Diarrhea in the postweaning period due to enterotoxigenic E. coli (ETEC) is an economically relevant disease in pig production worldwide. In Denmark, prevention is mainly achieved by zinc oxide administration (to be discontinued by 2022). In addition, a breeding program has been implemented that aims to reduce the prevalence of this illness. Treatment with antimicrobials contributes to the problem of antimicrobial resistance (AMR) development. As a novelty, this study aims to deeply understand the genetic population structure and variation among diarrhea-associated isolates by whole-genome sequencing characterization. ST100-F4ac is the dominant clonal group circulating in Danish herds and showed high similarity to ETEC ST100 isolates from China, the United States, and Spain. High rates of AMR and high diversity of virulence genes were detected. The characterization of diarrhea-related ETEC is important for understanding the disease epidemiology and pathogenesis and for implementation of new strategies aiming to reduce the impact of the disease in pig production.
KEYWORDS: pigs, diarrhea, enterotoxigenic E. coli, whole-genome sequencing, bioinformatics, enterotoxigenic, Escherichia coli
ABSTRACT
This study aimed to characterize in silico enterotoxigenic Escherichia coli F4- and F18-positive isolates (n = 90) causing swine postweaning diarrhea, including pathogenic potential, phylogenetic relationship, antimicrobial and biocide resistance, prophage content, and metal tolerance rates. F4 strains belonged mostly to the O149 and O6 serogroups and ST100 and ST48 sequence types (STs). F18 strains were mainly assigned to the O8 and O147 serogroups and ST10, ST23, and ST42. The highest rates of antimicrobial resistance were found against streptomycin, sulfamethoxazole, tetracycline, trimethoprim, and ampicillin. No resistance was found toward ciprofloxacin, cefotaxime, ceftiofur, and colistin. Genes conferring tolerance to copper (showing the highest diversity), cadmium, silver, and zinc were predicted in all genomes. Enterotoxin genes (ltcA, 100% F4, 62% F18; astA, 100% F4, 38.1% F18; sta, 18.8% F4, 38.1% F18; stb, 100% F4, 76.2% F18) and fimbria-encoding genes typed as F4ac and F18ac were detected in all strains, in addition to up to 16 other virulence genes in individual strains. Phage analysis predicted between 7 and 20 different prophage regions in each strain. A highly diverse variety of plasmids was found; IncFII, IncFIB, and IncFIC were prevalent among F4 isolates, while IncI1 and IncX1 were dominant among F18 strains. Interestingly, F4 isolates from the early 1990s belonged to the same clonal group detected for most of the F4 strains from 2018 to 2019 (ONT:H10-A-ST100-CH27-0). The small number of single-nucleotide polymorphism differences between the oldest and recent F4 ST100 isolates suggests a relatively stable genome. Overall, the isolates analyzed in this study showed remarkably different genetic traits depending on the fimbria type.
IMPORTANCE Diarrhea in the postweaning period due to enterotoxigenic E. coli (ETEC) is an economically relevant disease in pig production worldwide. In Denmark, prevention is mainly achieved by zinc oxide administration (to be discontinued by 2022). In addition, a breeding program has been implemented that aims to reduce the prevalence of this illness. Treatment with antimicrobials contributes to the problem of antimicrobial resistance (AMR) development. As a novelty, this study aims to deeply understand the genetic population structure and variation among diarrhea-associated isolates by whole-genome sequencing characterization. ST100-F4ac is the dominant clonal group circulating in Danish herds and showed high similarity to ETEC ST100 isolates from China, the United States, and Spain. High rates of AMR and high diversity of virulence genes were detected. The characterization of diarrhea-related ETEC is important for understanding the disease epidemiology and pathogenesis and for implementation of new strategies aiming to reduce the impact of the disease in pig production.
INTRODUCTION
Postweaning diarrhea (PWD) affects pigs after weaning, leading to significant economic costs for the pig industry due to weight loss and mortality as well as the cost of prevention (i.e., vaccination), treatment, and handling (1, 2). In addition to sudden death or profuse diarrhea, the disease is accompanied by growth retardation in surviving piglets (1, 3). During acute outbreaks, mortality due to PWD may reach 20 to 30% over a 1 to 2 months’ time span among infected pigs (3). As it is one of the most common reasons for the use of antimicrobials in the pig industry worldwide, PWD significantly contributes to the problem of antimicrobial resistance (AMR) development (2, 4, 5).
Enterotoxigenic Escherichia coli (ETEC) is the main etiological agent involved in PWD worldwide. The ETEC pathotypes in pigs are characterized by the expression of specific fimbrial adhesins, which mediate bacterial colonization of the gut mucosal surface. The most commonly detected types of fimbriae are F4 (previously termed K88) and F18 (F107, 2134P, and 8813). Both include different antigenic variants, three for F4 (ab, ac, and ad), with F4ac being the most prevalent, and two types for F18 (ab and ac), with F18ac being the main one associated with PWD (6, 7). Intestinal adhesion and subsequent colonization by ETEC depends on F4- or F18-specific receptors, the presence of which is essential for ETEC to cause disease (8). ETEC F4 is usually related to PWD of recently weaned piglets occurring 2 to 3 days after weaning (classical PWD), while F18 is commonly found associated with diarrhea 2 to 6 weeks after weaning. The age-dependent expression of F4 and F18 receptors in the small intestine might explain why ETEC F4 infection mainly takes place right after weaning as well as during the neonatal period, while ETEC F18 infection mainly occurs later in the postweaning period (5).
Once ETEC bacteria have adhered and colonized the small intestine, they can produce an enterotoxin(s) leading to diarrhea. Both ETEC F4 and F18 are reported to encode two classes of enterotoxins, heat-labile (LT) and heat-stable (Sta, Stb, and EAST1, for enteroaggregative heat-stable toxin 1) enterotoxins, which induce secretory diarrhea in the pigs (1, 2, 9–11). The predominant serogroup of ETEC associated with classical PWD in pigs worldwide is O149-F4 (1).
In Denmark and other countries, zinc oxide (ZnO) in therapeutic concentrations has been used in recent decades to prevent PWD in the first 14 days after weaning. ZnO has been found to improve growth performance and reduce scours (ETEC induced) in weaning piglets (12). Moreover, ZnO reduces bacterial adhesion and inflammatory cytokine expression and prevents the disruption of membrane integrity caused by ETEC (13). However, due to the environmental toxicity and potential coselection for AMR, the use of ZnO will be banned in pig production in the EU by 2022 (https://www.ema.europa.eu/en/medicines/veterinary/referrals/zinc-oxide). Other strategies that have been used to tackle PWD caused by ETEC include a breeding program (DanBred), which has been implemented in some Danish farms since 2003, and vaccines. The former consists of breeding pigs that do not express the F4ac-specific receptors on the intestinal mucosa, aiming at reducing the occurrence of diarrhea due to ETEC F4 (5). The live attenuated vaccine Coliprotec F4/F18 (https://www.ema.europa.eu/en/medicines/veterinary/EPAR/coliprotec-f4f18) has been developed to diminish the incidence of PWD caused by both ETEC F4 and F18 bacteria. Besides these preventive strategies, neomycin, apramycin, spectinomycin, tetracycline, amoxicillin, and sulfadiazine-trimethoprim are the antimicrobials commonly used to treat PWD in Denmark (14).
In the present work, we characterized a collection of Danish ETEC F4- and F18-positive strains through whole-genome sequencing (WGS) to analyze the pathogenic potential of the strains through the identification of relevant virulence factors and to determine the occurrence of AMR as well as biocide and metal resistances. Further, the analysis allowed us to understand the population genetic structure and variation among strains that are associated with PWD. Moreover, we also analyzed the phylogenetic relationship between the Danish strains under study and swine ETEC strains from other countries of the world.
RESULTS
Phylogroups, sequence types, clonotypes, and serotypes.
Most of the ETEC isolates belonged to phylogroup A (74 isolates, 82.2%), and the remaining were assigned to four different phylogroups, B1 (one isolate, 1.1%), C (nine isolates, 10%), D (five isolates, 5.5%), and E (one isolate, 1.1%). The strains displayed eight different sequence types (STs) by multilocus sequence typing (MLST) (ST10, ST23, ST42, ST48, ST90, ST100, ST155, and ST3524), with ST100 accounting for 56 (62.2%) isolates. Based on fumC-fimH allele combinations, eight clonotypes (CHs) were identified, with CH27-0 being the most prevalent type (56 isolates, 62.2%), corresponding to strains assigned to ST100 (see Table S2 in the supplemental material).
The SerotypeFinder tool detected six O serogroups (O6, O8, O29, O141, O147, and O149) and eight different H antigens (H4, H10, H12, H14, H16, H17, H19, and H30). It was not able to predict the O and H antigen in 14 and 8 isolates, respectively. Overall, 13 O:H combinations (serotypes) were found (Table S2), with O149:H10 being the most common serotype identified (49 isolates, 54.4%), followed by O6:H16 (11 isolates, 12.2%).
The association between clonal groups (defined by serotype, phylogroup, sequence type, and clonotype) and fimbrial type detected among the strains is shown in Table 1. Two main clonal groups were identified among the 69 F4-positive isolates, O149/ONT:H10/HNT-A-ST100-CH27-0 and O6:H16-A-ST48-CH11-34, representing 81.2% (56 strains) and 15.9% (11 strains) of the isolates, respectively. The remaining F4-positive isolates belonged to the O8:H19-C-ST90-CH4-54 clonal group (two isolates, 2.9%). Interestingly, five out of the six F4-positive isolates isolated in the early 1990s also belonged to the ONT:H10-A-ST100-CH27-0 clonal group shown for most of the F4 isolates collected during the period of 2018 to 2019 (Table S2). In contrast to the homogeneity observed among F4 isolates, the 21 F18-positive isolates showed a higher diversity and were assigned to six different clonal groups, of which O141/ONT:H4-A-ST10-CH11-24 (33.3%; seven isolates) and O8:H17-C-ST23-CH4-54 (28.6%; six isolates) were the predominant ones. The remaining F18-positive isolates were assigned to the O147/ONT:H14-D-ST42-CH-28-65 (five isolates, 23.8%), O29:H12-B1-ST155-CH4-121, ONT:H19-C-ST90-CH4-0 (E. coli Nysø), and O8:H31-E-ST3524-CH23-31 (one isolate each, 4.7%) clonal groups. F18-positive strains from the same herd belonged to the same clonal group, including seven, six, and five isolates from herd D, herd B, and herd C, respectively (Table S2).
TABLE 1.
Serotype, phylogroup, sequence type, and clonotypes associated with fimbrial antigens in the 90 ETEC isolates from pigs
| Clonal group (serotype-PG-ST-CH)a | No. (%) of isolates | Fimbrial antigen |
|---|---|---|
| O149:H10-A-ST100-CH27-0 | 49 (54.4) | F4 |
| ONT:H10-A-ST100-CH27-0 | 6 (6.7) | F4 |
| O149:HNT-A-ST100-CH27-0 | 1 (1.1) | F4 |
| O6:H16-A-ST48-CH11-34 | 11 (12.2) | F4 |
| O8:H19-C-ST90-CH4-54 | 2 (2.2) | F4 |
| O8:H17-C-ST23-CH4-54 | 6 (6.7) | F18 |
| ONT:H4-A-ST10-CH11-24 | 5 (5.6) | F18 |
| O141:H4-A-ST10-CH11-24 | 2 (2.2) | F18 |
| O147:H14-D-ST42-CH28-65 | 3 (3.3) | F18 |
| ONT:H14-D-ST42-CH28-65 | 2 (2.2) | F18 |
| O29:H12-B1-ST155-CH4-121 | 1 (1.1) | F18 |
| ONT:H19-C-ST90-CH4-0 | 1 (1.1) | F18 |
| O8:H31-E-ST3524-CH23-31 | 1 (1.1) | F18 |
PG, phylogroup; ST, sequence type; CH, clonotype.
Phylogeny analysis of ETEC isolates.
The raw read mapping of all 90 genomes to the reference E. coli K-12 genome showed that 3,475,685 out of 4,641,652 (74.8%) nucleotide positions in the reference genome were present in all of the analyzed genomes. A total of 42,172 variable nucleotide positions were detected in this core genome (Table S3).
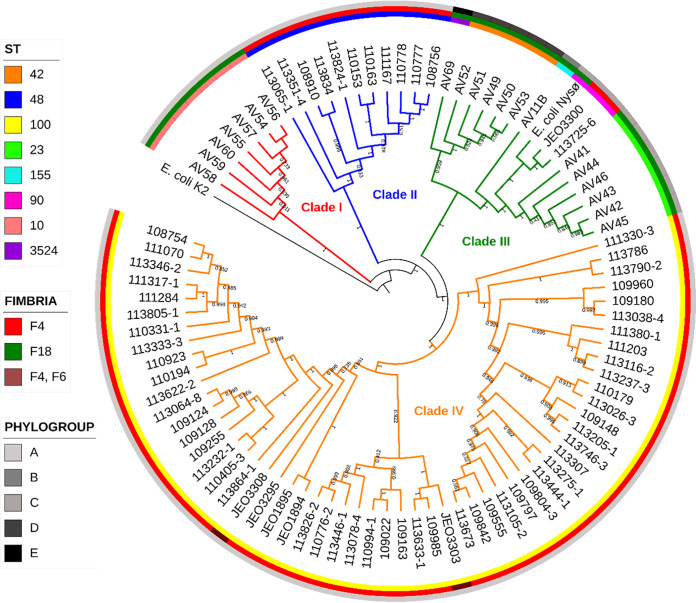
Isolates were clustered into four major clades (I, II, III, and IV) and grouped according to their ST (Fig. 1). Clade I included seven ST10-F18 isolates recovered from the same herd and showed 1 to 14 single-nucleotide polymorphism (SNP) differences. Isolates belonging to ST48 and encoding F4 fimbriae (11 isolates) were grouped in clade II and showed from 6 (between isolates from the same herd) to 633 SNP differences. Clade III encompassed isolates assigned to different STs (ST155, ST23, ST42, ST3524, and ST90), and all except two ST90 isolates were F18 positive. This cluster was divided into two well-defined subclades: subclade A (including five ST42 isolates from the same herd, with numbers of SNP differences ranging from 4 to 18, and one ST3524 isolate) and subclade B, containing six ST23 isolates (between 3 and 21 SNP differences), three ST90 (two of them were F4 positive) strains (between 104 and 556 SNP differences), and a single ST155 isolate. Lastly, clade IV consisted of F4-positive isolates belonging to ST100 (56 isolates) and was split into two subclades, with the number of different SNPs spanning from 16 to 866 among all genomes and from 4 to 15 among isolates belonging to the same herd. Numbers of SNP differences among the five F4-ST100 isolates from the 1990s and F4-ST100 strains recovered during 2018 to 2019 ranged from 130 to 699.
FIG 1.
SNP-based phylogeny of the 90 ETEC isolates from pigs. Colors in the outer ring correspond to phylogroups, the middle ring to fimbria type, and the inner ring to sequence type.
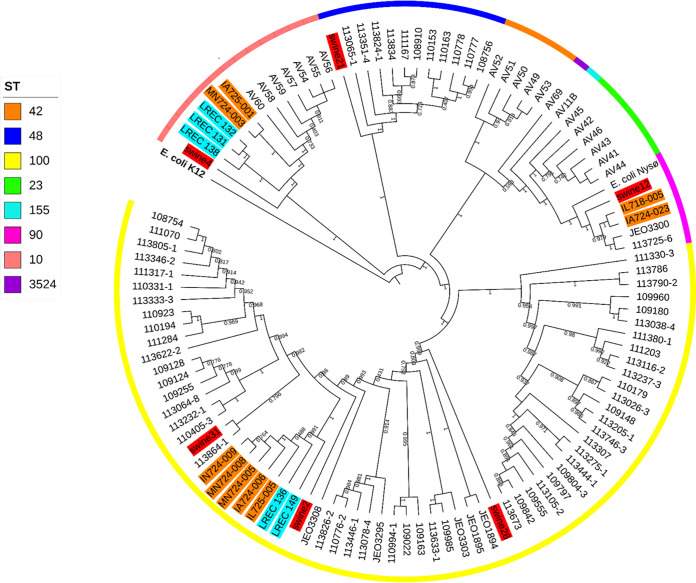
We investigated the relationship between our strains and porcine ETEC isolates from other countries (Fig. 2 and Table S11). Phylogenetic analysis based on SNP indicated that the Danish ST100 isolates are closely related to ST100 isolates from China (between 120 and 443 SNP differences), Spain (178 to 708), and the United States (196 to 466). The lowest numbers of SNP differences among ST10 isolates were detected versus ST10 strains from the United States (300 and 301), followed by Spain (1,779 to 1,852) and China (2,023 to 2,025). Regarding ST48, the single isolate from China, included for comparisons, showed between 4,163 and 4,291 SNP differences compared to the Danish ST48 isolates. Finally, our ST90 strains were more similar to the ST90 isolate from China (between 80 and 268 SNP differences) than to the ST90 strains from the United States (ranging from 388 to 476 SNP differences).
FIG 2.
SNP-based phylogeny of the 90 ETEC isolates from pigs in Denmark and the 20 porcine ETEC isolates from different countries. Colors in the ring correspond to sequence type. Isolates highlighted in red, orange, and blue correspond to isolates from China, the United States, and Spain, respectively.
Antimicrobial resistance phenotypes and genotypes.
The antimicrobial susceptibility testing revealed that 85 (94.4%) of the isolates were resistant to at least one of the antimicrobials investigated, 60 (66.7%) were multidrug resistant (MDR), and only five were susceptible to all antimicrobials. High rates of resistance were found against streptomycin (68.9% of the isolates), sulfamethoxazole (67.8%), tetracycline (56.7%), spectinomycin (55.6%), trimethoprim (53.3%), and ampicillin (48.3%). Importantly, none of the isolates was resistant to ceftiofur, cefotaxime, colistin, or ciprofloxacin, which are considered highly critical drugs in human medicine (15) (Table 2). In addition, one and eight F4-positive isolates showed resistance to amoxicillin-clavulanic acid and nalidixic acid, respectively, while none of the F18 isolates tested positive for these drugs (Table 2).
TABLE 2.
Prevalence of antimicrobial resistance among the 90 ETEC isolates from pigs
| Antimicrobiala | No. (%) of resistant isolates | No. (%) of isolates of each fimbrial typeb |
P value | ORc (95% CI) | |
|---|---|---|---|---|---|
| F4 | F18 | ||||
| Ampicillin | 47 (48.3) | 35 (50.7) | 12 (57.1) | 0.6282 | |
| Amoxicillin-clavulanic acid | 1 (1.1) | 1 (1.5) | 0 | >0.9999 | |
| Chloramphenicol-florfenicol | 15 (16.7) | 9 (13.4) | 6 (28.5) | 0.1064 | |
| Apramycin | 8 (8.9) | 7 (10.14) | 1 (4.7) | 0.6752 | |
| Gentamicin | 6 (6.7) | 8 (11.6) | 1 (4.7) | 0.6792 | |
| Neomycin | 23 (25.6) | 16 (23.2) | 7 (33.3) | 0.3964 | |
| Spectinomycin | 50 (55.6) | 35 (50.5) | 15 (71.42) | 0.1327 | |
| Streptomycin | 62 (68.9) | 53 (76.8) | 9 (42.9) | 0.0061 | 4.417 (1.491–12.48) |
| Sulfamethoxazole | 61 (67.8) | 46 (66.7) | 15 (71.4) | 0.7932 | |
| Tetracycline | 51 (56.7) | 42 (60.9) | 9 (42.8) | 0.2084 | |
| Trimethoprim | 48 (53.3) | 35 (50.7) | 13 (62) | 0.4568 | |
| Nalidixic acid | 8 (8.9) | 8 (11.6) | 0 | 0.1901 | |
The breakpoints used correspond to EUCAST epidemiological cutoff values (ECOFFs) for ampicillin (8 μg/ml), chloramphenicol-florfenicol (8 μg/ml), gentamicin (2 μg/ml), nalidixic acid (8 μg/ml), neomycin (8 μg/ml), spectinomycin (64 μg/ml), streptomycin (16 μg/ml), sulfamethoxazole (64 μg/ml), tetracycline (8 μg/ml), and trimethoprim (2 μg/ml). EUCAST clinical breakpoints for amoxicillin-clavulanic acid (R > 8 μg/ml) and DANMAP 2015 for apramycin (R > 32 μg/ml) were also used.
Percentage is estimated based on the total number of strains associated with each fimbrial type. Significant differences (P < 0.05) are indicated in boldface.
Odds ratio (OR) is indicated when the P value is <0.05; CI, confidence interval.
Results from kappa statistical analysis showed that there was an almost perfect agreement between phenotypic resistance and the in silico prediction of resistance genotype (Table 3), which identified a total of 39 different AMR genes (Table S2). The genes blaTEM-1B, tet(A), and dfrA1 were the most commonly detected among ampicillin-, tetracycline-, and trimethoprim-resistant isolates, respectively. Similarly, sul1 and sul2 were the predominant genes responsible for sulfonamide resistance. A total of 16 different genes encoding aminoglycoside-modifying enzymes were identified, with the aph [phosphotransferases, i.e., aph(3′)-Ia, aph(3′)-Ib, aph(3″)-Ib, aph(4)-Ia, and aph(6)-Id] and aadA (nucleotidyltranferases, i.e., aadA1, aadA2, aadA5, aadA11, aadA12, aadA17, aadA22, and aadA24) genes being the most common, consistent with the high resistance to spectinomycin and streptomycin, respectively. Regarding phenicols, catA1, cmlA1, and floR genes were found among the resistant isolates (Table 3 and Table S2). Susceptibility to macrolides and lincosamides was not phenotypically tested; however, genes conferring resistance to both classes of drugs were detected (Table 3 and Table S2).
TABLE 3.
Antimicrobial resistance genes detected among the 90 ETEC isolates from pigs
| Antimicrobial group and gene | Total no. (%) of isolates | No. (%) of isolates of each fimbrial typea |
Kappa | P valueb | |
|---|---|---|---|---|---|
| F4 | F18 | ||||
| Beta-lactams | |||||
| blaTEM-1A | 4 (4.4) | 4 (5.8) | 0 | 0.933 | 0.0001 |
| blaTEM-1B | 42 (46.7) | 30 (43.5) | 12 (57.1) | ||
| blaTEM-30 | 1 (1.1) | 1 (1.4) | 0 | ||
| Aminoglycosides | |||||
| aph (phosphotransferases) | 58 (64.4) | 50 (72.5) | 8 (38.1) | 0.927 | 0.0001 |
| aadA (nucleotidyltransferases) | 57 (63.3) | 42 (60.9) | 15 (71.4) | ||
| aac (acetyltransferases) | 9 (10) | 8 (11.6) | 1 (4.8) | ||
| Phenicols | |||||
| catA1 | 3 (3.3) | 3 (4.3) | 0 | 1.000 | 0.0001 |
| cmlA1 | 8 (8.9) | 2 (2.9) | 6 (28.6) | ||
| floR | 5 (5.6) | 5 (7.2) | 0 | ||
| Macrolides | |||||
| mdf(A) | 90 (100) | 69 (100) | 21 (100) | ND | ND |
| mph(A) | 8 (8.9) | 8 (11.6) | 0 | ||
| mph(B) | 7 (7.8) | 7 (10.1) | 0 | ||
| erm(B) | 9 (10) | 9 (13) | 0 | ||
| Lincosamides | |||||
| lnu(F) | 5 (5.6) | 5 (7.2) | 0 | ND | ND |
| lnu(G) | 5 (5.6) | 5 (7.2) | 0 | ||
| Sulfonamides | |||||
| sul1 | 30 (33.3) | 28 (40.6) | 2 (9.5) | 0.898 | 0.0001 |
| sul2 | 42 (46.7) | 35 (50.7) | 7 (33.3) | ||
| sul3 | 9 (10) | 3 (4.3) | 6 (28.6) | ||
| Tetracycline | |||||
| tet(A) | 40 (44.4) | 32 (46.4) | 8 (38.1) | 1.000 | 0.0001 |
| tet(B) | 13 (14.4) | 12 (17.4) | 1 (4.8) | ||
| tet(X) | 1 (1.1) | 1 (1.4) | 0 | ||
| Trimethoprim | |||||
| dfrA1 | 34 (37.8) | 27 (39.1) | 7 (33.3) | 0.978 | 0.0001 |
| dfrA5 | 2 (2.2) | 2 (2.9) | 0 | ||
| dfrA12 | 8 (8.9) | 2 (2.9) | 6 (28.6) | ||
| dfrA14 | 5 (5.6) | 5 (7.2) | 0 | ||
| dfrA17 | 2 (2.2) | 2 (2.9) | 0 | ||
Percentage is estimated based on the total number of strains associated with each fimbrial type.
A P value of <0.05 is considered significant.
Notably, none of the isolates was found to harbor genes that suggested extended-spectrum beta-lactamase (ESBL) production or transferrable colistin resistance (mcr-class genes).
The ResFinder bioinformatics tool also allows the identification of chromosomal mutations related to antimicrobial resistance. Eight isolates, phenotypically resistant to nalidixic acid, showed a single chromosomal mutation in the gyrA(S83L) gene. The substitution V161G, associated with colistin resistance, was also detected in the pmrB gene in five isolates; however, these isolates were phenotypically susceptible to the antimicrobial. In addition, one isolate had a nucleotide change in the ampC promoter (32T→A) that is associated with resistance to beta-lactams (Table S2).
Prediction of biocide and metal tolerance genes.
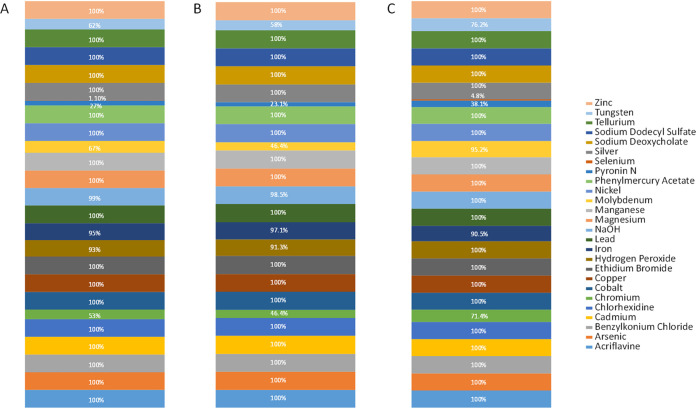
The ETEC isolates showed a large variety of biocide resistance and metal tolerance genes (Fig. 3 and Table S4). A total of 117 different genes were detected, and the number of genes per isolate ranged from 40 to 88. Eleven genes associated with metal tolerance and/or related to metal transport and metabolism as well as biocide resistance were shared by all of the strains in the collection: corA (magnesium [Mg], cobalt [Co], nickel [Ni], and manganese [Mn]), glpF (antimony [Sb] and arsenic [As]), mgtA (Co and Mg), zntR (yhdM) (zinc [Zn]), phoB (benzalkonium chloride and chlorhexidine), mdtF (yhiV), mdtK (ydhE) (several biocides, including benzalkonium chloride and ethidium bromide), ostA (lptD) (N-hexane), emrB (phenylmercury acetate, 2-chlorophenylhydrazine, and carbonylcyanide m-chlorophenyl hydrazine), and acrB and acrF (envD) (acriflavine). In addition, different genes conferring metal tolerance to cadmium (Cd), silver (Ag), mercury (Hg), lead (Pb), copper (Cu), and tellurium (Te) were predicted in each strain.
FIG 3.
Distribution of metal tolerance and biocide resistance among the 90 ETEC isolates from pigs (A), the F4-positive isolates (B), and the F18-positive isolates (C).
The highest diversity of genes conferring biocide resistance and metal tolerance were identified for copper (16 genes), hydrogen peroxide (15 genes), ethidium bromide (13 genes), sodium deoxycholate (13 genes), cadmium (12 genes), acriflavine (11 genes), and zinc (11 genes).
Genes involved in the uptake and transport of metals are ubiquitously found in bacteria, since metal ions play a role in many different biological processes and are essential for bacterial survival (16). All genes detected in the ETEC isolates were predicted in E. coli K-12, with the exception of those presumably conferring metal tolerance to Co (cuiD, pcoB, and pcoS), Hg (merA, merE, and merR), quaternary ammonium compounds (qacE, qacEdelta1, and qacF), Ag (silA, silE, and silP), and Te (terB, terC, terD, terW, and terZ), which were exclusively predicted in ETEC strains (data not shown).
Virulence genes carried by ETEC isolates.
The VirulenceFinder bioinformatics tool corroborated the presence of F4 and F18 fimbrial genes in 69 and 21 E. coli isolates, respectively, and further identified them as F4ac (100%) and F18ac (100%). Two isolates also were predicted to carry F6 type fimbria together with F4. WGS analysis also confirmed the presence of sta (21 isolates, 23.3%) and stb (85 isolates, 94.4%) heat-stable enterotoxins in those isolates positive for the toxins by PCR.
Apart from the fimbrial F4 and F18 and enterotoxin sta and stb genes, a total of 16 different virulence genes were predicted among the 90 E. coli isolates, such as ltcA, the gene associated with the heat-labile enterotoxin (LT) and present in most of the isolates (82 isolates, 91.1%). The gene astA (encoding the EAST-1 heat-stable toxin) and the gene iha (encoding a nonhemagglutinating adhesion protein [17]) were also present in 85% of the isolates (Table 4). All isolates carried at least five different virulence genes, and 60% of the isolates harbored eight or more virulence determinants.
TABLE 4.
Distribution of virulence genes (other than F4- and F18-encoding fimbria) among the 90 ETEC isolates
| Function and gene | Total no. (%) of isolates | No. (%) of isolates of each fimbrial typea
|
P value | ORb (95% CI) | |
|---|---|---|---|---|---|
| F4 | F18 | ||||
| Toxins | |||||
| astA | 77 (85.6) | 69 (100) | 8 (38.1) | <0.0001 | ∞ (25.71–∞) |
| ltcA | 82 (91.1) | 69 (100) | 13 (62) | <0.0001 | ∞ (9.402–∞ |
| sta | 21 (23.3) | 13 (18.8) | 8 (38.1) | 0.0820 | |
| stb | 85 (94.4) | 69 (100) | 16 (76.2) | 0.0005 | ∞ (5.465–∞) |
| Fimbriae | |||||
| lpfA | 15 (16.7) | 2 (2.9) | 13 (61.9) | <0.0001 | 0.01837 (0.003635–0.08) |
| fasA | 2 (2.2) | 2 (2.9) | 0 | >0.9999 | |
| Adhesion | |||||
| iha | 77 (85.6) | 57 (82.6) | 20 (95.2) | 0.2854 | |
| air | 4 (4.4) | 0 | 4 (19) | 0.0023 | 0 (0.0–0.02885) |
| Colicin | |||||
| cba | 33 (36.7) | 33 (47.8) | 0 | <0.0001 | ∞ (4.748–∞) |
| cma | 47 (52.2) | 42 (60.9) | 5 (23.8) | 0.0053 | 4.978 (1.631–13.29) |
| celB | 10 (11.1) | 8 (11.6) | 2 (9.5) | >0.9999 | |
| Microcin | |||||
| mchB | 9 (10) | 9 (13) | 0 | 0.1096 | |
| mchC | 9 (10) | 9 (13) | 0 | 0.1096 | |
| mchF | 9 (10) | 9 (13) | 0 | 0.1096 | |
| mcmA | 9 (10) | 9 (13) | 0 | 0.1096 | |
| Others | |||||
| capU | 55 (61.1) | 54 (78.3) | 1 (4.8) | <0.0001 | 72 (11.75–759.7) |
| gad | 40 (44.4) | 35 (50.7) | 5 (23.8) | 0.0439 | 3.294 (1.09–8.811) |
| sepA | 10 (11.1) | 10 (14.5) | 0 | 0.1088 | |
| iss | 20 (22.2) | 1 (1.4) | 19 (90.5) | <0.0001 | 0.001548 (0.0001506–0.0) |
| eilA | 5 (5.6) | 0 | 5 (23.8) | 0.0005 | 0 (0.0–0.1830) |
Percentage is estimated based on the total number of strains associated with each fimbria type. Significant differences (P < 0.05) are indicated in boldface.
The odds ratio (OR) is indicated when the P value is <0.05; CI, confidence interval; ∞, infinity.
Notably, statistically significant differences were detected in the distribution of virulence genes depending on the fimbria type (Table 4). While all F4-positive isolates harbored astA, ltcA, and stb genes, only a portion of F18-positive strains carried these genes (38%, 61%, and 76%, respectively). The genes gad (encoding a glutamate decarboxylase, enabling resistance to gastric acidity) (18) (35 isolates), capU (hexosyltransferase) (54 isolates), and the cba (33 isolates), cma (42 isolates), and celB (eight isolates) colicin-encoding genes (bacteriocins active against closely related E. coli bacteria or other members of the Enterobacteriaceae) (19) also were more frequently detected among F4- than among F18-positive strains. However, the genes lpfA (long polar fimbriae) (13 isolates) and iss (increased serum survival; role in extraintestinal pathogenic E. coli [ExPEC] virulence [20]) (19 isolates) were predominantly found in F18-positive isolates. In addition, microcin-encoding genes (nine isolates) and sepA (Shigella extracellular protein A, involved in tissue invasion) (10 isolates) (21) were exclusively identified in F4-positive strains, while air (enteroaggregative immunoglobulin protein with epithelial adhesion function) (four isolates) and eilA (Salmonella HilA homolog; transcriptional activator of SPI-1 genes) (five isolates) (22) were unique to F18-positive isolates. Hybrid variants VTEC (verotoxin-producing E. coli)/ETEC were not identified in the present work.
In silico plasmid detection and prediction of cooccurrence of AMR genes with virulence and metal tolerance genes.
Twenty-one different plasmid replicons were predicted among the E. coli strains. The most prevalent replicon types were IncFII (92.2% of the strains) and IncFIB (91.1%), followed by IncI1 (72.2%), IncFIC (54.4%), and IncX1 (36.7%), and the number of plasmid replicons per isolate ranged from three to eight (Table 5). At least one IncF replicon was detected in all the isolates.
TABLE 5.
Plasmid replicons predicted by PlasmidFinder among the 90 ETEC isolates
| Plasmid group and replicon | Total no. (%) of isolates | No. (%) of each fimbrial typea
|
P value | ORb (95% CI) | |
|---|---|---|---|---|---|
| F4 | F18 | ||||
| IncF | |||||
| FII | 83 (92.2) | 69 (100) | 14 (66.7) | <0.0001 | ∞ (7.269–∞) |
| FIB | 82 (91.1) | 69 (100) | 13 (61.9) | <0.0001 | ∞ (9.402–∞) |
| FIC | 49 (54.4) | 41 (59.4) | 8 (38.1) | 0.1323 | |
| FIA | 1 (1.1) | 1 (1.4) | 0 | >0.9999 | |
| IncI | |||||
| I1 | 65 (72.2) | 45 (65.2) | 20 (95.2) | 0.0057 | 0.09375 (0.0086–0.6311) |
| I2 | 12 (13.3) | 12 (17.4) | 0 | 0.0614 | |
| IncX | |||||
| X1 | 33 (36.7) | 12 (17.4) | 21 (100) | <0.0001 | 0.000 (0.000–0.06711) |
| X4 | 3 (3.3) | 3 (4.3) | 0 | >0.9999 | |
| IncQ | |||||
| Q1 | 21 (23.3) | 21 (30.4) | 0 | 0.0024 | ∞ (2.202–∞) |
| Col-like | |||||
| Col156 | 15 (16.7) | 13 (18.8) | 2 (9.5) | 0.5056 | |
| Col4400II | 2 (2.2) | 2 (2.9) | 0 | >0.9999 | |
| ColRNAI | 2 (2.2) | 2 (2.9) | 0 | >0.9999 | |
| IncHI | |||||
| 1A | 1 (1.1) | 1 (1.4) | 0 | >0.9999 | |
| 1B | 1 (1.1) | 1 (1.4) | 0 | >0.9999 | |
| 2 | 7 (7.7) | 7 (10.1) | 0 | 0.1933 | |
| 2A | 7 (7.7) | 7 (10.1) | 0 | 0.1933 | |
| IncN | 12 (13.3) | 12 (17.4) | 0 | 0.0614 | |
| IncB/O/Z | 10 (11.1) | 3 (4.3) | 7 (33.3) | 0.0012 | 0.09091 (0.02421–0.415) |
| IncY | 5 (5.5) | 5 (7.2) | 0 | 0.5869 | |
| p0111 | 21 (23.3) | 14 (20.3) | 7 (33.3) | 0.2450 | |
| pKPC | 3 (3.3) | 3 (4.3) | 0 | >0.9999 | |
Percentage is estimated based on the total number of strains associated with each fimbrial type. Significant differences (P < 0.05) are indicated in boldface.
The odds ratio (OR) is indicated when the P value is <0.05; CI, confidence interval.
With regard to fimbrial type, all F4-positive isolates contained replicons of both IncFII and IncFIB types, while all F18-positive strains harbored replicons of the IncX1 type and all but one carried IncI1 (95.2%). The latter was found in 65.2% of the F4-positive strains, and the IncI2 type was exclusively detected in F4 isolates. The replicon IncX1 also was found in all F18 isolates and only in 12 F4 isolates, while the replicon IncX4 was unique to three F4 isolates. In addition, IncHI, IncN, and IncQ replicons were only found in F4-positive isolates (Table 5).
According to the plasmidSPAdes and ResFinder bioinformatics tools, the majority of the AMR genes detected in the ETEC genomes were predicted to be plasmid located. Specifically, blaTEM, floR, catA, cmlA, and sul3 genes were always detected in plasmid contigs in all isolates. On the contrary, mdf(A) was predicted to be a chromosomal gene, since it was not detected in any plasmid component (Table S5).
Similarly, toxins were predicted to be located on plasmids in most of the strains: sta (71.4%, 15 out of 21 positive isolates), stb (83.5%, 71 out of 85 positive isolates), ltcA (87.8%, 72 out of 82 positive isolates), and astA (89.6%, 69 out of 77 positive isolates). In 58 out of the 90 genomes, the genes astA and stb were found in the same contigs, predicted to be a plasmid, and 22 of these also contained the gene ltcA in the same plasmid contig. Both F4 and F18 fimbrial genes appeared to be plasmid associated in all genomes too (Table S6). Interestingly, where both AMR and virulence genes were predicted to be carried by plasmids in a strain, these two types of genes were identified in the same plasmid component but not in the same plasmid contig.
Results from BacMet bioinformatics analysis were analyzed together with the plasmid bioinformatics tool; pitA and zintA (yodA) genes, conferring Zn tolerance, were predicted to be plasmid located in 4 and 16 out of 32 and 65 ETEC isolates that harbored these genes. This analysis also predicted that genes related to Cu tolerance were putatively plasmid located in some cases: cuiD in one out of six strains, cueO in 1 out of 27 strains, pcoS in four out of six strains, and pcoB in 34 out of 53 strains. Despite all of these Zn and Cu tolerance genes being predicted in the same plasmid component as certain AMR genes, cooccurrence in the same plasmid contig was not detected (Table S7). Additionally, most of the sil-like genes, mer-like genes, and ter-like genes encoding tolerance to Ag, Hg, and Te, respectively, were also predicted to be plasmid located and were also identified in the same plasmid component as several AMR genes (Table S8).
Prediction of prophage sequences in ETEC strains.
The in silico analysis of the 90 ETEC genomes with PHASTER resulted in the prediction of a minimum of 7 to a maximum of 20 different prophage regions in each strain (Table S9). The genome sizes of these prophage regions spanned from 2.7 to 107.9 kb, with a GC content ranging from 40.45 to 57.46%. This is very similar to the GC content of E. coli, that is, approximately 50% (23). Of the total predicted prophages, 39.1% were intact, whereas 48.2% were incomplete and the remaining 12.6% questionable (Fig. S1). The genome size, the GC content, and the number of prophage regions identified did not statistically differ between F4 and F18 strains. Most of the prophage regions identified in the ETEC genomes showed similarity to P88 (GenBank accession no. NC_026014) and PhiP27 (NC_003356) Enterobacteriaceae prophages (from 3.9% to 93.3% and from 4.8% to 60.9%, respectively). P88 is an inducible prophage of E. coli strain K88, able to lyse avian pathogenic E. coli strains (24), and the coliphage PhiP27 is an Stx2e-encoding phage, but since none of the ETEC isolates encoded Shiga toxins and the highest similarity is only 60.9%, the ETEC prophages could be part of the gene pool producing PhiP27 by recombination (25, 26).
DISCUSSION
The characterization of ETEC causing PWD is important for understanding disease epidemiology and pathogenesis and for the implementation of new strategies aiming to reduce the impact of the disease in the pig industry. In this study, we characterized in silico a collection of 90 ETEC F4- and F18-positive isolates to determine the levels of antimicrobial and biocide resistance, metal tolerance, and virulence genes associated with PWD. The antimicrobial resistance phenotype was also investigated. We also determined how many clonal groups are circulating in the production system as well as the phylogenetic relationships between ETEC strains currently causing PWD in Denmark and in other countries worldwide.
Overall, ETEC isolates showed significantly different genetic traits depending on the fimbria type. It should be mentioned that while the 69 F4-positive strains were recovered from at least 30 different herds, the 21 F18-positive isolates were collected from just five herds.
ETEC isolates causing PWD are commonly reported to belong to the serogroups O8, O138, O139 (often associated with edema disease), O141, 147, O149, and O157 (1, 27, 28), of which O8, O141, O147, and O149 were predicted to be among strains in the current investigation, together with O29 and O6, both commonly recognized serogroups associated with ETEC and enteroinvasive E. coli (EIEC) in humans (29). Differences were observed concerning serogroups between F4- and F18-positive isolates. O149 was the predominant serogroup among F4 isolates (72.46% versus 0%, P < 0.0001), while O8 was the most prevalent among F18 isolates (2.9% versus 33.33%, P < 0.0001). O149 and O138 (not detected here) have been the most frequent serogroups related to the classical ETEC F4 and F18, respectively, in Denmark (28) and other countries (30–32). As reported in Australia, the United States, Canada, Germany, and Thailand (32, 33), ST100 was the most frequent ST among the Danish porcine ETEC isolates, and ST48 and ST10 were also commonly observed. The latter STs belong to the largest clonal complex, 10, within E. coli, with isolates from both animal and human sources acting as commensals or as pathogens, and generally are associated with antimicrobial susceptibility and low virulence (34). ST10 is dominant in Spain among mcr-positive isolates and in China (35–37), followed by ST48 in the latter country (37).
To study the genetic relatedness of the collected E. coli isolates, phylogenetic analysis based on SNPs was performed. As expected, the isolates clustered depending on their ST in four well-defined clades, with three of them exclusively encompassing a single ST, while clade III was more diverse, including isolates belonging to different STs. Isolates recovered from the same herd showed the lowest number of different SNPs, suggesting that the same (or a very similar) ETEC strain circulates in a specific farm. F4-positive isolates appeared to be more closely related than F18-positive strains, since the maximum number of SNPs detected was lower in this group than among F18 strains (24,593 versus 45,742 SNPs), and the diversity of STs was lower among F4 isolates than F18 strains (three versus six). Results obtained from the phylogeny comparison, including isolates from four different countries, suggest that the ETEC ST100 isolates are closely related, since the SNP differences were relatively small among them. Interestingly, a relatively small number of SNPs (130 to 699) was identified between the F4-positive isolates from the 1990s and those recovered during 2018 to 2019, indicating that very similar clones have been circulating for at least 30 years. To confirm this hypothesis, a larger number of F4 strains from previous years (from the 1990s to now) should be analyzed. The clone O6:H16-ST48, which included 11 isolates which were recovered from five different herds and differed by 6 to 633 SNPs, was the second most prevalent among F4-positive isolates. Notably, O6 is one of the most important ETEC serogroups involved in human diarrhea globally, particularly among children under the age of five in developing countries (38), and it is not classically associated with PWD. A recent study based on the genomic characterization of 40 ETEC O6:H16/HNT human isolates collected during 1975 to 2016 showed significant genomic diversity among them, but none was assigned to ST48 (38). The occurrence of ETEC O6:H16 among pig ETEC isolates indicates zoonotic potential; however, the strains did not harbor known fimbrial genes involved in adhesion to the human intestine.
The World Health Organization has defined AMR as a global health issue in both humans and animals and has recommended surveillance for AMR bacteria in food-producing animals, such as pigs, as they represent a possible source and disseminator of AMR to humans (39). ETEC isolates from pigs are not considered zoonotic (the key virulence factors required to cause disease differ between pigs and humans) (40), but treatment with antimicrobials against PWD may select for AMR in commensal intestinal bacteria, and such bacteria may transfer critical resistances to humans via the food chain. To investigate this aspect, we determined, both phenotypically and in silico, the AMR levels among the ETEC isolates. The highest rates of AMR were found against aminoglycosides and sulfamethoxazole, the use of which has increased in livestock in Denmark during recent years (41). Resistance to tetracycline, trimethoprim, and ampicillin, which are among the antimicrobials frequently used for treatment of PWD in Denmark, was broadly detected. Notably, levels of tetracycline and ampicillin resistance were similar to those detected in a previous study on pathogenic E. coli from pigs in Denmark (42). All of the isolates were susceptible to ciprofloxacin, cefotaxime, ceftiofur, and colistin, which are critically important antimicrobials for human medicine (15), but treatment with other antimicrobial agents may allow their coselection. The absence of these resistances may be linked to the restricted use of these drug classes in the pig industry, where fluoroquinolones, cephalosporins, and colistin all bear a penalty of factor 10 in the herd-level registration scheme of the use of antimicrobials (the yellow-card scheme) in Danish pig production (41). Similar findings were described in a recent study in Denmark, where the highest proportions of AMR among ETEC isolates were found for ampicillin (60.7%), sulfamethoxazole (69.7%), tetracycline (47.2%), and trimethoprim (69.7%), while AMR to ciprofloxacin, ceftiofur, and colistin was not detected (43). MDR in the pig industry has been linked with the wide use of aminoglycosides and beta-lactams in veterinary medicine (44). Here, MDR was detected in 60 isolates (66.7%), far from the 94% detected among ETEC isolates carrying mcr-1 and causing PWD in Spain (36). This highlights that the in silico prediction of AMR genes showed an almost perfect agreement with the phenotypic analysis according to kappa statistical analysis. In addition, genes encoding macrolide (specifically for erythromycin via MdfA) or lincosamide resistance, which were not phenotypically tested, were also predicted. Both drug classes are commonly employed in Denmark and other countries for treatment against Lawsonia intracellularis, an intracellular pathogen causing enteric disease in pigs (45–47). The increase in the administration of macrolides during recent years and the steady use of lincosamides in the pig industry in Denmark (41) could have selected for resistance to the drugs in E. coli.
Studies of ETEC across several countries worldwide, including old studies from Denmark, describe ETEC F4 as the most common type associated with PWD, followed by F18 (48–52). However, in other countries, such as Poland, Cuba, Japan, and Spain (36, 53–55), the highest prevalence was found for ETEC F18. In a recent study in Denmark, the number of F4 and F18 strains detected was similar (annual report from 2018; https://diagnostik.dtu.dk/raadgivning/aarsrapporter-for-diagnostik_overvaagning/aarsrapporter-svin). As demonstrated in previous studies, the 90 strains under study were all F4ac or F18ac (6, 7). Further, Denmark has had a breeding strategy to reduce the susceptibility of pigs to ETEC F4ac. The strategy consisted of the inactivation (based on one SNP change) of the candidate gene of the F4ac receptor MUC4 (5, 56); however, according to our results, F4 strains of type ac are still recovered. Since information on the farms is confidential, we acknowledge whether the herds under study have this strategy implemented and/or piglets were vaccinated. Thus, the reason why F4ac is still being detected could be that some of the isolates tested were recovered from herds where the strategy and/or vaccination has not been applied. Some studies also suggest that MUC13 and not MUC4 is the most likely gene governing susceptibility to ETEC F4ac, and this might explain why ETEC F4ac is still the predominant causative agent of PWD right after weaning (57).
In two F4 isolates, F4 and F6 fimbrial genes were detected concurrently. ETEC isolates encoding more than one fimbria have been previously described (28, 48, 49, 58, 59), and such strains have been suggested to have a pathogenetic advantage (27). The most prevalent enterotoxin detected in our study was STb (85 isolates), consistent with previous studies performed in other countries (28, 48, 52). All F4-positive isolates carried astA, ltcA, and stb genes, in line with results of previous studies, where the F4 fimbria-encoding gene was strongly associated with lt and stb (28, 48, 60). However, in our study, only a portion of F18-positive isolates (n = 8) carried astA, ltcA, and stb genes, while the remaining strains harbored ltcA and/or stb and sta genes.
Additionally, 16 other virulence genes were predicted among strains, and their distribution was associated with the type of fimbria, suggesting that F4- and F18-positive isolates use different virulent strategies to cause disease. As suggested, ETEC strains harboring additional fimbrial adhesins may exploit alternative pathways for the colonization of the host (17). Overall, the analysis showed that ETEC isolates from Danish pigs harbor other virulence factors than their characteristic adhesins and toxins, but currently the role for such factors, if any, in intestinal disease is not known.
Plasmids play an important role in the spread and dissemination of both AMR and virulence genes (61). IncF, IncI, and IncX replicon types were the most prevalent, and at least one IncF replicon was detected in all strains, as previously reported in porcine ETEC isolates from Australia and Spain (32, 62). In general, F4 isolates showed more plasmid replicon diversity (21 different plasmid replicons) than the F18 isolates (eight different plasmid replicons). IncF plasmid type is the most commonly described in bacteria from humans and animals, particularly in E. coli, and is known to carry virulence and AMR genes (63, 64). IncI1 plasmids, the most dominant among F18 strains, were also detected in all O141-F18 isolates and in 87.1% of O149:F4 isolates from Australia (32). IncI1-type plasmids are often associated with AMR and known to be ESBL carriers (65); however, in the current study, ESBL were not detected, and the link to AMR was not investigated. IncX plasmids are narrow-host-range plasmids of Enterobacteriaceae, which are known to provide additional advantages commonly associated with AMR and biofilm formation (64, 66). Since ETEC fimbria and toxin genes often have been reported to be plasmid located (67), we investigated the putative plasmid localization of these genes and potential cooccurrence with AMR determinants. F4 and F18 fimbria-encoding genes were found to be plasmid located in all strains, and toxins were plasmid located in more than 70% of the isolates. In addition, the majority of the AMR genes were predicted to be plasmid located. Interestingly, AMR and virulence genes were often predicted to be part of the same plasmid component; however, further detailed studies are needed to confirm this, since the program used for predictions (plasmidSPAdes) does not separate plasmids of the same type present in a single strain.
Recent studies have reported that AMR might be associated with tolerance to heavy metals existing naturally or used in food animal production, such as zinc oxide and copper (41, 68–70). Here, we predicted that aminoglycoside, tetracycline, and ampicillin resistance genes and zinc or copper tolerance genes were located on the same plasmid component; however, as mentioned above for AMR and virulence genes, more detailed studies are needed to confirm this. Colocalization implies that the use of ZnO and Cu coselects for AMR. In addition to heavy metals, biocides, including disinfectants and antiseptics, widely used in farms, also could promote the spread of AMR (68). All isolates analyzed here were predicted to carry genes responsible for biocidal resistance, but cooccurrence with AMR was not investigated.
Each of the 90 isolates under study contained at least seven prophages, mostly similar to the coliphages P88 (from 3.9% to 93.3%) and Phi27 (from 4.8% to 60.9%). Although Phi27 is known to encode Stx2e, none of the ETEC isolates encoded Shiga toxins, indicating that, despite the similarity, it is not the same phage.
In conclusion, the current study showed a high clonal diversity among F18 isolates, while, in contrast, similar F4 clonal groups might be circulating in Danish herds. High rates of AMR against aminoglycosides, sulfamethoxazole, tetracycline, trimethoprim, and ampicillin were detected, as was the high diversity of virulence genes, including toxin genes (ltcA, astA, sta, and stb) and fimbria-encoding genes typed as F4ac and F18ac.
MATERIALS AND METHODS
Bacterial strains and PCR detection of fimbria types F4 and F18.
ETEC F4 isolates were collected from pigs with diarrhea in 2018 (n = 34, from 30 different farms), 2019 (n = 29, from 29 different farms), and 1989 to 1992 (n = 6, from six different farms). Presumptive ETEC F18 isolates (n = 20) were recovered from five different farms (collected at the same time point on each farm) in 2019. In addition, E. coli Nysø, a well-characterized ETEC strain recovered in the 1970s (71), was included as an F18 historical control. Strains were obtained during routine diagnostic procedures, and use for research purposes did not require ethical clearance as long as farm identity was not disclosed.
Strains were confirmed positive for F4 or F18 fimbriae using PCR with primers and conditions as previously reported (52).
Antimicrobial resistance phenotype.
MIC values for E. coli isolates were determined for amoxicillin (2 to 32 μg/ml)-clavulanic acid (1 to 16 μg/ml), ampicillin (1 to 32 μg/ml), apramycin (4 to 32 μg/ml), cefotaxime (0.125 to 4 μg/ml), ceftiofur (0.5 to 8 μg/ml), chloramphenicol (2 to 64 μg/ml), ciprofloxacin (0.015 to 4 μg/ml), colistin (1 to 16 μg/ml), florfenicol (2 to 64 μg/ml), gentamicin (0.5 to 16 μg/ml), nalidixic acid (4 to 64 μg/ml), neomycin (2 to 32 μg/ml), spectinomycin (16 to 256 μg/ml), streptomycin (8 to 128 μg/ml), sulfamethoxazole (64 to 1024 μg/ml), tetracycline (2 to 32 μg/ml), and trimethoprim (1 to 32 μg/ml) by the broth microdilution method using Sensititre microtiter trays (DKMVN4, Sensititre system; Thermo Fisher Scientific, West Sussex, United Kingdom). E. coli ATCC 25922 was used as a quality control. Results were interpreted according to EUCAST epidemiological cutoff values, EUCAST clinical breakpoints for amoxicillin-clavulanic acid (www.EUCAST.org), and DANMAP for apramycin (72). Isolates were defined as susceptible when classified as wild type and resistant when classified as not wild type. Multidrug-resistant (MDR) strains were those resistant to one agent from three or more different antimicrobial classes (73).
PCR for detection of sta and stb toxin genes.
E. coli DNA was extracted from a single overnight-grown colony by the boiling lysis method as reported previously (52), and the PCR amplification of sta and stb toxin genes was performed using the primers and PCR conditions previously described (52).
DNA extraction and whole-genome sequencing (WGS).
DNA was extracted using the Maxwell system (Promega) by following the instructions provided by the Maxwell RSC cultured cell DNA kit (Promega). The quality of the DNA was determined by a NanoDrop-1000 (Thermo Fisher Scientific), and DNA quantification was performed using a double-stranded DNA BR assay kit with a Qubit 2.0 fluorometer (Invitrogen, USA).
The libraries for sequencing were prepared using the Nextera DNA Flex library preparation kit (Illumina, Inc., San Diego, CA, USA) according to the manufacturer’s protocol and sequenced using Illumina NextSeq (Illumina). The paired-end raw reads were assembled using SPAdes Genome Assembler v.3.13.0 (74), and the quality of assembly was evaluated with QUAST v.5.0.2. (75).
Whole-genome characterization.
The assembled contigs, with genomic sizes between 5.1 and 5.7 Mbp (mean size, 5.4 Mbp) (see Table S1 in the supplemental material), were analyzed using the bioinformatics tools of the Center for Genomic Epidemiology (CGE) for the presence of antibiotic resistance (ResFinder v.3.2) (76), virulence genes (VirulenceFinder v.2.0) (77), and plasmid replicon types (PlasmidFinder v.2.1) (78), as well as identification of clonotypes (CHTyper v.1.0), sequence types (MLST v.2.0) (79), and serotypes (SerotypeFinder v.2.0) (75). All of the CGE predictions were called using default settings. The identification of antibacterial biocide and metal tolerance genes was assessed using BacMet-Scan v.2.0 (80). The ClermonTyping tool (http://clermontyping.iame-research.center/) and PHASTER webserver (http://phaster.ca/) were used to predict the phylogroups and putative prophage sequences in the bacterial genomes, respectively (81, 82).
In silico prediction of localization of antimicrobial resistance, virulence, and metal tolerance genes and potential cooccurrence on plasmids.
The putative localization of AMR and virulence genes (F4, F18, ltcA, astA, sta, and stb) was predicted using a combination of plasmidSPAdes v.3.13.0 (83), ResFinder, and VirulenceFinder tools. Briefly, plasmidSPAdes was used to identify contigs most likely belonging to plasmid DNA and to assign them to components. Each component is considered a putative plasmid consisting of one or more contigs. This tool is not able to separate similar plasmids (for example, similar plasmids of the same type present in a single strain); thus, their contigs may be assigned to the same identifier (ID) (same component). ResFinder and VirulenceFinder were used to analyze the presence of AMR and virulence genes in all contigs identified as putative DNA regions of plasmids. The output from both tools provides the contig ID and component on which the specific genes were located. Genes contained in the same contig and/or component were predicted to be plasmid located, while those antimicrobial and/or virulence genes not detected in plasmid DNA contigs were assumed to be chromosome located.
The BacMet database (BacMet-Scan v.2.0.), which includes metal tolerance and biocide resistance genes (metal tolerance genes include those genes that are indirectly related to metals) was used to investigate the presence of these genes in all of the genomes. The genome of the E. coli K-12 substrain MG1655 (GenBank accession number NC_000913.3) was also included in the analysis. Plasmid contigs identified and assigned to components with plasmidSPAdes v.3.13.0, as described above, were analyzed with BacMet-Scan v.2.0. Next, the plasmid contigs were manually inspected for the presence of metal tolerance genes (zinc, copper, silver, mercury, and tellurium) previously identified on the genome assembly by using the BacMet database (Table S4). The cooccurrence of AMR genes (detected as indicated above) and the metal tolerance genes on the same plasmid were predicted when they were found to belong to the same contig.
Phylogenetic analysis.
Phylogenetic relationships between the isolates were analyzed based on SNP trees constructed using the bioinformatics tool CSI Phylogeny v.1.4 (84), available from CGE. The genome of the E. coli K-12 substrain MG1655 was included as a reference strain, and CGE default parameters were used during SNP analysis. The phylogenetic tree was visualized and edited by using the bioinformatics tool iTOL v5 (85).
The ETEC isolates under study also were compared with 20 swine ETEC isolates from three different countries (China, the United States, and Spain), as mentioned above. The accession numbers and MLST types of the 20 ETEC strains used in this phylogenetic analysis are indicated in Table S10.
Statistical analysis.
Differences between F4- and F18-positive strains regarding serogroups, antimicrobial resistance, virulence gene content, and plasmid replicons were analyzed using two-tailed Fisher's exact test with GraphPad Prism version 8.3 software (GraphPad Inc.). P values of <0.05 were considered statistically significant.
Cohen’s kappa statistical analysis was used to analyze the correlation between phenotypic resistance and in silico gene predictions using SPSS, version 26 (IBM, USA). Kappa values of ≤0 indicate no agreement; 0.01 to 0.20, none to slight; 0.21 to 0.40, fair; 0.41 to 0.60, moderate; 0.61 to 0.80, substantial; and 0.81 to 1.00, almost perfect agreement (86).
Data availability.
The draft genome sequences of E. coli isolates from pigs in Denmark in this study are available in the European Nucleotide Archive (ENA) under the study accession number PRJEB38608.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Joakim Larsson and Johan Bengtsson-Palme for their assistance with the use of the Bac-Met database as well as K. Aagard and M. Carlsen for their technical assistance.
V. García acknowledges the Consellería de Cultura, Educación e Ordenación Universitaria, Xunta de Galicia, for her postdoctoral grant (grant number ED481B-2018/018). This work was supported by Innovationsfonden (grant number 8088-00032B) and the Danish Ministry of Food of Environment (VetforligIII).
We declare that we have no competing interests.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Fairbrother JM, Nadeau E, Gyles CL. 2005. Escherichia coli in postweaning diarrhea in pigs: an update on bacterial types, pathogenesis, and prevention strategies. Anim Health Res Rev 6:17–39. doi: 10.1079/ahr2005105. [DOI] [PubMed] [Google Scholar]
- 2.Luppi A. 2017. Swine enteric colibacillosis: diagnosis, therapy and antimicrobial resistance. Porcine Health Manag 3:16. doi: 10.1186/s40813-017-0063-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amezcua R, Friendship RM, Dewey CE, Gyles C, Fairbrother JM. 2002. Presentation of postweaning Escherichia coli diarrhea in southern Ontario, prevalence of hemolytic E. coli serogroups involved, and their antimicrobial resistance patterns. Can J Vet Res 66:73–78. [PMC free article] [PubMed] [Google Scholar]
- 4.Rhouma M, Fairbrother JM, Beaudry F, Letellier A. 2017. Post weaning diarrhea in pigs: risk factors and non-colistin-based control strategies. Acta Vet Scand 59:31. doi: 10.1186/s13028-017-0299-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luise D, Lauridsen C, Bosi P, Trevisi P. 2019. Methodology and application of Escherichia coli F4 and F18 encoding infection models in post-weaning pigs. J Anim Sci Biotechnol 10:53. doi: 10.1186/s40104-019-0352-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Westerman RB, Mills KW, Phillips RM, Fortner GW, Greenwood JM. 1988. Predominance of the ac variant in K88-positive Escherichia coli isolates from swine. J Clin Microbiol 26:149–150. doi: 10.1128/JCM.26.1.149-150.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rippinger P, Bertschinger HU, Imberechts H, Nagy B, Sorg I, Stamm M, Wild P, Wittig W. 1995. Designations F18ab and F18ac for the related fimbrial types F107, 2134P and 8813 of Escherichia coli isolated from porcine postweaning diarrhea and from edema disease. Vet Microbiol 45:281–295. doi: 10.1016/0378-1135(94)00141-i. [DOI] [PubMed] [Google Scholar]
- 8.Nagy B, Fekete PZ. 2005. Enterotoxigenic Escherichia coli in veterinary medicine. Int J Med Microbiol 295:443–454. doi: 10.1016/j.ijmm.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 9.Loos M, Geens M, Schauvliege S, Gasthuys F, van der Meulen J, Dubreuil JD, Goddeeris BM, Niewold T, Cox E. 2012. Role of heat-stable enterotoxins in the induction of early immune responses in piglets after infection with enterotoxigenic Escherichia coli. PLoS One 7:e41041. doi: 10.1371/journal.pone.0041041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagy B, Wilson RA, Whittam TS. 1999. Genetic diversity among Escherichia coli isolates carrying f18 genes from pigs with porcine postweaning diarrhea and edema disease. J Clin Microbiol 37:1642–1645. doi: 10.1128/JCM.37.5.1642-1645.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fairbrother J, Gyles C. 2012. Colibacillosis, p 723–749. In Zimmerman J, Karriker L, Ramirez A, Schwartz K, Stevenson G (ed), Diseases of swine, 10th ed Willey-Blackwell, Hoboken, NJ. [Google Scholar]
- 12.Li X, Yin J, Li D, Chen X, Zang J, Zhou X. 2006. Dietary supplementation with zinc oxide increases Igf-I and Igf-I receptor gene expression in the small intestine of weanling piglets. J Nutr 136:1786–1791. doi: 10.1093/jn/136.7.1786. [DOI] [PubMed] [Google Scholar]
- 13.Roselli M, Finamore A, Garaguso I, Britti M, Mengheri E. 2003. Zinc oxide protects cultures enterocytes from the damage induced by Escherichia coli. J Nutr 133:4077–4082. doi: 10.1093/jn/133.12.4077. [DOI] [PubMed] [Google Scholar]
- 14.DANMAP. 2017. Use of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, food and humans in Denmark The Danish Integrated Antimicrobial Resistance Monitoring and Research Programme, Copenhagen, Denmark. [Google Scholar]
- 15.WHO. 2019. Critically important antimicrobials for human medicine–6th rev. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 16.Porcheron G, Garenaux A, Proulx J, Sabri M, Dozois CM. 2013. Iron, copper, zinc, and manganese transport and regulation in pathogenic Enterobacteria: correlations between strains, site of infection and the relative importance of the different metal transport systems for virulence. Front Cell Infect Microbiol 3:90. doi: 10.3389/fcimb.2013.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu XY, Chapman T, Trott DJ, Bettelheim K, Do TN, Driesen S, Walker MJ, Chin J. 2007. Comparative analysis of virulence genes, genetic diversity, and phylogeny of commensal and enterotoxigenic Escherichia coli isolates from weaned pigs. Appl Environ Microbiol 73:83–91. doi: 10.1128/AEM.00990-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castanie-Cornet MP, Penfound TA, Smith D, Elliott JF, Foster JW. 1999. Control of acid resistance in Escherichia coli. J Bacteriol 181:3525–3535. doi: 10.1128/JB.181.11.3525-3535.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cascales E, Buchanan SK, Duche D, Kleanthous C, Lloubes R, Postle K, Riley M, Slatin S, Cavard D. 2007. Colicin biology. Microbiol Mol Biol Rev 71:158–229. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson TJ, Wannemuehler YM, Nolan LK. 2008. Evolution of the iss gene in Escherichia coli. Appl Environ Microbiol 74:2360–2369. doi: 10.1128/AEM.02634-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benjelloun-Touimi Z, Si Tahar M, Montecucco C, Sansonetti PJ, Parsot C. 1998. SepA, the 110 kDa protein secreted by Shigella flexneri: two-domain structure and proteolytic activity. Microbiology 144:1815–1822. doi: 10.1099/00221287-144-7-1815. [DOI] [PubMed] [Google Scholar]
- 22.Sheikh J, Dudley EG, Sui B, Tamboura B, Suleman A, Nataro JP. 2006. EilA, a HilA-like regulator in enteroaggregative Escherichia coli. Mol Microbiol 61:338–350. doi: 10.1111/j.1365-2958.2006.05234.x. [DOI] [PubMed] [Google Scholar]
- 23.Mann S, Chen YP. 2010. Bacterial genomic G+C composition-eliciting environmental adaptation. Genomics 95:7–15. doi: 10.1016/j.ygeno.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 24.Chen M, Zhang L, Xin S, Yao H, Lu C, Zhang W. 2017. Inducible prophage mutant of Escherichia coli can lyse new host and the key sites of receptor recognition identification. Front Microbiol 8:147. doi: 10.3389/fmicb.2017.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Recktenwald J, Schmidt H. 2002. The nucleotide sequence of Shiga toxin (Stx) 2e-encoding phage phiP27 is not related to other Stx phage genomes, but the modular genetic structure is conserved. Infect Immun 70:1896–1908. doi: 10.1128/iai.70.4.1896-1908.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith DL, Rooks DJ, Fogg PC, Darby AC, Thomson NR, McCarthy AJ, Allison HE. 2012. Comparative genomics of Shiga toxin encoding bacteriophages. BMC Genomics 13:311. doi: 10.1186/1471-2164-13-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagy B, Fekete PZ. 1999. Enterotoxigenic Escherichia coli (ETEC) in farm animals. Vet Res 30:259–284. [PubMed] [Google Scholar]
- 28.Frydendahl K. 2002. Prevalence of serogroups and virulence genes in Escherichia coli associated with postweaning diarrhea and edema disease in pigs and a comparison of diagnostic approaches. Vet Microbiol 85:169–182. doi: 10.1016/S0378-1135(01)00504-1. [DOI] [PubMed] [Google Scholar]
- 29.Nataro JP, Kaper JB. 1998. Diarrheagenic Escherichia coli. Clin Microbiol Rev 11:142–201. doi: 10.1128/CMR.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sting R, Stermann M. 2008. Duplex real-time PCR assays for rapid detection of virulence genes in E. coli isolated from post-weaning pigs and calves with diarrhea. Dtsch Tierarztl Wochenschr 115:231–238. [PubMed] [Google Scholar]
- 31.Barth S, Schwanitz A, Bauerfeind R. 2011. Polymerase chain reaction-based method for the typing of F18 fimbriae and distribution of F18 fimbrial subtypes among porcine Shiga toxin-encoding Escherichia coli in Germany. J Vet Diagn Investig 23:454–464. doi: 10.1177/1040638711403417. [DOI] [PubMed] [Google Scholar]
- 32.Abraham S, Trott DJ, Jordan D, Gordon DM, Groves MD, Fairbrother JM, Smith MG, Zhang R, Chapman TA. 2014. Phylogenetic and molecular insights into the evolution of multidrug-resistant porcine enterotoxigenic Escherichia coli in Australia. Int J Antimicrob Agents 44:105–111. doi: 10.1016/j.ijantimicag.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 33.Jiang F, Wu Z, Zheng Y, Frana TS, Sahin O, Zhang Q, Li G. 2019. Genotypes and antimicrobial susceptibility profiles of hemolytic Escherichia coli from diarrheic piglets. Foodborne Pathog Dis 16:94–103. doi: 10.1089/fpd.2018.2480. [DOI] [PubMed] [Google Scholar]
- 34.Manges AR, Johnson JR. 2012. Food-borne origins of Escherichia coli causing extraintestinal infections. Clin Infect Dis 55:712–719. doi: 10.1093/cid/cis502. [DOI] [PubMed] [Google Scholar]
- 35.García V, García-Meniño I, Mora A, Flament-Simon SC, Díaz-Jiménez D, Blanco JE, Alonso MP, Blanco J. 2018. Co-occurrence of mcr-1, mcr-4 and mcr-5 genes in multidrug-resistant ST10 enterotoxigenic and Shiga toxin-producing Escherichia coli in Spain (2006–2017). Int J Antimicrob Agents 52:104–108. doi: 10.1016/j.ijantimicag.2018.03.022. [DOI] [PubMed] [Google Scholar]
- 36.García-Meniño I, García V, Mora A, Diaz-Jiménez D, Flament-Simon SC, Alonso MP, Blanco JE, Blanco M, Blanco J. 2018. Swine enteric colibacillosis in Spain: pathogenic potential of mcr-1 ST10 and ST131 E. coli isolates. Front Microbiol 9:2659. doi: 10.3389/fmicb.2018.02659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang GY, Guo L, Su JH, Zhu YH, Jiao LG, Wang JF. 2019. Frequency of diarrheagenic virulence genes and characteristics in Escherichia coli isolates from pigs with diarrhea in China. Microorganisms 7:308. doi: 10.3390/microorganisms7090308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pattabiraman V, Katz LS, Chen JC, McCullough AE, Trees E. 2018. Genome wide characterization of enterotoxigenic Escherichia coli serogroup O6 isolates from multiple outbreaks and sporadic infections from 1975–2016. PLoS One 13:e0208735. doi: 10.1371/journal.pone.0208735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.WHO. 2014. Antimicrobial resistance global report on surveillance. http://www.who.int/drugresistance/documents/surveillancereport/en/.
- 40.Zhang W, Robertson DC, Zhang C, Bai W, Zhao M, Francis DH. 2008. Escherichia coli constructs expressing human or porcine enterotoxins induce identical diarrheal diseases in a piglet infection model. Appl Environ Microbiol 74:5832–5837. doi: 10.1128/AEM.00893-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DANMAP. 2018. Use of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, food and humans in Denmark. The Danish Integrated Antimicrobial Resistance Monitoring and Research Programme, Copenhagen, Denmark. [Google Scholar]
- 42.Holmer I, Salomonsen CM, Jorsal SE, Astrup LB, Jensen VF, Hog BB, Pedersen K. 2019. Antibiotic resistance in porcine pathogenic bacteria and relation to antibiotic usage. BMC Vet Res 15:449. doi: 10.1186/s12917-019-2162-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosager WN, Peter NJ, Erik Lind JS, Svend H, Matthew D, Steen PK. 2017. Comparison of antimicrobial resistance in E. coli isolated from rectal and floor samples in pens with diarrheic nursery pigs in Denmark. Prev Vet Med 147:42–49. doi: 10.1016/j.prevetmed.2017.08.007. [DOI] [PubMed] [Google Scholar]
- 44.Rhouma M, Beaudry F, Thériault W, Letellier A. 2016. Colistin in pig production: chemistry, mechanism of antibacterial action, microbial resistance emergence, and One Health perspectives. Front Microbiol 7:1789. doi: 10.3389/fmicb.2016.01789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pyorala S, Baptiste KE, Catry B, van Duijkeren E, Greko C, Moreno MA, Pomba MC, Rantala M, Ruzauskas M, Sanders P, Threlfall EJ, Torren-Edo J, Torneke K. 2014. Macrolides and lincosamides in cattle and pigs: use and development of antimicrobial resistance. Vet J 200:230–239. doi: 10.1016/j.tvjl.2014.02.028. [DOI] [PubMed] [Google Scholar]
- 46.Karuppannan AK, Opriessnig T. 2018. Lawsonia intracellularis: revisiting the disease ecology and control of this fastidious pathogen in pigs. Front Vet Sci 5:181. doi: 10.3389/fvets.2018.00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arnold M, Crienen A, Swam H, von Berg S, Jolie R, Nathues H. 2019. Prevalence of Lawsonia intracellularis in pig herds in different European countries. Porcine Health Manag 5:31. doi: 10.1186/s40813-019-0137-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luppi A, Gibellini M, Gin T, Vangroenweghe F, Vandenbroucke V, Bauerfeind R, Bonilauri P, Labarque G, Hidalgo A. 2016. Prevalence of virulence factors in enterotoxigenic Escherichia coli isolated from pigs with post-weaning diarrhea in Europe. Porcine Health Manag 2:20. doi: 10.1186/s40813-016-0039-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vu-Khac H, Holoda E, Pilipcinec E. 2004. Distribution of virulence genes in Escherichia coli strains isolated from diarrheic piglets in the Slovak Republic. J Vet Med B Infect Dis Vet Public Health 51:343–347. doi: 10.1111/j.1439-0450.2004.00769.x. [DOI] [PubMed] [Google Scholar]
- 50.Zajacova ZS, Konstantinova L, Alexa P. 2012. Detection of virulence factors of Escherichia coli focused on prevalence of EAST1 toxin in stool of diarrheic and non-diarrheic piglets and presence of adhesion involving virulence factors in astA positive strains. Vet Microbiol 154:369–375. doi: 10.1016/j.vetmic.2011.07.029. [DOI] [PubMed] [Google Scholar]
- 51.Smith MG, Jordan D, Chapman TA, Chin JJ, Barton MD, Do TN, Fahy VA, Fairbrother JM, Trott DJ. 2010. Antimicrobial resistance and virulence gene profiles in multi-drug resistant enterotoxigenic Escherichia coli isolated from pigs with post-weaning diarrhea. Vet Microbiol 145:299–307. doi: 10.1016/j.vetmic.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 52.Zhang W, Zhao M, Ruesch L, Omot A, Francis D. 2007. Prevalence of virulence genes in Escherichia coli strains recently isolated from young pigs with diarrhea in the US. Vet Microbiol 123:145–152. doi: 10.1016/j.vetmic.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 53.Osek J, Gallien P, Truszczyñski M, Protz D. 1999. The use of polymerase chain reaction for determination of virulence factors of Escherichia coli strains isolated from pigs in Poland. Comp Immunol Microbiol Infect Dis 22:163–174. doi: 10.1016/s0147-9571(98)00083-6. [DOI] [PubMed] [Google Scholar]
- 54.Blanco M, Lazo L, Blanco JE, Dahbi G, Mora A, López C, González EA, Blanco J. 2006. Serotypes, virulence genes, and PFGE patterns of enteropathogenic Escherichia coli isolated from Cuban pigs with diarrhea. Int Microbiol 9:53–60. [PubMed] [Google Scholar]
- 55.Kusumoto M, Hikoda Y, Fujii Y, Murata M, Miyoshi H, Ogura Y, Gotoh Y, Iwata T, Hayashi T, Akiba M. 2016. Emergence of a multidrug-resistant Shiga toxin-producing enterotoxigenic Escherichia coli lineage in diseased swine in Japan. J Clin Microbiol 54:1074–1081. doi: 10.1128/JCM.03141-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peng QL, Ren J, Yan XM, Huang X, Tang H, Wang YZ, Zhang B, Huang LS. 2007. The g.243A>G mutation in intron 17 of MUC4 is significantly associated with susceptibility/resistance to ETEC F4ab/ac infection in pigs. Anim Genet 38:397–400. doi: 10.1111/j.1365-2052.2007.01608.x. [DOI] [PubMed] [Google Scholar]
- 57.Ren J, Yan X, Ai H, Zhang Z, Huang X, Ouyang J, Yang M, Yang H, Han P, Zeng W, Chen Y, Guo Y, Xiao S, Ding N, Huang L. 2012. Susceptibility toward enterotoxigenic Escherichia coli F4ac diarrhea is governed by the MUC13 gene in pigs. PLoS One 7:e44573. doi: 10.1371/journal.pone.0044573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kwon D, Choi C, Jung T, Chung HK, Kim JP, Bae SS, Cho WS, Kim J, Chae C. 2002. Genotypic prevalence of the fimbrial adhesins (F4, F5, F6, F41 and F18) and toxins (LT, STa, STb and STx2e) in Escherichia coli isolated from postweaning pigs with diarrhea or edema disease in Korea. Vet Rec 150:35–37. doi: 10.1136/vr.150.2.35. [DOI] [PubMed] [Google Scholar]
- 59.Chen X, Gao S, Jiao X, Liu XF. 2004. Prevalence of serogroups and virulence factors of Escherichia coli strains isolated from pigs with postweaning diarrhea in eastern China. Vet Microbiol 103:13–20. doi: 10.1016/j.vetmic.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 60.Post K, Bosworth B, Knoth J. 2000. Frequency of virulence factors in Escherichia coli isolated from pigs with postweaning diarrhea and edema disease in North Carolina. Swine Health Production 8:119–120. [Google Scholar]
- 61.Carattoli A. 2009. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother 53:2227–2238. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.García-Meniño I, Díaz-Jiménez D, García V, de Toro M, Flament-Simon SC, Blanco J, Mora A. 2019. Genomic characterization of prevalent mcr-1, mcr-4, and mcr-5 Escherichia coli within swine enteric colibacillosis in Spain. Front Microbiol 10:2469. doi: 10.3389/fmicb.2019.02469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Villa L, Garcia-Fernandez A, Fortini D, Carattoli A. 2010. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J Antimicrob Chemother 65:2518–2529. doi: 10.1093/jac/dkq347. [DOI] [PubMed] [Google Scholar]
- 64.Rozwandowicz M, Brouwer MSM, Fischer J, Wagenaar JA, Gonzalez-Zorn B, Guerra B, Mevius DJ, Hordijk J. 2018. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J Antimicrob Chemother 73:1121–1137. doi: 10.1093/jac/dkx488. [DOI] [PubMed] [Google Scholar]
- 65.Wang J, Stephan R, Karczmarczyk M, Yan Q, Hächler H, Fanning S. 2013. Molecular characterization of blaESBL–harboring conjugative plasmids identified in multi-drug resistant Escherichia coli isolated from food-producing animals and healthy humans. Front Microbiol 4:188. doi: 10.3389/fmicb.2013.00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Johnson TJ, Bielak EM, Fortini D, Hansen LH, Hasman H, Debroy C, Nolan LK, Carattoli A. 2012. Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid 68:43–50. doi: 10.1016/j.plasmid.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 67.Dubreuil JD, Isaacson RE, Schifferli DM. 2016. Animal enterotoxigenic Escherichia coli. EcoSal Plus 7. doi: 10.1128/ecosalplus.ESP-0006-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cheng G, Ning J, Ahmed S, Huang J, Ullah R, An B, Hao H, Dai M, Huang L, Wang X, Yuan Z. 2019. Selection and dissemination of antimicrobial resistance in agri-food production. Antimicrob Resist Infect Control 8:158. doi: 10.1186/s13756-019-0623-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pal C, Asiani K, Arya S, Rensing C, Stekel DJ, Larsson DGJ, Hobman JL. 2017. Metal resistance and its association with antibiotic resistance. Adv Microb Physiol 70:261–313. doi: 10.1016/bs.ampbs.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 70.Rensing C, Moodley A, Cavaco LM, McDevitt SF. 19 April 2018. Resistance to metals used in agricultural production. Microbiol Spectr doi: 10.1128/microbiolspec.ARBA-0025-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Larsen J. 1981. Effect of pectin on secretion in pig jejunal loops challenged to enteropathogenic E. coli or enterotoxin (LT). A preliminary report. Nord Vet Med 33:218–223. [PubMed] [Google Scholar]
- 72.DANMAP. 2015. Use of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, food and humans in Denmark. The Danish Integrated Antimicrobial Resistance Monitoring and Research Programme, Copenhagen, Denmark. [Google Scholar]
- 73.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 74.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Joensen KG, Tetzschner AM, Iguchi A, Aarestrup FM, Scheutz F. 2015. Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J Clin Microbiol 53:2410–2426. doi: 10.1128/JCM.00008-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, Moller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Ponten T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pal C, Bengtsson-Palme J, Rensing C, Kristiansson E, Larsson DG. 2014. BacMet: antibacterial biocide and metal resistance genes database. Nucleic Acids Res 42:D737–D743. doi: 10.1093/nar/gkt1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Beghain J, Bridier-Nahmias A, Le Nagard H, Denamur E, Clermont O. 2018. ClermonTyping: an easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb Genom 4:e000192. doi: 10.1099/mgen.0.000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Arndt D, Marcu A, Liang Y, Wishart DS. 2019. PHAST, PHASTER and PHASTEST: tools for finding prophage in bacterial genomes. Brief Bioinform 20:1560–1567. doi: 10.1093/bib/bbx121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Antipov D, Hartwick N, Shen M, Raiko M, Lapidus A, Pevzner PA. 2016. plasmidSPAdes: assembling plasmids from whole genome sequencing data. Bioinformatics 32:3380–3387. doi: 10.1093/bioinformatics/btw493. [DOI] [PubMed] [Google Scholar]
- 84.Kaas RS, Leekitcharoenphon P, Aarestrup FM, Lund O. 2014. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS One 9:e104984. doi: 10.1371/journal.pone.0104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McHugh M. 2012. Interrater reliability: the kappa statistic. Biochem Med 22:276–282. doi: 10.11613/BM.2012.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The draft genome sequences of E. coli isolates from pigs in Denmark in this study are available in the European Nucleotide Archive (ENA) under the study accession number PRJEB38608.