SUMMARY
The dysregulation of the metabolic regulator TOR complex I (TORC1) contributes to a wide array of human pathologies. Tuberous sclerosis complex (TSC) is a potent inhibitor of TORC1. Here we demonstrate that the Rag GTPase acts in both the amino acid sensing and growth factor signaling pathways to control TORC1 activity through the regulation of TSC dynamics in HeLa cells and Drosophila. We find that TSC lysosomal/cytosolic exchange increases in response to both amino acid and growth factor restriction. Moreover, the rate of exchange mirrors TSC function, with depletions of the Rag GTPase blocking TSC lysosomal mobility and rescuing TORC1 activity. Finally, we show that the GATOR2 complex controls the phosphorylation of TSC2, which is essential for TSC exchange. Our data support the model that the amino acid and growth factor signaling pathways converge on the Rag GTPase to inhibit TORC1 activity through the regulation of TSC dynamics.
Keywords: TSC, TORC1, Rag GTPase, GATOR2, AKT1, lysosome, amino acid signaling, growth factor signaling, Drosophila, HeLa cell
eTOC Blurb
The metabolic regulator TORC1 responds to numerous upstream signals. Yang et al. report a unified model of TORC1 regulation in which the Rag GTPase integrates information from the amino acid and growth factor signaling pathways to control the activation of the TORC1 inhibitor TSC by regulating its lysosomal/cytosolic exchange rate.
INTRODUCTION
Cells must sense and adapt to a constantly changing external environment. Accordingly, in both single celled and multicellular organisms the ability to switch between anabolism, which supports growth, and catabolism which is activated in response to stress, is critical to the maintenance of metabolic homeostasis. The highly conserved Target Of Rapamycin (TOR) Complex 1 (TORC1) is central to the regulation of metabolism in eukaryotes (Dibble and Manning, 2013; Kim and Guan, 2019; Sabatini, 2017). TORC1 serves as a hub to integrate multiple upstream signaling inputs and regulates the execution of downstream metabolic pathways which control growth, proliferation and cell death (Laplante and Sabatini, 2012; Saxton and Sabatini, 2017). TORC1 consists of five subunits including mTOR, which functions as a serine threonine kinase, as well as Deptor, Raptor, mLST8 and PRAS40 (Aylett et al., 2016). In the presence of positive upstream inputs, TORC1 promotes anabolic metabolism by phosphorylating downstream effectors such as 4E-BP1 and S6K to increase mRNA translation and protein synthesis (Hara et al., 1998; Schalm et al., 2003). Concurrently, TORC1 inhibits catabolic pathways such as autophagy, by phosphorylating essential autophagic proteins including ULK1 and Atg13 (Ganley et al., 2009; Hosokawa et al., 2009). Thus, through the regulation of mTORC1 activity, cells can rapidly react to a diverse array of positive and negative environmental cues.
An important aspect of TORC1 regulation is its recruitment to and activation on lysosomes. These two processes require different Ras-related small GTPases, Rag and Rheb respectively. In the presence of amino acids, TORC1 is translocated from the cytosol to lysosomal membrane by the Rags (Sabatini, 2017; Sancak et al., 2010; Yang et al., 2017). Specifically, RagA or RagB dimerizes with RagC or RagD to form a heterodimeric complex that recruits TORC1 to lysosome by directly interacting with the Raptor subunit in an amino aciddependent manner (Kim et al., 2008; Sancak et al., 2008). Once on the lysosome, Rheb binds to the subunit mTOR resulting in a conformational change that exposes active site resides allowing TORC1 to bind and phosphorylate a wide array of substrates (Yang et al., 2017).
As small GTPases, the functions of Rags and Rheb are regulated by their guanine nucleotide binding cycle, which is tightly controlled by upstream signals (Avruch et al., 2006; Inoki et al., 2003; Lee et al., 2018; Nicastro et al., 2017; Roccio et al., 2006; Shen and Sabatini, 2018; Shen et al., 2019). The Rags switch their TORC1-recruiting function by changing their guanine nucleotide binding status. When cells lack access to appropriate levels of amino acids, the GTPase-activating protein toward Rags 1 (GATOR1) complex blocks the function of TORC1 by acting as a GTPase-activating protein (GAP) toward RagA/B (Bar-Peled et al., 2013; Panchaud et al., 2013a). Thus, under amino acid starvation the guanine nucleotide binding status of RagA/B changes to GDP inhibiting the ability of the Rag heterodimer to recruit TORC1 onto lysosomes (Bar-Peled et al., 2013). Another GATOR subcomplex GATOR2, functions to oppose GATOR1 activity and is composed of five highly conserved proteins (Mio, Wdr24, Wdr59, Seh1 and Sec13) several of which have been shown to localize to lysosomes (Cai et al., 2016; Dokudovskaya and Rout, 2011, 2015; Wei et al., 2014). In the presence of sufficient amino acids, the GATOR2 complex activates TORC1 activity by opposing the activity of the GATOR1 complex (Bar-Peled et al., 2013; Panchaud et al., 2013b; Wei et al., 2014). The mechanistic details of how the GATOR2 complex opposes the activity of the GATOR1 complex to activate TORC1 remain unclear.
The small GTPase Rheb is inhibited by a three subunit complex, called the Tuberous Sclerosis Complex (TSC), comprised of the proteins TSC2, TSC1 and TBC1D7 (Dibble and Manning, 2013). In the absence of insulin, TSC2 possesses a GAP activity toward Rheb (Inoki et al., 2003). TSC serves as a potent inhibitor of the TORC1 signaling pathway. Mutations or deletions in TSC subunits result in the growth of benign tumors, epilepsy and developmental delay (Curatolo et al., 2008). The upstream signals controlling TSC have been extensively studied (Manning and Toker, 2017). Essentially, growth factors including insulin, activate class I phosphatidylinositol-3-kinase (PI3K), which in turn stimulates the protein kinase AKT. AKT inhibits TSC by directly phosphorylating multiple serine residues in TSC2 (Inoki et al., 2002; Manning et al., 2002). Thus, growth factors and insulin positively regulate the TORC1 signaling pathway by preventing TSC from inhibiting the TORC1 activator Rheb.
While the model that TSC controls TORC1 activity through the inhibition of Rheb is well established, the detailed mechanism of how TSC is regulated remains poorly understood. It is widely accepted that TSC responds to insulin and other growth factors through a PI3K-AKT circuit (Cai et al., 2006; Inoki et al., 2002). However, the role of TSC in response to changes in amino acid levels is unclear. One possibility is that TORC1 is regulated by two independent pathways; the PI3K-AKT pathway which regulates the activity of the TORC1 inhibitor TSC in response to growth factors and the Rag GTPase pathway that responds to amino acid levels (Bar-Peled and Sabatini, 2014; Lawrence and Zoncu, 2019). However, multiple laboratories have reported an essential role for TSC in the response to amino acid starvation (Avruch et al., 2006; Demetriades et al., 2014; Gao et al., 2002; Smith et al., 2005; Wei et al., 2014). Additionally, it is unclear exactly how intracellular TSC dynamics impact the ability of TSC to inhibit TORC1. Several studies have shown that TSC shuttles between lysosomes and the cytoplasm in response to amino acid availability (Carroll et al., 2016; Demetriades et al., 2014), whereas others have reported that the lysosomal localization of TSC is independent of amino acid status with the TSC complex being released from lysosomes in response to high levels of insulin (Menon et al., 2014). Notably, in some cell types, such as HeLa cells, TSC is constitutively present on lysosomes even in the presence of physiological concentrations of insulin (Demetriades et al., 2016; Menon et al., 2014). Thus, whether the recruitment of TSC from the cytosol to the lysosomes is a common feature of TSC regulation in all cell types remains unclear. Finally, the detailed mechanism of how the phosphorylation of TSC2 by AKT affects TSC’s function is not well understood. Several studies proposed that TSC2 undergoes proteasomal degradation after phosphorylation by AKT (Cai et al., 2006; Dan et al., 2002; Plas and Thompson, 2003). However, more recent work suggests that TSC is not regulated at the level of protein stability but that the entire TSC complex is released from lysosomes upon phosphorylation of the TSC2 subunit by AKT (Menon et al., 2014). Thus, a full understanding of TSC regulation requires further exploration.
Here we demonstrate that the regulation of TSC lysosomal dynamics by the Rag GTPase is required for the full response to both amino acid and growth factor restriction. Moreover, using Fluorescence Recovery After Photobleaching (FRAP) and a photoconvertible fluorescently tagged TSC2 we demonstrate that in response to negative stimuli, the Rag GTPase drives the rapid cycling of TSC on and off lysosomes in both HeLa cells and Drosophila. Importantly, we find that GATOR2 and the Rag GTPase impact the lysosomal cycling of TSC2 by regulating its inhibitory phosphorylation by AKT. Our data support the model that the Rag GTPase works in concert with the PI3/AKT/TSC pathway to regulate TORC1 activity in response to both amino acid and growth factor restriction.
RESULTS
Knockdowns of the Rag GTPase increase TORC1 activity in GATOR2 mutant ovaries and HeLa cells
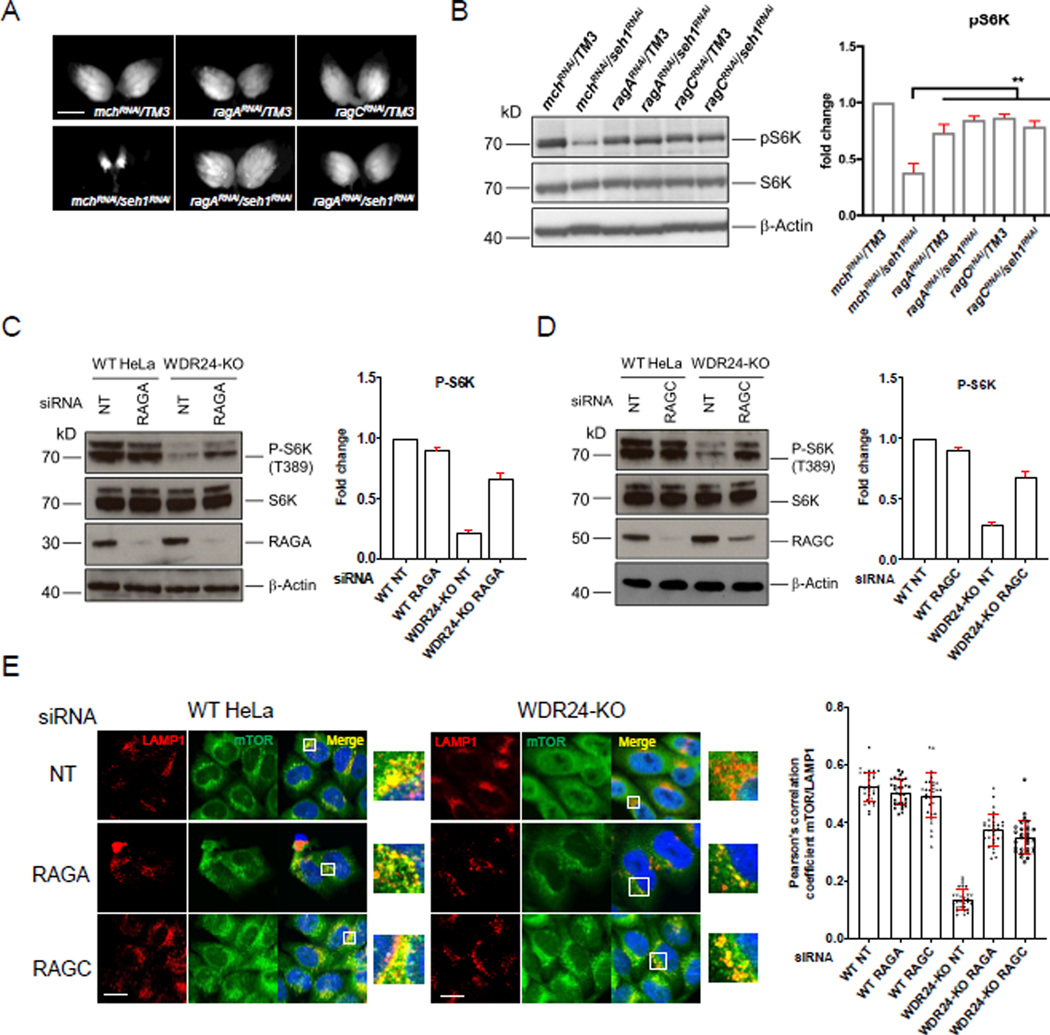
In order to identify additional genes that function in the GATOR pathway we performed a suppression screen in Drosophila. In null mutants of the GATOR2 component seh1, low TORC1 activity in the female germline results in a block to oocyte growth and development (Senger et al., 2011; Wei et al., 2014). Expressing a short hairpin RNA specifically against the germline seh1 transcript, recapitulates the seh1 mutant ovarian phenotype (Figure 1A). We previously demonstrated co-depleting the GATOR1 components nprl2, nprl3, and iml1 dramatically rescues the seh1RNAi ovarian phenotype, consistent with the model that a primary function of the GATOR2 component seh1 is to promote TORC1 activity through the inhibition of GATOR1 activity (Bar-Peled et al., 2013; Panchaud et al., 2013b; Wei et al., 2014). Using this epistatic relationship, we performed an RNAi based screen to identify additional genes that when co-depleted with seh1, rescued the seh1 ovarian phenotype. We predicted this screen would identify additional genes that act with GATOR1 to inhibit TORC1 activity. Surprisingly, we found that co-depleting the components of Rag GTPase, RagA and RagC, robustly rescued the seh1 ovarian growth deficit (Figure 1A). Moreover, the rescue of the seh1 ovarian growth phenotype was accompanied by a rescue of TORC1 activity (Figure 1B).
Figure 1. The Rag GTPase inhibits TORC1 in GATOR2 mutant Drosophila and HeLa cells.
(A) Co-depletions of ragA or ragC rescue the small ovary phenotype of seh1RNAi females. ragARNAi, ragCRNAi and seh1RNAi TRiP constructs were driven by the germline specific driver nanos-GAL4VP16. mchRNAi: mCherry-RNAi. Scale bar: 1 mm.
(B) Knockdowns of ragA or ragC increase pS6K levels in seh1RNAi ovaries, error bars in pS6K quantification graph represent standard deviation. Unpaired Student’s t-test was used to calculate p values. mchRNAi: mCherry-RNAi. **: p < 0.01.
(C) Immunoblot for P-S6K (T389) in WT and WDR24-KO HeLa cells treated with control (NT) or RAGA siRNAs. Error bars represent standard deviation.
(D) Immunoblot for P-S6K (T389) in WT and WDR24-KO HeLa cells treated with control (NT) or RAGC siRNAs.
(E) mTOR and LAMP1 immunostaining and colocalization analysis in WT and WDR24-KO HeLa cells treated with control (NT), RAGA or RAGC siRNA. Scale bar: 10 μm. The colocalization coefficient was calculated based on 30 cells per condition. Error bars represent standard deviation. See also Figure S1.
The results from our Drosophila RNAi screen indicated that the RagA/RagC heterodimer acts to inhibit TORC1 activity in seh1 mutant ovaries. To obtain a mechanistic understanding of this observation, we turned to a mammalian cell culture system. We previously determined that HeLa cell knockouts for the GATOR2 component WDR24 have decreased TORC1 activity relative to WT HeLa cells (Cai et al., 2016). Consequently, we utilized a WDR24 HeLa cell knockout (WDR24-KO) line to examine how depleting RAGA and RAGC impacts TORC1 activity when the GATOR2 complex is compromised (Cai et al., 2016). First, we explored how siRNA knockdowns of RAGA or RAGC affects TORC1 activity in WT and WDR24-KO cells by examining the phosphorylation status of the TORC1 downstream target S6K. Consistent with our results in the Drosophila ovary, siRNA knockdowns of RAGA (RAGA-KD) (Figure 1C) or RAGC (RAGC-KD) (Figure 1D) restored TORC1 activity in WDR24-KO, WDR59-KO and MIOS-KO cells as indicated by increased p-S6K levels (Figures 1C, D and S1). Interestingly, RAGA-KD or RAGC-KD in WT HeLa cells under fed conditions, had little effect on p-S6K levels. These data indicate that consistent with previous work, the Rags are dispensable for maintaining TORC1 activity under conditions of metabolic homeostasis (Demetriades et al., 2014; Jewell et al., 2015).
One explanation for how depleting the Rag GTPase might restore TORC1 activity in WDR24-KO cells is that knockdowns of the Rag GTPases allow for the return and activation of TORC1 on lysosomes in the WDR24-KO background. To test this hypothesis, we examined the localization of the TORC1 subunit mTOR and the lysosomal marker LAMP1 by immunofluorescence staining. We observed that under fed conditions, mTOR is largely colocalized with LAMP1 in WT HeLa cells while in WDR24-KO cells mTOR is diffusely localized in the cytosol (Bar-Peled et al., 2013) (Figure 1E). Importantly, knockdowns of either RAGA or RAGC in WDR24-KO cells significantly increased the colocalization between mTOR and LAMP1 (Figure 1E), indicating that TORC1 is recruited back to lysosomes in WDR24-KO cells in the absence of Rag GTPase. In contrast, consistent with the minimal impact on TORC1 activity, knockdowns of RAGA or RAGC had only a minor effect on the lysosomal localization of TORC1 in WT HeLa cells under fed condition. In summary, our data support the idea that in WDR24-KO HeLa cells, RAGA and RAGC act as inhibitors of TORC1 by preventing its activation on lysosomes.
The GATOR2 complex inhibits the dynamic recruitment of TSC to lysosomes
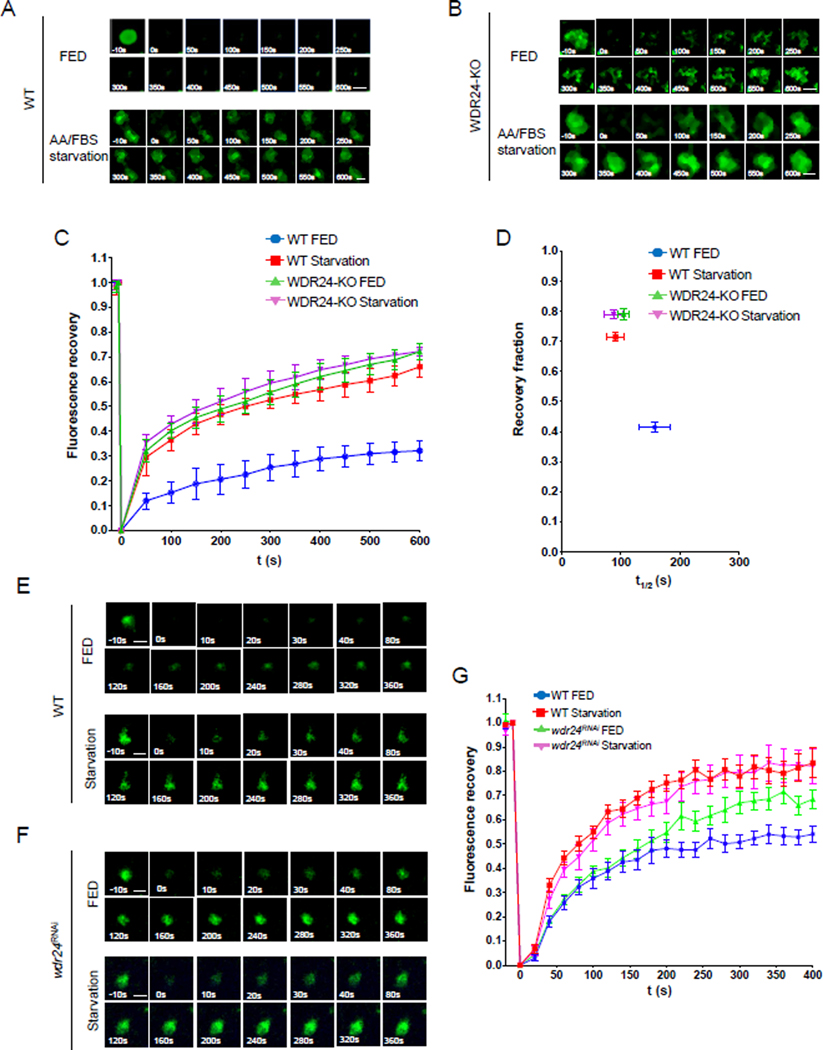
How might the Rag GTPase function as an inhibitor of TORC1 in the GATOR2 mutant background? In HeLa cells, deletions of GATOR2 components result in a dramatic decrease in TORC1 activity, presumably due to de-inhibition of the GATOR1 complex and conversion of the Rag GTPase into its “inactive” RAGAGDP form (Bar-Peled et al., 2013; Cai et al., 2016). Previous studies demonstrated that when amino acids are limited, the Rag GTPase, in its RAGAGDP: RAGCGTP form, recruits the TORC1 inhibitor TSC to lysosomal membranes (Demetriades et al., 2014). Thus, one possible explanation for our results, is that in the absence of the GATOR2 complex, knocking down components of the Rag GTPase prevents the recruitment of TSC to lysosomes, thus allowing TORC1 to bind directly to its activator Rheb (Demetriades et al., 2014). Whereas TSC shuttles on and off lysosomes in response to multiple stimuli in several cell lines, in HeLa cells, lysosomes retain a pool of TSC under conditions that favor growth (Figure S2) (Demetriades et al., 2016; Menon et al., 2014); However, analysis of steady-state distributions of proteins rarely inform whether they are static or dynamic. Therefore, we used an alternative technique to examine the regulation and functional importance of TSC lysosomal dynamics in the regulation of TORC1 activity in HeLa cells and Drosophila. Because of the known role of TSC in the regulation of TORC1 activity in response to multiple upstream inputs, we reasoned that the lysosomal pool of TSC in HeLa cells may mask changes in the rate of recruitment of TSC to lysosomes in response to upstream inputs (Demetriades et al., 2016). To test this hypothesis, we used the Fluorescence Recovery After Photobleaching (FRAP) (Lippincott-Schwartz et al., 2018) technique to examine the dynamic behavior of the TSC subunit TSC2 in various nutrient conditions and genetic backgrounds.
We tagged TSC2 with a Halo tag and labeled it with the fluorescence dye Halo Ligand-Oregon Green. First, we transiently expressed the Halo tagged TSC2 in HeLa cells. We confirmed that the Halo-TSC2 protein localizes to lysosomes in both WT and WDR24-KO cells using the lysosomal marker LAMP1 (Figure S3). To eliminate the effect of microtubule based lysosomal movement, we treated cells with nocodazole as suggested by a previous study (Lawrence et al., 2018). We then photobleached a single Halo-TSC2 labeled lysosome and used lysosomal FRAP to determine the rate of recruitment of TSC2 to lysosomes in WT versus WDR24-KO cells under fed versus starved conditions. First, we examined if nutrient conditions control TSC2 dynamics in WT HeLa cells (Figures 2A, C and D). In nutrient replete (+FBS, +AA) conditions photobleached lysosomes reached 40% of their original fluorescence fraction with a 150s half-time (t1/2) (Figure 2D). The partial recovery of Halo-TSC2 indicates that there is also a significant immobile fraction of TSC present on lysosomes under conditions that favor growth. Under nutrient-depleted conditions TSC2 had a higher exchange rate as indicated by the decreased half-time (around 100s). These results indicated that the rate of recruitment of TSC2 to lysosome dramatically increases upon nutrient limitation. Next, we used FRAP to examine the mobility of TSC2 in WDR24-KO HeLa cells (Figure 2B). We predicted that WDR24-KO cells cultured under fed conditions (+FBS, +AA) would have TSC2 dynamics similar to those observed in starved (−FBS, −AA) cells. Consistent with this prediction, TSC2 recovered to approximately 80% of pre-bleach fluorescence fraction with a half-time around 100s in both fed and starved conditions (Figure 2D). Thus, we conclude that in HeLa cells the exchange of TSC2 between the cytoplasm and the lysosome is increased in response to nutrient stress downstream of the GATOR2 component WDR24.
Figure 2. The GATOR2 complex inhibits the recruitment of TSC to lysosomes.
(A) Time point images from the Halo-TSC2 FRAP experiment in WT HeLa cells. The 0 s image represents photobleaching. Scale bar: 0.5 μm.
(B) Time point images from the Halo-TSC2 FRAP experiment in WDR24-KO cells.
(C) Fluorescence recovery versus time curves in A and B. A total of 30 lysosomes from different cells were used to plot the curves for each treatment. Error bars represent standard error.
(D) Plot showing the relation between the recovery fraction versus half time (t1/2) from curves in C. Error bars represents standard error.
(E) Time point pictures from the GFP-TSC1 FRAP experiment in WT Drosophila ovary.
(F) Time point pictures from the GFP-TSC1 FRAP experiment in wdr24RNAi Drosophila ovary. Scale bar: 2 μm.
(G) Fluorescence recovery versus time curves in E and F. A total of 10 lysosomes in different ovaries from each treatment were used in plotting the curve. Error bars represent standard error. See also Figure S2, S3 and S4.
Metabolism is often deregulated in cancer and cancer cell lines (Cornu et al., 2013; Efeyan and Sabatini, 2010; Mossmann et al., 2018). Thus, to complement our studies in HeLa cells, we examined TSC1-GFP dynamics in the Drosophila ovary. We noted that in the Drosophila ovary, consistent with what is observed in HeLa cells, lysosomes contain a stable pool of TSC1 under nutrient replete conditions (Figure S4). To define the dynamics of TSC1 in response to nutrient inputs we performed lysosomal FRAP of TSC1-GFP in the germline derived nurse cells of stage 6 egg chambers in cultured ovaries. We determined that, as we observed in HeLa cells, TSC1-GFP rate of recovery after lysosomal photobleaching increased in response to nutrient deprivation (Figures 2E and G). Additionally, depleting the GATOR2 component wdr24 in the female germline by RNAi partially phenocopied the response to nutrient stress (Figures 2F and G). Taken together these data demonstrate that in both HeLa cells and the Drosophila ovary, nutrient stress increases the dynamic recruitment of TSC to lysosomes. Moreover, in non-starvation conditions the GATOR2 component WDR24 opposes the dynamic exchange of TSC between the lysosome and the cytoplasm.
The GATOR2 complex acts through the Rag GTPase to regulate TSC recruitment to lysosomes
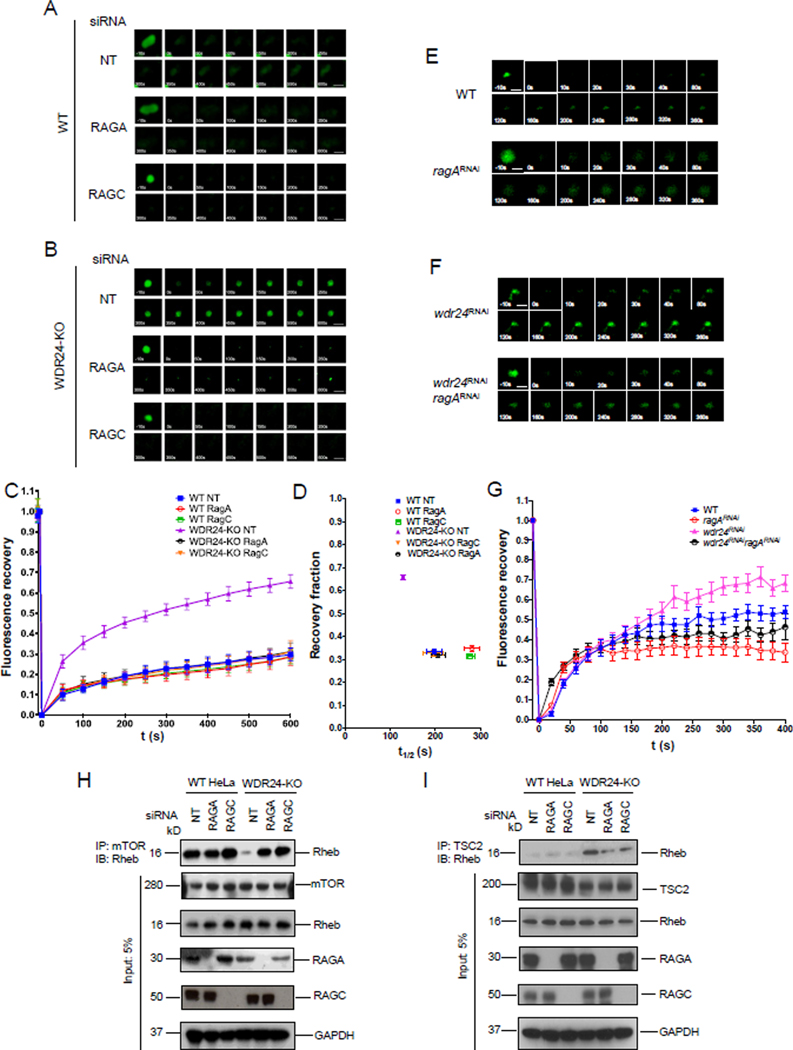
One model to explain our data is that the increased rate of recruitment of TSC to lysosomes observed during nutrient stress, and in WDR24-KO cells, requires the activity of the Rag GTPase. Consistent with this model, RAGA-KD or RAGC-KD in WDR24-KO cells, but not WT HeLa cells, significantly decreased the rate of fluorescence recovery of Halo-TSC2 after lysosomal FRAP relative to a control siRNA (mCh) (Figures 3A-D). Similarly, in the Drosophila ovary, co-depleting ragA with wdr24 in the female germline, reversed the rapid exchange of TSC1-GFP between the lysosome and the cytoplasm observed in wdr24RNAi ovaries (Figures 3E–G). These data suggest that the Rag GTPase drives the rapid lysosomal exchange of TSC in GATOR2 mutants. Notably, siRNA knockdowns of RAGA or RAGC did not affect the slow recruitment of TSC2 to lysosomes in WT HeLa cells cultured in nutrient replete condition (+FBS. +AA) (Figure 3A). This is consistent with RAGA and RAGC having minimal effect on TORC1 activity under fed (AA+, FBS+) conditions. Taken together our data demonstrate that the Rag GTPase promotes the recruitment of the TSC to lysosomes in the absence of GATOR2 activity.
Figure 3. The GATOR2 complex regulates the recruitment of TSC to lysosomes via the Rag GTPase.
(A) Time point images from the Halo-TSC2 FRAP experiment in WT HeLa cells treated with Non-targeting, RAGA and RAGC siRNAs. Scale bar: 0.5 μm.
(B) Time point images from the Halo-TSC2 FRAP experiment in WDR24-KO HeLa cells treated with Non-targeting, RAGA and RAGC siRNAs.
(C) Fluorescence recovery versus time curves in A and B. A total of 30 lysosomes in different cells were used to plot the curve for each treatment. Error bars represent standard error.
(D) Plot showing the relationship between the recovery fraction versus half time (t1/2) from the curves in C. Error bars represent standard error.
(E) Time point pictures from the GFP-TSC1 FRAP experiments in WT and ragARNAi Drosophila ovaries. Scale bar: 2 μm.
(F) Time point images from the GFP-TSC1 FRAP experiments in wdr24RNAi and wdr24RNAi ragARNAi Drosophila ovaries.
(G) Fluorescence recovery versus time curves in H and I. A total of 10 lysosomes from different ovaries were examined. Error bars represent standard error.
(H) Knockdowns of RAGA or RAGC increase the interaction between mTOR and Rheb in WDR24-KO cells. Cell lysates were immunoprecipitated by an anti-mTOR antibody and immunoblotted by an anti-Rheb antibody.
(I) Knockdowns of RAGA or RAGC decrease the interaction between TSC2 and Rheb in WDR24-KO HeLa cells. Cell lysates were immunoprecipitated by an anti-TSC2 antibody and immunoblotted by an anti-Rheb antibody.
TSC2 inhibits TORC1 by serving as a GAP for the TORC1 activator Rheb (Inoki et al., 2003). As presented above, the Rag GTPase promotes the dynamic recruitment of TSC to lysosomes in the absence of the GATOR2 component WDR24, a condition that mimics nutrient stress. We performed co-immunoprecipitations (co-IPs) to explore if the changes in TSC2 dynamics documented above, correlate with changes in the interaction of Rheb with TSC2 and mTOR. In WT HeLa cells expressing a non-targeting (NT) siRNA, Rheb readily co-IPs mTOR, but not the TORC1 inhibitor TSC2, both in the presence or absence of RAGA and RAGC (Figures 3H, I). These data are consistent with the ability of mTOR to directly bind Rheb in the absence of the Rag GTPase under nutrient replete conditions (Demetriades et al., 2014). Conversely, in WDR24-KO cells, Rheb co-IPs the TORC1 inhibitor TSC but not mTOR, consistent with the low TORC1 activity observed in WDR24-KO cells (Figures 3H, I). Moreover, knocking down RAGA or RAGC in WDR24-KO cells, restored the interaction between mTOR and Rheb while decreasing the interaction between TSC2 and Rheb (Figures 3H, I). These data support the model that in GATOR2 mutant cells, the Rag GTPase recruits TSC to lysosomes where it binds and inhibits Rheb. However, knockdowns of the Rag GTPase components RAGA or RAGC block recruitment of TSC2 to lysosomes thus allowing for the recruitment of TORC1 to lysosomes and rescue of the WDR24-KO phenotype.
The GTP/GDP status of RAGA and RAGC control the recruitment of TSC to lysosomes
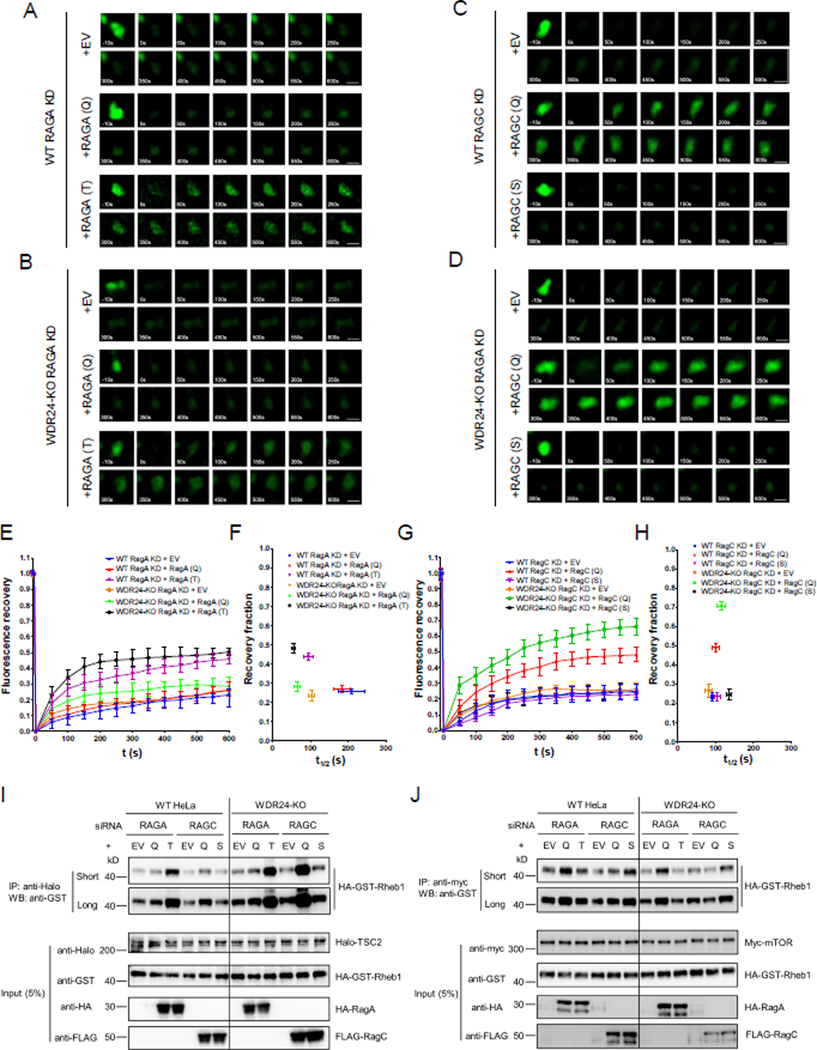
As small GTPases, the Rags are cycled between GTP-bound and GDP-bound forms. To formally test the hypothesis that GDP/GTP binding to the Rag GTPase affects TSC2 recruitment to lysosome, we knocked down endogenous RAGA and RAGC with siRNAs targeting their 3’ UTR (untranslated region), then used point mutations in RAGA and RAGC to lock the two proteins into either a GTP bound (RAGA-Q66L and RAGC-Q120L) or GDP bound (RAGA-T21N and RAGC-S75N) forms. Subsequently, the dynamics of TSC2 were examined by FRAP for each RAGA and RAGC point mutant. First, we examined if the Rag GTPase needs to be in the RAGAGDP: RAGCGTP configuration to facilitate the rapid recruitment of TSC2 to lysosomes. We found that in contrast to the empty vector (EV) control, expression of GDP-bound RAGA-T21N in WT and WDR24-KO HeLa cells increased the recovery rate of Halo-TSC2, while expression of the GTP-bound RAGA-Q66L had no effect (Figures 4A, B, E and F). From these data, we conclude that GDP bound RAGA facilitates the dynamic recruitment of TSC to lysosomes. Conversely with RAGC, which is in the GDP-bound form under nutrient replete conditions, we observed the opposite effect. Expression of the GTP-bound RAGC-Q120L increased the exchange rate of the Halo-TSC2 in both WT and WDR24-KO cells, while the GDP-bound RAGC-S75N had no effect (Figures 4C, D, G and H). These results demonstrate that the guanine nucleotide binding status of both RAGA and RAGC impacts the dynamic recruitment of TSC to lysosomes.
Figure 4. The guanine nucleotide binding status of RAGA and RAGC control the recruitment of TSC to lysosomes.
(A) Time point images from Halo-TSC2 FRAP experiment in WT HeLa cells transfected with GDP or GTP bound RAGA. Scale bar: 0.5 μm.
(B) Time point images from the Halo-TSC2 FRAP experiment in WDR24-KO HeLa cells transfected with GDP or GTP bound RAGA.
(C) Time point images from the Halo-TSC2 FRAP experiment in WT HeLa cells transfected with GDP or GTP bound RAGC.
(D) Time point pictures from the Halo-TSC2 FRAP experiment in WDR24-KO HeLa cells transfected with GDP or GTP bound RAGC.
(E) Fluorescence recovery versus time curves in A and B. A total of 30 lysosomes from 30 different cells were used to plot the curves for each treatment. Error bars represent standard error.
(F) Plot showing the relation between the recovery fraction versus half time (t1/2) from the curves in E. Error bars represent standard error.
(G) Fluorescence recovery versus time curves in C and D. A total of 30 lysosomes from different cells were used in plotting the curves for each treatment. Error bars represent standard error.
(H) Plot showing the relation between the recovery fraction versus half time (t1/2) from the curves in G. Error bars represent standard error.
(I) Co-IP experiments showing that GDP-bound RAGA and GTP-bound RAGC increased the interaction between Halo-TSC2 and HA-GST-Rheb. Cell lysates were immunoprecipitated by an anti-Halo antibody and immunoblotted by an anti-GST antibody. (Short: blot was exposed for 10 s; Long: 1 min)
(J) Co-IP experiment showing GTP-bound RAGA and GDP-bound RAGC increased the interaction between Myc-mTOR and HA-GST-Rheb. Cell lysates were immunoprecipitated by an anti-Myc antibody and immunoblotted by an anti-GST antibody.
Next we examined whether GDP/GTP binding of RAGA or RAGC affected the ability of TSC2 and TORC1 to associate with Rheb. As shown in Figure 4I, we determined that the GDP-bound RAGA-T21N and the GTP-bound RAGC-Q120L increased the interaction between TSC2 and Rheb in both WT and WDR24-KO HeLa cells (Figure 4I). In contrast, the interaction between mTOR and Rheb increased with the transfection of the GTP-bound RAGA-Q66L and the GDP-bound RAGC-S75N (Figure 4J). Thus as previously suggested (Demetriades et al., 2014), our data indicate that the guanine nucleotide binding status of the Rag GTPases regulates the dynamic recruitment of TSC to the lysosome and its ability to interact with and inhibit Rheb.
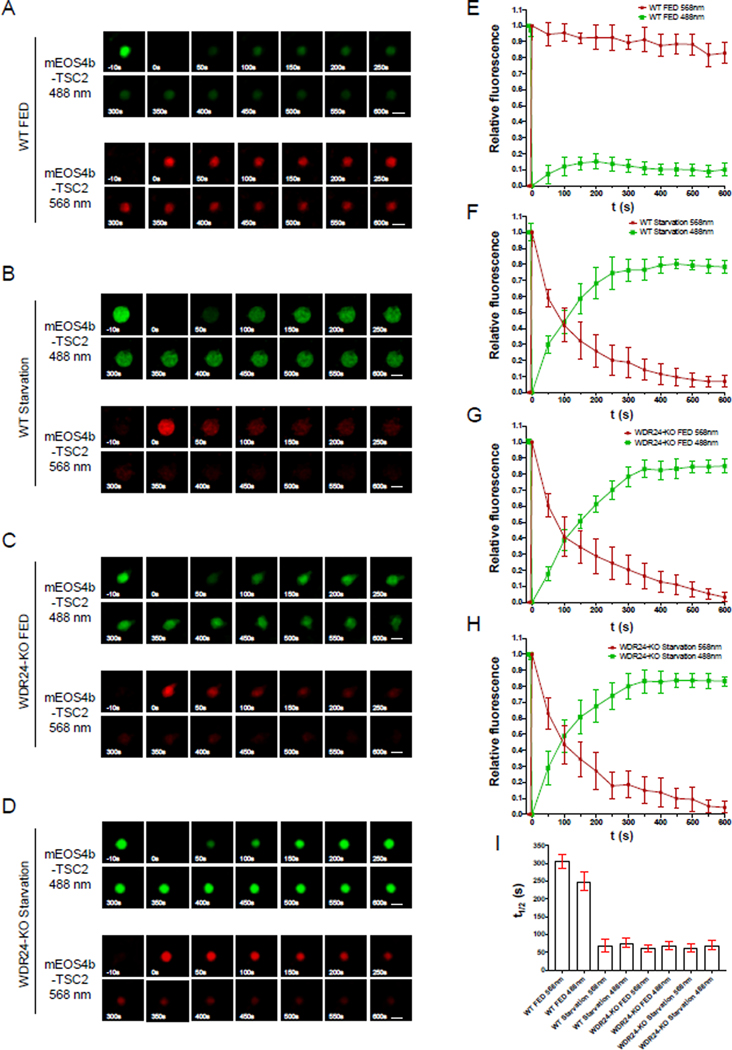
TSC2 rapidly cycles on and off lysosomes in GATOR2 mutant and starved cells
There are two models to explain the rapid association of TSC2 with lysosomes in WDR24-KO cells as well as in WT HeLa cells cultured in starved conditions. One model is that during starvation and in WDR24-KO cells, TSC2 is rapidly and stably recruited to lysosomes, where it remains and functions as a GAP for lysosomal Rheb. A second model posits that after acting as a GAP for lysosomal Rheb, TSC2 rapidly dissociates from the lysosome. In order to distinguish between these two models, we took advantage of the photoconvertible fluorescence tag mEOS4b (Paez-Segala et al., 2015). Before photoconversion, mEOS4b is detected at the wavelength of 488 nm, however after photoconversion with a 405 nm laser, mEOS4b emits at wavelength of 568 nm. By monitoring the changes in fluorescence signals at these two wavelengths after photoconversion, it is possible to determine both the “on-rate” and the “off-rate” of the mEOS4b tagged TSC2 on lysosomes in various nutrient conditions and genetic backgrounds.
First, we examined the behavior of lysosomal TSC2-mEOS4b in WT Hela cells cultured in nutrient replete (+FBS, +AA) conditions (Figures 5A, E). After photoconversion, the red fluorescence of the converted mEOS4b-TSC2 remained high, with minimal fluorescence loss over time. These data indicate that most of the TSC2 present on lysosomes is immobile under fed conditions. Additionally, consistent with our FRAP results, we observed minimal new recruitment of unconverted green mEOS4b-TSC2 to lysosomes from the cytoplasmic pool (Figure 5A, E). In contrast, nutrient depletion (−FBS, −AA) resulted in the rapid recruitment of the cytoplasmic green TSC2 to the lysosome, while the fluorescence of the converted “old” red TSC2 rapidly diminished (Figures 5B, F). The simplest explanation for these results is that under starvation conditions, TSC2 rapidly cycles on and off lysosomes. As predicted by our previous results, in WDR24-KO Hela cells TSC2-mEOS4b had on and off rates similar to WT HeLa cells cultured in starvation in both nutrient conditions (Figures 5C, D, G, H). In summary, by using a photoconvertible fluorescence protein, we demonstrated that TSC2 rapidly cycles between the cytoplasm and the lysosome in GATOR2 mutants and in WT cells subject to starvation.
Figure 5. TSC2 rapidly cycles on and off lysosomes in GATOR2 mutant and starved cells.
(A) Time point images from photoconversion experiment in WT Hela cells transfected with mEOS4b-TSC2 in nutrient replete conditions. The 0s image represents photoconversion. Scale bar: 0.5 μm.
(B) Time point images from photoconversion experiment in WT Hela cells transfected with mEOS4b-TSC2 under starvation condition.
(C) Time point images from photoconversion experiment in WDR24-KO Hela cells transfected with mEOS4b-TSC2 subjected to nutrient replete conditions.
(D) Time point images from photoconversion experiment in WDR24-KO Hela cells transfected with mEOS4b-TSC2 under starvation condition.
(E-H) Change in relative fluorescence of unconverted mEOS4b-TSC2 at 488 nm (green curve) and photo converted protein in 568 nm (red curve) over 600s period corresponding to the results shown in A-D. A total of 30 lysosomes in each condition were used. Error bars represent standard error.
(I) Comparisons of the half-time (t1/2) for mEOS4b-TSC2 at 488 nm and 568 nm in corresponding to panels A-D. Scale bars represent standard error.
The Rag GTPase controls the dynamic recruitment of TSC to lysosomes in response to both amino acid and growth factor restriction
The insulin-PI3K-AKT signaling pathway is a critical upstream regulator of TSC activity (Cai et al., 2006; Dan et al., 2002; Inoki et al., 2002). To obtain a better understanding of the regulation of TSC dynamics we examined the behavior of Halo-TSC2 in WT HeLa cells under either amino acid or growth factor (FBS) restriction using lysosomal FRAP. From these experiments, we determined that independently restricting either amino acids (−AA +dFBS) or serum (+AA −FBS) increased the rate of recruitment of Halo-TSC2 to lysosomes relative to complete media (WT FED) (NT images in Figures 6A, C, E). Additionally, in all three nutrient conditions depleting RAGA-KD significantly decreased the rate of TSC2 fluorescence recovery in WT HeLa cells (Figures 6C, E). These data indicate that the Rag GTPase regulates TSC dynamics downstream of both amino acid and growth factor restriction.
Figure 6. The Rag GTPase controls the dynamic recruitment of TSC to lysosomes in response to both amino acids and growth factor restriction.
(A) Time point images from the Halo-TSC2 FRAP experiment in WT HeLa cells treated with control or RAGA siRNA in FED condition (+AA, +FBS). Scale bar: 0.5 μm. Fluorescence recovery versus time curve was plotted by using a total of 30 lysosomes in cells. Error bars represent standard error.
(B) Time point images from the Halo-TSC2 FRAP experiment in WDR24-KO HeLa cells treated with control or RAGA siRNA in FED condition (+AA, +FBS).
(C) Time point images from the Halo-TSC2 FRAP experiment in WT HeLa cells treated with control or RAGA siRNA in amino acid starvation (-AA, +dFBS).
(D) Time point images from the Halo-TSC2 FRAP experiment in WDR24-KO HeLa cells treated with control or RAGA siRNA in amino acid starvation (-AA, +dFBS).
(E) Time point images from the Halo-TSC2 FRAP experiment in WT HeLa cells treated with control or RAGA siRNA in serum starvation (+AA, -FBS).
(F) Time point images from the Halo-TSC2 FRAP experiment in WDR24-KO HeLa cells treated with control or RAGA siRNA in serum starvation (+AA, -FBS).
As predicted by our previous results (Figure 3) WDR24-KO cells exhibited a high rate of Halo-TSC2 fluorescence recovery after lysosomal FRAP in both fed and nutrient depleted conditions (Figure 6B). Additionally, similar to what is observed in WT HeLa cells, knockdowns RAGA in individually restricted conditions (-AA +dFBS) and (+AA - FBS), impedes the recovery of TSC2 to lysosomes (Figures 6D, F).
Moreover, in amino acid or serum restriction (−AA +dFBS or +AA- FBS) conditions, removing RAGA in WT or WDR24-KO HeLa cells restored TORC1 activity as measured by the level of pS6K (Figure 7A). These results again indicate in nutrient or growth factor depletion conditions, the Rag GTPase prevents TORC1 activity. In summary, we find that both amino acids and growth factors individually impact the rate of exchange of TSC between the cytosol and lysosomes as well as TORC1 activity through the Rag GTPase.
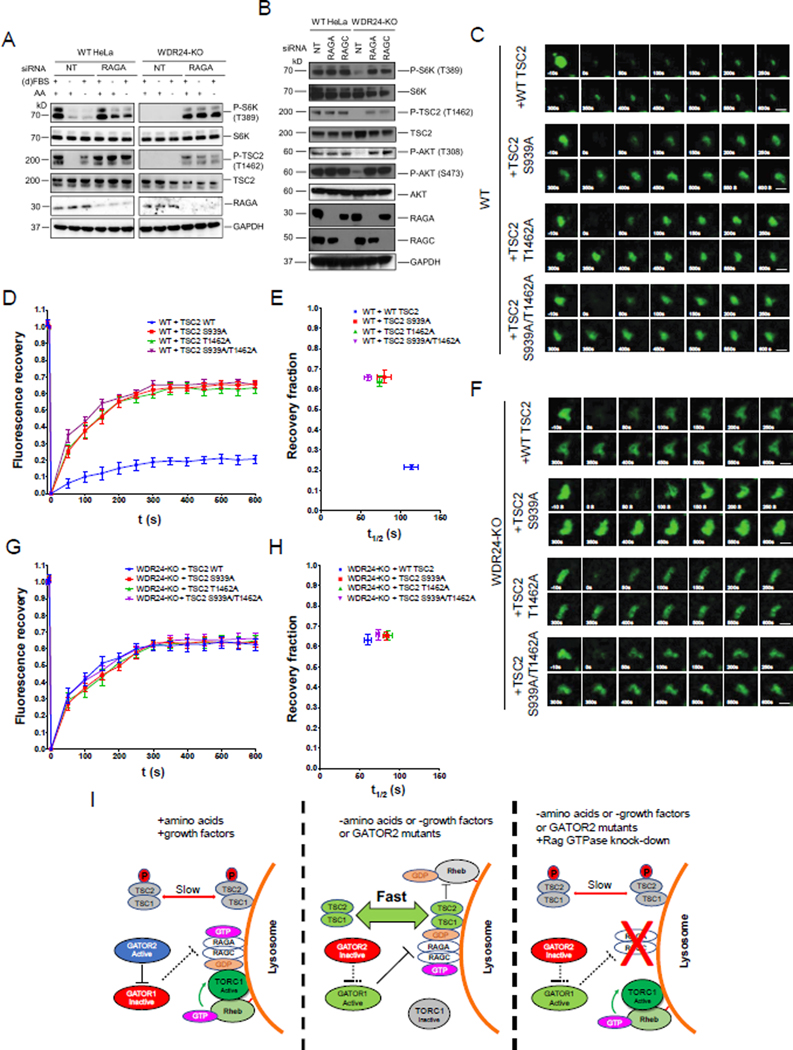
Figure 7. Phosphorylation by AKT decreases the lysosomal dynamics of TSC2.
(A) Immunoblots to detect the level of P-S6K (T389), P-TSC2 (T1462) in WT and WDR24-KO HeLa cells treated with control (NT) or RAGA siRNA, cultured in +AA +FBS, +AA -FBS and AA +dFBS conditions.
(B) Immunoblots show knockdowns of RAGA and RAGC in WDR24-KO cells increase the level of P-AKT (T308) and P-AKT (S473).
(C) Time point images from the Halo-TSC2 FRAP experiment in WT HeLa cells transfected with Halo-TSC2 T1462A, Halo-TSC2 S939A and the Halo-TSC2 T1462A/S939A. Scale bar: 0.5 μm.
(D) Fluorescence recovery versus time curves in C. A total of 30 lysosomes from 30 different cells were used in plotting the curve for each treatment. Error bars represent standard error.
(E) Plot showing the relation between the recovery fraction versus half time (t1/2) from the curves in D. Error bar represents standard error.
(F) Time point images from the Halo-TSC2 FRAP experiment in WDR24-KO HeLa cells transfected with Halo-TSC2 T1462A, Halo-TSC2 S939A and the Halo-TSC2 T1462A/S939A.
(G) Fluorescence recovery versus time curves in F. A total of 30 lysosomes from 30 different cells were used in plotting the curve for each treatment.
(H) Plot showing the relation between the recovery fraction versus half time (t1/2) from the curves in G.
(I) Proposed model: The GATOR complex regulates TORC1 activity by controlling the exchange rate of TSC between lysosome and cytosol through the Rag GTPase and TSC2 phosphorylation. Refer to Discussion for details. See also Figure S5.
WDR24 opposes the AKT-dependent inhibition of TSC activity and lysosomal dynamics
In the presence of adequate insulin, AKT phosphorylates TSC2 on multiple sites, including T1462, resulting in TSC inhibition and release from the lysosome (Menon et al., 2014). Thus, levels of P-TSC2 (T1462) reflect the functional status of TSC2. As reported by others (Inoki et al., 2002), we find that in WT HeLa cells TSC2-T1462 phosphorylation is dependent on the presence of FBS (Figure 7A). Additionally, we determined that pTSC2-T1462 levels were modestly diminished in HeLa cells grown under conditions of amino acid starvation relative to HeLa cells grow in complete media (Figure 7A). These data are consistent with previous work demonstrating a role for TSC in the response to amino acid stress (Demetriades et al., 2014). Surprisingly, we found that in WDR24-KO cells, pTSC2-T1462 levels were below detection in all three media conditions (+AA +FBS), (+AA -FBS) and (−AA +dFBS) (Figure 7A). Thus, TSC2-T1462 phosphorylation is dependent on the presence of the GATOR2 component WDR24, suggesting that the GATOR2 complex is an upstream inhibitor of TSC.
Next, we examined if the downregulation of TSC2-T1462 phosphorylation observed in WDR24-KO cells is dependent on the function of the Rag GTPase. First, we demonstrated that knocking down RAGA in wild-type HeLa cells had no effect on pTSC2-T1462 levels (Figure 7A). In contrast, levels of pTSC2-T1462, were nearly undetectable in WDR24-KO cells (Figure 7A). pTSC2-T1462 levels were restored in WDR24-KO cells depleted for RAGA regardless of nutrient inputs (Figure 7A). Similar to WDR24-KO, RagA is required for the reduction in TSC2 phosphorylation observed upon either amino acid or FBS removal (Figure 7A). Phosphorylation of pTSC2 on T1462 occurs via AKT (Dan et al., 2002; Inoki et al., 2002; Potter et al., 2002). AKT is activated by phosphorylation at its Thr308 and Ser473 sites. Interestingly, we found in WDR24-KO cells, the levels of both P-AKT T308 and S473 were lower than WT. Moreover, knockdowns of RAGA and RAGC restore AKT phosphorylation (Figure 7B). Thus, these data support the idea that in WDR24-KO cells the Rag GTPase prevents, either directly or indirectly, the AKT-dependent inhibitory phosphorylation of TSC2.
Conditions that favor the formation of the inactive pTSC2-T1462 correlate with increased mobility of TSC2 on lysosomes and decreased TORC1 activity (Figures 3, 6 and 7). Thus, we hypothesized that the phosphorylation of TSC2 by AKT decreases the lysosomal mobility of TSC. To test this model, we made point mutations of TSC2 that are resistant to AKT phosphorylation. AKT phosphorylates TSC2 at T1462 and S939 (Inoki et al., 2002). We mutated these two amino acids to alanine either together or individually and tagged each construct with a Halo tag. First, we confirmed that the three proteins mutants TSC2 T1462A, TSC2 S939A and TSC2 T1462A/S939A were localized on lysosomes by immunostaining, through colocalization with the lysosomal marker LAMP1 (Figure S5). Next, we performed FRAP to compare the dynamics of the Halo-TSC2 point mutants relative to control WT HaloTSC2. Consistent with our prediction, in WT HeLa cells cultured in nutrient replete conditions, all three Halo tagged TSC2 point mutants exhibited faster fluorescence recovery after lysosomal FRAP relative to WT TSC2 (Figures 7C, D and E). In contrast, the TSC2 point mutants had similarly high rates of fluorescence recovery relative to WT TSC2 in WDR24-KO HeLa cells (Figures 7F, G and H). In summary, our results suggest that AKT phosphorylation state correlates with TSC2 dynamics and that AKT may inhibit TSC2 function by opposing its dynamic exchange between lysosome and cytosol.
DISCUSSION
TSC is a critical inhibitor of TORC1 signaling (Dibble and Manning, 2013; Tee et al., 2002). Currently there are two working models for the role of TSC in the inhibition of TORC1 activity. The first model posits that TSC lies exclusively downstream of the PI3K-AKT growth factor signaling pathway while the second model proposes that TSC is a critical downstream effector of both the growth factor signaling and amino acid sensing pathways (Demetriades et al., 2014; Gao et al., 2002; Laplante and Sabatini, 2012; Roccio et al., 2006; Smith et al., 2005). Our findings on the function of the GATOR2 complex are consistent with the second model, which implicate different nucleotide states of the Rag GTPase in the recruitment of TORC1 versus TSC to lysosomes in response to amino acid starvation (Demetriades et al., 2014; Gao et al., 2002). Moreover, we find that the Rag GTPase, which has previously been thought to exclusively function in amino acid sensing, regulates the recruitment of TSC to lysosomes in response to growth factor restriction. Thus, our data support a model in which both the amino acid sensing pathway and growth factor signaling pathway converge on the Rag GTPase to recruit TSC to lysosomes in response to inhibitory signals. Notably, we found in both HeLa cells and Drosophila, the Rag GTPase promotes the rapid exchange of TSC between the lysosome and cytosol in response to negative inputs. Finally, demonstrating further integration of the amino acid sensing and growth factor signaling pathways, we show that the GATOR2 complex acts upstream of the Rag GTPase to promote both, the activating phosphorylation of AKT and the AKT-dependent inhibitory phosphorylation of TSC2.
An important outstanding question in the field of TORC1 regulation concerns the role of the Rag GTPase and TSC (Demetriades et al., 2014; Dibble and Manning, 2013).Our data indicate that in both Drosophila and HeLa cells, the GATOR-Rag GTPase axis inhibits TORC1 activity through the regulation of the dynamic behavior of TSC. We find that in cells without a functional GATOR2 complex, the Rag GTPase, locked in its RAGAGDP: RAGCGTP-bound form due to the activation of GATOR1, recruits TSC to lysosomes precluding the recruitment and activation of TORC1. This inhibited state is relieved by depleting RAGA or RAGC, resulting in the recruitment and activation of TORC1 on lysosomes. Mutations in GATOR2 components mimic amino acid starvation (Bar-Peled et al., 2013; Wei and Lilly, 2014). Thus, our results are consistent with reports from the Teleman laboratory indicating that amino acid starvation promotes the Rag GTPase-dependent recruitment of TSC to lysosomes (Demetriades et al., 2014). Moreover, our data confirm that under conditions of metabolic homeostasis TORC1 can be recruited to lysosomes and activated by Rheb in the absence of the Rag GTPase (Demetriades et al., 2014; Jewell et al., 2015).
Intriguingly, in budding yeast, which do not have TSC, the homolog of the Rag GTPase, GTRl1/GTRL2, has a similar dual role in the regulation of TORC1 activity with the verexpression of GTR1GTP promoting TORC1 activity while GTR1GDP is associated with low TORC1 activity (Binda et al., 2009). Thus, the Rag GTPase may have a conserved role in the inhibition of TORC1 activity that goes beyond the regulation of TSC dynamics.
Surprisingly, we also demonstrate a central role for the Rag GTPase in the recruitment of TSC to lysosomes in response to growth factor restriction. Specifically, the low TORC1 activity and reduced TSC mobility associated with growth factor depletion were rescued by depleting the Rag GTPase component RagA. Taken together, our data argue that the Rag GTPase and TSC are critical components of both the amino acid sensing and growth factor signaling pathways.
The idea that TSC acts downstream of the GATOR-Rag GTPase pathway is consistent with our previous observations in Drosophila showing that depleting components of TSC rescue the low TORC1 activity observed in GATOR2 mutant ovaries (Cai et al., 2016; Wei et al., 2014). Indeed, in Drosophila TSC is epistatic to the GATOR2 complex with respect to TORC1 activity, in that depleting TSC in GATOR2 mutant cells resulted in high TORC1 levels similar to those observed in TSC single mutants (Cai et al., 2016; Gao and Pan, 2001; Wei et al., 2014). As indicated above, our data are also consistent with several previous reports that TSC acts in the amino acid sensing pathway (Demetriades et al., 2014; Demetriades et al., 2016; Gao et al., 2002; Wei and Lilly, 2014).
Recent reports indicate that the recruitment of TSC to lysosomes is a common response to cellular stress in many mammalian cell types (Demetriades et al., 2014; Demetriades et al., 2016; Menon et al., 2014). However, these studies have not examined if TSC remains static on lysosomes or actively exchanges with the cytoplasmic pool during periods of TORC1 inhibition. Using FRAP and a photoconvertible TSC2 to follow the intracellular dynamics of TSC components, we determined that the rate of exchange of TSC between the lysosome and the cytosol increases in response to nutrient deprivation and growth factor restriction. This rapid increase in cycling on and off lysosomes requires the Rag GTPase in both HeLa cells and the Drosophila ovary. Notably, increased cycling of TSC correlates with its increased interaction with Rheb, and with a concomitant decrease in TORC1 activity. Depleting components of the Rag GTPase in cells grown in amino acid or growth factor restricted media dramatically reduced the lysosomal/cytosol TSC rate of exchange, decreased its binding with Rheb, and rescued TORC1 activity. Thus, our data support the model that the Rag GTPase increases the exchange rate of TSC between the lysosome and the cytoplasm in response to both amino acid starvation and growth factor restriction.
It has long been established that AKT controls TSC activity through the inhibitory phosphorylation of TSC2 (Inoki et al., 2002; Manning et al., 2002; Potter et al., 2002). More recent studies indicate that AKT controls TSC activity by regulating its association with Rheb on the surface of lysosomes (Menon et al., 2014). We find that in nutrient replete conditions, the inhibitory phosphorylation of TSC2 by AKT slows the rate of lysosomal/cytosolic exchange of TSC and allows for TORC1 activation. When TSC2 is rendered resistant to AKT-dependent phosphorylation, TSC rapidly cycled on and off lysosomes independent of growth factor and amino acid status. Importantly, cells expressing AKT resistant TSC have low TORC1 activity (Inoki et al., 2002; Menon et al., 2014). We determined that in WDR24-KO cells, in which the RAG GTPase is in the RAGAGDP: RAGCGTP state, the AKT dependent phosphorylation of TSC2 is strongly diminished, resulting in the increased mobility of the TSC complex on lysosomes and decreased TORC1 activity. Depleting components of the Rag GTPase in WDR24-KO cells, rescued these phenotypes resulting in increased AKT-dependent phosphorylation of TSC, decreased TSC lysosomal motility and increased TORC1 activity (Inoki et al., 2002; Menon et al., 2014). Further examination revealed that levels of activated AKT are kept low in WDR24-KO cells by a Rag GTPase dependent mechanism. These data are consistent with previous observations that RAGA-KO cells have increased levels of activated AKT while WDR24-KO cells fail to increase AKT activation after Sestrin2 overexpression (Efeyan et al., 2014; Kowalsky et al., 2020). Taken together, our data indicate that the GATOR2 complex promotes the activation of AKT, which facilitates the AKT-dependent inhibitory phosphorylation of TSC, upstream of the Rag GTPase. Moreover, they demonstrate that the rate of TSC cycling on and off lysosomes reflects TSC activity.
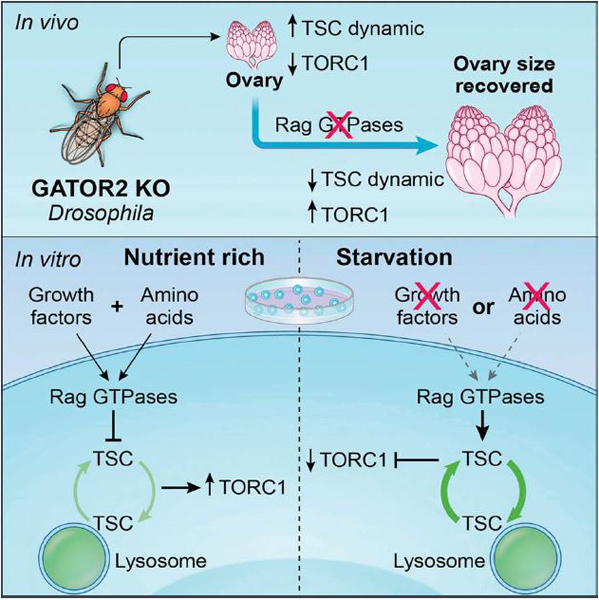
Based on our data, as well as data from others (Demetriades et al., 2014; Demetriades et al., 2016; Menon et al., 2014), we propose the following model (Figure 7I). Under conditions of amino acid and growth factor sufficiency GATOR2 inhibits GATOR1 resulting in the Rag GTPase adopting the RAGAGTP: RAGCGDP configuration which favors the recruitment and activation of TORC1 on lysosomes and results in the limited exchange of TSC between the lysosome and the cytoplasm. In contrast, under conditions of amino acid or growth factor depletion, or in GATOR2 mutant cells, GATOR1 is active, resulting in the Rag GTPase adopting the RAGAGDP: RAGCGTP configuration, which promotes the rapid exchange of TSC between the lysosome and the cytoplasm and decreases TORC1 activity. Under these restricted conditions (AA-, serum- or WDR24-KO), knockdowns of RAGA or RAGC prevent the rapid cycling of TSC allowing for the recovery of TORC1 activity due to the inherent ability of TORC1 to bind Rheb directly (Demetriades et al., 2014). Currently, whether the rapid cycling of TSC on and off lysosomes in response to upstream signals directly promotes TSC activity, or serves a regulatory role, has not been definitively established. In summary, our data support a model in which both the amino acid sensing and growth factor signaling pathways utilize the Rag GTPase to inhibit TORC1 activity through the regulation of TSC lysosomal dynamics.
A recent study reported that RAGA rapidly cycles between the lysosome and the cytosol in response to nutrients (Lawrence et al., 2018). Notably, RAGAGTP cycles on and of lysosomes while RAGAGDP remains more tightly associated with the Ragulator at the lysosomal surface. Intriguingly, we find that RAGAGDP promotes the rapid cycling of the TSC complex on and off the lysosome. Thus, in the future it will be important to determine precisely how the dynamic behavior of RAGA and TSC are coordinated to control TORC1 activity.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Mary Lilly (lillym@helix.nih.gov).
Materials Availability
Drosophila stocks, HeLa cell knockout lines and other reagents generated in this study will be available upon request from the Lead Contact Mary Lilly (lillym@helix.nih.gov).
Data and Code Availability
Original data are deposited in Mendeley Data: http://dx.doi.org/10.17632/n8gztd29sv.1
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell culture and treatments
All mammalian cells were maintained and cultured at 37°C, 5% CO2, in high-glucose Dulbecco’s modified Eagle’s medium GlutaMAX™-I with pyruvate, supplemented with 10% Fetal bovine serum (FBS) and 100 IU/ml Penicillin-Streptomycin (refer as the completed DMEM thereafter). For amino acid starvation, cells were cultured in the completed DMEM medium until 80% confluency, then were washed twice with 1x phosphate buffered saline (PBS), medium was changed to DMEM without amino acids (glucose was adjusted to 4.5g/l), with 10% dialyzed FBS for 2 hr. For FBS starvation, the medium was changed to DMEM without adding FBS, the cells were cultured for 16 hrs. Wild-type (WT) HeLa cell was obtained from ATCC. Its identity was verified and tested for mycoplasma.
Drosophila stocks
All Drosophila stocks were maintained at 25°C on JAZZ-mix Drosophila food (Fisher Scientific). The germline specific driver nanos-Gal4 was obtained from Ruth Lehmann (Van Doren et al., 1998). The TSC1-GFP fTRG transgenic line (VDRC #318377) was obtained from Vienna Drosophila Resource Center. The stocks UAS-Wdr24 RNAi (BDSC #62393), UAS-RagA RNAi (BDSC #34590), UAS-RagC RNAi (BDSC #32342) and UAS-mCherry RNAi (BDSC #35787) were obtained from Bloomington Stock Center. The seh1Δ15 line is made previously (Senger et al., 2011).
Generation MIOS and WDR59 knockout HeLa cells by CRISPR-Cas9
MIOS and WDR59 were knocked out in WT HeLa cells by using CRISPR/Cas9 based on the protocol by Ran et al., 2013 (Ran et al., 2013). Briefly the following sense or antisense nucleotide (sgDNA) encoding single guide RNAs (sgRNA) were assembled in vitro and cloned into the px459 (pSpCas9(BB)-2A-Puro) plasmid.
sgMIOS_1_For: CACCGTGTGATCAAAGGGTCCACTC sgMIOS_1_Rev: aaacGAGTGGACCCTTTGATCACAC
sgMIOS_2_For: CACCGCTAGCCAGTTACTATCCAG
sgMIOS_2_Rev: aaacCTGGATAGTAACTGGCTAGC
The combination of the sgMIOS_1 and 2 guides Cas9 to cut through the Exon 4 of the MIOS protein coding sequence including its start codon.
sgWDR59_1_For: CACCGACTCGGCCTCTAGCTCACCT sgWDR59_1_Rev: aaactccccAGTATCCCGGGACCGC
sgWDR59_2_For: CACCGACTCGGCCTCTAGCTCACCT sgWDR59_2_Rev: aaacAGGTGAGCTAGAGGCCGAGTC
The combination of the sgWDR59_1 and 2 guides Cas9 to cut through the Exon 1 of the WDR59 protein coding sequencing including its start codon.
The resulted px459 sgDNA constructs were verified by sequencing.
On day one, 1 million WT HeLa cells were seeded into 10-cm dish in the completed DMEM medium without antibiotics. After 24 hrs, a total of 500 ng px459 guide constructs (250 ng for each sgDNA assembly) were transfected into HeLa cells by using the Lipofectamine 3000 reagent. Untransfected cells were eliminated by puromycin (puromycin dihydrochloride, Thermo Fisher, A1113802) on the next day. 48 hrs later, the culture medium was changed into fresh completed DMEM without antibiotics. Cells were disassociated by trypsin and serial diluted to the concentration of 0.5 cells per 100 ul medium. The diluted cells were passed into 96-well plates in the amount of 100 ul per well. 1 week after plating, the plates were examined and wells with a single cell colony were marked. After expansion for another 2 weeks, genome of each cell colony was collected and their MIOS or WDR59 genome loci were analyzed by PCR. Cells with PCR products of the expected size after editing by Cas9 were collected and their genome were sent for further sequencing. Finally, the protein level of MIOS or WDR59 was verified by immunoblot using the antibodies against the two protein respectively. Those cells with undetectable level of MIOS or WDR59 were marked as MIOS-KO and WDR59-KO cells.
METHOD DETAILS
Immunofluorescence microscopy for mammalian cells
HeLa cells were seeded onto the 8-well Nunc™ Lab-Tek™ II Chamber Slide™ System (Thermo Fisher, 154534) coated with fibronectin in the density of 104 per well and cultured in the completed DMEM medium overnight. Cells were washed 3 times with 1 × PBS and fixed with 4% paraformaldehyde (PFA) in 1 × PBS at room temperature for 15 mins. Cells were washed 3 times with PBS and permeabilized with 0.2% Trition X-100 in PBS for 10 mins at room temperature. Then cells were blocked in blocking buffer (0.1% BSA, 0.2% Trition X-100 in PBS) for 1 hr at room temperature. The chamber slides were incubated overnight at 4°C in blocking buffer containing diluted primary antibody. Then they were washed 3 times in 1 × PBS and incubated in blocking buffer containing secondary antibody conjugated with fluorophore for 1 hr in dark at room temperature, following by a 10 mins’ incubation with 300 nM 4’,6-diamidino-2-phenylindole (DAPI). The chamber slides were washed 3 times in PBS and then mounted on slides using the ProLong™ Diamond Antifade solution (Thermo Fisher, P36970) after remove the chamber well. All confocal images were captured by a Zeiss 880 laser scanning microscopy with a Plan-Apochromat 63X/1.4 oil immersion objective. Pearson’s correlation coefficients of different fluorophores were calculated by using ImageJ with Fiji plugin. 30 cells per condition were analyzed. Each set of immunofluorescence experiments was repeated at least three times.
Fluorescence recovery after photobleaching (FRAP) and mEOS4b photoconversion
FRAP experiments were performed on HeLa cells transfected with plasmid constructs as descried in the “Plasmid transfections for mammalian cell” section, two days before imaging. 40 min before the experiment, HeLa cells were cultured in imaging buffer with 2.5 μg/ml nocodazole as indicated previously (Lawrence et al., 2018), with or without serum or amino acids. FRAP live cell images were acquired at 5 s intervals for 10 min after photobleaching a single lysosome to background fluorescence level, using a Zeiss 880 system with a 100% laser power. The FRAP data were processed in GraphPad Prism 8. The recovery fraction and half-time (t½) were calculated by fitting the FRAP fluorescence recovery curve into the model Y=Y0 + (Plateau-Y0)*(1-exp(−K*x)) through using one-phase association as described before (Sprague and McNally, 2005; Zheng et al., 2011). Photoconversion of mEos4b was performed by using a 405 nm laser. Red signal after conversion was acquired at 50 s intervals for 10 min. The kinetic of red decline (post-conversion) and green recovery (pre-conversion) on lysosome represent the off and on rate of protein respectively. The acquired images were processed by using Imaris 9.3 software.
Plasmid transfections for mammalian cell
Plasmid transfections into HeLa cells were performed by using the Xfect™ Transfection reagent, according to manufacturer’s protocol. A total of 5 μg plasmid was used in transfection for 0.8 million cells.
Mammalian cell siRNA transfections and knockdowns
siRNAs against RAGGA, RAGGC and LAMTOR1/p18 in HeLa cells were from the SMARTpool ON-TARGETplus siRNA set (GE Healthcare). All siRNAs were transfected by the Xfect™ RNA Transfection reagent, according to manufacturer’s protocol. An ON-TARGETplus Non-targeting Control siRNAs was used as the control. 0.3 million HeLa cells were seeded into 6-well plates and cultured in completed DMEM medium overnight. For next two days, each day a combination of 5 ul siRNA stock (20 μM) and 9 ul of the transfection reagent was added into culture medium per well. Cells were assayed in the third day for immunoblotting or imaging.
Mammalian protein extraction and immunoblot
HeLa cells were seeded into 6-well plates. 1.2 million HeLa cells from each well were washed twice by PBS. Cells were then covered by 600 ul of the M-PER mammalian protein extraction buffer plus proteinases inhibitor cocktail with or without phosphatases inhibitor cocktail, followed by gently shaking at room temperature for 5 mins. The solutions were collected, and the soluble parts were separated by centrifugation. Target proteins in the soluble part were detected by immunoblot using specific antibodies. To detect phosphorylated protein, Pierce™ Protein-Free T20 (TBS) Blocking Buffer was used to block the membrane and dilute the antibodies. HRP signals were visualized by using a Clarity™ Western ECL substrate kit (Biorad) and detected with a Biorad ChemiDoc™ MP imaging system. The grey scale of each band was quantified by Photoshop CC. Each set of immunoblot experiments was repeated at least three times. Representative examples are shown in each figure.
Mammalian protein co-immunoprecipitation
Proteins from HeLa cells were extracted in buffer with proteinase and phosphatase inhibitor cocktail as mentioned above. Antibody against endogenous TSC2 or mTOR was crosslinked to Dynabeads™ Protein A by disuccinimidyl suberate (DSS) according to manufacturer’s protocol. Protein extracts were incubated overnight at 4°C with the antibody linked protein A beads. After incubation beads were washed three times with protein extraction buffer, then proteins were eluted by heating the beads in protein sample buffer [60 mM Tris (pH 6.8), 10% glycerol, 100 mM DTT, 0.2% bromophenol blue, 2% SDS] at 70°C for 10 mins, followed by immunoblot. An anti- Rheb antibody was used to detect the interactions between TSC2- Rheb or mTOR- Rheb from different conditions. For pulling down the Myc tagged protein, Pierce™ c-Myc-Tag IP/Co-IP Kit from Thermo Fisher was used according to manufacturer’s instruction. For pulling down Halo tagged protein, the Halo-Trap kit from Chromotek was used based on manufacturer’s instruction.
Lysosomal FRAP assay in Drosophila germline
The FRAP experiments were performed on stage 6 egg chambers in Drosophila ovaries. Briefly, flies were cultured in standard media for 2–3 days before dissection. The ovaries were dissected in Schneider’s medium supplemented with 10% FBS, 10 μg/ml insulin,100 nm nocodazole and 1μg/ml Hoechst. For acute starvation, the ovaries were dissected in amino acid starvation media as previously described (Wilson et al., 2004) and suppled with 100 nM nocodazole and 1μg/ml Hoechst 33342. 100% laser was used for lysosomal photobleaching, images were acquired at 10 seconds intervals for 7 mins.
QUANTIFICATION AND STATISTICAL ANALYSIS
Sample Size and Statistical Analysis
Detailed sample size can be found in figure legends. All graphs report the mean ± SEM and represent data from three independent experiments. Unless otherwise indicates. Statistical comparisons were made using Unpaired Student’s t-test provided by GraphPad Prism 8 software.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| LAMP1 Mouse monoclonal, H4A3 | Developmental Studies Hybridoma Bank | Cat# H4A3, RRID:AB_2296838 |
| mTOR (7C10) Rabbit mAb | Cell Signaling Technology | Cat# 2983, RRID:AB_2105622 |
| RagA (D8B5) Rabbit mAb | Cell Signaling Technology | Cat# 4357, RRID:AB_10545136 |
| RagC (D31G9) XP Rabbit mAb | Cell Signaling Technology | Cat# 5466, RRID:AB_10692651 |
| RHEB (E1G1R) Rabbit mAb | Cell Signaling Technology | Cat# 13879, RRID:AB_2721022 |
| TSC2 (D93F12) XP Rabbit mAb | Cell Signaling Technology | Cat# 4308, RRID:AB_10547134 |
| Phospho-Tuberin/TSC2 (Thr1462) Antibody | Cell Signaling Technology | Cat# 3611, RRID:AB_329855 |
| P70 S6 kinase (49D7) Rabbit mAb | Cell Signaling Technology | Cat# 2708, RRID:AB_390722 |
| Phospho-p70 S6K (T389) Rabbit mAb | Cell Signaling Technology | Cat# 9025, RRID:AB_2734746 |
| Phospho-Drosophila p70 S6K (T389) Rabbit Ab | Cell Signaling Technology | Cat# 9209, RRID:AB_2269804 |
| GAPDH (14C10) Rabbit mAb | Cell Signaling Technology | Cat# 2118, RRID:AB_561053 |
| β-Actin (D6A8) Rabbit mAb | Cell Signaling Technology | Cat# 8457, RRID:AB_10950489 |
| Mios (D12C6) Rabbit mAb | Cell Signaling Technology | Cat# 13557, RRID:AB_2798254 |
| WDR59 (D4Z7A) Rabbit mAb | Cell Signaling Technology | Cat# 53385, RRID:AB_2799432 |
| HA-Tag (C29F4) Rabbit mAb | Cell Signaling Technology | Cat# 3724, RRID:AB_1549585 |
| DYKDDDDK Tag (D6W5B) Rabbit mAb | Cell Signaling Technology | Cat# 14793, RRID:AB_2572291 |
| Myc-Tag (9B11) Mouse mAb | Cell Signaling Technology | Cat# 2276, RRID:AB_331783 |
| GST (26H1) Mouse mAb | Cell Signaling Technology | Cat# 2624, RRID:AB_2189875 |
| Anti-Rabbit IgG, HRP-linked | Cell Signaling Technology | Cat# 7074, RRID:AB_2099233 |
| Anti-Mouse IgG, HRP-linked | Cell Signaling Technology | Cat# 7076, RRID:AB_330924 |
| Akt (pan) (C67E7) Rabbit mAb | Cell Signaling Technology | Cat# 4691, RRID:AB_915783 |
| Phospho-Akt (Thr308) (D25E6) XP Rabbit mAb | Cell Signaling Technology | Cat# 13038, RRID:AB_2629447 |
| Phospho-Akt (Ser473) (D9E) XP Rabbit mAb | Cell Signaling Technology | Cat# 4060, RRID:AB_2315049 |
| Anti HaloTag Mouse mAb | Promega | Cat# G9211, RRID:AB_2688011 |
| Anti-Rabbit IgG AlexaFluor 488 conjugated | Thermo Fisher | Cat# A-11008, RRID:AB_143165 |
| Anti-Rabbit IgG AlexaFluor 594 conjugated | Thermo Fisher | Cat# A-11037, RRID:AB_2534095 |
| Anti-Mouse IgG AlexaFluor 488 conjugated | Thermo Fisher | Cat# A-11029, RRID:AB_138404 |
| Anti-Mouse IgG AlexaFluor 594 conjugated | Thermo Fisher | Cat# A-11005, RRID:AB_141372 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Fibronectin human plasma | Sigma-Aldrich | Cat# F0895 |
| Paraformaldehyde | Electron Microscopy Sciences | Cat# 15714 |
| Triton X-100 | Sigma-Aldrich | Cat# T8787 |
| Disuccinimidyl suberate | Thermo Fisher | Cat# 21555 |
| Proteinases inhibitor cocktail | Thermo Fisher | Cat# 78430 |
| Phosphatases inhibitor cocktail | Thermo Fisher | Cat# 78420 |
| 4’,6-diamidino-2-phenylindole (DAPI) | Thermo Fisher | Cat# D1306 |
| Hoechst 33342 | Thermo Fisher | Cat# H3570 |
| Nocodazole | Sigma-Aldrich | Cat# M1404 |
| Critical Commercial Assays | ||
| Pierce™ c-Myc-Tag IP/Co-IP Kit | Thermo Fisher | Cat# 23630 |
| Halo-Trap kit | Chromotek | Cat# otak-20 |
| Experimental Models: Cell Lines | ||
| HeLa (Wild type) | ATCC | Cat# CCL-2 |
| HeLa, WDR24-KO | (Cai et al., 2016) | N/A |
| HeLa, WDR59-KO | This paper | N/A |
| HeLa, MIOS-KO | This paper | N/A |
| Experimental Models: Drosophila Stocks | ||
| seh1Δ15 | (Senger et al., 2011) | N/A |
| UAS-Wdr24 RNAi | Bloomington Drosophila Stock Center | Cat# 62393 |
| UAS-RagA RNAi | Bloomington Drosophila Stock Center | Cat# 34590 |
| UAS-RagC RNAi | Bloomington Drosophila Stock Center | Cat# 32342 |
| UAS-mCherry RNAi | Bloomington Drosophila Stock Center | Cat# 35787 |
| Nanos-Gal4 | (Van Doren et al., 1998) | N/A |
| TSC1-GFP | Vienna Drosophila Resource Center | Cat# 318377 |
| Lamp1–3xmCherry | (Hegedus et al., 2016) | N/A |
| Oligonucleotides | ||
| ON-TARGETplus Non-targeting siRNA control | GE Healthcare | D-001810-01-05 |
| SMARTpool: ON-TARGETplus LAMTOR1 siRNA | GE Healthcare | L-020916-02-0005 |
| SMARTpool: ON-TARGETplus RRAGA siRNA | GE Healthcare | L-016070-00-0005 |
| SMARTpool: ON-TARGETplus RRAGC siRNA | GE Healthcare | L-017822-01-0005 |
| Recombinant DNA | ||
| FLAG-TSC2 | (Tee et al., 2002) | Addgene plasmid #8996 |
| mPlum-LAMP1 | (Shaner et al., 2008) | Addgene plasmid #55982 |
| mEOS4b-TSC2 | This paper | N/A |
| Halo-TSC2 | This paper | N/A |
| Halo-TSC2 T1462A | This paper | N/A |
| Halo-TSC2 S939A | This paper | N/A |
| Halo-TSC2 T1462A/S939A | This paper | N/A |
| FLAG-RAGA(T21N) | (Shen et al., 2017) | Addgene plasmid #99715 |
| FLAG-RAGA(Q66L) | (Shen et al., 2017) | Addgene plasmid #99716 |
| FLAG-RAGC(Q120L) | (Shen et al., 2017) | Addgene plasmid #99725 |
| FLAG-RAGC(S75N) | (Shen et al., 2017) | Addgene plasmid #112757 |
| HA-GST-RHEB1 | (Sancak et al., 2007) | Addgene plasmid #14951 |
| Myc-mTOR | (Sarbassov et al., 2004) | Addgene plasmid #1861 |
| HA-RAGA | (Shen et al., 2017) | Addgene plasmid #99710 |
| FLAG-RAGC | (Shen et al., 2017) | Addgene plasmid #99723 |
| Software and Algorithms | ||
| Microsoft Excel | Microsoft | https://products.office.com/en-us/excel |
| GraphPad Prism 8 | GraphPad Software Inc. | https://www.graphpad.com/scientificsoftware/prism/ |
| Imaris 9.3.0 | Bitplane Inc. | https://imaris.oxinst.com/products/imaris-forcell-biologists |
| ImageJ | National Institutes of Health | https://imagej.nih.gov/ij/ |
| Photoshop CC | Adobe | https://www.adobe.com/products/photoshop.ht ml |
| Other | ||
| Dulbecco’s modified Eagle’s medium GlutaMAX™-I with pyruvate | Thermo Fisher | Cat# 10569010 |
| Fetal bovine serum | Thermo Fisher | Cat# A3160602 |
| Penicillin-Streptomycin | Thermo Fisher | Cat# 15140122 |
| DMEM without amino acids | MyBioSource | Cat# MBS653087 |
| Fetal bovine serum, dialyzed | Thermo Fisher | Cat# A3382001 |
| TrypLE™ Express | Thermo Fisher | Cat# 12605010 |
| Xfect™ Transfection reagent | Takara | Cat# 631318 |
| Xfect™ RNA Transfection reagent | Takara | Cat# 631450 |
| Dynabeads™ Protein A | Thermo Fisher | Cat# 1001D |
| M-PER mammalian protein extraction buffer | Thermo Fisher | Cat# 78503 |
| Pierce™ Protein-Free T20 (TBS) Blocking Buffer | Thermo Fisher | Cat# 37571 |
| Clarity™ Western ECL substrate | Biorad | Cat# 170–5060 |
Highlights.
The Rag GTPase recruits TSC to lysosomes in response to growth factor restriction
The Rag GTPase functions as an inhibitor of TORC1 in GATOR2 knockouts
GATOR2 opposes the Rag-GTPase-dependent cycling of TSC on and off lysosomes
GATOR2 promotes the phosphorylation of AKT upstream of the Rag GTPase
ACKNOWLEDGEMENTS
Multiple stocks used in this study were obtained from the Bloomington Drosophila Stock Center supported by NIH Grant P40OD018537. We thank Juan Bonifacino and Eric Snapp for comments on the manuscript. Research supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development Intramural Research program (to M.A.L., HD00163 16).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Avruch J, Hara K, Lin Y, Liu M, Long X, Ortiz-Vega S, and Yonezawa K. (2006). Insulin and amino-acid regulation of mTOR signaling and kinase activity through the Rheb GTPase. Oncogene 25, 6361–6372. [DOI] [PubMed] [Google Scholar]
- Aylett CH, Sauer E, Imseng S, Boehringer D, Hall MN, Ban N, and Maier T. (2016). Architecture of human mTOR complex 1. Science 351, 48–52. [DOI] [PubMed] [Google Scholar]
- Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, and Sabatini DM (2013). A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 340, 1100–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L, and Sabatini DM (2014). Regulation of mTORC1 by amino acids. Trends Cell Biol 24, 400–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binda M, Peli-Gulli MP, Bonfils G, Panchaud N, Urban J, Sturgill TW, Loewith R, and De Virgilio C. (2009). The Vam6 GEF controls TORC1 by activating the EGO complex. Mol Cell 35, 563–573. [DOI] [PubMed] [Google Scholar]
- Cai SL, Tee AR, Short JD, Bergeron JM, Kim J, Shen J, Guo R, Johnson CL, Kiguchi K, and Walker CL (2006). Activity of TSC2 is inhibited by AKT-mediated phosphorylation and membrane partitioning. J Cell Biol 173, 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W, Wei Y, Jarnik M, Reich J, and Lilly MA (2016). The GATOR2 Component Wdr24 Regulates TORC1 Activity and Lysosome Function. PLoS Genet 12, e1006036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll B, Maetzel D, Maddocks OD, Otten G, Ratcliff M, Smith GR, Dunlop EA, Passos JF, Davies OR, Jaenisch R, et al. (2016). Control of TSC2-Rheb signaling axis by arginine regulates mTORC1 activity. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornu M, Albert V, and Hall MN (2013). mTOR in aging, metabolism, and cancer. Curr Opin Genet Dev 23, 53–62. [DOI] [PubMed] [Google Scholar]
- Curatolo P, Bombardieri R, and Jozwiak S. (2008). Tuberous sclerosis. Lancet 372, 657–668. [DOI] [PubMed] [Google Scholar]
- Dan HC, Sun M, Yang L, Feldman RI, Sui XM, Ou CC, Nellist M, Yeung RS, Halley DJ, Nicosia SV, et al. (2002). Phosphatidylinositol 3-kinase/Akt pathway regulates tuberous sclerosis tumor suppressor complex by phosphorylation of tuberin. J Biol Chem 277, 35364–35370. [DOI] [PubMed] [Google Scholar]
- Demetriades C, Doumpas N, and Teleman AA (2014). Regulation of TORC1 in response to amino acid starvation via lysosomal recruitment of TSC2. Cell 156, 786–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demetriades C, Plescher M, and Teleman AA (2016). Lysosomal recruitment of TSC2 is a universal response to cellular stress. Nat Commun 7, 10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibble CC, and Manning BD (2013). Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat Cell Biol 15, 555–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokudovskaya S, and Rout MP (2011). A novel coatomer-related SEA complex dynamically associates with the vacuole in yeast and is implicated in the response to nitrogen starvation. Autophagy 7, 1392–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokudovskaya S, and Rout MP (2015). SEA you later alli-GATOR - a dynamic regulator of the TORC1 stress response pathway. J Cell Sci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efeyan A, and Sabatini DM (2010). mTOR and cancer: many loops in one pathway. Curr Opin Cell Biol 22, 169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley IG, Lam du H, Wang J, Ding X, Chen S, and Jiang X. (2009). ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem 284, 12297–12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, and Pan D. (2001). TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth. Genes Dev 15, 1383–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Zhang Y, Arrazola P, Hino O, Kobayashi T, Yeung RS, Ru B, and Pan D. (2002). Tsc tumour suppressor proteins antagonize amino-acid-TOR signalling. Nat Cell Biol 4, 699–704. [DOI] [PubMed] [Google Scholar]
- Hara K, Yonezawa K, Weng QP, Kozlowski MT, Belham C, and Avruch J. (1998). Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem 273, 14484–14494. [DOI] [PubMed] [Google Scholar]
- Hegedus K, Takats S, Boda A, Jipa A, Nagy P, Varga K, Kovacs AL, and Juhasz G. (2016). The Ccz1-Mon1-Rab7 module and Rab5 control distinct steps of autophagy. Mol Biol Cell 27, 3132–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, et al. (2009). Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell 20, 1981–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Li Y, Xu T, and Guan KL (2003). Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev 17, 1829–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Li Y, Zhu T, Wu J, and Guan KL (2002). TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol 4, 648–657. [DOI] [PubMed] [Google Scholar]
- Jewell JL, Kim YC, Russell RC, Yu FX, Park HW, Plouffe SW, Tagliabracci VS, and Guan KL (2015). Metabolism. Differential regulation of mTORC1 by leucine and glutamine. Science 347, 194–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Goraksha-Hicks P, Li L, Neufeld TP, and Guan KL (2008). Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol 10, 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, and Guan KL (2019). mTOR as a central hub of nutrient signalling and cell growth. Nat Cell Biol 21, 63–71. [DOI] [PubMed] [Google Scholar]
- Laplante M, and Sabatini DM (2012). mTOR signaling in growth control and disease. Cell 149, 274–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence RE, Cho KF, Rappold R, Thrun A, Tofaute M, Kim DJ, Moldavski O, Hurley JH, and Zoncu R. (2018). A nutrient-induced affinity switch controls mTORC1 activation by its Rag GTPase-Ragulator lysosomal scaffold. Nat Cell Biol 20, 1052–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence RE, and Zoncu R. (2019). The lysosome as a cellular centre for signalling, metabolism and quality control. Nat Cell Biol 21, 133–142. [DOI] [PubMed] [Google Scholar]
- Lee M, Kim JH, Yoon I, Lee C, Fallahi Sichani M, Kang JS, Kang J, Guo M, Lee KY, Han G, et al. (2018). Coordination of the leucine-sensing Rag GTPase cycle by leucyl-tRNA synthetase in the mTORC1 signaling pathway. Proc Natl Acad Sci U S A 115, E5279–E5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Snapp EL, and Phair RD (2018). The Development and Enhancement of FRAP as a Key Tool for Investigating Protein Dynamics. Biophys J 115, 1146–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning BD, Tee AR, Logsdon MN, Blenis J, and Cantley LC (2002). Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell 10, 151–162. [DOI] [PubMed] [Google Scholar]
- Manning BD, and Toker A. (2017). AKT/PKB Signaling: Navigating the Network. Cell 169, 381–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon S, Dibble CC, Talbott G, Hoxhaj G, Valvezan AJ, Takahashi H, Cantley LC, and Manning BD (2014). Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell 156, 771–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossmann D, Park S, and Hall MN (2018). mTOR signalling and cellular metabolism are mutual determinants in cancer. Nat Rev Cancer 18, 744–757. [DOI] [PubMed] [Google Scholar]
- Nicastro R, Sardu A, Panchaud N, and De Virgilio C. (2017). The Architecture of the Rag GTPase Signaling Network. Biomolecules 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez-Segala MG, Sun MG, Shtengel G, Viswanathan S, Baird MA, Macklin JJ, Patel R, Allen JR, Howe ES, Piszczek G, et al. (2015). Fixation-resistant photoactivatable fluorescent proteins for CLEM. Nat Methods 12, 215–218, 214 p following 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchaud N, Peli-Gulli MP, and De Virgilio C. (2013a). Amino Acid Deprivation Inhibits TORC1 Through a GTPase-Activating Protein Complex for the Rag Family GTPase Gtr1. Sci Signal 6, ra42. [DOI] [PubMed] [Google Scholar]
- Panchaud N, Peli-Gulli MP, and De Virgilio C. (2013b). Amino acid deprivation inhibits TORC1 through a GTPase-activating protein complex for the Rag family GTPase Gtr1. Sci Signal 6, ra42. [DOI] [PubMed] [Google Scholar]
- Plas DR, and Thompson CB (2003). Akt activation promotes degradation of tuberin and FOXO3a via the proteasome. J Biol Chem 278, 12361–12366. [DOI] [PubMed] [Google Scholar]
- Potter CJ, Pedraza LG, and Xu T. (2002). Akt regulates growth by directly phosphorylating Tsc2. Nat Cell Biol 4, 658–665. [DOI] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, and Zhang F. (2013). Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8, 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roccio M, Bos JL, and Zwartkruis FJ (2006). Regulation of the small GTPase Rheb by amino acids. Oncogene 25, 657–664. [DOI] [PubMed] [Google Scholar]
- Sabatini DM (2017). Twenty-five years of mTOR: Uncovering the link from nutrients to growth. Proc Natl Acad Sci U S A 114, 11818–11825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, and Sabatini DM. (2010). Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, and Sabatini DM (2008). The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, and Sabatini DM (2007). PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell 25, 903–915. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, and Sabatini DM (2004). Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol 14, 1296–1302. [DOI] [PubMed] [Google Scholar]
- Saxton RA, and Sabatini DM (2017). mTOR Signaling in Growth, Metabolism, and Disease. Cell 169, 361–371. [DOI] [PubMed] [Google Scholar]
- Schalm SS, Fingar DC, Sabatini DM, and Blenis J. (2003). TOS motif-mediated raptor binding regulates 4E-BP1 multisite phosphorylation and function. Curr Biol 13, 797–806. [DOI] [PubMed] [Google Scholar]
- Senger S, Csokmay J, Akbar T, Jones TI, Sengupta P, and Lilly MA (2011). The nucleoporin Seh1 forms a complex with Mio and serves an essential tissue-specific function in Drosophila oogenesis. Development 138, 2133–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL, Davidson MW, and Tsien RY (2008). Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods 5, 545–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K, Choe A, and Sabatini DM (2017). Intersubunit Crosstalk in the Rag GTPase Heterodimer Enables mTORC1 to Respond Rapidly to Amino Acid Availability. Mol Cell 68, 821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K, and Sabatini DM (2018). Ragulator and SLC38A9 activate the Rag GTPases through noncanonical GEF mechanisms. Proc Natl Acad Sci U S A 115, 9545–9550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K, Valenstein ML, Gu X, and Sabatini DM (2019). Arg-78 of Nprl2 catalyzes GATOR1-stimulated GTP hydrolysis by the Rag GTPases. J Biol Chem 294, 2970–2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EM, Finn SG, Tee AR, Browne GJ, and Proud CG (2005). The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. J Biol Chem 280, 18717–18727. [DOI] [PubMed] [Google Scholar]
- Sprague BL, and McNally JG (2005). FRAP analysis of binding: proper and fitting. Trends Cell Biol 15, 84–91. [DOI] [PubMed] [Google Scholar]
- Tee AR, Fingar DC, Manning BD, Kwiatkowski DJ, Cantley LC, and Blenis J. (2002). Tuberous sclerosis complex-1 and −2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc Natl Acad Sci U S A 99, 13571–13576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doren M, Williamson AL, and Lehmann R. (1998). Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr Biol 8, 243–246. [DOI] [PubMed] [Google Scholar]
- Wei Y, and Lilly MA (2014). The TORC1 inhibitors Nprl2 and Nprl3 mediate an adaptive response to amino-acid starvation in Drosophila. Cell Death Differ 21, 1460–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Reveal B, Reich J, Laursen WJ, Senger S, Akbar T, Iida-Jones T, Cai W, Jarnik M, and Lilly MA (2014). TORC1 regulators Iml1/GATOR1 and GATOR2 control meiotic entry and oocyte development in Drosophila. Proc Natl Acad Sci U S A 111, E5670–5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RI, Turner GC, and Laurent G. (2004). Transformation of olfactory representations in the Drosophila antennal lobe. Science 303, 366–370. [DOI] [PubMed] [Google Scholar]
- Yang H, Jiang X, Li B, Yang HJ, Miller M, Yang A, Dhar A, and Pavletich NP (2017). Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40. Nature 552, 368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng CY, Petralia RS, Wang YX, and Kachar B. (2011). Fluorescence recovery after photobleaching (FRAP) of fluorescence tagged proteins in dendritic spines of cultured hippocampal neurons. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original data are deposited in Mendeley Data: http://dx.doi.org/10.17632/n8gztd29sv.1