Wastewater treatment generates large amounts of waste-activated sludge (WAS), which consists mainly of recalcitrant microbial cells and particulate organic matter. Though WAS pretreatment is an effective way to release sludge organic matter for subsequent digestion, detailed information on the impact of the sludge pretreatment on the digestion sludge microbiome remains scarce. Our study provides unprecedented genome-centric metagenomic insights into how WAS pretreatments change the digestion sludge microbiomes, as well as their metabolic networks. Moreover, digestion sludge microbiomes could be a unique source for exploring microbial dark matter. These results may inform future optimization of methanogenic sludge digestion and resource recovery.
KEYWORDS: APAD, digestion sludge microbiome, metagenome, sludge pretreatment
ABSTRACT
Pretreatment of waste-activated sludge (WAS) is an effective way to destabilize sludge floc structure and release organic matter for improving sludge digestion efficiency. Nonetheless, information on the impact of WAS pretreatment on digestion sludge microbiomes, as well as mechanistic insights into how sludge pretreatment improves digestion performance, remains elusive. In this study, a genome-centric metagenomic approach was employed to investigate the digestion sludge microbiome in four sludge digesters with different types of feeding sludge: WAS pretreated with 0.25 mol/liter alkaline/acid (APAD), WAS pretreated with 0.8 mol/liter alkaline/acid (HS-APAD), thermally pretreated WAS (thermal-AD), and fresh WAS (control-AD). We retrieved 254 metagenome-assembled genomes (MAGs) to identify the key functional populations involved in the methanogenic digestion process. These MAGs span 28 phyla, including 69 yet-to-be-cultivated lineages, and 30 novel lineages were characterized with metabolic potential associated with hydrolysis and fermentation. Interestingly, functional populations involving carbohydrate digestion were enriched in APAD and HS-APAD, while lineages related to protein and lipid fermentation were enriched in thermal-AD, corroborating the idea that different substrates are released from alkaline/acid and thermal pretreatments. Among the major functional populations (i.e., fermenters, syntrophic acetogens, and methanogens), significant correlations between genome sizes and abundance of the fermenters were observed, particularly in APAD and HS-APAD, which had improved digestion performance.
IMPORTANCE Wastewater treatment generates large amounts of waste-activated sludge (WAS), which consists mainly of recalcitrant microbial cells and particulate organic matter. Though WAS pretreatment is an effective way to release sludge organic matter for subsequent digestion, detailed information on the impact of the sludge pretreatment on the digestion sludge microbiome remains scarce. Our study provides unprecedented genome-centric metagenomic insights into how WAS pretreatments change the digestion sludge microbiomes, as well as their metabolic networks. Moreover, digestion sludge microbiomes could be a unique source for exploring microbial dark matter. These results may inform future optimization of methanogenic sludge digestion and resource recovery.
INTRODUCTION
Waste-activated sludge (WAS) from wastewater treatment plants contains high levels of organic matter in the form of cells, extracellular polymeric substances (EPS), and macromolecules generated from cell lysis, as well as pathogens and other biohazards (1, 2). Anaerobic digestion is a sustainable sludge treatment technology that can convert these organic substances into biogas via a multiple-step process consisting of hydrolysis, fermentation, acetogenesis, and methanogenesis (3). The organic matter (e.g., microbial cells and EPS) in WAS form stable and complicated sludge flocs. These sludge flocs are recalcitrant to anaerobic digestion and consequently require sludge pretreatment to destabilize their structure and release organic matter for improving digestion efficiency (4). A variety of pretreatment techniques (e.g., thermal, ultrasonic, microwave, and alkaline pretreatments) have been developed to enhance digestion efficiency by releasing sludge organic matter (5–7). For example, thermal hydrolysis (>100°C) pretreatments can enhance solubilization of sludge organic matter, increasing sludge fermentation efficiencies by 4 to 20%, compared to non-pretreated controls (7). In our previous studies, a new alkaline/acid pretreatment and anaerobic digestion (APAD) process was developed, in which organic carbon removal reached 52.8% ± 1.7% (2). Further increasing alkaline/acid concentrations from 0.25 mol/liter (APAD) to 0.8 mol/liter (HS-APAD) did not show notable changes in digestion performance and community taxonomic composition in the sludge digesters (8). In contrast, the organic carbon removal rates were significantly lower in other digesters under the same operational conditions but with different influent-sludge-derived organic matter, i.e., 42.4% ± 1.6% in thermal-AD (i.e., WAS with thermal sludge pretreatment) and 30.9% ± 2.2% in control-AD (i.e., WAS without sludge pretreatment) (2). Therefore, dissolved organic compounds (DOC) derived from sludge pretreatments, rather than alkaline/acid pretreatment-derived salinity, could be a predominant selective pressure driving the performance and microbiome changes in sludge digesters (8). The mechanistic details of the digestion improvement and impact of WAS pretreatment on digestion sludge microbiomes remain unknown.
The conversion of sludge organic matter into biogas relies heavily on the complex and tightly coupled synergistic interactions of digestion sludge microbial populations (9). Previous studies based on 16S rRNA gene amplicon sequencing and metagenomic analyses showed that the relative abundances of bacteria and archaea in digestion sludge microbiomes were generally >95% and <5%, respectively (9–12). In terms of the bacterial community composition, Bacteroidetes, Proteobacteria, Spirochaetes, and Firmicutes were generally the dominant phyla in sludge digesters, most of which were fermentative bacteria with high compositional and functional redundancy (9, 13–16). Although a variety of factors (e.g., feeding substrates, pH, temperature, and ammonia) might change community composition and function of the digestion sludge microbiome (11, 12, 16, 17), the information generated from the 16S rRNA gene-based analyses was largely limited to community composition and succession (18).
Metagenome sequencing could theoretically obtain all microbial genome information within a sample, providing direct access to the metabolic potential and networks in the highly complex digestion sludge microbiome. Early metagenomic studies mainly relied on gene-centric analyses, which were biased toward existing databases (9, 19, 20). Current advances in both high-throughput sequencing technologies and population genome binning algorithms allowed the development of genome-centric approaches for the assessment of complex microbiomes (12, 18, 21). For example, genome-centric metagenomic analysis was employed to recover 101 population genomes and revealed their metabolic potential and interactions in a cellulose-degrading digester (16). Moreover, metagenomics changed the pace of virus discovery by enabling the accurate identification of viral genome sequences without requiring isolation of viruses (22, 23). In contrast to increasing metagenomic data on anaerobic digestion, no genome-centric metagenomic information was available on the impact of WAS pretreatment on digestion sludge microbiomes.
The genome-centric and strain-resolved metagenomic approaches could provide a systematic understanding of digestion sludge microbiomes, particularly the yet-to-be-elucidated impact of WAS pretreatment on digestion sludge microbiomes. In this study, we employed a metagenomic approach to explore prokaryotic and DNA viral community composition and the function of digestion sludge microbiomes in four sludge digesters (i.e., APAD, HS-APAD, thermal-AD, and control-AD). Coassembly of the four metagenomes followed by genomic binning resulted in the recovery of 254 population genomes that constituted the majority of the digestion sludge microbiome. Their metabolic potential and networks were further reconstructed to reveal the impact of WAS pretreatment on the digestion sludge microbiome. The results provided unprecedented genome-centric insights into how WAS pretreatment changes community composition and function, as well as metabolic networks of key players and their genomic traits in the digestion sludge microbiome.
RESULTS
Superkingdom community composition of digestion sludge microbiomes.
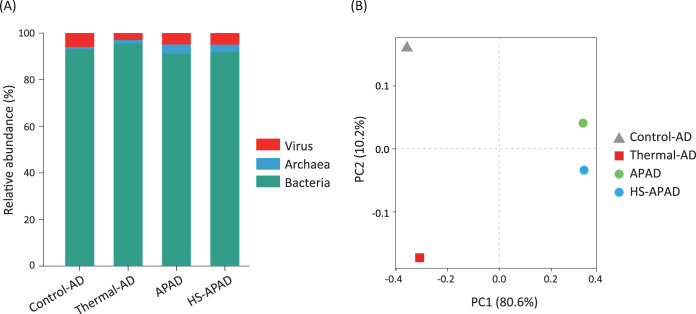
Four metagenomes (144 Gb total raw sequencing reads) from APAD, HS-APAD, thermal-AD, and control-AD were coassembled, generating 833,655 contigs with a combined length of 2,395 Mb (N50 = 3,658 bp) (see Table S1 in the supplemental material). Community composition investigation based on the relative number of microorganisms showed that Bacteria, Archaea, and DNA viruses were the three major taxonomic groups of the digestion sludge microbiome, exhibiting average relative abundances of 92.9%, 2.4%, and 4.7%, respectively (Fig. 1A). Notably, the lowest relative abundance of virus in thermal-AD (i.e., 2.9% in thermal-AD versus 5.9% in control-AD, 4.9% in APAD, and 4.9% in HS-APAD) might be due to the efficient removal of bacteriophages in thermal sludge pretreatment (24). Also, compared to control-AD, the higher abundance of methanogenic Archaea in APAD and HS-APAD corroborated their enhanced carbon removal and biogas generation (2, 8). Principal-coordinate analysis (PCoA) showed that thermal and alkaline/acid pretreatment had very different impacts on the digestion sludge microbiome. In contrast, APAD and HS-APAD exhibited similar microbial community patterns (Fig. 1B), consistent with their 16S rRNA gene-based community clustering (Fig. S1) (2, 8).
FIG 1.
Main taxonomic groups of the digestion sludge microbiome at the domain/kingdom level (A) and their principal-coordinate analysis (PCoA) (B).
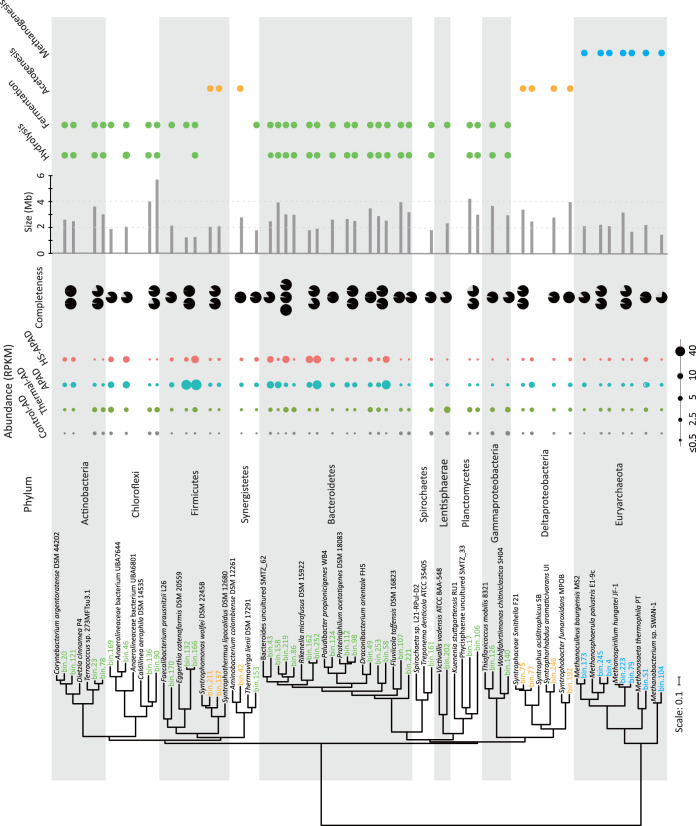
Population genome binning of the coassembled metagenomes enabled the recovery of 245 bacterial and 9 archaeal population genomes with >70% completeness and <5% contamination (Fig. 2; Table S2). Both bacterial and archaeal metagenome-assembled genomes (MAGs) represented major populations in digestion sludge microbiomes, i.e., 66.4%, 65.7%, 38.2%, and 30.2% of the total community in APAD, HS-APAD, thermal-AD, and control-AD, respectively, based on percentages of read mapping. These populations were phylogenetically diverse and belonged to 28 different phyla, of which 69 MAGs represented novel lineages at the order level and even higher taxonomic levels (Fig. S2). Major populations carrying out different steps in anaerobic sludge digestion (i.e., hydrolysis, fermentation, syntrophic acetogenesis, and methanogenesis) had significant differences in their abundances, normalized as reads per kilobase per million (RPKM), e.g., 31.4 to 200.9 for fermentative bacteria, 2.5 to 12.5 for syntrophic acetogens, and 0.75 to 8.3 for methanogens (Fig. 2), which was further corroborated by the 16S rRNA gene-based analyses (Fig. S1).
FIG 2.
Phylogeny, genome completeness, and abundance of retrieved MAGs which were the major populations involved in sludge digestion. The phylogenetic tree was constructed based on an alignment of concatenated ribosomal protein sequences from the MAGs and their related lineages. Phyla are distinguished by alternating background colors. The genome completeness of the MAGs is shown as pie charts. The right-hand columns indicated the presence (circles) and absence (blank) of MAG functions (i.e., hydrolysis, fermentation, syntrophic acetogenesis, and methanogenesis) in sludge digestion. See Fig. S2 for phylogenetic analysis of all 254 MAGs.
Metabolic networks in sludge digesters.
To construct the metabolic networks, the metabolic potential of major microorganisms (or MAGs) involving conversion of sludge organic matter into biogas was identified based on the carbon flow from organic macromolecules to methane. The predominant organic macromolecules to be degraded in the sludge digesters were polysaccharides, proteins, and lipids. These macromolecules could be converted into methane via mediation by three major groups of functional microorganisms, i.e., hydrolyzing and fermentative bacteria, syntrophic acetogenic bacteria, and methanogenic archaea (Table S3).
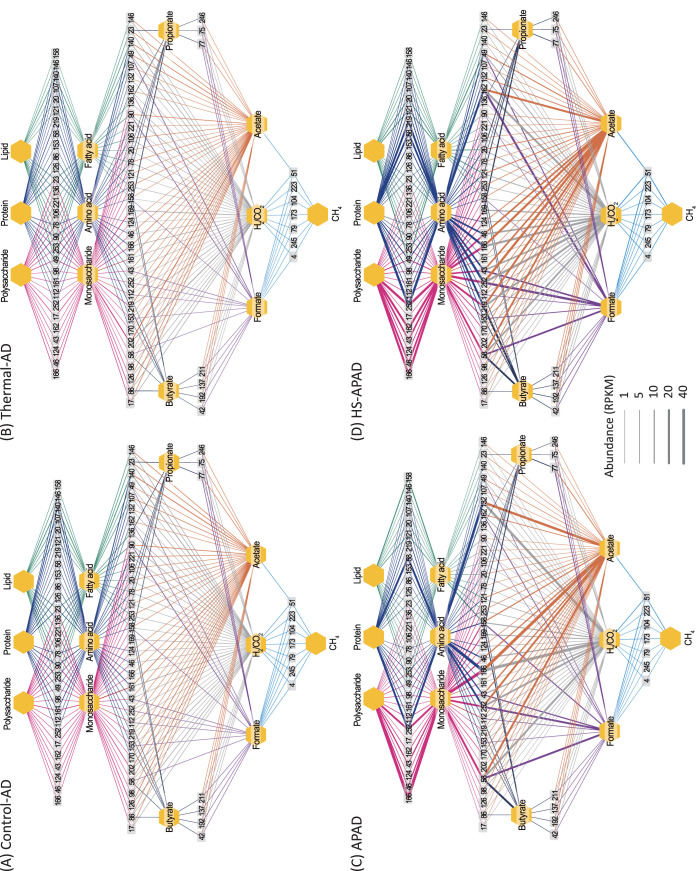
With regard to metabolic networks in the four sludge digesters, the presence of multiple populations capable of carrying out the same function (e.g., hydrolysis, fermentation, and methanogenesis) suggested high levels of functional redundancy (Fig. 3; Table S4). The high levels of functional redundancy were contributed by both core functional populations (major and essential functional microorganisms) and redundant functional populations (minor and nonessential functional populations), and the two different groups of functional populations could be interconvertible when operational conditions of sludge digestion were changed (25, 26). In line with the microbial community composition, the overall metabolic networks in APAD and HS-APAD were highly similar (P = 0.696) (Fig. 3C and D) but different from those in thermal-AD and control-AD (Fig. 3A and B). These distinct metabolic networks indicated different digestion substrates released from alkaline/acid and thermal sludge pretreatments, which was further corroborated by measured organic matter in their influent sludge (Table S5). Notably, the polysaccharide and protein metabolisms were significantly enriched in APAD and HS-APAD (Fig. 3C and D), suggesting effective destruction of both EPS and microbial cells to release polysaccharides and proteins in the alkaline/acid pretreatment. Accordingly, bacterial MAGs of Firmicutes (e.g., bin 132 and bin 166) and Bacteroidetes (e.g., bin 58 and bin 252) phyla were selectively enriched as core functional populations in APAD and HS-APAD. In addition, the predominant hydrolyzing and fermentative bacterial populations were different in thermal-AD and control-AD, e.g., bin 169 of Chloroflexi and bin 202 of Lentisphaerae in the thermal-AD, and bin 140 of Proteobacteria and bin 161 of Actinobacteria in the control-AD. In the syntrophic acetogenic process, propionate and butyrate were major volatile fatty acids (VFAs) as substrates for syntrophic acetogenic bacteria (e.g., bin 42 and bin 137 of the phyla Synergistetes and Firmicutes, respectively, for butyrate oxidation and bin 75 and bin 77 of the phylum Proteobacteria for propionate oxidation), which were significantly enriched as common and core syntrophic acetogens in sludge digesters fed with thermally pretreated or alkaline/acid-pretreated WAS (Fig. 3; Table S4). For methanogenesis, a variety of acetoclastic and hydrogenotrophic methanogens were observed, with a high level of functional redundancy in the four digesters. Particularly, members of Methanosaeta (bin 51), which are capable of producing methane from acetate, formate, and H2, were enriched as core methanogens in APAD and HS-APAD (Fig. 3C and D), which is in good agreement with their high methane production (2, 8).
FIG 3.
Metabolic networks based on the functional classification of all populations with abundances of >33.8 RPKM or the top seven most abundant lineages for methanogenic digestion in control-AD (A), thermal-AD (B), APAD (C), and HS-APAD (D). The colors correspond to conversion of different substrates, and the thickness of the lines is representative of the MAG abundance. See Table S4 for detailed abundances of the MAGs.
Microbial dark matter.
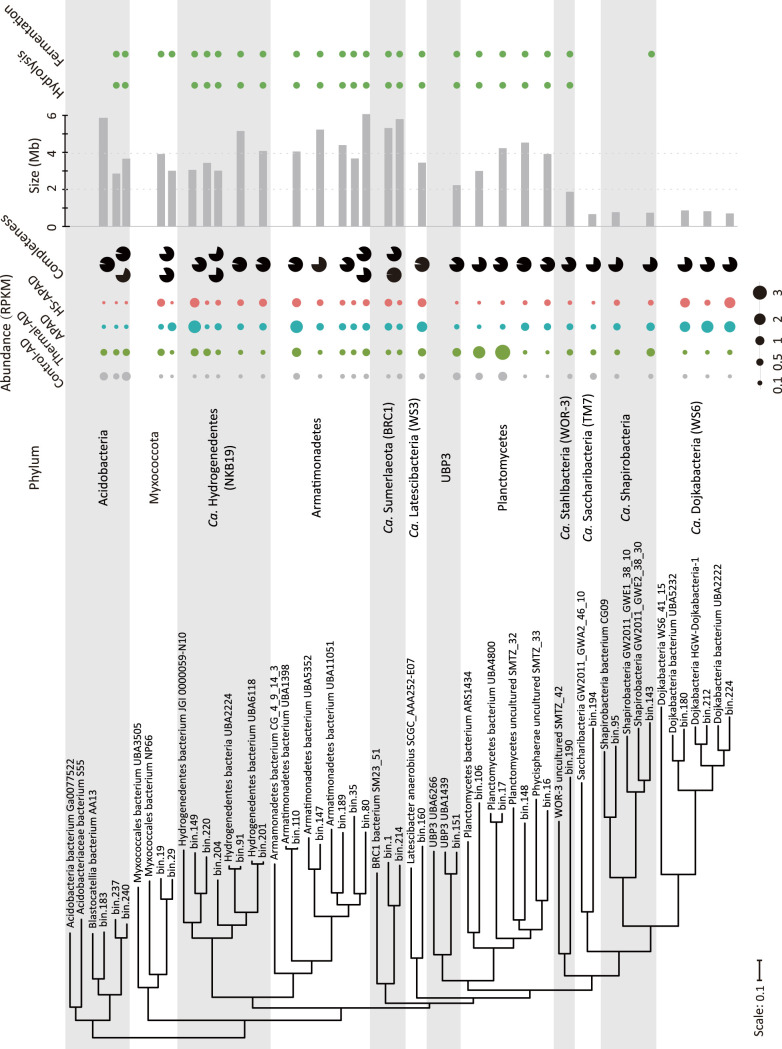
To elucidate the representative yet-to-be-cultivated lineages, or microbial dark matter, single MAGs were selected from each order or even higher taxonomic levels based on their genome completeness and relative abundance, which finally generated 30 out of the 69 uncultivated MAGs as representative microbial dark matter (Fig. 4; Table S6). Genome-predicted metabolic potential suggested that 24 of 30 MAGs were involved in hydrolysis and fermentation of cellular macromolecules in WAS. The abundances of these hydrolyzing and fermentative members of the microbial dark matter were generally <2.1 RPKM, with an exception of the abundant bin 17 and bin 106 populations in thermal-AD. Notably, several populations of candidate phyla with small genomes (<1 Mb), e.g., “Candidatus Stahlbacteria” (TM7), “Candidatus Shapirobacteria,” and “Candidatus Dojkabacteria,” were identified in the sludge microbiomes without functions directly linked to the major methanogenic digestion procedures (Fig. 4).
FIG 4.
Phylogeny, genome completeness, genome size, and abundance of 30 MAGs that were the representative microbial dark matter in the four digestion sludge microbiome. The phylogenetic tree was constructed based on an alignment of concatenated ribosomal protein sequences from the MAGs and their related lineages. Phyla are distinguished by alternating background colors. The genome completeness of the MAGs is shown as pie charts. The right-hand columns indicated the presence (circles) and absence (blank) of MAG functions (i.e., hydrolysis, fermentation, syntrophic acetogenesis, and methanogenesis) in sludge digestion.
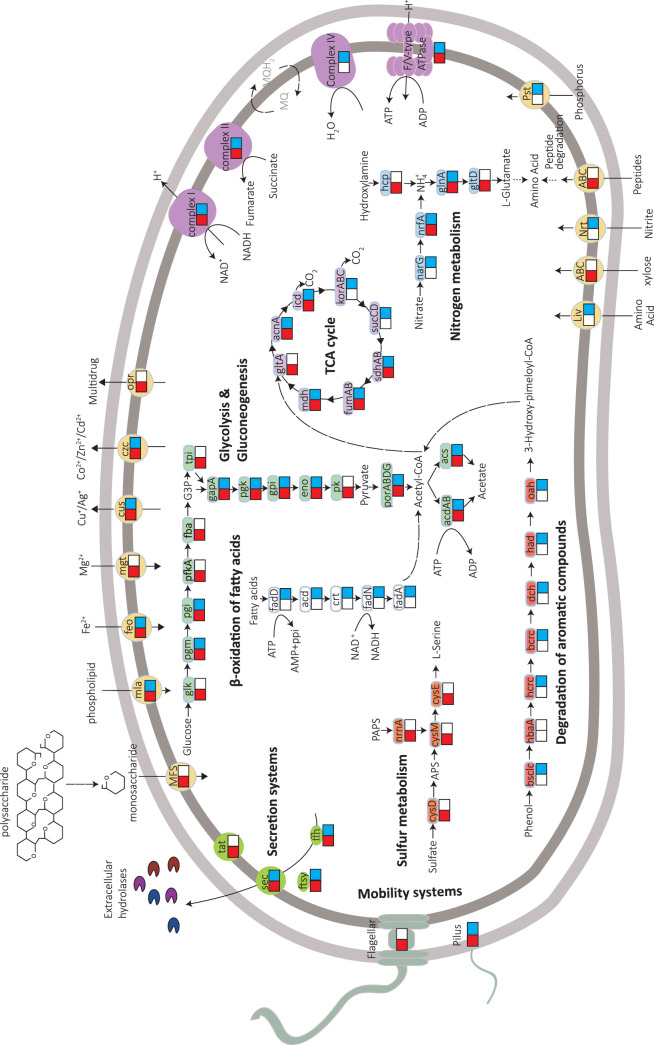
Of the 30 MAGs, the two most abundant populations (i.e., bin 17 and bin 106 with 89.2% and 76.6% genome completeness, respectively) belonging to the phylum Planctomycetes were chosen for subsequent metabolic reconstruction (Fig. 5; Table S7). Both genomes encoded enzymes of glycosylic hydrolase (GH) families for hydrolysis of cellulose, hemicellulose, and cellobiose, which could convert sludge polysaccharides into simple sugars for their subsequent fermentation and acetogenesis. In contrast to the nearly complete tricarboxylic acid (TCA) cycle in the bin 106 genome, genes encoding 2-oxoglutarate/2-oxoacid ferredoxin oxidoreductase and succinyl coenzyme A (succinyl-CoA) synthetase were absent in the bin 17 genome. Consequently, the partial TCA cycle might provide biosynthetic precursors only for anabolism of the bin 17 population. Notably, in addition to the glycolysis pathway, genes in the bin 106 genome also encoded enzymes for β-oxidation of fatty acids and phenol degradation, as well as a quinone-dependent electron transport chain containing a cbb3-type cytochrome-c oxidase with high O2 affinity. These potential metabolic traits implied a versatile lifestyle of the bin 106 population and its aerobic respiration capability under microaerobic conditions. In addition, the cell mobility of bin 17 and bin 106 populations might be different based on their gene contents. For example, genes encoding proteins for basal body, hook, and filament assembly, as well as type IV pilus, were detected in the bin 17 genome, suggesting its high mobility and capability to move toward a favorable growth environment. In contrast, only pilus-encoding genes were detected in the bin 106 genome, implying limited motility of the bin 106 population.
FIG 5.
Reconstructed metabolic capability of bin 17 and bin 106 populations. Pathways with genes detected in bin 17 and bin 106 genomes are in red and blue, respectively. Detailed information on genes assigned to specific metabolic pathways is available in Table S7.
Genomic traits of digestion sludge microbiomes.
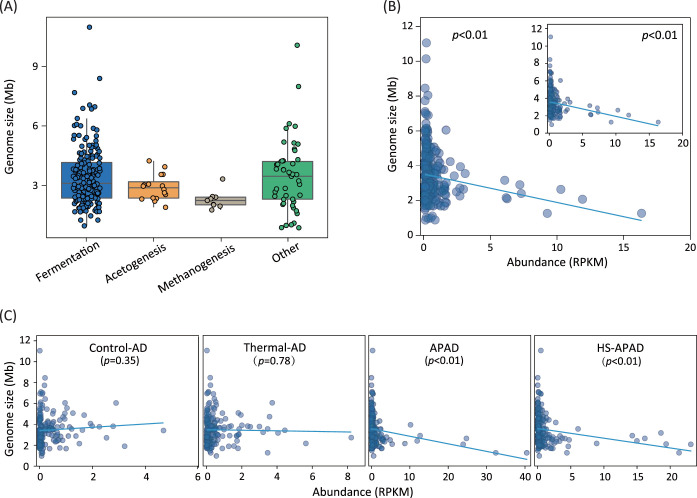
Genomic traits (e.g., genome size, coding sequence [CDS], and GC content) of microorganisms can be defining features of microbial cell growth and consequently associated with population abundance in a complex microbiome. In the four sludge digesters, the average genome size and number of CDSs of functional microorganisms (i.e., fermentative bacteria, acetogenic syntrophs, and methanogens) decreased with the successive methanogenic digestion procedures (Fig. 6A; Fig. S3), consistent with the Gibbs free energy changes of each redox reactions. Particularly, the genome size was shown to have a significant (P < 0.01) correlation with the abundance of overall digestion sludge microbiome (Fig. 6B). Of the three major functional groups of microorganisms, acetogenic syntrophs and methanogens mediated the rate-limiting steps (i.e., acetogenesis and methanogenesis), and their growth was substrate replete compared to that of fermentative bacteria. Accordingly, genome size could best predict the cell growth rate of the substrate-competitive fermentative bacteria (P < 0.01) (Fig. 6B; Fig. S4A and B). Interestingly, correlations between genome sizes of fermentative bacteria and their abundance were significant (P < 0.01) in APAD and HS-APAD, which have high carbon removal efficiencies (>50%), in contrast to their low correlations in control-AD (P = 0.35) and thermal-AD (P = 0.78; Fig. 6C). The low correlations in control-AD and thermal-AD might be due to their low efficiencies (30 to 40%) in destruction and removal of sludge flocs and associated microbial cells (2). The low efficiencies further resulted in high retention of cellular macromolecules (e.g., genomic DNA) of influent sludge cells (Fig. 6A; Fig. S4C) and consequent complication of the digestion sludge microbiome.
FIG 6.
Genome size distribution and relationship between genome size and abundance of MAGs. (A) Genome sizes of functional microbial groups in the coassembly of the four digestion sludge metagenomes. (B) Relationship between genome sizes and abundance of all retrieved MAGs; the inset shows the relationship between genome sizes and abundance of fermentative bacterial MAGs. The abundance is the average for the MAGs in the four digestion sludge metagenomes. (C) Relationships between genome sizes and abundance of all retrieved MAGs in the four sludge digesters.
DISCUSSION
This study provided genome-centric metagenomic insights into how WAS pretreatments changed the digestion sludge microbiome and subsequent digestion performance, as well as into the associated microbial dark matter. The highly complex digestion sludge microbiome guaranteed efficient conversion of organic polymers into biogas, and its performance could be changed by a variety of parameters, particularly influent-sludge composition and operational conditions (e.g., pH, temperature, and salinity) (27, 28). Nonetheless, impacts of these parameters on digestion sludge microbiomes were interconnected, and it was consequently challenging to identify the exact impacts of a specific parameter on the sludge microbiome and to differentiate the core functional populations from functionally redundant and to-be-digested microorganisms derived from influent WAS.
In view of the microbial community composition and function, prokaryotic populations in digestion sludge microbiomes could be classified into three major groups: group I consisted of core functional populations which were abundant and played essential roles in sludge digestion (e.g., bin 166, bin 252, and bin 58 as the predominant hydrolyzing and fermentative bacteria, bin 77 as syntrophic acetogens, and bin 51 as methanogenic archaeal in APAD and HS-APAD); group II included redundant functional populations which had functions parallel to those of the core functional populations but were minor and nonessential populations; and group III contained nonfunctional populations which entered digesters as substrates to be digested (e.g., aerobic bacteria derived from influent WAS). Recently, a study investigating the digestion efficiency in full-scale anaerobic sludge digesters estimated that 82% of microbial populations in feeding WAS were group III nonfunctional populations and could be digested (10). Most of the (group III) nonfunctional populations in feeding sludge were facultative aerobes or anaerobes, whose functions could barely be inferred from their 16S rRNA gene-based information and which could easily be confused with functional populations (group I and group II microorganisms) in digestion sludge microbiomes.
In this study, the composition and function of the three groups of microorganisms, particularly group I and group II populations, were clearly differentiated by employing genome-centric metagenomics and metabolic network reconstruction. Accordingly, several interesting observations were obtained: (i) WAS pretreatments changed group I assemblies (core functional populations) by releasing different digestion substrates from WAS, and (ii) group II microorganisms with high levels of functional redundancy provided a candidate pool for flexible and robust sludge digestion, and the group II minor and nonessential functional populations could be converted into group I predominant and functionally essential microorganisms when operational conditions were changed. These results might guide future optimization of sludge digestion by providing knowledge on the specific impact of WAS pretreatment on digestion sludge microbiomes.
The methanogenic sludge digesters with prolific nutrients, optimum temperature, and long retention time provided excellent habitats for an extremely wide range of anaerobic microorganisms, supporting diverse and complicated microbiomes for efficient sludge digestion. Of the digestion sludge microbiome, many populations have yet to be cultivated, due to their inability to grow in standard culture media (29). Recent advances in single-cell genomic and metagenomic techniques enabled researchers to bypass the complicated microbial cultivation and facilitated the discovery of numerous previously unknown, deep branches of the tree of life without cultivated representatives (30, 31). For example, a complete genome sequence of “Ca. Cloacamonas acidaminovorans” belonging to “Ca. Cloacimonetes” (formerly known as WWE1) was retrieved, which suggested that “Ca. Cloacamonas acidaminovorans” could derive most of its carbon and energy from the fermentation of amino acids (32).
In our study, several novel bacterial lineages were identified in the sludge microbiomes, including group I core functional populations (e.g., bin 17 and bin 106), group II functionally redundant populations (e.g., bin 160 and bin 214), and functionally unknown microorganisms (e.g., bin 180 and bin 224). All group I and group II functional microorganisms of novel lineages were identified to be fermentative bacteria, which suggested the high functional redundancy supported by the thermodynamically favorable fermentation, in contrast to the subsequent rate-limiting steps mediated by both acetogenic syntrophs and methanogens. Interestingly, energy metabolism of the functionally unknown lineages is not directly involved in the sludge digestion procedures. Consequently, these microorganisms might be second-hand metabolizers and their survival might depend on secreta of the group I and group II populations. Nonetheless, their detailed functional information awaited future in-depth analyses.
Conclusions.
This study provided unprecedented genome-centric metagenomic insights into how WAS pretreatments change digestion sludge microbiomes, as well as their metabolic networks. Results suggested that (i) WAS pretreatments govern the core functional population assemblies by changing digestion substrates, while redundant functional populations provide a candidate pool for flexible and robust sludge digestion; (ii) the genome-predicted metabolic potentials of microbial dark matter in digestion sludge microbiomes suggest their predominant roles in fermentation; and (iii) the genome sizes of the fermentative bacteria and their abundance are significantly correlated. These results may help guide future optimization of methanogenic sludge digestion and resource recovery.
MATERIALS AND METHODS
Characterization of organic matter in influent sludge.
Four mesophilic anaerobic sludge digesters were set up to treat fresh WAS (control-AD), thermally pretreated sludge (thermal-AD), and sludge pretreated with 0.25 mol/liter alkaline/acid (APAD) and 0.8 mol/liter alkaline/acid (HS-APAD), as described previously (2, 8). Influent-sludge samples were centrifuged and filtered through 0.45-μm filters to collect soluble fractions for measurement of dissolved organic matter. Concentrations of carbohydrates and proteins were quantified with the anthrone method and modified Lowry method, respectively, as described previously (33). DNA concentration was measured by using the diphenylamine colorimetric method (34). The total lipids were extracted from sludge with chloroform-methanol (2:1 [vol/vol]) (35), and the obtained mixtures were centrifuged. Then the lipid-containing organic phase was transferred to clean tubes. Total lipid content was determined by evaporating the organic solvents under nitrogen gas flushing, drying the mixture in the oven at 45°C for 15 min, and weighing (36).
Sample collection, DNA extraction, and metagenome sequencing.
Sludge samples for metagenomic sequencing were collected from steady-state digesters (sampling time points were set on day 192 for APAD, thermal-AD, and control-AD and on day 142 for HS-APAD), and their genomic DNA (gDNA) was extracted using a FastDNA spin kit for soil (MP Biomedicals, Carlsbad, CA, USA) (37). The quality and quantity of the DNA extract were evaluated with gel electrophoresis and a Quantus fluorometer (Promega, Madison, WI, USA). The DNA sequencing libraries and subsequent Illumina HiSeq sequencing services were provided by BGI (Shenzhen, China).
Metagenome assembly and genome binning.
All metagenomic sequencing raw data were filtered to remove low-quality bases/reads using Sickle (38), with the parameters set to “-q = 20” and “-l = 100.” Then the four metagenomes were combined and de novo coassembled using SPAdes (version 3.12.0) (39) with the following parameters: “-k 33,55,77” and “-meta.” To generate high-quality metagenome-assembled genomes (MAGs), contigs with lengths of <1,000 bp in the assembly were removed. Three binning methods, i.e., MetaBAT (40), Maxbin (41), and Concoct (42), were employed and compared for the binning of the contigs into population genomes as described previously (43). To improve binning quality, RefineM (44) was used to filter contigs with divergent genomic properties, incongruent taxonomic classification, and 16S rRNA genes. Manual curation of these population genomes was executed using single-copy genes, k-mer frequency distribution, contig coverage, and GC content. To obtain optimal genome quality, clean reads for each bin were recruited using BBMap (45) with following parameters: k = 15, minid = 0.9, build = 1. Then, genome bins were reassembled by SPAdes (version 3.12.0) (39) with the following parameters: --careful -k 21,33,55,77. The completeness, contamination, and strain heterogeneity of each bin were evaluated using CheckM (46). Finally, 254 bins with >70% completeness and <5% contamination were obtained for subsequent analyses.
Genome annotation and metabolic reconstruction.
Protein coding sequences (CDSs) were determined using prokka (47) with the “--quiet” option for all genome bins. Functional annotations were conducted based on comparisons with the KEGG (48), and eggnog (49) databases using DIAMOND (50) with an E value threshold of “<1e-5.” Carbohydrate-active enzymes were identified using the Carbohydrate-Active Enzymes (CAZy) database (51). A subset of CAZy genes involving in extracellular polysaccharide degradation pathways (52) was selected for further analysis. Peptidases were identified using MEROPS (53). To reconstruct the metabolic pathways, genome bins were uploaded to RAST (Rapid Annotation using Subsystem Technology) (54) for gene prediction and annotation, and their CDSs were annotated at the KEGG automatic annotation server (KAAS) (55). The metabolic potential (i.e., hydrolysis, fermentation, and syntrophic acetogenesis) of major populations of known lineages were further manually confirmed.
Genome tree phylogeny.
A total of 16 universally present and rarely horizontally transferred ribosomal proteins (i.e., L2, L3, L4, L5, L6, L14, L15, L16, L18, L22, L24, S3, S8, S10, S17, and S19) were selected to reconstruct the phylogenetic tree (56). Protein sequences were extracted separately using AMPHORA2 (57). Multiple-sequence alignments (MSAs) of the individual protein sequence were built using Mafft (58) (iterate 100 times). Poorly aligned regions were filtered by TrimAL (59) to remove columns containing >95% gap positions, and then the 16 filtered ribosomal protein sequences were concatenated for each genome. The phylogenomic tree was constructed using IQ-TREE (version 1.6.10) (60) with the following parameters: -mset JTT -mrate I,G,I+G. The resulting Newick tree file was uploaded to iTOL v4 (61) for visualization and formatting. To confirm their phylogeny, MAGs were further classified with GTDBtk (version 1.0.2, database release 89) (62).
Relative abundance and community composition.
Taxonomic classification of contigs was performed using Kraken 2 as described previously (63) with a database comprising 38,156 bacterial, 11,953 viral, and 535 archaeal complete reference genomes. The sequencing reads were then realigned back to the contigs using BBMap (45) to calculate their coverages. Taxon read hit counts were obtained by combining the realigned reads of all contigs belonging to the taxon, which were further normalized by genome size to calculate taxon abundance. For the relative abundance and diversity analysis, genomes with coverage less than 1 were removed to decrease the effect of low-abundance misclassification. The abundance of MAGs was calculated based on retrieved genome coverage and normalized as reads per kilobase per million (RPKM) as described previously (64, 65).
Data availability.
Raw Illumina HiSeq sequencing reads were deposited into European Nucleotide Archive (ENA) database with accession numbers ERR3454363 to ERR3454366.
Supplementary Material
ACKNOWLEDGMENTS
This study was financially supported by the National Natural Science Foundation of China (41671310) and Natural Science Foundation of Guangdong Province (2018B030314012).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Ju F, Li B, Ma L, Wang Y, Huang D, Zhang T. 2016. Antibiotic resistance genes and human bacterial pathogens: co-occurrence, removal, and enrichment in municipal sewage sludge digesters. Water Res 91:1–10. doi: 10.1016/j.watres.2015.11.071. [DOI] [PubMed] [Google Scholar]
- 2.Wang S, Yu S, Lu Q, Liao Y, Li H, Sun L, Wang H, Zhang Y. 2020. Development of an alkaline/acid pre-treatment and anaerobic digestion (APAD) process for methane generation from waste activated sludge. Sci Total Environ 708:134564. doi: 10.1016/j.scitotenv.2019.134564. [DOI] [PubMed] [Google Scholar]
- 3.Mao C, Feng Y, Wang X, Ren G. 2015. Review on research achievements of biogas from anaerobic digestion. Renew Sust Energ Rev 45:540–555. doi: 10.1016/j.rser.2015.02.032. [DOI] [Google Scholar]
- 4.Carrere H, Antonopoulou G, Affes R, Passos F, Battimelli A, Lyberatos G, Ferrer I. 2016. Review of feedstock pretreatment strategies for improved anaerobic digestion: from lab-scale research to full-scale application. Bioresour Technol 199:386–397. doi: 10.1016/j.biortech.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 5.Wang X, Duan X, Chen J, Fang K, Feng L, Yan Y, Zhou Q. 2016. Enhancing anaerobic digestion of waste activated sludge by pretreatment: effect of volatile to total solids. Environ Technol 37:1520–1529. doi: 10.1080/09593330.2015.1120783. [DOI] [PubMed] [Google Scholar]
- 6.Li X, Zhao J, Wang D, Yang Q, Xu Q, Deng Y, Yang W, Zeng G. 2016. An efficient and green pretreatment to stimulate short-chain fatty acids production from waste activated sludge anaerobic fermentation using free nitrous acid. Chemosphere 144:160–167. doi: 10.1016/j.chemosphere.2015.08.076. [DOI] [PubMed] [Google Scholar]
- 7.Gonzalez A, Hendriks ATWM, van Lier JB, de Kreuk M. 2018. Pre-treatments to enhance the biodegradability of waste activated sludge: elucidating the rate limiting step. Biotechnol Adv 36:1434–1469. doi: 10.1016/j.biotechadv.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 8.Lu Q, Yu Z, Yu S, Liang Z, Li H, Sun L, Wang S. 2019. Organic matter rather than salinity as a predominant feature changes performance and microbiome in methanogenic sludge digesters. J Hazard Mater 377:349–356. doi: 10.1016/j.jhazmat.2019.05.075. [DOI] [PubMed] [Google Scholar]
- 9.Guo J, Peng Y, Ni BJ, Han X, Fan L, Yuan Z. 2015. Dissecting microbial community structure and methane-producing pathways of a full-scale anaerobic reactor digesting activated sludge from wastewater treatment by metagenomic sequencing. Microb Cell Fact 14:33. doi: 10.1186/s12934-015-0218-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mei R, Narihiro T, Nobu MK, Kuroda K, Liu WT. 2016. Evaluating digestion efficiency in full-scale anaerobic digesters by identifying active microbial populations through the lens of microbial activity. Sci Rep 6:34090. doi: 10.1038/srep34090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mei R, Nobu MK, Narihiro T, Kuroda K, Sierra JM, Wu Z, Ye L, Lee PKH, Lee PH, van Lier JB, McInerney MJ, Kamagata Y, Liu WT. 2017. Operation-driven heterogeneity and overlooked feed-associated populations in global anaerobic digester microbiome. Water Res 124:77–84. doi: 10.1016/j.watres.2017.07.050. [DOI] [PubMed] [Google Scholar]
- 12.Campanaro S, Treu L, Rodriguezr LM, Kovalovszki A, Ziels RM, Maus I, Zhu X, Kougias PG, Basile A, Luo G, Schlüter A, Konstantinidis KT, Angelidaki I. 2019. The anaerobic digestion microbiome: a collection of 1600 metagenome-assembled genomes shows high species diversity related to methane production. bioRxiv doi: 10.1101/680553. [DOI]
- 13.Yang Y, Yu K, Xia Y, Lau FT, Tang DT, Fung WC, Fang HH, Zhang T. 2014. Metagenomic analysis of sludge from full-scale anaerobic digesters operated in municipal wastewater treatment plants. Appl Microbiol Biotechnol 98:5709–5718. doi: 10.1007/s00253-014-5648-0. [DOI] [PubMed] [Google Scholar]
- 14.Abendroth C, Vilanova C, Gunther T, Luschnig O, Porcar M. 2015. Eubacteria and archaea communities in seven mesophile anaerobic digester plants in Germany. Biotechnol Biofuels 8:87. doi: 10.1186/s13068-015-0271-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Westerholm M, Crauwels S, Van Geel M, Dewil R, Lievens B, Appels L. 2016. Microwave and ultrasound pre-treatments influence microbial community structure and digester performance in anaerobic digestion of waste activated sludge. Appl Microbiol Biotechnol 100:5339–5352. doi: 10.1007/s00253-016-7321-2. [DOI] [PubMed] [Google Scholar]
- 16.Vanwonterghem I, Jensen PD, Rabaey K, Tyson GW. 2016. Genome-centric resolution of microbial diversity, metabolism and interactions in anaerobic digestion. Environ Microbiol 18:3144–3158. doi: 10.1111/1462-2920.13382. [DOI] [PubMed] [Google Scholar]
- 17.Pervin HM, Dennis PG, Lim HJ, Tyson GW, Batstone DJ, Bond PL. 2013. Drivers of microbial community composition in mesophilic and thermophilic temperature-phased anaerobic digestion pre-treatment reactors. Water Res 47:7098–7108. doi: 10.1016/j.watres.2013.07.053. [DOI] [PubMed] [Google Scholar]
- 18.Knight R, Vrbanac A, Taylor BC, Aksenov A, Callewaert C, Debelius J, Gonzalez A, Kosciolek T, McCall LI, McDonald D, Melnik AV, Morton JT, Navas J, Quinn RA, Sanders JG, Swafford AD, Thompson LR, Tripathi A, Xu ZZ, Zaneveld JR, Zhu Q, Caporaso JG, Dorrestein PC. 2018. Best practices for analysing microbiomes. Nat Rev Microbiol 16:410–422. doi: 10.1038/s41579-018-0029-9. [DOI] [PubMed] [Google Scholar]
- 19.Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, Solovyev VV, Rubin EM, Rokhsar DS, Banfield JF. 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- 20.Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers YH, Smith HO. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 21.Turaev D, Rattei T. 2016. High definition for systems biology of microbial communities: metagenomics gets genome-centric and strain-resolved. Curr Opin Biotechnol 39:174–181. doi: 10.1016/j.copbio.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 22.Edwards RA, McNair K, Faust K, Raes J, Dutilh BE. 2016. Computational approaches to predict bacteriophage-host relationships. FEMS Microbiol Rev 40:258–272. doi: 10.1093/femsre/fuv048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blauwkamp TA, Thair S, Rosen MJ, Blair L, Lindner MS, Vilfan ID, Kawli T, Christians FC, Venkatasubrahmanyam S, Wall GD, Cheung A, Rogers ZN, Meshulam-Simon G, Huijse L, Balakrishnan S, Quinn JV, Hollemon D, Hong DK, Vaughn ML, Kertesz M, Bercovici S, Wilber JC, Yang S. 2019. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat Microbiol 4:663–674. doi: 10.1038/s41564-018-0349-6. [DOI] [PubMed] [Google Scholar]
- 24.Mocé-Llivina L, Muniesa M, Pimenta-Vale H, Lucena F, Jofre J. 2003. Survival of bacterial indicator species and bacteriophages after thermal treatment of sludge and sewage. Appl Environ Microbiol 69:1452–1456. doi: 10.1128/aem.69.3.1452-1456.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carballa M, Regueiro L, Lema JM. 2015. Microbial management of anaerobic digestion: exploiting the microbiome-functionality nexus. Curr Opin Biotechnol 33:103–111. doi: 10.1016/j.copbio.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 26.De Vrieze J, Christiaens ME, Walraedt D, Devooght A, Ijaz UZ, Boon N. 2017. Microbial community redundancy in anaerobic digestion drives process recovery after salinity exposure. Water Res 111:109–117. doi: 10.1016/j.watres.2016.12.042. [DOI] [PubMed] [Google Scholar]
- 27.Lefebvre O, Quentin S, Torrijos M, Godon JJ, Delgenes JP, Moletta R. 2007. Impact of increasing NaCl concentrations on the performance and community composition of two anaerobic reactors. Appl Microbiol Biotechnol 75:61–69. doi: 10.1007/s00253-006-0799-2. [DOI] [PubMed] [Google Scholar]
- 28.Shin J, Jang HM, Shin SG, Kim YM. 2019. Thermophilic anaerobic digestion: effect of start-up strategies on performance and microbial community. Sci Total Environ 687:87–95. doi: 10.1016/j.scitotenv.2019.05.428. [DOI] [PubMed] [Google Scholar]
- 29.Narihiro T, Nobu MK, Kim NK, Kamagata Y, Liu WT. 2015. The nexus of syntrophy-associated microbiota in anaerobic digestion revealed by long-term enrichment and community survey. Environ Microbiol 17:1707–1720. doi: 10.1111/1462-2920.12616. [DOI] [PubMed] [Google Scholar]
- 30.Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, Hedlund BP, Tsiamis G, Sievert SM, Liu WT, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM, Hugenholtz P, Woyke T. 2013. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 31.Doud DFR, Woyke T. 2017. Novel approaches in function-driven single-cell genomics. FEMS Microbiol Rev 41:538–548. doi: 10.1093/femsre/fux009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pelletier E, Kreimeyer A, Bocs S, Rouy Z, Gyapay G, Chouari R, Rivière D, Ganesan A, Daegelen P, Sghir A, Cohen GN, Médigue C, Weissenbach J, Le Paslier D. 2008. “Candidatus Cloacamonas acidaminovorans”: genome sequence reconstruction provides a first glimpse of a new bacterial division. J Bacteriol 190:2572–2579. doi: 10.1128/JB.01248-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frølund B, Palmgren R, Keiding K, Nielsen PH. 1996. Extraction of extracellular polymers from activated sludge using a cation exchange resin. Water Res 30:1749–1758. doi: 10.1016/0043-1354(95)00323-1. [DOI] [Google Scholar]
- 34.Sun Y, Clinkenbeard KD, Clarke C, Cudd L, Highlander SK, Dabo SM. 1999. Pasteurella haemolytica leukotoxin induced apoptosis of bovine lymphocytes involves DNA fragmentation. Vet Microbiol 65:153–166. doi: 10.1016/S0378-1135(98)00286-7. [DOI] [PubMed] [Google Scholar]
- 35.Folch J, Lees M, Stanley GS. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 226:497–509. [PubMed] [Google Scholar]
- 36.Adav SS, Lee DJ. 2008. Extraction of extracellular polymeric substances from aerobic granule with compact interior structure. J Hazard Mater 154:1120–1126. doi: 10.1016/j.jhazmat.2007.11.058. [DOI] [PubMed] [Google Scholar]
- 37.Xu G, Lu Q, Yu L, Wang S. 2019. Tetrachloroethene primes reductive dechlorination of polychlorinated biphenyls in a river sediment microcosm. Water Res 152:87–95. doi: 10.1016/j.watres.2018.12.061. [DOI] [PubMed] [Google Scholar]
- 38.Joshi NA, Fass JN. 2011. Sickle: a sliding-window, adaptive, quality-based trimming tool for FastQ files (version 1.33). https://github.com/najoshi/sickle.
- 39.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kang DD, Froula J, Egan R, Wang Z. 2015. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 3:e1165. doi: 10.7717/peerj.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu YW, Simmons BA, Singer SW. 2016. MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32:605–607. doi: 10.1093/bioinformatics/btv638. [DOI] [PubMed] [Google Scholar]
- 42.Alneberg J, Bjarnason BS, de Bruijn I, Schirmer M, Quick J, Ijaz UZ, Lahti L, Loman NJ, Andersson AF, Quince C. 2014. Binning metagenomic contigs by coverage and composition. Nat Methods 11:1144–1146. doi: 10.1038/nmeth.3103. [DOI] [PubMed] [Google Scholar]
- 43.Uritskiy GV, DiRuggiero J, Taylor J. 2018. MetaWRAP-a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome 6:158. doi: 10.1186/s40168-018-0541-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parks DH, Rinke C, Chuvochina M, Chaumeil PA, Woodcroft BJ, Evans PN, Hugenholtz P, Tyson GW. 2017. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat Microbiol 2:1533–1542. doi: 10.1038/s41564-017-0083-5. [DOI] [PubMed] [Google Scholar]
- 45.Bushnell B. 2014. BBMap: a fast, accurate, splice-aware aligner. Lawrence Berkeley National Laboratory, Berkeley, CA. [Google Scholar]
- 46.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 48.Kanehisa M, Goto S. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huerta-Cepas J, Szklarczyk D, Heller D, Hernandez-Plaza A, Forslund SK, Cook H, Mende DR, Letunic I, Rattei T, Jensen LJ, von Mering C, Bork P. 2019. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res 47:D309–D314. doi: 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 51.Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z, Busk PK, Xu Y, Yin Y. 2018. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 46:95–101. doi: 10.1093/nar/gky418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berlemont R, Martiny AC. 2013. Phylogenetic distribution of potential cellulases in bacteria. Appl Environ Microbiol 79:1545–1554. doi: 10.1128/AEM.03305-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rawlings ND, Barrett AJ, Finn R. 2016. Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res 44:D343–D350. doi: 10.1093/nar/gkv1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. 2007. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35:182–185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, Castelle CJ, Butterfield CN, Hernsdorf AW, Amano Y, Ise K, Suzuki Y, Dudek N, Relman DA, Finstad KM, Amundson R, Thomas BC, Banfield JF. 2016. A new view of the tree of life. Nat Microbiol 1:16048. doi: 10.1038/nmicrobiol.2016.48. [DOI] [PubMed] [Google Scholar]
- 57.Wu M, Scott AJ. 2012. Phylogenomic analysis of bacterial and archaeal sequences with AMPHORA2. Bioinformatics 28:1033–1034. doi: 10.1093/bioinformatics/bts079. [DOI] [PubMed] [Google Scholar]
- 58.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Letunic I, Bork P. 2019. Interactive Tree of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:256–259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil PA, Hugenholtz P. 2018. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol 36:996–1004. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 63.Wood DE, Lu J, Langmead B. 2019. Improved metagenomic analysis with Kraken2. Genome Biol 20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bischoff V, Bunk B, Meier-Kolthoff JP, Spröer C, Poehlein A, Dogs M, Nguyen M, Petersen J, Daniel R, Overmann J, Göker M, Simon M, Brinkhoff T, Moraru C. 2019. Cobaviruses—a new globally distributed phage group infecting Rhodobacteraceae in marine ecosystems. ISME J 13:1404–1421. doi: 10.1038/s41396-019-0362-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kruger K, Chafee M, Francis TB, Rio TG, Becher D, Schweder T, Amann RI, Teeling H. 2019. In marine Bacteroidetes the bulk of glycan degradation during algae blooms is mediated by few clades using a restricted set of genes. ISME J 13:2800–2816. doi: 10.1038/s41396-019-0476-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw Illumina HiSeq sequencing reads were deposited into European Nucleotide Archive (ENA) database with accession numbers ERR3454363 to ERR3454366.