Our in situ survey demonstrates that RNA-based sequencing of 16S rRNA amplicons is a reliable approach to qualitatively probe, at high taxonomic resolution, the changes triggered by new and existing cleaning/sanitizing strategies in the environmental microbiota in human-built environments. This approach could soon represent a fast tool to clearly define which routine sanitizing interventions are more suitable for a specific food processing environment, thus limiting the costs of special cleaning interventions and potential product loss.
KEYWORDS: gaseous ozone, RNA-based surveillance, meat processing plants, spoilage bacteria, environmental microbiota
ABSTRACT
Microbial complexity and contamination levels in food processing plants heavily impact the final product fate and are mainly controlled by proper environmental cleaning and sanitizing. Among the emerging disinfection technologies, ozonation is considered an effective strategy to improve the ordinary cleaning and sanitizing of slaughterhouses. However, its effects on contamination levels and environmental microbiota still need to be understood. For this purpose, we monitored the changes in microbiota composition in different slaughterhouse environments during the phases of cleaning/sanitizing and ozonation at 40, 20, or 4 ppm. Overall, the meat processing plant microbiota differed significantly between secondary processing rooms and deboning rooms, with a greater presence of psychrotrophic taxa in secondary processing rooms because of their lower temperatures. Cleaning/sanitizing procedures significantly reduced the contamination levels and in parallel increased the number of detectable operational taxonomic units (OTUs), by removing the masking effect of the most abundant human/animal-derived OTUs, which belonged to the phylum Firmicutes. Subsequently, ozonation at 40 or 20 ppm effectively decreased the remaining viable bacterial populations. However, we could observe selective ozone-mediated inactivation of psychrotrophic bacteria only in the secondary processing rooms. There, the Brochothrix and Pseudomonas abundances and their viable counts were significantly affected by 40 or 20 ppm of ozone, while more ubiquitous genera like Staphylococcus showed a remarkable resistance to the same treatments. This study showed the effectiveness of highly concentrated gaseous ozone as an adjunct sanitizing method that can minimize cross-contamination and so extend the meat shelf life.
IMPORTANCE Our in situ survey demonstrates that RNA-based sequencing of 16S rRNA amplicons is a reliable approach to qualitatively probe, at high taxonomic resolution, the changes triggered by new and existing cleaning/sanitizing strategies in the environmental microbiota in human-built environments. This approach could soon represent a fast tool to clearly define which routine sanitizing interventions are more suitable for a specific food processing environment, thus limiting the costs of special cleaning interventions and potential product loss.
INTRODUCTION
It is widely recognized that food processing plants are colonized by a broad variety of microorganisms. These environments are some of the main sources of food contamination, and their proper cleaning/sanitizing plays a key role in the subsequent product shelf life (1). This is particularly true for red meat processing; together with the endogenous microbiota of the cattle carcasses, the environmental microbiota of the slaughterhouse heavily impacts the bacterial contamination of the final product (2). Contamination of the meat occurs mainly during the deboning of quarters and the subsequent manipulations of primal cuts (cutting, mincing, and trimming) before packaging, which take place in the deboning rooms (DRs) and secondary processing rooms (PRs), respectively (3–5). During these manipulations, the bacteria (especially psychrotrophic) from human skin, cleaning waters, and meat cuts are collected on the surfaces of chopping boards, conveyor belts, and equipment. Accordingly, meat cross-contamination reaches the highest level when cleaning and sanitizing are improperly performed and meat exudates are available for bacterial growth (6–9).
It has been observed that the common core spoilage microbiota in different meat products and processing plants is mainly composed of Gram-negative bacteria derived from dust, cleaning water, or air, while the Gram-positive species of animal origin are usually less abundant and specific for each processing plant and meat product (6, 10). The spoilage microbiota housed in meat processing plants can be also divided into two groups, i.e., transient and resident populations. The latter group is mainly composed of bacteria capable of forming a biofilm, a multispecies interaction that promotes cell attachment on food contact surfaces and confers to microbes resistance to sanitizing treatments (11). It should be pointed out that, during red meat processing, psychrotrophic biofilm-forming genera like Pseudomonas, Psychrobacter, Acinetobacter, and Brochothrix represent the spoilage bacteria shared more between working surfaces and the meats (7).
Disinfection of food contact surfaces in such environments is a challenging task, and meat industries are constantly seeking new ecofriendly and economical solutions to optimize the cleaning and sanitizing process, which should be designed to guarantee a microbial reduction equal to the daily accumulation. In addition, such alternative treatments should be tested in order to verify reduction of pathogenic microorganisms (12). In this frame, periodic treatments with gaseous ozone are considered a complementary practice to improve the effectiveness of ordinary environmental cleaning and sanitizing, mainly inactivating the airborne microbiota (13). Indeed, ozone has been shown to inactivate bacteria (14, 15), filamentous fungi (16, 17), yeasts (18), viruses (16), and mycotoxin synthesis (19). Concerning bactericidal activity, molecular ozone and the derived free radical species operate first through oxidative lysis of the membranes and cells (20), while the definitive bacterial inactivation is mainly due to the damage of internal cell structures and the deactivation of oxidative stress-protective mechanisms of bacteria (21, 22). Unlike other chemical sanitizers used in the food processing plants, the main advantage of using gaseous ozone is its spontaneous decomposition to oxygen. However, its disinfecting capability needs to be assessed on site, taking into account any possible specific resistance of spoilage bacteria or pathogens (23).
Here, we aim to provide comprehensive monitoring of the environmental microbiota in red meat processing plants, as well as its compositional changes after routine cleaning/sanitizing procedures and after additional ozonation treatments at 4, 20, and 40 ppm. To accomplish this goal, we coupled the RNA-based amplicon sequencing of 16S rRNA with plate counts in order to map the viable microbiota and to define any specific bacterial group inactivation. This approach provides detailed scenarios of the environmental microbiota distribution in the processing plants and the changes caused by cleaning/sanitizing and ozonation steps.
RESULTS
Microbial landscape of the meat processing plants.
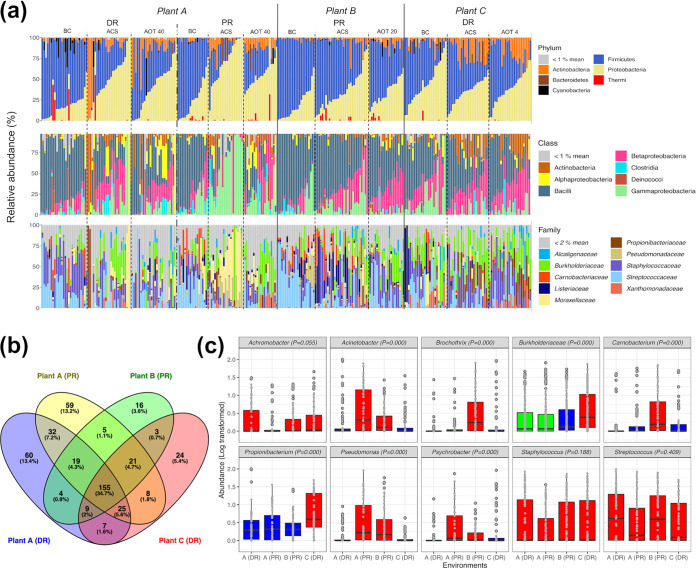
To explore the microbiota composition and distribution of three red meat processing plants, a total of 278 environmental swabs were collected during three temporal phases (Table 1), i.e., before cleaning (BC), after cleaning/sanitizing (ACS), and after three different ozone treatments (AOT4, AOT20, and AOT40). The environmental microbiota of plant A, which contains a DR and a secondary PR, and those of the single-room environments of plants B (PR) and C (DR) were mainly composed of Firmicutes, Proteobacteria, and Actinobacteria, while Bacteroidetes, Cyanobacteria, and Deinococcus-Thermus represented minor phyla (Fig. 1a). In all samples, Firmicutes comprised primarily Staphylococcaceae and Streptococcaceae, while Proteobacteria mainly contained members of Burkholderiaceae and Moraxellaceae.
TABLE 1.
Characteristics of the red meat processing plants and room types monitored in this study across three temporal phases
| Plant | Room type | Cleaning/sanitizing procedures | Phases | Ozone concentration (ppm) | Environment (vol [no. of treatments]) | Avg temperature (°C) | No. of samples fora
: |
||
|---|---|---|---|---|---|---|---|---|---|
| PTFE chopping boards | Conveyor belts | Steel equipment | |||||||
| A | DR adjoining secondary PR | Initial washing; foaming alkaline active chlorine detergent (pH 13; >350 ppm free chlorine); final rinse | BC, ACS, AOT40 | 40 | A-DR (990 m3 [2 treatments]) | 6.8 ± 3.6 | 48 | 24 | 24 |
| A-PR (980 m3 [2 treatments]) | 5.1 ± 1.0 | 12 | 24 | 6 | |||||
| B | Secondary PR | Initial washing; foaming alkaline active chlorine detergent (pH 11; >500 ppm free chlorine); final rinse | BC, ACS, AOT20 | 20 | B-PR (400 m3 [3 treatments]) | 4.5 ± 1.5 | 18 | 18 | 45 |
| C | DR | Initial washing; foaming alkaline active chlorine detergent (pH 11; >500 ppm free chlorine); final rinse | BC, ACS, AOT4 | 4 | C-DR (1500 m3 [2 treatments]) | 7.5 ± 1.3 | 36 | 36 | 6 |
Surface categories sampled (average area, ∼1 m2) at each sampling time are reported for each of the four environments (A-DR, A-PR, B-PR, and C-DR). Chopping boards were made of polytetrafluoroethylene (PTFE).
FIG 1.
(a) Stacked bar plots showing the microbiota composition (relative abundance) in phylum, class, and family taxon ranks, with a color coding key; samples are grouped according to the origin (plant and room type) and then according to the temporal phase (BC, ACS, or AOT). (b) Venn diagram displaying the number of shared OTUs at the genus level (or the highest taxonomic rank available when genus determination was not achievable) among the four environments considered (A-DR, A-PR, B-PR, and C-DR). (c) Box plots displaying the logarithmically transformed relative abundances of core OTUs in the four environments; Kruskal-Wallis test P values are displayed, and different box plot colors highlight significantly different abundances (P < 0.05 [FDR adjusted], Kruskal-Wallis and Wilcoxon tests).
Subsequently, we decided to explore the microbiota composition to the genus level (or to the highest taxonomic resolution available when genus identification was not achievable), since the numbers of operational taxonomic units (OTUs) classified to the species level were limited. Overall, we detected a total of 452 OTUs, excluding the singletons. Parametric permutational multivariate analysis of variance (PERMANOVA) performed on this OTU abundance matrix showed that the greatest portion of bacterial community variance was explained by temporal phases and environments (R2 = 0.055 and R2 = 0.052, respectively; P = 0.001), while processing plant and room type contributed to explain smaller portions of the overall variance (R2 = 0.029 and R2 = 0.023, respectively; P = 0.001) (see Table S1 in the supplemental material). Microbiota variance was not affected (P > 0.001) by the surface categories considered, which were chopping board (S1), conveyor belt (S2), and smooth steel surfaces (S3). Accordingly, we analyzed separately the microbiota of the four environments (A-DR, A-PR, B-PR, and C-DR) across the temporal phases considered, whereas data collected from the three surface categories were pooled for each of the four environments.
Regardless of the impact of cleaning/sanitizing and ozonations, 155 of these 452 OTUs were shared by the four environments (Fig. 1b). It is noteworthy that A-DR and A-PR showed greater numbers of environment-specific OTUs (60 and 59 OTUs, respectively) than shared OTUs (32 OTUs), although they were located in the same plant. Within the pool of 155 shared OTUs, the genera Streptococcus, Staphylococcus, Propionibacterium, Acinetobacter, Carnobacterium, Brochothrix, Pseudomonas, Achromobacter, and Psychrobacter, together with the family Burkholderiaceae, constituted the core microbiota of these processing plants, representing more than 50% of the relative abundance in 80% of the samples collected. As far as their spatial partitioning (Fig. 1c), Staphylococcus and Streptococcus were homogeneously distributed across the four environments, while Brochothrix-Carnobacterium and Propionibacterium-Burkholderiaceae were significantly (P < 0.001) more abundant in the environments B-PR and C-DR, respectively. By considering the two room types and thus the meat manipulation activities occurring inside, the OTUs identified as Acinetobacter, Psychrobacter, and Pseudomonas were significantly (P < 0.001) more abundant in the two secondary PR environments, regardless of the processing plant of origin.
Co-occurrence patterns at the sub-OTU level.
A total of 9,888,845 reads assigned to the 10 core OTUs were individually oligotyped in order to investigate their microdiversity at single-nucleotide resolution and to identify co-occurrence patterns at the sub-OTU level across the four environments. Of a total of 1,711 oligotypes, more than 93% were found in all four environments and 96% were homogenously distributed on the three meat contact surfaces considered (Fig. S1).
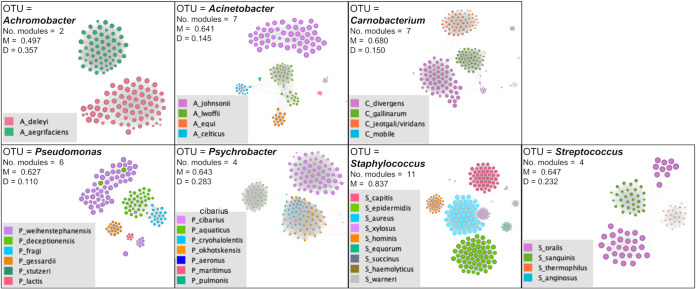
As far as the taxonomic classification of oligotypes is concerned, Burkholderiaceae, Brochothrix, and Propionibacterium encompassed single species, identified as Paraburkholderia fungorum, Brochothrix thermosphacta, and Propionibacterium cyclohexanicum, respectively (Table 2). The remaining seven OTUs harbored a minimum of two (Achromobacter) to a maximum of nine (Staphylococcus) different species. As a first step to investigate the oligotype distribution, we calculated for each OTU the checkboard (C) and togetherness (T) scores, which indicate an environmental pairwise partitioning of the oligotypes based on patterns of presence-absence (high C scores) or aggregation (high T scores). These two indices were negatively correlated with each other (Spearman's correlation, rho = −0.9; P < 0.01), but the C scores were positively correlated with the numbers of oligotypes (rho = 0.82; P < 0.05) and species (rho = 0.80; P < 0.05) found in each OTU.
TABLE 2.
Average C and T scores for oligotype occurrence matrices generated from the 10 selected core OTUs
| OTU identification | Higher taxon classification | Total no. of oligotypes | No. of species | Mean C score | Mean T score | Species (no. of oligotypes) |
|---|---|---|---|---|---|---|
| denovo307699 | Achromobacter | 126 | 2 | 0.58 | 0.14 | Achromobacter deleyi (69 oligotypes), Achromobacter aegrifaciens (57 oligotypes) |
| denovo443152 | Acinetobacter | 158 | 4 | 0.71 | 0.08 | Acinetobacter johnsonii (98 oligotypes), Acinetobacter lwoffii (51 oligotypes), Acinetobacter equi (6 oligotypes), Acinetobacter celticus (3 oligotypes) |
| denovo84598 | Brochothrix | 78 | 1 | 0.46 | 0.18 | Brochothrix thermosphacta |
| denovo440404 | Burkholderiaceae | 71 | 1 | 0.22 | 0.18 | Paraburkholderia fungorum |
| denovo191751 | Carnobacterium | 238 | 4 | 0.82 | 0.02 | Carnobacterium divergens (103 oligotypes), Carnobacterium gallinarum (57 oligotypes), Carnobacterium jeotgali/viridans (56 oligotypes), Carnobacterium mobile (22 oligotypes) |
| denovo629236 | Propionibacterium | 197 | 1 | 0.71 | 0.06 | Propionibacterium cyclohexanicum |
| denovo245059 | Pseudomonas | 155 | 6 | 0.67 | 0.08 | Pseudomonas weihenstephanensis (60 oligotypes), Pseudomonas deceptionensis (39 oligotypes), Pseudomonas fragi (21 oligotypes), Pseudomonas gessardii (19 oligotypes), Pseudomonas stutzeri (9 oligotypes), Pseudomonas lactis (7 oligotypes) |
| denovo210339 | Psychrobacter | 192 | 7 | 0.73 | 0.06 | Psychrobacter cibarius (61 oligotypes), Psychrobacter aquaticus (48 oligotypes), Psychrobacter cryohalolentis (36 oligotypes), Psychrobacter okhotskensis (35 oligotypes), Psychrobacter aeronus (8 oligotypes), Psychrobacter pulmonis (2 oligotypes), Psychrobacter maritimus (2 oligotypes) |
| denovo294860 | Staphylococcus | 349 | 9 | 0.85 | 0.03 | Staphylococcus capitis (59 oligotypes), Staphylococcus epidermidis (56 oligotypes), Staphylococcus aureus (55 oligotypes), Staphylococcus xylosus (50 oligotypes), Staphylococcus hominis (41 oligotypes), Staphylococcus equorum (34 oligotypes), Staphylococcus succinus (29 oligotypes), Staphylococcus haemolyticus (19 oligotypes), Staphylococcus warneri (6 oligotypes) |
| denovo518842 | Streptococcus | 99 | 4 | 0.61 | 0.10 | Streptococcus oralis (36 oligotypes), Streptococcus sanguinis (32 oligotypes), Streptococcus thermophilus (27 oligotypes), Streptococcus anginosus (4 oligotypes) |
Subsequently, the significant positive pairwise correlations existing among oligotypes (Spearman's correlation, rho > 0.6; P [false discovery rate {FDR}] < 0.001) were used to infer a correlation network for each OTU and to visualize subnetwork groups of oligotypes with high rates of co-occurrence, defined as modules. Interestingly, in the Staphylococcus, Streptococcus, Carnobacterium, Acinetobacter, and Achromobacter networks, the oligotypes co-occurred in relation to the species of belonging, while lower correspondence among covarying modules and oligotype species was observed for Pseudomonas and Psychrobacter (Fig. 2). To exclude the possibility that these sub-OTU correlation patterns were simply determined by temporal succession of species during the three phases (BC, ACS, and AOT) or different distributions in the four environments, we assessed the numbers of pairwise correlations existing among oligotypes of the same species in all environments in the three temporal phases. In accordance with the network modules, the majority of positive pairwise correlations (Spearman's correlation, rho > 0.6; P [FDR] < 0.001) occurred among oligotypes of the same species for Staphylococcus, Streptococcus, Acinetobacter, and Achromobacter, highlighting within these genera a spatial species segregation independent of environment and time (Fig. S2); that is, in each of the four environments, the microorganisms of the same species shared the same sampling area during the three temporal phases (BC, ACS, and AOT), excluding the simultaneous presence of other species of the same genus.
FIG 2.
Co-occurrence networks of bacterial oligotypes belonging to Achromobacter, Acinetobacter, Psychrobacter, Pseudomonas, Propionibacterium, Carnobacterium, Streptococcus, and Staphylococcus. Each network is based on the log-transformed relative abundance matrix of each genus, and the edges represent significant positive correlations (co-occurrence) between the oligotypes (nodes) by means of Spearman’s correlations (rho > 0.6; FDR, P < 0.001). Nodes were made proportional to the weighted degree (total occurrence of an oligotype in the whole data set) and are colored in relation to the species of belonging. Edges were made proportional to the Spearman’s rho value. The M and D values shown in the box represent the modularity (clustering coefficient) and density (calculated using the ratio of the number of edges) of each genus-based network.
Effectiveness and selectivity of cleaning/sanitizing procedures and ozonation.
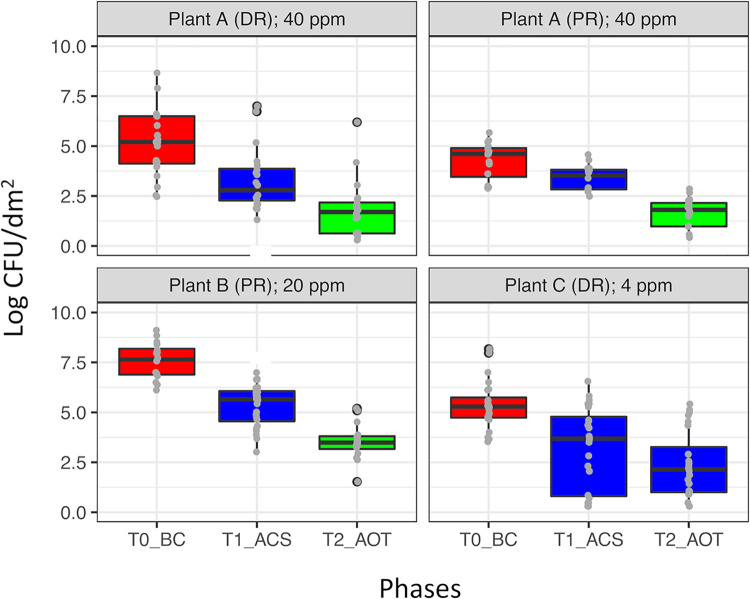
In order to assess the effectiveness of cleaning/sanitizing procedures and the subsequent ozone treatments, the total viable counts (TVCs) of mesophilic bacteria were determined in each of the four environments (A-DR, A-PR, B-PR, and C-DR) after each phase. The TVCs before cleaning ranged from 4 to 8 log CFU/dm2, and the B-PR environment showed the highest counts in all phases (BC, ACS, and AOT). The cleaning/sanitizing procedures reduced significantly (P < 0.01) the TVCs in all environments, whereupon only the ozone treatments at 40 and 20 ppm significantly decreased the remaining TVCs (P < 0.01) (Fig. 3). Taking into account TVC dynamics and the compensatory properties of relative abundance, we regarded the reduction of OTU abundances across the phases as meaningful indices to explain the impact of cleaning/sanitizing and ozonation on the microbiota composition.
FIG 3.
TVCs detected in the four environments (A-DR, A-PR, B-PR, and C-DR) across the three temporal phases, i.e., BC, ACS, and AOT performed at 40 ppm (A-DR and A-PR), 20 ppm (B-PR), or 4 ppm (C-DR) of ozone (concentrations used are displayed for each environment). Box plot colors highlight significant differences (P < 0.05 [FDR adjusted], Kruskal-Wallis and Wilcoxon tests) among the TVCs of the three temporal phases (BC, ACS, and AOT).
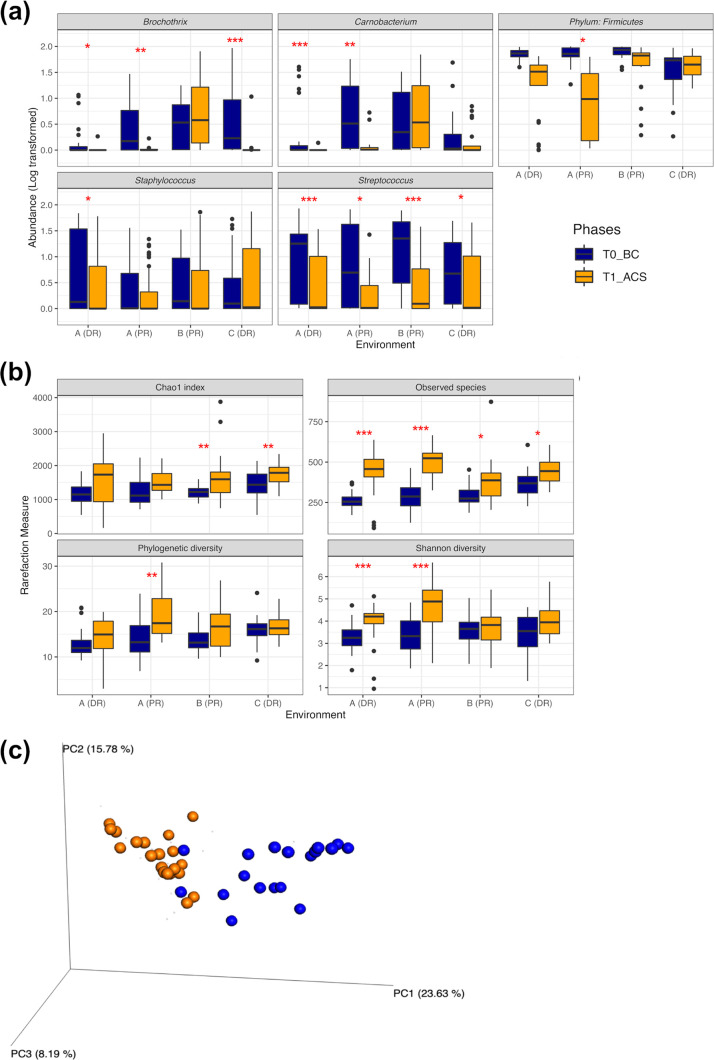
In this regard, after cleaning/sanitizing, the relative abundance of Firmicutes decreased significantly (P < 0.01) in the environments A-DR, A-PR, and B-PR but not in the environment C-DR. Depending on the environments considered, the cleaning/sanitizing procedures determined significant (P < 0.05) reductions of Streptococcus (A-DR, A-PR, and B-PR), Carnobacterium (A-DR and A-PR), Brochothrix (A-DR, A-PR, and C-DR), and Staphylococcus (A-PR) abundances (Fig. 4a). In parallel with these abundance reductions, the numbers of observed species increased significantly (P < 0.05) after these procedures in all environments (Fig. 4b), resulting in a marked increase of Shannon’s diversity (P < 0.001) in the environments of plant A and in the Chao1 index (P < 0.01) in the environments B-PR and C-DR. However, in A-PR only, the microbiota before (BC) and after (ACS) cleaning/sanitizing represented two phylogenetically distinct communities (Fig. 4c), in accordance with both α-diversity (phylogenetic diversity [PD] whole tree, P < 0.01) and β-diversity (weighted UniFrac distance, Adonis results, and analysis of similarities [ANOSIM], P < 0.001) parameters.
FIG 4.
(a and b) Box plots displaying abundance decreases of the core OTUs (a) and the variations of α-diversity measures (b) that occurred in each environment before (BC) (blue) and after (ACS) (orange) the cleaning/sanitizing phase. Asterisks highlight significant differences (FDR-adjusted P value from Wilcoxon’s test); *, 0.05; **, 0.01; ***, 0.001. (c) PCoA chart displaying the weighted UniFrac distance matrix (β-diversity) of the environment PR of plant A before (BC) and after (ACS) cleaning/sanitizing procedures; BC and ACS are different communities (P < 0.001 [FDR adjusted], ANOSIM and Adonis tests).
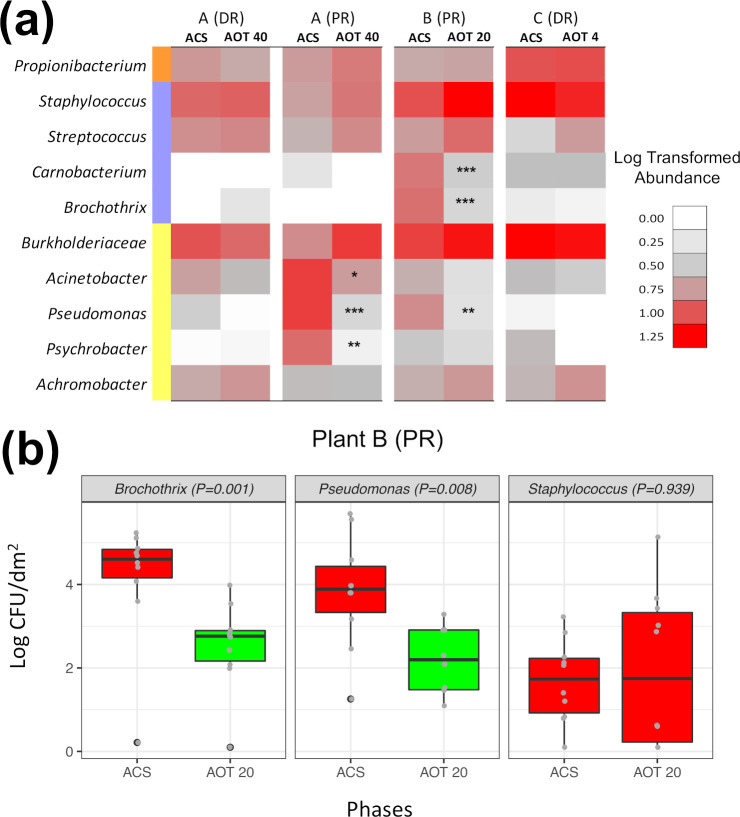
The 40-, 20-, and 4-ppm ozonation treatments did not additionally modify the α- and β-diversity metrics observed after cleaning/sanitizing of plants A, B, and C. In terms of relative abundance variations, Proteobacteria decreased significantly (P < 0.05) after ozonation at 40 ppm performed in the environment A-PR but not in the environment A-DR (data not shown). At the genus level, we observed significant (P < 0.01) decreases of Carnobacterium and Pseudomonas abundances in both secondary PRs treated with 40 and 20 ppm of ozone. In addition, Brochothrix, for which OTU abundance was still marked after cleaning/sanitizing of B-PR, was significantly affected by 20-ppm ozonation carried out in this environment (Fig. 5a). Among the other abundant OTUs, the genera Staphylococcus, Streptococcus, and Propionibacterium and the Burkholderiaceae family did not seem affected by the ozone treatments. To elucidate the extent by which a significant abundance decrease of a given genus was representative of its actual sensitivity to the treatments, microbial counts of Brochothrix, Pseudomonas, and Staphylococcus were determined in the environment B-PR in parallel with the RNA-based surveillance (Fig. 5b). In accordance with the sequencing data, we observed a significant (P < 0.05) decrease of viable counts for Brochothrix and Pseudomonas and a steady Staphylococcus population before and after 20-ppm ozonation (P > 0.05).
FIG 5.
(a) Pseudo-heatmap summarizing the abundance variations of the 10 core OTUs (>50% of the total abundance in >80% of the samples) occurred during the ozone treatments. Asterisks highlight significant decreases in relative abundances in each environment after ozone treatment at 40, 20, or 4 ppm (FDR-adjusted P value from Wilcoxon’s test). *, 0.05; **, 0.01; ***, 0.001. (b) Viable counts of Brochothrix, Pseudomonas, and Staphylococcus before (ACS) and after (AOT 20) the 20-ppm ozonation carried out in the environment B-PR; box plot colors highlight significant differences between ACS and AOT 20 counts (P < 0.05 [FDR adjusted], Wilcoxon’s test).
DISCUSSION
Our study provides the landscape of potentially active microbiota in red meat processing plants and highlights the main shifts in the core microbiota composition occurring after cleaning/sanitizing and ozonation. This RNA-based surveillance was carried out on meat contact surfaces, which are useful indicators for the control of the procedures of cleaning and disinfection (24). The majority of bacterial sequences collected from these surfaces belonged to Actinobacteria, Firmicutes, and Proteobacteria, which are recognized as the dominant phyla in indoor environments with food processing activities (8, 25). Going more deeply in the microbiota composition, meat processing plants showed a greater number of OTUs (400 to 500 OTUs at the genus/species level) than did dairies and meal cooking centers (24, 26, 27), confirming that they harbored more complex microbiota than similar food processing plants (7–9).
Unlike findings observed in ecological surveys of breweries, wineries, cooking centers, and kitchens (24, 25, 28–30), the microbiota composition of each environment did not vary across the surface categories in relation to their structure, layout, or material. We can speculate that here the daily cleaning with pressure washing yields intensive bacterial dispersion and determines a homogeneous distribution of the microbiota across chopping boards, conveyor belts, and smooth steel surfaces of tables and machines. Instead, significant variations of microbiota composition were observed mainly among the environments and the temporal phases, as well as in relation to the type of room. In this regard, the greater abundance and persistence of psychrotrophic taxa (Acinetobacter, Psychrobacter, Pseudomonas, and Brochothrix) in secondary PRs is likely correlated with the lower temperatures of these environments, in comparison with those of the DRs (12). Except for this spatial partitioning, the other abundant OTUs were ubiquitously distributed in all environments.
However, oligotyping analysis and co-occurrence networks highlighted, in each environment and regardless of temporal phase, a meaningful spatial segregation of species belonging to Staphylococcus, Streptococcus, Acinetobacter, and Achromobacter genera. In this frame, intragenus species competition has been already observed for Streptococcus and Staphylococcus on human skin, mouth, and nostrils (31–33), and it is likely that genetically closest ecotypes tend to compete for the same niches also in meat processing plants. Since the extent to which positive correlations among abundances reflects their ecological interactions is still unclear (34, 35), the intragenus species competition hypothesized here needs further investigations, which are beyond the scope of this observational exploratory study. In addition, by oligotyping the core microbiota we investigated to a deeper taxonomic level the potential presence of pathogens in the processing plants, such as in the case of the Burkholderiaceae family, for which all oligotypes were identified as Paraburkholderia fungorum by excluding the presence of the Burkholderia cepacia pathogen complex (36–38).
As far as the two temporal phases considered, cleaning/sanitizing reduced the bacterial counts in all environments. Moreover, in this phase the physical removal of the most abundant human/animal-derived Firmicutes strains increased the number of detectable OTUs, which were previously masked on uncleaned surfaces (39). The bacterial population that survived to cleaning/sanitizing was further reduced by ozone treatments at 40 and 20 ppm, concentrations at which efficacy has been proven for surfaces artificially contaminated with foodborne pathogens (40, 41). The ineffectiveness of the 4-ppm ozone treatment is in contrast to its in vitro efficacy for shorter treatment times (41). The complexity of treated surfaces (roughness, porosity, and notches), the high contamination level, and the presence of highly oxidizable residues can limit the bacterial inactivation and killing capabilities of ozone, which would explain the differences between the previous in vitro outcomes and our in situ observations (13, 42, 43). Taking into account the low temperature of these environments, we must also consider that cold stresses can increase bacterial resistance to ozone (44). All together, these observations pointed out the need to confirm on site, in each food processing environment, the efficacy of any ozone treatment previously observed in vitro.
Focusing on ozone selectivity, since the N-acetylglucosamine constituting the peptidoglycan layer is poorly oxidizable, Gram-positive organisms are generally considered more resistant than Gram-negative organisms (13, 20, 45). The abundance decreases of Proteobacteria observed in one of the two environments treated with ozone at 40 ppm seem to confirm this assumption. In parallel, the discrepancies in bacterial inactivation produced by the same ozonation protocol in two different environments highlight how the selectivity of any antimicrobial treatment is dependent on the initial composition of the treated microbiota, both in foods and in processing environments (46, 47). Accordingly, only in the two secondary PRs (A-PR and B-PR) did the higher concentrations of ozone (40 and 20 ppm) show a selective inactivation capability toward bacteria, as already observed in several survival assays (14, 15, 41, 48). In terms of relative abundance, the psychrotrophic genera associated with these environments (Pseudomonas, Psychrobacter, Brochothrix, and Carnobacterium) showed significant sensitivity to ozone, while a remarkable resistance was observed for the ubiquitous Staphylococcus, Streptococcus, Proteobacterium, and Burkholderiaceae. We are aware that RNA-based amplicon sequencing has a limited quantitative capability. However, the rRNA abundance of a given taxon can indicate its ecological potential for growth and acclimation in response to external stimuli like ozone with greater reliability than other sequencing methods (49, 50). This was confirmed in the present study; concordance was observed in the trend of the viable counts and the relative abundance by rRNA amplicon sequencing for Brochothrix, Pseudomonas, and Staphylococcus. Focusing on the survival capability of these genera with ozone, the literature data are often discordant and direct comparisons are difficult or pointless in relation to different treatment times, ozonation modes, and detection methods, as well as the aforementioned discrepancies between in vitro and in situ conditions (14, 15, 23, 41, 48, 51, 52). Beside the comparison, it is noteworthy that high ozone concentrations selectively inactivated Brochothrix and Pseudomonas, which survived to cleaning/sanitizing and are commonly responsible of meat spoilage during the product shelf life (53–55).
In summary, we showed that high-concentration ozonations are effective antimicrobial treatments to be used in addition to and complementing current cleaning/sanitizing procedures in meat processing plants. The selective bacterial inactivation observed after single treatments undoubtedly needs additional investigations, in order to clearly define the effects of prolonged and routine ozone exposures on the resident microbiota of these and other food processing plants. Although the practical conclusion of this survey can be dichotomously summarized as effectiveness or ineffectiveness of a given treatment, based on microbial count, the complementary 16S rRNA metataxonomic analysis performed here allowed us to understand how the layout and organization of a food processing environment impact the microbial distribution and thus affect the sanitizing interventions. Similar approaches will be largely used by food industries to make final decisions regarding sanitizing aspects and product shelf life.
MATERIALS AND METHODS
Ozonation treatments, sampling procedures, and microbiological analysis.
Three red meat processing environments located in the same geographical area (Piedmont, northwest Italy) were treated for 12 h with gaseous ozone. Briefly, the environment A, which composed of two rooms (PR and DR), was treated with 40 ppm of ozone, while the single rooms of the environments B (PR) and C (DR) were treated with 20 and 4 ppm of ozone, respectively (Table 1). A minimum of two independent treatments were performed in each plant (A, B, and C), at 3-week intervals, from January to March 2018. The environmental ozone treatments were carried out during the weekly closing days of the slaughterhouses after the completion of their routine cleaning and sanitizing procedures. The gas was produced by using an ozone generator (model Power-H; Industrie De Nora SpA, MI, Italy) fed by pure oxygen, with a nominal production capacity of 100 g O3/h, and inflated into sealed rooms, and the effective concentrations were constantly monitored by the recirculation of ozone-enriched air from the chamber through a BMT 964 UV-photometric ozone analyzer (BMT Messtechnik Gmbh, Germany), which controlled the ozone generator output. In each room, up to 11 environmental samples (9,118 ± 215 cm2, on average) were chosen from three different categories of meat contact surfaces, i.e., (i) chopping boards (Teflon); (ii) conveyor belts (Teflon and rubber); and (iii) machines, equipment, and tables (smooth steel surfaces). Knives and small equipment (saws, slicer blades, and internal mincer parts) were not sampled since they undergo specific cleaning/sanitizing procedures and are often removed from the environment at the end of butchering activities. The same areas were sampled at three different time points, i.e., before the cleaning procedures (BC); 1 h after cleaning and sanitizing (ACS); and after the end of ozone treatments (AOT), which included the 12-h treatment and the further gas decay (from 8 to 24 h; final concentration, <1 ppm). The surface samples were aseptically collected with sponges (sterile sample bags with specimen sponges; VWR International, Leuven, Belgium) that had been previously hydrated with 10 ml of buffered peptone water (BPW; Sigma), stored at 5°C, and analyzed.
After a further addition of 5 ml of BPW, the sponges were manually massaged for 2 min and squeezed, yielding approximately 10 ml of suspension. An aliquot of 5 ml was centrifuged, and the pellet was collected and stored with RNA-later (Ambion, Thermo Scientific, Milan, Italy) at − 80°C until RNA extraction and amplicon-based sequencing analysis were performed.
The remaining 5 ml was added to 45 ml of BPW and used to perform microbiological analyses by counting TVCs of mesophilic bacteria on plate count agar (Biolife s.p.a., Milan, Italy) incubated at 30°C for 72 h, Brochothrix spp. on streptomycin-thallous acetate-actidione medium (Biolife) incubated at 25°C for 48 h, Pseudomonas spp. on Pseudomonas agar base with cetrimide-fucidin-cephaloridine selective supplement (Biolife) incubated at 25°C for 48 h, and Staphylococcaceae on mannitol salt agar (Oxoid) incubated at 30°C for 48 h. Microbiological data were expressed and statistically analyzed as log-transformed CFU per square decimeter.
RNA extraction and cDNA synthesis.
Total RNA was extracted using the Master Pure complete DNA and RNA purification kit (Epicentre, Madison, WI, USA) according to the manufacturer’s instructions. Three microliters of Turbo DNase (Ambion) was added to digest the DNA in the RNA samples, with an incubation of 3 h at 37°C. The complete denaturation of genomic DNA was confirmed by PCR amplification of the partial 16S rRNA gene, using forward primer FD1 and reverse primer RD1 (56). The quality of the extracted RNA was evaluated and quantified using a NanoDrop spectrophotometer (Thermo Scientific, Milan, Italy). The cDNA was synthesized from 2 μg of RNA with the Moloney murine leukemia virus reverse transcriptase system (Promega, Milan, Italy), as described previously (46).
Amplification, sequencing, and data processing.
V3 to V4 region libraries were constructed from the 16S rRNA gene region by using primers and conditions described previously (46). The PCR products were purified by means of an Agencourt AMPure kit (Beckman Coulter, Milan, Italy), and the resulting products were tagged with sequencing adapters using the Nextera XT library preparation kit (Illumina, San Diego, CA), according to the manufacturer’s instructions. Sequencing was performed using an Illumina MiSeq system with v3 chemistry, which generated 2 × 250-bp paired-end reads. MiSeq Control Software v2.3.0.3, RTA v1.18.42.0, and CASAVA v1.8.2 were used for the base calling and Illumina barcode demultiplexing processes.
Paired-end reads were first assembled using FLASH software (57) with default parameters. Joint reads were further quality filtered (at Phred scores of <Q20) using QIIME v1.9.0 software (58), and short reads (<250 bp) were discarded through Prinseq (59). Chimera filtering was performed with USEARCH software v8.1 (60). OTUs were picked at a 97% similarity threshold by UCLUST (60) algorithms, and centroid sequences of each cluster were matched to the Greengenes 16S rRNA gene database v2013.
After sequencing, a total of 18,952,580 raw reads (2 × 250 bp) were obtained from 297 samples. After merging, a total of 13,643,420 reads passed the filters applied by QIIME, with an average value of 48,802 reads/sample and a sequence length of 460 bp. In order to avoid biases due to the different sequencing depths, OTU tables were rarefied to 5,000 reads/sample by limiting the further analysis to 278 samples. The rarefaction analysis and Good’s coverage percentages indicated an average value of 94.1%. When the taxonomy assignment was not able to reach the genus, the highest taxonomic level available was displayed.
Oligotyping.
Within-OTU diversity was resolved using the oligotyping procedure described previously (61). Briefly, all reads assigned to the 10 most abundant OTUs were extracted, and Shannon’s entropy analysis was performed in order to identify positional variations at the single-nucleotide level that explained the biological diversity within each OTU. The list of OTUs oligotyped, taxon assignments, initial numbers of reads, average Shannon’s entropy values, high-entropy positions chosen (“C” option), and minimum substantive abundance values (“M” option) are summarized in Table S2 in the supplemental material. Moreover, each oligotype was required to appear in at least 10 samples at 1.0% abundance (“A” option), to reduce the noise generated by low-abundance oligotypes. BLASTn (https://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to query the representative oligotype sequences against the NCBI database, and the top hits (higher percent identity values) were considered for taxonomic assignment.
Statistical analysis.
Statistical analyses and data plotting were performed using the R program for Statistical Computing v3.6.0 (http://www.r-project.org) unless otherwise stated. In detail, the normality and homogeneity of the data (log-transformed abundances and counts) were checked by means of the Shapiro-Wilk W test and Levene’s tests, respectively. Kruskal–Wallis nonparametric tests were used to assess the overall variation and differences between the multiple groups. Comparisons between individual groups were performed with Wilcoxon’s test. Non-normally distributed variables were presented in box plots as medians (with the interquartile range, minimum and maximum values, and outliers).
PERMANOVA was conducted to quantitatively evaluate the effects of each categorical variable (plant, environment, room type, surface category, and temporal phase) or their interactions on the variations in bacterial communities (OTUs abundances) using the Adonis function based on 999 permutations and Brey-Curtis dissimilarity distances. Distribution of the OTUs and oligotypes along the environments or surface categories was visualized with a Venn diagram by using Venny v2.1 (http://bioinfogp.cnb.csic.es/tools/venny).
The α-diversity was assessed by analyzing the number of observed species, the Chao1 index, the Shannon-Wiener diversity index H′, and the PD whole tree. All parameters were calculated using the diversity function of the vegan package (62). The β-diversity among bacterial communities was visualized by plotting the principal-coordinate analysis (PCoA) of the weighted UniFrac distance matrix with EMPeror (63); Adonis and ANOSIM statistical tests were used to detect significant differences among the microbial communities, as considered by using the weighted UniFrac distance matrices.
For each OTU analyzed, we calculated the mean checkerboard (C) and togetherness (T) scores among oligotypes using the R package bipartite. The co-occurrence among oligotypes was investigated by analyzing the log-transformed abundances with Spearman's rank correlation, which is recommended as a robust method for nonparametric data (64); positive correlations were considered significant with Spearman's rho values of >0.6 and P values [FDR] of <0.001. Significant correlations were used to infer co-occurrence networks and to detect the presence of recurrent subnetwork modules (groups of oligotype that covary) through the algorithm described by Blondel and colleagues (65). Oligotype networks were visualized using the program Gephi v0.9.2-beta (https://gephi.org).
Data availability.
Sequencing data were deposited in the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA629433.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by the Piedmont Region, Italy, through the P.O.R. FESR 2014–2020 financing system, under European Commission decision C (2017) 6892.
We thank Maria Chiara Cavallero (Polo AGRIFOOD, Dronero, Italy) and Simonetta Riva (Veterinary Food Safety, Salmour, Italy) for invaluable help during the experiments, as well as the staffs of the participating abattoirs for their fruitful collaboration, in particular, Cristina Candela, Barbara Rossi, and Gianluca Scrivano. We are also grateful to Cristian Carboni and the staffs of ozone technology facilities (Industrie De Nora S.p.a and Emporio Enologico Albese s.r.l.) for their help and technical support, as well as to Elisabetta Genta (Laemmegroup s.r.l., Italy) for preliminary discussion of the experimental plan.
Laemmegroup s.r.l. is a Tentamus Company.
C.B., L.C., and K.R. designed and conducted the research. C.B. and A.P. conducted the microbiological analysis. C.B. and I.F. performed the RNA extraction and cDNA synthesis. I.F. carried out the 16S rRNA gene sequencing and the bioinformatic analyses. C.B. performed the statistical analysis, generated the manuscript figures, and analyzed the data. C.B. wrote the main manuscript text. I.F. supervised the data analysis and contributed to manuscript preparation. All authors read and approved the final manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Doyle CJ, O'Toole PW, Cotter PD. 2017. Metagenome-based surveillance and diagnostic approaches to studying the microbial ecology of food production and processing environments. Environ Microbiol 19:4382–4391. doi: 10.1111/1462-2920.13859. [DOI] [PubMed] [Google Scholar]
- 2.Yang X, Tran F, Youssef MK, Gill CO. 2015. Determination of sources of Escherichia coli on beef by multiple-locus variable-number tandem repeat analysis. J Food Prot 78:1296–1302. doi: 10.4315/0362-028X.JFP-15-014. [DOI] [PubMed] [Google Scholar]
- 3.Williams GD, Keener KM. 2013. Design considerations for the construction and operation of meat and poultry processing facilities. ASABE Meet, paper 082382. doi: 10.13031/2013.24882. [DOI]
- 4.Nel S, Lues JFR, Buys EM, Venter P. 2004. Bacterial populations associated with meat from the deboning room of a high throughput red meat abattoir. Meat Sci 66:667–674. doi: 10.1016/S0309-1740(03)00187-6. [DOI] [PubMed] [Google Scholar]
- 5.Shale K, Jacoby A, Plaatjies Z. 2006. The impact of extrinsic sources on selected indicator organisms in a typical deboning room. Int J Environ Health Res 16:263–272. doi: 10.1080/09603120600734162. [DOI] [PubMed] [Google Scholar]
- 6.Kaur M, Bowman JP, Porteus B, Dann AL, Tamplin M. 2017. Effect of abattoir and cut on variations in microbial communities of vacuum-packaged beef. Meat Sci 131:34–39. doi: 10.1016/j.meatsci.2017.04.021. [DOI] [PubMed] [Google Scholar]
- 7.Stellato G, La Storia A, De Filippis F, Borriello G, Villani F, Ercolini D. 2016. Overlap of spoilage microbiota between meat and meat processing environment in small-scale 2 vs large-scale retail distribution. Appl Environ Microbiol 82:4045–4054. doi: 10.1128/AEM.00793-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hultman J, Rahkila R, Ali J, Rousu J, Björkroth KJ. 2015. Meat processing plant microbiome and contamination patterns of cold-tolerant bacteria causing food safety and spoilage risks in the manufacture of vacuum-packaged cooked sausages. Appl Environ Microbiol 81:7088–7097. doi: 10.1128/AEM.02228-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Filippis F, La Storia A, Villani F, Ercolini D. 2013. Exploring the sources of bacterial spoilers in beefsteaks by culture-independent high-throughput sequencing. PLoS One 8:e70222. doi: 10.1371/journal.pone.0070222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaillou S, Chaulot-Talmon A, Caekebeke H, Cardinal M, Christieans S, Denis C, Desmonts MH, Dousset X, Feurer C, Hamon E, Joffraud J-J, La Carbona S, Leroi F, Leroy S, Lorre S, Macé S, Pilet M-F, Prévost H, Rivollier M, Roux D, Talon R, Zagorec M, Champomier-Vergès M-C. 2015. Origin and ecological selection of core and food-specific bacterial communities associated with meat and seafood spoilage. ISME J 9:1105–1118. doi: 10.1038/ismej.2014.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giaouris E, Heir E, Hébraud M, Chorianopoulos N, Langsrud S, Møretrø T, Habimana O, Desvaux M, Renier S, Nychas GJ. 2014. Attachment and biofilm formation by foodborne bacteria in meat processing environments: causes, implications, role of bacterial interactions and control by alternative novel methods. Meat Sci 97:289–309. doi: 10.1016/j.meatsci.2013.05.023. [DOI] [PubMed] [Google Scholar]
- 12.Møretrø T, Langsrud S. 2017. Residential bacteria on surfaces in the food industry and their implications for food safety and quality. Compr Rev Food Sci Food Saf 16:1022–1041. doi: 10.1111/1541-4337.12283. [DOI] [PubMed] [Google Scholar]
- 13.Khadre MA, Yousef AE, Kim J-G. 2001. Microbiological aspects of ozone applications in food: a review. J Food Sci 66:1242–1252. doi: 10.1111/j.1365-2621.2001.tb15196.x. [DOI] [Google Scholar]
- 14.Fontes B, Cattani Heimbecker AM, de Souza Brito G, Costa SF, van der Heijden IM, Levin AS, Rasslan S. 2012. Effect of low-dose gaseous ozone on pathogenic bacteria. BMC Infect Dis 12:358–357. doi: 10.1186/1471-2334-12-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Candia S, Morea M, Baruzzi F. 2015. Eradication of high viable loads of Listeria monocytogenes contaminating food-contact surfaces. Front Microbiol 6:733. doi: 10.3389/fmicb.2015.00733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hudson JB, Sharma M. 2009. The practical application of ozone gas as an anti-fungal (anti-mold) agent. Ozone Sci Eng 31:326–332. doi: 10.1080/01919510903043996. [DOI] [Google Scholar]
- 17.Iacumin L, Manzano M, Comi G. 2012. Prevention of Aspergillus ochraceus growth on and Ochratoxin A contamination of sausages using ozonated air. Food Microbiol 29:229–232. doi: 10.1016/j.fm.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 18.Bellincontro A, Catelli C, Cotarella R, Mencarelli F. 2017. Postharvest ozone fumigation of Petit Verdot grapes to prevent the use of sulfites and to increase anthocyanin in wine. Aust J Grape Wine Res 23:200–206. doi: 10.1111/ajgw.12257. [DOI] [Google Scholar]
- 19.Freitas-Silva O, Venâncio A. 2010. Ozone applications to prevent and degrade mycotoxins: a review. Drug Metab Rev 42:612–620. doi: 10.3109/03602532.2010.484461. [DOI] [PubMed] [Google Scholar]
- 20.Thanomsub B, Anupunpisit V, Chanphetch S, Watcharachaipong T, Poonkhum R, Srisukonth C. 2002. Effects of ozone treatment on cell growth and ultrastructural changes in bacteria. J Gen Appl Microbiol 48:193–199. doi: 10.2323/jgam.48.193. [DOI] [PubMed] [Google Scholar]
- 21.Girgin Ersoy Z, Barisci S, Dinc O. 2019. Mechanisms of the Escherichia coli and Enterococcus faecalis inactivation by ozone. LWT 100:306–313. doi: 10.1016/j.lwt.2018.10.095. [DOI] [Google Scholar]
- 22.Patil S, Valdramidis VP, Karatzas KAG, Cullen PJ, Bourke P. 2011. Assessing the microbial oxidative stress mechanism of ozone treatment through the responses of Escherichia coli mutants. J Appl Microbiol 111:136–144. doi: 10.1111/j.1365-2672.2011.05021.x. [DOI] [PubMed] [Google Scholar]
- 23.Alexander J, Knopp G, Dötsch A, Wieland A, Schwartz T. 2016. Ozone treatment of conditioned wastewater selects antibiotic resistance genes, opportunistic bacteria, and induce strong population shifts. Sci Total Environ 559:103–112. doi: 10.1016/j.scitotenv.2016.03.154. [DOI] [PubMed] [Google Scholar]
- 24.Stellato G, La Storia A, Cirillo T, Ercolini D. 2015. Bacterial biogeographical patterns in a cooking center for hospital foodservice. Int J Food Microbiol 193:99–108. doi: 10.1016/j.ijfoodmicro.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 25.Flores GE, Bates ST, Caporaso JG, Lauber CL, Leff JW, Knight R, Fierer N. 2013. Diversity, distribution and sources of bacteria in residential kitchens. Environ Microbiol 15:588–596. doi: 10.1111/1462-2920.12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pothakos V, Stellato G, Ercolini D, Devlieghere F. 2015. Processing environment and ingredients are both sources of Leuconostoc gelidum, which emerges as a major spoiler in ready-to-eat meals. Appl Environ Microbiol 81:3529–3541. doi: 10.1128/AEM.03941-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stellato G, De Filippis F, La Storia A, Ercolini D. 2015. Coexistence of lactic acid bacteria and potential spoilage microbiota. Appl Environ Microbiol 81:7893–7904. doi: 10.1128/AEM.02294-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bokulich NA, Bergsveinson J, Ziola B, Mills DA. 2015. Mapping microbial ecosystems and spoilage-gene flow in breweries highlights patterns of contamination and resistance. eLife 4:e04634. doi: 10.7554/eLife.04634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bokulich NA, Mills DA. 2013. Facility-specific “house” microbiome drives microbial landscapes of artisan cheesemaking plants. Appl Environ Microbiol 79:5214–5223. doi: 10.1128/AEM.00934-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bokulich NA, Ohta M, Richardson PM, Mills DA. 2013. Monitoring seasonal changes in winery-resident microbiota. PLoS One 8:e66437. doi: 10.1371/journal.pone.0066437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kreth J, Merritt J, Shi W, Qi F. 2005. Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J Bacteriol 187:7193–7203. doi: 10.1128/JB.187.21.7193-7203.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tristan A, Vandenesch F, Lina G, Boutite F, Bes M, Etienne J. 2003. Bacterial competition for human nasal cavity colonization: role of staphylococcal agr alleles. Appl Environ Microbiol 69:18–23. doi: 10.1128/aem.69.1.18-23.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Canovas J, Baldry M, Bojer MS, Andersen PS, Grzeskowiak PK, Stegger M, Damborg P, Olsen CA, Ingmer H. 2016. Cross-talk between Staphylococcus aureus and other staphylococcal species via the agr quorum sensing system. Front Microbiol 7:1733. doi: 10.3389/fmicb.2016.01733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berry MA, White JD, Davis TW, Jain S, Johengen TH, Dick GJ, Sarnelle O, Denef VJ. 2017. Are oligotypes meaningful ecological and phylogenetic units? A case study of microcystis in freshwater lakes. Front Microbiol 8:365. doi: 10.3389/fmicb.2017.00365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Freilich MA, Wieters E, Broitman BR, Marquet PA, Navarrete SA. 2018. Species co-occurrence networks: can they reveal trophic and non-trophic interactions in ecological communities? Ecology 99:690–699. doi: 10.1002/ecy.2142. [DOI] [PubMed] [Google Scholar]
- 36.Eberl L, Vandamme P. 2016. Members of the genus Burkholderia: good and bad guys. F1000Res 5:1007. doi: 10.12688/f1000research.8221.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morici L, Torres AG, Titball RW. 2019. Novel multi-component vaccine approaches for Burkholderia pseudomallei. Clin Exp Immunol 196:178–188. doi: 10.1111/cei.13286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sass A, Slachmuylders L, Van Acker H, Vandenbussche I, Ostyn L, Bové M, Crabbé A, Chiarelli LR, Buroni S, Van Nieuwerburgh F, Abatih E, Coenye T. 2019. Various evolutionary trajectories lead to loss of the tobramycin-potentiating activity of the quorum-sensing inhibitor baicalin hydrate in Burkholderia cenocepacia biofilms. Antimicrob Agents Chemother 63:e02092-18. doi: 10.1128/AAC.02092-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang H, He A, Yang X. 2018. Dynamics of microflora on conveyor belts in a beef fabrication facility during sanitation. Food Control 85:42–47. doi: 10.1016/j.foodcont.2017.09.017. [DOI] [Google Scholar]
- 40.Nicholas R, Dunton P, Tatham A, Fielding L. 2013. The effect of ozone and open air factor on surface-attached and biofilm environmental Listeria monocytogenes. J Appl Microbiol 115:555–564. doi: 10.1111/jam.12239. [DOI] [PubMed] [Google Scholar]
- 41.Marino M, Maifreni M, Baggio A, Innocente N. 2018. Inactivation of foodborne bacteria biofilms by aqueous and gaseous ozone. Front Microbiol 9:2024. doi: 10.3389/fmicb.2018.02024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Megahed A, Aldridge B, Lowe J. 2018. The microbial killing capacity of aqueous and gaseous ozone on different surfaces contaminated with dairy cattle manure. PLoS One 13:e0196555. doi: 10.1371/journal.pone.0196555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller FA, Silva CLM, Brandao TRS. 2013. A review on ozone-based treatments for fruit and vegetables preservation. Food Eng Rev 5:77–106. doi: 10.1007/s12393-013-9064-5. [DOI] [Google Scholar]
- 44.Wani S, Barnes J, Singleton I. 2016. Investigation of potential reasons for bacterial survival on “ready-to-eat” leafy produce during exposure to gaseous ozone. Postharvest Biol Technol 111:185–190. doi: 10.1016/j.postharvbio.2015.08.007. [DOI] [Google Scholar]
- 45.Moore G, Griffith C, Peters A. 2000. Bactericidal properties of ozone and its potential application as a terminal disinfectant. J Food Prot 63:1100–1106. doi: 10.4315/0362-028X-63.8.1100. [DOI] [PubMed] [Google Scholar]
- 46.Botta C, Ferrocino I, Cavallero MC, Riva S, Giordano M, Cocolin L. 2018. Potentially active spoilage bacteria community during the storage of vacuum packaged beefsteaks treated with aqueous ozone and electrolyzed water. Int J Food Microbiol 266:337–345. doi: 10.1016/j.ijfoodmicro.2017.10.012. [DOI] [PubMed] [Google Scholar]
- 47.Ferrocino I, Greppi A, La Storia A, Rantsiou K, Ercolini D, Cocolin L. 2016. Impact of nisin-activated packaging on microbiota of beef burgers during storage. Appl Environ Microbiol 82:549–559. doi: 10.1128/AEM.03093-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tachikawa M, Yamanaka K, Nakamuro K. 2009. Studies on the disinfection and removal of biofilms by ozone water using an artificial microbial biofilm system. Ozone Sci Eng 31:3–9. doi: 10.1080/01919510802586566. [DOI] [Google Scholar]
- 49.Blazewicz SJ, Barnard RL, Daly RA, Firestone MK. 2013. Evaluating rRNA as an indicator of microbial activity in environmental communities: limitations and uses. ISME J 7:2061–2068. doi: 10.1038/ismej.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li R, Tun HM, Jahan M, Zhang Z, Kumar A, Fernando D, Farenhorst A, Khafipour E. 2017. Comparison of DNA-, PMA-, and RNA-based 16S rRNA Illumina sequencing for detection of live bacteria in water. Sci Rep 7:5752. doi: 10.1038/s41598-017-02516-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Restaino L, Frampton EW, Hemphill JB, Palnikar P. 1995. Efficacy of ozonated water against various food-related microorganisms. Appl Environ Microbiol 61:3471–3475. doi: 10.1128/AEM.61.9.3471-3475.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Choudhury B, Portugal S, Mastanaiah N, Johnson JA, Roy S. 2018. Inactivation of Pseudomonas aeruginosa and methicillin-resistant Staphylococcus aureus in an open water system with ozone generated by a compact, atmospheric DBD plasma reactor. Sci Rep 8:17573. doi: 10.1038/s41598-018-36003-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Casaburi A, De Filippis F, Villani F, Ercolini D. 2014. Activities of strains of Brochothrix thermosphacta in vitro and in meat. Food Res Int 62:366–374. doi: 10.1016/j.foodres.2014.03.019. [DOI] [Google Scholar]
- 54.Papadopoulou OS, Doulgeraki AI, Botta C, Cocolin L, Nychas G-JE. 2012. Genotypic characterization of Brochothrix thermosphacta isolated during storage of minced pork under aerobic or modified atmosphere packaging conditions. Meat Sci 92:735–738. doi: 10.1016/j.meatsci.2012.06.030. [DOI] [PubMed] [Google Scholar]
- 55.De Filippis F, La Storia A, Villani F, Ercolini D. 2018. Strain-level diversity analysis of Pseudomonas fragi after in situ pangenome reconstruction shows distinctive spoilage-associated metabolic traits clearly selected by different storage conditions. Appl Environ Microbiol 85:e02212-18. doi: 10.1128/AEM.02212-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 1991. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Magoč T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eren AM, Maignien L, Sul WJ, Murphy LG, Grim SL, Morrison HG, Sogin ML. 2013. Oligotyping: differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol Evol 4:1111–1119. doi: 10.1111/2041-210X.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dixon P. 2003. VEGAN, a package of R functions for community ecology. J Veg Sci 14:927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x. [DOI] [Google Scholar]
- 63.Vázquez-Baeza Y, Pirrung M, Gonzalez A, Knight R. 2013. EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience 2:16. doi: 10.1186/2047-217X-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weiss S, Van Treuren W, Lozupone C, Faust K, Friedman J, Deng Y, Xia LC, Xu ZZ, Ursell L, Alm EJ, Birmingham A, Cram JA, Fuhrman JA, Raes J, Sun F, Zhou J, Knight R. 2016. Correlation detection strategies in microbial data sets vary widely in sensitivity and precision. ISME J 10:1669–1681. doi: 10.1038/ismej.2015.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blondel VD, Guillaume JL, Lambiotte R, Lefebvre E. 2008. Fast unfolding of communities in large networks. J Stat Mech 2008:P10008. doi: 10.1088/1742-5468/2008/10/P10008. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data were deposited in the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA629433.