Abstract
Background
The Chromobox (CBX) protein family, which is a crucial part of the epigenetic regulatory complex, plays an important role in the occurrence and development of cancer; however, the function and prognostic value of CBX family members in gastric cancer is not clear.
Methods
we investigated the relationship between CBX members and gastric cancer using a range of tools and databases: Oncomine, Kaplan–Meier plotter, cBioPortal, ULCAN, Metascape, and GEPIA.
Results
The results showed that, relative to normal gastric tissue, mRNA expression levels of CBX1-6 were significantly higher in gastric cancer tissue, whereas the level of CBX7 was significantly lower. Furthermore, overexpression of CBX3-6 and underexpression of CBX7 mRNAs was significantly related to the poor prognosis and survival of gastric cancer patients, making these CBX family members useful biomarkers. Finally, overexpression of CBX1 mRNA was significantly related to the poor prognosis of gastric cancer patients treated with adjuvant 5-fluorouracil-based chemotherapy.
Conclusions
The members of the CBX family can be used as prognosis and survival biomarkers for gastric cancer and CBX1 may be a biomarker for choosing the chemotherapy regimen of gastric cancer patients.
Keywords: Chromobox (CBX) protein, gastric cancer, prognosis
Introduction
Globally, gastric cancer is the third leading cause of cancer death with ~1 million new gastric cancer cases reported annually worldwide (1). The known risk factors for gastric cancer include Helicobacter pylori infection, the intake of salt and pickled foods, and a lack of dietary fruits and vegetables (2). The occurrence of gastric cancer is a complex, multi-step pathological process involving multiple, interacting factors, e.g., Helicobacter pylori infection, diet, and heredity (3). Currently, the mechanism of gastric cancer formation remains to be fully elucidated (4); therefore, it is necessary to develop our understanding of the pathogenesis of gastric cancer in order to identify biomarkers of treatment and prognosis.
Recent studies have shown that abnormal epigenetic regulation plays an important role in the occurrence and development of gastric cancer (5). Polycomb Repressor Complex 1 (PCR1) is an epigenetic regulatory complex; its main function is to suppress and thereby silence target genes (6). Chromobox (CBX) family proteins are the core members of PCR1; these proteins can regulate the occurrence and development of many types of cancer by promoting the self-renewal of tumor stem cells (7). Eight CBX proteins have been identified in the human genome, with links primarily to heterochromatin, gene expression, apoptosis, and the regulation of development (6).
Members of the CBX family are known to be abnormally overexpressed in tumorigenesis and affect tumor prognosis. For example, overexpression of CBX8 in hepatocellular carcinoma patients is related to short overall survival time (OS) (8). In addition, CBX7 can be activated by the AKT pathway and downregulated by the tumor suppressor protein p16 in gastric cancer, which plays an important role in maintaining the stem cell-like characteristics of gastric cancer cells (9). However, the roles of different CBX family members in the occurrence and development of gastric cancer remain unclear. Therefore, in the present study, we used a range of tools and databases to investigate the relationship between CBX family members and gastric cancer. Specifically, we used Oncomine, Kaplan-Meier plotter, cBioPortal, Metascape, and GEPIA (Gene Expression Profiling Interactive Analysis) to analyze the expression, mutation, and clinical relationships of CBX proteins in gastric cancer patients and to predict their potential functions and prognostic values.
We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/jgo-20-223).
Methods
Oncomine database
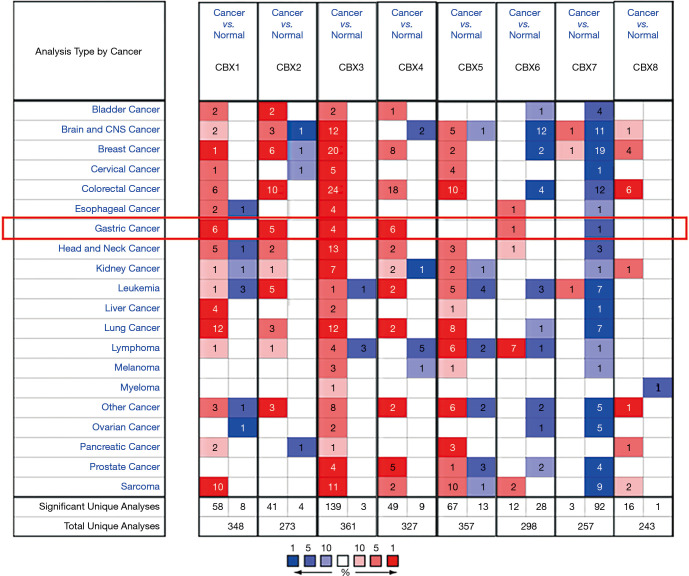
As shown in Table 1, we analyzed the mRNA expression of eight CBX family members in various gastric cancer types using data from four studies, Cho et al. (10), Chen et al. (11), D’Errico et al. (12), and Wang et al. (13), and Oncomine (www.oncomine.org), an integrated online cancer microarray database for DNA or RNA sequence analysis that facilitates discovery from gene-wide expression analyses (14). The expression of CBX mRNAs in clinical tumors and normal control tissues was compared by Student’s t-test. Sufficient fold change and significant p values were defined as 1.5 and 0.01, respectively.
Table 1. Significant changes in Chromobox (CBX) family transcription levels in gastric cancer tissues relative to normal gastric tissue.
| Cancer types | Fold change | P value | t-test | Source data (reference) | |
|---|---|---|---|---|---|
| CBX1 | Gastric adenocarcinoma | 2.415 | 4.52E-06 | 6.913 | Cho et al. (10) |
| Gastric mixed adenocarcinoma | 1.660 | 2.25E-06 | 7.026 | Chen et al. (11) | |
| Gastric intestinal-type adenocarcinoma | 1.613 | 2.38E-13 | 9.531 | Chen et al. (11) | |
| Gastric intestinal-type adenocarcinoma | 2.116 | 2.21E-13 | 10.157 | D’Errico et al. (12) | |
| Diffuse gastric adenocarcinoma | 1.516 | 2.83E-08 | 6.996 | Chen et al. (11) | |
| Diffuse gastric adenocarcinoma | 1.730 | 5.00E-03 | 3.756 | D’Errico et al. (12) | |
| CBX2 | Gastric cancer | 2.501 | 2.00E-03 | 3.330 | Wang et al. (13) |
| Gastric mixed adenocarcinoma | 2.077 | 3.75E-04 | 4.349 | Cho et al. (10) | |
| Gastric intestinal-type adenocarcinoma | 1.603 | 1.00E-03 | 3.303 | Cho et al. (10) | |
| Gastric intestinal-type adenocarcinoma | 4.485 | 1.70E-09 | 7.310 | D’Errico et al. (12) | |
| Diffuse gastric adenocarcinoma | 2.290 | 6.01E-09 | 6.862 | Cho et al. (10) | |
| CBX3 | Gastric cancer | 1.736 | 6.79E-04 | 3.719 | Wang et al. (13) |
| Gastric mixed adenocarcinoma | 1.998 | 1.62E-07 | 8.875 | Chen et al. (11) | |
| Gastric intestinal-type adenocarcinoma | 1.878 | 1.13E-16 | 11.061 | Chen et al. (11) | |
| Gastric intestinal-type adenocarcinoma | 3.014 | 6.64E-14 | 9.795 | D’Errico et al. (12) | |
| CBX4 | Gastric mixed adenocarcinoma | 1.625 | 7.18E-04 | 3.633 | Cho et al. (10) |
| Gastric mixed adenocarcinoma | 1.955 | 3.03E-06 | 8.080 | Chen et al. (11) | |
| Gastric mixed adenocarcinoma | 3.314 | 2.29E-06 | 6.444 | D’Errico et al. (12) | |
| Gastric intestinal-type adenocarcinoma | 1.783 | 2.55E-17 | 10.753 | Chen et al. (11) | |
| Diffuse gastric adenocarcinoma | 1.730 | 4.23E-04 | 4.295 | Chen et al. (11) | |
| Diffuse gastric adenocarcinoma | 2.466 | 2.45E-05 | 4.862 | D’Errico et al. (12) | |
| CBX5 | NA | NA | NA | NA | NA |
| CBX6 | Diffuse gastric adenocarcinoma | 1.758 | 8.38E-05 | 4.643 | Chen et al. (11) |
| CBX7 | Diffuse gastric adenocarcinoma | –1.656 | 9.09E-05 | –4.072 | Cho et al. (10) |
| CBX8 | NA | NA | NA | NA | NA |
Analyses were conducted using Oncomine.
GEPIA
GEPIA (http://gepia.cancer-pku.cn/) is an interactive web server for analyzing the RNA sequencing expression data of 9,736 tumors and 8,587 normal samples, obtained from The Cancer Genome Atlas (TCGA) and the Genotype-Tissue Expression projects, using a standard processing pipeline (15). We used GEPIA to analyze the relationship between the expression of CBX mRNAs and the clinicopathological stage of cancer. We also identified genes that were similar to the CBX family members.
Kaplan-Meier plotter
The Kaplan-Meier plotter (http://kmplot.com/analysis/) can be used to assess the effects of 54,000 genes on survival in 21 cancer types; primarily, the tool is used for meta-analysis-based discovery and validation of survival biomarkers (16). The incorporated datasets include gastric cancer data (n=1,440). We used the Kaplan-Meier plotter to analyze the relationship between mRNA expression of CBX family members and OS, including survival associated with various cancer treatments. Differences were considered to be statistically significant when P<0.05.
cBioPortal
cBioPortal (www.cbioportal.org) is a comprehensive web resource used to visualize and analyze multidimensional cancer genomics data (17). We used cBioPortal to analyze the relationships among CBX gene mutations, OS, and disease-free survival (DFS) in patients with gastric cancer. We obtained mRNA expression z-8-13scores (RNA-Seq V2 RSEM) using a z-score threshold of ±2.0.
Metascape
Metascape (http://metascape.org) is a tool for gene annotation and gene list enrichment analysis (18). We used Metascape to enrich CBX genes and similar genes via the “CustomAnalysis” module.
UALCAN
UALCAN (http://ualcan.path.uab.edu/analysis.html), a comprehensive web resource, provides analyses based on The Cancer Genome Atlas (TCGA) and MET500 cohort data (19). In this study, UALCAN was used to analyze Relationships between mRNA expression levels of 8 Chromobox (CBX) family members and the grades of gastric cancer.
Statistical analysis
Transcriptional expressions of 8 different CBXs members between different cancer tissues and their corresponding adjacent normal control samples were got from ONCOMINE database. The expression of CBX mRNAs in clinical tumors and normal control tissues was compared by Student’s t-test. Sufficient fold change and significant p values were defined as 1.5 and 0.01. UALCAN and GEPIA were used to analyze the mRNA expressions of 8 CBXs family members in gastric cancer and their association with clinicopathologic parameters. Difference of mRNA expression was compared by students’ t test and P<0.05 was considered as statistically significant. Survival curves were generated by Kaplan-Meier plots. Differences were considered to be statistically significant when P<0.05.
Results
Expression of CBX family members in patients with gastric cancer
As shown in Figure 1 and Table 1, we analyzed the mRNA expression of eight CBX family members in 20 types of tumor and various gastric cancers using Oncomine. Figure 1 shows that, in analysis of gastric cancer tissues from multiple datasets, the expression of CBX1/2/3/4/6 mRNA was significantly increased whereas the expression of CBX7 mRNA was significantly decreased. Table 1 shows that, relative to normal tissues, CBX1 mRNA was overexpressed in gastric adenocarcinoma, diffuse gastric adenocarcinoma, gastric mixed adenocarcinoma, and gastric intestinal-type adenocarcinoma [11-13] with fold changes of 1.660–2.415. Similarly, CBX2 mRNA was overexpressed in diffuse gastric adenocarcinoma, gastric mixed adenocarcinoma, and gastric intestinal-type adenocarcinoma [11-13], as well as gastric cancer in general (13), with fold changes of 1.603–4.485. CBX3 mRNA was overexpressed in gastric mixed adenocarcinoma, gastric intestinal-type adenocarcinoma, and gastric cancer in general (13) with fold changes of 1.736–3.014. CBX4 mRNA was overexpressed in gastric intestinal-type adenocarcinoma, gastric mixed adenocarcinoma, and diffuse gastric adenocarcinoma [11-13] with fold changes of 1.625–3.314. CBX6 (11) and CBX7 (10) mRNAs were overexpressed in diffuse gastric adenocarcinoma with fold changes of 1.758 and 1.656, respectively.
Figure 1.
Transcriptional expression of Chromobox (CBX) family members in 20 types of cancer. Analyses were conducted in Oncomine. Red box shows data for gastric cancer.
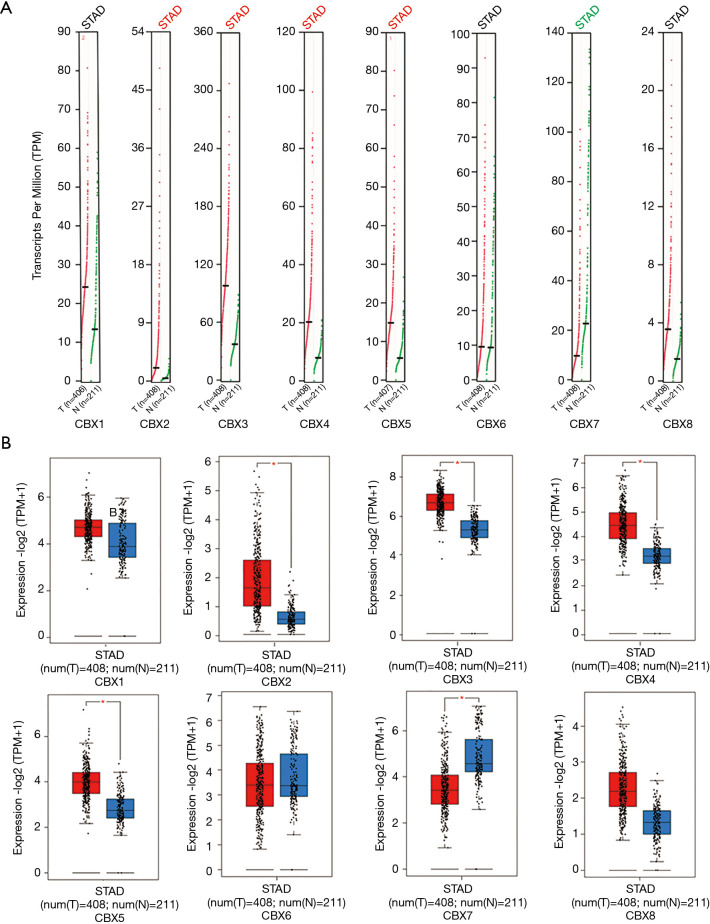
We also used GEPIA to compare the expression of CBX mRNA between tumor and normal tissues (this analysis differs from the Oncomine analysis because the data in GEPIA comes from TCGA). As shown in Figure 2, the expression of CBX2-5 mRNAs was significantly higher in tumor tissues than in normal tissues, whereas the expression of CBX7 mRNA was significantly lower in tumor tissues than in normal tissues.
Figure 2.
mRNA expression of eight Chromobox (CBX) family members in gastric cancer and normal tissues. Analyses were conducted in GEPIA. (A) Scatter diagram of CBX mRNA expression. Red: overexpressed in tumor tissue; green: underexpressed in tumor tissue; black: no significant difference in expression (P>0.05). (B) Box plots of CBX mRNA expression. Red: tumor tissue; blue: normal tissue; *, P<0.05.
In summary, CBX1-6 are apparently overexpressed in gastric cancer, whereas CBX7 is underexpressed.
Relationship between CBX family mRNA expression levels and clinicopathological parameters
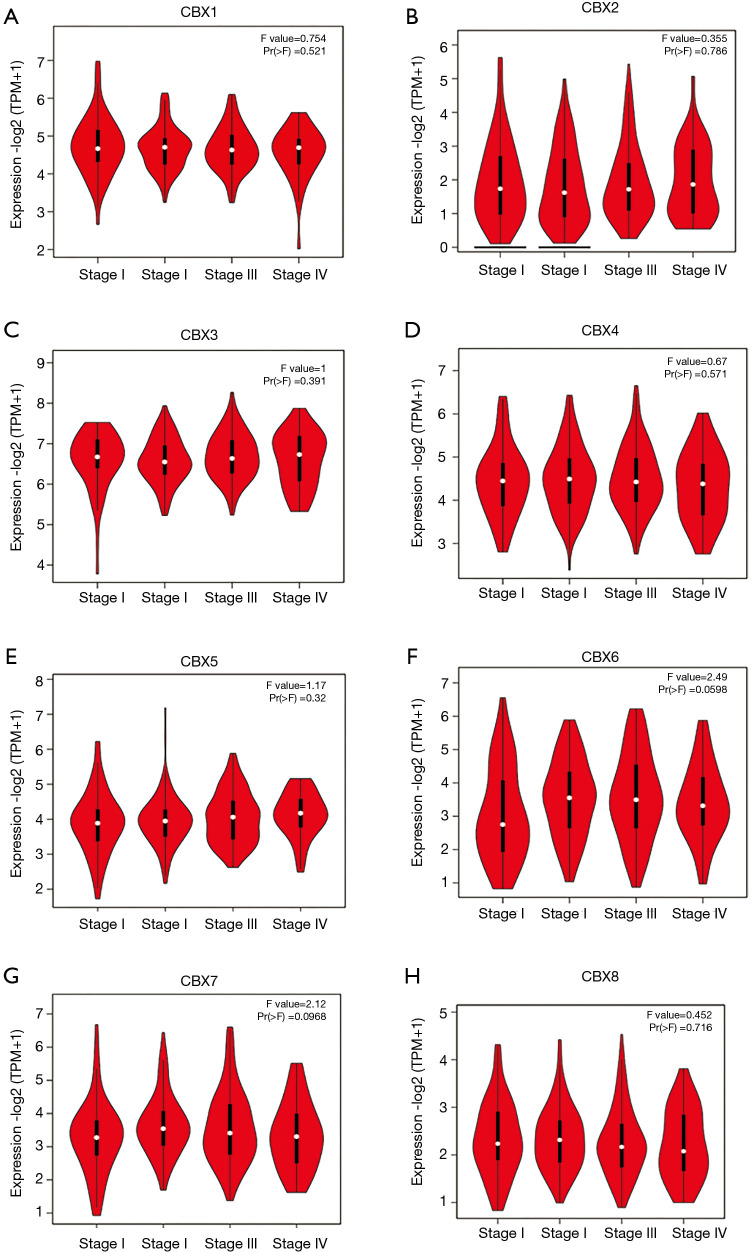
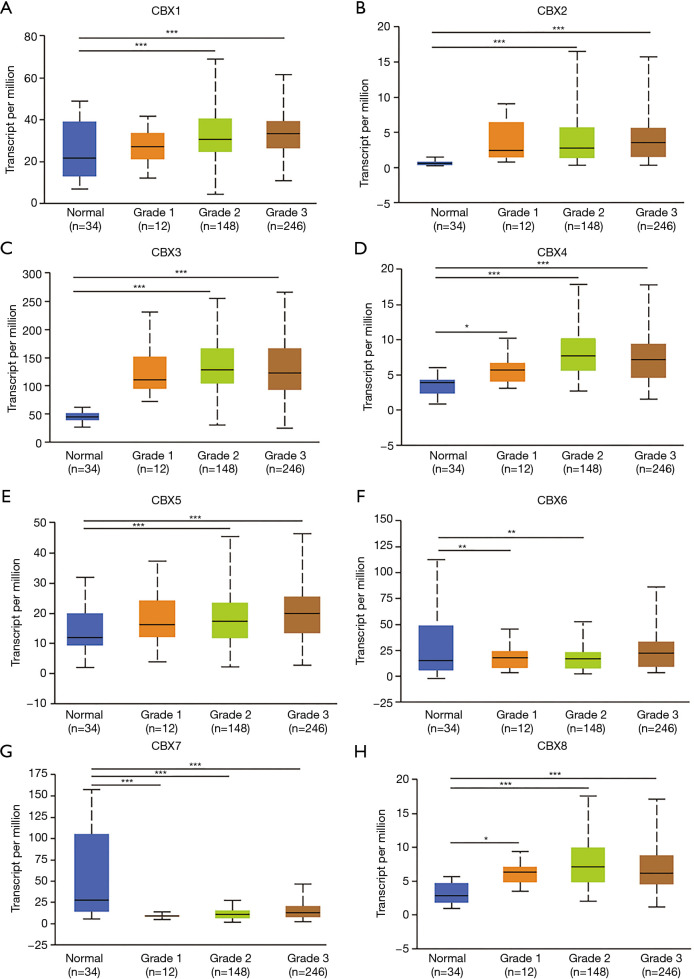
As shown in Figure 3, according to GEPIA analysis, there were no significant relationships between CBX1-8 mRNA levels and the tumor stages of gastric cancer (P>0.05). As shown in Figure 4, the mRNA expressions of 8 CBXs family members were significantly related to tumor grades. The mRNA expression of CBX1/2/3/4/5/6/8 tended to be higher as tumor grade increased. The highest mRNA expressions of CBX1/2/5/6 were found in tumor grade 3 (A-B, E-F), while the highest mRNA expressions of CBX3/4/8 were found in grade 2 (C-D, H). However, the highest mRNA expression of CBX7 was found in normal, and as tumor grade increased, the mRNA expression of CBX7 tended to be lower.
Figure 3.
Relationships between mRNA expression levels of eight Chromobox (CBX) family members and the clinicopathological stages of gastric cancer. Analyses were conducted using GEPIA.
Figure 4.
Relationships between mRNA expression levels of eight Chromobox (CBX) family members and the grades of gastric cancer. Analyses conducted using the UALCAN. *, P<0.05; **, P<0.01; ***, P<0.001.
Relationship between CBX family mRNA expression levels and prognosis of gastric cancer patients
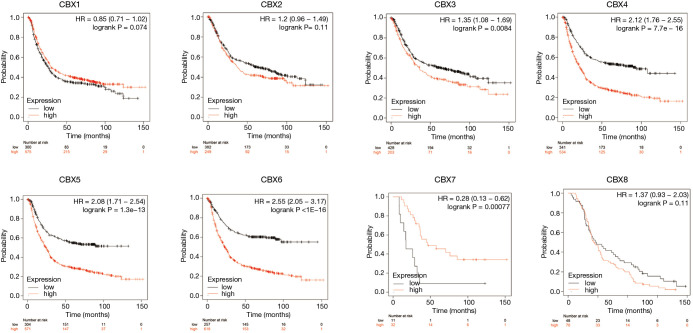
As shown in Figure 5, Kaplan–Meier plotter analysis showed that overexpression of CBX3 (HR =1.35, P=0.0084), CBX4 (HR =2.21, P=7.7−16), CBX5 (HR =2.08, P=1.3−13), and CBX6 (HR =2.55, P<1−16) mRNA, as well as underexpression of CBX7 mRNA (HR =0.28, P=0.00077), was significantly correlated with shorter OS in gastric cancer patients. Thus, the expression of CBX3-7 mRNA is closely related to the prognosis of gastric cancer patients and could potentially be used as a predictive biomarker.
Figure 5.
Relationships between mRNA expression levels of eight Chromobox (CBX) family members and the prognosis of patients with gastric cancer. Analyses were conducted using the Kaplan-Meier plotter.
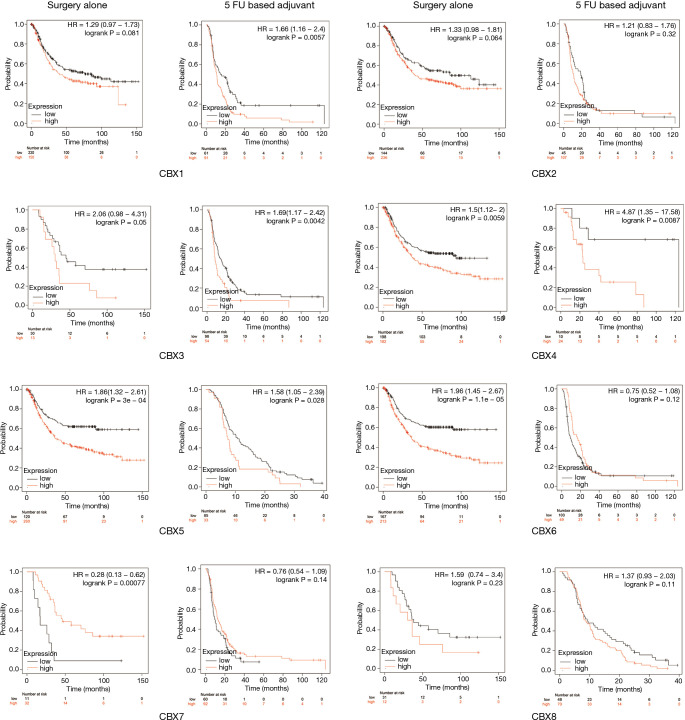
Relationship between CBX family mRNA expression levels and prognosis of gastric cancer patients undergoing different treatments
Similar to the analysis in section 3.3, we also used the Kaplan-Meier plotter to assess the relationship between CBX family mRNA expression and the prognosis of gastric cancer patients receiving differing treatment regimes. Figure 6 shows that the overexpression of CBX3 (HR =2.06, P=0.05), CBX4 (HR =1.5, P=0.0059), CBX5 (HR =1.86, P=0.0003), and CBX6 (HR =1.96, P=0.0001.1) mRNAs in gastric cancer patients that received surgery was significantly correlated with shorter OS, whereas the overexpression of CBX7 mRNA (HR =0.28, P=0.00077) was significantly correlated with longer OS. In addition, the overexpression of CBX1 (HR =1.66, P=0.0057), CHX3 (HR =1.69, P=0.0042), CBX4 (HR =4.87, P=0.0087), and CBX5 (HR =1.58, P=0.028) mRNAs in adjuvant 5-fluorouracil-based chemotherapy-treated patients was significantly correlated with shorter OS.
Figure 6.
Relationships between the mRNA expression levels of eight Chromobox (CBX) family members and the prognosis of patients given differing treatments. Treatments were surgery (alone, i.e., with no other treatment) or 5-fluorouracil (5 FU)-based adjuvant chemotherapy. Analyses were conducted using the Kaplan-Meier plotter.
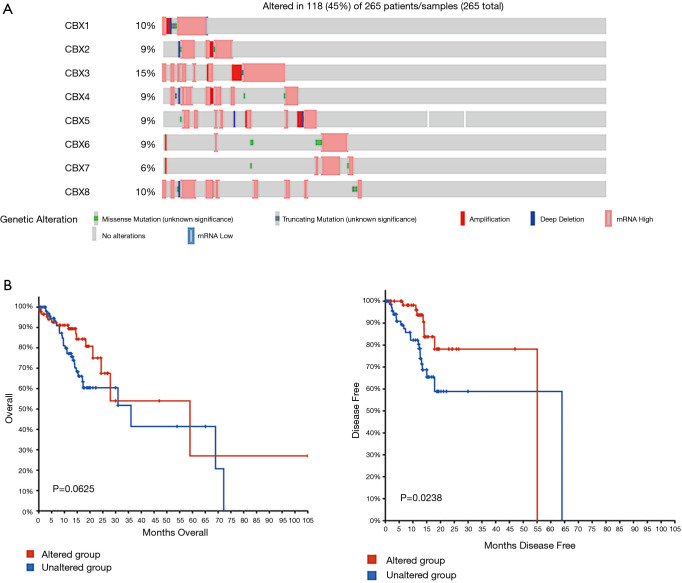
Relationships among CBX gene alterations, OS, and DFS in gastric cancer patients
As shown in the cBioPortal analysis in Figure 7A, 118 CBX gene alterations were observed in 265 patients with an alteration rate of 45%. Figure 7B shows the analysis of the relationship between CBX gene changes and OS or DFS: CBX gene alterations were significantly associated with shorter DFS (P=0.0238) but not OS (P=0.0625).
Figure 7.
Relationships between Chromobox (CBX) gene mutations and survival in gastric cancer patients. (A) The CBX gene mutations [118] observed in gastric cancer patients [265]. (B) Overall survival time (OS, left) and disease-free survival (DFS, right) in gastric cancer patients. Analyses were conducted in cBioPortal.
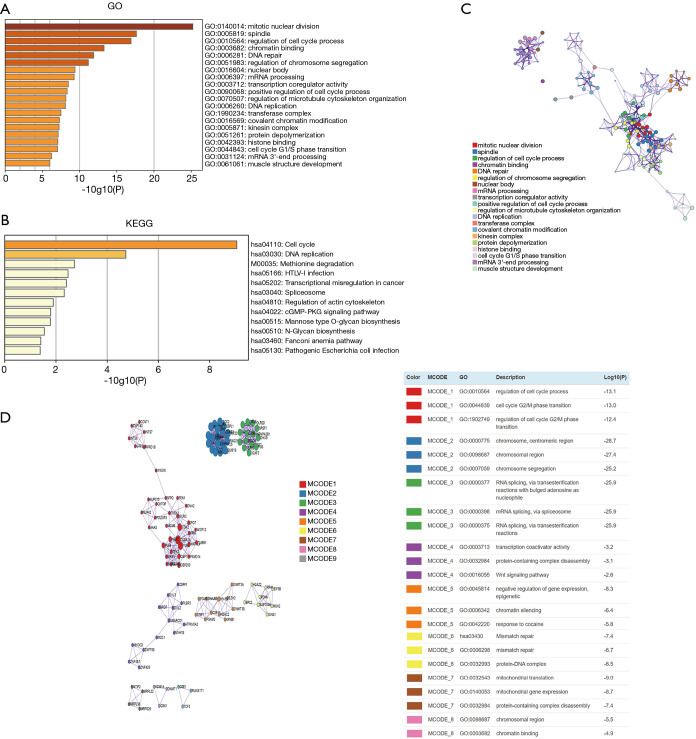
Predicting the functions and pathways of CBX genes and similar genes in gastric cancer patients
First, we used GEPIA to find CBX-like genes in gastric cancer tissues; we used the first 100 similar genes for each CBX gene. Next, we used Metascape for Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genome (KEGG), and protein-protein interaction (PPI) enrichment analyses. The predicted functions and pathways of CBX genes and similar genes are shown in Figure 8A,B,C. These genes were involved in the range of biological processes (13 GO terms), cellular components (5 terms), and molecular functions (3 terms). Most prominently the genes were enriched with the following GO terms: mitotic nuclear division, spindle, regulation of cell cycle process, chromatin binding, DNA repair, and regulation of chromosome segregation. The genes were related to 12 KEGG pathways, including cell cycle, DNA replication, and transcriptional misregulation in cancer (which is specifically related to the development of cancer). Figure 8D shows the PPI network related to the genes; it was mainly associated with mitotic nuclear division, the Wnt signaling pathway, chromatin silencing, cell division, negative regulation of gene expression, and epigenetics.
Figure 8.
The predicted functions, pathways, and protein-protein interactions of CBX genes and similar genes. (A) Gene Ontology (GO) analysis, (B) Kyoto Encyclopedia of Genes and Genome (KEGG) analysis, and (C) Network of enriched terms of CBX genes and similar genes. (D) Protein-protein interaction (PPI) enrichment analysis of these genes and MCODE (Molecular Complex Detection) components identified in the gene lists. Analyses were conducted in Metascape.
Discussion
Our results showed that CBX family members were differentially expressed in gastric cancer tissues: CBX1-6 mRNAs were overexpressed in gastric cancer tissues whereas CBX7 mRNA was underexpressed. Furthermore, overexpression of CBX3-6 mRNAs and CBX7 mRNA was significantly correlated with shorter and longer OS in gastric cancer patients, respectively. This suggests that CBX3-7 could be used as prognostic markers for patients with gastric cancer. In addition, we found a significant relationship between CBX gene mutations and shorter DFS, which further highlights the importance of this family in gastric cancer.
Previously, we found that CBX1 showed carcinogenic activity in hepatocellular carcinoma and predicted poor prognosis (20). In addition, overexpression of CBX1 in prostate cancer and breast cancer are significantly correlated with shorter OS (21,22). The present study showed that the mRNA and protein levels of CBX1 in gastric cancer tissues were significantly higher than those in normal tissues, which is consistent with the results of previous studies. Furthermore, although there was no significant relationship between CBX1 mRNA expression and prognostic OS in gastric cancer patients treated with surgery alone, there was a significant correlation between the overexpression of CBX1 mRNA and shorter prognostic OS in gastric cancer patients treated with 5-fluorouracil-based adjuvant chemotherapy. The expression of heterochromatin protein 1 (HP1) has been shown to enhance the stability of heterochromatin structure and reduce the cytotoxicity of chemotherapeutic drugs (23). Interestingly, CBX1 is one of the core proteins in HP1 (24). Therefore, we speculate that the overexpression of CBX1 leads to the expression of HP1, which reduces the cytotoxicity of chemotherapeutic drugs and thereby reduces the prognosis OS. Moreover, CBX1 may be a novel biomarker for choosing appropriate chemotherapy regimens for gastric cancer patients.
CBX2 is overexpressed in breast cancer and can promote the progression of this disease through the PI3K/AKT signal pathway (25). Furthermore, CBX2 can regulate the proliferation and apoptosis of hepatocellular carcinoma cells in patients by phosphorylating YAP (26). In patients with gastric cancer, overexpression of CBX2 in tumor tissues has previously been related to tumor metastasis but shown to have no significant relationship with OS (27). In the present study, consistent with this previous study, CBX2 mRNA and protein levels were significantly increased in gastric cancer tissues, but these levels were not significantly associated with the prognostic OS of patients.
CBX3 has been found to regulate glycolysis and promote proliferation by inhibiting fructose-1,6-bisphosphatase 1 in pancreatic cancer (28). In glioma patients, CBX3 is overexpressed in tumor tissue, which can stimulate the growth of glioma U87 cells by targeting CDKN1A; CBX3 overexpression also predicts the poor prognosis of glioma patients (29). Moreover, in patients with liver and breast cancers, CBX3 overexpression promotes the proliferation of tumor cells and predicts poor prognoses (21,30). In our results, CBX3 mRNA and protein levels were also significantly increased in gastric cancer tissues, and CBX3 mRNA overexpression was significantly correlated with a short OS prognosis in patients. Thus, CBX3 may be a biomarker of prognosis and survival in gastric cancer patients.
In renal clear cell carcinoma, CBX4 plays a carcinogenic role by inhibiting tumor suppressor KLF6 via an interaction with histone deacetylase (31). CBX4 also promotes the proliferation and metastasis of lung cancer cells by regulating the expression of BMI-1, and it is a prognostic indicator in lung cancer (32). Furthermore, CBX4 promotes the development of breast cancer through the mir137-mediated Notch1 signaling pathway and can be used as a biomarker of prognosis in the disease (33). Additionally, CBX4 is overexpressed in hepatocellular carcinoma and significantly related to the growth and prognosis of tumor cells (34). In the present study, CBX4 mRNA and protein levels increased significantly in gastric cancer tissues, and the overexpression of CBX4 mRNA was significantly correlated with a short OS prognosis in patients. Similar to CBX3, CBX4 may be a biomarker of prognosis and survival in gastric cancer patients.
CBX5 is overexpressed in breast and lung cancers; it promotes tumor cell proliferation and metastasis, and is significantly associated with poor prognosis (35,36). In a previous study of gastric cancer, CBX5 was overexpressed in cancer tissues; microRNA758-3p, which is a tumor suppressor, was found to play an important role in inhibiting the proliferation, migration, and invasion of gastric cancer by targeting CBX5 (37). Here, consistent with this previous study, we showed that CBX5 mRNA was overexpressed in gastric cancer and that overexpression of CBX5 mRNA was significantly correlated with the short OS prognosis of patients. We therefore postulate that CBX5 plays an important role in the occurrence and development of gastric cancer and could be used as a biomarker to predict patient prognosis and survival.
CBX6 is overexpressed in hepatocellular carcinoma and promotes the growth of hepatocellular carcinoma cells; this has an important effect on the disease by regulating the S100A9/NF-κB/MAPK pathway (38). CBX6 is also overexpressed in glioblastoma and is significantly associated with poor prognosis (39). In the present study, CBX6 mRNA was overexpressed in gastric cancer and this overexpression was significantly correlated with short OS prognosis in gastric cancer patients, making CBX6 another potential biomarker of patient prognosis and survival.
Many studies have shown that CBX7 may be a tumor suppressor. For example, in ovarian cancer, CBX7 combines with E-box to inhibit TWIST1 function, and thereby inhibits tumor growth and metastatic potential (40). The expression of CBX7 in lung cancer tissue has been shown to be lower than that in normal tissue, and the tumor inhibits the activity of CBX7 (41). In addition, CBX7 expression in gastric cancer, colorectal cancer, and hepatocellular carcinoma is known to lower than that in normal tissues (42). In pancreatic cancer, CBX7 is downregulated in tumor tissue and is an important tumor suppressor, which negatively regulates PTEN/AKT signal transduction during pancreatic tumorigenesis (43). Finally, CBX7 inhibits the occurrence of breast adenocarcinoma by increasing the expression of the Wnt antagonist DKK-1 and thereby inhibiting the Wnt/β-catenin pathway (44). We also found that the expression of CBX7 in tumor tissues was lower than that in normal tissues, and underexpression of CBX7 mRNA was significantly correlated with short OS prognosis. Therefore, as well as being an important tumor suppressor factor in gastric cancer, CBX7 may be a biomarker of patient prognosis and survival in this disease.
CBX8 plays a contradictory role in esophageal squamous cell carcinoma: it inhibits metastasis by inhibiting Snail while also promoting cell proliferation (45). In bladder cancer, CBX8 overexpression promotes the proliferation of bladder transitional cell carcinoma cells by inhibiting the p53 pathway, which plays an important role in the invasiveness of bladder transitional cell cancer cells (46). However, in our study, we did not find a significant difference between CBX8 mRNA and protein levels in gastric cancer and normal tissues.
B11r results, overexpression of CBX3-6 mRNA and underexpression of CBX7 mRNA in gastric cancer could be used as biomarkers of patient prognosis and survival; however, we note the following limitations in our study. First, our data were all taken from databases; thus, further research will be required to verify our results in vivo and in vitro. In addition, we discussed the prognostic value of CBX family members only; our study lacked research on the related mechanisms. Finally, due to the limitations of our data, we did not conduct grouping analysis.
In conclusion, we found that the overexpression and underexpression of CBX3-6 mRNA and CBX7 mRNA, respectively, are significantly related to the poor prognosis and survival of gastric cancer patients. These members of the CBX family can therefore be used as prognosis and survival biomarkers for this disease. Furthermore, the overexpression of CBX1 mRNA is significantly related to the poor prognosis of gastric cancer patients treated with adjuvant 5-fluorouracil-based chemotherapy; thus, CBX1 could be a biomarker for choosing chemo3-1therapy treatments in these patients (47).
Acknowledgments
Funding: This work was supported by the Science and Technology Planning Project of Guangdong Province (grant number.2017A020215036).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
Footnotes
Reporting Checklist: The authors present the study in accordance with the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/jgo-20-223
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jgo-20-223
Peer Review File: Available at http://dx.doi.org/10.21037/jgo-20-223
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jgo-20-223). The authors have no conflicts of interest to declare.
References
- 1.Ferro A, Peleteiro B, Malvezzi M, et al. Worldwide trends in gastric cancer mortality (1980-2011), with predictions to 2015, and incidence by subtype. Eur J Cancer 2014;50:1330-44. 10.1016/j.ejca.2014.01.029 [DOI] [PubMed] [Google Scholar]
- 2.Karimi P, Islami F, Anandasabapathy S, et al. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev 2014;23:700-13. 10.1158/1055-9965.EPI-13-1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strong VE. Progress in gastric cancer. Updates Surg 2018;70:157-9. 10.1007/s13304-018-0543-3 [DOI] [PubMed] [Google Scholar]
- 4.Goral V. Etiopathogenesis of Gastric Cancer. Asian Pac J Cancer Prev 2016;17:2745-50. [PubMed] [Google Scholar]
- 5.Fu DG. Epigenetic alterations in gastric cancer (Review). Mol Med Rep 2015;12:3223-30. 10.3892/mmr.2015.3816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma RG, Zhang Y, Sun TT, et al. Epigenetic regulation by polycomb group complexes: focus on roles of CBX proteins. J Zhejiang Univ Sci B 2014;15:412-28. 10.1631/jzus.B1400077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klauke K, Radulović V, Broekhuis M, et al. Polycomb Cbx family members mediate the balance between haematopoietic stem cell self-renewal and differentiation. Nat Cell Biol 2013;15:353-62. 10.1038/ncb2701 [DOI] [PubMed] [Google Scholar]
- 8.Gao SB, Sun SL, Zheng QL, et al. Genetic alteration and misexpression of Polycomb group genes in hepatocellular carcinoma. Am J Cancer Res 2015;5:2969-79. [PMC free article] [PubMed] [Google Scholar]
- 9.Ni SJ, Zhao LQ, Wang XF, et al. CBX7 regulates stem cell-like properties of gastric cancer cells via p16 and AKT-NF-κB-miR-21 pathways. J Hematol Oncol 2018;11:17. 10.1186/s13045-018-0562-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho JY, Lim JY, Cheong JH, et al. Gene expression signature-based prognostic risk score in gastric cancer. Clin Cancer Res 2011;17:1850-7. 10.1158/1078-0432.CCR-10-2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen X, Leung SY, Yuen ST, et al. Variation in gene expression patterns in human gastric cancers. Mol Biol Cell 2003;14:3208-15. 10.1091/mbc.e02-12-0833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D'Errico M, de Rinaldis E, Blasi MF, et al. Genome-wide expression profile of sporadic gastric cancers with microsatellite instability. Eur J Cancer 2009;45:461-9. 10.1016/j.ejca.2008.10.032 [DOI] [PubMed] [Google Scholar]
- 13.Wang Q, Wen YG, Li DP, et al. Upregulated INHBA expression is associated with poor survival in gastric cancer. Med Oncol 2012;29:77-83. 10.1007/s12032-010-9766-y [DOI] [PubMed] [Google Scholar]
- 14.Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 2004;6:1-6. 10.1016/S1476-5586(04)80047-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. 10.1093/nar/gkx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagy Á, Lanczky A, Menyhart O, et al. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep 2018;8:9227. 10.1038/s41598-018-27521-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis ofsystems-level datasets. Nat Commun 2019;10:1523. 10.1038/s41467-019-09234-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. 10.1016/j.neo.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang YF, Pan YH, Tian QH, et al. CBX1 Indicates Poor Outcomes and Exerts Oncogenic Activity in Hepatocellular Carcinoma. Transl Oncol 2018;11:1110-8. 10.1016/j.tranon.2018.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang YK, Lin H, Chen C, et al. Prognostic values of distinct CBX family members in breast cancer. Oncotarget 2017;8:92375-87. 10.18632/oncotarget.21325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ngollo M, Lebert A, Daures M, et al. Global analysis of H3K27me3 as an epigenetic marker in prostate cancer progression. BMC Cancer 2017;17:261. 10.1186/s12885-017-3256-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mojardín L, Botet J, Moreno S, et al. Chromosome segregation and organization are targets of 5'-Fluorouracil in eukaryotic cells. Cell Cycle 2015;14:206-18. 10.4161/15384101.2014.974425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vincenz C, Kerppola TK. Different polycomb group CBX family proteins associate with distinct regions of chromatin using nonhomologous protein sequences. Proc Natl Acad Sci U S A 2008;105:16572-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng S, Lv P, Su J, et al. Overexpression of CBX2 in breast cancer promotes tumor progression through thePI3K/AKT signaling pathway. Am J Transl Res 2019;11:1668-82. [PMC free article] [PubMed] [Google Scholar]
- 26.Mao J, Tian Y, Wang C, et al. CBX2 Regulates Proliferation and Apoptosis via the Phosphorylation of YAP in Hepatocellular Carcinoma. J Cancer 2019;10:2706-19. 10.7150/jca.31845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He YL, Zhang B. Clinical significance of expression of CBX2 in gastric cancer. World Chinese Journal of Digestology 2019;27:872-7. 10.11569/wcjd.v27.i14.872 [DOI] [Google Scholar]
- 28.Chen LY, Cheng C, Qu C, et al. CBX3 promotes proliferation and regulates glycolysis via suppressing FBP1 in pancreatic cancer. Biochem Biophys Res Commun 2018;500:691-7. 10.1016/j.bbrc.2018.04.137 [DOI] [PubMed] [Google Scholar]
- 29.Zhao SP, Wang F, Yang M, et al. CBX3 promotes glioma U87 cell proliferation and predicts an unfavorable prognosis. J Neurooncol 2019;145:35-48. 10.1007/s11060-019-03286-w [DOI] [PubMed] [Google Scholar]
- 30.Zhong X, Kan A, Zhang W, et al. CBX3/HP1gamma promotes tumor proliferation and predicts poor survival inhepatocellular carcinoma. Aging 2019;11:5483-97. 10.18632/aging.102132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang N, Niu G, Pan Y, et al. CBX4 transcriptionally suppresses KLF6 via interaction with HDAC1 to exert oncogenic activities in clear cell renal cell carcinoma. EBioMedicine 2020;53:102692. 10.1016/j.ebiom.2020.102692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu C, Zhang Q, Tang Q, et al. CBX4 promotes the proliferation and metastasis via regulating BMI-1 in lung cancer. J Cell Mol Med 2020;24:618-31. 10.1111/jcmm.14771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeng JS, Zhang ZD, Pei L, et al. CBX4 exhibits oncogenic activities in breast cancer via Notch1 signaling. Int J Biochem Cell Biol 2018;95:1-8. 10.1016/j.biocel.2017.12.006 [DOI] [PubMed] [Google Scholar]
- 34.Wang B, Tang J, Liao D, et al. Chromobox homolog 4 is correlated with prognosis and tumor cell growth in hepatocellular carcinoma. Ann Surg Oncol 2013;20 Suppl 3:S684-92. 10.1245/s10434-013-3171-7 [DOI] [PubMed] [Google Scholar]
- 35.Yu YH, Chiou G, Huang P, et al. Network biology of tumor stem-like cells identified a regulatory role of CBX5 in lung cancer. Sci Rep 2012;2:584. 10.1038/srep00584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vad-Nielsen J, Jakobsen KR, Daugaard TF, et al. Regulatory dissection of the CBX5 and hnRNPA1 bi-directional promoter in human breast cancer cells reveals novel transcript variants differentially associated with HP1α down-regulation in metastatic cells. BMC Cancer 2016;16:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo J, Zhang Z, Pan L, et al. Identification of miR-758-3p as Potential Modulator of CBX5 Expression in Gastric Cancer. Technol Cancer Res Treat. 2018;17:1533033818816061. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Zheng H, Jiang W, Tian T, et al. CBX6 overexpression contributes to tumor progression and is predictive of a poor prognosis in hepatocellular carcinoma. Oncotarget 2017;8:18872-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li G, Warden C, Zou Z, et al. Altered expression of polycomb group genes in glioblastoma multiforme. PLoS One 2013;8:e80970. 10.1371/journal.pone.0080970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J, Alvero AB, Nuti S, et al. CBX7 binds the E-box to inhibit TWIST-1 function and inhibit tumorigenicity and metastatic potential. Oncogene 2020;39:3965-79. 10.1038/s41388-020-1269-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Forzati F, Federico A, Pallante P, et al. Tumor suppressor activity of CBX7 in lung carcinogenesis. Cell Cycle 2012;11:1888-91. 10.4161/cc.20022 [DOI] [PubMed] [Google Scholar]
- 42.Guan ZP, Gu LK, Xing BC, et al. Downregulation of chromobox protein homolog 7 expression in multiple humancancer tissues. Zhonghua Yu Fang Yi Xue Za Zhi 2011;45:597-600. [PubMed] [Google Scholar]
- 43.Ni S, Wang H, Zhu X, et al. CBX7 suppresses cell proliferation, migration, and invasion through theinhibition of PTEN/Akt signaling in pancreatic cancer. Oncotarget 2017;8:8010-21. 10.18632/oncotarget.14037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim HY, Park JH, Won HY, Lee JY, Kong G. CBX7 inhibits breast tumorigenicity through DKK-1-mediated suppression of the Wnt/β-catenin pathway. FASEB J 2015;29:300-13. 10.1096/fj.14-253997 [DOI] [PubMed] [Google Scholar]
- 45.Wang G, Tang J, Zhan W, et al. CBX8 Suppresses Tumor Metastasis via Repressing Snail in Esophageal Squamous Cell Carcinoma. Theranostics 2017;7:3478-88. 10.7150/thno.20717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuan GJ, Chen X, Lu J, et al. Chromobox homolog 8 is a predictor of muscle invasive bladder cancer and promotes cell proliferation by repressing the p53 pathway. Cancer Sci 2017;108:2166-75. 10.1111/cas.13383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gu X, Wang L, Boldrup L, et al. AP001056.1, A Prognosis-Related Enhancer RNA in Squamous Cell Carcinoma of the Head and Neck. Cancers (Basel) 2019;11:347. 10.3390/cancers11030347 [DOI] [PMC free article] [PubMed] [Google Scholar]