Tissue engineering offers tremendous potential to overcome issues with organ transplant shortage, drug screening, and to study molecular phenomena that are involved in biological functions such as tissue morphogenesis, wound healing, and inflammatory reactions.[1] However, conventional planar fabrication techniques are unable to create the complex multi-scale architectures such as the interface between the vascular endothelium and surrounding connective tissue and parenchymal cells. This may limit the applications of these structures for studying basic functions of the respective tissues.
A recent interesting development in the field of tissue engineering is the use of three dimensional (3D) bioprinting technologies to merge the use of cells, biomaterials, and microfabrication.[2–4] The selection of a proper biomaterial as the ink is important for 3D bioprinting of constructs with biomimetic architectures and properties. In the past few years, different biomaterials, including synthetic polymers,[5, 6] natural polymers,[7, 8] and decellularized extracellular matrix (ECM),[9] have been engineered as bioinks to print complex 3D structures such as perfusable vascular networks. For example, in a recent study, Dvir and colleagues 3D bioprinted a decellularized ECM hydrogel into a vascularized heart model.[10] However, the construct was not perfused long term in vitro, and the viability and function of the encapsulated cells were not reported. Alternatively, different techniques such as casting[11] and sacrificial ink-writing,[12] that involve extra processing steps, have been combined with 3D bioprinting to mimic vasculatures. The fabrication of complex biomimetic structure that are entirely based on 3D bioprinting is still challenging primarily due to the lack of suitable bioinks with high printability, biocompatibility, biomimicry, and proper mechanical properties. Therefore, new biomaterial-based approaches are needed to address the limitations of currently available bioinks.
Here, we present a highly biocompatible recombinant human tropoelastin-based bioink for 3D bioprinting of complex soft tissues. The engineered bioink is composed of two ECM-based biopolymers gelatin methacryloyl (GelMA) and methacryloyl-substituted recombinant human tropoelastin (MeTro) which can be crosslinked with visible light. ECM materials have been widely used in tissue engineering field given that they provide a cell the environment similar to its origin. However, tropoelastin as one of the main components of connective tissues has not yet been studied for 3D bioprinting of soft tissues. Our recombinant human tropoelastin is identical to the naturally secreted human form. Elastic fibers are integral to vertebrate tissues such as blood vessels, skin, lung, and heart, where strength and elasticity are required for regular tissue function (i.e. stretching and contracting). Elasticity of tropoelastin originates from its intermolecular alignment governed by a modular structure of alternating hydrophobic and hydrophilic domains including lysine residues (Figure 1A). In addition to intrinsic elasticity and resilience, tropoelastin provides cell binding motifs and cell signaling pathways.[13] Though the binding site or mechanism of bonding are not clearly revealed, tropoelastin have been demonstrated to support endothelial cell recruitment and growth and to play a significant role in migration and angiogenesis.[14, 15] In addition to tropoelastin, gelatin was selected as a base material in bioink formulation due to its properties. Gelatin is an irreversibly hydrolyzed form of collagen, yet possesses many characteristics of collagen (i.e. cell-binding sites, and matrix-metalloproteinase (MMP) degradation sites).[16] While elastin is synthesized in the body by enzymatic cross-linking of lysine residues of tropoelastin, recombinant human tropoelastin can be chemically modified with methacryloyl groups to synthesize MeTro, which can be photopolymerized to form highly elastic hydrogel (Figure 1B).[17] Similarly, gelatin backbone can be also chemically functionalized with methacryloyl groups to form GelMA (Figure 1C).[18] While GelMA in the bioink supports an amenable cell culture environment,[19] the incorporation of MeTro enhances elasticity and mechanical stability for the printed structures.[20] An optimized formulation of bioink based on testing different ratios of GelMA/MeTro was used as a versatile platform for 3D bioprinting of soft tissues.
Figure 1. Synthesis and mechanical characterization of GelMA/MeTro composite hydrogel.
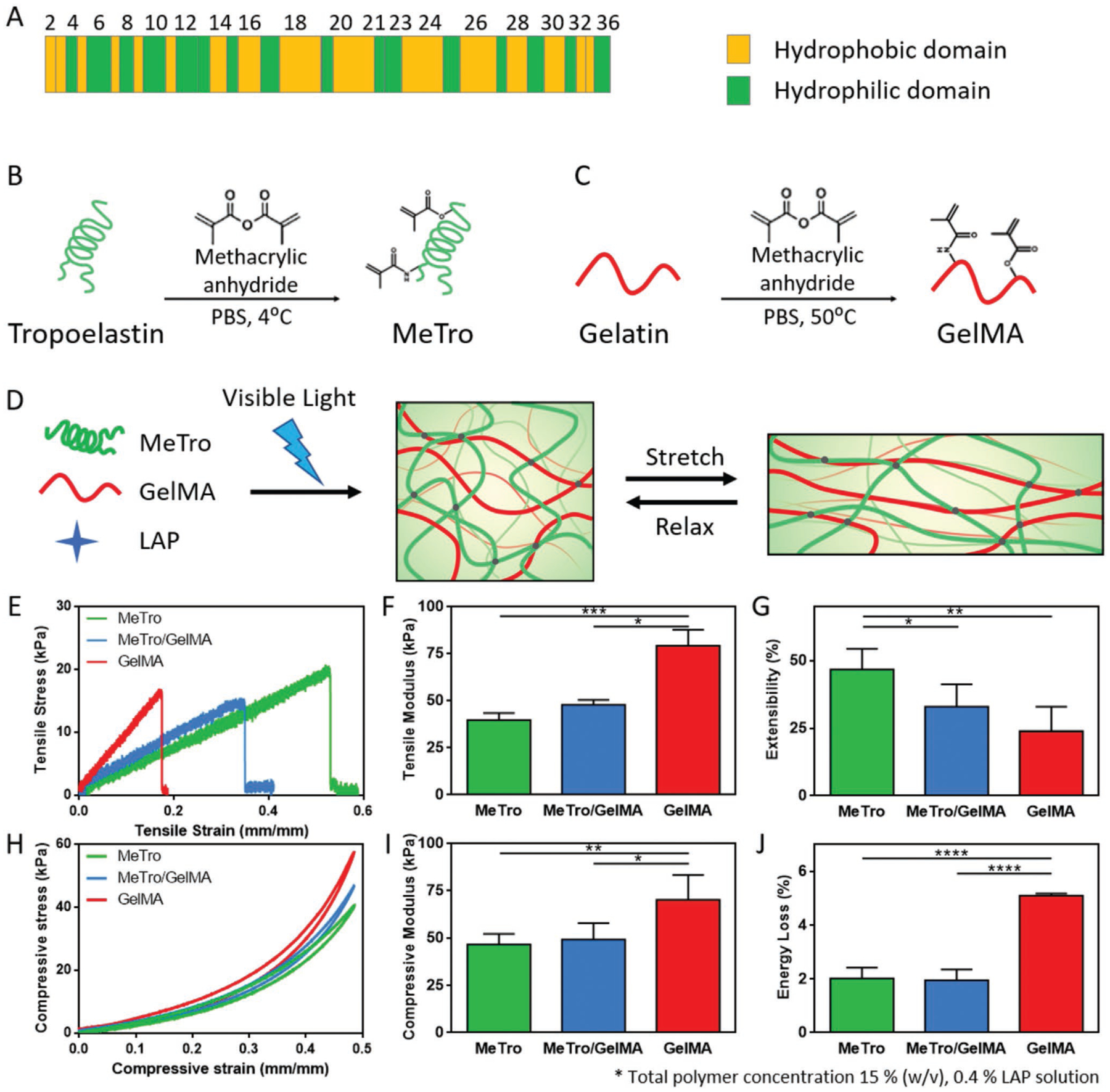
(A) Domain map of human tropoelastin. Methacrylation of (B) tropoelastin and (C) gelatin. (D) A schematic to describe the formation of MeTro/GelMA hydrogels. GelMA and MeTro polymers are covalently crosslinked upon exposure to visible light in the presence of LAP photoinitiator to form a highly elastic hydrogel network. Mechanical properties of MeTro, GelMA and MeTro/GelMA composite hydrogels, showing (E) representative tensile stress-strain curves; (F) tensile moduli; (G) extensibility; (H) representative compressive stress-strain curves; (I) compressive moduli; and (J) energy loss. (* p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001)
In our previous work, we engineered GelMA/MeTro composite hydrogels for different tissue engineering applications such as wound healing[21] and nerve tissue regeneration[22]. Here, we aim to explore the use of GelMA/MeTro as ink for 3D bioprinting of vascularized soft tissues such as cardiac tissue. Due to the hydrophobic hydration of tropoelastin, water molecules around tropoelastin are expelled at temperatures above a characteristic transition temperature, which results in coacervation of tropoelastin. Therefore, solutions of tropoelastin or MeTro, whose solubility is identical with tropoelastin, in this study were handled in temperature around 8 °C. We previously reported that the Young’s moduli of the visible light crosslinked MeTro/GelMA hydrogels, using Eosin Y/triethanolamine (TEA)/N-vinylcaprolactam (VC) photoinitiation system, was in the range of 4.05–10.25 kPa,[23] which was comparatively lower than those for UV crosslinked MeTro/GelMA hydrogels using Irgacure 2959 (16.5–49.8 kPa).[24] Although the UV crosslinked hydrogels were highly elastic, the procedure required exposure to UV light for a few min, which can be harmful for the encapsulated cells by damaging their DNA.[25] To eliminate the negative effects of UV irradiation, in this study, we investigated another photoinitiator, lithium phenyl-2,4,6-trimethylbenzoylphos-phinate (LAP), to photopolymerize the bioprinted structures containing different type of cells (Figure 1D). LAP can initiate photocrosslinking around 405 nm and demonstrated remarkable advantages as compared to other photoinitiators, including high water solubility and low cytotoxicity.[26] To verify the crosslinking within the hydrogels, the 1H NMR (400 MHz) spectra of the MeTro/GelMA hydrogels was compared with MeTro and GelMA prepolymers (Figure S1). The results demonstrated that 87.7 ± 8.6% of the methacryloyl groups in the MeTro/GelMA prepolymers were involved in the formation of the hydrogel network.
Tensile tests demonstrated statistically significant differences in the mechanical properties between the composites, pure GelMA and MeTro hydrogels. Specifically, the tensile modulus of MeTro/GelMA hydrogel was calculated to be 47.9 ± 2.6 kPa, which was higher than the value for pure MeTro (39.7 ± 3.8 kPa), and lower than pristine GelMA hydrogel (79.2 ± 8.5 kPa) (Figure 1E, F). In contrast, an opposite trend was observed for the extensibility and ultimate stress values (Figures 1G and S2). As expected, pure MeTro hydrogel had extensibility around 2-fold higher than pure GelMA hydrogel and the MeTro/GelMA exhibited extensibility of 33.1 ± 8.3 %. Cyclic compression test showed similar trend where MeTro/GelMA hydrogels presented compressive modulus of 49.2 ± 8.7 kPa between the modulus of pure GelMA (46.6 ± 5.6 kPa) and pure MeTro hydrogels (70.2 ± 13.2 kPa) (Figure 1H, I). In addition, energy loss calculation proved that the addition of MeTro increased the resilience of the composite hydrogel by showing a reduced energy loss for both MeTro and MeTro/GelMA hydrogels as compared to pristine GelMA hydrogel (Figure 1J). Biodegradable hydrogels are the most promising bioinks for 3D bioprinting as they mimic salient features of ECMs, can be modified to achieve mechanics similar to soft tissues, and can support cell adhesion and proliferation. However, many of these hydrogel precursors shear easily and have a low resistance to deformation, which limits their utilization as bioinks for 3D printing purpose. To address this limitation, rheology modifiers such as glycerol,[27] cellulose nanofibers,[28, 29] or silicate nanoparticles[5, 29–31] have been used to endow the enhanced printability to the existing bioinks. However, these additional bioink components can alter the mechanical and physiological properties of the resulting printed structures, which can negatively affect the cell growth and differentiation within the materials. Therefore, sacrificial materials, such as alginate[32] and poloxamer 407[33] have been used to improve printability of hydrogel-based bioinks. In this study, cold water fish gelatin was used as a sacrificial material to enhance the printability of the designed bioink (Figure 2A).
Figure 2. Optimization of 3D printing parameters.
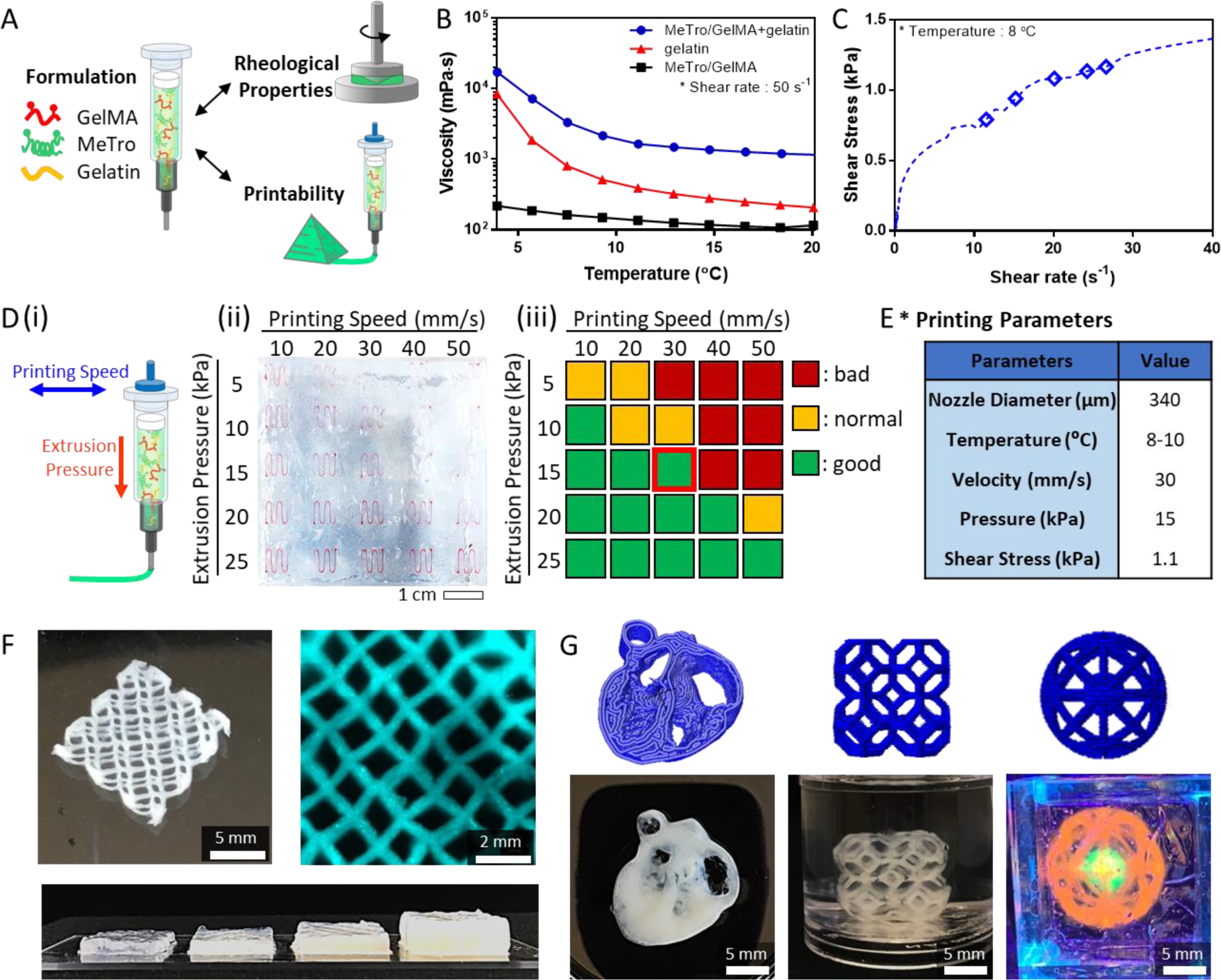
(A) Bioink formulation for 3D bioprinting. (B) Viscosity of MeTro/GelMA/gelatin, gelatin, and MeTro/GelMA as a function of temperature at shear rate 50 s−1. (C) Shear stress of MeTro/GelMA bioink measured as a function of shear rate. The diamond points indicate the actual shear stresses on the cells encapsulated in the bioink exiting the nozzle under the extrusion pressures 5, 10, 15, 20, and 25 kPa, respectively (from left to right). (D) Optimization of the printing speed and extrusion pressure: (i) a schematic to illustrate printing speed and extrusion pressure; (ii) MeTro/GelMA bioink filaments printed into the support bath at different printing pressures and speeds; (iii) qualitative evaluation of the MeTro/GelMA bioink printability at different printing pressures and speeds. (E) Optimized printing parameters for MeTro/GelMA bioink. (F) A lattice-shaped construct printed up to 16 layers to form constructs with a linear relationship between the number of layers and the height of the construct. (G) Various 3D constructs printed with MeTro/GelMA bioink (from left to right: heart slice, lattice cube and cat toy).
The MeTro/GelMA solution originally exhibited a viscosity (η) of ~162.6 mPa·s at shear rate of ~50 s−1 and temperature of ~8 °C which were close to the conditions used during printing (Figure 2B). The addition of gelatin transforms the solution into a viscoelastic fluid and imparts the shear thinning behavior under 10 °C due to the reversible gelation of gelatin (Figure S3A, B). The resulting material, referred to MeTro/GelMA bioink, possesses a viscosity that exceeded 3000 mPa·s which was 20-fold higher than the composite bioink without gelatin. The non-Newtonian behavior of this bioink was also evidenced by its high storage modulus (G′) over the loss modulus (G″) (Figure S3C).
Although the use of gelatin increases the rheological properties, the uncured ink lacks the ability to provide mechanical stability and immediately deforms upon exiting the nozzle. Thus, we utilized a freeform reversible embedding of suspended hydrogels (FRESH) printing technique to enable the fabrication of complex structures.[34] Carbopol bath was used due to its biocompatibility and ease of processing. Our biocompatible support bath is mainly composed of crosslinked poly(acrylic acid) particles, which can be deswollen post printing and removed from the structure permanently. Following photopolymerization of the printed construct, Carbopol bath was liquefied by addition of monovalent cations such as Dulbecco’s phosphate-buffered saline (DPBS) and the print was released from the Carbopol.
Next, extrusion pressure and printing speed for 3D printing of MeTro/GelMA bioink were optimized to achieve the optimal printing time, provide structural integrity to the printed constructs, and reduce stress to the cells during the printing process. We indirectly measured actual shear stress on the cells to study the impact of the extrusion pressure on the cells encapsulated within the bioink. The results revealed that the shear stress increased from 0.79 to 1.17 kPa, when the extrusion pressure increased from 5 kPa to 25 kPa (Figures 2C and S3D). To qualitatively assess the printability of MeTro/GelMA bioink, curved-line structures were printed under different printing speeds from 10 mm/s to 50 mm/s and extrusion pressures from 5 kPa to 25 kPa (Figure 2D). The printing conditions were categorized into good, normal and bad. Based on the results, the final printing condition was selected to be 15 kPa pressure and 30 mm/s speed at 8–10 °C which had 1.08 kPa of shear stress on the cells in MeTro/GelMA bioink (Figure 2E).
The flow of MeTro/GelMA bioink exiting the nozzle was smooth without clogging and the resulting constructs after crosslinking were mechanically stable and robust, allowing to print multi-layered constructs. Lattice constructs were printed up to 16 layers (corresponding to 5 mm) and a linear relationship was obtained between the number of layers and the height of the construct (Figures 2F and S4). To ensure reliable printing fidelity of our bioink and printing system, constructs with complex architecture were designed and printed with different sizes and heights including a heart slice, a lattice cube, and a cat toy (Figure 2G).
To test the stability of the 3D printed MeTro/GelMA lattice constructs in aqueous solution, the weight and microscopic structure of the constructs were monitored during a week of incubation in DPBS at 37 °C (Figure S5). The results showed more than 30 %(w/w) of the original weight of the printed structures decreased in a day with no significant changes afterwards, implying the gelatin and residual Carbopol gel in the 3D printed constructs were removed during the first 24 h.
The enzymatic degradation of 3D printed constructs was characterized using collagenase type II solution in DPBS for up to 14 days (Figure S6). Since we observed significant amount of the construct weight was lost due to the removal of gelatin and Carbopol, the printed constructs were incubated in DPBS at 37 °C in order to remove the residual gelatin and Carbopol for 3 days prior to the degradation test. Results showed 12.7 ± 2.5 % degradation of the construct during the first 24 h, however, the degradation rate slowed down and reached 17.9 ± 3.5 % at day 14.
Cytocompatibility of MeTro/GelMA bioink was studied using different cell types. First, 3T3 cells were used for 2D cell seeding on the printed structure as well as for bioprinting 3D cell-laden structures. In both 2D and 3D culture conditions, high cell viabilities (> 90 %) were achieved (Figures S7 and S8). To evaluate the suitability of our engineered bioinks for printing cardiovascular tissues, we next encapsulate cardiomyocytes (CMs), cardiac fibroblasts (CFs) and human umbilical vein endothelial cells (HUVECs) within the MeTro/GelMA bioink to 3D bioprint constructs containing multiple cell types. A CM/CF-laden bioink and a HUVEC-laden bioink were loaded to separate syringes and sequentially co-printed into a heterogeneous lattice construct (Figure 3A). For 3D bioprinting, duration of the time during which cells were encapsulated in the bioink at 8°C was around 30 minutes. The cold shock responses of mammalian cells generally involve suppression of transcription and subsequent translation, modulation of the cell cycle and reduction in metabolism. However, no significant responses of cells to cold stress were observed in our in vitro experiments, suggesting that the exposure to 8°C for this relatively short period of time had negligible effects on cells. All cells showed over 85 % of cell viability during the 7 days of culture (Figures S9). In addition, the result of immunostaining analysis confirmed the expression of sarcomeric α-actinin by the CMs and CD31 by HUVECs on day 7 (and 3B). These results demonstrate that our printing approach supports cellular growth and proliferation without affecting the cells’ phenotype.
Figure 3. 3D bioprinting of cell-laden elastic constructs using MeTro/GelMA bioink.
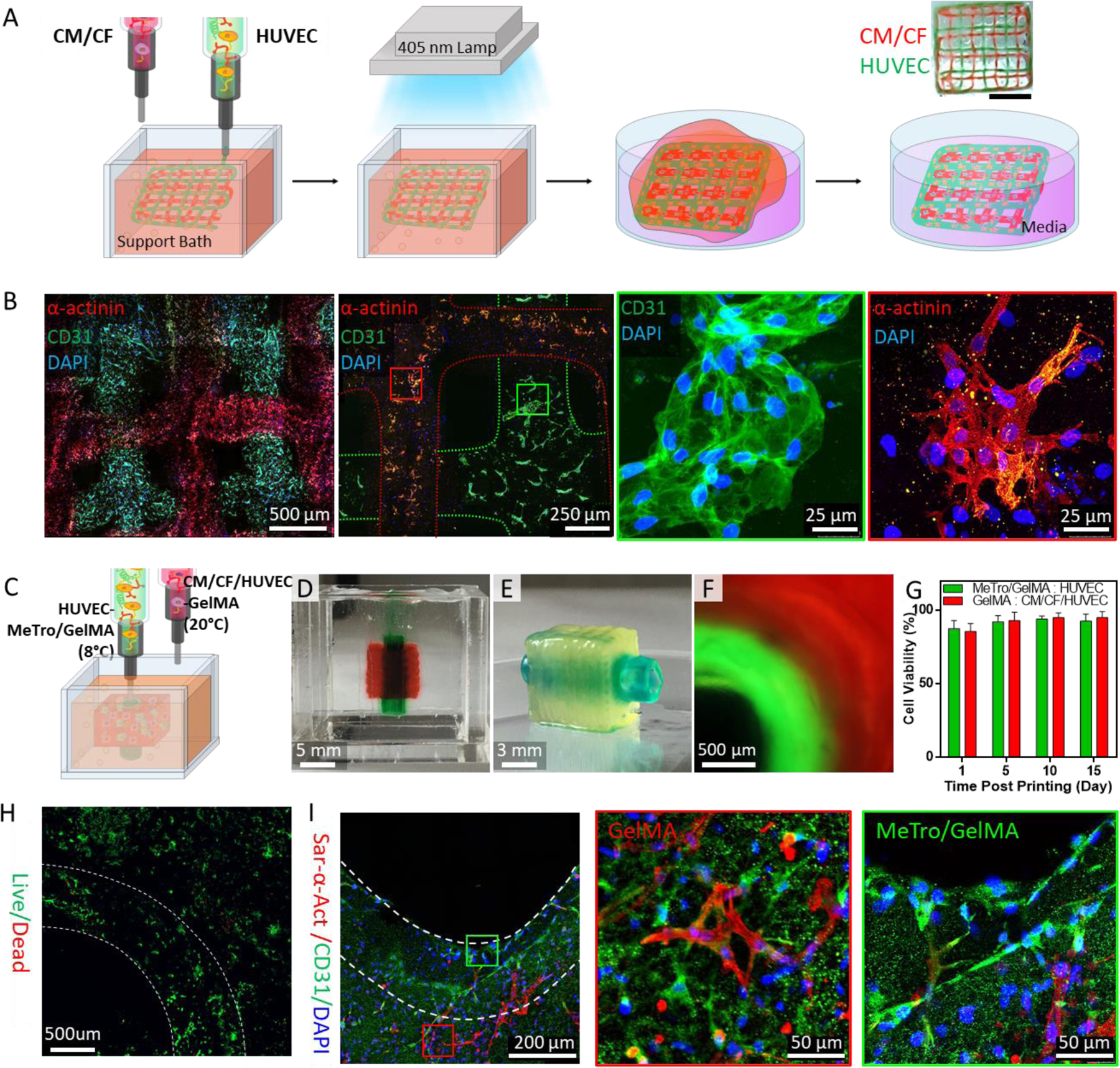
(A) A schematic illustration of 3D bioprinting of lattice scaffolds using HUVECs- and CMs/CFs-laden MeTro/GelMA bioinks. Green and red food colors were used to distinguish the HUVECs- and CMs/CFs-laden inks, respectively, only for imaging experiments. (B) Immunostaining of the lattice structure against sarcomeric α-actinin (red), CD31 (green), and DAPI (blue) at day 7 post bioprinting. Printed CMs/CFs and HUVECs are marked with red and green boxes, respectively. (C) A schematic to describe 3D bioprinting vascularized cardiac constructs with HUVECs-laden MeTro/GelMA bioink and CMs/CFs/HUVECs-laden GelMA bioink. (D) A vascularized cardiac construct in the support bath right after printing process and (E) the construct after the photocrosslinking and washing steps. Green and red food colors were used to distinguish the MeTro/GelMA and GelMA bioinks, respectively, only for imaging experiments. (F) A cross-sectional fluorescence image of the vascularized cardiac construct. Fluorescein and rhodamine dyes were added to the MeTro/GelMA and GelMA bioinks, respectively. (G) Viability of HUVECs (in MeTro/GelMA bioink) and CMs/CFs/HUVECs (in GelMA bioink) within vascularized cardiac tissue constructs. (H) Live/dead staining of the vascularized cardiac construct at day 10 post bioprinting. (I) Immunostaining of the vascularized cardiac construct against sarcomeric α-actinin (red), CD31 (green), and DAPI (blue) at day 10 post bioprinting. HUVECs (in MeTro/GelMA bioink) and CMs/CFs/HUVECs (in GelMA bioink) are marked with green and red boxes, respectively.
In human body, blood vessels have essential roles in constant nutrient-waste exchange between the blood and tissue as well as in homeostasis and regulation of the human body system. Likewise, 3D bioprinted tissue constructs require vascular systems in order to circumvent necrosis and mimic native tissue function. Connective tissues such as cartilage and bone have been successfully printed without vasculature,[35, 36] however, the creation of human-sized soft tissues for clinical translation remains challenging due to slow vascularization rate in tissue constructs. One solution for this challenge is to create vascular channel within artificial tissues to speed up the vascularization process when implanted in vivo,[10, 11] which requires higher standards on printing speed, accuracy, processability and structural heterogeneity compared to printing simple structure like lattice. Therefore, in our second in vitro model, we aimed to use our elastin-based bioink to print a vascularized cardiac tissue as a proof of concept.
To this end, two bioinks were prepared to engineer a vascularized cardiac tissue model (Figure 3C): i) GelMA bioink loaded with CFs/CMs and HUVECs to represent parenchymal cardiac tissue; ii) MeTro/GelMA bioink loaded with HUVECs for vessel formation. Prior to 3D bioprinting, rheological properties and printing conditions for GelMA bioink were optimized using a similar method as described for MeTro/GelMA bioink (Figures S10 and S11). In the degradation study, 3D printed GelMA constructs showed faster degradation rate than MeTro/GelMA constructs due to the high MMP-sensitivity of GelMA (Figure S12).
Our cardiac tissue model was composed of 67 layers with 7.2 mm in length and width, and 16.5 mm in height (Figure S13). The bioprinting process took around 11 min and the bioprinted structure was photocrosslinked upon exposure to visible light for 3 min (Figure 3D–F, Movies 1 and 2). For the in vitro analysis, the printed constructs were sectioned into 3 mm slices with a razor blade. The viabilities of CFs/CMs and HUVECs in the printed structure were above 85% up to 15 days of culture (Figure 3G, H and S14). In addition, the printed cardiac tissue constructs demonstrated high retention of cells’ phenotype as confirmed by immunofluorescent staining for sarcomeric α-actinin, CD31 and DAPI at day 10 (Figure 3I). The endothelial cells proliferated in both GelMA and MeTro/GelMA hydrogels, as confirmed by CD31 expression throughout the whole constructs. Also, we observed that the CMs were elongated and branched in every direction to connect to each other.
The layer of endothelial cells in human blood vessel is essential to form a semi-permeable barrier that regulates the transport of water, proteins and blood cells between the blood and interstitial fluids, which is important for normal blood vessel function.[37] In our study, to quantify the barrier properties imparted by HUVECs in vascular construct, we measured the diffusional permeability of fluorescein isothiocyanate-conjugated dextran (FITC-Dex) (Figure S15A). The results revealed that compared to the acellular construct (1.4 ± 0.4 ×10−3 cm/s), the HUVECs-laden structure (0.8 ± 0.3 ×10−3 cm/s) showed a 2-fold reduction in diffusional permeability, confirming the barrier function of the endothelium (Figures 4A–C and S15B).
Figure 4. Evaluation of in vitro function and in vivo biocompatibility of the 3D printed vascularized cardiac constructs.
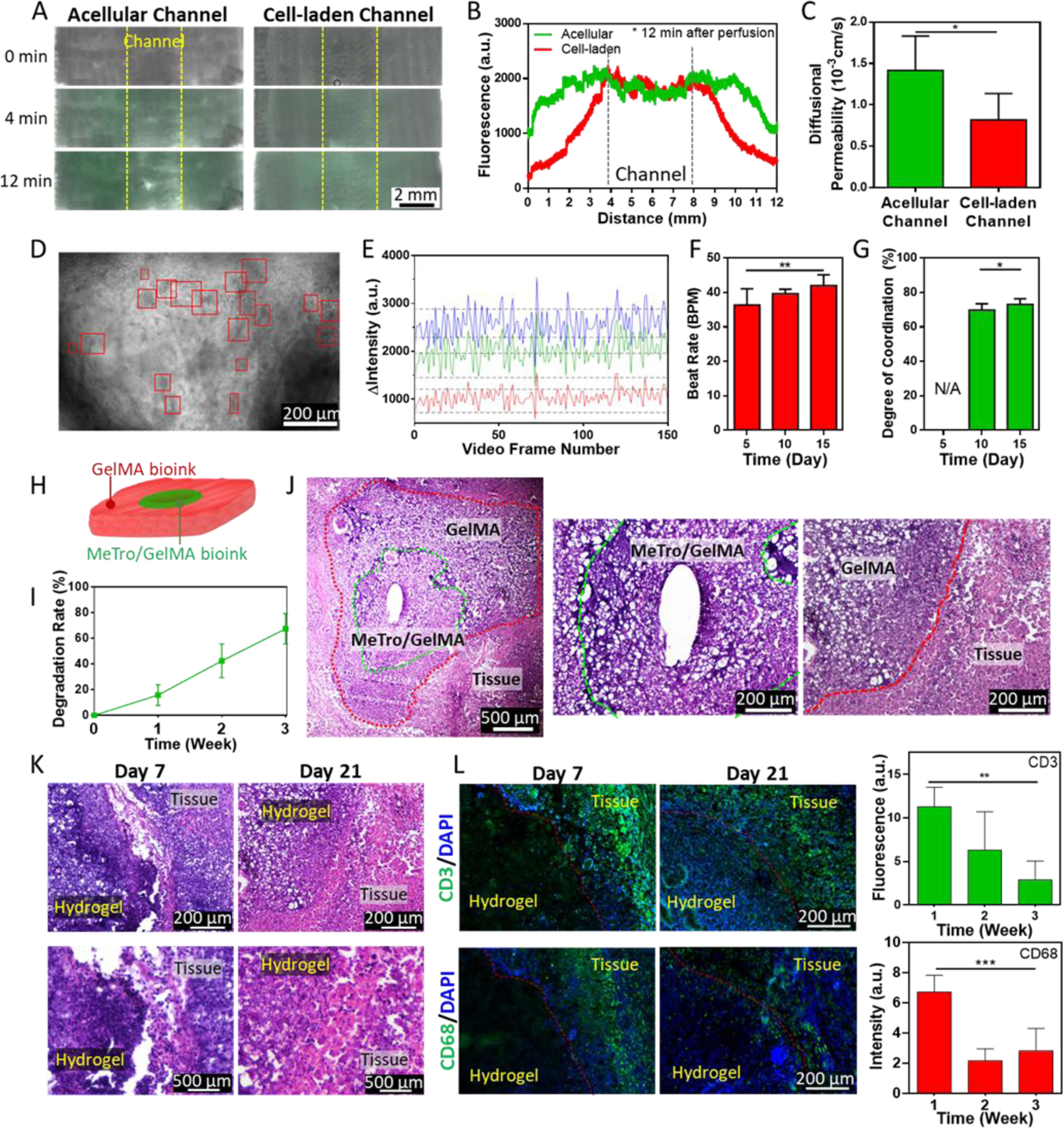
(A) Representative images of the cardiac constructs with acellular and HUVECs-laden channel 0, 4, and 12 after infusion of FITC-Dex to quantify the barrier properties imparted by HUVECs in the printed construct. (B) FITC-Dex distribution within the constructs plotted as a function of distance from vasculature. (C) Diffusional permeability of FITC-Dex in acellular and HUVECs-laden channels. (D) A representative image of the vascularized cardiac construct at day 15 post printing. Beating CMs were identified and marked with red boxes as region of interests (ROIs). (E) Beating quantification of the CMs in vascularized cardiac construct at day 15 post bioprinting. In the representative plot, beating was recorded as change in pixel intensity within three different ROIs. (F) Beat rate and (G) degree of coordination measured for the beating CMs. (H) A schematic of the 3D printed implant structure. (I) In vivo biodegradation rate of the 3D printed constructs. (J) H&E staining of the 3D printed constructs explanted at day 21 post implantation. (K) H&E staining images showing the interface between implanted hydrogel constructs and tissues at day 7 and 21 post implantation at 10 × (top) and 40 × (bottom) magnifications. (L) IHC staining against CD3, (top, green) and CD68 markers (bottom, green) and DAPI (blue) in the explanted 3D printed constructs at day 7 and 21 post implantation. Inflammatory response was quantified by calculating fluorescence intensity from IHC images. (* p<0.05, ** p<0.01, *** p<0.001)
Cardiac excitation-contraction coupling (EC coupling) describes a series of events from the production of an electrical impulse to the contraction of muscles in the heart.[38] In a native heart tissue, when more than two independently beating cardiomyocytes are joined, the cell with the highest inherent rate sets the pace. Cardiomyocytes are interconnected with gap junctions and the electrical impulses to stimulate contractions spread from the fastest one to the rest, resulting in synchronized beating. In this study, we evaluated EC coupling of the 3D bioprinted cardiac tissue constructs via video microscopy and a custom algorithm to quantify beat frequency and degree of coordination as previously described.[26] Beating cardiac cells were identified within 3D constructs based upon inclusion criteria (i.e., signal-to-noise ratio, peak-to-peak frequency) and the dynamic beating behavior of individual cells was quantitatively analyzed. The contracting CMs were first noticed at day 5 post bioprinting. During the 15 days of culture, the contractions became more synchronous with increased interconnection between striated muscles (Figures 4D–G and S16, Movies 3–5), as confirmed by immunofluorescent staining of sarcomeric α actinin-positive myofibrils. The beating rate and the coordination reached 42.0 ± 3.1 bpm and 73.1 ± 3.2 % at day 15 (The degree of coordination for native heart is considered 100 %). This result implies that the cells were tightly interconnected through gap junctions, which contributed to the rapid transmission of the electric impulse between cells and enabled the synchronized contraction.
To assess in vivo degradation and biocompatibility of our bioink, the acellular constructs that were 3D printed with MeTro/GelMA and GelMA bioinks were subcutaneously implanted in rats (Figure 4H). The samples were explanted on days 7, 14, and 21 in order to obtain the degradation rate and perform histological examination. The average biodegradation of the implanted samples increased from 15.8 ± 8.1 % at day 7 to 67.4 ± 11.9 % at day 21 post-implantation (Figure 4I). Similarly, the hematoxylin and eosin (H&E) staining of the samples revealed effective biodegradation and significant integration between engineered constructs and surrounding tissue (Figure 4J). This result suggests that the bioink can be potentially used for repair and replacement of the damaged or diseased soft tissues due to fast degradation and bio-integration. A fibrous capsule was observed around the explanted samples at day 7 but disappeared after 14 and 21 days, indicating minimal inflammatory response after implantation of the constructs (Figures 4K and S17). In addition, the immunohistochemical (IHC) staining for macrophages (CD68) and T-lymphocytes (CD3) was performed on the cryo-sectioned samples and analyzed both qualitatively and quantitatively to evaluate biocompatibility of the constructs (Figure 4L). The results showed infiltration of CD3 and CD68 antigens on day 7 due to natural foreign body response. However, this response was not observed after 21 days, showing significant reduction of both markers, which proves high biocompatibility of the printed constructs. Similar to H&E stain results, a significant cell infiltration was observed on day 21 post implantation, confirming the construct was well-integrated with the host tissue.
In this work, we demonstrate, for the first time, the use of recombinant human tropoelastin combined with gelatin as an elastic bioink for 3D bioprinting. The synergistic association of two biopolymers allowed a high-resolution printing with great cell viability. As a proof of concept to fabricate an entirely 3D-printed artificial tissue, vascularized cardiac tissue constructs were 3D printed and characterized both in vitro and in vivo. The printed constructs presented endothelium barrier function and spontaneous beating of cardiac cells which are important functions of cardiac tissue in vivo. Furthermore, the printed construct elicited minimal inflammatory responses, and were shown to be efficiently biodegraded in vivo when implanted subcutaneously in rats. Taken together, our results demonstrate the potential of MeTro/GelMA bioinks for printing complex 3D functional cardiac tissues which could eventually be used for cardiac replacement.
3D printing holds great promise for engineering whole organs, but there are still challenges remaining. These include the needs for a physiologically relevant number of cells and scalable approaches to print complex human-scale tissues with hierarchical vascular network. The vascularized cardiac construct presented in this work has proven that we can fabricate small-scaled constructs that recapitulate the architectural and functional properties of native tissues. For clinical translation, therapeutic value of cell-laden cardiac tissue should be evaluated by using a relevant animal model (e.g. myocardial infarction model). We envision that integrating our approach with modular tissue design will enable fabricating a complex multifunctional tissue at a clinically relevant scale.
Experimental Section
Hydrogel preparation:
MeTro and GelMA were synthesized as described previously (more details: Methods Section, Supporting Information).[16, 23] To form MeTro/GelMA hydrogels, different concentrations of MeTro (0, 7.5 and 15 % (w/v)) and GelMA (0, 7.5 and 15 % (w/v)) were dissolved in DPBS with 0.4 % (w/v) Lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) as a photoinitiator at 4 °C. The precursor solutions were then placed in polydimethylsiloxane (PDMS) molds and photocrosslinked using a LED light (405 nm, 10 W) for 120 sec.
Bioink preparation:
Gelatin from cold water fish skin (Sigma-Aldrich) were used to enhance the printability of the both MeTro/GelMA and GelMA bioinks. After evaluating the extrudability and the cell viability, the final concentrations of gelatin were determined for each bioinks. For MeTro/GelMA bioinks, 7.5 % (w/v) of MeTro, 7.5 % (w/v) of GelMA, 20 % (w/v) gelatin and 0.4 % (w/v) LAP were dissolved in cell culture media at around 10 °C and the pH was adjusted to 7. For GelMA bioinks, 10 % (w/v) of GelMA, 23 % (w/v) gelatin and 0.4 % (w/v) LAP were dissolved in cell culture media and the pH was adjusted to 7.
To make cell-laden bioinks, each step was carefully conducted under sterile conditions and appropriate number of cells were mixed with the solutions at the last step. The lattice structures were printed with MeTro/GelMA bioink with 2×107 cells/ml of HUVECs and MeTro/GelMA bioink with 2×107 cells/ml of CMs and 1×107 cells/ml of CFs. For the vascularized cardiac constructs, MeTro/GelMA bioink with 2×107 cells/ml of HUVECs and GelMA bioink with 2×107 cells/ml of CMs, 1×107 cells/ml of CFs and 5×106 cells/ml of HUVECs were used.
Support bath preparation:
Carbopol gel support baths were prepared as described previously with some modification (Supporting Information, Methods).[39]
Mechanical properties:
The tensile and compressive properties of the biomaterials were evaluated using an Instron 5542 mechanical tester, as described before (Supporting Information, Methods).[40]
Rheological properties:
A rheometer (MCR 92, Anton Paar) equipped with a parallel plate with a gap size of 1 mm and a diameter of 8 mm was used to characterize the rheological properties different solutions including MeTro/GelMA pre-polymers, gelatin solution, MeTro/GelMA bioinks and GelMA bioinks. Viscosity and dynamic modulus of the solutions were determined as a function of temperature. Viscosity and shear stress of both bioinks were measured as a function of shear rate from 0.1 to 1000 s−1. The shear force applied to cells during bioprinting was measured indirectly by calculating the shear rate during printing and matching it to shear stress-shear rate data. More details on rheological properties are outlined in Methods Section, Supporting Information.
3D printing and bioprinting:
Acellular and cell-laden bioinks were printed into different structures using an INKREDIBLE+ bioprinter (Cellink®) as detailed in Methods Section, Supporting Information. The printed structures were then exposed to light (405 nm) to crosslink up to 3 min depending on the shape and the size of the structures, carefully removed from the support bath, then washed with warm DPBS or cell culture media.
Enzymatic degradation:
Enzymatic degradation of 3D printed constructs were assessed as described before (Supporting Information, Methods).[41]
Cell culture and isolation:
Human umbilical vein endothelial cells (HUVECs, passage 6) were obtained from Lonza and cultured in endothelial growth BulletKit (EGM-2, Lonza) at 37 °C in a humidified atmosphere containing 5% CO2. Neonatal ventricular rat cardiomyocytes and cardiac fibroblasts were isolated by the UCLA NRVM core facility. (more details: Methods Section, Supporting Information).
Cell viability and immunofluorescence analysis:
Cell viability was evaluated using a calcein AM/ethidium homodimer-1 live/dead kit (Invitrogen) as described previously.[40] For immunofluorescence analysis, samples were stained against rabbit polyclonal anti-CD31 (ab32457, Abcam), mouse monoclonal anti-sarcomeric α-actinin (ab9465, Abcam), and goat polyclonal anti-connecxin-43 (ab11370, Abcam) antibodies as described elsewhere.[42]
Cardiac beating quantification:
Beat frequency and degree of coordination of cardiac cells were assessed using video microscopy and a custom algorithm as previously described (more details: Methods Section, Supporting Information).[26]
Evaluation of endothelium barrier function:
The barrier function of the printed vasculature, and diffusional permeability was quantified by perfusing culture media with FITC-conjugated 70-kDa dextran (Sigma) in the vascular channel (Methods Section, Supporting Information).[12]
Dorsal subcutaneous implantation of hydrogels:
The cardiac constructs 3D printed with MeTro/GelMA and GelMA bioinks were subcutaneously implanted in rats. (Methods Section, Supporting Information).
Histological and immunohistochemical analysis:
Histological and immunohistochemical analysis were performed as described previously. (Methods Section, Supporting Information).[40]
Statistical analysis:
All values are presented as mean ± standard deviation. Statistical differences between sample means at each condition were evaluated with two-way ANOVA tests using GraphPad (Software Inc., CA, USA) as P-values were defined as *< 0.05, **< 0.01, ***< 0.001, and ****< 0.0001.
Supplementary Material
Acknowledgements
N.A. acknowledges the support from American Heart Association (AHA) (DE02750601) and National Institutes of Health (NIH) (R01EB023052; R01HL140618). A.S.W. acknowledges support from the National Health and Medical Research Council. Confocal laser scanning microscopy was performed at the Advanced Light Microscopy/Spectroscopy Laboratory and the Leica Microsystems Center of Excellence at the California NanoSystems Institute at UCLA with funding support from NIH Shared Instrumentation Grant S10OD025017 and NSF Major Research Instrumentation grant CHE-0722519. The authors acknowledge Jonathan Soucy for producing the MATLAB code for cardiac cell analysis.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Andrew R. Spencer, Holistick Medical, Paris 75003, France
Yvonne Guan, Department of Chemical and Biomolecular Engineering, University of California- Los Angeles, Los Angeles, California 90095, USA.
Anthony S. Weiss, School of Life and Environmental Sciences, Charles Perkins Centre, University of Sydney, NSW, Australia Charles Perkins Centre, University of Sydney, NSW, Australia; Bosch Institute, University of Sydney, NSW, Australia.
Nasim Annabi, Department of Chemical and Biomolecular Engineering, University of California- Los Angeles, Los Angeles, California 90095, USA; Center for Minimally Invasive Therapeutics (C-MIT), California NanoSystems Institute (CNSI), University of California – Los Angeles, Los Angeles, CA 90095, USA.
Reference
- [1].Murphy SV, Atala A, Nature biotechnology 2014, 32, 773. [DOI] [PubMed] [Google Scholar]
- [2].Murphy SV, De Coppi P, Atala A, Nature biomedical engineering 2019, 1. [DOI] [PubMed] [Google Scholar]
- [3].Park JH, Jang J, Lee J-S, Cho D.-W. J. A. o. b. e., 2017, 45, 180. [DOI] [PubMed] [Google Scholar]
- [4].Derby BJS, 2012, 338, 921. [DOI] [PubMed] [Google Scholar]
- [5].Hong S, Sycks D, Chan HF, Lin S, Lopez GP, Guilak F, Leong KW, Zhao X, Advanced materials 2015, 27, 4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Highley C, Rodell C, Burdick J, “Direct 3D printing of shear-thinning hydrogels at high resolution using open-source technologies”, presented at Front. Bioeng. Biotechnol. Conference Abstract: 10th World Biomaterials Congress. doi: 10.3389/conf FBIOE, 2016. [DOI] [Google Scholar]
- [7].Włodarczyk-Biegun MK, del Campo A, Biomaterials 2017, 134, 180. [DOI] [PubMed] [Google Scholar]
- [8].Cui X, Boland T, Biomaterials 2009, 30, 6221. [DOI] [PubMed] [Google Scholar]
- [9].Ma X, Yu C, Wang P, Xu W, Wan X, Lai CSE, Liu J, Koroleva-Maharajh A, Chen S, Biomaterials 2018, 185, 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Noor N, Shapira A, Edri R, Gal I, Wertheim L, Dvir T, Advanced Science 2019, 1900344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lee A, Hudson A, Shiwarski D, Tashman J, Hinton T, Yerneni S, Bliley J, Campbell P, Feinberg A, Science 2019, 365, 482. [DOI] [PubMed] [Google Scholar]
- [12].Kolesky DB, Homan KA, Skylar-Scott MA, Lewis JA, Proceedings of the national academy of sciences 2016, 113, 3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wise SG, Yeo GC, Hiob MA, Rnjak-Kovacina J, Kaplan DL, Ng MK, Weiss AS, Acta biomaterialia 2014, 10, 1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Williamson MR, Shuttleworth A, Canfield AE, Black RA, Kielty CM, Biomaterials 2007, 28, 5307. [DOI] [PubMed] [Google Scholar]
- [15].Yin Y, Wise SG, Nosworthy NJ, Waterhouse A, Bax DV, Youssef H, Byrom MJ, Bilek MM, McKenzie DR, Weiss AS, Biomaterials 2009, 30, 1675. [DOI] [PubMed] [Google Scholar]
- [16].Sani ES, Lara RP, Aldawood Z, Bassir SH, Nguyen D, Kantarci A, Intini G, Annabi NJM, 2019. [DOI] [PMC free article] [PubMed]
- [17].Annabi N, Mithieux SM, Zorlutuna P, Camci-Unal G, Weiss AS, Khademhosseini A, Biomaterials 2013, 34, 5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Noshadi I, Hong S, Sullivan KE, Shirzaei Sani E, Portillo-Lara R, Tamayol A, Shin SR, Gao AE, Stoppel WL, Black Iii LD, Khademhosseini A, Annabi N, Biomater Sci 2017. [DOI] [PMC free article] [PubMed]
- [19].Yue K, Trujillo-de Santiago G, Alvarez MM, Tamayol A, Annabi N, Khademhosseini A, Biomaterials 2015, 73, 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yeo GC, Keeley FW, Weiss AS, Advances in colloid and interface science 2011, 167, 94. [DOI] [PubMed] [Google Scholar]
- [21].Annabi N, Zhang Y-N, Assmann A, Sani ES, Cheng G, Lassaletta AD, Vegh A, Dehghani B, Ruiz-Esparza GU, Wang X. J. S. t. m., 2017, 9, eaai7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Soucy JR, Shirzaei Sani E, Portillo Lara R, Diaz D, Dias F, Weiss AS, Koppes AN, Koppes RA, Annabi N, Tissue Engineering Part A 2018. [DOI] [PMC free article] [PubMed]
- [23].Annabi N, Rana D, Sani ES, Portillo-Lara R, Gifford JL, Fares MM, Mithieux SM, Weiss AS, Biomaterials 2017, 139, 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Assmann A, Vegh A, Ghasemi-Rad M, Bagherifard S, Cheng G, Sani ES, Ruiz-Esparza GU, Noshadi I, Lassaletta AD, Gangadharan S, Tamayol A, Khademhosseini A, Annabi N, Biomaterials 2017, 140, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sinha RP, Häder D-P, Photochemical & Photobiological Sciences 2002, 1, 225. [DOI] [PubMed] [Google Scholar]
- [26].Soucy JR, Askaryan J, Diaz D, Koppes AN, Annabi N, Koppes RA, Biofabrication 2019, 12, 015014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yeh Y-C, Highley CB, Ouyang L, Burdick JA, Biofabrication 2016, 8, 045004. [DOI] [PubMed] [Google Scholar]
- [28].Shin S, Park S, Park M, Jeong E, Na K, Youn HJ, Hyun J, BioResources 2017, 12, 2941. [Google Scholar]
- [29].Jungst T, Smolan W, Schacht K, Scheibel T, Groll J. r., Chemical reviews 2015, 116, 1496. [DOI] [PubMed] [Google Scholar]
- [30].Yang F, Tadepalli V, Wiley BJ, ACS Biomaterials Science & Engineering 2017, 3, 863. [DOI] [PubMed] [Google Scholar]
- [31].Chimene D, Peak CW, Gentry JL, Carrow JK, Cross LM, Mondragon E, Cardoso GB, Kaunas R, Gaharwar AK, ACS applied materials & interfaces 2018, 10, 9957. [DOI] [PubMed] [Google Scholar]
- [32].Compaan AM, Christensen K, Huang Y, ACS Biomaterials Science & Engineering 2016, 3, 1519. [DOI] [PubMed] [Google Scholar]
- [33].Kraut G, Yenchesky L, Prieto F, Tovar GE, Southan A, Journal of Applied Polymer Science 2017, 134, 45083. [Google Scholar]
- [34].Hinton TJ, Jallerat Q, Palchesko RN, Park JH, Grodzicki MS, Shue H-J, Ramadan MH, Hudson AR, Feinberg AW, Science advances 2015, 1, e1500758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Nguyen D, Hägg DA, Forsman A, Ekholm J, Nimkingratana P, Brantsing C, Kalogeropoulos T, Zaunz S, Concaro S, Brittberg M, Scientific reports 2017, 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kang H-W, Lee SJ, Ko IK, Kengla C, Yoo JJ, Atala A, Nature biotechnology 2016, 34, 312. [DOI] [PubMed] [Google Scholar]
- [37].Malik AB, Lynch JJ, Cooper JA, Journal of Investigative Dermatology 1989, 93, S62. [DOI] [PubMed] [Google Scholar]
- [38].Santana LF, Cheng EP, Lederer WJ, Journal of molecular and cellular cardiology 2010, 49, 901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Spencer A, Shirzaei Sani E, Soucy JR, Corbet CC, Primbetova A, Koppes RA, Annabi N, ACS applied materials & interfaces 2019. [DOI] [PMC free article] [PubMed]
- [40].Shirzaei Sani E, Portillo-Lara R, Spencer A, Yu W, Geilich BM, Noshadi I, Webster TJ, Annabi N, ACS Biomaterials Science & Engineering 2018. [DOI] [PMC free article] [PubMed]
- [41].Sani ES, Kheirkhah A, Rana D, Sun Z, Foulsham W, Sheikhi A, Khademhosseini A, Dana R, Annabi N, Science Advances 2019, 5, eaav1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Walker BW, Lara RP, Yu C, Sani ES, Kimball W, Joyce S, Annabi NJB, 2019. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


