Abstract
Cytokines are key factors affecting the fate of intestinal stem cells (ISCs) and effective reagents to manipulate ISCs for research purpose. Tumor necrosis factor alpha (TNFα) is a cytokine produced primarily by monocytes and macrophages. It can induce apoptotic cell death and inflammation, and to inhibit tumorigenesis and viral replication. Additionally, TNFα has been shown to play a critical role in the pathogenesis of inflammatory bowel disease (IBD). It is therefore important to identify the mechanism by which individual cytokines affect particular cell types. For this purpose, we used both conventional (CONV) and altered Schaedler flora (ASF) C3H/HeN mice to elucidate the effect of different microbial populations (complex versus defined) on growth of miniguts derived from two different intestinal environments. Furthermore, we studied the effects of different concentrations of TNFα extracted from the lymph and spleen on the growth and viability of ISCs recovered from mice bearing the ASF or CONV microbiota. The effect of TNFα on miniguts growth depends not only on the source and concentration, but also on the intestinal microenvironment from which the ISCs were derived. The findings suggest that TNFα influenced the proliferation of miniguts derived from ISCs and, therefore, modulates mucosal homeostasis of the host.
Keywords: TNFα, Microbiota, Intestinal organoids
1. Introduction
As an important component of the mucosal barrier, the intestinal epithelium plays a key role in the pathophysiology of gastrointestinal (GI) disorders as well as non-enteric diseases[1-6]. Historically, research in this area has been limited by the structural complexity of the small intestinal epithelium, the multiple signalling pathways involved in controlling the functional specificity, as well as the lack of facile culture methods and appropriate animal models for comparative studies[7-10]. Moreover, materials, substrates, pathways and growth factors involved in the differentiation of GI stem cells into specific lineages is not fully understood[11, 12]. The intestinal crypt culture system, which was established in 2009[13], has enabled the development and adoption of intestinal organoid cultures, resulting in a dramatic increase in the availability of tools and model systems to study pathophysiologic mechanisms involving the intestinal epithelium. This discovery has led to numerous advances[14-17] in these fields, including the development of culture conditions for human organoids derived from different gastrointestinal (GI) tissues such as primary colonic tissue[18] and human pluripotent stem cells[19]. The widespread adoption of experimental techniques and analysis tools[20-23] to interrogate minigut cultures has greatly advanced the application of organoids in cell biology[24, 25], disease modelling[26, 27], as well as, drug development and screening[28, 29]. Among these studies, the complex and dynamic interactions between the host’s epithelium and GI microorganisms is an integral component in the investigation of diverse GI diseases including inflammatory bowel diseases[30-33].
The intestinal microbiota, which represents the most complex ecosystem in the body, comprises a highly diverse abundance of species. Several factors impact gut microbiota structure, including host age, host genotype, postnatal environment, diet, and antibiotic use. Gut microbiota contribute to disease pathogenesis in a variety of ways: the presence or high abundance of specific bacteria may initiate inflammatory diseases such as inflammatory bowel disease, and the metabolites of the intestinal microbiota have been implicated in obesity and colon cancer by affecting host signalling pathways[34]. Gut microbes communicate to the central nervous system through nervous, endocrine, and immune signalling mechanisms that were involved in the community structure and function of the gut microbiota[35]. It is increasingly evident that the development of targeted strategies influencing the regulation of intestinal microbiota is essential for the prevention and treatment of disease[36].
The taxonomic composition of the microbiota has important scientific and clinical significance as different microbial species have been linked to several aspects of host health and disease. The effect of the microbiota, however, on cultured miniguts remains poorly understood. The altered Schaedler flora (ASF) mouse represents a powerful animal model because it possesses a highly simplified microbiota that supports normal growth and development of the mice similar to that observed for conventionally reared (CONV) mice. The gut microbiota of ASF mice is comprised of a limited community of eight bacterial species that originate from CONV mice[37-39]. The ASF mouse offers significant advantages over the CONV animals for the study of epithelium-microbiota interact ons[40, 41] by greatly simplifying the complexity of microbiota-host interactions compared with a vastly more complex, conventional microbiota[42].
In addition, cytokines are key factors affecting the fate of ISCs and effective reagents to manipulate ISCs for research and clinical purpose. Tumor necrosis factor alpha (TNFα) is a cytokine produced primarily by monocytes and macrophages. It can induce apoptotic cell death and inflammation, and to inhibit tumorigenesis and viral replication. Additionally, TNFα has been shown to play a critical role in the pathogenesis of IBD. In the in vivo environment, ISCs are exposed to complicated niches where their self-renewal, proliferation, and differentiation are influenced by combinatorial signals generated by multiple cytokines, growth factors, and interactions with other types of cells. It is, therefore, important to clarify the mechanism by which individual cytokines effect particular cell types.
Due to advances in the identification and culture of specific adult stem cells, ex vivo models of organoids have been recently described [43]. The generated multicellular intestinal organoid structures include all cell types presented in the intestine epithelium including enterocytes, Paneth cells, goblet cells, enteroendocrine cells, tuft cells, and M cells [44]. The intestinal organoids can be passaged weekly for at least 1.5 year and the phenotype and karyotype remain unchanged. Moreover, the self-renewing kinetics and multicellular structures of organoids closely resemble the in vivo situation. In addition, intestinal organoid orthotopic transplantation has the potential to facilitate regenerative therapy for inflammatory bowel disease [17]. The engrafted intestinal organoids have been shown to seamlessly integrate into the colonic tissue providing a single-layered patch that is indiscernible from adjacent epithelium.
The capability of intestinal organoids to demonstrate long term (6 months) proliferation and renewal without changing their histological characteristics provides evidence of their value to regenerative medicine. This is further supported by the ability of ex vivo cultured ISCs to functionally engraftment into the gut while maintaining histological functional features of intestinal epithelium. Intestinal organoids derived from mouse intestinal tissue are amenable to similar in vitro experimental manipulations used for cell lines. They can be studied by mass spectrometry, gene expression analysis, live cell imaging, confocal immunofluorescence microscopy and immunohistochemistry. Intestinal organoids can also be monitored in real time facilitating a detailed evaluation of any consequential phenotypic changes. The general plasticity of ISCs present in intestinal organoids endow this intestinal epithelial model with the ability to rapidly adjust cellular and tissue-related homeostasis in response to local stimuli. All these examples demonstrate the feasibility and flexibility of intestinal organoids to investigate the impact of the luminal microbial environment on gut function.
In the present study, as depicted in Figure 1, organoids grown from ISCs isolated from small intestinal tissue recovered from ASF and CONV mice were cultured successfully ex vivo. The cell morphology and growth characteristics of the organoids were found to remain unaltered after multiple passages. Comparison of organoid morphology, as well as, growth and histological characteristics were performed to elucidate the impact of different microbial populations (conventional versus ASF) on the growth characteristics of organoids derived from two different intestinal environments. We also studied the effects of host cell-derived cytokines on the growth and viability of ISCs recovered from mice harbouring the ASF or a conventional microbiota.
Figure 1.
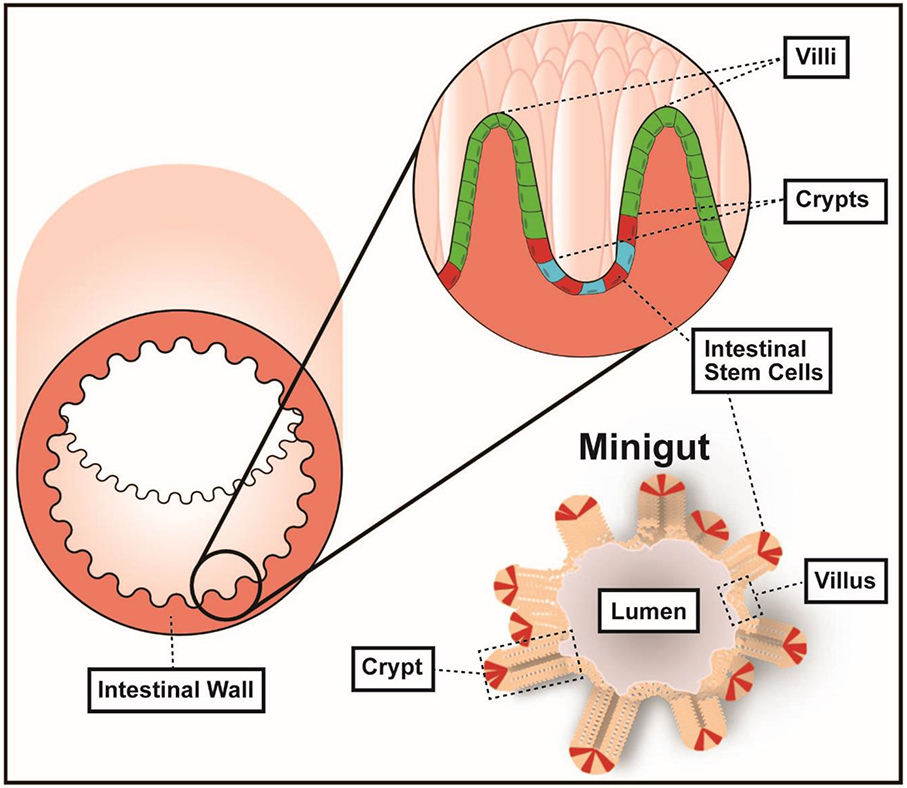
Intestinal organoids, miniguts, derived from intestinal stem cells from mice with both defined and conventional microbiota.
2. Methods
2.1. Materials
Conventional C3H/HeN CONV mice and C3H/HeN ASF mice were obtained from the College of Veterinary Medicine of Iowa State University. All animal experiments were performed with the permission of the Iowa State University Institutional Animal Care and Use Committee (IACUC # 9-04-5755-M). All methods and procedures in the experiments were performed in full compliance with the Committee's guidelines and regulations. Epidermal Growth Factor, Noggin, and R-spondin-1were purchased from PeproTech Inc. and Matrigel from BD Biosciences. LIVE/DEAD Reduced Biohazard Viability/ Cytotoxicity Kit (L7010) was purchased from Thermo Fisher Scientific Inc. All other reagents were from Life Technologies and used as received.
2.2. Preparation of ISC Culture Solution
The basal medium was prepared from Advanced DMEM/F-12 containing GlutaMax (2mM), Hepes (10 mM), penicillin (100 Units/mL), and streptomycin (100 μg/mL). The integrated ISC culture medium was prepared by mixing the basal culture medium with N2 supplement (1X), B27 supplement (1X), Epidermal Growth Factor (50 ng/mL), R-spondin-l(1 μg/mL), N-acetylcysteine (1 mM), and Noggin (100 ng/mL).
2.3. Isolation and culture of Primary ISCs
The CONV and ASF C3H/HeN mice were euthanized by CO2 asphyxiation. The proximal half of the small intestine of mice was isolated and longitudinally opened. The intestine was cut into small pieces (about 2 mm) using scissors and transferred into 50 mL tubes after thoroughly rinsing using pre-chilled (~ 4 °C) PBS (0.05 M, pH 7.2). The fragments were washed with 30 mL of ice-cold PBS by gently aspirating into a 10 mL pipette. The supernatant was removed once the fragments settled to the bottom of the conical tube. This wash step was repeated several times until the supernatant was completely clear. Then, 30 mL of ice-cold crypt isolation buffer (containing 2mM EDTA in PBS) was added, and the tube was gently agitated on its side at 4 °C for 30 min. Once the fragments settled, the supernatant was discarded.
Next, 30 mL of ice-cold PBS was added, and the tube was agitated gently by hand to remove the remaining EDTA. This was followed by the addition of 30 mL of ice-cold PBS; the tube was then mixed using a 10 mL pipette to release the crypts into solution. The solution was filtered with a 70 μm cell strainer, and the supernatant containing crypt fragments was collected in a tube pre-coated with 1% BSA. The crypt fractions were centrifuged at 300 × g for 5 min. Next, the pellet was transferred into a 15 mL tube pre-coated with 1% BSA by re-suspending in 10 mL of ice-cold basal medium. The tube was then centrifuged at a lower speed (200 × g, 2 min) to eliminate single cells (mostly lymphocytes). The cell pellet was re-suspended in 1 mL of basal medium in a microfuge tube and centrifuged at 300 × g for 3 min; then, the supernatant was decanted. This was followed by the addition of 250 μL of Matrigel using a pre-cooled tip. The solution was mixed gently and kept on ice to avoid the formation of air bubbles. Then, 38 μL of Matrigel was pipetted into each well of a 24-well culture dish pre-incubated in a CO2 incubator (5% CO2, 37°C), followed by an additional incubation for 20 min in an incubator (5% CO2, 37°C). Next, 500 μL of complete ISC culture medium was added into wells containing the Matrigel - intestinal cells mixture.
2.4. Live/dead viability
Viability Kit (L7013) was thawed to room temperature before use. A diluted working solution was prepared by pipetting SYTO 10 solution (1 ×) and ethidium homodimer-2 solution (1 ×) into HBSS (500 ×) and mixing thoroughly by vortexing. The original culture medium was removed and 0.5 mL HBSS was added. Next, the HBSS was decanted and replaced with 0.5 mL of the live/dead working solution. The solution was incubated in the dark at 25 °C for 20 min. The working solution was drawn and the miniguts were washed with an equal volume of HBSS.
2.5. Haematoxylin and eosin Staining
Samples of miniguts were fixed in neutral buffered formalin (4% formaldehyde in 0.1 M phosphate buffer) for several days at 4 °C. First, miniguts were dehydrated in an ethanol gradient (30%-100%); then, the miniguts were cleared in xylene, penetrated, and re-embedded with melted paraffin 3 times. Next, 8 μm sections were cut using an A/O 820 rotary microtome and fixed on to glass slides. Next, the slides were deparaffinized using an automated system and haematoxylin and eosin staining was performed. The samples were then dehydrated, cleared, and sealed with a coverslip.
2.6. Preparation of Cytokines and Treatment ex vivo
Single suspensions of lymph node or spleen cells were generated from tissue harvested from ASF mice previously colonized with Escherichia coli (E. coli) LF82. Lymph node or spleen cells were cultured at 1-2 X 106 cells/mL in cell culture medium (DMEM 4.5g/L) supplemented with HEPES, streptomycin, penicillin, glutamine, pyruvate, and 2-mercaptoethanol. In addition, lymph node and spleen cells were stimulated with anti-CD3 (10 μg/mL) plus anti-CD28 (0.5 μg/mL) for 72 hours in 12 well flat bottom plates. The culture supernatants were harvested, clarified by centrifugation, and stored frozen at −80 °C until used. As a relative measure of the cytokine content, the concentration of TNFα in the culture supernatant was measured by a multiplex bead-based assay employing Bio-Plex® MAGPIX™ Multiplex Reader. Standard curves were generated using Bio-Plex Manager Software Version 6.0 (Bio-Rad Laboratories, Inc., Carlsbad, CA, USA).
TNFα-containing supernatants were added to the culture medium after intestinal miniguts had been cultured for 7 days, then added fresh culture medium was added every 2 days to keep the total amount of cytokines constant. To measure the proliferation of ISCs, photographic images were captured using a LeicaDMi1 inverted microscope. The total area of the organoids calculated using ImageJ. Each data point is the average value obtained by tracking the change in size of 10 miniguts/ treatment group.
2.7. Statistical Analysis
For statistical analysis, statistical software JMP® was used with a regression modelling approach. The data were presented as means ± SD. The associated p-values were reported; p < 0.05 indicated a statistically significant difference; p < 0.01 represented a higher significant difference.
2.8. Data Calculation and Date Availability Statement
The area of the miniguts was calculated with ImageJ which was download from the open platform of the ImageJ official website https://imagej.net/.
The average growth rate of the miniguts was calculated using the following formula.
All data generated or analysed during this study are included in this published article.
3. Results and Discussions
3.1. Comparison of morphology of ISCs isolated from ASF and CONV mice
The miniguts derived from CONV and ASF mice exhibited a self-organized morphology with continuous expansion in the size/volume of the individual miniguts (Figure 2). After 3 days, the miniguts generated asymmetric structures, and efficiently formed large organoids with a central lumen within 14 days. The bottom 4 rows of photomicrographs depicted in Figure 2 show the change in size of a single isolated miniguts generated from the small intestines of CONV and ASF mice over time. In both cases, crypts were formed from the self-organizing ISCs differentiating into an organoid. The lumen of the organoid filled with sloughed epithelial cells. Comparison of the change of size of the miniguts derived from the two different set of mice showed that the organoids from the ASF mice grew faster than those from CONV mice.
Figure 2.
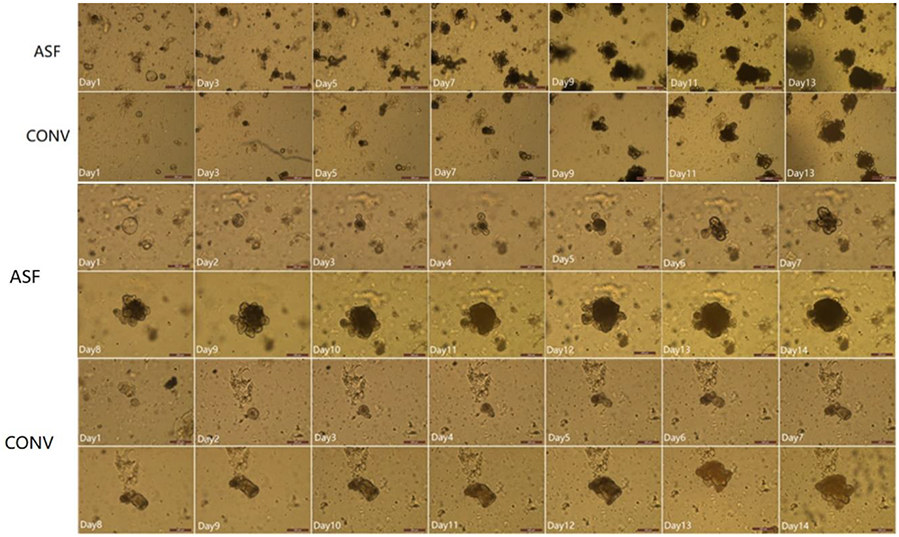
Morphological features of miniguts derived from ASF (top row) and CONV (2nd row) mice. Numbers in the images represent the number of days since the intestinal stem cell cultures were initiated. The scale bars in 1st and 2nd rows represent 500 μm. Time course for the change in size of a single miniguts derived from ASF (the 3rd and 4th rows) or CONV (the 5th and 6th rows) mice. Numbers in the images represent the number of days since the intestinal stem cell cultures were initiated. The scale bars in 3rd, 4th, 5th and 6th rows represent 200 μm.
3.2. Comparison of average growth rate
Growth rate (y) continuously increases with time (x), i.e., days, and thus, a regression modelling approach was used to determine this relationship for CONV (C) and ASF (A) mice. The general least squares regression model used in this work is:
| (1) |
where yi = ith measured response, xi is the ith measured time, θ is a vector of unknown model parameters, f(xi; θ) = μY∣xi is the true unknown expectation function for yi, and εi random deviation from Y∣xi that is an independent normally distributed random variable with mean 0 and unknown and constant variance σ2 for i = 1, …, n, the sample size, i.e.,
| (2) |
The modelling objective is to estimate the unknown quantities and functions in compliance with the modelling assumptions to make sound formal statistical inference regarding the comparative growth behaviour of C and A. More specifically, the statistical inference goal is to accurately estimate Y∣xi for C and A and then statistically assess their significant difference using 100(1-95)% confidence intervals (CIs) for Y∣xi at all experimental value of i, which are called “confidence bands.”
The scatter plots of both data sets are given in Fig. 4, where very similar monotonically exponential growth behaviour is observed. To avoid the complexities of nonlinear regression, in particularly, for inference, the class for f(xi;θ) was restricted to linear regression forms, i.e., forms that are linear in model parameters. Based on the response behaviour, a second order (i.e., quadratic) linear regression structure was fit to the data. The residual plot for C for its fit is given in Fig. 5, where the ith residual, , and , is the ith estimate of μY∣xi. The residual plot against time reveals the unacceptability of this structure very clearly as the residuals increase as time (xi) increases. This structure leads to highly biased estimates because their values are more strongly dependent on the data at the later times. Unbiased estimation requires an equal weighing of the all the data over the input range. To accomplish this objective, the following structure was used to fit the data:
| (3) |
Figure 4.
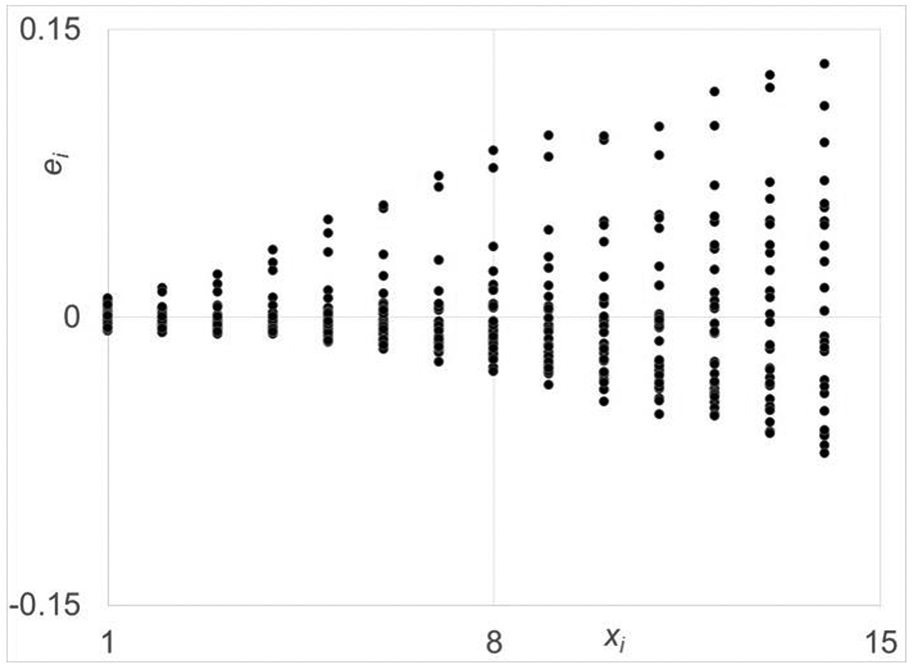
Residual plot for a quadratic linear regression function of the data set derived from the miniguts of CONV mouse miniguts. The spread in the residuals increase as days increase and will result in very biased estimation of the model parameters.
Figure 5.
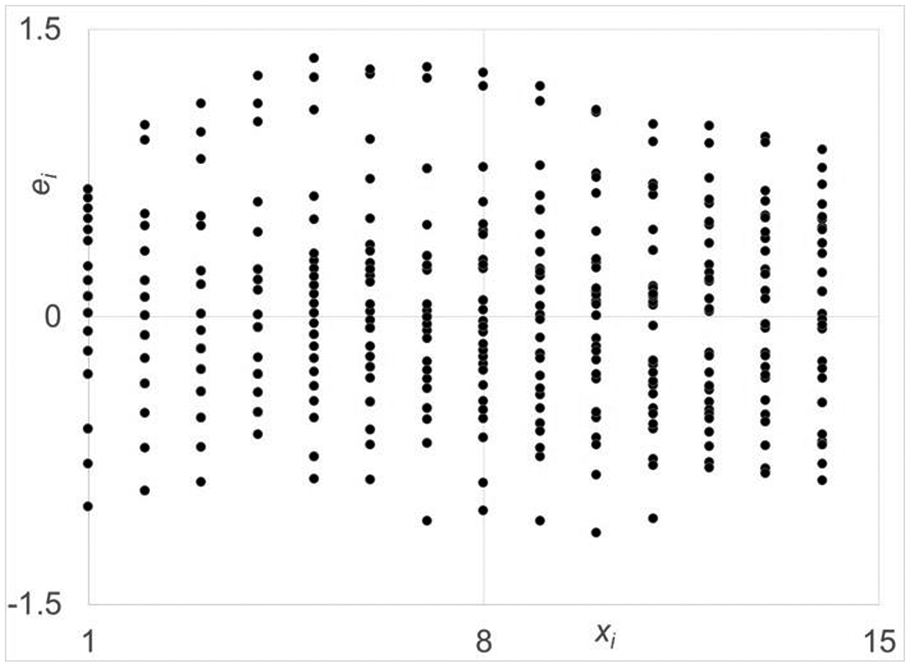
Residual plot for proposed log transformed data set for the miniguts from CONV mice. The spread in the residuals are fairly uniform over the days and will result in only slightly biased estimation of the model parameters.
The residual plot against time for the C fit using this structure is shown Fig. 6. The one for A is very similar. As shown, the spread of Eq. 3 residuals is nearly uniform over , the ln (Days). Upon meeting this criterion, this model structure was fully evaluated for compliance to all assumptions, i.e., independence and normality also, which were met quite well for both cases, but not shown for space considerations. In addition, the coefficient of determination for model fits for A and C were 0.75 and 0.71, respectively. These values are quite good given the very large spread in the responses at each time point of data collection.
Figure 6.
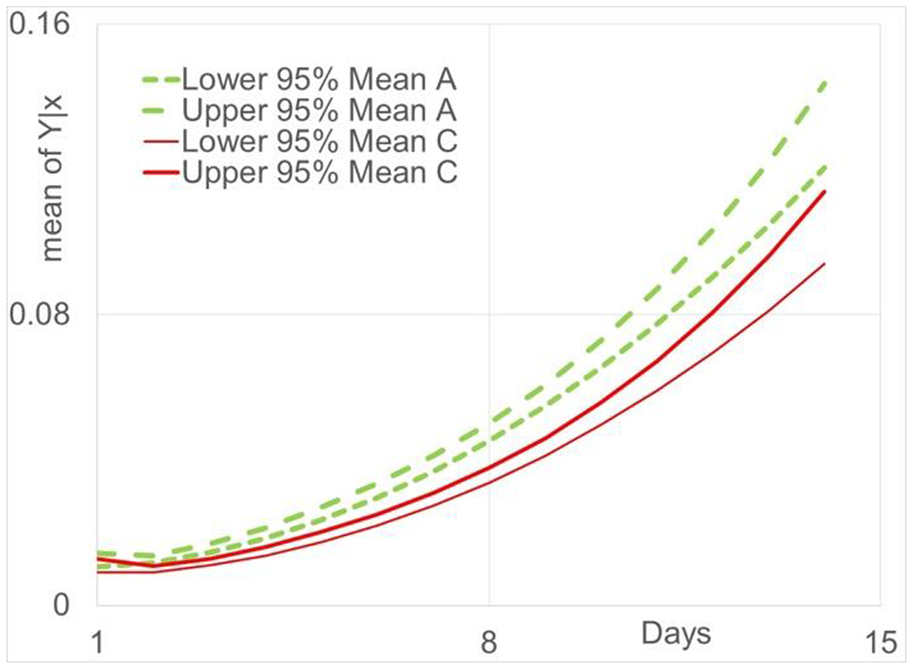
95% confidence bands for Y∣xi to miniguts derived from ASF (green lines) and CONY (red lines) mice.
The efficacies of the fitted models are supported and evaluated for statistical inference using 95% confidence bands for Y∣xi and 95% prediction bands for individual values spread about μY∣xi. Figure 3 gives the estimated curves for μY∣xi, i.e., , and the lower and upper 95% prediction bands for C and A, respectively. Each data set consists of the same number of samples at each time point for a total of 420 values. On the average, 95% prediction bands should contain 95% of the measured values. Given the size of this data set, it is informative to evaluate the efficacy of these fits informally. For C and A, 93% and 94% of the values are in the 95% prediction bands, respectively, and thus, supporting the accuracy and soundness of our approach. Figure 4 shows the residual plot for a quadratic linear regression function for the CONV data set. The spread in the residuals increase as days increase and will result in very biased estimation of the model parameters. Figure 5 shows the residual plot for proposed log transformed fit for the CONV data set. The spread in the residuals are fairly uniform over the days and will result in only slightly biased estimation of the model parameters. The 95% confidence bands for both cases are shown in Figure 6. Overlap of the intervals indicate no statistically significant differences in the estimated means, and no overlap indicates statistically significant differences at the specified significance level. As shown in Figure 6, at the earliest time points, there is overlap but this quickly changes and there is no more overlap of the confidence bands, with A being greater than C. Therefore, A appears to have significantly greater growth rate than C over most of the days. This further confirms that this data set is informative to evaluate the efficacy of our approach fits informally.
Figure 3.
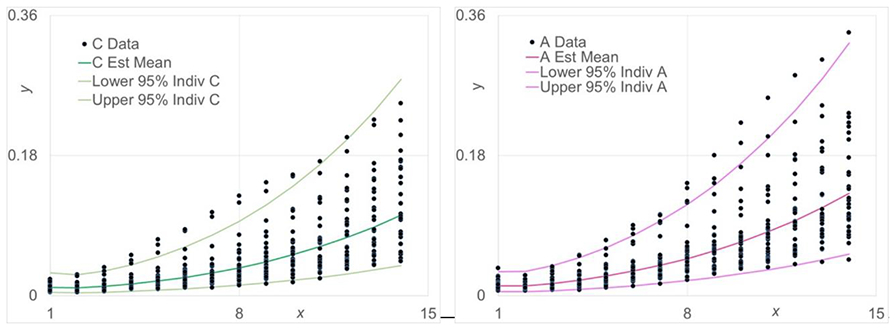
Scatter plots for change in the size of organoid derived from CONV (left) or ASF (right) mice. Data represents the estimated mean (central line) and the 95 % confidence intervals (upper and lower lines).
3.3. Viability and Haematoxylin and eosin staining
The viability assay, which enables the determination of cell viability based on two-colour fluorescence, was performed using a fluorescence microscope. Green high-permeability dye marks all cells, while red dye enables the specific identification of damaged cells owing to its poor cell permeability. Under conditions of equal dye concentrations and balanced relative affinities, cells are differentially stained because of their differential permeability. Live cells appear fluorescent green and dead cells, fluorescent red, enabling the qualitative determination of live and dead of cells. Comparison of the fluorescence micrographs, as shown in Figure 7, indicates that the red area of CONV miniguts at Day 5 and Day 9 is stronger in intensity versus that of the ASF miniguts at the same time points suggesting that the miniguts from the ASF mice are healthier and less prone to cell death. This result is consistent with the findings showing that miniguts derived from ASF mice grow more rapidly than those derived from CONV mice. The growth of miniguts includes both stem cell proliferation and differentiation.
Figure 7.
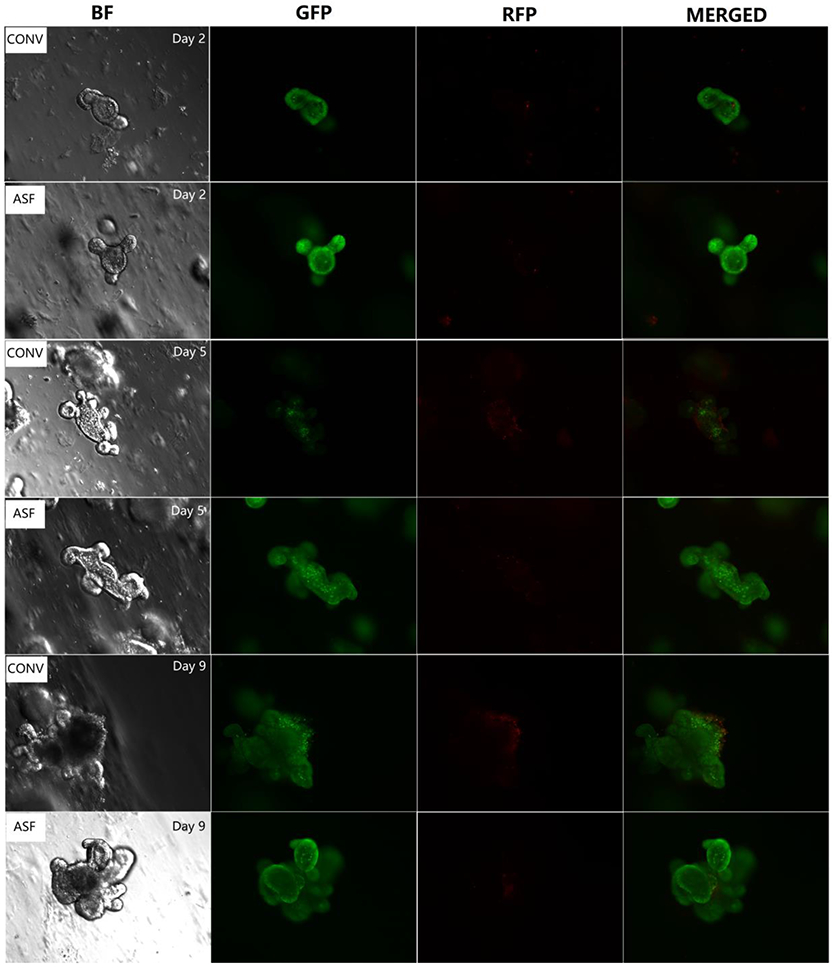
Assessment of live (green) and dead (red) cells of miniguts cultured after 2, 5, and 9 days in culture.
The use of various immunohistochemical dyes to stain specific organoid components enables the analysis of their differences by microscopic evaluation. Haematoxylin and eosin staining images of the miniguts is shown in Figure 8. Because of the effects of the fixative and inhibition of infiltration, breakage of tissues occurred, as shown in the images. However, it is still clear that miniguts derived from ASF mice are more structured and organized compared to those from CONV mice especially on day 5.
Figure 8.
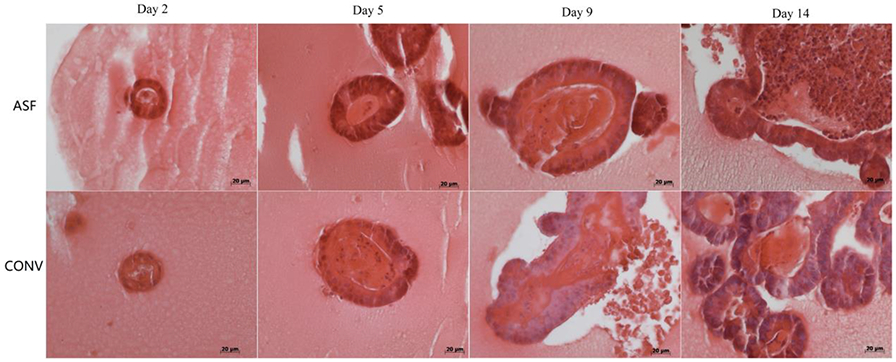
Representative haematoxylin & eosin staining of miniguts after days 2, 5, 9, and 14 in culture.
Under identical culture conditions, miniguts derived from ASF mice grow faster than those derived from CONV mice. These data indicate that miniguts derived from ISC recovered from mice harbouring complex microbiota are differentially programmed, as evidenced by their slower proliferation rate. The presence of diverse microorganisms in a complex intestinal environment may inhibit the proliferation of miniguts as shown in this study. Other potential mechanisms are apoptosis.
3.4. Effect of Cytokine Treatment on ex vivo Growth
It was found that culture supernatants high in TNFα derived from different tissue sources has different effects on intestinal organoids’ growth. As shown in Figure 9a, compared with the control group, at the same concentration (based on level of TNFα = 0.02 ng/mL), the culture supernatants produced by splenocytes stimulated with anti-CD3 plus anti-CD28 inhibited the growth of ISCs from ASF mice while similarly induced supernatants from lymph node cells promoted organoid growth.
Figure 9.
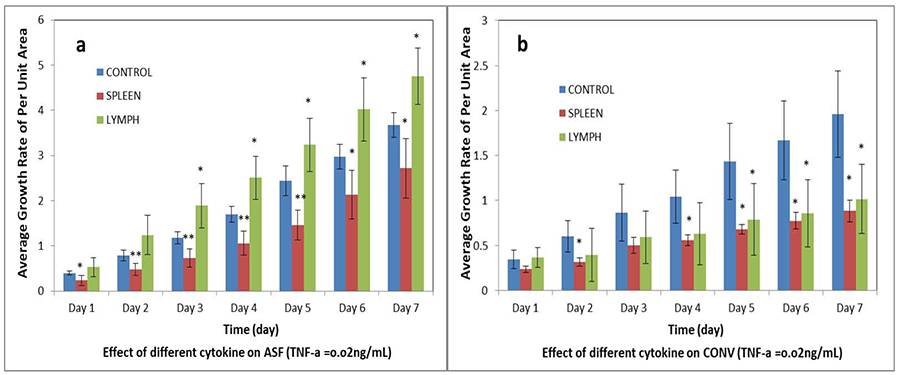
Effect of cytokines in culture supernatants on the change in size of intestinal organoids. a: intestinal organoids derived from the small intestine of ASF mice. b: intestinal organoids derived from the small intestine of CONV. The culture supernatants containing the cytokines including TNFα were generated by stimulating spleen cells (red) or lymph nodes cells (green) with anti-CD3 plus anti-CD28 as described in the Methods section. N = 20; *p < 0.05, **p < 0.01.
Compared to non-treated control organoids, supernantants from stimulated spleen cells significantly (p < 0.05) inhibited organoid growth as measured at days two to seven. However, growth of ISCs from CONV mice were significantly (p < 0.05) inhibited by supernatants from both stimulated splenocytes and lymph node cells (Figure 9b). We also found that effects of different concentrations of cytokines, based on the concentration of TNFα, on ISCs from ASF or CONV mice were also different. As shown in Figure 10a, ISCs from ASF mice were significantly inhibited (p < 0.05) by a lower concentration of cytokines compared to organoids from CONV mice. As the concentration of supernatants was increased, the inhibitory effect became less obvious, with evidence of growth promotion as shown in Figure 9a. While evaluating ISCs from CONV mice (Figure 10b), regardless of the concentration of cytokines, the growth of the miniguts was significantly (p < 0.05) inhibited. We conclude that the effect of TNFα on ISCs replication and organoid growth is subtle and complex. These studies demonstrate that the complexities affecting organoid development depend not only on the source and concentration of TNFα, but also on the microenvironment (i.e., microbial composition) of the GI tract from which the miniguts were derived.
Figure 10.
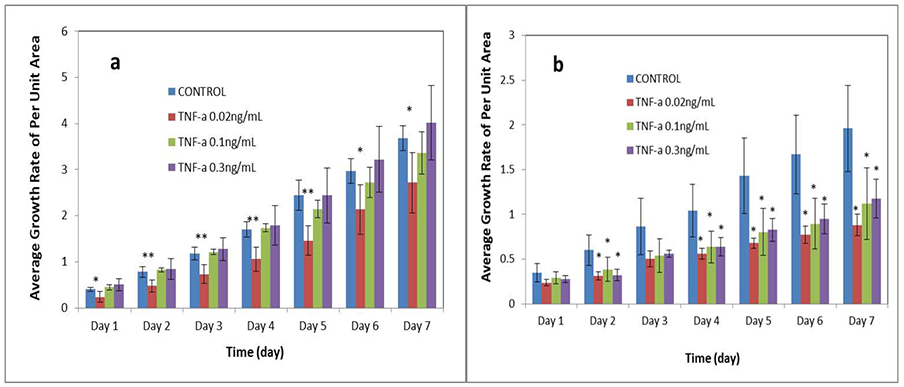
Effect of the concentration of stimulated spleen cell culture supernatants on the change in size of miniguts (based on TNF-α concentration in supernatants of anti-CD3 + anti-CD28 stimulated spleen cells). a: intestinal organoids isolated from ASF. b: intestinal organoids isolated from CONY. N = 20; *p < 0.05, **p < 0.01
Acknowledgements
Dr. Wang is grateful for the support from Crohn’s & Colitis Foundation of America (CCFA) Career Award (No. 348137), PhRMA Foundation Research Starter Award (No. RSGTMT17), and McGee-Wagner Interdisciplinary Research Foundation. Dr. Wang and Dr. Mansell acknowledge support from National Institutes of Health (R21GM131189) grant. Drs. Phillips and Wannemuehler likewise acknowledge NIH support (R01GM099537).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
The authors declare no competing interests.
References
- [1].Egberts H, Koninkx J, Van Dijk J, Mouwen J, Biological and pathobiological aspects of the glycocalyx of the small intestinal epithelium. A review, Veterinary Quarterly 6(4) (1984) 186–199. [DOI] [PubMed] [Google Scholar]
- [2].Groschwitz KR, Hogan SP, Intestinal barrier function: molecular regulation and disease pathogenesis, Journal of Allergy and Clinical Immunology 124(1) (2009) 3–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Clevers H, The intestinal crypt, a prototype stem cell compartment, Cell 154(2) (2013) 274–284. [DOI] [PubMed] [Google Scholar]
- [4].Sato T, Clevers H, Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications, Science 340(6137) (2013) 1190–1194. [DOI] [PubMed] [Google Scholar]
- [5].Leushacke M, Barker N, Ex vivo culture of the intestinal epithelium: strategies and applications, Gut 63(8) (2014) 1345–1354. [DOI] [PubMed] [Google Scholar]
- [6].Wilson SS, Tocchi A, Holly MK, Parks WC, Smith JG, A small intestinal organoid model of non-invasive enteric pathogen-epithelial cell interactions, Mucosal Immunology 8(2) (2015) 352–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bowen JM, Gibson RJ, Stringer AM, Chan TW, Prabowo AS, Cummins AG, Keefe DM, Role of p53 in irinotecan-induced intestinal cell death and mucosal damage, Anti-cancer drugs 18(2) (2007) 197–210. [DOI] [PubMed] [Google Scholar]
- [8].Evans GS, Flint N, Somers A, Eyden B, Potten CS, The development of a method for the preparation of rat intestinal epithelial cell primary cultures, Journal of Cell Science 101(1) (1992) 219–231. [DOI] [PubMed] [Google Scholar]
- [9].Whitehead RH, DemmLer K, Rockman SP, Watson NK, Clonogenic growth of epithelial cells from normal colonic mucosa from both mice and humans, Gastroenterology 117(4) (1999) 858–865. [DOI] [PubMed] [Google Scholar]
- [10].Fukamachi H, Proliferation and differentiation of fetal rat intestinal epithelial cells in primary serum-free culture, Journal of Cell Science 103(2) (1992) 511–519. [DOI] [PubMed] [Google Scholar]
- [11].Wang Q, Cheng H, Peng H, Zhou H, Li PY, Langer R, Non-genetic engineering of cells for drug delivery and cell-based therapy, Advanced Drug Delivery Reviews 91 (2015) 125–140. [DOI] [PubMed] [Google Scholar]
- [12].Hu X, Tang Y, Wang Q, Li Y, Yang J, Du Y, Kennedy JF, Rheological behaviour of chitin in NaOH/urea aqueous solution, Carbohydrate Polymers 83(3) (2011) 1128–1133. [Google Scholar]
- [13].Sato T, Vries RG, Snippert HJ, Van De Wetering M, Barker N, Stange DE, Van Es JH, Abo A, Kujala P, Peters PJ, Single Lgr5 stem cells build crypt villus structures in vitro without a mesenchymal niche, Nature 459(7244) (2009) 262–265. [DOI] [PubMed] [Google Scholar]
- [14].Peng H, Wang C, Xu X, Yu C, Wang Q, An intestinal Trojan horse for gene delivery, Nanoscale 7(10) (2015) 4354–4360. [DOI] [PubMed] [Google Scholar]
- [15].Peng H, Poovaiah N, Forrester M, Cochran E, Wang Q, Ex vivo culture of primary intestinal stem cells in collagen gels and foams, ACS Biomaterials Science & Engineering 1(1) (2014)37–42. [DOI] [PubMed] [Google Scholar]
- [16].Sala FG, Matthews JA, Speer AL, Torashima Y, Barthel ER, Grikscheit TC, A multicellular approach forms a significant amount of tissue-engineered small intestine in the mouse, Tissue Engineering Part A 17(13–14) (2011) 1841–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Yui S, Nakamura T, Sato T, Nemoto Y, Mizutani T, Zheng X, Ichinose S, Nagaishi T, Okamoto R, Tsuchiya K, Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5+ stem cell, Nature Medicine 18(4) (2012) 618–623. [DOI] [PubMed] [Google Scholar]
- [18].Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van Den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD, Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium, Gastroenterology 141(5) (2011) 1762–1772. [DOI] [PubMed] [Google Scholar]
- [19].Spence JR, Mayhew CN, Rankin SA, Kuhar MF, Vallance JE, Tolle K, Hoskins EE, Kalinichenko VV, E Wells S, Zorn AM, Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro, Nature 470(7332) (2011) 105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Andersson-Rolf A, Fink J, Mustata RC, Koo B-K, A video protocol of retroviral infection in primary intestinal organoid culture, Journal of visualized experiments: JoVE (90) (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Fujii M, Shimokawa M, Date S, Takano A, Matano M, Nanki K, Ohta Y, Toshimitsu K, Nakazato Y, Kawasaki K, A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis, Cell Stem Cell 18(6) (2016) 827–838. [DOI] [PubMed] [Google Scholar]
- [22].Matano M, Date S, Shimokawa M, Takano A, Fujii M, Ohta Y, Watanabe T, Kanai T, Sato T, Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids, Nature Medicine 21(3) (2015) 256–262. [DOI] [PubMed] [Google Scholar]
- [23].Yin Y, Bijvelds M, Dang W, Xu L, van der Eijk AA, Knipping K, Tuysuz N, Dekkers JF, Wang Y, de Jonge J, Modeling rotavirus infection and antiviral therapy using primary intestinal organoids, Antiviral Research 123 (2015) 120–131. [DOI] [PubMed] [Google Scholar]
- [24].Gregorieff A, Liu Y, Inanlou MR, Khomchuk Y, Wrana JL, Yap-dependent reprogramming of Lgr5+ stem cells drives intestinal regeneration and cancer, Nature 526(7575) (2015)715–718. [DOI] [PubMed] [Google Scholar]
- [25].Oudhoff MJ, Braam MJ, Freeman SA, Wong D, Rattray DG, Wang J, Antignano F, Snyder K, Refaeli I, Hughes MR, SETD7 controls intestinal regeneration and tumorigenesis by regulating Wnt/μ-catenin and hippo/YAP signaling, Developmental Cell 37(1) (2016) 47–57. [DOI] [PubMed] [Google Scholar]
- [26].Wiegerinck CL, Janecke AR, Schneeberger K, Vogel GF, van Haaften–Visser DY, Escher JC, Adam R, Thöni CE, Pfaller K, Jordan AJ, Loss of syntaxin 3 causes variant microvillus inclusion disease, Gastroenterology 147(1) (2014) 65–68.elO. [DOI] [PubMed] [Google Scholar]
- [27].Zietek T, Rath E, Haller D, Daniel H, Intestinal organoids for assessing nutrient transport, sensing and incretin secretion, Scientific Reports 5 (2015) 16831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Koppens MA, Bounova G, Comelissen-Steijger P, de Vries N, Sansom OJ, Wessels LF, van Lohuizen M, Large variety in a panel of human colon cancer organoids in response to EZH2 inhibition, Oncotarget 7(43) (2016) 69816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Boehnke K, Iversen PW, Schumacher D, Lallena MJ, Haro R, Amat J, Haybaeck J, Liebs S, Lange M, Schäfer R, Assay establishment and validation of a high-throughput screening platform for three-dimensional patient-derived colon cancer organoid cultures, Journal of Biomolecular Screening 21(9) (2016) 931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Mueller K, Ash C, Pennisi E, Smith O, The gut microbiota, Science 336(6086) (2012) 1245–1245. [DOI] [PubMed] [Google Scholar]
- [31].Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S, Hostgut microbiota metabolic interactions, Science 336(6086) (2012) 1262–1267. [DOI] [PubMed] [Google Scholar]
- [32].Roeselers G, Ponomarenko M, Lukovac S, Wortelboer HM, Ex vivo systems to study host–microbiota interactions in the gastrointestinal tract, Best Practice & Research Clinical Gastroenterology 27(1) (2013) 101–113. [DOI] [PubMed] [Google Scholar]
- [33].Trosvik P, Muinck EJ, Ecology of bacteria in the human gastrointestinal tract—identification of keystone and foundation taxa, Microbiome 3(1) (2015) 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Putignani L, Del Chierico F, Petrucca A, Vemocchi P, Dallapiccola B, The human gut microbiota: a dynamic interplay with the host from birth to senescence settled during childhood, Pediatric Research 76(1) (2014) 2–10. [DOI] [PubMed] [Google Scholar]
- [35].De Palma G, Collins SM, Bercik P, Verdu EF, The microbiota–gut–brain axis in gastrointestinal disorders: stressed bugs, stressed brain or both?, The Journal of Physiology 592(14) (2014) 2989–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Marchesi JR, Adams DH, Fava F, Hermes GD, Hirschfield GM, Hold G, Quraishi MN, Kinross J, Smidt H, Tuohy KM, The gut microbiota and host health: a new clinical frontier, Gut (2015) gutjnl-2015-309990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wymore Brand M, Wannemuehler MJ, Phillips GJ, Proctor A, Overstreet A-M, Jergens AE, Orcutt RP, Fox JG, The altered Schaedler flora: continued applications of a defined murine microbial community, ILAR Journal 56(2) (2015) 169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Alexander AD, Orcutt RP, Henry JC, Baker J, Bissahoyo AC, Threadgill DW, Quantitative PCR assays for mouse enteric flora reveal strain-dependent differences in composition that are influenced by the microenvironment, Mammalian Genome 17(11) (2006) 1093–1104. [DOI] [PubMed] [Google Scholar]
- [39].Stehr M, Greweling MC, Tischer S, Singh M, Blöcker H, Monner DA, Müller W, Charles River altered Schaedler flora (CRASF®) remained stable for four years in a mouse colony housed in individually ventilated cages, Laboratory Animals 43(4) (2009) 362–370. [DOI] [PubMed] [Google Scholar]
- [40].Stecher B, Chaffron S, Käppeli R, Hapfelmeier S, Freedrich S, Weber TC, Kirundi J, Suar M, McCoy KD, von Mering C, Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria, PLoS Pathogens 6(1)(2010)e1000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Jergens AE, Wilson-Welder JH, Dom A, Henderson A, Liu Z, Evans RB, Hostetter J, Wannemuehler MJ, Helicobacter bilis triggers persistent immune reactivity to antigens derived from the commensal bacteria in gnotobiotic C3H/HeN mice, Gut 56(7) (2007) 934–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lyte J, Proctor A, Phillips G, Lyte M, Wannemuehler M, Altered Schaedler flora mice: A defined microbiota animal model to study the microbiota-gut-brain axis, Behavioural Brain Research 356 (2019) 221–226. [DOI] [PubMed] [Google Scholar]
- [43].Scoville DH, Sato T, He XC, Li L, Current view: intestinal stem cells and signaling, Gastroenterology 134(3) (2008) 849–864. [DOI] [PubMed] [Google Scholar]
- [44].Van Der Flier LG, Clevers H, Stem cells, self-renewal, and differentiation in the intestinal epithelium, Annual Review of Physiology 71 (2009) 241–260. [DOI] [PubMed] [Google Scholar]


