Abstract
Background
Chromobox (CBX) family proteins are a class of transcriptional repressors involved in epigenetic regulation and developmental processes of various tumors, including gastric cancer. However, the function and prognosis of different CBXs in gastric cancer remain unknown.
Methods
This study addresses this issue by synthesizing several mainstream databases (Oncomine, GEPIA2, cBioportal, and Kaplan-Meier plotter, among others) that currently contain many tumor samples and provide very reliable analysis results, investigating the role of CBXs in the prognosis of gastric cancer.
Results
The mRNA of CBX1/2/3/4/5/8 was highly expressed in gastric cancer, the mRNA of CBX7 was lowly expressed in gastric cancer, and the mRNA expression of CBX6 was not significantly different in CRC. Besides, high and low CBXs mRNA expression correlated with cancer stage, node metastasis status, H. pylori infection status, and tumor grade in CRC patients. We found that high mRNA expression of CBX4/5/6/7/8 was significantly associated with worse overall survival (OS), progression-free survival (FP), and post-progression survival (PPS) in a large number of CRC patients. High mRNA expression of CBX3 was significantly associated with better OS and FP. We also found that none of the eight CBXs family genes had a mutation rate of less than 5% in gastric cancer, and the highest mutation rate was in CBX3 (14%).
Conclusions
These results suggest that CBX3/4/5/6/7/8 could be a prognostic biomarker in gastric cancer patients.
Keywords: Gastric cancer, chromobox (CBX), prognosis, biomarkers, Kaplan-Meier plotter
Introduction
The CBXs family is a class of genes that can encode transcription factors (TFs) that regulate the development and progression of various cancers by inhibiting cell differentiation and self-renewal of cancer stem cells (1). To date, eight CBXs (CBX1–8) family proteins have been identified in the human genome. They are all involved in the regulation of heterochromatin, gene expression, and developmental processes. According to the molecular structure of CBXs family proteins, they can be divided into two groups: the HP1 group (CBX1/3/5) and the PC group (CBX2/4/6/7/8). The functions of different CBXs family proteins are associated with different chromatin regions and show non-overlap in mammalian embryonic stem cells (2-4). Current studies have reported that CBXs are dysregulated in a variety of tumors, including gastric cancer (5), lung cancer (6), cervical cancer (7), liver cancer (8,9), breast cancer (10,11), pancreatic cancer (12), esophageal squamous cell carcinoma (13) and high grade serous ovarian carcinoma (14) and so on.
Gastric cancer is the fourth most common cancer and the second leading cause of cancer death worldwide. Despite significant advances in understanding epidemiology, pathology, molecular mechanisms, and treatment options and strategies, the burden is still substantial, and surgical resection is still the most effective means of treating gastric cancer. Continued research into the molecular mechanisms of gastric cancer is urgently needed to develop more effective and unique treatments (15). Although some members of the CBXs family have been identified to function in a variety of tumors, including gastric cancer, to our knowledge, bioinformatics analysis has not been applied to explore the role of CBXs in gastric cancer. New biomarkers as prognostic indicators are needed in this field to enhance prognosis and individualize treatment effectively. A comprehensive study of different CBXs family members in gastric cancer will help reveal the molecular mechanisms of GC development and may reveal new prognostic and therapeutic targets.
In this study, we analyzed the expression and mutation of different CBXs family members and their association with clinical parameters in gastric cancer patients to determine the expression patterns, potential functions, and different prognostic values of CBXs in gastric cancer. We also analyzed the predicted functions and pathways of CBXs mutations and the genes with which they have co-expression relationships.
We present the following article in accordance with the Materials Design Analysis Reporting (MDAR) reporting checklist (available at http://dx.doi.org/10.21037/jgo-20-208).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study did not involve animal or human experiments and did not require ethical approval or patient consent.
Oncomine analysis
Oncomine datasets (https://www.oncomine.org/) were used to analyze the transcription levels of CBXs in different cancers. Student’s t-test was used to compare the differences in transcript levels. The critical values of P value and fold change are defined as 0.01 and 2.
GEPIA2 database
GEPIA2 (http://gepia2.cancer-pku.cn/) is a web server containing RNA sequencing expression data of 9,736 tumors and 8,587 normal samples from the TCGA and GTEx projects, all of which were analyzed by standard processing pipelines (16). The RNA-Seq datasets were mainly based on the UCSC Xena project (http://xena.ucsc.edu). We sequentially analyzed the expression differences of each gene of the CBXs family between gastric cancer and healthy gastric tissues. Statistical analysis is based on the limma (17) package of R software. Wilcox test is used to calculate the P value. Only when |Log2FC| >1 and P<0.01, the difference in the expression of the target gene in tumor and healthy tissue has significant statistical differences.
Association of CBXs family proteins with clinicopathological features
UALCAN (http://ualcan.path.uab.edu/) is a comprehensive interactive web server for analyzing cancer OMICS data. UALCAN provides easy access to public cancer OMICS data (TCGA and MET500) and allows users to identify biomarkers or perform computer verification of potential genes of interest. This study analyzed the mRNA expression of eight CBXs family members in gastric cancer and their relationship with clinicopathological parameters. Similarly, the Student t-test was used to verify expression differences. P<0.01 indicates that the difference is statistically significant. Due to the lack of typical clinical information in UALCAN, we downloaded transcriptome data (381 tumor samples and 32 matched paracancerous tissues) and corresponding clinical information for gastric cancer from the TCGA database. After deleting some samples with incomplete clinical information, we analyzed the remaining 349 tumor samples and clinical information for statistical associations between CBX family proteins and various clinicopathological characteristics. The study was conducted using the survival (https://cran.r-project.org/web/packages/survival/index.html) package of R software, univariate and multivariate Cox analysis was performed separately to construct the proportional hazards model. P<0.05 indicates a statistically significant difference in results.
Human Protein Atlas
The Human Protein Atlas (https://www.proteinatlas.org/) is an online tool that contains transcriptome profiling data and immunohistochemistry profiling data for more than 8,000 patients and 17 major cancer types, allowing users to directly profile the protein expression patterns of specific genes in specified tumors (18). In this study, we obtained immunohistochemical images in this tool to directly compare the protein expression of different CBXs family members in human healthy and gastric cancer tissues and analyze the CBXs family protein expression patterns in depth.
The Kaplan-Meier plotter
The Kaplan Meier plotter (http://kmplot.com/analysis/) is an online tool, capable of assessing the impact of 54K genes on survival in 21 cancer types. This database contains many gastric cancer samples (n=1,440) and can analyze the effect of each gene of the CBXs family on the survival time of gastric cancer patients. By dividing the median expression values of CXBs family genes in gastric cancer patients into two groups (high vs. low expression), the results were presented by plotting survival curves and simultaneously analyzing hazard ratio (HR) with 95% confidence intervals (CIs) and log-rank P value. The JetSet Best Probe Set of CBXs was performed on to Obtain Kaplan-Meier plots. The Kaplan-Meier method was used to analyze the relationship between target gene expression and patient survival, and the significance index P value was calculated using the log-rank test. P<0.05 indicated significant statistical significance. The tool automatically treats genes as risk factors and other pathological indicators and calculates HR values for high expression of target genes based on COX regression.
The Cancer Genome Atlas Data and cBioPortal
The Cancer Genome Atlas (https://www.cancer.gov/tcga), a landmark cancer genome project, matched sequencing data to pathological data for normal samples covering 33 cancer types. The cBioPortal (https://www.cbioportal.org/) database includes all data in TCGA and is continuously updated (19,20). We used the Stomach Adenocarcinoma (TCGA, PanCancer Atlas) dataset (21) to analyze the mutation information of CBXs family genes in gastric cancer and corrected each gene of CBXs family to analyze the correlation between each gene. We used the corrplot (https://cran.r-project.org/web/packages/corrplot/index.html) package to analyze the association between the eight gene expressions, and the difference significance index was calculated using the Spearman’s correlation test. P<0.05 indicates a statistically significant difference between the two gene expressions.
GO and KEGG pathway analyses
DAVID (https://david.ncifcrf.gov/) (22,23), a compelling database capable of functional enrichment and pathway annotation for a range of genes, we analyzed the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of these genes by annotating the genes in the resulting CBXs family co-expression network in STRING. The functions and underlying molecular mechanisms of the CBXs family in gastric cancer were further analyzed. Due to the output of the limited pathway in the DAVID database, we also introduced Reactome (https://reactome.org/) databases. The Reactome is open-source, open access, manual management, and peer-reviewed access database that references more than 100 different online bioinformatics resources (including NCBI Gene, Ensembl, and UniProt databases, UCSC Genome Browser, ChEBI small molecule database, and PubMed Literature databases, etc.). Researchers can use this tool to uncover reliable pathways from genomic (24). We analyzed the possible molecular mechanisms involved in the role of CXBs in gastric cancer accordingly.
Results
Transcriptional levels of CBXs in patients with gastric cancer
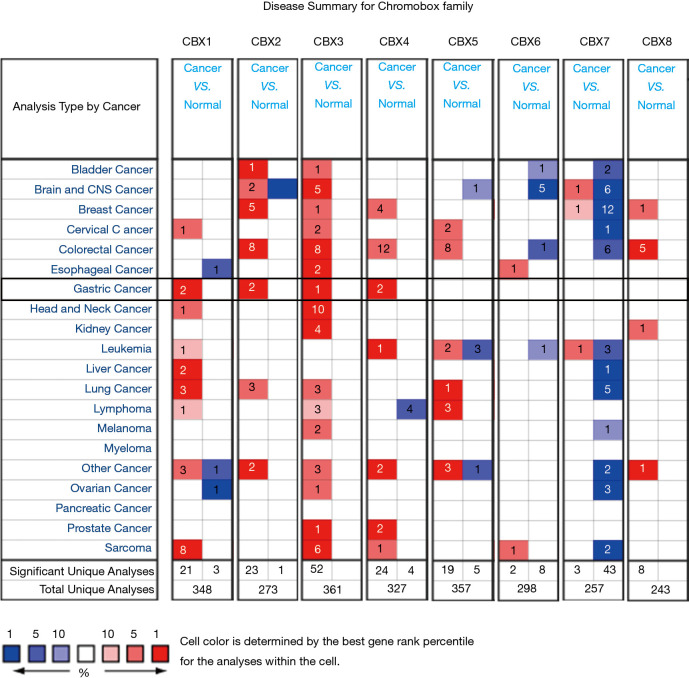
We identified eight CBXs expressed in different diseases and gastric cancer in the Oncomine and GEPIA2 databases. In the Oncomine database, we found that CBXs family members are highly expressed in various tumors (Figure 1) and that there are multiple CBXs with higher transcript levels in gastric cancer (Table 1). Regarding CBX1, we found high expression in the Cho gastric statistics dataset (FC =2.415, P=4.52E-06) and also in the Derrico gastric statistics dataset (FC =2.116, P=2.21E-13). Regarding CBX2, high mRNA expression levels were also found in the Cho Gastric Statistics (FC =2.290, P=6.01e-9) and Derrico Gastric Statistics (FC =4.485, P=1.70e-09) datasets, respectively. In addition, we also found high CBX3 expression in the DeRrico Gastric Statistics dataset (FC =3.014, P=6.64E-14). In the Derrico Gastric Statistics dataset, differential expression contrast between Diffuse Gastric Adenocarcinoma and Normal samples revealed that CBX4 was more highly expressed in tumors (FC =2.466, P=2.45E-05). Moreover, differential expression between Gastric Mixed Adenocarcinoma and Normal samples revealed that CBX4 was more highly expressed in tumors (FC =3.314, P=2.29E-6). However, no differential expression information of CBX5-8 in gastric cancer was found in Oncomine dataset. Subsequently, we further measured the transcript levels of eight CBXs family members in the GEPIA2 (RNA-Seq and clinical Data of 31 cancer types based on the TCGA database) database (Figure 2). Compared with normal samples, the mRNA expression of CBX1/2/3/4/5/8 was significantly up-regulated in gastric cancer tissues (P<0.05), the mRNA expression of CBX7 was significantly downregulated in gastric cancer tissues (P<0.05), and no expression difference was found for CBX6.
Figure 1.
The transcription levels of CBXs in 20 different types of cancers (Oncomine). Difference of transcriptional expression was compared by Students’ t-test. The cut-off of parameters: P<0.01; fold change: 1.5; gene rank: 10%.
Table 1. The significant changes of CBX expression between different types of gastric cancer and normal gastric tissues (Oncomine Database).
| Chromobox family | Type of gastric cancer versus normal tissue | Fold change | P value | t-test | Source and/or reference |
|---|---|---|---|---|---|
| CBX1 | Gastric adenocarcinoma vs. normal | 2.415 | 4.52E-06 | 6.913 | Cho gastric statistics |
| Gastric intestinal type adenocarcinoma vs. normal | 2.116 | 2.21E-13 | 10.157 | DErrico gastric statistics | |
| CBX2 | Diffuse gastric adenocarcinoma vs. normal | 2.290 | 6.01E-9 | 6.862 | Cho gastric statistics |
| Gastric intestinal type adenocarcinoma vs. normal | 4.485 | 1.70E-09 | 7.310 | DErrico gastric statistics | |
| CBX3 | Gastric intestinal type adenocarcinoma vs. normal | 3.014 | 6.64E-14 | 9.795 | DErrico gastric statistics |
| CBX4 | Diffuse gastric adenocarcinoma vs. normal | 2.466 | 2.45E-05 | 4.862 | DErrico gastric statistics |
| Gastric mixed adenocarcinoma vs. normal | 3.314 | 2.29E-6 | 6.444 | DErrico gastric statistics |
Cho Gastric Statistics: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE13861; DErrico Gastric Statistics: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE13911; CBX5-8: not available.
Figure 2.
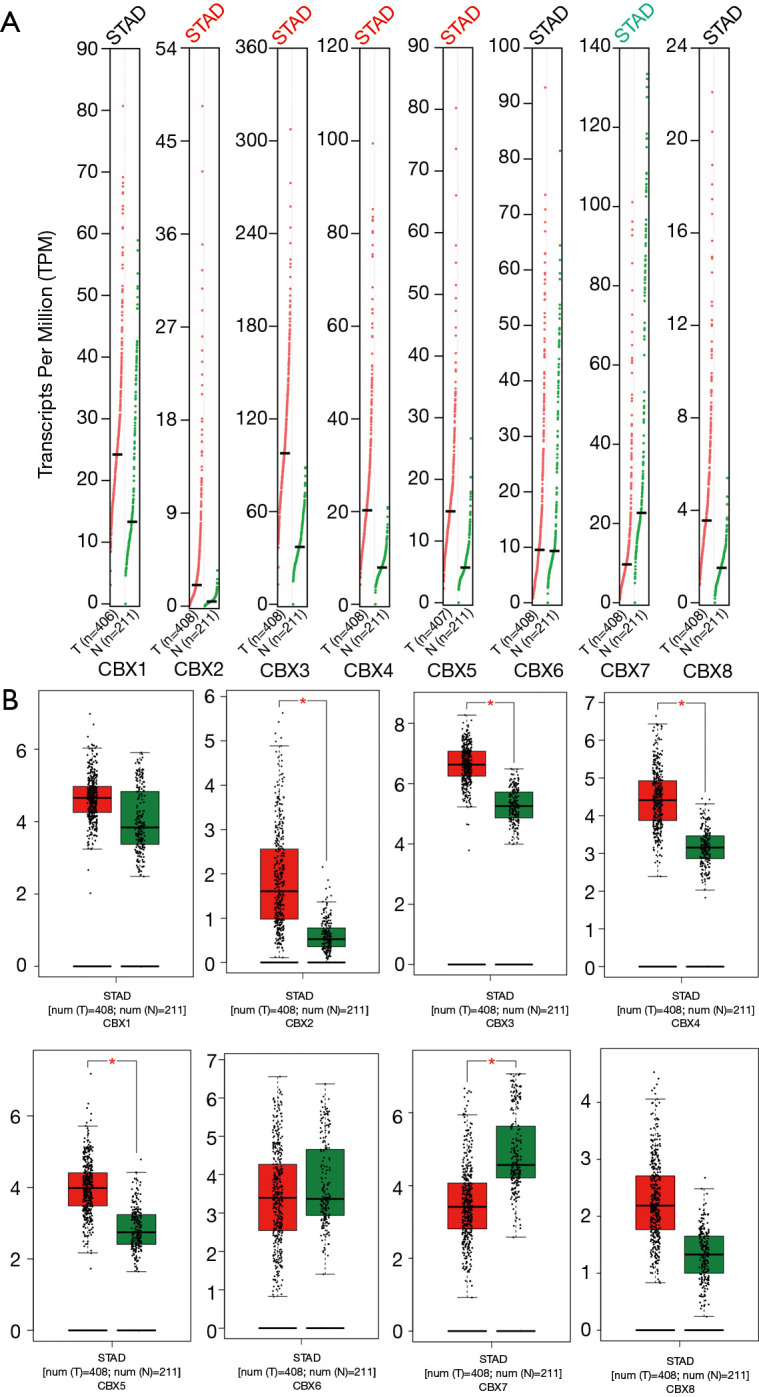
The mRNA expression of CBXs in gastric cancer (GEPIA2). The mRNA transcript levels of eight CBXs family genes in gastric cancer and normal gastric tissues (A: scatter plot; B: box plot). *, P<0.05.
Relationship to clinicopathological parameters
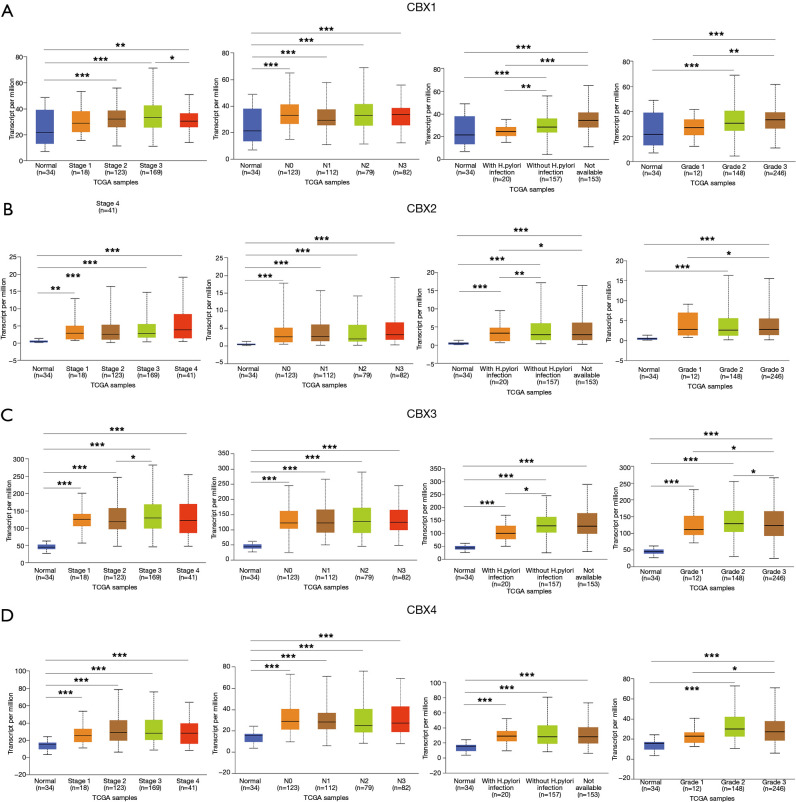
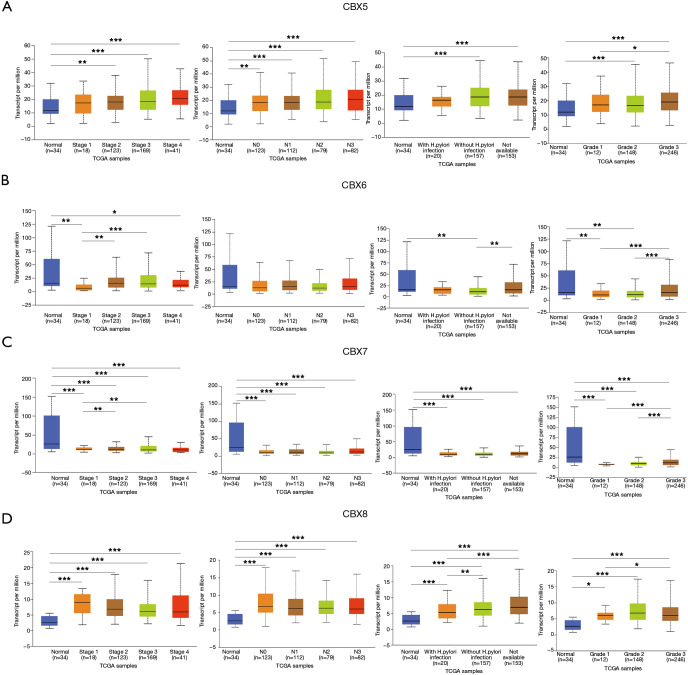
After determining the expression of each CBXs member in gastric cancer, the relationship between mRNA expression of different CBXs family members and clinicopathological parameters of gastric cancer patients such as cancer stage, node metastasis status, H. pylori infection status, and tumor grade was then analyzed in the UALCAN database (TCGA data processed by unified standards) (Figures 3,4). We found a specific correlation between the mRNA expression of the 8 CBXs family members and each clinicopathological parameter of gastric cancer. Statistically, most of the mRNA expression of CBX1-6 tended to be higher in more advanced tumors. However, the mRNA expression of CBX7/8 showed a decreasing trend with increasing tumor stage, with the difference that CBX7 was down-regulated in tumors, whereas CBX8 was up-regulated. However, no significant trend was found regarding the relationship between mRNA expression of CBXs family members and node metastasis status.
Figure 3.
Association of CBX1-4 transcript levels with clinical pathology. Relationship between mRNA expression levels of CBX1-4 and cancer stage, node metastasis status, H. pylori infection status and tumor grade (*, P<0.05; **, P<0.01, ***, P<0.001).
Figure 4.
Association of CBX5-8 transcript levels with clinical pathology. Relationship between mRNA expression levels of CBX5-8 and cancer stage, node metastasis status, H. pylori infection status and tumor grade (*, P<0.05; **, P<0.01, ***, P<0.001).
Regarding the relationship between mRNA expression of CBXs family members and H. pylori infection status, we statistically found that CBX2 had higher mRNA expression in the presence of H. pylori infection in tumor samples. However, the mRNA expression of CBX1/3/8 was higher in the absence of H. pylori infection, and the mRNA expression of CBX4/5/6/7 was not associated with the presence of H. pylori infection. In the statistics of the association between tumor grade and CBXs family mRNA expression was found to have an upward trend roughly with grade stage, the mRNA expression of CBX1/2/5/6/7 was the highest in grade III, while the mRNA expression of CBX3/4/8 was the highest in grade II. In conclusion, the above statistical analysis indicated that mRNA expression of 8 CBXs family members was significantly correlated with clinicopathological parameters in gastric cancer patients.
Proportional risk models for CBX family proteins and clinicopathological factors
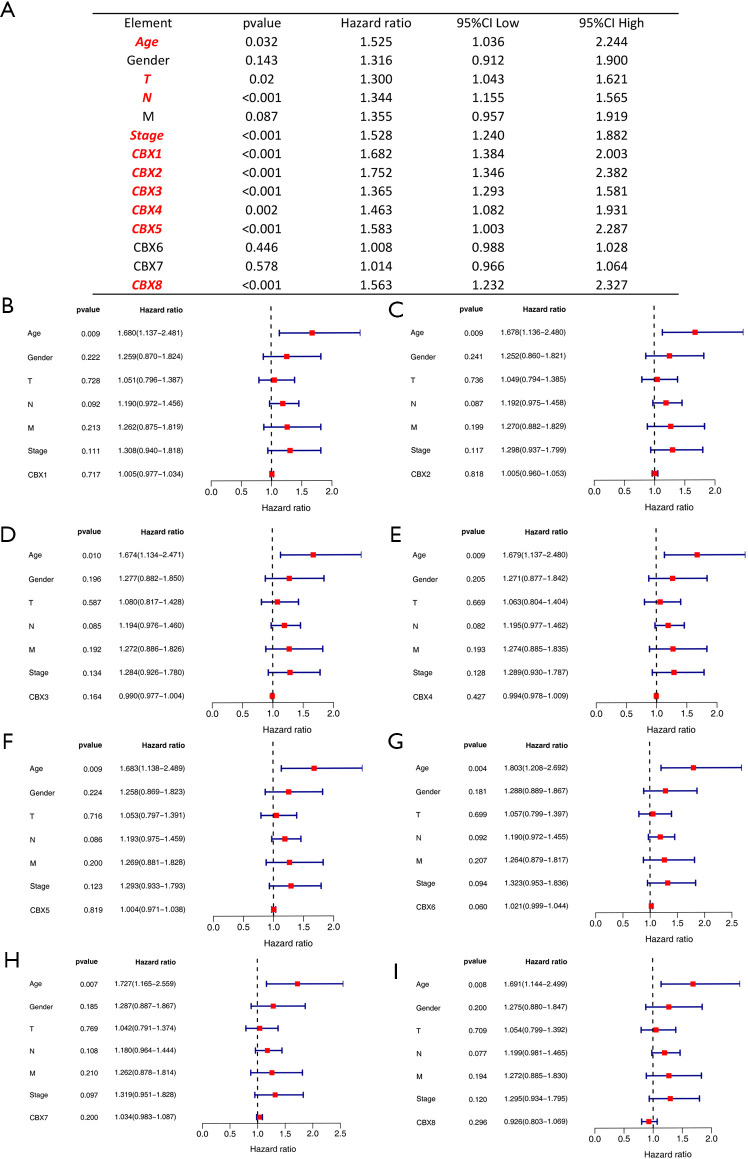
We extracted transcriptomic and clinical data from 349 gastric cancer patients containing complete pathological parameters. Based on the expression of CBXs family proteins and tumor patients’ age, sex, TNM stage, cancer stage, and other variables, followed by single- and multifactor Cox analysis (Figure S1). The results of the single-factor Cox analysis indicated patient age, tumor T and N grade, tumor stage, and CBX1/2/3/4/5/8 were all independent high-risk factors for survival of gastric cancer patients. In the multifactorial Cox that comprehensively analyzed the association between these factors and the prognosis of gastric cancer patients, we found that the patients' age was still high-risk. However, the results for other pathological parameters and CBXs family proteins were not statistically different (P>0.05).
Figure S1.
Proportional risk models for CBX family proteins and clinicopathological factors. Univariate and multivariate Cox analyses were performed on 349 gastric cancer tumor samples in TCGA. Univariate analysis (A) showed age, T, N, tumor stage, and CBX1/2/3/4/5/8 was an independent high-risk factor for patients with gastric cancer. Multivariate analysis (B,C,D,E,F,G,H,I) showed age as an independent high-risk factor for gastric cancer patients, but the remaining risk factors were not statistically different (P>0.05).
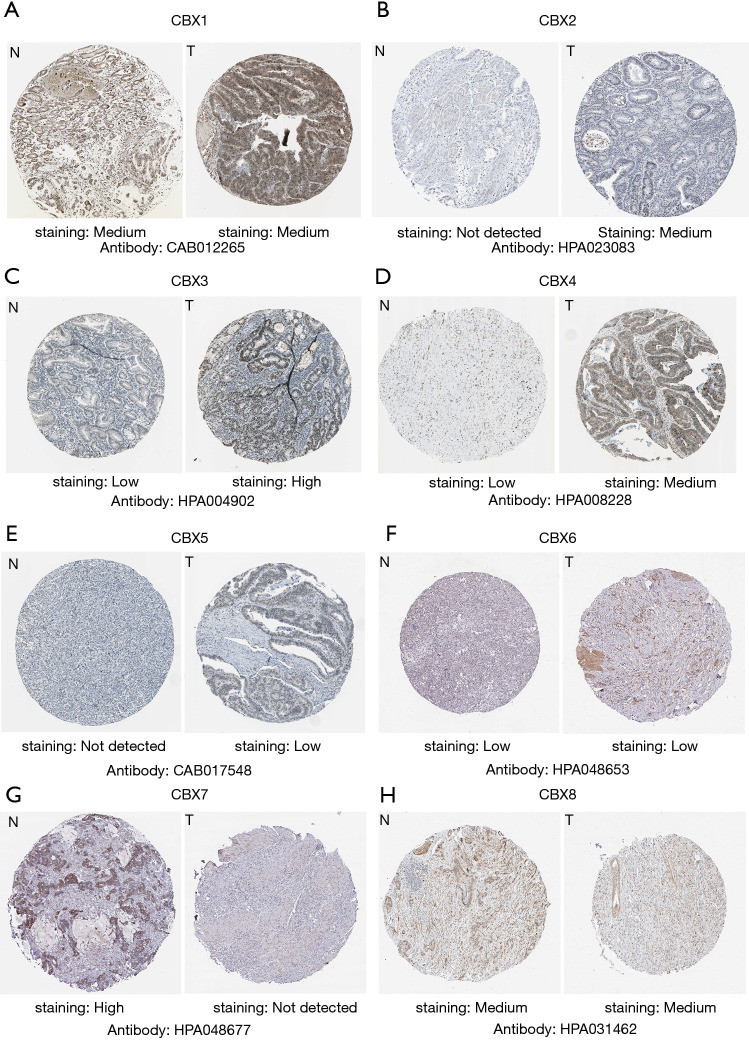
Protein expression of CBXs family members
We explored the protein expression patterns of CBXs in the Human Protein Atlas (Figure 5) and found that CBX2/3/4/5 protein expression was higher in tumor tissues, but CBX7 protein expression was significantly higher in healthy gastric tissues. Meanwhile, we found that CBX1/6/8 protein was not significantly differentially expressed between gastric cancer tissues and healthy gastric tissues. Except for CBX1/8, these results are approximately the same trend as CBXs family mRNA expression.
Figure 5.
Immunohistochemical images of CBXs family in Human Protein Atlas. To predict the protein expression pattern of CBXs family, 8 CBXs family members in gastric cancer and normal gastric tissue typical protein expression in immunohistochemical map. The legend for each image is 200 µm.
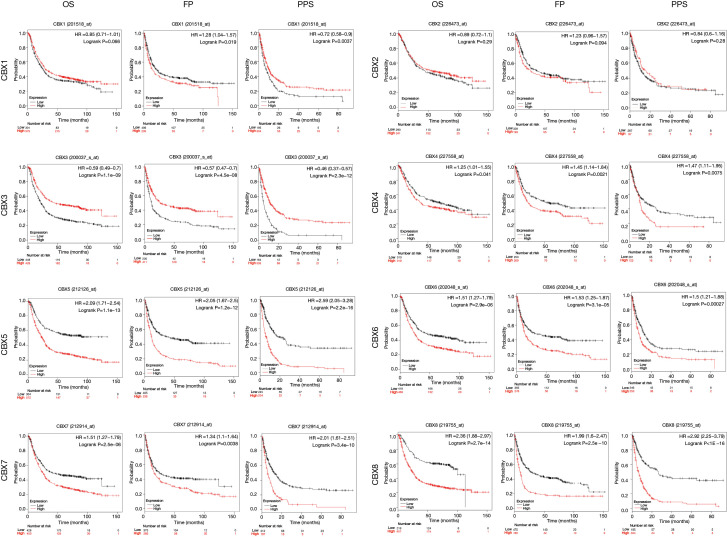
Prognostic value of the CBXs family
We analyzed the relationship between the mRNA expression of CBXs family and the survival time of gastric cancer patients in the Kaplan-Meier plotter to determine the prognostic value of each CBXs. The result showed that most members of the CBXs family were significantly correlated with the prognosis of patients with gastric cancer (Figure 6). Kaplan-Meier curve and log-rank test analysis showed higher CBX4/5/6/7/8 mRNA expression and poor OS, FP, and PPS was significantly correlated (P<0.05). However, higher CBX3 mRNA expression was significantly associated with better OS, FP, and PPS. There was no significant correlation between CBX2 mRNA expression and survival in patients with gastric cancer. High CBX1 mRNA expression was significantly associated with worse FP and better PPS, but not with OS. It shows that CBX3/4/5/6/7/8 is significantly related to the prognosis of gastric cancer patients and can be used as a useful biomarker to predict gastric cancer patients’ survival rate.
Figure 6.
The Prognostic value of mRNA expression of CBXs family members in gastric cancer patients (Kaplan-Meier plotter). Association between mRNA expression of eight CBXs family members and OS, FP and PPS in gastric cancer patients. The samples of gastric cancer patients were obtained from the datasets of GSE14210 (n=146), GSE15459 (n=200), GSE2237 (n=43), GSE29272 (n=268), GSE51105 (n=94), and GSE62254 (n=300) in GEO database which were processed by the same standard.
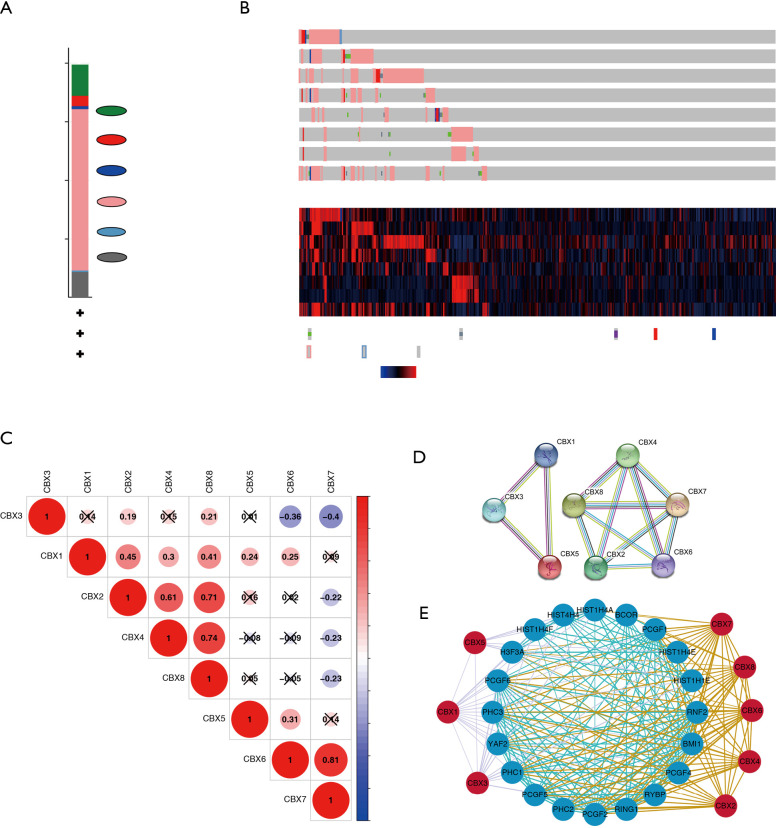
CBXs gene mutations and co-expressed genes of the CBX family
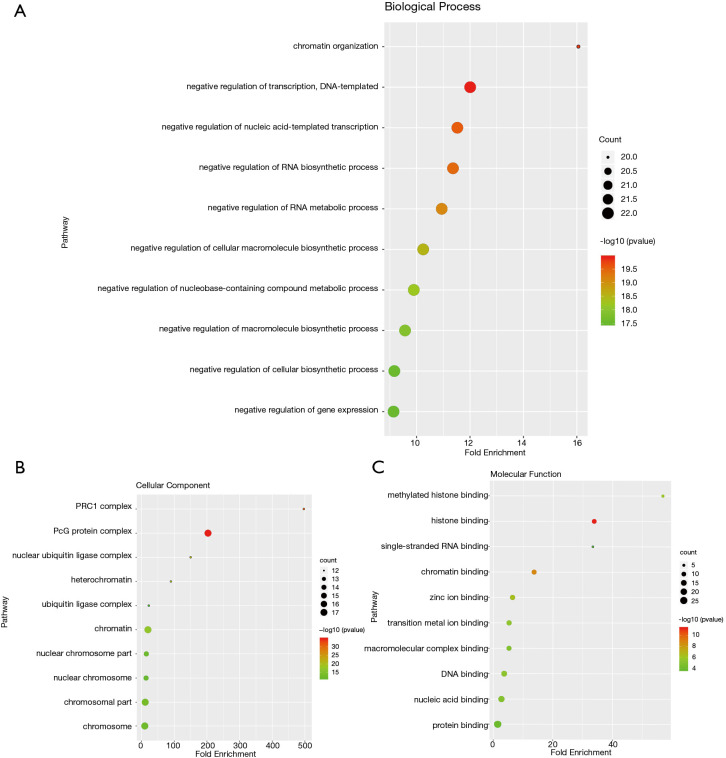
We analyzed the mutation of CBXs gene in gastric cancer patients in the cBioPortal online tool and observed that CBXs gene was mutated in 160 patients from 407 gastric cancer patients with a mutation rate of 39% (Figure 7A,B). We also calculated the correlation among CBXs by analyzing their mRNA expression, and Pearson’s correction was included. The results are shown in Figure 7C, where several genes with the most significant positive correlations are present: CBX2 with CBX4 and CBX8; CBX4 with CBX8; CBX6 with CBX7. We constructed the interaction network of these eight CBXs family proteins and the genes with which they have co-expression relationships in STRING (Figure 7D,E). The functions of CBXs and their co-expressed genes and the molecular mechanisms by which CBXs regulate tumor were subsequently predicted in DAVID6.8 (Figure 8). Gene ontology (GO) includes biological process (BP), cellular component (CC), and molecular function (MF). By annotating, we found that CBXs and their co-expressed genes are mainly enriched in negative regulation of transcription, DNA-templated, chromatin organization; polycomb group (PcG) protein complex, PRC1 complex; histone binding, chromatin binding and so on. The KEGG pathways involved in these genes were subsequently analyzed. Furthermore, because few pathways were involved, and most of them did not have related CBXs family genes, we simultaneously analyzed the pathways involved in regulating tumors by CBXs in Reactome. The results are shown in Table 2. We found that these genes were mainly enriched in signaling pathways of stem cells, viral carcinogenesis, regulation of PTEN gene transcription, and oxidative stress-induced senescence and associated with the function of CBXs mutations in gastric cancer.
Figure 7.
Gene expression and mutation analysis of CBXs in gastric cancer. (A,B) CBX gene expression and mutation analysis in gastric cancer (cBioPortal). (C) PPI network of CBXs family proteins. (D) Correction between different CBXs in gastric cancer. (E) CBXs family co-expression network diagram. Blue nodes indicate proteins that have co-expression relationships with CBXs family proteins. A thicker line between each node indicates a stronger co-expression relationship.
Figure 8.
The gene ontology (GO) analysis of CBXs and genes significantly associated with CBX. (A) Gene ontology inclusion biological process, (B) cellular component and (C) molecular function are represented in the figures as circles, triangles and squares, respectively.
Table 2. The functions of CBXs and genes significantly associated with CBX alterations were predicted by the analysis of pathway in regulating gastric cancer.
| Category | Term | Count | P value/false discovery rate | Genes |
|---|---|---|---|---|
| KEGG_Pathway | Systemic lupus erythematosus | 5 | P=1.61E-05 | HIST4H4, HIST1H4A, HIST1H4E, H3F3A, HIST1H4F |
| Signaling pathways regulating pluripotency of stem cells | 5 | P=1.91E-05 | BMI1, PCGF5, PCGF2, PCGF6, PCGF1 | |
| Alcoholism | 5 | P=4.83E-05 | HIST4H4, HIST1H4A, HIST1H4E, H3F3A, HIST1H4F | |
| Viral carcinogenesis | 4 | P=0.001918327 | HIST4H4, HIST1H4A, HIST1H4E, HIST1H4F | |
| Reactome_Pathway | RUNX1 interacts with co-factors whose precise effect on RUNX1 targets is not known | 4 | False discovery rate =2.90E-08 | CBX2, CBX4, CBX6, CBX8 |
| SUMOylation of chromatin organization proteins | 4 | False discovery rate =6.00E-08 | CBX2, CBX4, CBX5, CBX8 | |
| Regulation of PTEN gene transcription | 4 | False discovery rate =6.98E-08 | CBX2, CBX4, CBX6, CBX8 | |
| Oxidative Stress Induced Senescence | 4 | False discovery rate =2.83E-07 | CBX2, CBX4, CBX6, CBX8 | |
| SUMOylation of DNA methylation proteins | 3 | False discovery rate =2.83E-07 | CBX2, CBX4, CBX8 | |
| SUMOylation of transcription cofactors | 3 | False discovery rate =1.83E-06 | CBX2, CBX4, CBX8 | |
| SUMOylation of RNA binding proteins | 3 | False discovery rate =1.83E-06 | CBX2, CBX4, CBX8 | |
| Generic Transcription Pathway | 6 | False discovery rate =2.07E-06 | CBX2, CBX3, CBX4, CBX5, CBX6, CBX8 | |
| SUMOylation of DNA damage response and repair proteins | 3 | False discovery rate =5.11E-06 | CBX2, CBX4, CBX8 | |
| Transcriptional Regulation by E2F6 | 2 | False discovery rate =0.00012 | CBX3, CBX5 |
Discussion
It has been shown that members of the CBXs family play a role in a variety of tumors, including gastric cancer, but the differential roles of this family in gastric cancer remain elucidated. PcG proteins are a class of epigenetic regulators that play a crucial role in stem cell recognition, differentiation, and disease. They always play a role in Polycomb inhibitory complexes (including PRC1 and PRC2) and exert their functions in regulating cell proliferation, aging, and tumorigenesis through normal growth-regulating pathways (25). The CBXs family proteins are the normative component of PRC1, responsible for targeting PRC1 to chromatin (26). After the regulatory relationship between the CBXs family proteins and some interacting proteins is clarified, its role in gastric cancer can be analyzed through normal pathways. According to our understanding, this study is the first time to use bioinformatics to analyze the mRNA expression, mutation, and prognostic value of CBXs family in gastric cancer patients (OS, FP, PPS). We hoped that our findings would help broaden the existing knowledge on gastric cancer, improve or propose new treatment designs, and improve the accuracy of prognosis of gastric cancer patients.
Chromobox 1 (CBX1) belongs to the heterochromatin protein 1 (HP1) family, also known as HP1β. Yang et al. found CBX1 was significantly up-regulated in HCC tissues and cell lines and was significantly associated with larger tumor volumes, distant metastases, poor tumor differentiation, and tumor vessel infiltration through the standardized cell and tissue experiments. Mechanistically, it was found that the over-expressed CBX1 interacts with HMGA2 to trigger the Wnt/β-Catenin signaling pathway and enhance liver cancer cell proliferation and migration (27). The high mobility group protein A2 (HMGA2) is an architectural transcription factor that plays a role in the development of a variety of tumors. Zha et al. found that the protein and mRNA levels of HMGA2 were up-regulated in gastric cancer cells and tissues by immunohistochemistry and cytology. That overexpression of HMGA2 could enhance the oncogenicity of cell lines of gastric epithelial origin, and enhance migration and invasion in vitro and in vivo. They studied molecular biological changes in gastric cancer cells and changes in HMGA2 target genes after HMGA2 overexpression and knockdown. The results show that HMGA2 could not only translocate β-catenin through TWIST1 but also protect β-catenin from phosphorylation and degradation by inhibiting the transcription of AXIN1, thereby activating the Wnt/β-catenin pathway and causing EMT (Epithelial-Mesenchymal Transition) to improve the progression of gastric cancer (28). Other studies have shown that in pancreatic cancer, the WNT pathway has a significant role in promoting an aggressive tumor phenotype (29).
CBX1 is also up-regulated in Prostate cancer tissues, and higher CBX1 expression can enhance androgen receptor pathway activity, thereby regulating prostate cancer growth (30). In patients with thyroid tumors, the results vary, with CBX1 downregulated in thyroid cancer tissue and potentially enhancing the malignant and metastatic potential of thyroid cancer. It may be related to the inhibitory effect of miRNAs that are highly expressed in thyroid cancer (miR-205, etc.) (31). Also, CBX1 expression increases in breast cancer and is associated with poor prognosis in breast cancer patient (32). At present, there is no report on the role of CBX1 in various tumors, including gastric cancer. Our study found that CBX1 was up-regulated in gastric cancer, and its expression was correlated with clinical parameters such as cancer stage and tumor grade. We also found that CBX1 has regulatory relationships with H3F3A, PCGF6, PHC3, YAF2, and PHC1 protein, which may provide directions for future studies on the function and molecular mechanisms of CBX1 in gastric cancer. We also found that the high expression of CBX1 was associated with worse FP and better PPS in gastric cancer patients. The results demonstrated that CBX1 might be a potential molecular marker of gastric cancer, and its specific function and deeper mechanism in gastric cancer need further study.
Clermont et al. found high CBX2 mRNA expression in breast, lung, colorectal, prostate, brain, and hematopoietic tumors associated with metastasis and worse OS in breast cancer (33). Besides, it has been shown that CBX2 is highly expressed in high grade serous ovarian carcinoma (HGSOC). Meanwhile, up-regulated CBX2 promotes HGSOC by inducing a stem-like transcriptional profile and suppressing imbalance, which is associated with worse survival time (14). No studies have reported the role of CBX2 in gastric cancer. In this study, we found that CBX2 was significantly highly expressed in gastric cancer. We also found that the expression of CBX2 was correlated with the presence of H. pylori infection and cancer grade. Therefore, CBX2 can be a potential molecular marker of gastric cancer, which can be a more in-depth study of gastric cancer to define the function of CBX in gastric cancer.
CBX3 is highly expressed in gliomas. In glioma patients, up-regulated CBX3 is indicative of worse relapse-free survival (RFS) and OS (34). Other studies have shown that CBX3 is highly expressed in osteosarcoma, which is associated with worse disease-free survival (DFS) and OS in patients with osteosarcoma, large tumor volumes, and high distant metastasis rates, and high clinical staging (35). Chen et al. found that CBX3 overexpression in pancreatic adenocarcinoma (PAAD) tissues induced PAAD cells to proliferate in vitro, anchor-free growth, migration, and invasion, which were related to poor OS and DFS in PAAD patients (12). Some studies show that CBX3 plays a role in the Tongue Squamous Cell Carcinoma (36) and vascular smooth muscle cells and Vascular Smooth Muscle Cell (37,38). No studies have yet demonstrated the role played by CBX3 in gastric cancer. In our study, CBX3 was highly expressed in gastric cancer. Furthermore, CBX3 expression was higher in higher tumor stages and tumor tissues with H. pylori invasion. The OS, FP, and PPS regarding prognosis were better in gastric cancer patients with high CBX3 expression. It is suggested that CBX3 may be a potential biomarker for gastric cancer, and maybe an excellent prognostic marker for gastric cancer patients, and guide clinical treatment.
The role of CBX4 in a variety of cancers has been thoroughly studied. Hu et al. found that CBX4 is highly expressed in lung cancer and can significantly promote the proliferation and migration of cancer cells by regulating the expression of BMI-1 (6). CBX4 can be up-regulated by HIF-1α signaling activation in osteosarcoma. The high expression of CBX4 is related to the late clinical-stage, high malignancy, and low tumor necrosis rate, and it plays a pro-cancer role in osteosarcoma (39). CBX4 increases angiogenesis and metastasis of hepatocellular carcinoma (HCC) through its SUMOylating effect on hypoxia-inducible factor-1α. It was also associated with histological grade, tumor size, pathological differentiation, lymph node metastasis, microvessel density, distant metastasis, and hematogenous metastasis of HCC. High expression of CBX4 predicted shorter OS and RFS in HCC patients (40-42). The highly expressed CBX4 also plays a cancer-promoting role in breast cancer by promoting breast cancer through miR-137-mediated Notch1 signaling and can contribute to the poor prognosis of breast cancer patients (43).
Similarly, specific studies on the role of CBX4 in gastric cancer are still lacking, so our study is instructive for developing the role of CBX4 in gastric cancer. In our study, CBX4 was highly expressed in gastric cancer. Higher CBX4 expression in gastric cancer patients predicted worse OS, FP, and PPS. These results indicate that CBX4 might be an independent prognostic factor for gastric cancer. So new therapies for gastric cancer can be developed and directed in the future.
CBX5, also named heterochromatic protein 1α (HP1α), is closely related to CBX1 (HP1β) and CBX3 (HP1γ) and is involved in gene silencing, genome stability and chromosome segregation (44). Previous studies have shown that CBX5 is highly expressed in gastric cancer, can promote the proliferation, migration, and invasion of gastric cancer cells in vitro, and can be inhibited by highly expressed microRNA-758-3p (45). CBX5 is highly expressed in glioma cells, which is conducive to cell proliferation and survival (46). CBX5 expression is reduced only in metastatic carcinomas and poorly differentiated thyroid carcinomas and contributes to tumorigenesis. In lung cancer patients, CBX5 mRNA expression was significantly increased in tumor samples and metastatic lesions associated with poorer OS. The knockdown of CBX5 significantly inhibited the migration of CD133+ cancer stem-like cells (TSLCs) in vitro and the tumorigenic engraftment, tumor growth rate, and propensity for lung metastasis caused by lung CD133+ TSLCs in vivo (47). In breast cancer cells, the expression of HP1α protein is down-regulated. It is an inhibitor of invasive breast cancer cell migration and invasion. CBX5 is mainly down-regulated at the transcription level, and E2F transcription factors can regulate CBX5 transcription. Other studies have shown that decreased expression of YY1 in breast cancer cells may help breast cancer cells acquire an aggressive phenotype by increasing cell migration and decreasing CBX5 expression (48,49). Our study also demonstrated that CBX5 is up-regulated in gastric cancer. High CBX5 expression in gastric cancer patients predicted worse OS, FP, and PPS. These results suggest that CBX5 might be a prognostic marker for gastric cancer patients. It is worthwhile to further study the specific mechanism of CBX5 in gastric cancer to improve the understanding of gastric cancer and develop new therapeutic means.
The role of CBX6 in a variety of tumors has been initially identified. CBX6 was found to be down-regulated in glioblastoma multiforme (GBM) cells and glioma tissues. The induced over-expression of CBX6 inhibited the proliferation of glioblastoma cells (50). Other studies have shown that CBX6 is up-regulated in both HCC cells and tissues. The role of CBX6 in HCC is mediated by S100A9/NF-κB/MAPK pathway. Up-regulation of CBX6 in liver cancer is associated with the promotion of in vitro and in vivo growth of cancer cells and poor prognosis (51,52). There is no clear evidence that shows that CBX6 is significantly down-regulated in breast cancer. Over-expression of CBX6 inhibits breast cancer cells' ability to proliferate and metastasize and can inhibit the growth of breast cancer (10). Up to now, no study has reported the role of CBX6 in gastric cancer. According to our study, CBX6 was not differentially expressed in gastric cancer tissue and healthy gastric tissue. However, gastric cancer patients with high CBX6 expression showed worse OS, FP, and PPS compared to gastric cancer patients with low CBX6 expression according to the results in Kaplan-Meier Plotter. These results indicate that CBX6 can be a potential prognostic marker in gastric cancer.
In the functional summary of CBX7 by Pallante et al., we can learn that CBX7 is down-regulated in various human malignancies, such as carcinoma of the bladder, thyroid, colorectal, breast, pancreas, lung carcinoma and glioma. Moreover, a decreased expression of CBX7 was significantly associated with higher invasiveness and poorer prognosis of tumors. For specific mechanisms, multiple miRNAs (including miR-9, miR-421, miR-181b, miR-182, and miR-183) and proteins (HMGA1 and MYC) are directly or indirectly involved in the repression of CBX7 expression. CBX7 can also be highly expressed in prostate cancer, ovarian cancer, and lymphoma, with oncogenic effects. In ovarian cancer patients, we found that patients with CBX7 over-expression had reduced OS and PFS rates (53,54). Li et al. found that CBX7 was up-regulated in cervical cancer, and high expression of CBX7 could inhibit p65, induce E-cadherin expression, inhibit cell proliferation, induce apoptosis, inhibit cell migration and invasion. They also found that the knockdown of CBX7 promoted the growth and migration as well as invasion of cervical cancer cells by molecular means, which showed that CBX7 is a potential tumor suppressor for cervical cancer (7). Other studies have shown that CBX7 expression is significantly lower in HCC tissues than in adjacent non-tumor tissues. It is associated with cirrhosis. Down-regulation of CBX7 in HCC patients predicts poor prognosis (55). Other relevant mechanisms of CBX7 carcinogenesis may interact with CBX7 with histone-modifying enzymes such as HDAC2 and PRMT1 to enhance E-cadherin expression. Disruption of this balance may induce impaired E-cadherin expression, increase cell migration, and lead to EMT, leading to cancer progression (56).
Nguyen et al. found that in hepatocellular carcinoma, CBX family proteins, particularly CBX7, are associated with KRAS-dependent oncogenic driving mechanisms (57). In gastric cancer, Ni et al. confirmed that CBX7 was highly expressed in gastric cancer cells by standardized cell experiments and that highly expressed CBX7 enhanced gastric cancer cell proliferation and colony-forming ability, also enhanced cell migration and invasion, resulting in reduced sensitivity to gastric cancer cellularization. Immunohistochemical experiments on 95 gastric cancer tissues showed that CBX7 was highly expressed in 40 gastric cancer tissues and positively correlated with regional lymph node metastasis and TNM stage. Besides, CBX7 expression was positively correlated with the expression of the stem cell markers OCT-4 and CD133, indicating that it could positively regulate the gastric cancer stem cell phenotype. In a study of the molecular mechanisms of CBX7 regulation of gastric cancer, show that CBX7 promotes cancer stem cell properties of gastric cancer cells by downregulating p16INK4a/ARF. CBX7 overexpression promoted AKT and ERK activation, whereas CBX7 depletion downregulated the levels of p-AKT and p-ERK in gastric cancer cells, and CBX7 regulated the stem cell properties of gastric cancer cells by activating the expression of miR-21 through activation of the AKT-NF-κB pathway (5).
CBX7 can also modulate the stem cell-like properties of gastric cancer cells in an immunodeficiency model, and immunodeficiency is significantly associated with a high incidence of gastrointestinal manifestations and an increased risk of gastric cancer and lymphoma (including genetics, immune disorders, and pyloric screw Chronic infection of bacteria) (58). These studies provide a decent idea for studying the role and molecular mechanism of other proteins of the CBXs in gastric cancer. By combining our findings, we can carry out available research. Sorafenib is an oral kinase inhibitor found to have promising applications in several specific clinical scenarios (liver and gastric cancer) (59,60), which could be instructive for future targeted therapy of related tumors. CBX7 mRNA expression was downregulated in gastric cancer in our study, which is contrary to the existing conclusion. Nevertheless, immunohistochemical plots showed that CBX7 protein expression was significantly elevated in tumor tissues, consistent with the current study. We found that relatively high CBX7 expression in gastric cancer patients means worse OS, FP, and PPS, suggesting that CBX7 is also a poor prognostic indicator of gastric cancer. CBX7 is strongly linked to PHC family proteins and PCGF family proteins, and the mechanism by which CBX7 is linked to these proteins can be investigated in gastric cancer to guide future therapy.
CBX8 can regulate cell differentiation, senescence, and cell cycle progression in many cancers. It has been shown to play different roles in a variety of tumors. CBX8 is highly expressed in HCC and interacts with YBX1 to regulate the cell cycle. Overexpression of CBX8 promotes HCC cell growth. At the same time, it is associated with poor prognosis of HCC patients. The related molecular mechanism may be since CBX8 promotes the proliferation of HCC cells through YBX1-mediated cell cycle processes. In a study on the role of CBX8 in HCC by Tang et al., they found that up-regulation of CBX8 in HCC patients and could enhance cancer stem-like and metastatic ability. High CBX8 expression in HCC tissues predicted a lower survival rate of HCC patients. Further research indicates that these functions of CBX8 are mainly exerted by H3K27me3’s control of the transcriptional expression of bone morphogenetic protein 4 (BMP4) and regulation of Smads and mitogen-activated protein kinase (MAPK) signaling pathways. It has also been shown that CBX8 exhibits oncogenic activity by activating the Akt/β-catenin signaling pathway in HCC (9,61,62).
CBX8 is also up-regulated in bladder cancer tissues and is associated with progression to T, N, and M stages. Promote the proliferation of cancer cells by inhibiting the p53 pathway. Simultaneously, it also leads to a poor prognosis for patients with muscular-invasive bladder cancer (63). CBX8 is over-expressed in both CRC tissues and cell lines and promotes CRC proliferation by inhibiting p53, but interestingly low-expression CBX8 is associated with poor DFS and OS of CRC patients (64,65). After M6A methylation-induced upregulation, CBX8 interacts with KMT2B and Pol II to promote LGR5 expression, which helps increase the dryness of cancer and reduces the sensitivity of CRC to chemotherapy. CBX8 interacts with KMT2B and Pol II and promotes the expression of LGR5 in an atypical manner in colon cancer after induction of upregulation by m6A methylation, leading to increase cancer stemness and reduce chemosensitivity in colon cancer (66). In addition to the diseases mentioned above, CBX8 also plays a crucial role in esophageal squamous cell carcinoma (ESCC) (67), lymphoma (68), breast cancer (69,70), esophageal cancer (71), and other diseases. So far, there is no precise study shows the exact role of CBX8 in gastric cancer. Our study showed that CBX8 was over-expressed in gastric cancer and correlated with clinicopathological parameters such as tumor grade and whether there was H. pylori infection. Besides, we analyzed the relationship between gastric cancer patients' survival time and the expression of CBX8. The high expression of CBX8 in gastric cancer represents worse OS, FP, and PPS. These results indicate that CBX8 can serve as a potential molecular marker and a prognostic and therapeutic target for gastric cancer.
Conclusions
In this study, we systematically analyzed the expression and prognostic value of CBXs in gastric cancer, opening broad prospects for the potential of CBXs as diagnostic and prognostic markers of gastric cancer. We also found that CBXs genes can associate with each other by regulating one or more genes (some H4 Histone Family proteins, PCGF1/2/4/5/6 of the Polycomb Group Ring Finger family, or PHC1/2/3 of the Polycomb group of gene family) to function together in tumors. It may provide a reliable molecular basis for future comprehensive studies on the function and molecular mechanisms of the CBXs family in gastric cancer. However, our study also has some known limitations. For instance, our large number of samples were derived from online databases. Furthermore, a larger sample size is needed to validate our results. Some members of the CBXs family proteins as potential markers and prognostic indicators of gastric cancer have not been validated experimentally nor in-depth mechanistic studies.
Nevertheless, we endeavored to provide reliable results. Our results indicated increased expression of CBX1/2/3/4/5/8 might profoundly affect the prognosis of gastric cancer patients. The high expression of CBX4/5/6/7/8 in gastric cancer patients is a potential indicator of poor prognosis in gastric cancer patients. High mRNA expression of CBX3 may be a beneficial prognostic factor in gastric cancer patients. Genetic alterations in CBXs are associated with poor prognosis in gastric cancer patients. In conclusion, we have shown that CBX3/4/5/6/7/8 can be used as a prognostic biomarker in gastric cancer patients.
Acknowledgments
The data presented here is derived, in part or whole, from the TCGA Research Network (https://www.cancer.gov/tcga). The CBXs expression and gastric cancer patient survival association results are based on data generated by the GEO datasets (https://www.ncbi.nlm.nih.gov/geo/) and Kaplan Meier plotter (http://kmplot.com/analysis/index.php?p=service&cancer=gastric).
Funding: This study was supported by grants from the National Natural Science Foundation of China (No. 881560389), the Project of the Jiangxi Provincial Department of Science and Technology (No. 20181BBG70015), and the Jiangxi Provincial Health Department (No. 20171068).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study did not involve animal or human experiments and did not require ethical approval or patient consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
Footnotes
Reporting Checklist: The authors have completed the Materials Design Analysis Reporting (MDAR) reporting checklist. Available at http://dx.doi.org/10.21037/jgo-20-208
Peer Review File: Available at http://dx.doi.org/10.21037/jgo-20-208
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jgo-20-208). The authors have no conflicts of interest to declare.
References
- 1.Klauke K, Radulović V, Broekhuis M, et al. Polycomb Cbx family members mediate the balance between haematopoietic stem cell self-renewal and differentiation. Nat Cell Biol 2013;15:353-62. 10.1038/ncb2701 [DOI] [PubMed] [Google Scholar]
- 2.Morey L, Pascual G, Cozzuto L, et al. Nonoverlapping functions of the Polycomb group Cbx family of proteins in embryonic stem cells. Cell Stem Cell 2012;10:47-62. 10.1016/j.stem.2011.12.006 [DOI] [PubMed] [Google Scholar]
- 3.Vincenz C, Kerppola TK. Different polycomb group CBX family proteins associate with distinct regions of chromatin using nonhomologous protein sequences. Proc Natl Acad Sci U S A 2008;105:16572-7. 10.1073/pnas.0805317105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wotton D, Merrill JC. Pc2 and SUMOylation. Biochem Soc Trans 2007;35:1401-4. 10.1042/BST0351401 [DOI] [PubMed] [Google Scholar]
- 5.Ni SJ, Zhao LQ, Wang XF, et al. CBX7 regulates stem cell-like properties of gastric cancer cells via p16 and AKT-NF-κB-miR-21 pathways. J Hematol Oncol 2018;11:17. 10.1186/s13045-018-0562-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu C, Zhang Q, Tang Q, et al. CBX4 promotes the proliferation and metastasis via regulating BMI-1 in lung cancer. J Cell Mol Med 2020;24:618-31. 10.1111/jcmm.14771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li R, Yan Q, Tian P, et al. CBX7 Inhibits Cell Growth and Motility and Induces Apoptosis in Cervical Cancer Cells. Mol Ther Oncolytics 2019;15:108-16. 10.1016/j.omto.2019.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mao J, Tian Y, Wang C, et al. CBX2 Regulates Proliferation and Apoptosis via the Phosphorylation of YAP in Hepatocellular Carcinoma. J Cancer 2019;10:2706-19. 10.7150/jca.31845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang CZ, Chen SL, Wang CH, et al. CBX8 Exhibits Oncogenic Activity via AKT/β-Catenin Activation in Hepatocellular Carcinoma. Cancer Res 2018;78:51-63. 10.1158/0008-5472.CAN-17-0700 [DOI] [PubMed] [Google Scholar]
- 10.Deng H, Guan X, Gong L, et al. CBX6 is negatively regulated by EZH2 and plays a potential tumor suppressor role in breast cancer. Sci Rep 2019;9:197. 10.1038/s41598-018-36560-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng S, Lv P, Su J, et al. Overexpression of CBX2 in breast cancer promotes tumor progression through the PI3K/AKT signaling pathway. Am J Transl Res 2019;11:1668-82. [PMC free article] [PubMed] [Google Scholar]
- 12.Chen LY, Cheng CS, Qu C, et al. Overexpression of CBX3 in Pancreatic Adenocarcinoma Promotes Cell Cycle Transition-Associated Tumor Progression. Int J Mol Sci 2018;19:1768. 10.3390/ijms19061768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Chen H, Zhu H, et al. CBX8 promotes tumorigenesis and confers radioresistance in esophageal squamous cell carcinoma cells through targeting APAF1. Gene 2019;711:143949. 10.1016/j.gene.2019.143949 [DOI] [PubMed] [Google Scholar]
- 14.Wheeler LJ, Watson ZL, Qamar L, et al. CBX2 identified as driver of anoikis escape and dissemination in high grade serous ovarian cancer. Oncogenesis 2018;7:92. 10.1038/s41389-018-0103-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Cutsem E, Sagaert X, Topal B, et al. Gastric cancer. Lancet 2016;388:2654-64. 10.1016/S0140-6736(16)30354-3 [DOI] [PubMed] [Google Scholar]
- 16.Tang Z, Kang B, Li C, et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 2019;47:W556-60. 10.1093/nar/gkz430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uhlen M, Zhang C, Lee S, et al. A pathology atlas of the human cancer transcriptome. Science 2017;357:eaan2507. [DOI] [PubMed]
- 19.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoadley KA, Yau C, Hinoue T, et al. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell 2018;173:291-304.e6. 10.1016/j.cell.2018.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 23.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1-13. 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jassal B, Matthews L, Viteri G, et al. The reactome pathway knowledgebase. Nucleic Acids Res 2020;48:D498-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Croce L, Helin K. Transcriptional regulation by Polycomb group proteins. Nat Struct Mol Biol 2013;20:1147-55. 10.1038/nsmb.2669 [DOI] [PubMed] [Google Scholar]
- 26.Ma RG, Zhang Y, Sun TT, et al. Epigenetic regulation by polycomb group complexes: focus on roles of CBX proteins. J Zhejiang Univ Sci B 2014;15:412-28. 10.1631/jzus.B1400077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang YF, Pan YH, Tian QH, et al. CBX1 Indicates Poor Outcomes and Exerts Oncogenic Activity in Hepatocellular Carcinoma. Transl Oncol 2018;11:1110-8. 10.1016/j.tranon.2018.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zha L, Zhang J, Tang W, et al. HMGA2 elicits EMT by activating the Wnt/β-catenin pathway in gastric cancer. Dig Dis Sci 2013;58:724-33. 10.1007/s10620-012-2399-6 [DOI] [PubMed] [Google Scholar]
- 29.Argentiero A, De Summa S, Di Fonte R, et al. Gene Expression Comparison between the Lymph Node-Positive and -Negative Reveals a Peculiar Immune Microenvironment Signature and a Theranostic Role for WNT Targeting in Pancreatic Ductal Adenocarcinoma: A Pilot Study. Cancers (Basel) 2019;11:942. 10.3390/cancers11070942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shiota M, Song Y, Yokomizo A, et al. Human heterochromatin protein 1 isoform HP1beta enhances androgen receptor activity and is implicated in prostate cancer growth. Endocr Relat Cancer 2010;17:455-67. 10.1677/ERC-09-0321 [DOI] [PubMed] [Google Scholar]
- 31.Tretiakova MS, Bond SD, Wheeler D, et al. Heterochromatin protein 1 expression is reduced in human thyroid malignancy. Lab Invest 2014;94:788-95. 10.1038/labinvest.2014.68 [DOI] [PubMed] [Google Scholar]
- 32.Lee YH, Liu X, Qiu F, et al. HP1β is a biomarker for breast cancer prognosis and PARP inhibitor therapy. PLoS One 2015;10:e0121207. 10.1371/journal.pone.0121207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clermont PL, Sun L, Crea F, et al. Genotranscriptomic meta-analysis of the Polycomb gene CBX2 in human cancers: initial evidence of an oncogenic role. Br J Cancer 2014;111:1663-72. 10.1038/bjc.2014.474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao SP, Wang F, Yang M, et al. CBX3 promotes glioma U87 cell proliferation and predicts an unfavorable prognosis. J Neurooncol 2019;145:35-48. 10.1007/s11060-019-03286-w [DOI] [PubMed] [Google Scholar]
- 35.Ma C, Nie XG, Wang YL, et al. CBX3 predicts an unfavorable prognosis and promotes tumorigenesis in osteosarcoma. Mol Med Rep 2019;19:4205-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang H, Chen W, Fu X, et al. CBX3 promotes tumor proliferation by regulating G1/S phase via p21 downregulation and associates with poor prognosis in tongue squamous cell carcinoma. Gene 2018;654:49-56. 10.1016/j.gene.2018.02.043 [DOI] [PubMed] [Google Scholar]
- 37.Xiao Q, Wang G, Yin X, et al. Chromobox protein homolog 3 is essential for stem cell differentiation to smooth muscles in vitro and in embryonic arteriogenesis. Arterioscler Thromb Vasc Biol 2011;31:1842-52. 10.1161/ATVBAHA.111.230110 [DOI] [PubMed] [Google Scholar]
- 38.Zhang C, Chen D, Maguire EM, et al. Cbx3 inhibits vascular smooth muscle cell proliferation, migration, and neointima formation. Cardiovasc Res 2018;114:443-55. 10.1093/cvr/cvx236 [DOI] [PubMed] [Google Scholar]
- 39.Yang J, Cheng D, Zhu B, et al. Chromobox Homolog 4 is Positively Correlated to Tumor Growth, Survival and Activation of HIF-1α Signaling in Human Osteosarcoma under Normoxic Condition. J Cancer 2016;7:427-35. 10.7150/jca.13749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiao HK, Xu Y, Li J, et al. Prognostic significance of Cbx4 expression and its beneficial effect for transarterial chemoembolization in hepatocellular carcinoma. Cell Death Dis 2015;6:e1689. 10.1038/cddis.2015.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mei Z, Jiao H, Wang W, et al. Polycomb chromobox 4 enhances migration and pulmonary metastasis of hepatocellular carcinoma cell line MHCC97L. Sci China Life Sci 2014;57:610-7. 10.1007/s11427-014-4663-9 [DOI] [PubMed] [Google Scholar]
- 42.Wang B, Tang J, Liao D, et al. Chromobox homolog 4 is correlated with prognosis and tumor cell growth in hepatocellular carcinoma. Ann Surg Oncol 2013;20 Suppl 3:S684-92. 10.1245/s10434-013-3171-7 [DOI] [PubMed] [Google Scholar]
- 43.Zeng JS, Zhang ZD, Pei L, et al. CBX4 exhibits oncogenic activities in breast cancer via Notch1 signaling. Int J Biochem Cell Biol 2018;95:1-8. 10.1016/j.biocel.2017.12.006 [DOI] [PubMed] [Google Scholar]
- 44.De Koning L, Savignoni A, Boumendil C, et al. Heterochromatin protein 1alpha: a hallmark of cell proliferation relevant to clinical oncology. EMBO Mol Med 2009;1:178-91. 10.1002/emmm.200900022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo J, Zhang Z, Pan L, et al. Identification of miR-758-3p as Potential Modulator of CBX5 Expression in Gastric Cancer. Technol Cancer Res Treat 2018;17:1533033818816061. 10.1177/1533033818816061 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Lai X, Deng Z, Guo H, et al. HP1α is highly expressed in glioma cells and facilitates cell proliferation and survival. Biochem Biophys Res Commun 2017;490:415-22. 10.1016/j.bbrc.2017.06.056 [DOI] [PubMed] [Google Scholar]
- 47.Yu YH, Chiou GY, Huang PI, et al. Network biology of tumor stem-like cells identified a regulatory role of CBX5 in lung cancer. Sci Rep 2012;2:584. 10.1038/srep00584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lieberthal JG, Kaminsky M, Parkhurst CN, et al. The role of YY1 in reduced HP1alpha gene expression in invasive human breast cancer cells. Breast Cancer Res 2009;11:R42. 10.1186/bcr2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomsen R, Christensen DB, Rosborg S, et al. Analysis of HP1α regulation in human breast cancer cells. Mol Carcinog 2011;50:601-13. 10.1002/mc.20755 [DOI] [PubMed] [Google Scholar]
- 50.Li G, Warden C, Zou Z, et al. Altered expression of polycomb group genes in glioblastoma multiforme. PLoS One 2013;8:e80970. 10.1371/journal.pone.0080970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mei S, Qin Q, Wu Q, et al. Cistrome Data Browser: a data portal for ChIP-Seq and chromatin accessibility data in human and mouse. Nucleic Acids Res 2017;45:D658-D662. 10.1093/nar/gkw983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng H, Jiang WH, Tian T, et al. CBX6 overexpression contributes to tumor progression and is predictive of a poor prognosis in hepatocellular carcinoma. Oncotarget 2017;8:18872-84. 10.18632/oncotarget.14770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pallante P, Forzati F, Federico A, et al. Polycomb protein family member CBX7 plays a critical role in cancer progression. Am J Cancer Res 2015;5:1594-601. [PMC free article] [PubMed] [Google Scholar]
- 54.Scott CL, Gil J, Hernando E, et al. Role of the chromobox protein CBX7 in lymphomagenesis. Proc Natl Acad Sci U S A 2007;104:5389-94. 10.1073/pnas.0608721104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu X, Qin M, Li C, et al. Downregulated Expression of Chromobox Homolog 7 in Hepatocellular Carcinoma. Genet Test Mol Biomarkers 2019;23:348-52. 10.1089/gtmb.2018.0293 [DOI] [PubMed] [Google Scholar]
- 56.Federico A, Sepe R, Cozzolino F, et al. The complex CBX7-PRMT1 has a critical role in regulating E-cadherin gene expression and cell migration. Biochim Biophys Acta Gene Regul Mech 2019;1862:509-21. 10.1016/j.bbagrm.2019.02.006 [DOI] [PubMed] [Google Scholar]
- 57.Nguyen AT, Emelyanov A, Koh CHV, et al. A high level of liver-specific expression of oncogenic Kras(V12) drives robust liver tumorigenesis in transgenic zebrafish. Dis Model Mech 2011;4:801-13. 10.1242/dmm.007831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leone P, Vacca A, Dammacco F, et al. Common Variable Immunodeficiency and Gastric Malignancies. Int J Mol Sci 2018;19:451. 10.3390/ijms19020451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brunetti O, Gnoni A, Licchetta A, et al. Predictive and Prognostic Factors in HCC Patients Treated with Sorafenib. Medicina (Kaunas) 2019;55:707. 10.3390/medicina55100707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun W, Powell M, O'Dwyer PJ, et al. Phase II study of sorafenib in combination with docetaxel and cisplatin in the treatment of metastatic or advanced gastric and gastroesophageal junction adenocarcinoma: ECOG 5203. J Clin Oncol 2010;28:2947-51. 10.1200/JCO.2009.27.7988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tang B, Tian Y, Liao Y, et al. CBX8 exhibits oncogenic properties and serves as a prognostic factor in hepatocellular carcinoma. Cell Death Dis 2019;10:52. 10.1038/s41419-018-1288-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xiao L, Zhou Z, Li W, et al. Chromobox homolog 8 (CBX8) Interacts with Y-Box binding protein 1 (YBX1) to promote cellular proliferation in hepatocellular carcinoma cells. Aging 2019;11:7123-49. 10.18632/aging.102241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yuan GJ, Chen X, Lu J, et al. Chromobox homolog 8 is a predictor of muscle invasive bladder cancer and promotes cell proliferation by repressing the p53 pathway. Cancer Sci 2017;108:2166-75. 10.1111/cas.13383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Song X, Tang T, Li C, et al. CBX8 and CD96 Are Important Prognostic Biomarkers of Colorectal Cancer. Med Sci Monit 2018;24:7820-7. 10.12659/MSM.908656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tang J, Wang G, Zhang M, et al. Paradoxical role of CBX8 in proliferation and metastasis of colorectal cancer. Oncotarget 2014;5:10778-90. 10.18632/oncotarget.2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Y, Kang M, Zhang B, et al. m6A modification-mediated CBX8 induction regulates stemness and chemosensitivity of colon cancer via upregulation of LGR5. Mol Cancer 2019;18:185. 10.1186/s12943-019-1116-x [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67.Wang G, Tang J, Zhan W, et al. CBX8 Suppresses Tumor Metastasis via Repressing Snail in Esophageal Squamous Cell Carcinoma. Theranostics 2017;7:3478-88. 10.7150/thno.20717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Béguelin W, Teater M, Gearhart MD, et al. EZH2 and BCL6 Cooperate to Assemble CBX8-BCOR Complex to Repress Bivalent Promoters, Mediate Germinal Center Formation and Lymphomagenesis. Cancer Cell 2016;30:197-213. 10.1016/j.ccell.2016.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chung CY, Sun Z, Mullokandov G, et al. Cbx8 Acts Non-canonically with Wdr5 to Promote Mammary Tumorigenesis. Cell Rep 2016;16:472-86. 10.1016/j.celrep.2016.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee SH, Um SJ, Kim EJ. CBX8 suppresses Sirtinol-induced premature senescence in human breast cancer cells via cooperation with SIRT1. Cancer Lett 2013;335:397-403. 10.1016/j.canlet.2013.02.051 [DOI] [PubMed] [Google Scholar]
- 71.Xiao W, Ou C, Qin J, et al. CBX8, a novel DNA repair protein, promotes tumorigenesis in human esophageal carcinoma. Int J Clin Exp Pathol 2014;7:4817-26. [PMC free article] [PubMed] [Google Scholar]