Abstract
Adhesive pili in Gram-positive bacteria represent a variety of extracellular multi-protein polymers that mediate bacterial colonization of specific host tissues and associated pathogenesis. Pili are assembled in two distinct but coupled steps, an orderly crosslinking of pilin monomers and subsequent anchoring of the polymer to peptidoglycan, catalyzed by two transpeptidase enzymes – the pilus-specific sortase and the housekeeping sortase. Here, we review this biphasic assembly mechanism from studies of two prototypical models, the heterotrimeric pili in Corynebacterium diphtheriae and the heterodimeric pili in Actinomyces oris, highlighting some newly emerged basic paradigms. The disparate mechanisms of protein ligation mediated by the pilus-specific sortase and the spatial positioning of adhesive pili on cell surface modulated by the housekeeping sortase are amongst the notable highlights.
Keywords: Pili, Sortase, Pilus assembly, Transpeptidation, Protein ligation, Coaggregation, Virulence
INTRODUCTION
Fiber-like appendages called pili or fimbriae are microscopic structures present on the cell surface of both Gram-negative and Gram-positive bacteria. They are involved in a wide range of cellular activities, including adherence, motility, conjugation, and virulence [1–3]. Among these, the only pilus form known to date in which individual subunits are covalently bonded, is the pili that are assembled by the action of sortase enzymes conserved in Gram-positive bacteria [4], but not in Gram-negative bacteria that produce pili in which the monomer subunits are joined via protein-protein interaction. The various sortases found thus far are broadly grouped into six classes (SrtA-F), based on sequence alignment and substrate preference [5, 6]; of these, only members of the class C and class E sortases are shown to catalyze the two distinct steps of Gram-positive pilus assembly, pilus polymerization and anchoring of pili to the cell wall, respectively (Box 1) [7].
Box 1: Overview of class A, C, and E sortases.
Staphylococcus aureus SrtA is the prototype class A sortase [61], which recognizes a LPXTG motif, preceding a hydrophobic domain and a positively charged tail that together constitutes a “cell wall sorting signal” (CWSS) located at the C-terminus of a sortase substrate [62]. As a transpeptidase, SrtA catalyzes cell wall anchoring of surface proteins harboring the tripartite sorting signal, by first hydrolyzing the peptide bond between threonine and glycine and then covalently joining the cleaved threonine residue to the pentaglycine peptide of lipid II in the cell wall [63]. Structurally, the class A sortase harbors a unique β-barrel fold, with the catalytic site containing the sole cysteine residue [64], which is absolutely required for sortase activity [65].
Class C sortases, or pilus-specific sortases, are found in bacterial species that produce covalently linked pili [5]. They are structurally similar to class A sortases with the 8-stranded β-barrel fold encapsulating the active site [66]. Unique to class C sortases is a flexible N-terminal hydrophobic “lid” which covers the catalytic pocket and has been proposed to play a role in substrate recognition and sortase stability [6, 67–70].
Present in various Gram-positive bacterial species and abundant in Actinobacteria [5, 71], class E sortase enzymes recognize a distinct LAXTG sorting motif [5]. Compared to sortases of class A and C, structures of class E sortases have been less well studied, with only two available sortase structures from Streptomyces coelicolor and A. oris [60, 72]. While both harbor the conserved 8-stranded β-barrel fold without the aforementioned lid, they contain a conserved tyrosine residue within the β3/β4 sheet that appears to be involved in the recognition of the LAXTG sorting motif [60, 72].
Historically, the connection between sortase and pilus polymerization was somewhat serendipitous based on two types of genetic observations. First, it was recognized that the protein sequences of different fimbrial subunits in the actinobacterium Actinomyces naeslundii harbor the same C-terminal cell wall sorting signal (CWSS) as the classically defined cell surface protein, the protein A of Staphylococcus aureus [8, 9] (Box 1). Secondly, many sets of genes coding for sortase enzymes and surface proteins with the LPXTG motif are clustered in the same operons in the actinobacterium Corynebacterium diphtheriae [10]. Indeed, immuno-electron microscopic analysis using antibodies against some of these surface proteins revealed the presence of distinct classes of pili on the surface of C. diphtheriae [10]. Since the first demonstration of the essential function of specific sortases in pilus assembly in C. diphtheriae [3, 10, 11], the past decade has seen the extensive investigations of pilus assembly in many other Gram-positive bacteria including Bacillus cereus, Enterococcus faecalis, and streptococci [12–18]. Because both Actinomyces and C. diphtheriae continue to serve as excellent models in the studies of the genetic, biochemical and structural mechanisms of Gram-positive pilus assembly, here we focus our review on these two actinobacterial species only to highlight the recent advances in the field. For a more comprehensive description of pilus assembly in Gram-positive bacteria, we refer the reader to several excellent publications elsewhere [19–21].
Corynebacterium diphtheriae pili offer a new paradigm for protein ligation
Assembly of the SpaA pilus
The causative agent of human diphtheria C. diphtheriae is among the earliest bacterial species where pili were identified [22, 23]. As in many Gram-positive bacteria [4], the C. diphtheriae genes coding for distinct pilin subunits and dedicated pilus-specific sortases, which are all class C sortases, are organized into three operons [10]. Together, they encode three distinct pilus types specified by their major subunits, i.e. SpaA-type, SpaD-type and SpaH-type pili [10, 24, 25]. Pili constitute one of the major virulence factors in C. diphtheriae: A mutant devoid of all pilins is highly attenuated in virulence in a mouse model of diphtheritic toxemia [26]. Each type of pili of C. diphtheriae are heterotrimeric, meaning that each type of pili are made of three distinct pilin subunits; however, it is important to note further that the various molecules of the same pilus type (SpaA-, SpaD- or SpaH-type) vary in their length. Of these, the most highly studied is the SpaA pilus, which is composed of the shaft pilin SpaA, the tip adhesin SpaC and the base pilin SpaB anchored to the cell wall; assembly of this pilus requires the cognate sortase SrtA, a class C sortase [10] (Box 1). While all three pilins contain the CWSS, only SpaA has a recognizable pilin motif with a conserved lysine residue that serves as a nucleophile essential for sortase-mediated cross-linking of pilin monomers. In this reaction that can occur repeatedly, the pilus-specific sortase SrtA catalyzes hydrolysis of the LPXTG motif in a pilin subunit and links the cleaved threonine residue to the lysine residue within the pilin motif of another pilin that are adjoining each other on the bacterial membrane (Fig. 1). Because SpaC resides at the pilus tip (the tip first rule), the first transpeptidation reaction must occur between SpaC and SpaA, linking the threonine residue of the SpaC LPXTG motif to the reactive lysine residue within the pilin motif of SpaA (Fig. 1). Subsequently, pilus elongation ensues whereby SpaA pilins are added to the growing chain of pilus polymers. Ultimately, pilus polymerization is terminated with addition of SpaB (the base pilin), which is then anchored to peptidoglycan by the housekeeping sortase SrtF (class E sortase), whose gene is not genetically linked (or in proximity) to any of the three pilus gene clusters [27] (Fig. 1). This biphasic mechanism of pilus assembly appears to be universally applicable to other Gram-positive bacterial pili so far studied [12, 28–30].
Figure 1: Assembly of the heterotrimeric SpaA pili in Corynebacterium diphtheriae.
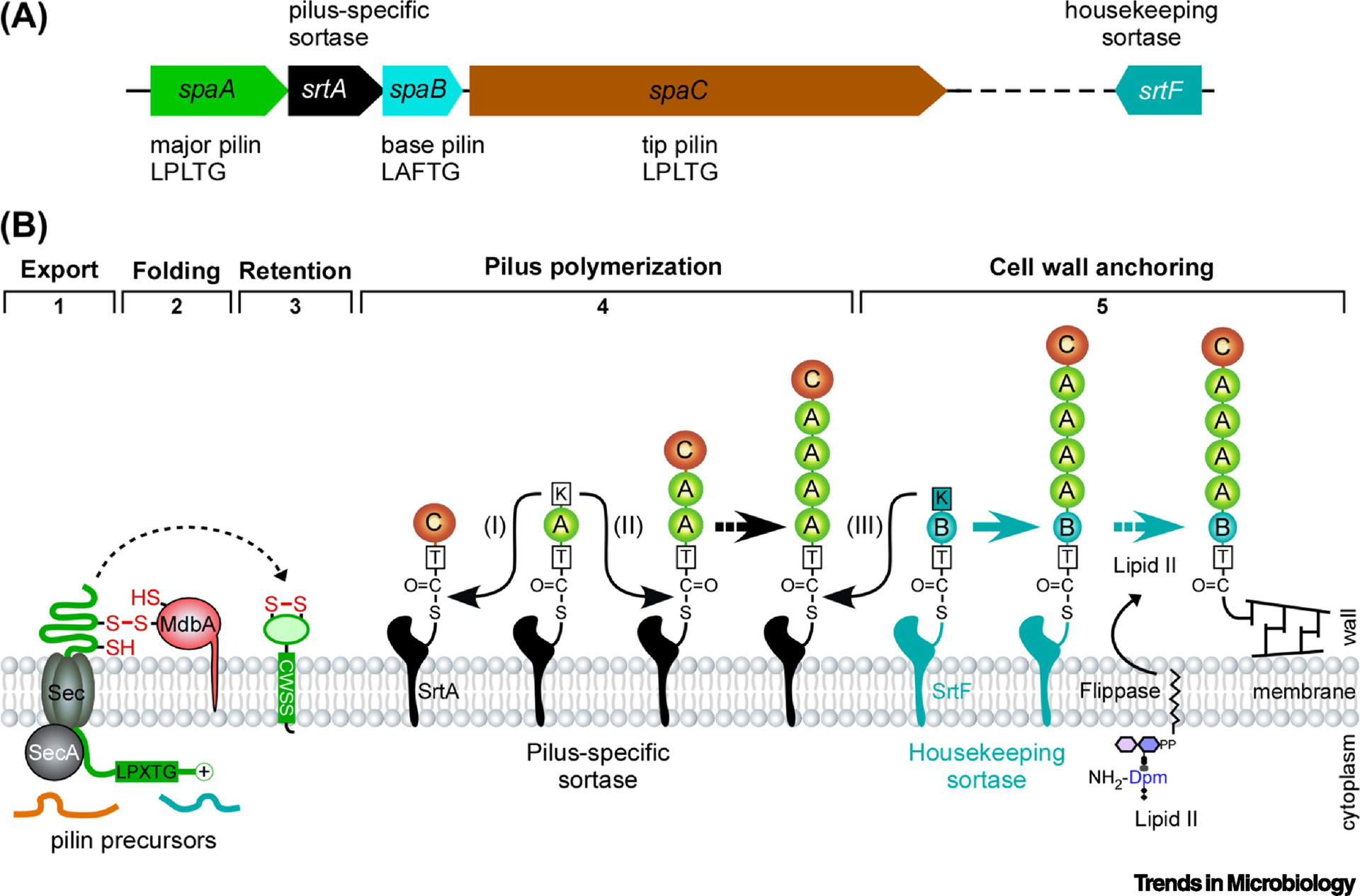
(A) Shown is the spaA pilus gene cluster in strain NCTC13129, encoding the pilus shaft SpaA, pilus base SpaB, and pilus tip SpaC, as well as the pilus-specific sortase SrtA. The SpaABC pilins contain a cell wall sorting signal with the LPLTG or LAFTG motif. The housekeeping sortase gene srtF is located elsewhere in the bacterial chromosome. (B) A the biphasic assembly mechanism of Gram-positive pili is depicted here with the SpaA-type pili. Pilin precursors are secreted in an unfolded state across the cytoplasmic membrane through the Sec translocon (Step 1). The membrane-bound thiol-disulfide oxidoreductase MdbA mediates disulfide bond formation and folding of pilin subunits (Step 2) prior to their insertion into the membrane (Step 3). The pilus-specific sortase SrtA catalyzed pilus polymerization into pilus fibers through successive lysine-transpeptidase reactions (Step 4). Incorporation of SpaB to the base of the pilus signals cell wall anchoring of the pilus by housekeeping sortase SrtF (Step 5). The order of the described transpeptidation reactions is marked by roman numerals (I-III); reproduced from Siegel et al [7].
Genetic and biochemical studies together have provided groundbreaking evidence to support the various steps of the aforementioned model (Fig. 1). Importantly, X-ray crystallographic studies of the SpaA pilin led to the discovery of an intramolecular disulfide bond and the role of a disulfide bridge-forming machine, termed MdbA, which is critically involved in mediating the post-translocational folding of the SpaA precursor pilin prior to its polymerization by pilus-specific sortase SrtA in the exoplasmic environment [26]. As expected from the model, deletion of srtA completely abolishes SpaA pilus polymerization [10], as does the genetic replacement of the nucleophilic lysine residue of the pilin motif (lysine to alanine substitution) that prevents pilus crosslinking [11]. The complete loss of SpaA pilus assembly in vivo in the absence of SrtA also demonstrates the substrate specificity of the pilus-specific sortase enzyme SrtA as other pilus-specific sortases expressed in vivo cannot substitute for SrtA function. When srtF or spaB is absent, however, abundant amounts of the generated pilus polymers are secreted into the culture medium, supporting the model that the pilus polymerization phase precedes the cell wall anchoring phase and that SpaB incorporation acts a pilus termination switch in this biphasic mode of pilus assembly [27, 31].
Recently, in vitro reconstitution of a Gram-positive pilus assembly system described for the first time that utilizes recombinant sortase enzyme and pilin substrate proteins has provided yet another foundational support for the biphasic model described above. This remarkable success in biochemical reconstitution was facilitated by structural genetic studies of the pilus-specific sortase SrtA, which uncovered novel structural features of the enzyme on the one hand, and also led to the engineering of a robust protein-polymerizing machine on the other hand. As we outlined in Box 1, the pilus-specific sortase SrtA harbors a structural lid that appears to occlude the enzyme’s catalytic pocket. Mutations of this lid could unmask the active site, thereby amplifying the net rounds of SpaA polymerization by this mutant enzyme (CdSrtA2M) to a level that was never observed with the wild-type enzyme, under comparable conditions [32].
The in vitro pilus assembly reaction contained the recombinant SrtA sortase truncated for its membrane localization domain and a SpaA protein devoid of its hydrophobic domain and charged tail [32], generating substantial amounts of pilus polymers within 24h that were easily detected by SDS-PAGE and Coomassie staining. Importantly, electron microscopic sampling revealed SpaA polymers with many >1 micron long fibers, while mass spectrometry authenticated the isopeptide linkage connecting individual subunits. Polymerization was abolished by a catalytic site mutation (C222A) as well as a pilin motif mutation (K190A); furthermore, the formation of an acyl-enzyme intermediate between SpaA and sortase, as the model predicted, was also observed. Remarkably, when SpaB or SrtF protein was added to the reaction, pilus polymerization was terminated, another prediction of the model [32]. The fact that SpaA polymers are formed without the presence of the tip pilin SpaC confirms the previous genetic observation that SpaC is dispensable for pilus assembly [10]. In essence, this test tube version of the reaction recapitulates much of the pilus assembly process that is observed in C. diphtheriae cells.
Protein ligation with pilus-specific sortase
The ability of a sortase to ligate proteins or peptides has significant implications in protein engineering, cell biology and biomedicine. Indeed, prior to the work with a pilus-specific sortase described above, Mao and coworkers have creatively utilized the most active recombinant sortase enzyme studied to date, i.e. S. aureus SrtA (SaSrtA), in protein ligation [33]. In this study, the recombinant staphylococcal SrtA (rSa-SrtA) is capable of joining one substrate protein that is C-terminally tagged with the LPXTG peptide with another substrate N-terminally tagged with the Gn peptide (n= 1–5). Ploegh and colleagues further advanced this method for protein labeling in living cells [34]. Dubbed “sortagging”, this method appears to be a promising new engineering tool, which has been further optimized for site-specificity and the ability to covalently-link peptides to a variety of non-peptide substrates, including folate, amino-terminated or glycine-tagged polyethylene glycol, and beads [33, 35]. Sortagging was also utilized to create peptide nucleic acid cell-penetrating peptide conjugants, thus providing an exciting new tool for designing highly specific cell-permeable drug therapies [36]. Importantly, the high affinity of sortase for the LPXTG motif has yielded yet another protein capture method in which LPXTG motif containing peptides can be efficiently and specifically captured from complex cell lysates [35, 37]. The ability of sortase to mediate the stable anchoring of proteins to surfaces for microarray based protein activity assays has also been explored as staphylococcal SrtA has been used to covalently attach LPXTG-containing proteins to planar surfaces, such as glass coverslips, following treatment of the surface with aminosilane and oligoglycine [37]. Sortase protein labelling technology is not limited to protein extract applications as staphylococcal SrtA was used to efficiently ligate a modified pentaglycine probe to the surface of the transmembrane protein CD40L in live HEK 293T cells [34]. Finally, Tanaka and colleagues successfully used sortagging as a method of protein specific fluorescent labelling by conjugated glycine-containing biotin, EGFP, and Alexa-fluor probes to the transmembrane protein osteoclast differentiation factor (ODF) in HEK 293T cells, without inducing any toxic phenotype to the cell culture [38].
The protein ligation reaction catalyzed by S. aureus SrtA is limited to the N-terminal to C-terminal protein joining involving two defined substrates. By comparison, a variety of pilus-specific sortase enzymes identified to date offer the unique advantage as a bioconjugation tool via the formation of an isopeptide bond that is mechanically stable and less susceptible to proteolytic cleavage [39, 40]. Recently, McConnell and colleagues generated a recombinant C. diphtheriae sortase enzyme termed CdSrtA3M that is more reactive than CdSrtA2M described above. When a substrate protein containing the pilin motif was incubated with green fluorescent protein harboring a C-terminal LPLTG motif in the presence of CdSrtA3M, the mutant enzyme catalyzed the covalent joining of the two recombinant proteins [41]. Furthermore, these authors demonstrated that both sortases SaSrtA and CdSrtA3M can be utilized in sequential transpeptidation reactions to modify a protein of interest at distinct sites and with high specificity. Using a Small Ubiquitin-like Modifier (SUMO) engineered to contain an N-terminal pentaglycine peptide and a C-terminal pilin motif, CdSrtA3M was first used to catalyze the addition of a FITC tag harboring the LPLTG motif to the lysine residue of the pilin motif. SaSrtA was then used to conjugate Alexa-fluor 546 harboring the LPATG motif to SUMO via pentaglycine [41]. Because of high degree of specificity for the ε-amine nucleophile within the pilin motif, protein ligation using pilus-specific sortase enzyme may provide selective labeling [41].
Actinomyces fimbriae – a paradigm of tissue tropism, hijacking of pilus machinery and spatial positioning of pili
The heterodimeric fimbriae of Actinomyces oris
Actinomyces are one of the most dominant and earliest colonizing genera of microbes present in the human oral cavity, with Actinomyces oris (formerly called Actinomyces naeslundii) detected in children as young as one year old [42, 43]. A. oris is a major contributor to dental plaque through its ability for coaggregation with other microbial species and thus a key to the genesis of complex biofilms on the surface of teeth and the mucosal epithelia [44–46]. This intrinsic adherence property of A. oris is largely attributed to the presence of two distinct fimbrial types, i.e. type 1 and type 2 fimbriae. A. oris has served as a pioneering model of tissue tropism mediated by Gram-positive pili as type 1 fimbriae mediate bacterial adherence to the salivary proline-rich proteins normally coating the tooth enamel [47], whereas type 2 fimbriae promote bacterial binding to receptor polysaccharides present on the surface of oral streptococci and various host cells [48–51]. Unlike C. diphtheriae and many other Gram-positive bacteria, A. oris fimbriae are heterodimeric, containing a tip component and another entity forming the pilus shaft. In the case of type 1 fimbriae, FimP forms the pilus shaft with FimQ located at the tip, and their assembly requires the pilus-specific sortase SrtC1, whose genes are all tightly linked together on the Actinomyces genome [52]. The specificity of Actinomyces sortases appears to be strict as the pilus-specific sortase SrtC2 is selectively required for formation of type 2 fimbriae only, which consist of the shaft FimA and tip FimB [53]. Since there are only two components in each fimbria, the last subunit of the shaft pilins should be the pilus base. This has raised an intriguing question of how pilus polymerization in Actinomyces or in any other two-component pilus systems, such as B. cereus [12] and Streptococcus suis [54], is terminated.
Using the type 2 fimbriae of A. oris as a prototype, the model of pilus assembly in Actinomyces is illustrated in Figure 2 [55]. Similar to what is described above for C. diphtheriae, pilin precursors translated in the cytoplasm are transported across the cytoplasmic membrane by the Sec translocon, and post-translocational protein folding is mediated by the disulfide bond machine MdbA, permitting membrane insertion of the pilin precursors [56]. When both tip and shaft pilins are available on the membrane, pilus polymerization is catalyzed by the pilus-specific sortase SrtC2 [57]. Finally, cell wall anchoring of the type 2 fimbriae is mediated by the housekeeping sortase SrtA, a class E sortase [58]. Surprisingly, unlike all known sortases studied to date, A. oris srtA is an essential gene. Genetic and biochemical studies to reveal the basis of srtA essentiality (Box 2) have serendipitously uncovered the molecular basis of regulated pilus polymerization and spatial positioning of pilus adhesins in Actinomyces that could not have been envisioned before (see below).
Figure 2: Assembly of the heterodimeric type 2 fimbrae in Actinomyces oris.
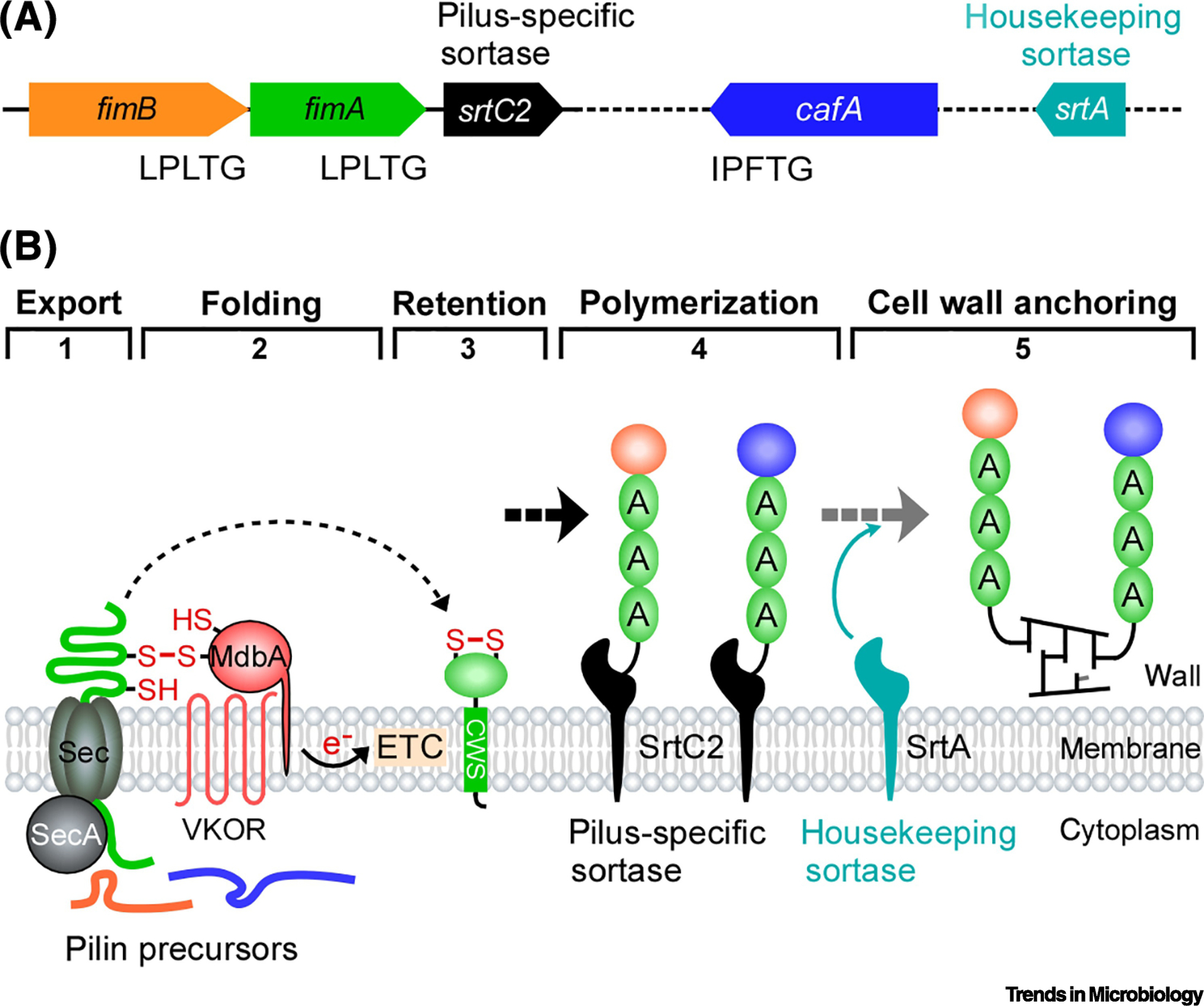
(A) The type 2 fimbriae are encoded by the three-gene operon. Genes encoding the coaggregation factor CafA and the housekeeping sortase SrtA are located elsewhere. (B) Similar to the assembly mechanism of the SpaA pili, assembly of the type 2 fimbriae begins with translocation (Step 1) and post-translocational folding of the shaft pilin FimA and tip pilin FimB mediated by the disulfide bond-forming machine MdbA/VKOR coupled to the electron transport chain (ETC) (Step 2). Membrane-insertion (Step 3) of these pilins permits pilus polymerization catalyzed by the pilus-specific sortase SrtC2 (Step 4), followed by cell wall anchoring of the pilus polymer catalyzed by the housekeeping sortase SrtA; reproduced from Sanchez et al. [55].
Box 2: Molecular basis of srtA essentiality in A. oris.
Unlike all other sortases studied to date, A. oris srtA is an essential gene as deletion of srtA has proven to be lethal [58]. A genetic suppressor screen – by Tn5 transposon mutagenesis – subsequently revealed that srtA essentiality is linked to the toxic accrual of a normally cell wall-anchored glycoprotein GspA, a SrtA substrate harboring a cell wall sorting signal with the LAXTG motif. In the absence of srtA, glycosylated GspA accumulates in the cytoplasmic membrane, causing lethal glyco-stress accompanied with expansion of the cell envelope and cell growth arrest [58]. A lcp mutant devoid of the glycosyltransferase LcpA and unable to glycosylate GspA [73] is one of 13 identified suppressor mutants, so is a gspA mutant lacking the cell wall sorting signal that permits membrane insertion prior to cell wall anchoring [58]. This illustrates the power of forward genetic analysis and the continued utility of isolating genetic suppressors in unveiling the intricacies of microbial genetic mechanisms.
The coaggregation factor CafA illustrates pilus hijacking in Gram-positive bacteria
The discovery that CafA is the coaggregation factor in A. oris has several significant implications, one that provides a paradigm of a surface protein hijacking the pilus assembly machine for pilus display and another a concept of spatial positioning of pilus adhesins for biological functions (see below). As mentioned above, type 2 fimbriae are essential for A. oris interactions, or coaggregation, with other oral bacteria, esp. oral streptococci as deletion of fimA abrogates coaggregation with Streptococcus oralis [57]. Surprisingly, A. oris coaggregation could not be blocked by polyclonal antibodies against FimA; nor the process was affected by a deletion of fimB, the gene that encodes the type 2 fimbrial tip pilin FimB [59]. Thus, a hunt was on for discovering the responsible adhesive principle that defied molecular genetic identification strategies taken so far. One potential scenario was that the FimA shaft contains some other protein to mediate co-aggregation. This prompted a systematic elimination of each individual LPXTG-containing surface proteins encoded in the Actinomyces genome. Indeed, among the fourteen candidate surface protein-encoding genes successfully deleted, one displayed a clear-cut co-aggregation defect on its own [59]. The implicated gene product, thus named CafA, was subsequently proved to be the long sought-after coaggregation factor by biochemical experiments: antibodies against CafA captured type 2 fimbriae and blocked bacterial coaggregation. Electron microscopic analyses revealed that CafA localizes at the pilus tip, forming a distinct pilus structure with shaft pilin FimA. Intriguingly, the CWSS sequence of CafA is strikingly similar to that of FimB, leading to the hypothesis that some Gram-positive surface proteins may hijack a pilus assembly machine via molecular mimicry to be displayed at the pilus tip [59]. As significant as this may be for advances in oral bacterial biology and therapeutic intervention, the broader implication of whether the general mechanism of pilus hijacking is more widespread or not still remains to be investigated.
Spatial positioning of pilus adhesins
Over a decade ago, it was speculated that Gram-positive adhesins appended at the pilus tip mediate the initial bacterial encounter with host cells due to the extended nature of pili [3]. Because pilus lengths vary greatly within individual pilus types as well as among various types of pili, it is important to know whether pilus-mediated adhesion processes depend on pilus length and whether and how pilus length is controlled in Gram-positive bacteria. A breakthrough on this problem came from the observation that A. oris mutants lacking the housekeeping sortase srtA produced exceedingly long pili, as might be expected, but surprisingly, they failed to adhere to oral streptococci [58]. This was puzzling since the coaggregation factor CafA was still abundantly detected at the tip of these long pili [60]. Through a series of probing experiments, Chang and colleagues demonstrated that as the pilus length was shortened by inducing expression of srtA, coaggregation could be restored, supporting the notion that the enzymatic activity of the housekeeping sortase SrtA is a key determinant of pilus length modulation. This new insight leads to the important question of whether SrtA activity is subject to regulation functionally or genetically. X-ray crystallization revealed that SrtA harbors two structural elements – a conserved tyrosine residue Y131 and a GVN tripeptide loop that may be of regulatory significance. Indeed, alanine-substitution of Y131 residue resulted in the production of shorter pili and defective coaggregation by A. oris, whereas mutations of the GVN loop led to assembly of extremely long pili and no coaggregation by the mutant bacteria [60], the phenotype similar to srtA depletion [58].
These results led to a mechanistic model that Y131 mutations alter the preference of the class E sortase SrtA, which normally recognizes the LAXTG motif, toward the LPXTG motif present in the FimA pilin subunits. As a result, pilus polymerization is terminated by SrtA-catalyzed cell wall anchoring of the FimA polymer, leading to the display of short pili on cell surface. Conversely, in the case of the GVN motif, its mutations diminished SrtA’s preference for the LAXTG motif, making SrtA’s capacity limited for cell wall anchoring. As a consequence, pilus polymerization continues unperturbed, leading to extremely long pili (Fig. 3). Consistent with this model, the deletion of gspA – which codes for one of the most abundant SrtA substrates with the LAXTG motif – resulted in normal assembly of FimA pili in the GVN mutation background and enabled positive-coaggregation by the mutant strain. Together, these structural genetic findings provide compelling grounds to posit that the housekeeping sortase functions as a molecular ruler for pilus polymerization and as such a positive effector of bacterial coaggregation and virulence.
Figure 3: A model of pilus length modulated by sortase enzymes in Actinomyces oris.
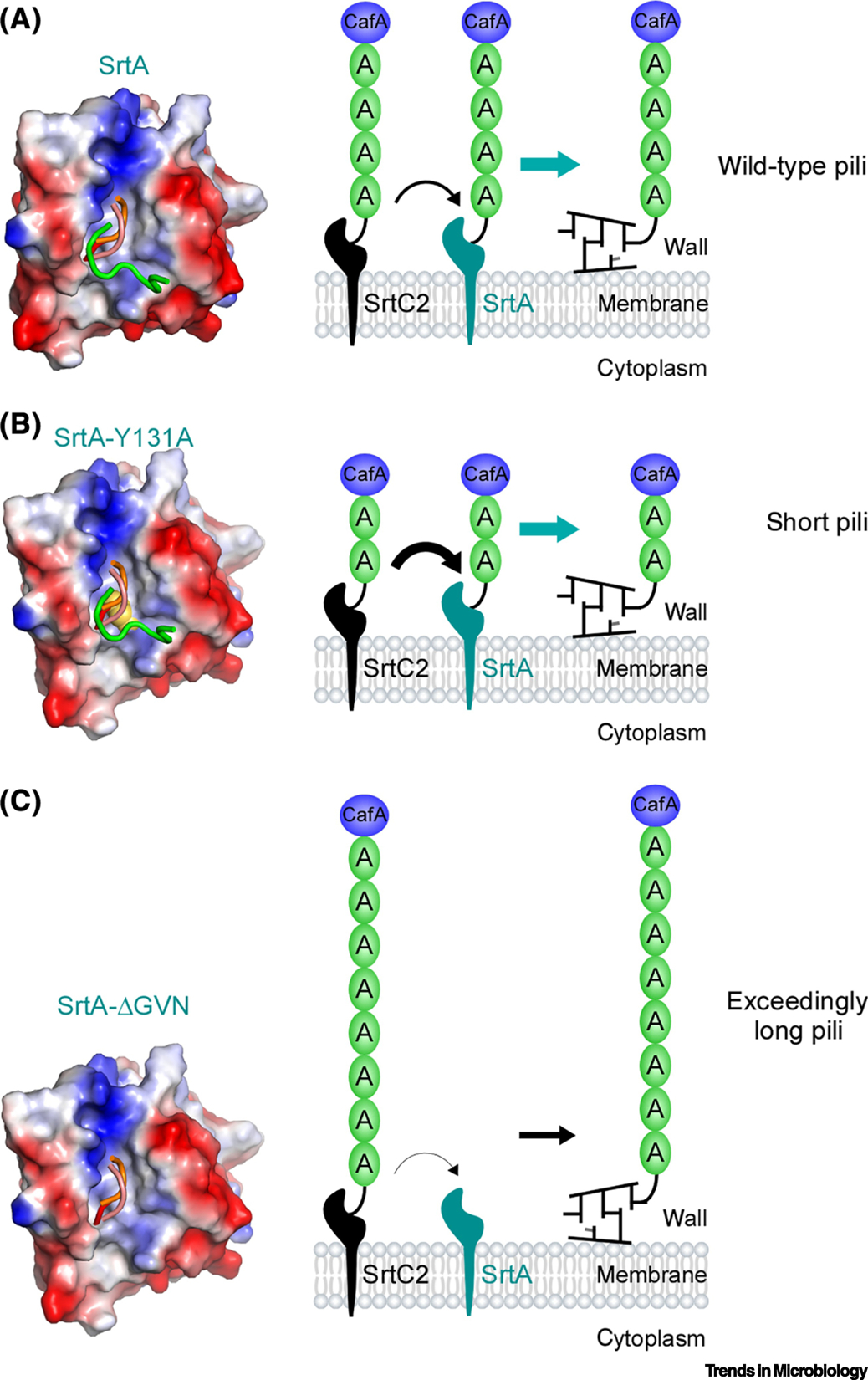
X-ray crystallization revealed the structural features of the housekeeping sortase SrtA: The tyrosine residue Y131 and the tripeptide loop GVN. (A) In the wild-type MG1, balanced activities of the pilus-specific sortase SrtC2 and the housekeeping sortase SrtA produce type 2 fimbriae with typical length. (B) Alanine-substitution of Y131 enhance SrtA sortase activity for the LPLTG motif of FimA, interfering with pilus polymerization, resulting in premature cell wall anchoring of short pili. (C) Mutation or deletion of the GVN loop reduces SrtA preference for the LPLTG motif of FimA, leading to continuous polymerization by SrtC2 and generating exceeding long fimbriae that can be anchored to the cell wall by this pilus-specific sortase enzyme; reproduced from Chang et al [60].
Concluding Remarks
Collective efforts during the last decade dissected the molecular assembly mechanisms of Gram-positive pili and probed their roles in bacterial pathogenesis as well as their utilization in the development of vaccines. While pilus vaccines have yet to emerge in the clinical arena, we now have made great strides in the basic biology and have a clearer view of pilus biogenesis in Gram-positive bacteria. A common feature in these monoderms is the biphasic mode of pilus assembly by distinct steps of enzymatic catalysis involving two sortases, whereby pilus polymerization catalyzed by pilus-specific sortase is followed by cell wall anchoring of pili promoted by the housekeeping sortase. Regardless of the sortase enzymes involved, the basic principle of these transpeptidation reactions in the polymerization phase is the same: The enzymatic cleavage of a substrate and covalent linkage of the cleaved substrate to a nucleophilic acceptor. This transpeptidation reaction generates an isopeptide bond that is mechanically strong and can resist a potential unfolding force up to 690 pN [39]. This unique property of isopeptide bonding, via lysine and threonine residues, is protease resistant and offers a versatile tool in protein engineering and bio-conjugation [41].
Where do we go from here? In spite of differences with their Gram-negative counterparts in the manner of assembly, the heteromeric pili of Gram-positive bacteria play significant roles in bacterial physiology and virulence as those of Gram-negative bacteria. Today, however, many fundamental questions regarding this still await to be addressed (Box 3). Given the importance of these questions, and the genetic and biochemical versatility of C. diphtheriae and A. oris as model organisms and their importance in significant human conditions, we believe these two systems will continue as fertile and attractive experimental models of pilus biogenesis in Gram-positive bacteria for some time to come.
Box 3: Outstanding Questions.
In the Actinobacteria C. diphtheriae and A. oris, the membrane-bound disulfide bond-forming machine MdbA promotes post-translocational folding of pilins. Given that no major protein folding machines are linked to pilus assembly in Firmicutes, how do these organisms solve the protein-folding problem in pilus assembly?
In a heterodimeric pilus system like A. oris, the last shaft pilin acts as pilus base and stop signal, and the housekeeping sortase appears to control pilus polymerization, hence pilus length. How does the housekeeping sortase indiscriminately recognize this base pilin from the rest? This raises an intriguing possibility that pilus polymerization and termination may depend on the stoichiometric availability of pilin substrates and sortase enzymes at the pilusosome. If so, does this require additional factors?
How are the tip pilins FimB and CafA in A. oris involved in pilus assembly? What mechanisms govern how a tip pilin is spatiotemporally incorporated only the tip of pili?
How do surface proteins with the LPXTG motif like CafA in A. oris hijack the sortase machine to be incorporated into the pilus tip?
Pili are found in the culture medium, esp. in late log-phase and stationary phase. What are the roles of secreted pili or are they products of cell wall turnover?
Highlights.
Covalently-linked pili are assembled on the cell surface of many Gram-positive bacteria via a biphasic mechanism, whereby pilus polymerization is catalyzed by the pilus-specific sortase followed by cell wall anchoring of pili by the housekeeping sortase.
Pilus-mediated adhesion depend on pilus length, which is modulated by the housekeeping sortase via unique structural features.
Some Gram-positive surface proteins with the LPXTG motif may hijack a pilus assembly machine via molecular mimicry to be displayed at the pilus tip.
Pilus-specific sortase enzymes provide a bioconjugation tool via the formation of an isopeptide bond that is mechanically stable and less susceptible to proteolytic cleavage.
ACKNOWLEDGEMENTS
We thank our laboratory members for critical review of the manuscript and discussion. Work in the Ton-That lab related to Gram-positive pili has been supported by the National Institute of Dental and Craniofacial Research (NIDCR)/NIH under the Award Numbers DE025015 and DE017382 (to H. T-T).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Kline KA et al. (2010) A tale of two pili: assembly and function of pili in bacteria. Trends Microbiol 18 (5), 224–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thanassi DG et al. (2012) Surface organelles assembled by secretion systems of Gram-negative bacteria: diversity in structure and function. FEMS Microbiol Rev 36 (6), 1046–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mandlik A et al. (2008) Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol 16 (1), 33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ton-That H and Schneewind O (2004) Assembly of pili in Gram-positive bacteria. Trends Microbiol 12 (5), 228–34. [DOI] [PubMed] [Google Scholar]
- 5.Spirig T et al. (2011) Sortase enzymes in Gram-positive bacteria. Mol Microbiol 82 (5), 1044–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dramsi S et al. (2005) Sorting sortases: a nomenclature proposal for the various sortases of Gram-positive bacteria. Res Microbiol 156 (3), 289–97. [DOI] [PubMed] [Google Scholar]
- 7.Siegel SD et al. (2016) Biogenesis of the Gram-positive bacterial cell envelope. Curr Opin Microbiol 34, 31–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeung MK et al. (1998) Identification of a gene involved in assembly of Actinomyces naeslundii T14V type 2 fimbriae. Infect Immun 66 (4), 1482–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yeung MK and Ragsdale PA (1997) Synthesis and function of Actinomyces naeslundii T14V type 1 fimbriae require the expression of additional fimbria-associated genes. Infect Immun 65 (7), 2629–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ton-That H and Schneewind O (2003) Assembly of pili on the surface of Corynebacterium diphtheriae. Mol Microbiol 50 (4), 1429–38. [DOI] [PubMed] [Google Scholar]
- 11.Ton-That H et al. (2004) Sortases and pilin elements involved in pilus assembly of Corynebacterium diphtheriae. Mol Microbiol 53 (1), 251–61. [DOI] [PubMed] [Google Scholar]
- 12.Budzik JM et al. (2007) Assembly of pili on the surface of Bacillus cereus vegetative cells. Mol Microbiol 66 (2), 495–510. [DOI] [PubMed] [Google Scholar]
- 13.Nallapareddy SR et al. (2006) Endocarditis and biofilm-associated pili of Enterococcus faecalis. J Clin Invest 116 (10), 2799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lauer P et al. (2005) Genome analysis reveals pili in Group B Streptococcus. Science 309 (5731), 105. [DOI] [PubMed] [Google Scholar]
- 15.Dramsi S et al. (2006) Assembly and role of pili in group B streptococci. Mol Microbiol 60 (6), 1401–13. [DOI] [PubMed] [Google Scholar]
- 16.Manetti AG et al. (2007) Streptococcus pyogenes pili promote pharyngeal cell adhesion and biofilm formation. Mol Microbiol 64 (4), 968–83. [DOI] [PubMed] [Google Scholar]
- 17.Falker S et al. (2008) Sortase-mediated assembly and surface topology of adhesive pneumococcal pili. Mol Microbiol 70 (3), 595–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turroni F et al. (2013) Role of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in modulating bacterium-host interactions. Proc Natl Acad Sci U S A 110 (27), 11151–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Telford JL et al. (2006) Pili in Gram-positive pathogens. Nat Rev Microbiol 4 (7), 509–19. [DOI] [PubMed] [Google Scholar]
- 20.Danne C and Dramsi S (2012) Pili of gram-positive bacteria: roles in host colonization. Res Microbiol 163 (9–10), 645–58. [DOI] [PubMed] [Google Scholar]
- 21.Hendrickx AP et al. (2011) Architects at the bacterial surface - sortases and the assembly of pili with isopeptide bonds. Nat Rev Microbiol 9 (3), 166–76. [DOI] [PubMed] [Google Scholar]
- 22.Rogers EA et al. (2011) Adhesion by pathogenic corynebacteria. Adv Exp Med Biol 715, 91–103. [DOI] [PubMed] [Google Scholar]
- 23.Yanagawa R and Honda E (1976) Presence of pili in species of human and animal parasites and pathogens of the genus corynebacterium. Infect Immun 13 (4), 1293–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaspar AH and Ton-That H (2006) Assembly of distinct pilus structures on the surface of Corynebacterium diphtheriae. J Bacteriol 188 (4), 1526–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swierczynski A and Ton-That H (2006) Type III pilus of corynebacteria: Pilus length is determined by the level of its major pilin subunit. J Bacteriol 188 (17), 6318–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reardon-Robinson ME et al. (2015) A thiol-disulfide oxidoreductase of the Gram-positive pathogen Corynebacterium diphtheriae is essential for viability, pilus assembly, toxin production and virulence. Mol Microbiol 98 (6), 1037–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swaminathan A et al. (2007) Housekeeping sortase facilitates the cell wall anchoring of pilus polymers in Corynebacterium diphtheriae. Mol Microbiol 66 (4), 961–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nobbs AH et al. (2008) Sortase A utilizes an ancillary protein anchor for efficient cell wall anchoring of pili in Streptococcus agalactiae. Infect Immun 76 (8), 3550–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nielsen HV et al. (2013) Pilin and sortase residues critical for endocarditis- and biofilm-associated pilus biogenesis in Enterococcus faecalis. J Bacteriol 195 (19), 4484–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sillanpaa J et al. (2013) Contribution of individual Ebp Pilus subunits of Enterococcus faecalis OG1RF to pilus biogenesis, biofilm formation and urinary tract infection. PLoS One 8 (7), e68813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mandlik A et al. (2008) The molecular switch that activates the cell wall anchoring step of pilus assembly in gram-positive bacteria. Proc Natl Acad Sci U S A 105 (37), 14147–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang C et al. (2018) In vitro reconstitution of sortase-catalyzed pilus polymerization reveals structural elements involved in pilin cross-linking. Proc Natl Acad Sci U S A 115 (24), E5477–E5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mao H et al. (2004) Sortase-mediated protein ligation: a new method for protein engineering. J Am Chem Soc 126 (9), 2670–1. [DOI] [PubMed] [Google Scholar]
- 34.Popp MW et al. (2007) Sortagging: a versatile method for protein labeling. Nat Chem Biol 3 (11), 707–8. [DOI] [PubMed] [Google Scholar]
- 35.Parthasarathy R et al. (2007) Sortase A as a novel molecular “stapler” for sequence-specific protein conjugation. Bioconjug Chem 18 (2), 469–76. [DOI] [PubMed] [Google Scholar]
- 36.Pritz S et al. (2007) Synthesis of biologically active peptide nucleic acid-peptide conjugates by sortase-mediated ligation. J Org Chem 72 (10), 3909–12. [DOI] [PubMed] [Google Scholar]
- 37.Chan L et al. (2007) Covalent attachment of proteins to solid supports and surfaces via Sortase-mediated ligation. PLoS One 2 (11), e1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanaka T et al. (2008) Site-specific protein modification on living cells catalyzed by Sortase. Chembiochem 9 (5), 802–7. [DOI] [PubMed] [Google Scholar]
- 39.Echelman DJ et al. (2016) CnaA domains in bacterial pili are efficient dissipaters of large mechanical shocks. Proc Natl Acad Sci U S A 113 (9), 2490–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mishra A et al. (2011) Two autonomous structural modules in the fimbrial shaft adhesin FimA mediate Actinomyces interactions with streptococci and host cells during oral biofilm development. Mol Microbiol 81 (5), 1205–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McConnell SA et al. (2018) Protein Labeling via a Specific Lysine-Isopeptide Bond Using the Pilin Polymerizing Sortase from Corynebacterium diphtheriae. J Am Chem Soc 140 (27), 8420–8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Papaioannou W et al. (2009) The microbiota on different oral surfaces in healthy children. Oral Microbiol Immunol 24 (3), 183–9. [DOI] [PubMed] [Google Scholar]
- 43.Sarkonen N et al. (2000) Oral colonization with Actinomyces species in infants by two years of age. J Dent Res 79 (3), 864–7. [DOI] [PubMed] [Google Scholar]
- 44.Cisar JO et al. (1979) Specificity of coaggregation reactions between human oral streptococci and strains of Actinomyces viscosus or Actinomyces naeslundii. Infect Immun 24 (3), 742–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skopek RJ et al. (1993) Dental plaque development on defined streptococcal surfaces. Oral Microbiol Immunol 8 (1), 16–23. [DOI] [PubMed] [Google Scholar]
- 46.Zijnge V et al. (2010) Oral biofilm architecture on natural teeth. PLoS One 5 (2), e9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gibbons RJ et al. (1988) Adsorbed salivary proline-rich protein 1 and statherin: receptors for type 1 fimbriae of Actinomyces viscosus T14V-J1 on apatitic surfaces. Infect Immun 56 (11), 2990–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McIntire FC et al. (1978) Mechanism of coaggregation between Actinomyces viscosus T14V and Streptococcus sanguis 34. Infect Immun 21 (3), 978–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stromberg N and Karlsson KA (1990) Characterization of the binding of Actinomyces naeslundii (ATCC 12104) and Actinomyces viscosus (ATCC 19246) to glycosphingolipids, using a solid-phase overlay approach. J Biol Chem 265 (19), 11251–8. [PubMed] [Google Scholar]
- 50.Ruhl S et al. (2000) Identification of polymorphonuclear leukocyte and HL-60 cell receptors for adhesins of Streptococcus gordonii and Actinomyces naeslundii. Infect Immun 68 (11), 6346–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruhl S et al. (1996) Recognition of immunoglobulin A1 by oral actinomyces and streptococcal lectins. Infect Immun 64 (12), 5421–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mishra A et al. (2007) Sortase-catalyzed assembly of distinct heteromeric fimbriae in Actinomyces naeslundii. J Bacteriol 189 (8), 3156–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu C et al. (2012) Structural determinants of Actinomyces sortase SrtC2 required for membrane localization and assembly of type 2 fimbriae for interbacterial coaggregation and oral biofilm formation. J Bacteriol 194 (10), 2531–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Okura M et al. (2011) The Minor Pilin Subunit Sgp2 Is Necessary for Assembly of the Pilus Encoded by the srtG Cluster of Streptococcus suis. J Bacteriol 193 (4), 822–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sanchez BC et al. (2017) Electron Transport Chain Is Biochemically Linked to Pilus Assembly Required for Polymicrobial Interactions and Biofilm Formation in the Gram-Positive Actinobacterium Actinomyces oris. mBio 8 (3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reardon-Robinson ME et al. (2015) A Disulfide Bond-forming Machine Is Linked to the Sortase-mediated Pilus Assembly Pathway in the Gram-positive Bacterium Actinomyces oris. J Biol Chem 290 (35), 21393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mishra A et al. (2010) The Actinomyces oris type 2 fimbrial shaft FimA mediates co-aggregation with oral streptococci, adherence to red blood cells and biofilm development. Mol Microbiol 77 (4), 841–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu C et al. (2014) Lethality of sortase depletion in Actinomyces oris caused by excessive membrane accumulation of a surface glycoprotein. Mol Microbiol 94 (6), 1227–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reardon-Robinson ME et al. (2014) Pilus hijacking by a bacterial coaggregation factor critical for oral biofilm development. Proc Natl Acad Sci U S A 111 (10), 3835–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chang C et al. (2019) Cell-to-cell interaction requires optimal positioning of a pilus tip adhesin modulated by gram-positive transpeptidase enzymes. Proc Natl Acad Sci U S A 116 (36), 18041–18049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mazmanian SK et al. (1999) Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285 (5428), 760–3. [DOI] [PubMed] [Google Scholar]
- 62.Navarre WW and Schneewind O (1999) Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol Mol Biol Rev 63 (1), 174–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ton-That H et al. (1999) Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc Natl Acad Sci U S A 96 (22), 12424–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ilangovan U et al. (2001) Structure of sortase, the transpeptidase that anchors proteins to the cell wall of Staphylococcus aureus. Proc Natl Acad Sci U S A 98 (11), 6056–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ton-That H et al. (2002) Anchoring of surface proteins to the cell wall of Staphylococcus aureus. Cysteine 184 and histidine 120 of sortase form a thiolateimidazolium ion pair for catalysis. J Biol Chem 277 (9), 7447–52. [DOI] [PubMed] [Google Scholar]
- 66.Khare B and S VLN (2017) Pilus biogenesis of Gram-positive bacteria: Roles of sortases and implications for assembly. Protein Sci 26 (8), 1458–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Khare B et al. (2011) The crystal structure analysis of group B Streptococcus sortase C1: a model for the “lid” movement upon substrate binding. J Mol Biol 414 (4), 563–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Khare B et al. (2011) Structural differences between the Streptococcus agalactiae housekeeping and pilus-specific sortases: SrtA and SrtC1. PLoS One 6 (8), e22995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Persson K (2011) Structure of the sortase AcSrtC-1 from Actinomyces oris. Acta Crystallographica Section D Biological Crystallography 67, 212–217. [DOI] [PubMed] [Google Scholar]
- 70.Manzano C et al. (2009) Sortase activity is controlled by a flexible lid in the pilus biogenesis mechanism of Gram-positive pathogens. Biochemistry 48, 10549–10557. [DOI] [PubMed] [Google Scholar]
- 71.Comfort D and Clubb RT (2004) A comparative genome analysis identifies distinct sorting pathways in gram-positive bacteria. Infect Immun 72 (5), 2710–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kattke MD et al. (2016) Crystal Structure of the Streptomyces coelicolor Sortase E1 Transpeptidase Provides Insight into the Binding Mode of the Novel Class E Sorting Signal. PLoS One 11 (12), e0167763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Siegel SD et al. (2019) Structure and Mechanism of LcpA, a Phosphotransferase That Mediates Glycosylation of a Gram-Positive Bacterial Cell Wall-Anchored Protein. mBio 10 (1), pii: e01580–18. [DOI] [PMC free article] [PubMed] [Google Scholar]


