Abstract
Background
This study analyzed the effect of circular RNA (circRNA), TBL1XR1, on the malignant progression of colon cancer cells and explored its possible molecular mechanism.
Methods
The expression levels of circTBL1XR1 in colorectal cancer and paracancerous tissues were detected by quantitative real-time polymerase chain (qRT-PCR). Changes in circTBL1XR1 expression in normal intestinal epithelial cells and colon cancer cell lines were detected at the cellular level. LoVo and SW620 cells with the highest circTBL1XR1 expression were transfected with circTBL1XR1, and the transfection efficiency was verified by qRT-PCR. The effects of circTBL1XR1 on the proliferation, migration, and invasion of colon cancer cells were detected by MTT, Transwell migration, and invasion assay, and a subcutaneous tumor model. Target genes of circTBL1XR1 were verified by luciferase reporter assay. Expression levels of circTBL1XR1 target genes were detected by qRT-PCR and Western blotting.
Results
circTBL1XR1 was highly expressed in colon cancer patients. circTBL1XR1 expression in colon cancer cells was also higher than that in normal intestinal cells. In vivo and in vitro experimental results showed that overexpression of circTBL1XR1 enhanced the proliferation, migration, and invasion of colon cancer cells. After lowering circTBL1XR1, the ability of migration and invasion of colon cancer cells was significantly reduced. Bioinformatics retrieval and luciferase reporter gene assay confirmed that circTBL1XR1 can bind to microRNA-424 (miR-424) and that the Smad7 gene is the target gene of miR-424.
Conclusions
circTBL1XR1 was highly expressed in colon cancer, and miR-424 was poorly expressed in colon cancer cells. circTBL1XR1 regulates the expression of Smad7 through miR-424, thereby affecting the malignant progression of colorectal cancer.
Keywords: circTBL1XR1, miR-424, Smad7, colorectal cancer, epithelial-mesenchymal transition (EMT)
Introduction
Colorectal cancer is a common tumor with a high mortality rate. Although the overall mortality rate of colorectal cancer has decreased with the improvement of early diagnosis technology and multidisciplinary comprehensive treatment, the incidence rate of colorectal cancer is increasing in more and more young people (1,2). In terms of treatment, stage III colorectal cancer is often treated with radical resection of colorectal cancer and postoperative adjuvant chemoradiotherapy, has good efficacy (3,4). However, due to the obvious heterogeneity of colorectal tumors and the lack of early diagnosis markers and effective intervention measures, the 5-year survival period of colorectal cancer is still poor (5,6), so it is particularly important to find early diagnosis markers and effective therapeutic targets (7-10).
In recent years, a large number of studies have confirmed that competing endogenous RNA (ceRNA) molecules are involved in the progress of colorectal cancer. These ceRNA molecules may be early diagnostic markers and therapeutic targets for colorectal cancer. Micro RNA (miRNA) can complementarily bind to the three prime untranslated region (3'UTR) of related genes, reduce the stability of messenger RNA (mRNA) or inhibit the translation of mRNA, and negatively regulate the expression of target genes. The 3'UTR of the target gene contains miRNA recognition elements (MREs), which can be complementary to the miRNA in forming a RNA-induced silencing complex. Related enzymes will destroy the structure of the target gene mRNA with the RNA-induced silencing complex or inhibit its translation. circRNA can be used as ceRNA to inhibit the malignant development of colorectal cancer. Qu et al. found that hsa_circ_001569 can sponge miR-145 to regulate the E2F5, BAG4, and FMNL2 genes, thereby promoting the proliferation and invasion of colorectal cancer (11). miRNA is abnormally expressed in most malignant tumors, occupies a role similar to proto-oncogenes and tumor suppressor genes, and contributes significantly to the occurrence and development of tumors. With this knowledge, it was found that miRNA can be used as a specific target for targeted therapy to guide the treatment of various malignant tumors in clinic (12).
Smad7 is located on chromosome 18q21 and is 3,111 bp in length. As a suppressor of the TGF-β/Smads signaling pathway (13-15), Smad7 plays a role in both the nucleus and cytoplasm. and thereby maintains the balance of the channel (16). Smad7 can play a role in both the nucleus and cytoplasm. Smad7 can inhibit the phosphorylation of R-Smads by blocking the receptor, suppress the polymerization of R-Smads and Co-Smads, and cause the ligand-receptor complex, thereby inhibiting the conduction of TGF-β/Smads signaling pathway (17). Inactivation of any component in this pathway will lead to accelerated cell growth and dysregulation of apoptosis signals, resulting in uncontrolled growth and differentiation of cells, which in turn induces cancerous cells (18,19).
In this study, we found that circTBL1XR1 was involved in the malignant progression of rectal cancer. CircTBL1XR1 could directly regulate the expression of miR-424, and silencing circTBL1XR1 could promote tumor proliferation, invasion and metastasis by upregulating miR-424. We further found that Smad7 is a target of miR-424, and miR-424 is involved in the regulation of Smad7 expression. In short, circTBL1XR1 can regulate the malignant progression of colorectal cancer by targeting the miR-424/Smad7 pathway. Our data provide new insights into the interactions between circRNA and miRNA in tumor molecular regulation.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/jgo-20-395).
Methods
Patient and specimen collection
Samples of colorectal cancer and paracancerous normal intestinal mucosal tissues were selected from 24 patients diagnosed with adenocarcinoma of colon by pathology who were admitted to the gastrointestinal surgery center of the First Hospital of Xi’an Jiaotong University from March to May, 2018. Among the 24 patients, there were 13 males and 11 females. The median age was 65 (range, 34–75 years). None of the patients had received preoperative radiotherapy, chemotherapy, combined treatment of Traditional Chinese and western medicine and immunotherapy, and no other tumors, autoimmune diseases or infectious diseases were associated. The tumor tissues and adjacent tissues were obtained and washed clean with running water, and 0.5 cm3 specimens were cut and immersed in RNAlater solution (Thermo Fisher Scientific, USA). The stored liquid volume was no less than 5 times that of the sample volume. After being immersed, the samples were immediately transported on ice and transferred to the refrigerator at −80 °C for storage within 30 minutes. When obtaining tumor tissues, the necrotic and organic parts of the tissue center were avoided. The adjacent tissues that were obtained were normal intestinal mucosal tissues that were farther than 5 cm away from the tumor tissues. All patients signed informed consent. This experiment received approval from the Ethics Committee of First Hospital of Xi’an Jiaotong University and was conducted according to the tenets of the Declaration of Helsinki (as revised in 2013).
Cell culture and transfection
LoVo, SW620, IEC-6, HCT 116 and SW480 used in this study were purchased from the American Germplasm Storage Center (ATCC) and cultivated according to ATCC-recommended conditions and methods. For LoVo cells, RPMI-1640 medium was used as the basic medium. The complete medium was composed of 10% fetal bovine serum (FBS, Gibco, USA) mixed with each basic medium; the passage was 1 mL 0.25% (w/v) pancreatin −0.53 mM EDTA solution per well, and the cells are digested at 37 °C. After 2 to 4 minutes, the digestion was terminated with 10 mL of complete medium, the cell suspension was centrifuged at 1,500 rpm, the supernatant was discarded, the complete medium was added to resuspend the cells, and the cells were seeded after counting using a hemocytometer. Culturing and proliferation was completed in a 6-well plate until 70% of the bottom area of the culture plate was covered, and the Lipofectamine2000 kit (Thermo Fisher Scientific, USA) was used to dissolve the small interfering RNA (siRNA) sequence (Ruibo, China) at a ratio of 10 ng: 1 µL. The plasmid was transfected using Lipofectamine3000 kit (Thermo Fisher Scientific, USA). According to the manufacturer’s instructions, 2 µg/µL of overexpression plasmid DNA (Ruibo, China) and 4 µL/µL of stem reagent were dissolved in the basic medium and incubated for 15 minutes at room temperature. After transfection, the cells were cultured at 37 °C under 5% CO2.
Establishment of animal tumor model
Nude mice, aged 5–6 weeks old were used in this study. A colon cancer cell line that stably expresses circTBL1XR1 lentivirus was selected. The cell concentration was adjusted to 2×107 cells/mL. The cells were injected subcutaneously into the armpit of the mouse, 0.1 mL was inoculated, and the tumor size was observed and measured every day. The tumor volume was calculated according to the following formula: V = Length × Width2 × 0.5. After the experiment, the mice were euthanized. After the tumors in each group were peeled off, they were photographed and recorded. This experiment received approval from the Ethics Committee of First Hospital of Xi’an Jiaotong University.
Cell proliferation experiment
The Cell Counting Kit-8 (CCK-8) test was performed according to the manufacturer’s instructions (Dongren, Japan). The cells were seeded at a rate of 5,000/cm2 into a 96-well cell culture plate (Corning, USA), and 10 µL of CCK-8 working solution was added after 72 hours of cultivation. After an incubation of 4 hours at °C, the absorbance optical density (OD) value was measured in a microplate reader (BioTek Instruments Inc., USA) at a wavelength of 450 nM.
Cell scratch test
Cells of each group at 24 hours after transfection were seeded in 6-well plates at 1×106/well. When the confluence of cells reached 90%, straight lines were drawn with a 10 µL sterile pipette tip in the vertical direction, and washed three times with phosphate-buffered saline (PBS) to remove shed cells. Then, 2 mL of fresh medium without serum were added, and pictures were taken under the microscope at 0 and 48 hours. The migration distance was measured with ImageJ software, and the relative distance of scratches of each group of cells was calculated according the following formula: relative distance of scratches (%) =48 hours scratches width/0 hours scratch width ×100%. The experiment was repeated three times.
Cell transwell experiment
Matrigel was diluted with a prechilled culture solution (dilution ratio 9:1), 40 µL of Matrigel dilution was added to each well of the upper chamber of the 24-well plate Transwell chamber, and the 24-well plate was placed in a 37 °C 5% CO2 incubator for 5 hours. Colorectal cancer cells in logarithmic growth phase were taken, digested with 0.25% trypsin and adjusted to a cell density of 5×104 cells/mL. 200 µL single cell suspension was added to the upper chamber of Transwell chamber, incubated at 37 °C for 24 hours. After that, the cells were washed with PBS 3 times × 5 (1) minutes, and fixed with paraformaldehyde solution for 10 minutes. Stain the fixed cells with 0.1% crystal violet staining solution for 10 minutes. Five fields were randomly selected for observation under a microscope to determine the number of invading cells.
Double luciferase reporter gene detection experiment
The dual luciferase reporter experiment with the StarBase database was used to analyze the miRNA bound to circTBL1XR1, and it was found that circTBL1XR1 and Smad7 had binding sites with miR-424. A dual luciferase report experiment was used to verify the targeted regulation of circTBL1XR1 on miR-424. Meanwhile, the targeting effect of miR-424 on Smad7 was verified. The wild-type (WT) circTBL1XR1 luciferase recombinant reporter vector (PmirGLO-circTBL1XR1-WT) was named the circTBL1XR1-WT vector. Based on the circTBL1XR1-WT vector, a mutant (MUT) recombination was constructed using the site-directed mutation method. The PmirGLO-circTBL1XR1-MUT vector was named the circTBL1XR1-MUT vector. The above recombinant vector was synthesized by Shanghai Jima Pharmaceutical Technology Co., Ltd. HEK293T cells were seeded in 96-well plates at a density of 3×104 cells/well. After 24 hours, they were transfected with miR-424 mimics or miR-NC-mimics and circTBL1XR1-WT or circTBL1XR1-MUT plasmids (combination of two pairs for transfection). After 48 hours, cells of each group were collected. After treatment with cell lysate, 20 µL of supernatant was centrifuged, 50 µL of the prepared reaction solution and stop solution were added, and the light signal value was measured with a GloMax20/20 luminescence detector (Promega Corporation, USA).
Quantitative real-time fluorescence polymerase chain reaction (qRT-PCR)
qRT-PCR was conducted using TRIzol (Invitrogen, USA), Tri Pure Isolation Reagent Kit (Roche, USA), and GeneJET RNA Purification Lit (Invitrogen) to collect cell and tissue RNA,. For circular RNA and mRNA, the study used β-actin levels as internal controls. The following primers were used: β-actin, F: 5'-CCTGTACGCCAACACAGTGC-3', R: 5'-ATACTCCTGCTTGCTGATCC-3'; the U6 primer sequences were the following: F: 5'-GCTTCGGCAGCACATATACTAAAAT-3', R: 5'-CGCTTCACGAATTTGCGTGTCAT-3'. Bcl-2, F: ATGCCTTTGTGGAACTATATGGC, R: GGTATGCACCCAGAGTGATGC; Bax, F: TGAGACAGGGGCCTTTTCC, CC: Random primers and complementary DNA (cDNA) were mixed, incubated at 70 °C for 5 minutes, and then quickly cooled on ice for 5 minutes. Afterwards, buffer, reverse transcriptase, nucleotides, and magnesium ions were added, annealed at 25 °C for 15 minutes, and extended at 42 °C for 60 minutes. The reaction conditions were as follows: pre-denaturation at 95 °C for 5 minutes; denaturation at 95 °C for 1 minute, annealing at 60 °C for 30 seconds, and extension at 70 °C for 30 seconds, for a total of 45 cycles. KAPA SYBR FAST qPCR Master Mix (Kapa Biosystems, USA) and CFX96 (Waveguide, USA) qRT-PCR were used to detect the amplified fluorescence intensity, and the 2−△△CT method was used to calculate the gene expression level.
Western blotting
The cells were lysed with 100 µL of RIPA lysate per well on ice for 30 minutes. The cells were scraped off with a cell scraper and centrifuged and stored. The bicinchoninic acid (BCA) method was used to determine the protein concentration of the sample. The protein was denatured at 100 °C before running the gel, the sample was loaded with 50 µg of protein per well, and separated by marker electrophoresis at 120 V of constant pressure. Cross-flow of the membrane occurred at 395 mA in an ice bath for 180 minutes. After blocking with 5% skimmed milk, the PVDF membrane was blocked with E-cadherin (Abcam, 1:1,000) and Vimentin (Abcam, 1:1,000) polyclonal antibody or anti-GAPDH monoclonal antibody (Biyuntian, China, 1:1,000) and shaken at 4 °C overnight. And the PVDF membrane was incubated with Goat anti-rabbit fluorescent labeled antibody (Biyuntian, China, 1:500) for 2 hours, immersed in luminous liquid for 2 minutes, and photographed with indoor exposure of the illuminometer.
Immunohistochemistry
The tumor tissue from patients and nude mice was collected and fixed with 4% paraformaldehyde for 48 h. Paraffin sections with a thickness of 4 µm were made and routinely dewaxed. The slices were incubated with 3% H2O2 at 37 °C for 5-10 minutes. The slices were blocked with 5% serum. Anti-smad7 antibody (1:100; Santa Cruz) and Anti-Ki67 antibody (1:300; Santa Cruz) were added onto the section and cultuerd at 4 °C overnight. Then the sections were incubated with Secondary Antibody (1:500; Santa Cruz) at 37 °C for 2 hours and stained with DAB. Finally, the slides were observed under a normal microscope (Nikon Corporation).
Statistical analysis
SPSS software (Version 20.0, SPSS Inc., USA) was used for calculation. The continuous variable comparison between the two groups was analyzed by t-test, the multiple group continuous variable comparison was analyzed by single factor analysis of variance, and the correlation between the two groups of continuous variables was analyzed by Pearson’s correlation analysis. The results are reported as mean ± standard deviation. A P value <0.05 was considered statistically significant.
Results
Expression of circTBL1XR1 in CRC tissues and cell lines
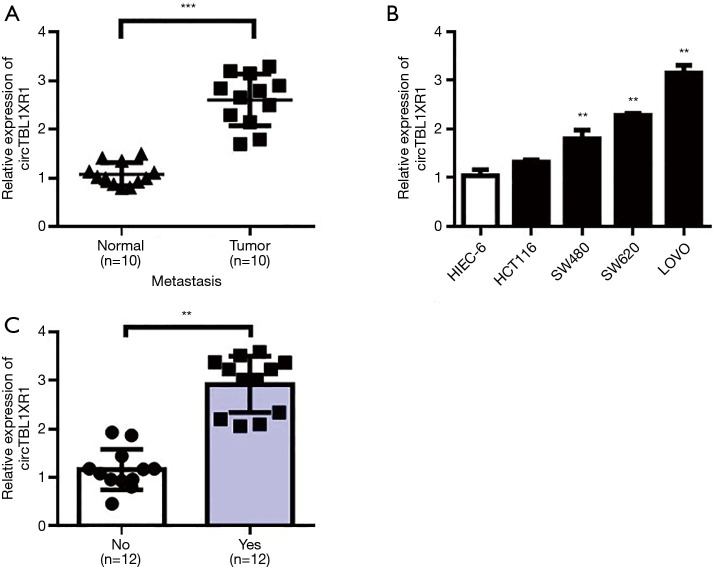
Expression differences of circTBL1XR1 in CRC and paracancerous tissues were detected by RT-qPCR. The results showed that the expression level of circTBL1XR1 in CRC tissues was significantly higher than that in paracancerous tissues (P<0.001) (Figure 1A). The expression levels of circTBL1XR1 in CRC cell lines LoVo, SW620, and SW480 were significantly higher than that of hiec-6 in human colorectal cells (all P values <0.01, Figure 1B). Further analysis showed that the expression of circTBL1XR1 in patients with colorectal cancer metastasis was higher than that in patients without metastasis (Figure 1C).
Figure 1.
Expression of circTBL1XR1 in colorectal cancer and adjacent tissues and an analysis of patients’ clinical information. (A) CircTBL1XR1 levels in adjacent tissues and colorectal cancer tissues; (B) CircTBL1XR1 expression levels in normal intestinal epithelial cells and colorectal cancer cells; (C) the level of circTBL1XR1 in patients with colorectal cancer in metastatic and non-metastatic patients. **, P<0.01; ***, P<0.001.
Overexpression of circTBL1XR1 promoted the proliferation, invasion and migration of CRC cells
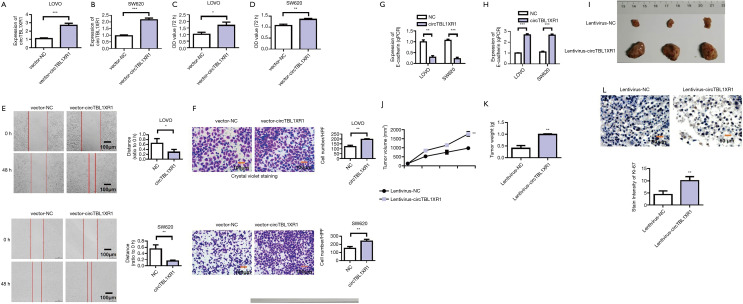
After transfection with the overexpression recombinant vector of circTBL1XR1 in LoVo and SW620 cells, the results showed that, compared with the NC group, the expression level of circTBL1XR1 in LoVo and SW620 cells was significantly upregulated (Figure 2A,B). The results of CCK-8 assay (Figure 2C,D) showed that after circTBL1XR1 was transferred into LoVo and SW620 cells for 72 hours, the proliferation capacity of the cells in the transfected group with the overexpressed recombinant vector circTBL1XR1 was significantly enhanced compared with that in the control group. These results suggest that the overexpression of circTBL1XR1 can promote the proliferation of CRC cells. The results of cell scratch and Transwell assay (Figure 2E,F) showed that after the transfection of circTBL1XR1 into LoVo and SW620 cells, the invasion and migration capacity of the cells in the transfected group with the overexpressed recombinant vector circTBL1XR1 were significantly enhanced compared with that in the control group. The detection results of E-cadherin and Vimentin showed that after the transfer of circTBL1XR1 into LoVo and SW620 cells, the expression of E-cadherin, an epithelial marker, was decreased in the transfected cells with the overexpression of the recombinant vector, while the expression of mesenchymal marker protein Vimentin was increased, indicating that circTBL1XR1 promoted the epithelial-mesenchymal transition (EMT) ability of CRC cells (Figure 2G,H). Animal experiments showed that overexpression of circTBL1XR1 promoted tumor proliferation (Figure 2I). The tumor volume test results showed that with the extension of tumor growth time, circTBL1XR1 significantly promoted the tumor proliferation (Figure 2J). Tumor weight test results also revealed that overexpression of circTBL1XR1 increased tumor weight (Figure 2K). Ki-67 immunohistochemical staining indicated changes in tumor cell activity and proliferation. The staining results showed that Ki-67 expression increased after overexpression of circTBL1XR1. This indicates that overexpression of circTBL1XR1 can promote tumor activity and proliferation (Figure 2L).
Figure 2.
CircTBL1XR1 promoted colorectal cancer proliferation and metastasis. (A) Detection of circTBL1XR1 transfection efficiency in LoVo cells; NC: negative control; (B) detection of circTBL1XR1 transfection efficiency in SW620 cells; (C) overexpression of circTBL1XR1 promoted the cell proliferation ability of LoVo cells; (D) overexpression of circTBL1XR1 promoted the cell proliferation ability of in SW620 cells; (E) overexpression of circTBL1XR1 promoted colon cancer cell migration ability in LoVo and SW620 cells; (F) overexpression of circTBL1XR1 promoted colon cancer cell invasion in LoVo and SW620 cells; Crystal violet staining for tumor cells: 0.1% crystal violet solution (Beyotime, Shanghai, China) was used for staining for 20 min. Next, gently wipe off the upper unmigrated cells with a cotton swab. Wash it with PBS three times. (G) overexpression of circTBL1XR1 inhibited the expression of E-cadherin; (H) overexpression of circTBL1XR1 promoted Vimentin expression; (I) overexpression of circTBL1XR1 promoted tumor proliferation; (J) the tumor volume test results indicated that circTBL1XR1 promoted tumor proliferation; (K) tumor weight test results after overexpression of circTBL1XR1; (L) Ki-67 immunohistochemical staining (magnification ×200). *, P<0.01; **, P<0.01; ***, P<0.001.
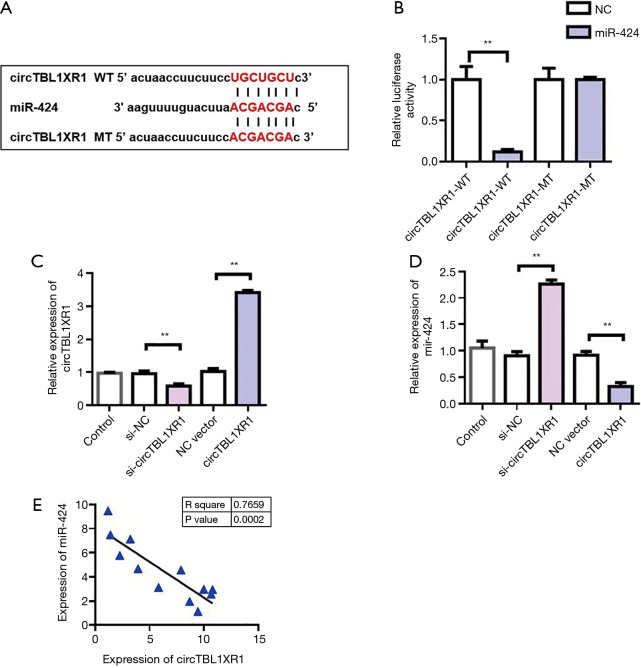
MiR-424 was a target gene for circTBL1XR1
To further investigate the mechanism of circTBL1XR1 in colorectal cancer, we first analyzed the miRNA profile of circTBL1XR1 binding. Through StarBase database analysis of miRNAs bound to circTBL1XR1, it was found that there was a binding site between circTBL1XR1 and miR-424 (Figure 3A). The circTBL1XR1 sequence was cloned into the luciferase reporter gene downstream of the reporter vector to obtain the wild-type circTBL1XR1-WT reporter vector. Meanwhile, the mutant reporting vector circTBL1XR1-MUT was constructed. The cotransfection of miR-196b-5p-mimics/miR-NC-mimics and circTBL1XR1-wt/circTBL1XR1-mut into HEK293T cells was carried out, and then the effect of miR-424 on luciferase activity of the reporter vector was detected using the dual luciferase reporter system. The results showed that miR-424 mimics significantly inhibited the luciferase activity of the circTBL1XR1-WT reporter vector, but had no effect on luciferase activity of the circTBL1XR1-MUT reporter vector. These results suggest that circTBL1XR1 can be targeted to bind to miR-424 (Figure 3B). To verify the regulatory effect of circTBL1XR1 on miR-424, we overexpressed and downregulated circTBL1XR1 (Figure 3C), respectively. The experimental results showed that the overexpression plasmid and siRNA could effectively regulate the expression of circTBL1XR1. The detection results of the expression level of miR-424 showed that overexpression of circTBL1XR1 could inhibit the expression of miR-424, while the reduction of circTBL1XR1 could promote the expression of miR-424 (Figure 3D). Further, we detected the co-expression correlation between circTBL1XR1 and miR-424. Pearson’s correlation analysis results (Figure 3E) showed that the expression level of circTBL1XR1 in CRC tissues was negatively correlated with miR-424.
Figure 3.
CircTBL1XR1 regulated the detection of miR-424. (A) Information maps of the binding sites of circTBL1XR1 and miR-424; (B) dual luciferase report assay detected the binding of circTBL1XR1 to miR-424; NC: negative control, WT: wild type, MT: mutant; (C) detection of transfection efficacy of circTBL1XR1 overexpression and knockdown; (D) CircTBL1XR1 inhibited miR-424 expression; (E) correlation analysis of circTBL1XR1 and miR-424 co-expression. **, P<0.01.
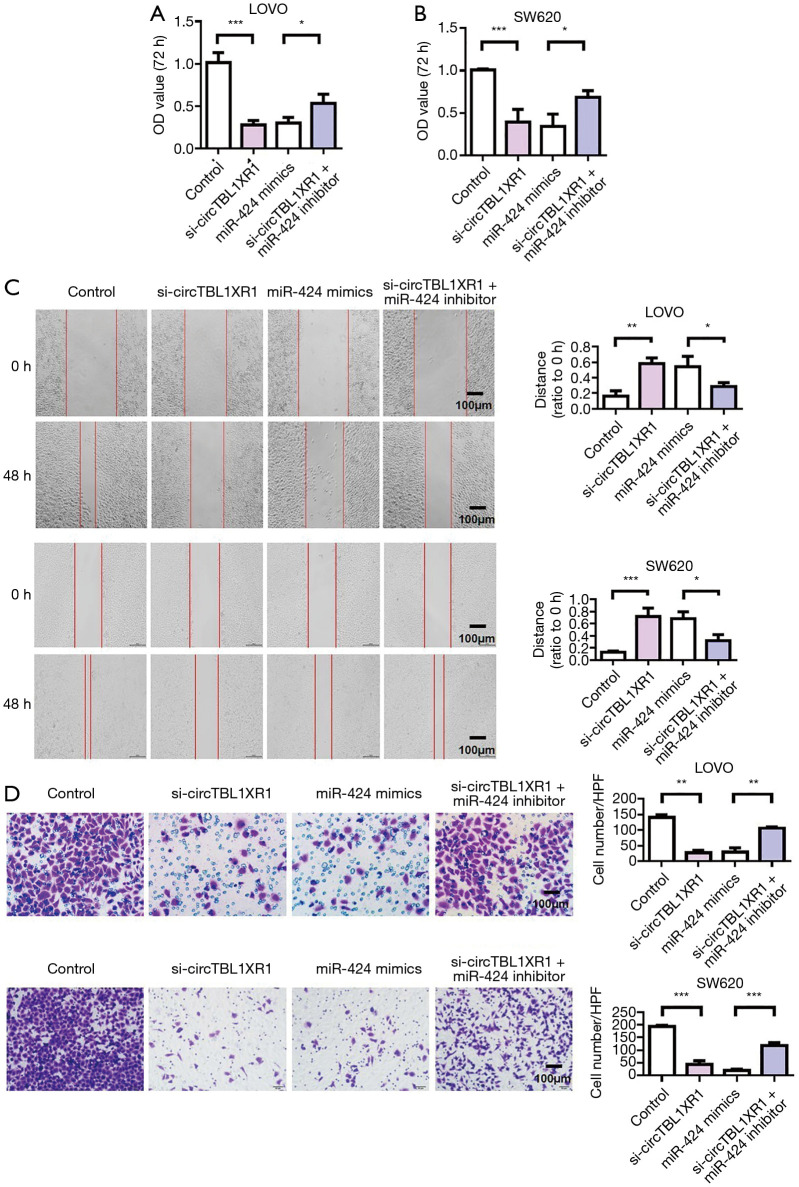
MiR-424 reversed the carcinogenic effect of circTBL1XR1
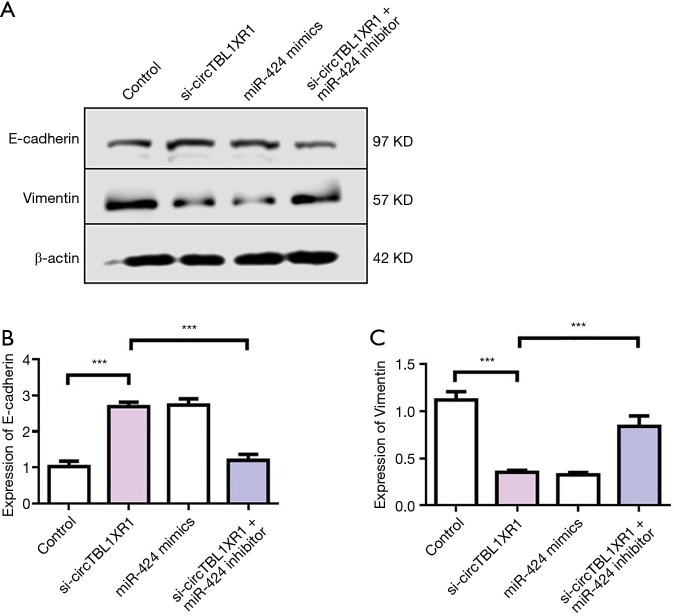
The results of cell proliferation detection showed that si-circTBL1XR1 could inhibit the proliferation of colorectal cancer cells. Similarly, miR-424 mimic also inhibits cell proliferation. After cotransfection with miR-424 inhibitor in the circTBL1XR1 knockout LoVo and SW620 cells, the proliferation of cells in the si-circTBL1XR1 + miR-424 inhibitor group was improved compared with that in the circTBL1XR1 knockout group, suggesting that the inhibition of cell proliferation by circTBL1XR1 knockout was partially reversed after the addition of miR-424 inhibitor (Figure 4A,B). Transwell and scratch detection results showed that si-circTBL1XR1 could inhibit the invasion and migration of colorectal cancer cells. Similarly, miR-424 mimic also inhibits cell invasion and migration. However, miR-424 can inhibit reverse si-circTBL1XR1 inhibition (Figure 4C,D). The results of E-cadherin expression detection showed that the knockdown of circTBL1XR1 and inhibition of miR-424 could upregulate the expression of E-cadherin (Figure 5A,B), while the knockdown of circTBL1XR1 and inhibition of miR-424 could downregulate the expression of Vimentin (Figure 5C). The experimental results showed that circRNA promoted the malignant progression of colorectal cancer by sponge adsorption of miR-424.
Figure 4.
CircTBL1XR1 adsorbed miR-424 to increase the expression of Smad7 through sponge action to promote the malignant evolution of colorectal cancer cells. (A,B) Cell proliferation detection of LoVo and SW620 cells; (C) cell migration detection; (D) detection of cell invasion. Crystal violet staining for tumor cells: 0.1% crystal violet solution (Beyotime, Shanghai, China) was used for staining for 20 min. Next, gently wipe off the upper unmigrated cells with a cotton swab. Wash it with PBS three times. Magnification ×200. *, P<0.01; **, P<0.01; ***, P<0.001.
Figure 5.
CircTBL1XR1 promoted EMT of colorectal cancer cells. (A) Western blot detection of E-cadherin and Vimentin expression; (B) detection of E-cadherin expression; (C) detection of Vimentin expression. ***, P<0.001.
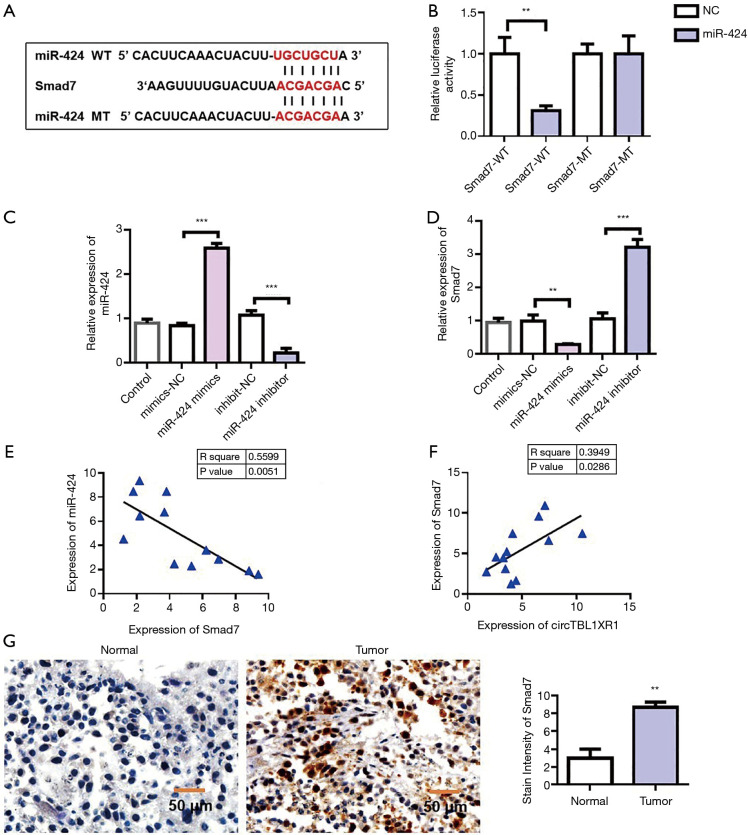
Smad7 was the target gene of miR-424
To further investigate the mechanism of miR-424 in colorectal cancer, we first analyzed the miRNA target genes. Target genes binding to miR-424 were analyzed by TargetScan database, and it was found that Smad7 had binding sites with miR-424 (Figure 6A). The Smad7 sequence was cloned into luciferase reporter gene downstream of the reporter vector to obtain the wild-type Smad7-wt reporter vector. Meanwhile, the mutant reporting vector Smad7-mut was constructed. The cotransfection of miR-196b-5p-mimics/miR-NC-mimics and Smad7-wt/Smad7-mut into HEK293T cells was carried out, and the effect of miR-424 on luciferase activity of the reporter vector was detected using the dual luciferase reporter system. The results showed that miR-424-mimics significantly inhibited luciferase activity of the Smad7-wt reporter vector, but had no effect on luciferase activity of Smad7-mut reporter vector. This result suggested that Smad7 could be targeted by miR-424 (Figure 6B). To verify the regulatory effect of miR-424 on Smad7, we overexpressed and downregulated miR-424, respectively (Figure 6C). Detection results of Smad7 expression showed that overexpression of miR-424 could inhibit the expression of Smad7, while knockdown of miR-424 could promote the expression of Smad7 (Figure 6D). Furthermore, we detected the co-expression correlation between Smad7 and miR-424. Pearson’s correlation analysis results (Figure 6E) showed that the expression level of Smad7 in CRC tissues was negatively correlated with miR-424 (Figure 6E). The co-expression results of circTBL1XR1 and Smad7 showed that there was a positive co-expression correlation between circTBL1XR1 and Smad7 (Figure 6F). Immunohistochemical results showed that Smad7 was highly expressed in tumor tissues (Figure 6G).
Figure 6.
MiR-424 inhibited Smad7 expression. (A) Information maps of Smad7 and miR-424 binding sites; (B) dual luciferase report detected the binding of Smad7 to miR-424; NC: negative control, WT: wild type, MT: mutant; (C) detection of transfection efficacy of miR-424 overexpression and knockdown; (D) the expression of Smad7 decreased with the addition of miR-424 mimics to colorectal cancer cell lines. Adding miR-424 inhibitor increased the expression of Smad7; (E) correlation analysis of Smad7 and miR-424 co-expression; (F) correlation analysis of circTBL1XR1 and Smad7 co-expression; (G) Smad7 immunohistochemical staining. **, P<0.01; ***, P<0.001.
Overexpression of Smad7 reversed the antitumor effects of miR-424
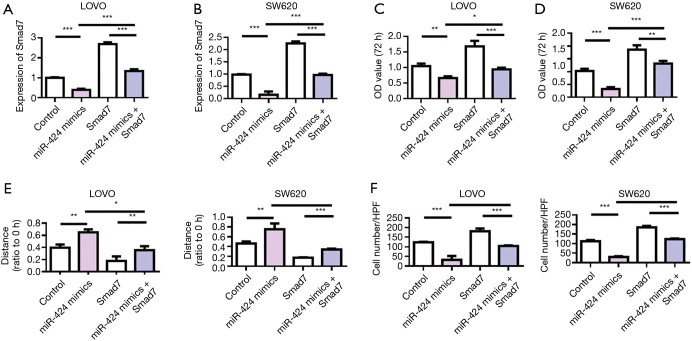
We further used rescue experiment to investigate the regulatory effect of miR-424 on Smad7. The detection results of Smad7 expression showed that miR-424 mimics could effectively inhibit the expression of Smad7. Smad7-overexpressed plasmid could effectively upregulate the expression of Smad7 (Figure 7A,B). Cell proliferation assay results showed that miR-424 mimics inhibited LoVo and SW620 cell proliferation, while overexpression of Smad7 promoted tumor cell proliferation, compared with the control group. Meanwhile, overexpression of Smad7 reversed the antitumor effect of miR-424 (Figure 7C,D). The scratch and Transwell results of LOVO and SW620 cells showed that miR-424 mimics inhibited the migration and invasion of cells, while overexpression of Smad7 promoted the migration and invasion of tumor cells, and thus reversed the antitumor effect of miR-424 (Figure 7E,F).
Figure 7.
Overexpression of Smad7 reversed the antitumor effects of miR-424. (A) Detection of Smad7 expression in LoVo cells; (B) detection of Smad7 expression in SW620 cells; (C,D) LoVo and SW620 cell proliferation detection; (E) detection of cell migration ability; (F) detection of cell invasion ability. *, P<0.01; **, P<0.01; ***, P<0.001.
Discussion
The incidence and mortality of colorectal cancer seriously endanger public health and are major concerns in tumor research (20-22). With the development of medical technology, many methods have been innovated for the treatment of colorectal cancer; however, the malignant progress of the tumor remains an unsolved problem, and colorectal cancer patients often die from other sites of metastasis. If we can understand the mechanism underlying the development of colorectal cancer, the survival and quality of life of colorectal cancer patients will likely be improved (23-25). The purpose of this study was to describe the role of the circTBL1XR1/miR-424 axis in the malignant progression of colorectal cancer and to describe the cellular/molecular mechanisms of the axis’s function.
circRNAs are closely related to the malignant progression of colorectal cancer and may be used as a molecular target for the treatment of colorectal cancer. In this study, our experiment produced several new findings. First, the expression of circTBL1XR1 was negatively correlated with that of miR-424, with the former upregulated and the latter downregulated in colorectal cancer. Secondly, overexpression of circTBL1XR1 significantly promoted tumor proliferation, invasion and metastasis. Recently, other circRNAs have been reported in colorectal cancer. For instance, Han et al. found that Hsa_circ_0020397 regulated the proliferation activity, apoptosis, and invasion of colorectal cancer by promoting the expression of miR-138 target genes (26). Downregulated circITCH inhibited Wnt/-catenin pathway and promoted the expression of ITCH, while upregulated circITCH inhibited proliferation (27). Other studies have found that has_circ_0000069 can promote the proliferation, migration, and invasion of colorectal cancer, which may be a potential target for diagnosis and treatment of colorectal cancer (28). CircCCDC66 may protect MYC mRNA as a miRNA sponge and promote the proliferation, migration, and metastasis of colorectal cancer (29). ciRS-7 is highly expressed in colorectal cancer, and high expression of ciRS-7 is associated with poor clinical prognosis. With regard to its mechanism, ciRS-7 acts as ceRNA of epidermal growth factor receptor (EGFR) and RAF1, while sponge adsorption of miR-7 promotes the malignant biological behavior of colorectal cancer (30).
Studies have shown that the expression of Smad7 protein is closely related to Dukes stage, the depth of colorectal cancer invasion, and lymph node metastasis, and is positively correlated with the expression in colorectal cancer (31,32). Myatt et al. found that in the precancerous lesions of colorectal cancer, such as in colorectal polyps and ulcerative colitis, the expression of Smad7 was significantly increased compared with that in the normal intestinal mucosa, which indicated that the high expression of Smad7 was closely related to the occurrence and development of colorectal cancer (33). In addition, Smad7 single nucleotide polymorphisms are associated with the development of colorectal cancer. In the study of the Smad7 gene, Shukla et al. found that there was a G>C single nucleotide polymorphism in the transcription factor binding region at about the 44703563bp gene fragment, which could lead to the abnormal increase of Smad7 protein expression, which was considered to be a susceptibility factor for colorectal cancer (34). Broderick et al. found that single nucleotide polymorphisms in rs4464148, rs4939827, and rs12953717 of Smad7 genes can increase genetic susceptibility to colorectal cancer (31). Kleeff et al. transfected Smad7 gene into pancreatic cancer cells of nude mice, and found that their malignancy to be significantly increased, with Smad7 protein being highly expressed in pancreatic cancer tissues. This thus indicated that an excessive expression of Smad7 can promote the development of tumor (13).
In the present study, Smad7, as the target gene of miR-424, was found to mediate the oncogenic effect of the circTBL1XR1/miR-424 axis. circRNA was found to act as a ceRNA through sequence complementation, limiting the functional availability of miR-424. Furthermore, circTBL1XR1 inhibited the anticancer effect of miR-424. Therefore, our study revealed new aspects of the cellular function and pathophysiological role of circTBL1XR1 and miR-424, both of which can be considered as potential molecular targets for the treatment of colorectal cancer.
Conclusions
Our data suggest that circTBL1XR1 is up-regulated in colon cancer tissues and cells and plays a procarcinogenic role by enhancing the malignant behavior of colon cancer cells and promoting the metastasis of colon cancer. In addition, circTBL1XR1 targeted miR-424 and inhibited the tumor inhibition of miR-424, while Smad7 effectively promoted the malignant behavior of colon cancer cells. In addition, the overexpression of circTBL1XR1 enhanced the malignant progression of colon cancer in vivo. Therefore, the circTBL1XR1/miR-424/Smad7/MAPK8 pathway may be critical for regulating the development and progression of colon cancer and may be a therapeutic target for intervention in colon cancer. Therefore, our findings may provide new insights into the pathogenesis of colon cancer.
Acknowledgments
Funding: None.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All patients signed informed consent. This experiment received approval from the Ethics Committee of First Hospital of Xi’an Jiaotong University and was conducted according to the tenets of the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
Reporting Checklist: The author has completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/jgo-20-395
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jgo-20-395
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jgo-20-395). The author has no conflicts of interest to declare.
(English Language Editor: J. Gray)
References
- 1.Roselló S, Simón S, Cervantes A. Programmed colorectal cancer screening decreases incidence and mortality. Transl Gastroenterol Hepatol 2019;4:84. 10.21037/tgh.2019.12.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klaassen CH, Jeunink MA, Prinsen CF, et al. Quantification of Human DNA in Feces as a Diagnostic Test for the Presence of Colorectal Cancer. Clin Chem 2003;49:1185-7. 10.1373/49.7.1185 [DOI] [PubMed] [Google Scholar]
- 3.Valentini AM, Cavalcanti E, Di Maggio M, et al. RAS-expanded Mutations and HER2 Expression in Metastatic Colorectal Cancer: A New Step of Precision Medicine. Appl Immunohistochem Mol Morphol 2018;26:539-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tilg H, Adolph TE, Gerner RR, et al. The Intestinal Microbiota in Colorectal Cancer. Cancer Cell 2018;33:954-64. 10.1016/j.ccell.2018.03.004 [DOI] [PubMed] [Google Scholar]
- 5.Kanaan Z, Rai SN, Eichenberger MR, et al. Plasma miR-21: a potential diagnostic marker of colorectal cancer. Ann Surg 2012;256:544-51. 10.1097/SLA.0b013e318265bd6f [DOI] [PubMed] [Google Scholar]
- 6.Grizzi F, Basso G, Borroni EM, et al. Evolving notions on immune response in colorectal cancer and their implications for biomarker development. Inflamm Res 2018;67:375-89. 10.1007/s00011-017-1128-1 [DOI] [PubMed] [Google Scholar]
- 7.Shi X, Gao K, Huang H, et al. Pretargeted Immuno-PET Based on Bioorthogonal Chemistry for Imaging EGFR Positive Colorectal Cancer. Bioconjug Chem 2018;29:250-4. 10.1021/acs.bioconjchem.8b00023 [DOI] [PubMed] [Google Scholar]
- 8.Schulz-Heddergott R, Stark N, Edmunds SJ, et al. Therapeutic Ablation of Gain-of-Function Mutant p53 in Colorectal Cancer Inhibits Stat3-Mediated Tumor Growth and Invasion. Cancer Cell 2018;34:298-314.e7. 10.1016/j.ccell.2018.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meng J, Ai X, Lei Y, et al. USP5 promotes epithelial-mesenchymal transition by stabilizing SLUG in hepatocellular carcinoma. Theranostics 2019;9:573-87. 10.7150/thno.27654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H, Zhong W, Zhao J, et al. Oleanolic Acid Inhibits Epithelial-Mesenchymal Transition of Hepatocellular Carcinoma by Promoting iNOS Dimerization. Mol Cancer Ther 2019;18:62-74. 10.1158/1535-7163.MCT-18-0448 [DOI] [PubMed] [Google Scholar]
- 11.Qu S, Yang X, Li X, et al. Circular RNA: A new star of noncoding RNAs. Cancer Lett 2015;365:141-8. 10.1016/j.canlet.2015.06.003 [DOI] [PubMed] [Google Scholar]
- 12.Fesler A, Liu H, Ju J. Modified miR-15a has therapeutic potential for improving treatment of advanced stage colorectal cancer through inhibition of BCL2, BMI1, YAP1 and DCLK1. Oncotarget 2017;9:2367-83. 10.18632/oncotarget.23414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kleeff J, Ishiwata T, Maruyama H, et al. The TGF-beta signaling inhibitor Smad7 enhances tumorigenicity in pancreatic cancer. Oncogene 1999;18:5363-72. 10.1038/sj.onc.1202909 [DOI] [PubMed] [Google Scholar]
- 14.Kaur G, Li CG, Chantry A, et al. SMAD proteins directly suppress PAX2 transcription downstream of transforming growth factor-beta 1 (TGF-β1) signalling in renal cell carcinoma. Oncotarget 2018;9:26852-67. 10.18632/oncotarget.25516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mota MSV, Jackson WP, Bailey SK, et al. Deficiency of tumor suppressor Merlin facilitates metabolic adaptation by co-operative engagement of SMAD-Hippo signaling in breast cancer. Carcinogenesis 2018;39:1165-75. 10.1093/carcin/bgy078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen W, Fu X, Sheng Z. Review of current progress in the structure and function of Smad proteins. Chin Med J (Engl) 2002;115:446-50. [PubMed] [Google Scholar]
- 17.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature 2003;425:577-84. 10.1038/nature02006 [DOI] [PubMed] [Google Scholar]
- 18.Blume JE, Wilhelmsen M, Benz RW, et al. Discovery and Validation of Plasma-Protein Biomarker Panels for the Detection of Colorectal Cancer and Advanced Adenoma in a Danish Collection of Samples from Patients Referred for Diagnostic Colonoscopy. J Appl Lab Med 2019;1:181-93. 10.1373/jalm.2016.020271 [DOI] [PubMed] [Google Scholar]
- 19.Duffy MJ. Carcinoembryonic Antigen as a Marker for Colorectal Cancer: Is It Clinically Useful? Clin Chem 2001;47:624-30. 10.1093/clinchem/47.4.624 [DOI] [PubMed] [Google Scholar]
- 20.Bychkov D, Linder N, Turkki R, et al. Deep learning based tissue analysis predicts outcome in colorectal cancer. Sci Rep 2018;8:3395. 10.1038/s41598-018-21758-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xi X, Liu N, Wang Q, et al. ACT001, a novel PAI-1 inhibitor, exerts synergistic effects in combination with cisplatin by inhibiting PI3K/AKT pathway in glioma. Cell Death Dis 2019;10:757. 10.1038/s41419-019-1986-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao T, Zhong W, Zhao J, et al. Polyphyllin I suppresses the formation of vasculogenic mimicry via Twist1/VE-cadherin pathway. Cell Death Dis 2018;9:906. 10.1038/s41419-018-0902-5 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Chubak J, Boudreau DM, Rulyak SJ, et al. Colorectal cancer risk in relation to antidepressant medication use. Int J Cancer 2011;128:227-32. 10.1002/ijc.25322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen L, Pan X, Hu XH, et al. Gene expression differences among different MSI statuses in colorectal cancer: Gene expression differences among MSI statuses. Int J Cancer 2018;143:1731-40. 10.1002/ijc.31554 [DOI] [PubMed] [Google Scholar]
- 25.Zhong W, Yang W, Qin Y, et al. 6-Gingerol stabilized the p-VEGFR2/VE-cadherin/beta-catenin/actin complex promotes microvessel normalization and suppresses tumor progression. J Exp Clin Cancer Res 2019;38:285. 10.1186/s13046-019-1291-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han B, Chao J, Yao H. Circular RNA and its mechanisms in disease: From the bench to the clinic. Pharmacol Ther 2018;187:31-44. 10.1016/j.pharmthera.2018.01.010 [DOI] [PubMed] [Google Scholar]
- 27.Xie H, Ren X, Xin S, et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget 2016;7:26680-91. 10.18632/oncotarget.8589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang XL, Xu LL, Wang F. Hsa_circ_0020397 regulates colorectal cancer cell viability, apoptosis, and invasion by promoting the expression of the miR-138 targets TERT and PD-L1. Cell Biol Int 2017;41:1056-64. 10.1002/cbin.10826 [DOI] [PubMed] [Google Scholar]
- 29.Huang G, Zhu H, Shi Y, et al. cir-ITCH Plays an Inhibitory Role in Colorectal Cancer by Regulating the Wnt/β-Catenin Pathway. PLoS One 2015;10:e0131225. 10.1371/journal.pone.0131225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo JN, Li J, Zhu CL, et al. Comprehensive profile of differentially expressed circular RNAs reveals that hsa_circ_0000069 is upregulated and promotes cell proliferation, migration, and invasion in colorectal cancer. Onco Targets Ther 2016;9:7451-8. 10.2147/OTT.S123220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Broderick P, Carvajal-Carmona L, Pittman AM, et al. A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat Genet 2007;39:1315-7. 10.1038/ng.2007.18 [DOI] [PubMed] [Google Scholar]
- 32.Dowdy SC, Mariani A, Reinholz MM, et al. Overexpression of the TGF-β antagonist Smad7 in endometrial cancer. Gynecol Oncol 2005;96:368-73. 10.1016/j.ygyno.2004.10.006 [DOI] [PubMed] [Google Scholar]
- 33.Myatt N, Aristodemou P, Neale MH, et al. Abnormalities of the transforming growth factor‐beta pathway in ocular melanoma. J Pathol 2000;192:511-8. [DOI] [PubMed] [Google Scholar]
- 34.Shukla A, Yang Y, Madanikia S, et al. Elevating CLIC4 in Multiple Cell Types Reveals a TGF-β Dependent Induction of a Dominant Negative Smad7 Splice Variant. PLoS One 2016;11:e0161410. 10.1371/journal.pone.0161410 [DOI] [PMC free article] [PubMed] [Google Scholar]