Abstract
Methylmercury (MeHg) can elicit cognitive and motor deficits due to its developmental neuro- and myotoxic properties. While previous work has demonstrated that Nrf2 antioxidant signaling protects from MeHg toxicity, in vivo tissue-specific studies are lacking. In Drosophila, MeHg exposure shows greatest developmental toxicity in the pupal stage resulting in failed eclosion (emergence of adults) and an accompanying ‘myosphere’ phenotype in indirect flight muscles (IFMs). To delineate tissue-specific contributions to MeHg-induced motor deficits, we investigated the potential of Nrf2 signaling in either muscles or neurons to moderate MeHg toxicity. Larva were exposed to various concentrations of MeHg (0–20μM in food) in combination with genetic modulation of the Nrf2 homolog cap-n-collar C (CncC), or its negative regulator Keap1. Eclosion behavior was evaluated in parallel with the morphology of two muscle groups, the thoracic IFMs and the abdominal dorsal internal oblique muscles (DIOMs). CncC signaling activity was reported with an antioxidant response element construct (ARE-GFP). We observed that DIOMs are distinguished by elevated endogenous ARE-GFP expression, which is only transiently seen in the IFMs. Dose-dependent MeHg reductions in eclosion behavior parallel formation of myospheres in the DIOMs and IFMs, while also increasing ARE-GFP expression in the DIOMs. Modulating CncC signaling via muscle-specific Keap1 knockdown and upregulation gives a rescue and exacerbation, respectively, of MeHg effects on eclosion and myospheres. Interestingly, muscle-specific CncC upregulation and knockdown both induce lethality. In contrast, neuron-specific upregulation of CncC, as well as Keap1 knockdown, rescued MeHg effects on eclosion and myospheres. Our findings indicate that enhanced CncC signaling localized to either muscles or neurons is sufficient to rescue muscle development and neuromuscular function from a MeHg insult. Additionally, there may be distinct roles for CncC signaling in myo-morphogenesis.
Keywords: Methylmercury, Nrf2 signaling, neuromuscular development, myotoxicity, Drosophila
1. INTRODUCTION
Methylmercury (MeHg) is a pervasive global pollutant that can accumulate in fish and seafood, which may in turn pose risks of toxicity for humans. (Chen et al. 2008; Cunningham et al. 2003; Kim and Zoh 2012). Historical accidental poisonings have shown that the developing fetus is an especially sensitive MeHg target (Bakir et al. 1973; Bose-O’Reilly et al. 2010; Harada 1995). At a sufficient prenatal dose, MeHg can elicit developmental deficits, including mental retardation, ataxia, muscle weakness, decreased muscle tone, and altered reflexes (Bakir et al. 1973; Harada 1995; McKeown-Eyssen et al. 1983). These clinical features are consistent with the multiple neurotoxic mechanisms ascribed to MeHg including inhibiting neuroblast proliferation and differentiation (Burke et al. 2006; Fujimura et al. 2012; Fujimura and Usuki 2015a; Rodier et al. 1984; Sager et al. 1984), neurite outgrowth (Fujimura and Usuki 2015b; Fujimura et al. 2016), and neurotransmission (Aschner et al. 2007; Carratu et al. 2006). However, accumulating evidence from cell culture and small organism models such as Drosophila melanogaster and zebrafish, as well as rodents, demonstrates MeHg can act directly on developing skeletal muscle. Indeed, MeHg is reported to inhibit myobast differentiation (Culbreth and Rand 2020; Prince and Rand 2018), alter muscle-specific gene expression (Prince and Rand 2017), pertub myotendonous juction formation (Peppriell et al. 2020), alter muscle fiber diameter (Usuki et al. 1998), and disrupt mitochondrial organization and enzyamtic activity (Cambier et al. 2009). Reduced performance in motor behaviors with developmental MeHg exposures in animal models has been reported numerous times (Dare et al. 2003; Fujimura et al. 2012; Spyker et al. 1972). Yet, the relative contribution of distinct neurotoxic and myotoxic mechanisms to these motor deficits remains to be characterized.
Studies using Drosophila have implicated mechanisms of MeHg susceptibility in the developing neuromuscular system. Prior investigations aimed at identifying a genetic basis for susceptibility to MeHg toxicity have screened for effects on eclosion behavior, the neuromuscular act of the adult fly emerging from the pupal case at the completion of metamorphosis (Magnusson and Ramel 1986; Montgomery et al. 2014). Parallel studies have reported developmental phenotypes in neurons and muscles in MeHg exposed fly embryos and pupae (Engel and Rand 2014; Prince and Rand 2017; Rand et al. 2009). A genome wide association study (GWAS) has identified muscle and neuromuscular development gene ontology categories that associate with MeHg tolerance (Montgomery et al. 2014). This GWAS study was also the first to describe a myo-morphogenesis defect whereby MeHg exposure causes the developing indirect flight muscles (IFMs) to condense into rounded “myospheres”. Subsequent studies have used transgenic expression systems to upregulate the multi-drug resistance like protein dMRP, and enzymes related to glutathione synthesis (glutamyl-cysteine ligase catalytic subunit, GCLc) and metabolism (glutathione-S-transferases (GSTs)) in a muscle-specific manner to rescue MeHg effects on eclosion behavior while also preventing MeHg-induced myospheres in the IFMs (Prince and Rand 2017; Rand et al. 2019; Vorojeikina et al. 2017). Interestingly, overexpression of GCLc or dMRP in a neural-specific manner shows moderate to no rescue of MeHg effects on eclosion (Prince and Rand 2017; Rand et al. 2019). These findings may underscore differences in MeHg susceptibility between nascent muscles and neurons. Intriguingly, they also point to the potential regulation of MeHg toxicity via the antioxidant response element (ARE) pathway, given that these genes are regulated by the Nrf2 transcription factor (Hayes and Dinkova-Kostova 2014). Consistent with this notion, global Nrf2 overexpression in Drosophila embryos can rescue their development from a toxic MeHg exposure (Rand et al. 2009).
The Nrf2 pathway is a highly conserved, inducible antioxidant response mechanism. Under basal conditions, the Nrf2 transcription factor is sequestered to the cytoplasm and constitutively targeted for degradation via interaction with its negative regulator, Keap1. Xenobiotic- and oxidative stress-dependent modification of Keap1 cysteine residues decreases Nrf2 turnover, facilitating Nrf2 accumulation and translocation to the nucleus where it subsequently binds to ARE sequences upstream of cytoprotective genes, including heme oxygenase-1 (Ho), GCL subunits, GSTs, and MRPs (Kumagai et al. 2013; Wakabayashi et al. 2004). Nrf2 signaling is conserved in Drosophila; it is encoded by the cap-n’-collar C (CncC) gene. The CncC protein partners with Drosophila Keap1 for negative regulation, and responds to oxidants by activating genes via binding to conserved sequences of the ARE (Chatterjee and Bohmann 2012; Pitoniak and Bohmann 2015; Sykiotis and Bohmann 2008). MeHg activates the mammalian Nrf2 pathway both in vitro and in vivo, to increase Nrf2-target genes such as heme oxygenase-1, NAD(P)H quinone dehydrogenase-1, GCLc, and Nrf2 itself (Culbreth et al. 2017; Feng et al. 2017; Ni et al. 2011; Wang et al. 2009). Nrf2-MeHg interaction studies have been mostly limited to cell culture systems, where Nrf2 activity can elicit a protective effect (Ni et al. 2010; Toyama et al. 2007). Studies in vivo also indicate that Nrf2 signaling protects neuromotor behaviors and lifespan from MeHg toxicity. (Akiyama et al. 2019; Toyama et al. 2011). However, these findings rely on an experimental design of global Nrf2 knockout, which limits the understanding of a tissue-specific role for Nrf2 signals in moderating MeHg toxicity during development.
In this study, we use Drosophila to investigate a role for Nrf2 signaling, here in after referred to as CncC signaling, to moderate MeHg toxicity explicitly at the level of the developing muscle and/or neuron. Examining combinations of MeHg exposure and genetic modulation of CncC signaling, we evaluate outcomes in the morphology of two distinct muscle groups in the developing fly: the IFMs in the thorax and the dorsal internal oblique muscles (DIOMs) in the developing adult abdomen. These morphological effects are correlated with eclosion behavior. We find that the CncC activity can rescue muscle development and neuromuscular function from MeHg toxicity, which can be initiated independently by CncC activity localized to the muscle or the neural component of the motor unit. Furthermore, attributes of CncC activity in the developing muscle highlight a novel role in muscle development and morphogenesis.
2. MATERIALS AND METHODS
Drosophila Stocks
Targeted manipualtions of gene expression were carried out using the conventional GAL4>UAS heterologous gene expression system in Drosophila (Brand and Perrimon 1993). The numerous Drosophila strains utilized in this study with their corresponding purpose and source are summarized in in Table S1 in Online Resource 2. In general, targeted manipulations of expression utilized GAL4 driver lines under the control of either a muscle specific promoter/enhancer (e.g. mycocyte enhancer factor 2 (Mef2) or myosin heavy chain (MHC)) or a neural specific promoter/enhancer (Elav or OK371 GAL4 drivers; See Table S1). CncC/Nrf2 related activity was modulated with upstream activating sequence (UAS) directed expression of either CncC or Keap1 or the corresponding RNAi hairpin construct for knockdown of expression ((Sykiotis and Bohmann 2008) and see Table S1). Additional fly lines used include a Cnc-GAL4 line (Mi{Trojan-GAL4.1cnc}MI12862-TG4.1), which was generated with a Minos-mediated integration cassette (MiMIC) within an intron of the cnc gene (MI12862) that was exchanged with a SA-T2A-GAL4-polA-containing cassette through PhiC31 integrase-mediated recombinase cassette exchange (Diao et al. 2015), personal communication with J. Wang of the Bellen lab (Baylor College of Medicine). The Mef2-RFP-GAL4, Cnc-RFP-GAL4, and Cnc-GFP-GAL4 recombinant strains were created by inducing recombination at 29°C with the respective GAL4 and UAS reporter strains. The antioxidant response element reporter (ARE-GFP) strain contains four tandem ARE sequences upstream of GFP with a nuclear localization sequences (NLS) (Chatterjee and Bohmann 2012). A recombinant strain carrying Mef2GAL4, ARE-GFP and a temperature sensitive GAL80 gene (GAL80TS) for temperature regulated expression, was created by and gifted from the Bohmann lab (University of Rochester). Flies were maintained on a 12/12-hour light-dark cycle in a 25°C humidified chamber on standard fly food made of cornmeal, molasses, yeast, and agar.
To achieve CncC/Nrf2 related gene expression modualtion various combinations of muscle- and neural-specific GAL4 drivers were crossed with UAS responder lines constructs. Flies carrying ARE-GFP were used independently to assess MeHg effects or in conjunction with various GAL4>UAS combinations to assess transgene modulation of CncC and Keap1 expression on endogenous CncC pathway activity. To demonstrate transgene activation of the endoegnous CncC pathway, ARE-GFP; Mef2-GAL4, Tub-GAL80TS virgins were crossed with various UAS-CncC or UAS-Keap1 constructs in males to generate indicated genotypes. Development of F1 progeny occurred at 25°C to maintain the GAL80TS repression. Pupae were staged at 0 hours after puparium formation (h APF) and maintained at 25°C until 12h APF, which is the average time for head eversion to occur. These pupae were then transferred to 29°C and incubated for 36 hours to relieve the GAL80TS repression of the Mef2-GAL4 driver and induce expression of the UAS construct. Imaging of the ARE-GFP fluorescence was performed and, in parallel, 10 pupa were pooled for protein extraction and Western blot analyses. Staged-matched controls reared at 25°C for each genotype were also imaged and collected for protein extraction. GFP protein quantification was done via SDS-PAGE and Western blotting methods (see below).
Eclosion Assays
MeHg tolerance in the various Drosophila genotypes were assayed using the previously described eclosion assay (Mahapatra et al. 2010; Rand et al. 2014). To generate F1 progeny of indicated genotypes, mating populations were composed of virgin GAL4 females (~150) and UAS males (~50) and placed in small mating chambers with exchangeable grape-agar plates. Grape-agar plates were replaced approximately every 12 hours to capture embryos. F1 embryos were allowed to develop to first instar (L1) stage at 25°C. L1 larvae were seeded at a density of 50 L1 larvae/vial of food (Jazz Mix, Fisher Scientific, #AS153) containing 0, 5, 10, 15, or 20 μM MeHg (methylmercury chloride, Sigma-Aldrich, #215465), and allowed to develop for 13 days at 25°C. Doses were chosen to remain in context with previous work and to demonstrate a reproducible dose-response effect on eclosion ability. Technical triplicates for each MeHg concentration was accomplished by collecting L1 larvae from three separate embryo-containing grape-agar plates from the same mating population. After 13 days, the number of successful eclosed (complete emergence from puparium) flies were counted, with the maximum being 150 flies. Biological triplets were completed by obtaining L1 larvae from three independent mating populations and seeding on three independent preparations of MeHg food. Viability eclosion assays were completed by using the above protocol, expect with the use of standard fly food containing no MeHg.
Pupal Staging and Harvesting
For MeHg experiments, pupae were staged based on the presence of physical characteristics that appear throughout metamorphosis, described in (Bainbridge and Bownes 1981), rather than based on hours APF as MeHg treatment slows development (unpublished observations). For experiments independent of MeHg, pupae were staged at 0h APF, such that subsequent imaging and sample collection would occur at the same developmental stage. Generally, p5, p6, p7, p8, and p9 pupal stages corresponds to 12–25, 25–43, 43–47, 47–57, 57–69h APF, respectively. Imaging, RT-qPCR, and Western blot samples were typically collected during p6-p8 stages.
Imaging and Muscle Phenotype Quantification
After larval exposure to MeHg, pupae were dissected from the puparium at the designated developmental stage or hours APF and placed dorsal up on double-stick tape on a glass microscope slide. Pupae were live imaged on a Nikon AZ100 Multizoom microscope (MVI, Avon MA). Within each experiment, at least 15 individuals per genotype per treatment of the same pupal stage or hours APF were imaged. Most experiments concerning muscle morphology used 0, 5, and 10 μM MeHg, as a rescue or exacerbation in eclosion ability is typically diminished by 15 μM MeHg treatment. Quantification of IFM phenotypes was based on one of two scoring criteria: the number of myospheres (“muscle ball”) per thorax or counting the number of missing dorsoventral muscles (DVMs) and dorsal lateral muscles (DLMs) per thorax. When imaging from the dorsal side of the pupa, the number of DVMs and DLMs that are visible are 4 and 2, respectively. Quantification of DIOMs myospheres focused on the most medial DIOMs in abdominal segments A2-A5, causing the maximum number of potential myospheres per abdomen to be 8. To reduce bias in scoring muscle dysmorphology, images were blinded and randomized. Data is represented as the mean number of myospheres or muscles missing per body region of interest.
ImageJ (NIH) was used for the determination of DIOMs nuclei fluorescence intensity in ARE-GFP pupa. Exposure to MeHg begun at L1 stage. Development occurred at 25°C. p5 ARE-GFP pupa were placed on double stick tape on a microscope slide, dissected from their puparium, and imaged on a Nikon AZ100 Multizoom microscope (MVI, Avon MA). Each image was captured at the same exposure and step size. TIF images were converted to 16-bit images. For each image, a single region of interest (ROI) composed of at least 100 individual DIOMs nuclei was measured, with measurement parameters including area, standard deviation, and integrated density. Selected nuclei were generally from the 10 most medial DIOMs in A1-A5 abdominal segments. An ROI composed of a cytosolic region in 3 random DIOMs determined an average background fluorescence of the DIOMs for each image. Corrected Total Cell Fluorescence (CTCF) equation was used to calculate the mean nuclei fluorescence, with the equation being CTCF = Intergrated Density − (Area of Nuceli ROI × Mean DIOMs Background Fluorescence). At least 5 abdomens per MeHg concentration were analyzed.
RT-qPCR
To measure MeHg-dependent pupal CncC pathway activation, 50 L1 Canton S larvae were seeded onto food containing either 0, 5, or 15 μM MeHg. Development occurred at 25°C. Pooled samples consisted of 10 p6 pupa for each MeHg treatment. Biological triplicates were completed from three different mating populations of Canton S and MeHg food preparations. Pooled pupae were homogenized in Trizol (Invitrogen) using a Kimble™ Kontes™ Pellet Pestle™ cordless motor (Fisher Scientific, #K749540–0000). Total RNA was extracted using Trizol and chloroform (Fisher Scientific, #BP1145–1). Total RNA was converted to cDNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, #4368813), according to manufacturer’s protocol. RT-qPCR was done using iTaq Universal SYBR® Green Supermix (Bio-Rad, #1725121) with forward and reverse primers specific for each gene of interest (Online Resource 3, represented 5’/3’), according to manufacturer’s protocol. Bio-Rad CFX96™ RealTime PCR System was programmed according to manufacturer instructions. Fold change was calculated using the 2-ΔΔCt method (Livak and Schmittgen 2001). The inter-sample ribosomal protein RP49 (L32) Cq value was subtracted from the gene of interest Cq value to calculate Delta Ct (ΔCt) values. RP49 is a commonly used reference gene for Drosophila and other insect gene expression analyses using qPCR (Deng and Kerppola 2013; Teng et al. 2012).
Western Blot
For quantification of GFP protein in ARE-GFP pupa after MeHg exposure, 50 L1 ARE-GFP larvae were seeded onto food containing 0, 5, 10, or 15 μM MeHg. Development occurred at 25°C. Pooled samples of 10 p6 pupa from each MeHg treatment were lysed in 4°C protein lysis buffer (50 mM Tris HCl, pH 7.4, 150 mM NaCl, 1% IGEPAL® CA-630 (MP Biomedicals, #198596)) and Halt Protease Inhibitor Cocktail EDTA-free (Thermo #87785) using a Kimble™ Kontes™ Pellet Pestle™ cordless motor (Fisher Scientific, #K749540–0000). Samples were frozen at −80°C until sample preparation. Biological triplicates were obtained by exposing ARE-GFP L1 larvae from three different mating populations to three separate preparations of MeHg food. Western blot samples were prepared in 2X Laemmli sample buffer (Bio-Rad #161–0737) and 2-mercaptoethanol (Fisher #BP176) with a final protein concentration of 1 mg/ml. Samples were boiled at 100°C. Samples (10 μl) and the Precision Plus Protein™ Kaleidoscope™ (Bio-Rad #161–0375) ladder (5 μl) were run on 12% polyacrylamide gels (30% acrylamide, 1.5 M Tris HCl, 10% SDS, ddH2O, 10% APS, TEMED). Gel transfer occurred at 4°C for overnight onto polyvinylidene difluoride (PVDF, Millipore #IPVH00010) membranes. Membranes were blocked with 1:1 Aquablock (EastCoast Bio, #PP82):1x PBS for 1 hour, then incubated overnight at 4°C with specific primary antibodies for protein of interest. Mouse anti-Actin (Developmental Studies Hybridoma Bank, #JLA20) antibody concentration was 0.5 μg/ml and rabbit anti-GFP (Torrey Pines Biolabs, #TP401) antibody dilution was 1:1000. Membranes were washed and then incubated for 1 hour at room temperature with appropriate secondary antibodies (dilution 1:5000), peroxidase-conjugated goat anti-mouse secondary (Jackson ImmunoResearch, #115–035-146) for actin and peroxidase-conjugated goat anti-rabbit secondary (Jackson ImmunoResearch, #111–005-003) for GFP. Protein bands were visualized with Clarity™ Western ECL Substrate (Bio-Rad #170–5060) with subsequent imaging on the Bio-Rad Chemi-Doc™ MP Imaging System. ImageJ (NIH) was used to quantify Western blot band intensity, with GFP band intensity normalized to respective MeHg treatment or genotype actin band prior to comparison between MeHg treatments and other genotypes. Sample preparation, blotting procedure, and analysis completed for the ARE-GFP; Mef2-GAL4, Tub-GAL80TS experiment followed the same protocol as above.
Statistical Analyses
All statistical analyses were completed in JMP Pro 14. Quantification of IFM and DIOMs dysmorphology were analyzed by either Mann-Whitney or Kruskal-Wallis nonparametric test with Dunn’s post-hoc for multiple comparisons between each genotype within a single MeHg treatment. At least 10 pupae per genotype and/or treatment was examined. For eclosion assays, a two-tailed Z-test within each MeHg concentration was performed categorically, due to the percent of flies successfully eclosed is a non-continuous value reaching 0% and 100% at the minima and maxima, respectively. The impact of MeHg on GFP protein levels and CncC pathway gene expression was analyzed by ANOVA with Tukey’s post hoc test for pair-wise comparisons. The impact of transgenic alterations in CncC and Keap1 expression GFP protein levels was analyzed by ANOVA with Tukey’s post-hoc with experimental crosses being compared to respective temperature control (w1118) cross. All experiments, except for IFM and DIOMs dysmorphology experiments, were done in biological triplicates. Data is presented as mean ± standard deviation of the mean (s.d.m.). Significance was defined as p ≤ 0.05.
3. RESULTS
Modulation of endogenous CncC pathway activity in DIOMs.
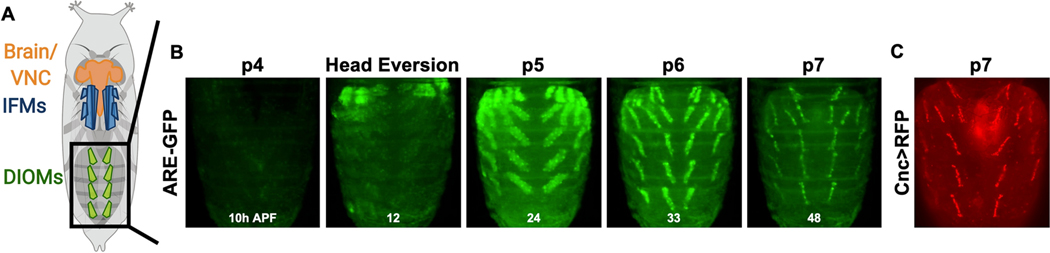
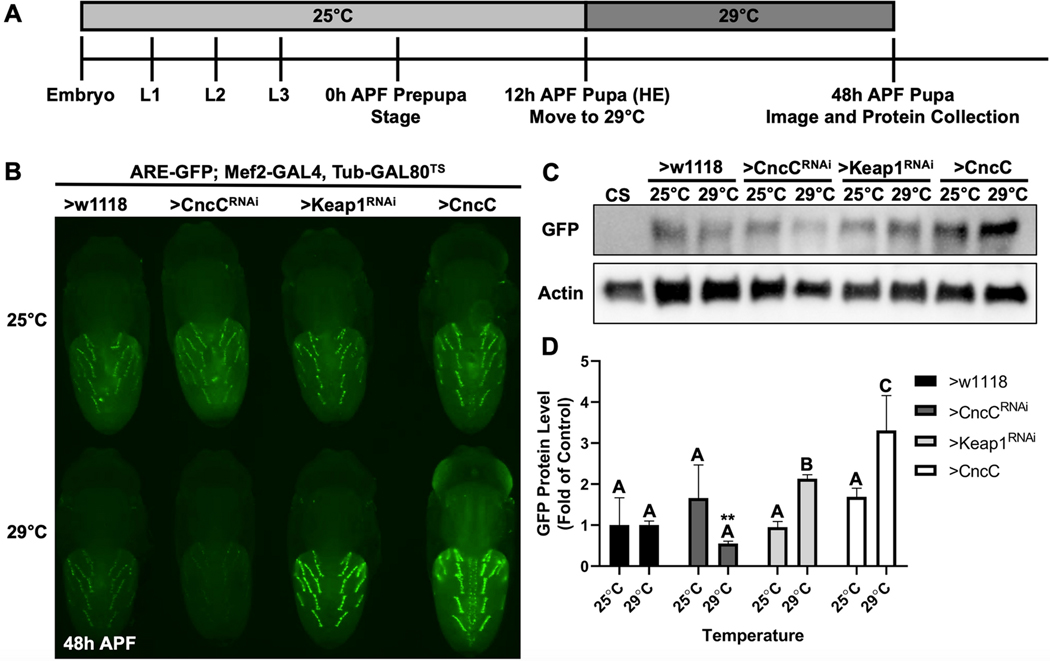
To characterize MeHg toxicity effects, we sought to define a system where muscle morphogensis could be evaluated in parallel with a motor behavior. We therefore focused on the DIOMs as these abdominal muscles are reported to be involved in eclosion behavior (Kimura and Truman 1990; Miller 1965). In a survey of reporters lines we unexpectedly found an ARE-GFP construct for the antioxidant response element (ARE) that localizes primarily to the DIOMs (Figure 1A, B; Online Resource 1, Movie S1). As a first approach to characterizing this system, we evaluated the developmental progression of DIOMs using time lapse microscopy, which revealed strong GFP expression that emerges in the DIOMs spanning the stages p4 to p7 (~10–48 h APF) (Figure 1A, B, Online Resource 1, Movie S1). While lower levels of GFP expression are seen in the larval template myotubes of the IFM precursors, this expression appears to be transient (Online Resource 1, Movie S1). DIOM GFP intensity is seen to peak at stages p5–6, highlighting progression of these muscles into a ‘remodelling’ phase to assume their mature anterior-posterior orientation, with stereotypically aligned nuclei at p7. This ARE expression profile was indictive of endogenous CncC signaling in these muscles. Consistent with this profile, the expression of a Cnc-GAL4>UAS-RFP reporter combination demonstrated a DIOM-restricted pattern (Figure 1C). To confirm if CncC activity is faithfully reported by the ARE-GFP construct, we modulated CncC and Keap1 expression levels in pupae via a temperature sensitive (TS) induction of a pan-muscle driver (Mef2-GAL4, TubGAL80TS) in developing flies carrying the ARE-GFP reporter (Figure 2). In control conditions, without induction of Keap or CncC modulation, the ARE-GFP reporter levels were nearly equal across all combinations (Figure 2B, top panel). Following induction of CncC knockdown, the fluorescence intensity of GFP decreased in the DIOMs, whereas with either Keap1 knockdown or CncC overexpression GFP intensity increased (Figure 2B, bottom panel). Western blotting for the GFP protein demonstrated a similar overall pattern as the fluorescence. CncC knockdown caused an apparent 50% reduction in GFP protein levels (Figure 2C, D; not significant by ANOVA, p<0.05 by Student’s t test with w1118 temperaturecontrol), while Keap1 knockdown and CncC overexpression caused significant increases of 2- and 3.5-fold in GFP protein, respectively (Fig 2C, D). Altogether, these data confirm that the ARE-GFP reporter effectively responds to CncC activity levels.
Fig. 1. Endogenous CncC signaling in DIOMs.
a A schematic of the pupal body plan. The brain and ventral nerve cord (VNC) are located in the head and extend into the thorax. The indirect flight muscles (IFMs) are located dorsally in the thorax. The dorsal internal oblique muscles (DIOMs) form parallel configurations in A1-A5 abdominal segments. b Expression of the ARE-GFP reporter in the DIOMs from stage p4 through p7 (~10–48h APF) (Also see Movie S1). Hours after puparium formation (APF) indicated at the bottom of each image. c Expression of Cnc-GAL4>UAS-RFP at stage p7.
Fig. 2. An in vivo ARE activity reporter is responsive to CncC and Keap1 expression.
a A schematic for inducing gene expression using the ARE-GFP; Mef2-GAL4, Tub-GAL80TS strain. b Representative images of non-induced and induced 48h APF pupae. Top row is of age-matched, temperature control pupa of each genotype. The bottom row is of pupa after a 36-hour 29°C induction starting at head eversion (HE). c Representative Western blot of GFP and actin protein from whole pupa protein extracts from each genotype and temperature. The Canton S (CS) wild-type strain acts as a negative control for GFP protein. d Quantification of GFP band intensities. CncC and Keap1 experimental groups are normalized to respective w1118 temperature controls (n = 3, mean ± s.d.m., One-way ANOVA with Tukey’s HSD, p<0.05, significant differences indicated by different letter labels). Comparison of 29°C w1118 and CncCRNAi band intensities by Student’s t test is statistically significant (**p<0.01).
MeHg disrupts DIOMs morphogenesis and activates the CncC pathway.
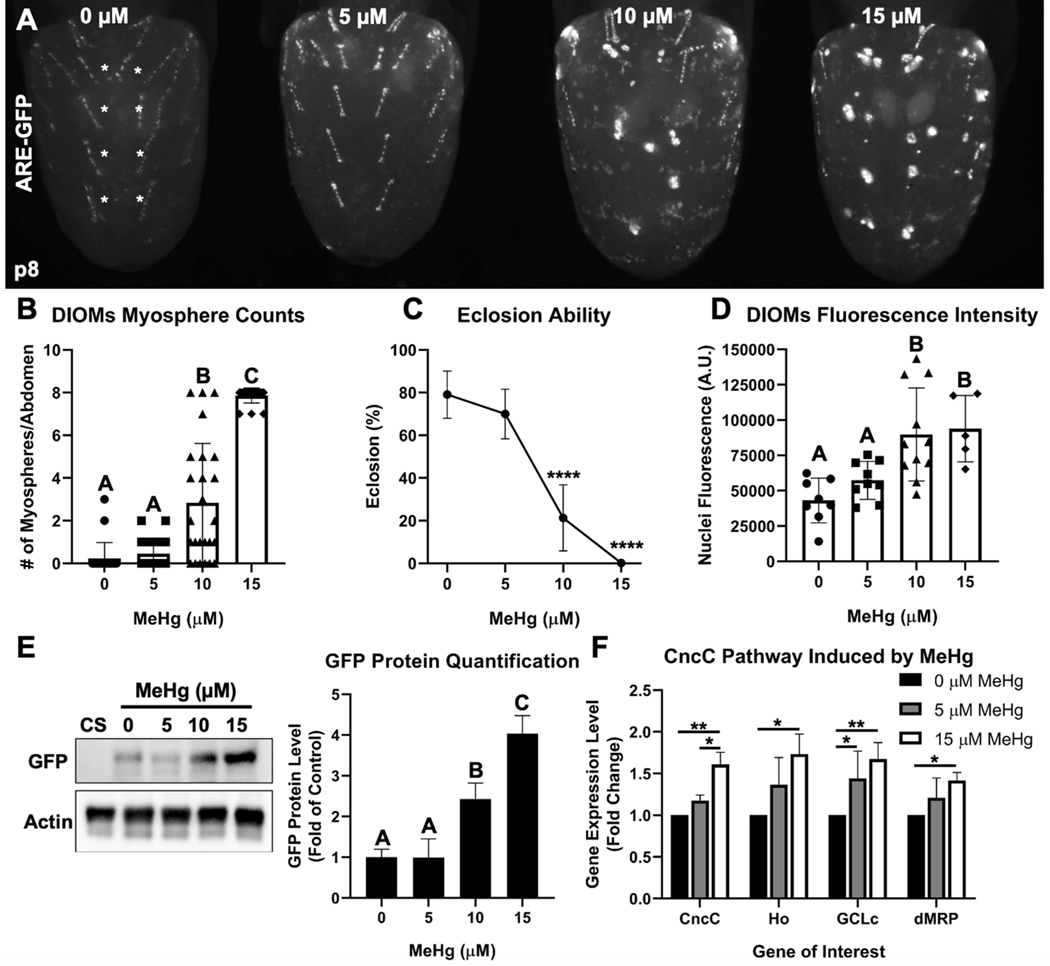
We next characterized the impact of MeHg exposure on DIOM morphology and CncC pathway activation. A dose-dependent increase in the number of myospheres in the DIOMs (Figure 3A, B) was seen to coincided with a decrease in eclosion ability (Figure 3C). Furthermore, we observed a significant dose-dependent increase in GPF intensity in nuclei at 10 and 15μM MeHg (Figure 3D, Online Resource 4) which corresponded with a 2.4- and 4-fold increase in GFP protein levels, respectively (Figure 3E). To further validate that CncC signaling is activated by MeHg, we quantified the transcript levels of CncC target genes Heme oxygenase (Ho), GCLc, dMRP, and CncC itself (Figure 3F). Whereas GCLc gene expression was significantly elevated at 5 μM MeHg, increased expression of all of these genes was observed at the 15 μM MeHg exposure. These data demonstrate that MeHg can compromise DIOM morphology to form myospheres which parallels a reduction in eclosion ability and an activation of the CncC pathway.
Fig. 3. MeHg induces DIOM myospheres and activates the CncC pathway.
a Representative image of DIOM morphology in ARE-GFP pupa after larval MeHg exposure. b Quantification of myospheres in most medial DIOMs in A2-A5 (denoted by the white asterisks) in ARE-GFP pupa upon larval MeHg exposure (n ≥ 22 abdomens/treatment, Kruskal-Wallis test with Dunn’s post-hoc for multiple comparisons, p<0.05, significant differences indicated by different letter labels). c Eclosion ability of ARE-GFP pupa upon larval MeHg exposure (n = 3, 450 flies/treatment, mean ± s.d.m., Z-test between each treatment group to 0 μM MeHg, ****p<0.0001). d Mean nuclei GFP fluorescence intensity (arbitrary units) of DIOMs upon larval MeHg exposure (100 individual nuclei/abdomen, n ≥ 5 abdomens/treatment). e Representative Western blot of GFP and actin protein bands from whole ARE-GFP pupa protein extracts upon larval MeHg exposure. The Canton S (CS) wild-type strain is a negative control for GFP protein. Quantification of GFP protein band intensities (normalized to actin) is represented as a bar graph (n = 3, d, e mean ± s.d.m., One-way ANOVA with Tukey’s HSD, p<0.05, significant differences indicated by different letter labels). f Gene expression by qPCR of CncC and CncC-regulated genes in Canton S pupa after larval exposure to 0, 5, or 15 μM MeHg (n = 3, mean ± s.d.m., One-way ANOVA with Tukey’s HSD, *p<0.05, **p<0.01).
Muscle-specific CncC rescue of eclosion and DIOM morphology from MeHg toxicity.
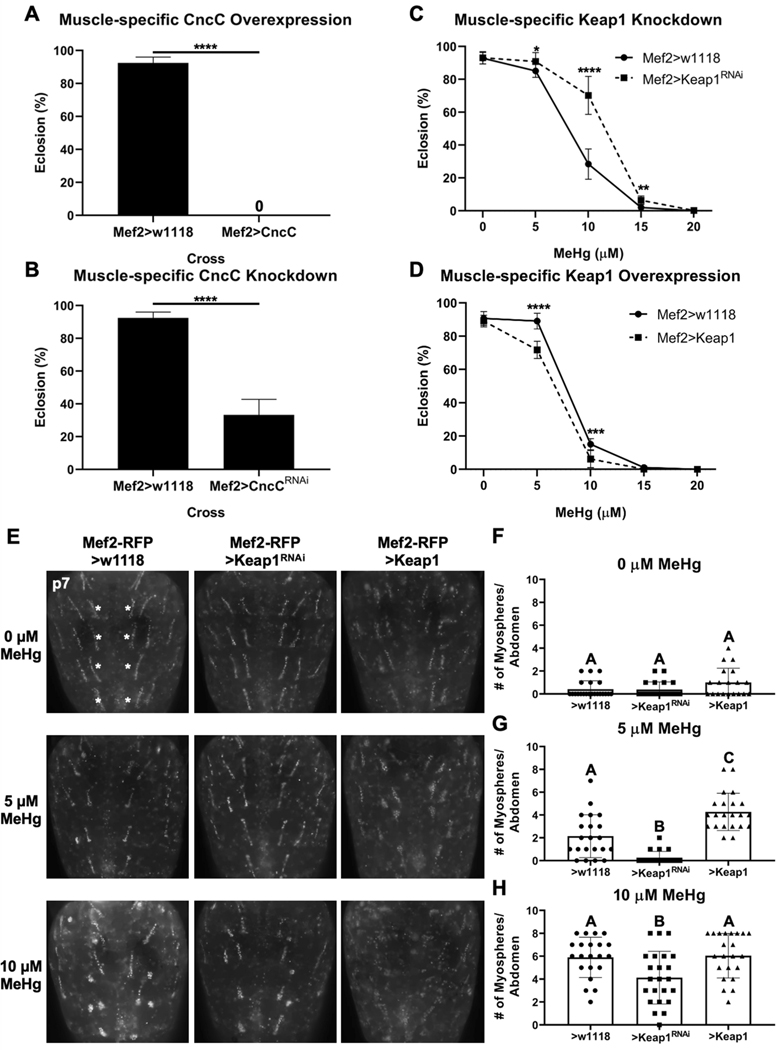
The above findings indicated that despite CncC pathway activation, DIOM morphogenesis and eclosion ability remain compromised following expoure to MeHg. We therefore investigated the possibility that muscle-specific enhancement of CncC signaling could counter the effects of MeHg. Following pan-muscle upregulation of CncC, we observed 100% lethality at the larval/early pupal stage that precluded eclosion (Figure 4A). Pan-muscle knockdown of CncC also proved toxic reducing eclosion ability by 70% (Figure 4B). To evaluate more moderate muscle-specific modulation of CncC, we targeted the expression of Keap1. Keap1 knockdown showed a rescue in eclosion ability, whereas and Keap1 overexpression slightly exacerbated eclosion defects following MeHg exposure (Figure 4C-D). We next examined if Keap1-mediated effects on eclosion paralleled changes in DIOM morphogenesis. Keap1 upregulation or knockdown alone did not alter the number of myospheres compared to control (Figure 4E top row, F). Exposure to 5 μM MeHg resulted in an average of 2.1 myospheres per abdomen (control), which was reduced with Keap1 knockdown (0.3 myospheres per abdomen) and increased with Keap1 overexpression (4.3 myospheres per abdomen) (Figure 4E middle row, G). A similar pattern was seen with 10 μM MeHg, with significant rescue of DIOM morphology with Keap1 knockdown, and a trend toward a more severe myosphere phenotype with Keap1 overexpression (Figure 4E bottom row, H). These effects of Keap1 on modulating the MeHg effect on DIOM morphology were also seen when expression was targeted to the DIOMs using the Cnc-GAL4 driver (Online Resource 5).
Fig. 4. Muscle-specific CncC signaling modulation of MeHg effects on eclosion ability and DIOM morphology.
a, b Effect of muscle-specific CncC overexpression (Mef2>CncC) and knockdown (Mef2>CncCRNAi) on eclosion. c, d Effect of Keap1 knockdown (>Keap1RNAi) and overexpression (>Keap1) on eclosion compared to control (Mef2>w1118) (a-d n = 3, 450 flies/genotype/treatment, mean ± s.d.m., Z-test, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001). e Representative images of MeHg effect on DIOM morphology without (>w1118 control) or with knockdown (>Keap1RNAi) or overexpression (>Keap1) of Keap1. Number of myospheres originating from the most medial DIOMs in the A2-A5 abdominal segments (indicated by white asterisks) were quantified: (f) 0 μM, (g) 5 μM, and (h) 10 μM MeHg (f-h n ≥ 20 abdomens/genotype/treatment, mean ± s.d.m., Kruskal-Wallis test with Dunn’s post-hoc for multiple comparisons, p<0.05, significant differences indicated by different letter labels).
The DIOMs are not required for eclosion.
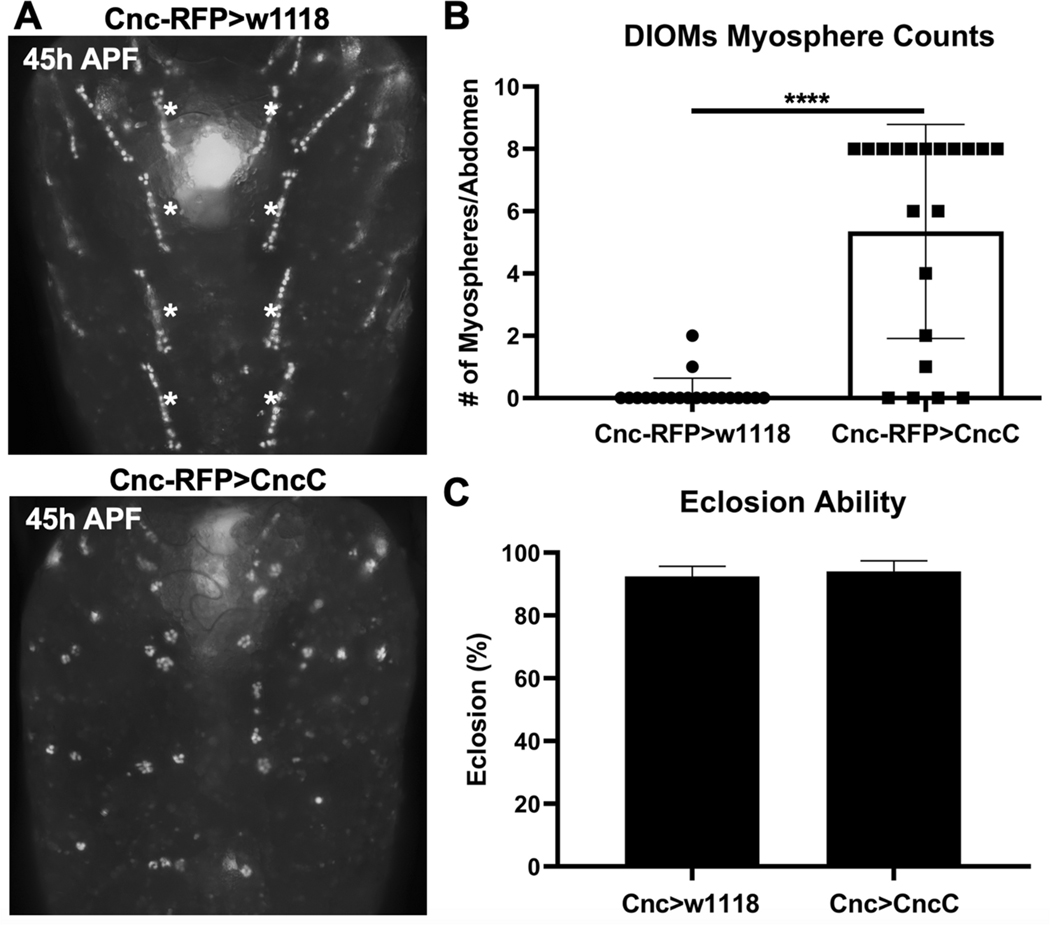
Since CncC overexpression with Mef2-GAL4 was early-pupal lethal (Figure 4A), we could not assess relevant myogenic phenotypes in the DIOMs. We therefore restricted CncC overexpression to the DIOMs using the Cnc-RFP-GAL4 driver to simultaneously overexpress CncC and visualize effects in DIOMs. CncC overexpression resulted in multiple myospheres (5.4 myospheres per abdomen) compared to controls (Figure 5A, B). Unexpectedly, DIOM perturbation through CncC overexpression did not alter eclosion ability (Figure 5D). This result demonstrated that while DIOMs may support eclosion behavior, they are not required as previously suggested (Kimura and Truman 1990). We therefore sought to investigate if the perturbations in DIOMs occurred more generally across developing muscle groups in the fly and turned our attention to the well characterized model of the IFMs.
Fig. 5. The DIOMs are not required for eclosion.
a Representative images of 45h APF (~p7) DIOMs morphology with control (CncC RFP>w1118) and CncC expression (CncC RFP>CncC) directed by the CncC Gal4 driver. b The number of myospheres originating from the most medial DIOMs in the A2-A5 abdominal segments (indicated by white asterisks) were quantified (n = 20 abdomens/genotype, mean ± s.d.m., Mann-Whitney test, ****p<0.0001). c Eclosion ability (n = 3, 450 flies/genotype, mean ± s.d.m, Z test).
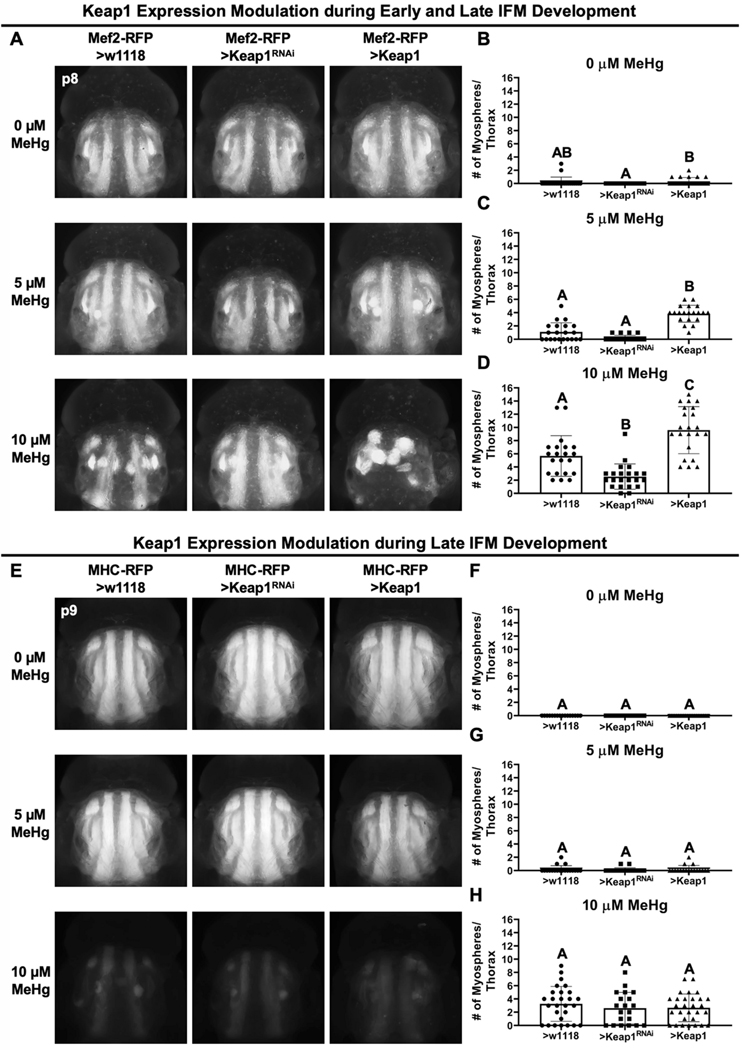
CncC signaling protects IFM development from MeHg toxicity.
MeHg exposure produced a dose-dependent increase in myospheres in the IFM (Figure 6A, B left panels and bars). Keap1 knockdown or overexpression, alone, did not significantly change the number of myospheres present per thorax compared to control (Figure 6A top row, B). Following 10 μM MeHg exposure, the number IFM myospheres observed in genotype controls (5.7 myospheres per thorax) was reduced with Keap1 knockdown (2.5 myospheres per thorax), whereas Keap1 overexpression increased the number of myospheres (9.6 myospheres per thorax) (Figure 6A bottom row, D). A similar profile was seen with the intermediate 5μM MeHg exposure (Figure 6A middle row, C).
Fig. 6. Muscle-specific CncC signaling modulation of MeHg effects on IFM morphology.
a Representative images of MeHg effects on IFM morphology with either no change (>w1118), knockdown (>Keap1RNAi), or overexpression (>Keap1) of Keap1 with the Mef2-RFP-GAL4 driver. b-d Number of myospheres in each thorax were quantified at indicated MeHg treatment (n ≥ 20/genotype/treatment, mean ± s.d.m., Kruskal-Wallis test with Dunn’s post-hoc for multiple comparisons, p<0.05, significant differences indicated by different letter labels). e Representative images of IFM morphology with either no change (>w1118), knockdown (>Keap1RNAi), or overexpression (>Keap1) of Keap1 with the MHC-RFP-GAL4 driver. f-h Number of myospheres in each thorax were quantified at indicated MeHg treatment (n ≥ 15/genotype/treatment, mean ± s.d.m., Kruskal-Wallis test with Dunn’s post-hoc for multiple comparisons, p<0.05, significant differences indicated by different letter labels).
To understand the sensitivity of the IFMs over the course of muscle development (i.e. early versus late), we examined the ability Keap1 expression levels to moderate MeHg toxicity using a late-stage myosin heavy chain driver, MHC-RFP-GAL4, which initiates expression at approximately p7 of IFM development (unpublished observations). With this reporter/driver combination MeHg effects on the IFMs are readily apparent at 10 μM MeHg exposure, where the most obvious phenotype is a reduced red fluorescence, which is attributed to the failure of IFM fibers to reach late stage development where MHC gene expression is initiated. Myospheres could nonetheless be detected and quantified. This reduction in IFM RFP fluorescence with MeHg exposure was, qualitatively, unaffected by Keap1 knockdown or overexpression driven by MHC-RFP-GAL4. Furthermore, the myospheres that were observed did not change in number (Figure 6E bottom row, H). These data point to a requirement for CncC signaling earlier in IFM development in order to protect against MeHg.
Neural-specific CncC rescue of eclosion and muscle morphology from MeHg toxicity.
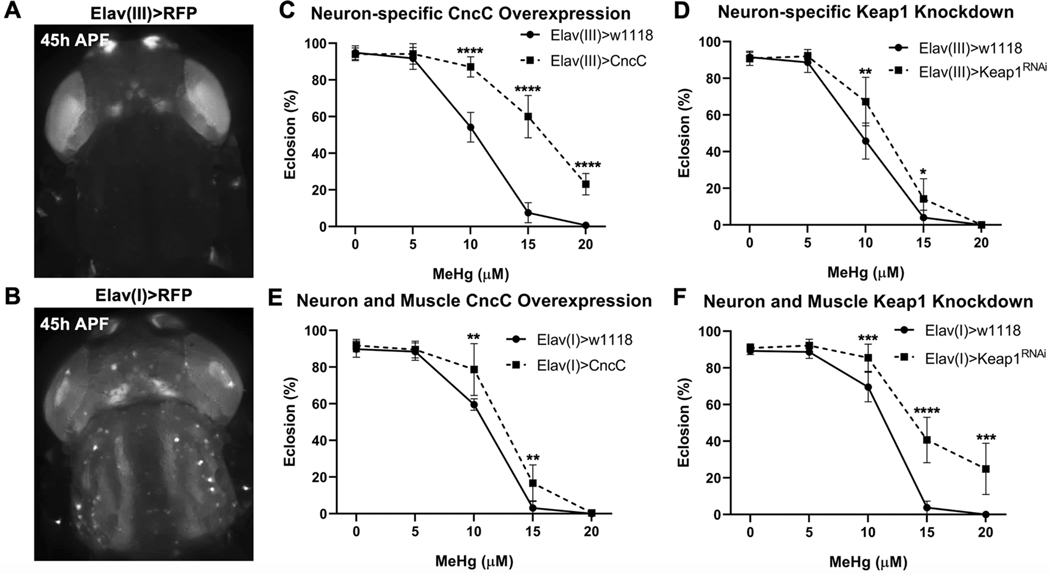
The above findings suggest that CncC can play an autonomous role in moderating MeHg toxicity in developing muscle. Nonetheless, neural inputs to the developing motor unit present an additional source of support and guidance for muscle development, particularly for IFM development. Thus, we investigated if CncC protective mechanisms within developing neurons act in parallel to support muscle morphogenesis using two approaches. Our first approach used two versions of a pan-neural driver, Elav(I)-GAL4 and Elav(III)-GAL4, which drive expression from enhancer elements of the neuron-associated ELAV gene, localized on either the first (I) or third (III) chromosome, respectively. Elav(I)-GAL4 is expressed in neural structures of the developing pupae such as the ommatidia of the eye, central brain, center brain, and antennae, as well as a ‘cross-over’ expression in the IFM fibers (Figure 7B). Elav(III)-GAL4 is expressed strictly in neural structures (Figure 7A and 8A). CncC overexpression in developing neurons via Elav(I) or Elav(III) is not pupal lethal and furthermore rescues eclosion ability with MeHg (Figure 7C, 7E). Moderate enhancement of CncC signaling in neurons via Keap1 knockdown also rescues eclosion ability with MeHg (Figure 7D).
Fig. 7. Neural-specific CncC signaling modulation of MeHg effects on eclosion.
a Expression domain of the Elav(III)-GAL4 driver. b Expression domain of Elav(I)-GAL4 driver. c-f Eclosion ability with CncC overexpression (>CncC) or Keap1 knockdown (>Keap1RNAi) under control of the Elav(III)-GAL4 driver (c, d) or Elav(I)-GAL4 driver (e, f) (n = 3, 450 flies/genotype/treatment, mean ± s.d.m., Z-test, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001).
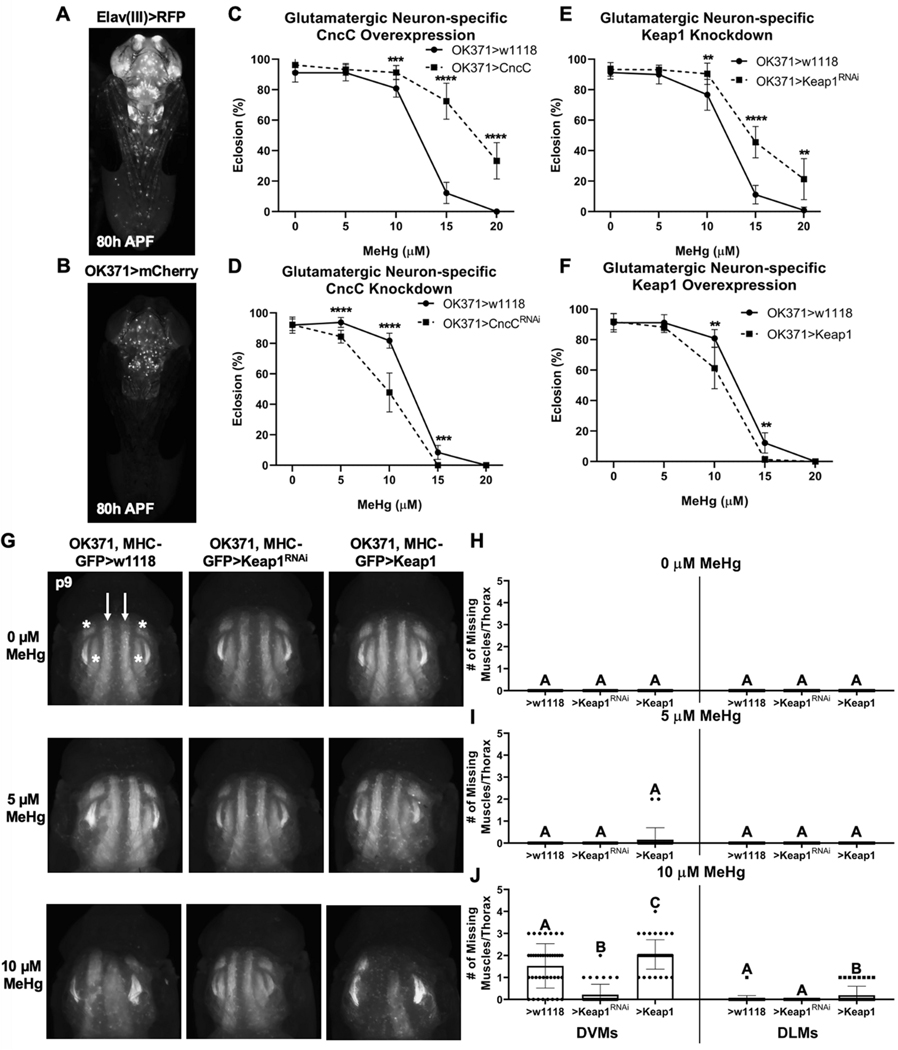
Fig. 8. Glutamatergic neuron-specific CncC signaling modulation of MeHg effects on eclosion.
a, b Expression pattern of the Elav(III)-GAL4 and OK371-GAL4 drivers (ventral view). c, d Eclosion ability with OK371-GAL4 driven CncC overexpression (>CncC) and knockdown (>CncCRNAi) compared to control (>w1118). e, f Eclosion ability with OK371-GAL4 driven Keap1 knockdown (>Keap1RNAi) and overexpression (>Keap1) compared to control (>w1118) (c-f n = 3, 450 flies/genotype/treatment, mean ± s.d.m., Z-test, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001). g Representative images of MeHg effects on IFM morphology with either no change (>w1118), knockdown (>Keap1RNAi), or overexpression (>Keap1) of Keap1 in glutamatergic neurons. Number of missing dorsoventral muscles (DVMs, indicated by white asterisks) and dorsal lateral muscles (DLMs, indicated by white arrows) in each thorax were quantified at indicated MeHg treatment (h-j n ≥ 20/genotype/treatment, mean ± s.d.m., Kruskal-Wallis test with Dunn’s post-hoc for multiple comparisons within same muscle group, p<0.05, significant differences indicated by different letter labels).
To further explore the neural specificity of MeHg-protective CncC activity, we restricted CncC modulations to glutamatergic neurons, which encompasses all motor neurons, using the OK371-GAL4 driver (Figure 8B). OK371-GAL4 demonstrates a pattern of limited expression in motor neurons of the ventral ganglion relative to pan-neural Elav(III) expression (Figure 8A). Overexpression and knockdown of CncC in glutamatergic neurons is seen to rescue and diminish, respectively, the MeHg dose-responsive eclosion abilities (Figure 8C, D). Glutamatergic neuron-restricted Keap1 knockdown moderately rescues eclosion while Keap1 overexpression decreases the MeHg dose-responsive eclosion abilities (Figure 8E, F). Finally, we examined if altering Keap1 expression strictly in the neuron can influence the impact of MeHg on IFM development. To do this, we used a strain that contains both the OK371-GAL4 and the myosin heavy MHC promotor controlling GFP expression such that all mature muscles fluoresce green. Exposure up to 10 μM MeHg to this strain was not seen to result in overt myospheres, yet the GFP intensity is noticeably reduced (Figure 8), similar to the response observed with the MHC-RFP reporter above (Figure 6E, bottom panels). Furthermore, in several intances missing IFM muscles were noted (Figure 8G bottom row), consistent with an overall lack of maturation of myofibers. We therefore quantified the number of missing IFMs. This missing muscle phenotype was rescued with motor neuron driven Keap1 knockdown and exacerbated with Keap1 overexpression (Figure 8G bottom row, J). Altering Keap1 in motor neurons did not influence IFM development at 0 or 5 μM MeHg (Figure 8G top and middle row, H, I).
4. DISCUSSION
We have capitalized on the strengths of the Drosophila model to characterize a role for tissue-specific Nrf2 signaling to moderate MeHg toxicity in the developing neuromuscular system, revealing that both developing muscles and neurons contribute targets for the apparent MeHg-induced motor deficits. Dose-dependent myotoxicity was apparent in two muscle groups; both the DIOMs and IFMs displayed myosphere phenotypes that correlated with reduced eclosion ability. Although elevations in CncC signaling occurred in parallel with these phenotypes, the antioxidant response was apparently insufficient to counter MeHg toxicity. However, we were able to reduce Keap1 levels in a muscle-restricted fashion, which achieved a moderate enhancement of CncC signaling and was sufficient to attenuate MeHg myotoxicity, as evident by improvement in eclosion ability and reduction in myospheres. In contrast, both strong enhancement and knockdown of CncC signaling in a pan-muscle-restricted fashion incurs lethality. These findings suggest that developing muscle is exquisitely sensitive to a narrow range of CncC signaling to afford protection against MeHg. However, different muscle groups may utilize endogenous CncC signals at various levels. Indeed, strong ARE reporter expression was observed across morphogenesis of the DIOMs but not the IFMs, indicating elevated basal CncC activity in the former. Nevertheless, MeHg exposure results in a similar dose-responsive occurrence of myospheres in both muscle groups, which can be offset by Keap1 reduction. On the other hand, restricting CncC overexpression to developing neurons yielded a robust rightward shift in the MeHg dose response for eclosion, and Keap1 knockdown in neurons resulted in a more moderate rescue, altogether suggesting that neurons accommodate a broader range of CncC signals than muscle. Furthermore, we see that MeHg phenotypes in IFM morphology are rescued by modulating CncC signaling in neurons, pointing to a non-autonomous protection of muscle development from MeHg toxicity by neuronal counterparts. Altogether, these findings demonstrate that motor deficits stemming from developmental MeHg exposure are due to the sensitivity of the neuromuscular unit, with both muscle and neuronal targets contributing to the susceptibility. The specificity of MeHg toxicity in this context of neuromuscular development and CncC signaling will require future studies examining a broader array of toxicants.
Enhanced CncC signaling could be protecting muscle morphogenesis and neuromotor function in several ways, including enhancing MeHg excretion or mitigating MeHg-mediated ROS production. These effects can be mediated by Nrf2 target genes such as GCLc and MRP (Unoki et al. 2018; Vollrath et al. 2006), which has been demonstrated in cell culture systems (Ni et al. 2010; Toyama et al. 2007; Wang et al. 2009). These effects are strengthened by in vivo studies in Drosophila demonstrating that transgenic GCLc upregulation can lead to an increase in GSH, reduced response to MeHg-induced oxidative stress and a decrease in mercury body burden which parallels rescue in eclosion (Rand et al. 2019). Consistent with findings here, it was previously seen that muscle- and neuron-specific GCLc upregulation rescued MeHg-inhibited eclosion (Rand et al. 2019). Yet, upregulation of MRP has shown to be effective only in muscle, with no rescue of MeHg-inhibited eclosion when upregulated in neurons (Prince and Rand 2017). This differential profile of MeHg-protective effects of CncC downstream genes in muscle versus neurons may reflect an overall intrinsic difference in CncC signaling between these two tissues. Previous in vitro and Drosophila studies have highlighted the toxicity accompanying hyperactivation of Nrf2/CncC signaling in developing muscle (Rajasekaran et al. 2020; Tsakiri et al. 2019). Neuron-specific enhancement of CncC induces more moderate dysfunction (Sykiotis and Bohmann 2008; Tsakiri et al. 2019), but does not impair major neuromuscular events or lifespan (Spiers et al. 2019). Future studies are needed to examine the array of target genes that are responsive to Nrf2 upregulation in developing muscle versus neuronal cells.
This study has afforded new insights into DIOM function, the generality of MeHg myotoxicity, and muscle morphogenesis. Firstly, we establish that the DIOMs are not required for eclosion. This is confirmed with our result with CncC overexpression in the DIOMs, via the Cnc-GAL4 driver, that causes a severe myosphere phenotype while eclosion is unaffected. The presumption that DIOMs are needed for eclosion is based solely on their temporal existence, since they persist only until 1–2 days after adult emergence, when they then histolyze (Kimura and Truman 1990). Our results support prior work which showed a DIOM dysmorphology that did not impact eclosion (Kuleesha et al. 2016a). Further studies are required to understand the purpose of the DIOMs during metamorphosis and eclosion. Although eclosion has been reported to rely on muscle groups in the thorax, the IFMs reportedly do not play a role in eclosion (Miyan 1989). Nonetheless, the fact that two distinct muscle groups show similar myosphere phenotypes following MeHg exposure allows us to surmise that MeHg perturbs myo-morphogenesis generally, including those muscle that are required for eclosion. Further evaluations will require unambiguous identification of the Drosophila eclosion muscles. Furthermore, distinct roles for the CncC pathway during different myo-morphogenesis processes are implicated by the restricted and elevated cnc gene transcription and ARE activity in DIOMs compared to the IFM. Muscle morphogenesis is imperative for proper muscle development and neuromuscular function (Goody et al. 2015; Schulman et al. 2015; Turner 1986). The myo-morphogenesis of the DIOMs differ from the IFM over the course of metamorphosis. In the IFMs, development consists of iterative myoblast fusion events to form multi-nucleated syncytia, followed by subsequent growth and characteristic myofibrillogenesis steps of muscle development (Fernandes et al. 1991; Spletter et al. 2018). In contrast, DIOM maturation involves a unique set of larval muscles that persist through histolysis and remodel into temporary adult muscles (Chinta et al. 2012; Kimura and Truman 1990; Miller 1965). DIOM remodeling involves autophagy-mediated atrophy (Kuleesha et al. 2016b). Both the mammalian Nrf2 and Drosophila CncC pathway regulate expression of autophagy-related genes and overexpression of CncC can induce excessive autophagy (Pajares et al. 2016; Yamada et al. 2019). Thus, the fundamental differences in IFM development versus DIOM maturation could explain why the DIOMs have a higher basal level of CncC signaling; the DIOMs uniquely require CncC signaling for their maturation, which involves myogenic features that are not shared with the IFM such as autophagy-mediated atrophy. Future studies are needed to investigate the potential for CncC/Nrf2 signaling to control aspects of muscle morphogenesis.
Remarkably, we see that elevated CncC activity in neurons can protect developing muscle from MeHg toxicity non-autonomously. This finding suggests that the intimate interplay of muscle and neurons during morphogenesis and establishment of the neuromuscular unit constitute a sensitive MeHg target. Neuron-muscle cross talk during morphogenesis uses conserved processes in mammals and Drosophila (Chakkalakal et al. 2010; Chen et al. 2018; Fernandes and Keshishian 1998; Fernandes and Keshishian 2005; Lin et al. 2008). The developing IFM of the pupa rely on the intersegmental motor neuron (ISN) to direct appropriate spatio-temporal myogenic events such as myoblast fusion, and ablation of the ISN drastically reduces IFM formation (Fernandes and Keshishian 1998). We predict that MeHg directly affects the ISN as well as the developing myotubes and myofibers. In accordance with this, we observed reduction in the MeHg effects on IFM morphology and eclosion after upregulating CncC in motor neurons, pointing to a critical neuronal ‘cue’ in muscle morphogenesis that is labile to MeHg. While the morphology of the ISN following MeHg exposure remains to be investigated in pupae, our prior study of MeHg exposure to embryos revealed dysmorphology of both the developing ISN and the larval lateral transverse muscles (Engel and Rand 2014; Prince and Rand 2017; Rand et al. 2009). Nevertheless, these data indicate that muscle development can be rescued through enhanced neuron-specific CncC signaling. Future studies will explore the potential for neuron-specific CncC to protect neuron morphology and its influence on myogenic processes.
In summary, this study is the first to demonstrate tissue-specific roles for Nrf2 activity in protecting against the developmental toxicity of MeHg. Our results also highlight the importance of neuron-muscle interaction during development for sustaining a MeHg insult. Finally, we demonstrate a unique pattern of endogenous Nrf2 signaling localized to remodeling muscle that implicates additional roles for this pathway in tissue morphogenesis.
Supplementary Material
Acknowledgements:
We would like to thank University of Rochester Drosophila researchers for communal fly husbandry materials. We thank I. Krout for the critical feedback during the course of the study. We thank J. Wang of the Bellen lab (Baylor College of Medicine) for providing the Cnc-T2AGAL4 stock description. We are especially grateful to D. Bohmann and R. Hoff (University of Rochester) for providing many of the fly strains and for useful discussions of this study.
Declarations:
Funding: This study was supported by National Institute of Environmental Health Sciences (R01 ES025721 (PI; M.D.R.), R01 ES010219 (co-I; M.D.R.), and P30 ES001247 (co-I; M.D.R.)) and the University of Rochester Environmental Health Sciences Center (T32 207026 (J.G.)).
Footnotes
Code availability: Not applicable.
Conflict of interests/Competing interests: The authors declare that they have no conflict of interest.
Availability of data and materials: All supplemental data will be available in supplemental files. Fly strains can be provided upon request.
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
5. REFERENCES
- Akiyama M, Unoki T, Shinkai Y, et al. (2019) Environmental Electrophile-Mediated Toxicity in Mice Lacking Nrf2, CSE, or Both. Environ Health Perspect 127(6):67002 doi: 10.1289/EHP4949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschner M, Syversen T, Souza DO, Rocha JB, Farina M (2007) Involvement of glutamate and reactive oxygen species in methylmercury neurotoxicity. Braz J Med Biol Res 40(3):285–91 doi: [DOI] [PubMed] [Google Scholar]
- Bainbridge SP, Bownes M (1981) Staging the metamorphosis of Drosophila melanogaster. J Embryol Exp Morphol 66:57–80 [PubMed] [Google Scholar]
- Bakir F, Damluji SF, Amin-Zaki L, et al. (1973) Methylmercury poisoning in Iraq. Science 181(4096):230–41 [DOI] [PubMed] [Google Scholar]
- Bose-O’Reilly S, McCarty KM, Steckling N, Lettmeier B (2010) Mercury exposure and children’s health. Curr Probl Pediatr Adolesc Health Care 40(8):186–215 doi: 10.1016/j.cppeds.2010.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118(2):401–15 [DOI] [PubMed] [Google Scholar]
- Burke K, Cheng Y, Li B, et al. (2006) Methylmercury elicits rapid inhibition of cell proliferation in the developing brain and decreases cell cycle regulator, cyclin E. Neurotoxicology 27(6):970–81 doi: 10.1016/j.neuro.2006.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambier S, Benard G, Mesmer-Dudons N, et al. (2009) At environmental doses, dietary methylmercury inhibits mitochondrial energy metabolism in skeletal muscles of the zebra fish (Danio rerio). Int J Biochem Cell Biol 41(4):791–9 doi: 10.1016/j.biocel.2008.08.008 [DOI] [PubMed] [Google Scholar]
- Carratu MR, Borracci P, Coluccia A, et al. (2006) Acute exposure to methylmercury at two developmental windows: focus on neurobehavioral and neurochemical effects in rat offspring. Neuroscience 141(3):1619–29 doi: 10.1016/j.neuroscience.2006.05.017 [DOI] [PubMed] [Google Scholar]
- Chakkalakal JV, Nishimune H, Ruas JL, Spiegelman BM, Sanes JR (2010) Retrograde influence of muscle fibers on their innervation revealed by a novel marker for slow motoneurons. Development 137(20):3489–99 doi: 10.1242/dev.053348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee N, Bohmann D (2012) A versatile PhiC31 based reporter system for measuring AP-1 and Nrf2 signaling in Drosophila and in tissue culture. PLoS One 7(4):e34063 doi: 10.1371/journal.pone.0034063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Amirbahman A, Fisher N, et al. (2008) Methylmercury in marine ecosystems: spatial patterns and processes of production, bioaccumulation, and biomagnification. Ecohealth 5(4):399–408 doi: 10.1007/s10393-008-0201-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HH, Tsai LK, Liao KY, et al. (2018) Muscle-restricted nuclear receptor interaction protein knockout causes motor neuron degeneration through down-regulation of myogenin at the neuromuscular junction. J Cachexia Sarcopenia Muscle 9(4):771–785 doi: 10.1002/jcsm.12299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinta R, Tan JH, Wasser M (2012) The study of muscle remodeling in Drosophila metamorphosis using in vivo microscopy and bioimage informatics. BMC Bioinformatics 13 Suppl 17:S14 doi: 10.1186/1471-2105-13-S17-S14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbreth M, Rand MD (2020) Methylmercury modifies temporally expressed myogenic regulatory factors to inhibit myoblast differentiation. Toxicol In Vitro 63:104717 doi: 10.1016/j.tiv.2019.104717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbreth M, Zhang Z, Aschner M (2017) Methylmercury augments Nrf2 activity by downregulation of the Src family kinase Fyn. Neurotoxicology 62:200–206 doi: 10.1016/j.neuro.2017.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham P, Cooter W, Sullivan E (2003) Mercury in Marine Life Database. In: Agency USEP (ed). U.S. EPA, p 1–226 [Google Scholar]
- Dare E, Fetissov S, Hokfelt T, Hall H, Ogren SO, Ceccatelli S (2003) Effects of prenatal exposure to methylmercury on dopamine-mediated locomotor activity and dopamine D2 receptor binding. Naunyn Schmiedebergs Arch Pharmacol 367(5):500–8 doi: 10.1007/s00210-003-0716-5 [DOI] [PubMed] [Google Scholar]
- Deng H, Kerppola TK (2013) Regulation of Drosophila metamorphosis by xenobiotic response regulators. PLoS Genet 9(2):e1003263 doi: 10.1371/journal.pgen.1003263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao F, Ironfield H, Luan H, et al. (2015) Plug-and-play genetic access to drosophila cell types using exchangeable exon cassettes. Cell Rep 10(8):1410–21 doi: 10.1016/j.celrep.2015.01.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel GL, Rand MD (2014) The Notch target E(spl)mdelta is a muscle-specific gene involved in methylmercury toxicity in motor neuron development. Neurotoxicol Teratol 43:11–8 doi: 10.1016/j.ntt.2014.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Xu Z, Wang F, et al. (2017) Sulforaphane Prevents Methylmercury-Induced Oxidative Damage and Excitotoxicity Through Activation of the Nrf2-ARE Pathway. Mol Neurobiol 54(1):375–391 doi: 10.1007/s12035-015-9643-y [DOI] [PubMed] [Google Scholar]
- Fernandes J, Bate M, Vijayraghavan K (1991) Development of the indirect flight muscles of Drosophila. Development 113(1):67–77 [DOI] [PubMed] [Google Scholar]
- Fernandes JJ, Keshishian H (1998) Nerve-muscle interactions during flight muscle development in Drosophila. Development 125(9):1769–79 [DOI] [PubMed] [Google Scholar]
- Fernandes JJ, Keshishian H (2005) Motoneurons regulate myoblast proliferation and patterning in Drosophila. Dev Biol 277(2):493–505 doi: 10.1016/j.ydbio.2004.09.038 [DOI] [PubMed] [Google Scholar]
- Fujimura M, Cheng J, Zhao W (2012) Perinatal exposure to low-dose methylmercury induces dysfunction of motor coordination with decreases in synaptophysin expression in the cerebellar granule cells of rats. Brain Res 1464:1–7 doi: 10.1016/j.brainres.2012.05.012 [DOI] [PubMed] [Google Scholar]
- Fujimura M, Usuki F (2015a) Low concentrations of methylmercury inhibit neural progenitor cell proliferation associated with up-regulation of glycogen synthase kinase 3beta and subsequent degradation of cyclin E in rats. Toxicol Appl Pharmacol 288(1):19–25 doi: 10.1016/j.taap.2015.07.006 [DOI] [PubMed] [Google Scholar]
- Fujimura M, Usuki F (2015b) Methylmercury causes neuronal cell death through the suppression of the TrkA pathway: in vitro and in vivo effects of TrkA pathway activators. Toxicol Appl Pharmacol 282(3):259–66 doi: 10.1016/j.taap.2014.12.008 [DOI] [PubMed] [Google Scholar]
- Fujimura M, Usuki F, Cheng J, Zhao W (2016) Prenatal low-dose methylmercury exposure impairs neurite outgrowth and synaptic protein expression and suppresses TrkA pathway activity and eEF1A1 expression in the rat cerebellum. Toxicol Appl Pharmacol 298:1–8 doi: 10.1016/j.taap.2016.03.002 [DOI] [PubMed] [Google Scholar]
- Goody MF, Sher RB, Henry CA (2015) Hanging on for the ride: adhesion to the extracellular matrix mediates cellular responses in skeletal muscle morphogenesis and disease. Dev Biol 401(1):75–91 doi: 10.1016/j.ydbio.2015.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada M (1995) Minamata disease: methylmercury poisoning in Japan caused by environmental pollution. Crit Rev Toxicol 25(1):1–24 doi: 10.3109/10408449509089885 [DOI] [PubMed] [Google Scholar]
- Hayes JD, Dinkova-Kostova AT (2014) The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem Sci 39(4):199–218 doi: 10.1016/j.tibs.2014.02.002 [DOI] [PubMed] [Google Scholar]
- Kim MK, Zoh KD (2012) Fate and transport of mercury in environmental media and human exposure. J Prev Med Public Health 45(6):335–43 doi: 10.3961/jpmph.2012.45.6.335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura KI, Truman JW (1990) Postmetamorphic cell death in the nervous and muscular systems of Drosophila melanogaster. J Neurosci 10(2):403–1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleesha Y, Puah WC, Wasser M (2016a) Live imaging of muscle histolysis in Drosophila metamorphosis. BMC Dev Biol 16:12 doi: 10.1186/s12861-016-0113-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleesha Y, Puah WC, Wasser M (2016b) A model of muscle atrophy based on live microscopy of muscle remodelling in Drosophila metamorphosis. R Soc Open Sci 3(2):150517 doi: 10.1098/rsos.150517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai Y, Kanda H, Shinkai Y, Toyama T (2013) The role of the Keap1/Nrf2 pathway in the cellular response to methylmercury. Oxid Med Cell Longev 2013:848279 doi: 10.1155/2013/848279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Landmann L, Ruegg MA, Brenner HR (2008) The role of nerve- versus muscle-derived factors in mammalian neuromuscular junction formation. J Neurosci 28(13):3333–40 doi: 10.1523/JNEUROSCI.5590-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25(4):402–8 doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Magnusson J, Ramel C (1986) Genetic variation in the susceptibility to mercury and other metal compounds in Drosophila melanogaster. Teratog Carcinog Mutagen 6(4):289–305 doi: 10.1002/tcm.1770060405 [DOI] [PubMed] [Google Scholar]
- Mahapatra CT, Bond J, Rand DM, Rand MD (2010) Identification of methylmercury tolerance gene candidates in Drosophila. Toxicol Sci 116(1):225–38 doi: 10.1093/toxsci/kfq097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown-Eyssen GE, Ruedy J, Neims A (1983) Methyl mercury exposure in northern Quebec. II. Neurologic findings in children. Am J Epidemiol 118(4):470–9 doi: 10.1093/oxfordjournals.aje.a113652 [DOI] [PubMed] [Google Scholar]
- Miller A (1965) The internal anatomy and histology of the imago of Drosophila melanogaster In: Demerec M (ed) The Biology of Drosophila. Hafner Publishing Company, New York, p 420–534 [Google Scholar]
- Miyan JA (1989) The thoracic mechanism for eclosion and digging during the extrication behaviour of Diptera. Physiological Entomology 14:309–317 doi: 10.1111/j.1365-3032.1989.tb01098.x [DOI] [Google Scholar]
- Montgomery SL, Vorojeikina D, Huang W, Mackay TF, Anholt RR, Rand MD (2014) Genome-wide association analysis of tolerance to methylmercury toxicity in Drosophila implicates myogenic and neuromuscular developmental pathways. PLoS One 9(10):e110375 doi: 10.1371/journal.pone.0110375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Li X, Yin Z, et al. (2010) Methylmercury induces acute oxidative stress, altering Nrf2 protein level in primary microglial cells. Toxicol Sci 116(2):590–603 doi: 10.1093/toxsci/kfq126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Li X, Yin Z, et al. (2011) Comparative study on the response of rat primary astrocytes and microglia to methylmercury toxicity. Glia 59(5):810–20 doi: 10.1002/glia.21153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajares M, Jimenez-Moreno N, Garcia-Yague AJ, et al. (2016) Transcription factor NFE2L2/NRF2 is a regulator of macroautophagy genes. Autophagy 12(10):1902–1916 doi: 10.1080/15548627.2016.1208889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peppriell A, Gunderson J, Vorojeikina D, Rand DM (2020) Methylmercury myotoxicity targets formation of myotendinous junction. Toxicology (In review) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitoniak A, Bohmann D (2015) Mechanisms and functions of Nrf2 signaling in Drosophila. Free Radic Biol Med 88(Pt B):302–313 doi: 10.1016/j.freeradbiomed.2015.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince LM, Rand MD (2017) Notch Target Gene E(spl)mdelta Is a Mediator of Methylmercury-Induced Myotoxicity in Drosophila. Front Genet 8:233 doi: 10.3389/fgene.2017.00233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince LM, Rand MD (2018) Methylmercury exposure causes a persistent inhibition of myogenin expression and C2C12 myoblast differentiation. Toxicology 393:113–122 doi: 10.1016/j.tox.2017.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasekaran NS, Shelar SB, Jones DP, Hoidal JR (2020) Reductive stress impairs myogenic differentiation. Redox Biol 34:101492 doi: 10.1016/j.redox.2020.101492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand MD, Dao JC, Clason TA (2009) Methylmercury disruption of embryonic neural development in Drosophila. Neurotoxicology 30(5):794–802 doi: 10.1016/j.neuro.2009.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand MD, Montgomery SL, Prince L, Vorojeikina D (2014) Developmental toxicity assays using the Drosophila model. Curr Protoc Toxicol 59:1 12 1–20 doi: 10.1002/0471140856.tx0112s59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand MD, Vorojeikina D, Pepriell P, Gunderson J, Prince L (2019) Drosophotoxicology: Elucidating Kinetic and Dynamic Pathways of Methylmercury Toxicity in a Drosophila Model. Front Genet 10(666) doi:10.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodier PM, Aschner M, Sager PR (1984) Mitotic arrest in the developing CNS after prenatal exposure to methylmercury. Neurobehav Toxicol Teratol 6(5):379–85 [PubMed] [Google Scholar]
- Sager PR, Aschner M, Rodier PM (1984) Persistent, differential alterations in developing cerebellar cortex of male and female mice after methylmercury exposure. Brain Res 314(1):1–11 doi: 10.1016/0165-3806(84)90170-6 [DOI] [PubMed] [Google Scholar]
- Schulman VK, Dobi KC, Baylies MK (2015) Morphogenesis of the somatic musculature in Drosophila melanogaster. Wiley Interdiscip Rev Dev Biol 4(4):313–34 doi: 10.1002/wdev.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiers JG, Breda C, Robinson S, Giorgini F, Steinert JR (2019) Drosophila Nrf2/Keap1 Mediated Redox Signaling Supports Synaptic Function and Longevity and Impacts on Circadian Activity. Front Mol Neurosci 12:86 doi: 10.3389/fnmol.2019.00086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spletter ML, Barz C, Yeroslaviz A, et al. (2018) A transcriptomics resource reveals a transcriptional transition during ordered sarcomere morphogenesis in flight muscle. Elife 7 doi: 10.7554/eLife.34058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spyker JM, Sparber SB, Goldberg AM (1972) Subtle consequences of methylmercury exposure: behavioral deviations in offspring of treated mothers. Science 177(4049):621–3 doi: 10.1126/science.177.4049.621 [DOI] [PubMed] [Google Scholar]
- Sykiotis GP, Bohmann D (2008) Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev Cell 14(1):76–85 doi: 10.1016/j.devcel.2007.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng X, Zhang Z, He G, Yang L, Li F (2012) Validation of reference genes for quantitative expression analysis by real-time rt-PCR in four lepidopteran insects. J Insect Sci 12:60 doi: 10.1673/031.012.6001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama T, Shinkai Y, Yasutake A, Uchida K, Yamamoto M, Kumagai Y (2011) Isothiocyanates reduce mercury accumulation via an Nrf2-dependent mechanism during exposure of mice to methylmercury. Environ Health Perspect 119(8):1117–22 doi: 10.1289/ehp.1003123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama T, Sumi D, Shinkai Y, et al. (2007) Cytoprotective role of Nrf2/Keap1 system in methylmercury toxicity. Biochem Biophys Res Commun 363(3):645–50 doi: 10.1016/j.bbrc.2007.09.017 [DOI] [PubMed] [Google Scholar]
- Tsakiri EN, Gumeni S, Iliaki KK, et al. (2019) Hyperactivation of Nrf2 increases stress tolerance at the cost of aging acceleration due to metabolic deregulation. Aging Cell 18(1):e12845 doi: 10.1111/acel.12845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner DC (1986) Cell-cell and cell-matrix interactions in the morphogenesis of skeletal muscle. Dev Biol (N Y 1985) 3:205–24 doi: 10.1007/978-1-4684-5050-7_11 [DOI] [PubMed] [Google Scholar]
- Unoki T, Akiyama M, Kumagai Y, et al. (2018) Molecular Pathways Associated With Methylmercury-Induced Nrf2 Modulation. Front Genet 9:373 doi: 10.3389/fgene.2018.00373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usuki F, Yasutake A, Matsumoto M, Umehara F, Higuchi I (1998) The effect of methylmercury on skeletal muscle in the rat: a histopathological study. Toxicol Lett 94(3):227–32 [DOI] [PubMed] [Google Scholar]
- Vollrath V, Wielandt AM, Iruretagoyena M, Chianale J (2006) Role of Nrf2 in the regulation of the Mrp2 (ABCC2) gene. Biochem J 395(3):599–609 doi: 10.1042/BJ20051518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorojeikina D, Broberg K, Love TM, Davidson PW, van Wijngaarden E, Rand MD (2017) Editor’s Highlight: Glutathione S-Transferase Activity Moderates Methylmercury Toxicity During Development in Drosophila. Toxicol Sci 157(1):211–221 doi: 10.1093/toxsci/kfx033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi N, Dinkova-Kostova AT, Holtzclaw WD, et al. (2004) Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proc Natl Acad Sci U S A 101(7):2040–5 doi: 10.1073/pnas.0307301101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Jiang H, Yin Z, Aschner M, Cai J (2009) Methylmercury toxicity and Nrf2-dependent detoxification in astrocytes. Toxicol Sci 107(1):135–43 doi: 10.1093/toxsci/kfn201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Iwata M, Warabi E, Oishi H, Lira VA, Okutsu M (2019) p62/SQSTM1 and Nrf2 are essential for exercise-mediated enhancement of antioxidant protein expression in oxidative muscle. FASEB J:fj201900133R doi: 10.1096/fj.201900133R [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.