expressing live cells also exist. SigX, the sigma factor essential for lytF expression, also induces the expression of genes involved in genetic competence. Therefore, since the bottom of the biofilm is expected to have abundant eDNA and competent cells, it has the potential to be a site where horizontal gene transfer occurs. Analyzing the heterogeneity in biofilms from the functional aspect is a future challenge.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains used in this study are listed in Table 1. The bacterial cells were grown in an aerobic atmosphere containing 5% CO2 in brain heart infusion (BHI) broth (Difco Laboratories, Detroit, MI) at 37°C.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant propertiesa | Source or reference |
|---|---|---|
| Strains | ||
| S. mutans | ||
| UA159 | Wild type ermS kanS specS | Wild type |
| YN102 | UA159 SMU_1405c::Pldh-ZsGreen ermBP | (56) |
| ΔcomDE | UA159 comDE deletion mutant; ermBP | This study |
| ΔcipB | UA159 cipB deletion mutant; ermBP | This study |
| ΔcomR | UA159 comR deletion mutant; ermBP | This study |
| ΔcomS | UA159 comS deletion mutant; ermBP | This study |
| ΔcomX | UA159 comX deletion mutant; ermBP | This study |
| ΔlytF | UA159 lytF deletion mutant; ermBP | This study |
| lytF comp.b | UA159 lytF deletion mutant; ermBP, SMU_437c::PlytF lytF aph3 | This study |
| PlytF reporter | UA159 SMU_437c::PlytF-mScarlet-I aph3 | This study |
| Pldh reporter | UA159 SMU_1405c::Pldh-mScarlet-I ermBP | This study |
| PcipB reporter | UA159 SMU_437c::PcipB-mScarlet-I aph3 | This study |
| Pldh and PlytF dual reporter | UA159 SMU_437c::PlytF-mScarlet-I aph3, SMU_1405c::Pldh-mNeonGreen ermBP | This study |
| ΔlytF Pldh reporter | UA159 lytF deletion mutant; aad9, SMU_1405c::Pldh-mScarlet-I ermBP | This study |
| lytF comp. Pldh reporter | UA159 lytF deletion mutant; aad9, SMU_437c::PlytF lytF aph3, SMU_1405c::Pldh-mScarlet-I ermBP | This study |
| ΔgtfB PlytF reporter | UA159 gtfB deletion mutant; ermBP, SMU_437c::PlytF-mScarlet-I aph3 | This study |
| ΔluxS PlytF reporter | UA159 luxS deletion mutant; ermBP, SMU_437c::PlytF-mScarlet-I aph3 | |
| B. subtilis | ||
| TAY3203 | trpC2 lys1 ΔaprE3 nprE18 nprR2 ΔydiR-ydjA ΔyqpP-yodU (ΔSPβ) amyE::nonA-spoVG 3′ UTR aph3 | (58) |
| Plasmids | ||
| pJIR418 | ermBP | (54) |
| pEX-A2J1-mScarlet-I | mScarlet-I | Eurofins genomics |
| pEX-A2J1-mNG | mNeonGreen | Eurofins genomics |
| pDL278 | aad9 | (55) |
ermS, erythromycin susceptibility; kanS, kanamycin susceptibility; specS, spectinomycin susceptibility.
comp., complemented.
Construction of deletion mutants.
The primers used for the construction of deletion mutant strains are listed in Table S1. Sequence information was obtained from the KEGG (http://www.genome.jp/kegg/) and NCBI (https://www.ncbi.nlm.nih.gov/) databases. The deletion mutant strains were constructed by replacing the target gene with an erythromycin resistance gene (ermBP) or spectinomycin resistance gene (aad9) using a DNA fragment constructed by overlap extension PCR. The upstream and downstream sequences of the target genes were amplified from the genomic DNA of S. mutans UA159. The ermBP and aad9 genes were amplified from pJIR418 and pDL278, respectively (54, 55). Nested PCR was performed as necessary. The amplicons were connected by overlap extension PCR and introduced into S. mutans UA159. To induce genetic competence, 1 μM sCSP was added to the S. mutans UA159 culture grown in BHI at early log phase. The transformants were screened on Mitis Salivarius (MS) agar (Difco Laboratories, Detroit, MI) plates with erythromycin (10 μg/ml) or spectinomycin (200 μg/ml). Insertion into the target region of genomic DNA was confirmed by colony PCR and DNA sequencing.
Construction of promoter reporter strains.
The primers used for the construction of promoter reporter strains are listed in Table S2. The promoter reporter strains were constructed by inserting the promoter sequence, fluorescent protein gene (mScarlet-I or mNeonGreen), and drug resistance marker (ermBP or aph3) at the loci of SMU_437c, a pseudogene, and/or SMU_1405c, a gene that does not affect the typical phenotype (56). The adjacent sequences of insertion loci, ermBP, and promoter sequences were amplified from the genomic DNA of S. mutans UA159 or YN102 (56). We used the ldh promoter as a constitutive reporter (57). The mScarlet-I and mNeonGreen genes were artificially synthesized (Eurofins Genomics, Tokyo, Japan) and amplified by PCR. A kanamycin resistance gene (aph3) was amplified from Bacillus subtilis TAY3203 (58). The DNA fragments were connected by overlap extension PCR and introduced into S. mutans UA159, as previously described (56). The transformants were screened on MS agar plates with erythromycin (10 μg/ml) and/or kanamycin (900 μg/ml). Insertion into the target region of genomic DNA was confirmed by colony PCR and DNA sequencing.
Construction of a complemented strain.
The primers used for the construction of the lytF-complemented strain (lytF comp) are listed in Table S3. We constructed the complemented strain by introducing lytF transcribed from the native promoter into the SMU_437c locus. The adjacent region of SMU_437c, the lytF promoter and the lytF gene were amplified by PCR from the genome of S. mutans UA159. aph3 from B. subtilis TAY3203 was connected downstream of lytF as a selection marker (58). DNA fragments were ligated by overlap extension PCR. The DNA fragment was introduced into ΔlytF. The transformants were screened on MS agar (Difco Laboratories, Detroit, MI) plates with kanamycin (900 μg/ml). Insertion into the target region of genomic DNA was confirmed by colony PCR and DNA sequencing.
Microfluidic experiment.
The microfluidic device was made from polydimethylsiloxane (PDMS) from a single mold and sealed with a cover glass (59). High-aspect ratio quasi-two-dimensional microfluidic chambers for two-dimensional bacterial growth were made using the mold previously reported by our group (59). We used the Sylgard 184 silicone elastomer kit (Dow Chemical Company, Midland, MI). The silicone elastomer base component and curing agent were mixed at a ratio of 10:1. After degassing, the mixture was poured into the mold. To solidify the PDMS, the mixture was heated to 70°C for 12 h. We cut out the device from the mold and punched holes to connect fluoropolymer tubing (FEP tube, Junkosha, Ibaraki, Japan). The device was sealed by permanently bonding it to cover glass (Matsunami, Osaka, Japan) by treating it in O2 plasma for 20 s at 50 W using a plasma cleaner (Cute, Femto Science, Gyeonggi, South Korea).
Bacterial cells in the mid-log phase were inoculated into the flow cell chamber filled with BHI or BHI with 1 μM CSP supplemented with 1.25 μM SYTOX Green and cultured at 37°C. The medium was infused at a constant rate (150 μl/h) using a syringe pump. We acquired images every 10 min using an Axio observer Z1 (Carl Zeiss, Oberkochen, Germany) with a Plan-Apochromat 100×/1.46 oil-immersion objective lens.
eDNA quantification.
We diluted the overnight culture to an optical density at 600 nm (OD600) of 0.05 with fresh medium with or without 1 μM sCSP and grew the cells in an aerobic atmosphere containing 5% CO2 at 37°C for 6 h. The cells and supernatant were separated by centrifugation, and the DNA was extracted from 500 μl of the supernatant by using cetyltrimethylammonium bromide (CTAB) and phenol. The supernatant was mixed with 500 μl of CTAB solution and incubated at 65°C for 15 min. An equal amount of phenol-chloroform-isoamyl alcohol (Nippon Gene, Tokyo, Japan) was added. After centrifugation, we collected the aqueous phase, and the DNA was precipitated using isopropanol. The DNA pellet was washed with 70% ethanol and dissolved in TE buffer (10 mM Tris-HCl, 1 mM EDTA). We quantified the DNA with a plate reader (Varioskan Flash, Thermo Fisher Scientific, Waltham, MA) using a Quant-iT PicoGreen dsDNA assay kit (Thermo Fisher Scientific, Waltham, MA). The results were standardized by OD600 of the 6-h culture solution.
Flow cytometry.
We diluted overnight culture to an OD600 of 0.05 with fresh BHI with or without 1 μM sCSP and grew the cells in an aerobic atmosphere containing 5% CO2 at 37°C for 6 h. For biofilm induction, we used BHI supplemented with 1% (wt/vol) sucrose (BHIs). After 6 h of incubation, the culture medium was removed and cells were washed twice with sterile phosphate-buffered saline (PBS). The biofilm cells were collected using a cell scraper and sonicated for 10 s at 25 W to disperse aggregates. We stained the dead cells with 500 nM SYTOX Green (Thermo Fisher Scientific, Waltham, MA) for 15 min. The cells were diluted with filtered PBS. The bacterial suspension was analyzed with a cell sorter (SH800Z, Sony, Japan). For SYTOX Green fluorescence, we used a laser excitation of 488 nm and detected the fluorescence with 525/±25 and 487.5 LP filters. For mScarlet-I fluorescence, we used a laser excitation of 561 nm and detected the fluorescence with 600/±30 and 561 LP sequential filters. The boundaries between the positive and negative fluorescence were set using nonfluorescent cells and single fluorescent cells (Fig. S1).
Fluorescence microscopy.
We diluted overnight culture to an OD600 of 0.05 with fresh medium with or without 1 μM sCSP and grew the cells in an aerobic atmosphere containing 5% CO2 at 37°C for 6 h. Dead cells were stained with 500 nM SYTOX Green for 15 min. The cells were placed on 0.8% (wt/vol) agarose and pressed with a cover glass to prevent the cells from moving during observation. We acquired images using an Axio observer Z1 with an AxioCam 506 mono camera or an AxioCam MRm camera and a Plan-Apochromat 63×/1.40 oil-immersion or an EC Plan-Neofluar 100×/1.30 oil-immersion objective lens.
Biofilm formation.
We diluted overnight culture to an OD600 of 0.05 with BHIs with or without 1 μM sCSP and grew the cells in a 6-well plate (Iwaki, Shizuoka, Japan) or glass-bottom dish (Matsunami, Osaka, Japan) in an aerobic atmosphere containing 5% CO2 at 37°C for 6 h. To remove the planktonic cells, the biofilms were washed twice with PBS. We stained the cells with 5 μM SYTO 59 (Thermo Fisher Scientific, Waltham, MA) for 30 min. The stained biofilms were washed 2 times with PBS. We acquired 3D images by using an upright confocal laser scanning microscope (LSM880, Carl Zeiss, Oberkochen, Germany) with an IR-Achroplan 40×/0.8 water-immersion objective lens or inverted confocal laser scanning microscope (LSM780, Carl Zeiss, Oberkochen, Germany) with an Apochromat 40×/1.1 water-immersion objective lens. Z-stacks were acquired at 0.5 μm intervals. mNeonGreen, mScarlet-I, and SYTO 59 were excited by 488 nm, 543 nm, and 633 nm lasers, respectively, and the emissions were detected (481 to 570 nm for mNeonGreen, 589 to 624 nm for mScarlet-I, and 642 to 695 nm for SYTO 59). For image analysis, we calculated the area where SYTO 59 and mScarlet-I fluorescence were detected using ImageJ and Fiji (60, 61).
We used a 96-well plate (Sumitomo Bakelite, Tokyo, Japan) to perform a biofilm inhibition assay by DNase I (Roche Diagnostics GmbH, Germany). We diluted overnight culture to an OD600 of 0.05 with BHIs with 1 μM sCSP and added DNase I to the medium at 50 U/ml. The cells were incubated in an aerobic atmosphere containing 5% CO2 at 37°C for 6 h. After incubation, the medium was removed, and the wells were washed 2 times with PBS. We stained the biofilms with 0.1% (wt/vol) crystal violet (CV) for 15 min. The wells were washed 2 times with PBS to remove the excess CV. CV was extracted from the biofilms with 100 μl of 70% ethanol, and the absorbance at 595 nm was measured with a plate reader.
To test the effect of DNase I on the preformed biofilm, we used the 6-h biofilm formed in a 96-well plate. Before staining with CV, the biofilms were treated with incubation buffer (40 mM Tris-HCl, 10 mM NaCl, 6 mM MgCl2, 1 mM CaCl2, pH 7.9) with or without DNase I (50 U/ml) at 37°C for 6 h. The wells were washed 2 times with PBS. We quantified the remaining biofilm using CV by the same procedure as above.
Attachment assay.
We diluted overnight culture to an OD600 of 0.05 with fresh BHI with 1 μM sCSP and grew the cells in an aerobic atmosphere containing 5% CO2 at 37°C for 6 h. The culture solution was placed in a 96-well plate (100 μl/well) at 4°C for 6 h. The adherent cells were stained with CV for 15 min. The wells were washed twice with PBS. CV was extracted from the biofilms with 100 μl of 70% ethanol and the absorbance was measured at 595 nm with a plate reader. The results were standardized by OD600 of the 6-h culture solution.
Statistics.
The differences were analyzed for statistical significance with Student’s t test or one-way analysis of variance and Bonferroni’s posttest (IBM SPSS statistics 24, IBM Corporation, Armonk, NY). In this study, three independent experiments were performed in duplicate or triplicate. A P value of <0.05 was considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant-in-aid for Scientific Research (18J21373) from the Japanese Society for the Promotion of Sciences and ERATO (JPMJER1502) from the Japan Science and Technology Agency.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Okshevsky M, Regina VR, Meyer RL. 2015. Extracellular DNA as a target for biofilm control. Curr Opin Biotechnol 33:73–80. doi: 10.1016/j.copbio.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 2.Petersen FC, Tao L, Scheie AA. 2005. DNA binding-uptake system: a link between cell-to-cell communication and biofilm formation. J Bacteriol 187:4392–4400. doi: 10.1128/JB.187.13.4392-4400.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas VC, Thurlow LR, Boyle D, Hancock LE. 2008. Regulation of autolysis-dependent extracellular DNA release by Enterococcus faecalis extracellular proteases influences biofilm development. J Bacteriol 190:5690–5698. doi: 10.1128/JB.00314-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qin Z, Ou Y, Yang L, Zhu Y, Tolker-Nielsen T, Molin S, Qu D. 2007. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology (Reading) 153:2083–2092. doi: 10.1099/mic.0.2007/006031-0. [DOI] [PubMed] [Google Scholar]
- 5.Regev-Yochay G, Trzcinski K, Thompson CM, Malley R, Lipsitch M. 2006. Interference between Streptococcus pneumoniae and Staphylococcus aureus: in vitro hydrogen peroxide-mediated killing by Streptococcus pneumoniae. J Bacteriol 188:4996–5001. doi: 10.1128/JB.00317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng L, Chen Z, Itzek A, Ashby M, Kreth J. 2011. Catabolite control protein A controls hydrogen peroxide production and cell death in Streptococcus sanguinis. J Bacteriol 193:516–526. doi: 10.1128/JB.01131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olwal CO, Ang’ienda PO, Onyango DM, Ochiel DO. 2018. Susceptibility patterns and the role of extracellular DNA in Staphylococcus epidermidis biofilm resistance to physico-chemical stress exposure. BMC Microbiol 18:40. doi: 10.1186/s12866-018-1183-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gödeke J, Paul K, Lassak J, Thormann KM. 2011. Phage-induced lysis enhances biofilm formation in Shewanella oneidensis MR-1. ISME J 5:613–626. doi: 10.1038/ismej.2010.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carrolo M, Frias MJ, Pinto FR, Melo-Cristino J, Ramirez M. 2010. Prophage spontaneous activation promotes DNA release enhancing biofilm formation in Streptococcus pneumoniae. PLoS One 5:e15678. doi: 10.1371/journal.pone.0015678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagasawa R, Sato T, Senpuku H. 2017. Raffinose induces biofilm formation by Streptococcus mutans in low concentrations of sucrose by increasing production of extracellular DNA and fructan. Appl Environ Microbiol 83:e00869-17. doi: 10.1128/AEM.00869-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawarai T, Narisawa N, Suzuki Y, Nagasawa R, Senpuku H. 2016. Streptococcus mutans biofilm formation is dependent on extracellular DNA in primary low pH conditions. J Oral Biosci 58:55–61. doi: 10.1016/j.job.2015.12.004. [DOI] [Google Scholar]
- 12.Perry JA, Jones MB, Peterson SN, Cvitkovitch DG, Lévesque CM. 2009. Peptide alarmone signalling triggers an auto-active bacteriocin necessary for genetic competence. Mol Microbiol 72:905–917. doi: 10.1111/j.1365-2958.2009.06693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Senadheera DB, Cordova M, Ayala EA, Chávez de Paz LE, Singh K, Downey JS, Svensäter G, Goodman SD, Cvitkovitch DG. 2012. Regulation of bacteriocin production and cell death by the VicRK signaling system in Streptococcus mutans. J Bacteriol 194:1307–1316. doi: 10.1128/JB.06071-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liao S, Klein MI, Heim KP, Fan Y, Bitoun JP, Ahn S-J, Burne RA, Koo H, Brady LJ, Wen ZT. 2014. Streptococcus mutans extracellular DNA is upregulated during growth in biofilms, actively released via membrane vesicles, and influenced by components of the protein secretion machinery. J Bacteriol 196:2355–2366. doi: 10.1128/JB.01493-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dufour D, Lévesque CM. 2013. Cell death of Streptococcus mutans induced by a quorum-sensing peptide occurs via a conserved streptococcal autolysin. J Bacteriol 195:105–114. doi: 10.1128/JB.00926-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mashburn-Warren L, Morrison DA, Federle MJ. 2010. A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol Microbiol 78:589–606. doi: 10.1111/j.1365-2958.2010.07361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reck M, Tomasch J, Wagner-Döbler I. 2015. The alternative sigma factor SigX controls bacteriocin synthesis and competence, the two quorum sensing regulated traits in Streptococcus mutans. PLoS Genet 11:e1005353. doi: 10.1371/journal.pgen.1005353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leung V, Dufour D, Lévesque CM. 2015. Death and survival in Streptococcus mutans: differing outcomes of a quorum-sensing signaling peptide. Front Microbiol 6:1176. doi: 10.3389/fmicb.2015.01176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khan R, Rukke HV, Høvik H, Åmdal HA, Chen T, Morrison DA, Petersen FC. 2016. Comprehensive transcriptome profiles of Streptococcus mutans UA159 map core streptococcal competence genes. mSystems 1:e00038-15. doi: 10.1128/mSystems.00038-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaspar J, Underhill SAM, Shields RC, Reyes A, Rosenzweig S, Hagen SJ, Burne RA. 2017. Intercellular communication via the comX-inducing peptide (XIP) of Streptococcus mutans. J Bacteriol 199:e00404-17. doi: 10.1128/JB.00404-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Underhill SAM, Shields RC, Kaspar JR, Haider M, Burne RA, Hagen SJ. 2018. Intracellular signaling by the comRS system in Streptococcus mutans genetic competence. mSphere 3: e00444-18. doi: 10.1128/mSphere.00444-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perry JA, Cvitkovitch DG, Lévesque CM. 2009. Cell death in Streptococcus mutans biofilms: a link between CSP and extracellular DNA. FEMS Microbiol Lett 299:261–266. doi: 10.1111/j.1574-6968.2009.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hossain MS, Biswas I. 2012. An extracelluar protease, SepM, generates functional competence-stimulating peptide in Streptococcus mutans UA159. J Bacteriol 194:5886–5896. doi: 10.1128/JB.01381-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dufour D, Cordova M, Cvitkovitch DG, Lévesque CM. 2011. Regulation of the competence pathway as a novel role associated with a streptococcal bacteriocin. J Bacteriol 193:6552–6559. doi: 10.1128/JB.05968-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Son M, Ahn SJ, Guo Q, Burne RA, Hagen SJ. 2012. Microfluidic study of competence regulation in Streptococcus mutans: environmental inputs modulate bimodal and unimodal expression of comX. Mol Microbiol 86:258–272. doi: 10.1111/j.1365-2958.2012.08187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lemme A, Gröbe L, Reck M, Tomasch J, Wagner-Döbler I. 2011. Subpopulation-specific transcriptome analysis of competence-stimulating-peptide-induced Streptococcus mutans. J Bacteriol 193:1863–1877. doi: 10.1128/JB.01363-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao J, Klein MI, Falsetta ML, Lu B, Delahunty CM, Yates JR, Heydorn A, Koo H. 2012. The exopolysaccharide matrix modulates the interaction between 3D architecture and virulence of a mixed-species oral biofilm. PLoS Pathog 8:e1002623. doi: 10.1371/journal.ppat.1002623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hwang G, Liu Y, Kim D, Sun V, Aviles-Reyes A, Kajfasz JK, Lemos JA, Koo H. 2016. Simultaneous spatiotemporal mapping of in situ pH and bacterial activity within an intact 3D microcolony structure. Sci Rep 6:32841. doi: 10.1038/srep32841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang C, Hou J, van der Mei HC, Busscher HJ, Ren Y. 2019. Emergent properties in Streptococcus mutans biofilms are controlled through adhesion force sensing by initial colonizers. mBio 10:e01908-19. doi: 10.1128/mBio.01908-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wenderska IB, Lukenda N, Cordova M, Magarvey N, Cvitkovitch DG, Senadheera DB. 2012. A novel function for the competence inducing peptide, XIP, as a cell death effector of Streptococcus mutans. FEMS Microbiol Lett 336:104–112. doi: 10.1111/j.1574-6968.2012.02660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Das T, Sehar S, Manefield M. 2013. The roles of extracellular DNA in the structural integrity of extracellular polymeric substance and bacterial biofilm development. Environ Microbiol Rep 5:778–786. doi: 10.1111/1758-2229.12085. [DOI] [PubMed] [Google Scholar]
- 32.Qi F, Kreth J, Lévesque CM, Kay O, Mair RW, Shi W, Cvitkovitch DG, Goodman SD. 2005. Peptide pheromone induced cell death of Streptococcus mutans. FEMS Microbiol Lett 251:321–326. doi: 10.1016/j.femsle.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 33.Stewart PS, Franklin MJ. 2008. Physiological heterogeneity in biofilms. Nat Rev Microbiol 6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 34.Das T, Sharma PK, Busscher HJ, van der Mei HC, Krom BP. 2010. Role of extracellular DNA in initial bacterial adhesion and surface aggregation. Appl Environ Microbiol 76:3405–3408. doi: 10.1128/AEM.03119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ye J, Shao C, Zhang X, Guo X, Gao P, Cen Y, Ma S, Liu Y. 2017. Effects of DNase I coating of titanium on bacteria adhesion and biofilm formation. Mater Sci Eng C Mater Biol Appl 78:738–747. doi: 10.1016/j.msec.2017.04.078. [DOI] [PubMed] [Google Scholar]
- 36.DiPeso L, Ji DX, Vance RE, Price JV. 2017. Cell death and cell lysis are separable events during pyroptosis. Cell Death Discov 3:17070. doi: 10.1038/cddiscovery.2017.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moscoso M, Claverys J-P. 2004. Release of DNA into the medium by competent Streptococcus pneumoniae: kinetics, mechanism and stability of the liberated DNA. Mol Microbiol 54:783–794. doi: 10.1111/j.1365-2958.2004.04305.x. [DOI] [PubMed] [Google Scholar]
- 38.Eldholm V, Johnsborg O, Haugen K, Ohnstad HS, Håvarstein LS. 2009. Fratricide in Streptococcus pneumoniae: contributions and role of the cell wall hydrolases CbpD, LytA and LytC. Microbiology (Reading) 155:2223–2234. doi: 10.1099/mic.0.026328-0. [DOI] [PubMed] [Google Scholar]
- 39.Wei H, Håvarstein LS. 2012. Fratricide is essential for efficient gene transfer between pneumococci in biofilms. Appl Environ Microbiol 78:5897–5905. doi: 10.1128/AEM.01343-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cullin N, Redanz S, Lampi KJ, Merritt J, Kreth J. 2017. Murein hydrolase LytF of Streptococcus sanguinis and the ecological consequences of competence development. Appl Environ Microbiol 83: e01709-17. doi: 10.1128/AEM.01709-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu Y, Kreth J. 2013. Role of LytF and AtlS in eDNA release by Streptococcus gordonii. PLoS One 8:e62339. doi: 10.1371/journal.pone.0062339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guiral S, Mitchell TJ, Martin B, Claverys J-P. 2005. Competence-programmed predation of noncompetent cells in the human pathogen Streptococcus pneumoniae: genetic requirements. Proc Natl Acad Sci U S A 102:8710–8715. doi: 10.1073/pnas.0500879102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Håvarstein LS, Martin B, Johnsborg O, Granadel C, Claverys J-P. 2006. New insights into the pneumococcal fratricide: relationship to clumping and identification of a novel immunity factor. Mol Microbiol 59:1297–1307. doi: 10.1111/j.1365-2958.2005.05021.x. [DOI] [PubMed] [Google Scholar]
- 44.Johnsborg O, Eldholm V, Bjørnstad ML, Håvarstein LS. 2008. A predatory mechanism dramatically increases the efficiency of lateral gene transfer in Streptococcus pneumoniae and related commensal species. Mol Microbiol 69:245–253. doi: 10.1111/j.1365-2958.2008.06288.x. [DOI] [PubMed] [Google Scholar]
- 45.Berg KH, Ohnstad HS, Håvarstein LS. 2012. LytF, a novel competence-regulated murein hydrolase in the genus Streptococcus. J Bacteriol 194:627–635. doi: 10.1128/JB.06273-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carniello V, Peterson BW, van der Mei HC, Busscher HJ. 2018. Physico-chemistry from initial bacterial adhesion to surface-programmed biofilm growth. Adv Colloid Interface Sci 261:1–14. doi: 10.1016/j.cis.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 47.Davies DG, Geesey GG. 1995. Regulation of the alginate biosynthesis gene algC in Pseudomonas aeruginosa during biofilm development in continuous culture. Appl Environ Microbiol 61:860–867. doi: 10.1128/AEM.61.3.860-867.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harapanahalli AK, Chen Y, Li J, Busscher HJ, van der Mei HC. 2015. Influence of adhesion force on icaA and cidA gene expression and production of matrix components in Staphylococcus aureus biofilms. Appl Environ Microbiol 81:3369–3378. doi: 10.1128/AEM.04178-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jenkinson HF, Demuth DR. 1997. Structure, function and immunogenicity of streptococcal antigen I/II polypeptides. Mol Microbiol 23:183–190. doi: 10.1046/j.1365-2958.1997.2021577.x. [DOI] [PubMed] [Google Scholar]
- 50.Mieher JL, Larson MR, Schormann N, Purushotham S, Wu R, Rajashankar KR, Wu H, Deivanayagam C. 2018. Glucan binding protein C of Streptococcus mutans mediates both sucrose-independent and sucrose-dependent adherence. Infect Immun 86: e00146-18. doi: 10.1128/IAI.00146-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bitoun JP, Liao S, Yao X, Ahn SJ, Isoda R, Nguyen AH, Brady LJ, Burne RA, Abranches J, Wen ZT. 2012. BrpA is involved in regulation of cell envelope stress responses in Streptococcus mutans. Appl Environ Microbiol 78:2914–2922. doi: 10.1128/AEM.07823-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Do H, Makthal N, VanderWal AR, Rettel M, Savitski MM, Peschek N, Papenfort K, Olsen RJ, Musser JM, Kumaraswami M. 2017. Leaderless secreted peptide signaling molecule alters global gene expression and increases virulence of a human bacterial pathogen. Proc Natl Acad Sci U S A 114:E8498–E8507. doi: 10.1073/pnas.1705972114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prudhomme M, Berge M, Martin B, Polard P. 2016. Pneumococcal competence coordination relies on a cell-contact sensing mechanism. PLoS Genet 12:e1006113. doi: 10.1371/journal.pgen.1006113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sloan J, Warner TA, Scott PT, Bannam TL, Berryman DI, Rood JI. 1992. Construction of a sequenced Clostridium perfringens-Escherichia coli shuttle plasmid. Plasmid 27:207–219. doi: 10.1016/0147-619x(92)90023-4. [DOI] [PubMed] [Google Scholar]
- 55.LeBlanc DJ, Lee LN, Abu-Al-Jaibat A. 1992. Molecular, genetic, and functional analysis of the basic replicon of pVA380-1, a plasmid of oral streptococcal origin. Plasmid 28:130–145. doi: 10.1016/0147-619x(92)90044-b. [DOI] [PubMed] [Google Scholar]
- 56.Nakanishi Y, Yamamoto T, Obana N, Toyofuku M, Nomura N, Kaneko A. 2018. Spatial distribution and chemical tolerance of Streptococcus mutans within dual-species cariogenic biofilms. Microbes Environ 33:455–458. doi: 10.1264/jsme2.ME18113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Merritt J, Kreth J, Qi F, Sullivan R, Shi W. 2005. Non-disruptive, real-time analyses of the metabolic status and viability of Streptococcus mutans cells in response to antimicrobial treatments. J Microbiol Methods 61:161–170. doi: 10.1016/j.mimet.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 58.Yamamoto T, Obana N, Yee LM, Asai K, Nomura N, Nakamura K. 2014. SP10 infectivity is aborted after bacteriophage SP10 infection induces nonA transcription on the prophage SPβ region of the Bacillus subtilis genome. J Bacteriol 196:693–706. doi: 10.1128/JB.01240-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kunoh T, Morinaga K, Sugimoto S, Miyazaki S, Toyofuku M, Iwasaki K, Nomura N, Utada AS. 2020. Polyfunctional nanofibril appendages mediate attachment, filamentation, and filament adaptability in Leptothrix cholodnii. ACS Nano 14:5288–5297. doi: 10.1021/acsnano.9b04663. [DOI] [PubMed] [Google Scholar]
- 60.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.