Abstract
Human natural killer (NK) cells develop in tonsils through discrete NK cell developmental intermediates (NKDIs), yet the mechanistic regulation of this process is unclear. We demonstrate that Notch activation in human tonsil-derived stage 3 (CD34−CD117+CD94−NKp80−) and 4A (CD34−CD117+/−CD94+NKp80−) NKDIs promoted non-NK innate lymphoid cell (ILC) differentiation at the expense of NK cell differentiation. In contrast, stage 4B (CD34−CD117+/−CD94+NKp80+) NKDIs were NK cell lineage committed despite Notch activation. Interestingly, while NK cell functional maturation from stage 3 and 4A NKDIs was independent of Notch activation, the latter was required for high NKp80 expression and a stage 4B-like phenotype by the NKDI-derived NK cells. The Notch-dependent effects required simultaneous engagement with OP9 stromal cells and were also stage-specific, with NOTCH1 and NOTCH2 receptors regulating stage 3 NKDIs, and NOTCH1 primarily regulating stage 4A NKDIs. These data establish stage-specific and stromal-dependent roles for Notch in regulating human NK cell developmental plasticity and maturation.
Keywords: NK cell development, Notch, ILC development, plasticity
INTRODUCTION
Natural killer (NK) cells are cytotoxic Group 1 innate lymphoid cells (ILCs) that can recognize and directly kill infected or malignant cells and also modulate immune responses through the production of chemokines and cytokines such as interferon-gamma (IFN-γ). NK cells develop from multipotent bone marrow hematopoietic stem cells and further restricted CD34+CD45RA+α4β7 integrin+ progenitor cells that traffic to secondary lymphoid tissues (SLTs) including lymph nodes and tonsils (1, 2). Once in these tissues, the progenitors lose CD34 expression and proceed through successive stages of NK cell developmental intermediates (NKDIs) that can be identified based on their differential expression of functional surface markers including CD117, CD94, NKp80, and CD16. Accordingly, stage 3 NKDIs (CD34−CD117+CD94−NKp80−CD16−) are the immediate precursors to stage 4 NKDIs (CD34−CD117+/−CD94+NKp80+/−CD16−), which in turn are the precursors to stage 5 NKDIs (CD34−CD117−CD94+/−NKp80+CD16+). The stage 4 population in SLTs was recently shown to be heterogeneous and can be further subdivided based on the expression of NKp80 into a “stage 4A” subset (CD34−CD117+/−CD94+NKp80−CD16−) that is non-cytotoxic and cannot produce IFN-γ, as well as a “stage 4B” subset (CD34−CD117+/−CD94+NKp80+CD16−) that is cytotoxic and can produce IFN-γ.
More recent work has discovered that the linear pathway of human NK cell development described above intersects with other pathways of ILC differentiation. Stage 3 NKDIs include the recently described population of common ILC precursors (ILCPs) that can generate Group 1–3 ILCs but no other leukocyte populations (3, 4). While it is known that the potential for NKDIs to diverge from NK cell development toward other pathways (i.e. developmental plasticity) becomes increasingly restricted (4, 5), it is not yet clear at which developmental stage NK cells become fully committed. Moreover, the mechanisms that regulate NK cell and ILC differentiation and maturation remain to be elucidated.
Previous work has implicated the Notch signaling pathway in regulating numerous aspects of lymphopoiesis including ILC differentiation and NK cell development (6–8). Canonical Notch signaling becomes activated when a Notch ligand such as Delta-like-1 (DL1) binds in trans to one of four Notch receptors expressed on the surface of an adjacent cell. Following receptor proteolysis, the Notch intracellular domain (NICD) is released to the nucleus where it interacts with the RBPJ-κ transcription factor and recruits co-activator proteins, thereby controlling the transcription of target genes such as HES1 (9). To that end, Notch has been previously studied for its role in NK cell developmental biology. For example, human cord blood-derived CD34+ progenitors can develop into functional CD56+ NK cells in the presence of Notch ligand (10, 11). Notch signaling in combination with IL-15 was also shown to upregulate killer immunoglobulin-like receptor (KIR) expression by human cord-blood CD34+ progenitor cell-derived NK cells (12). However, what remains unknown is how Notch signaling regulates NK cell plasticity and commitment at distinct intermediate developmental stages that have only been recently described in SLTs.
In this study, we demonstrate the stage-specific effects of different Notch receptors impacting both the developmental plasticity and maturation of tonsil-derived NKDIs. We observed that interactions with a stromal cell microenvironment were necessary for the Notch-dependent effects, and we also identified the specific stage of development at which NK cells were committed to the NK cell lineage. These data refine our current model of NK cell development establishing stage-specific requirements for both stromal cells and Notch ligands for the synchronous phenotypic and functional maturation of human NK cells.
MATERIALS AND METHODS
Human Tissue Samples
Human tissues were collected and used in accordance with protocols approved by The Ohio State University Institutional Review Board (OSU IRB). Donor consent was acquired when deemed appropriate according to the approved OSU IRB protocol. Human pediatric tonsils were obtained fresh following same-day surgery via the NCI-approved Cooperative Human Tissue Network (CHTN) from Nationwide Children’s Hospital, Columbus, OH. Leukocyte-depleted red blood cells were commercially obtained from the American Red Cross and Versiti.
Methods Details
Cell Isolation and Fluorescence-Activated Cell Sorting
Human ILCs were enriched from fresh tonsil tissue specimens using previously reported protocols (13). Briefly, single cell suspensions from fresh human pediatric tonsils were generated by dissociation via a GentleMACS Dissociator (Miltenyi Biotec) according to manufacturer’s instructions. Cells were resuspended in fetal bovine serum (FBS, Sigma-Aldrich) with leukocyte-depleted red blood cells (RBCs) and a custom human NK cell enrichment RosetteSep reagent containing a cocktail of bivalent antibodies against glycophorin A and CD3, CD4, CD19, CD20, CD36, CD66b, and CD123 (STEMCELL Technologies). Cells were mixed with RBCs and RosetteSep reagent, incubated on a nutator for 30 min at room temperature, diluted in PBS (Thermo Fisher Scientific), layered over Ficoll-Paque PLUS (GE Healthcare), and centrifuged at 1800 rpm for 30 min at room temperature with the brake off. The monolayers were harvested and residual RBCs were lysed using eBioscience 1x RBC Lysis Buffer (Thermo Fisher Scientific). From the tonsillar ILC-enriched fractions, NK cell developmental stages were sorted to >99% purity using a FACSAriaII cell sorter (BD Biosciences). Stage 3 cells were sorted as live Lineage− (Lin = CD3, CD4, CD5, CD14, CD19, CD20, CD123, FcεR1α, TCRαβ, TCRδγ) CD34−CD117+CD94−NKp80−CD16−KIR2D−KIR3DL1/2−NKG2C−NKp44−CD294−KLRG1− lymphocytes. Stage 4A cells were sorted as live Lin−CD34−CD117+/−CD200R+CD94+NKp80−CD16−KIR2D−KIR3DL1/2−NKG2C−CD294−KLRG1− lymphocytes. Stage 4B cells were sorted as live Lin−CD94+NKp80+CD16− lymphocytes. Stage 5 cells were sorted as live Lin−NKp80+CD16+ lymphocytes.
In Vitro Cell Cultures
All in vitro cell culture experiments were incubated at 37°C in 5% CO2 atmosphere. For bulk culture experiments, FACS-purified tonsil-derived ILCs were seeded at 1000 cells per well and cultured with IL-7 (10 ng/ml; Miltenyi) for 14–28 days in 200 μl culture media per well in 96-well flat-bottom plates (TrueLine). IL-15 and Stem cell factor (SCF) (10 ng/ml each) were used in experiments where indicated. For single cell clonal assays, FACS-purified populations were sorted directly into 96-well round-bottom plates, using established laboratory protocols for single cell clonal developmental assays (4, 14). Briefly, cells were monitored via light microscope for visible identification of viable cell populations and subsequent harvest for analysis by flow cytometry. Clones with fewer than 10 live CD45+ events were excluded. The numeric distribution of live CD45+ events resulting from stage 4A-derived clonal populations was mean = 135 events; median = 63 events. The numeric distribution of live CD45+ events resulting from stage 4B-derived clonal populations was mean = 242 events; median = 147 events. The clones were analyzed from among live CD3−CD14−CD45+ lymphocytes based on surface expression: stage 3/ILC3 like: CD94−NKp44+ lymphocytes; stage 4A-like: CD94+NKp80− lymphocytes; stage 4B-like: CD94+NKp80+ lymphocytes. The culture media for in vitro development experiments contained DMEM and F12 (2:1 ratio) supplemented with 1% antibiotic/antimycotic (Thermo Fisher Scientific), 20 mg/ml ascorbic acid, 24 μM 2-mercaptoethanol, 0.05 mg/ml sodium selenite (Sigma), and 10% heat-inactivated human AB serum (Valley Biomedical). For experiments with stromal feeder cells, OP9 or OP9-DL1 cells were obtained from the lab of Dr. Juan Carlos Zuniga-Pflucker (15). Stromal cells were maintained in MEM-α + Glutamax media (Thermo Fisher Scientific) with 10% fetal bovine serum and 1% antibiotic/antimycotic (Thermo Fisher Scientific). One day prior to culture of NKDIs, OP9 or OP9-DL1 cells were seeded at a density of 1000 cells per well (in a 96 well flat-bottom plate) and incubated at 37°C for cell adhesion. Stromal cell media was removed immediately prior to addition of FACS-sorted ILCs. Fresh media and cytokines were replenished twice per week. For intracellular cytokine analysis, cells were resuspended in 250 μl RPMI-1640 media and stimulated for 4 hr with either IL-2 (10 ng/ml, PeproTech) plus a mixture of 81 nM phorbol 12-myristate 13-acetate (PMA) and 1.34 μM ionomycin (Cell Stimulation Cocktail, Thermo Fisher Scientific); or IL-12 (Miltenyi), IL-15 (Miltenyi), and IL-18 (R&D Systems) (10 ng/ml each); or IL-2, IL-1β (Miltenyi), and IL-23 (Miltenyi) (10 ng/ml each). A mixture of 10.6 μM brefeldin A and 2 μM monensin (Protein Transport Inhibitor Cocktail, Thermo Fisher Scientific) was also added for 4 hr prior to analysis by flow cytometry. For K562 activation assay, in vitro-derived ILCs were incubated for 4 hr with 50,000 non-irradiated K562 cells with Protein Transport Inhibitor Cocktail in a 96 well V-bottom plate. For recombinant Notch ligand protein experiments, goat anti-human IgG Fc (10 μg/ml, R&D Systems) in PBS was immobilized in a 96 well flat-bottom plate overnight at 4°C. After washing twice with PBS, human IgG Fc control (4 μg/ml, Abcam), recombinant human DLL1 protein (4 μg/ml, Adipogen), or recombinant human DLL4 protein (4 μg/ml, Abcam) in PBS was added to each well and incubated at room temperature for 2 hr. Each well was washed twice with PBS prior to addition of ILCs. Specific human antibodies against the NOTCH1 or NOTCH2 negative regulatory regions (anti-NRR1, anti-NRR2; used at 5 μg/ml in vitro) were provided by Dr. Christian Siebel (Genentech Inc.) (16).
Flow Cytometry
Ex vivo and in vitro-derived ILC populations were stained using antibodies directed against surface or intracellular proteins according to manufacturer’s instructions. Where appropriate, isotype-matched or unstimulated controls were used to determine nonspecific staining. LIVE/DEAD Fixable Aqua Dead Cell Stain Kit and SYTOX Blue Dead Cell Stain (Thermo Fisher Scientific) viability dyes were used to exclude non-viable cells in the analyses. Intracellular staining was performed using Cytofix and Cytoperm Fixation and Permeabilization Solution Kit (BD Biosciences) for cytokine analysis, or Foxp3 Transcription Factor Staining Buffer Set (Thermo Fisher Scientific) for transcription factor analysis. For Notch receptor surface staining, the following antibodies (clone #, manufacturer) were used: NOTCH1 (REA849, Miltenyi); NOTCH2 (MHN2–25, Miltenyi); NOTCH3 (MHN3–21, Miltenyi); NOTCH4 (MHN4–2, Biolegend). Data were acquired on an LSRII cytometer (BD Biosciences) and analyzed using FlowJo (BD Biosciences) software.
Real-Time RT-PCR
From FACS-purified primary tonsil-derived NK cell populations, mRNA was isolated and purified using the Total RNA Purification Kit (Norgen Biotek) according to manufacturer’s instructions, and cDNA was synthesized using Superscript IV VILO Master Mix (Thermo Fisher Scientific). Standard quantitative real-time RT-PCR reactions were performed on a Viia7 Real-Time PCR System (Life Technologies) using PowerUp SYBR Green Master Mix (Thermo Fisher Scientific) and primer sequences obtained from published reports (17). Gene expression was normalized to the 18S mRNA internal control: ΔCt = Ct(gene of interest) – Ct(18S). Relative mRNA expression for each gene was calculated as 2^(-ΔCt).
Quantification and Statistical Analysis
Data are represented as mean ± SEM. Sample sizes for each experiment are included in the figure legends. Most of the experiments used cells from the same donors to compare outcomes that were generated from different treatment conditions or from different stages of NK cell development. The measured outcomes include, but are not limited to, the percentages of different cell lineages generated from the same cell culture, or a specified cell lineage generated under different treatment conditions, or gene expression measured using RT-PCR. For the RT-PCR data, the raw Ct values of the target genes were first normalized to the Ct values of the 18S internal controls. All comparisons were performed with linear mixed effects models to account for the correlation of observations from the same donor. The change in Notch receptor gene expression levels during NK cell development from stage 3 to stage 5 (trend analysis) was tested from the linear mixed effects models. Holm’s procedure was used to control for multiple comparisons. Adjusted p values < 0.05 were considered significant. All analyses were performed using SAS9.4 (SAS Institute, Inc.).
RESULTS
Generation of ILC2s and ILC3s, but not NK cells, requires Notch activation and stroma.
We previously demonstrated that human tonsil-derived lineage (“Lin”; B/T/myeloid)-negative, CD34−CD117+ ILCPs, or referred to here as “stage 3” NKDIs, were capable of developing into CD294+CD94−IL-13+ ILC2s, NKp44+CD94−IL-22+ ILC3s, and CD94+IFN-γ+ NK cells in the presence of recombinant human interleukin (IL)-7 and OP9-DL1 cells, a murine bone marrow-derived stromal cell line (OP9) transduced to constitutively express the human Notch ligand Delta-like-1 (DL1) (4). Here we first sought to determine the individual requirements for IL-7, stroma, and Notch ligands for differentiation. IL-7 was required for cell viability in vitro, as most stage 3 cells died when cultured in the absence of IL-7 (data not shown). Nonetheless, IL-7 was not sufficient to promote CD94 acquisition and/or to generate IFN-γ-producing cells indicative of NK cell differentiation (Figure 1A–1D, left panels; S1A). Furthermore, although small numbers of stage 3 cells from some donors acquired low amounts of surface CD294 or NKp44 expression following in vitro culture, these minor populations produced negligible IL-13 or IL-22, suggesting a lack of functionality (Figure 1A–1D; S1A). Importantly, fresh tonsil-derived mature ILC3s and NK cells retained not only their surface expression of NKp44 and CD94, respectively, they also retained their capacities for IL-22 or IFN-γ production when similarly cultured in IL-7 alone (Figure S1B), indicating that the latter in vitro condition was not inhibitory to mature ILCs. Therefore we concluded that IL-7 alone was not sufficient to support the differentiation of functional NK cells, ILC2s, and ILC3s from stage 3 cells.
Figure 1.
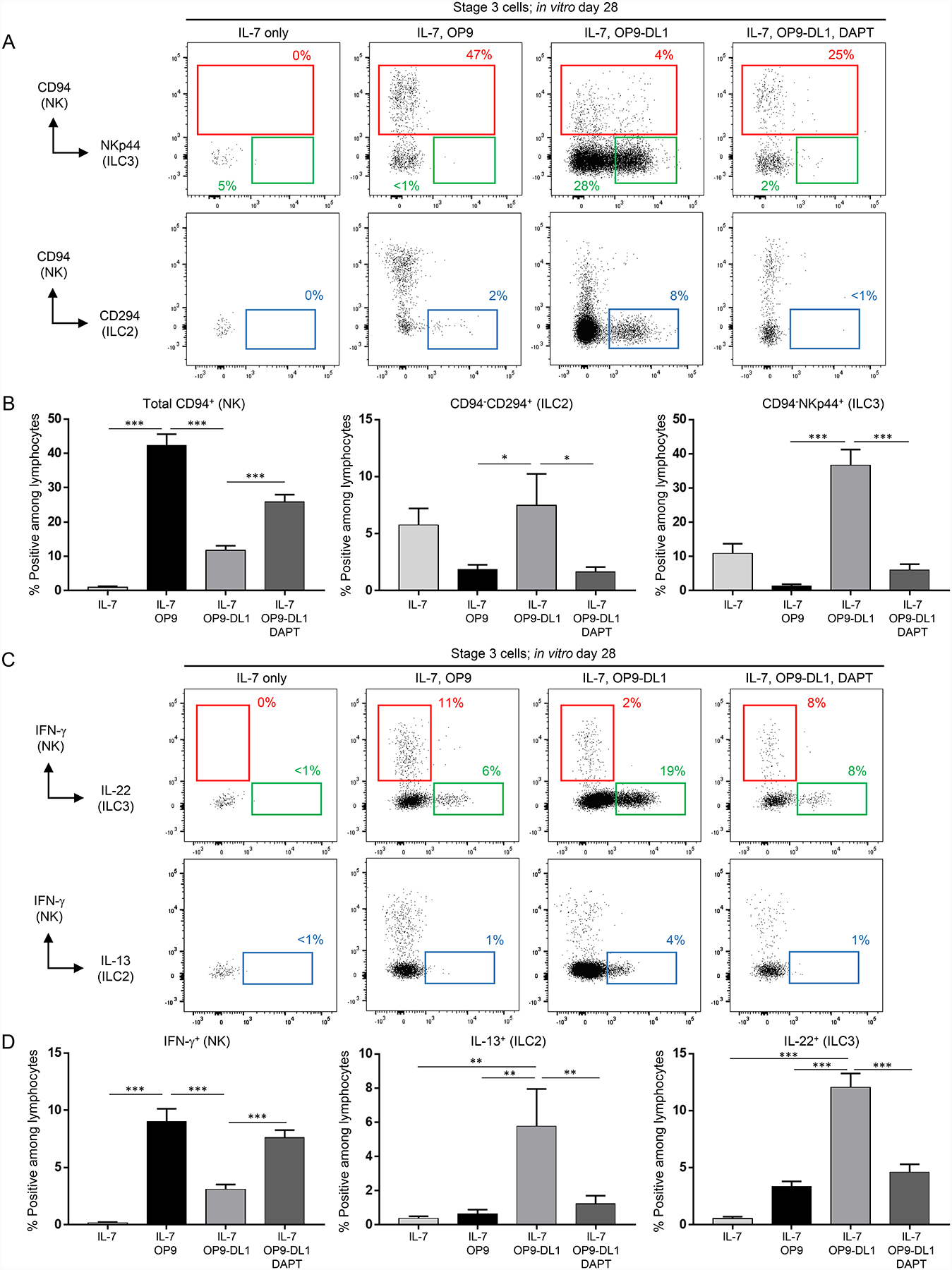
OP9-DL1 stroma support the differentiation of functional ILC2s, ILC3s, and NK cells from stage 3 cells. (A) Representative (n = 16; 5 independent experiments) surface flow cytometry analyses of NK cells (CD94+), ILC2s (CD94−CD294+), and ILC3s (CD94−NKp44+) generated in vitro following 28 day culture of freshly purified tonsil-derived stage 3 cells with recombinant human IL-7 (10 ng/ml for all experiments) alone, IL-7 + OP9 cells, or IL-7 + OP9-DL1 cells and treated with either vehicle control or DAPT (10 μM for all experiments). Dot plots in this and all successive figures are gated on live CD3−CD14−CD45+ lymphocytes unless otherwise labeled. Percentages shown in this and all successive figures represent each population relative to total in vitro-derived live CD3−CD14−CD45+ lymphocytes unless otherwise stated. (B) Quantification of NK cells, ILC2s, and ILC3s generated from stage 3 cells in the conditions described in (A). Data are represented as mean ± SEM. * p < 0.05; *** p < 0.001. (C) Representative (n = 23; 7 independent experiments) intracellular flow cytometry analyses of IFN-γ+, IL-22+, and IL-13+ cells generated in vitro following 28 day culture of freshly purified tonsil-derived stage 3 cells with IL-7 alone, IL-7 + OP9 cells, or IL-7 + OP9-DL1 cells and treated with either vehicle or DAPT. ILCs were stimulated with PMA, ionomycin, and IL-2 (10 ng/ml) for 4 hr prior to analysis of cytokine production. (D) Quantification of IFN-γ+, IL-22+, and IL-13+ cells generated from stage 3 cells in the conditions described in (C). Data are represented as mean ± SEM. ** p < 0.01; *** p < 0.001.
To determine if interactions with stromal cells were required to promote differentiation, we next co-cultured stage 3 cells with IL-7 and the OP9 stromal cell line (without overexpressed DL1). We observed significantly more CD94+IFN-γ+/− cells produced in the presence of OP9 cells compared to cultures with IL-7 alone (Figure 1A–1D, second panels from left; S1A). However, in these conditions, still only rare ILC2s or ILC3s were produced. Thus, while IL-7 alone did not support functional NK cell development, the addition of OP9 stromal cells was sufficient to generate CD94+IFN-γ+/− NK cells from stage 3 cells. Direct contact with the OP9 cells was required for stroma-mediated NK cell differentiation, as stage 3 cells cultured with OP9 cells separated by a transwell did not differentiate into NK cells (Figure S1C). We also tested the addition of the cytokines IL-15 and Stem cell factor (SCF), as we previously reported that IL-15 added with or without SCF in the absence of stroma can induce low amounts of CD94 acquisition (5, 18). Culture of stage 3 cells with IL-7 plus either IL-15 or SCF or both (in the absence of stroma) produced a marginal increase in CD94+ cells (not statistically significant) compared to IL-7 alone, yet significantly fewer than was observed in the presence of stroma. In addition the few CD94+ cells produced in the presence of IL-15 and/or SCF produced very little IFN-γ when stimulated, further suggesting the importance of stroma in promoting functional NK cell development (Figure S1D and data not shown).
Having established that the presence of stromal cells promoted differentiation of NK cells, but not of ILC2s or ILC3s, we next sought to determine the impact of Notch activation in the presence of stromal cells. In contrast to the results of co-culturing stage 3 cells with IL-7 and OP9 stroma (without overexpressed DL1), and consistent with our previous findings (4), stage 3 cells differentiated into not only NK cells but also into distinct populations of cytokine-producing ILC2s and ILC3s in the presence of OP9-DL1 stroma at day 28 (Figure 1A–1D, third panels from left; S1A). To confirm that this time point does indeed allow for accurate representation of ILC2, ILC3, and NK cell differentiation, we performed a time course experiment and observed that robust populations of ILC2s, ILC3s, and NK cells were obtained from stage 3 cells cultured with IL-7 and OP9-DL1 stroma following 28 days in culture. Although some variation in the kinetics of ILC differentiation was noted among different donor-derived stage 3 cell cultures, 28 days was the most consistent time point to ensure differentiation of NK cells, ILC2s, and ILC3s, supporting the use of this time point for these studies and also consistent with our previously reported culture experiments using a similar OP9-DL1-mediated in vitro development assay (4) (Figure S1E). These effects on ILC2 and ILC3 differentiation were abrogated in the presence of N-[N-(3,5-Difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester (DAPT), a global inhibitor of Notch receptor activation (Figure 1A–1D, right panels; S1A) (19). On the other hand, NK cell differentiation was not inhibited in the presence of DAPT. Just as was seen with the OP9 cultures, NK cell differentiation predominated in the presence of DAPT likely due to the absence of skewing stage 3 cells away from ILC2s or ILC3s. We also observed that while immobilized plate-bound Notch ligands induced downstream HES1 activation in Notch receptor-expressing THP1 cells (Figure S1F), plate-bound Notch ligands were insufficient to promote functional ILC differentiation from human tonsil-derived stage 3 cells. Rather, concurrent interactions with both stromal cells and Notch ligands (via the OP9-DL1 cells) were required for development of functional ILC2s and ILC3s (Figure S1G). Collectively, these data showed that Notch activation in the presence of IL-7 and stroma was necessary to generate functional ILC2s and ILC3s, whereas only IL-7 and co-culture with OP9 stroma were required for the generation of functional NK cells from stage 3 precursors.
Stage 4A cells retain ILC3 differentiation potential whereas stage 4B cells are committed.
Having established the requirements for IL-7, stromal cells, and Notch ligands in promoting ILC2 and ILC3 differentiation from stage 3 cells, we next sought to determine the non-NK cell developmental potentials of later stage NKDIs. To address this, FACS-purified tonsillar stage 4A or 4B cells were co-cultured with IL-7 plus OP9-DL1 stroma; stage 3 cells were cultured in parallel as positive control. In comparison to stage 3 cells, which developed into ILC2s, ILC3s, and NK cells (Figures 2A–2C and S2A, left panels), stage 4A cells cultured in the same conditions gave rise to ILC3s and NK cells but to only negligible amounts of ILC2s as shown by both surface phenotype and cytokine production (Figures 2A–2C and S2A, middle panels). In contrast, stage 4B cells maintained a stable functional NK cell phenotype (CD94+IFN-γ+/-) and did not develop into either ILC2s or ILC3s (Figures 2A–2C and S2A, right panels).
Figure 2.
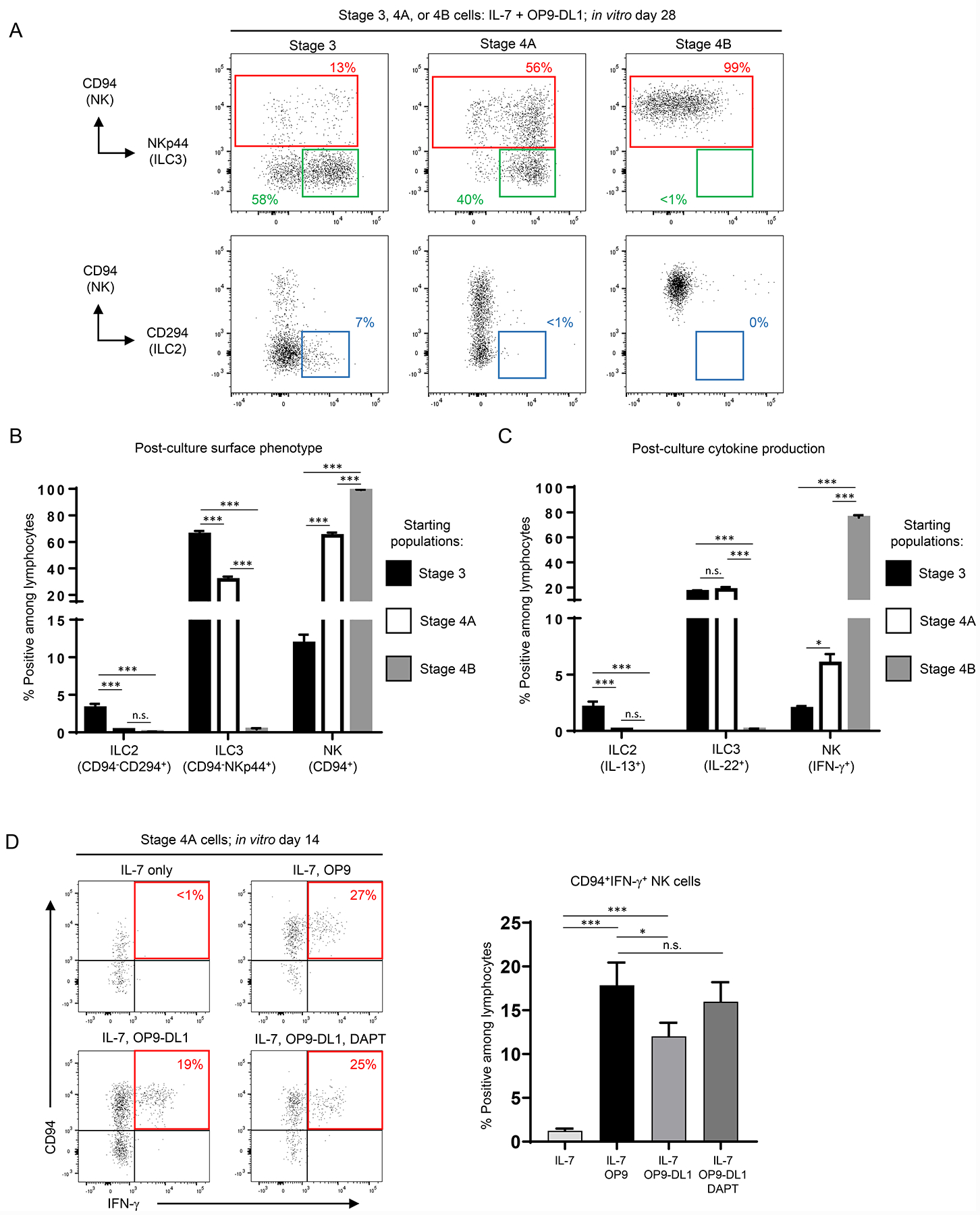
Stage 4A and 4B cells have decreased ILC plasticity compared to stage 3 cells. (A) Representative (n = 15; 4 independent experiments) surface flow cytometry analyses of NK cells (CD94+), ILC3s (CD94−NKp44+), and ILC2s (CD94−CD294+) generated in vitro following 28 day culture of the indicated freshly purified tonsil-derived populations with OP9-DL1 cells and IL-7. (B) Quantification of ILC2s, ILC3s, and NK cells generated in vitro following 28 day culture of the indicated freshly purified tonsil-derived populations with OP9-DL1 cells and IL-7. Data are represented as mean ± SEM. n.s. = not significant (p > 0.05); *** p < 0.001. (C) Quantification of IL-13+, IL-22+, and IFN-γ+ cells generated in vitro following 28 day culture of the indicated freshly purified tonsil-derived populations with OP9-DL1 cells and IL-7. ILCs were stimulated with PMA, ionomycin, and IL-2 for 4 hr following in vitro culture prior to analysis. Data are represented as mean ± SEM. n.s. = not significant (p > 0.05); * p < 0.05; *** p < 0.001. (D) Representative (n = 10; 3 independent experiments) surface flow cytometry analyses (left) and quantification (right) of ILCs generated in vitro following 14 day culture of freshly purified tonsil-derived stage 4A cells with IL-7 alone, IL-7 + OP9 cells, or IL-7 + OP9-DL1 cells and treated with either vehicle or DAPT. Cells were stimulated with PMA, ionomycin, and IL-2 for 4 hr prior to analysis of cytokine production. Data are represented as mean ± SEM. n.s. = not significant (p > 0.05); * p < 0.05; *** p < 0.001.
Similar to our findings with fresh tonsil-derived stage 3 cells, we observed that the differentiation of stage 4A cells into IFN-γ-producing NK cells required IL-7 and OP9 stroma but not Notch (Figure 2D). In contrast, differentiation into an ILC3 phenotype was dependent on IL-7 in the presence of Notch ligands and stroma. Indeed, compared to culture in IL-7 alone or IL-7 plus OP9 stroma, stage 4A cells cultured with IL-7 plus OP9-DL1 stroma had greater production of Lin−CD45+NKp44+CD94− ILC3s, and this was inhibited in the presence of DAPT (Figure S2B). In vitro CD94−NKp44+ cells derived from stage 4A cells produced IL-22 and expressed RORγt, consistent with a functional and transcriptional ILC3 profile (Figure S2A–S2C) (20). Collectively, these data showed that first ILC2 and then ILC3 developmental potentials were successively lost through sequential stages of NK cell development and that tonsil-derived stage 4B cells appeared to be NK lineage committed. Moreover, the capacity for stage 4A cells to downregulate CD94 and differentiate into ILC3s was largely dependent upon Notch signaling.
NK cell acquisition of NKp80 requires Notch ligands and stroma.
The findings above indicated that the differentiation of stage 3 and stage 4A NKDIs into CD94+IFN-γ+ NK cells required stroma but was independent of Notch. However, because previous studies had implicated Notch in the regulation of CD16 and KIR expression from cord blood CD34+ progenitor-derived NK cells (12), we hypothesized that Notch still influenced the phenotype of NK cells generated from tonsil-derived NKDIs. To address this, we cultured stage 3 cells for four weeks with IL-7 and OP9 or OP9-DL1 cells +/− DAPT and evaluated surface antigen expression by flow cytometry. The CD94+ NK cells derived on either OP9 or OP9-DL1 stroma expressed NKG2A, which is typically co-expressed with CD94 by ex vivo stage 4A cells (18). However, surface markers associated with terminally differentiated peripheral blood NK cells (CD16, CD57, KIR2D, KIR3DL1/2, and NKG2C) were low or absent in each of the culture conditions (21). We also measured the expression of CD56 and CD127, which have been used to identify NK cell subsets in the blood and other tissues (22, 23). Stage 3 cells cultured with IL-7 and OP9-DL1 stroma generated CD94+ NK cells that were positive/bright for CD56. However, CD127 (IL-7 receptor) expression was not detected, consistent with what other groups have reported when tonsil-derived precursor cells are cultured in the presence of IL-7 (Figure S2D) (23). In contrast, expression of the stage 4B-associated marker, NKp80, was significantly increased following culture in the presence of Notch signaling (Figure 3A). Culture of stage 3 cells with OP9 stroma did generate minor populations of NKp80+ cells (albeit dimly positive). Alternatively, co-culture with OP9-DL1 stroma resulted in distinct populations of CD94+NKp80+ NK cells, which closely recapitulated the surface expression pattern of fresh tonsil-derived stage 4B cells (Figure 3B and data not shown). Indeed, not only were the percentages of NKp80+ cells (among culture-derived CD94+ cells) and absolute numbers of CD94+NKp80+ cells significantly higher in the presence of Notch, the mean fluorescence intensities (MFIs) of NKp80 among these cells were also significantly increased (Figure 3C, S2E). Treatment with DAPT in the OP9-DL1 co-culture condition significantly decreased both the percentages and absolute numbers of NKp80+ cells and MFIs of NKp80 to amounts seen with OP9 co-culture (Figure 3C). Culture of stage 3 cells with IL-15 and/or SCF in addition to IL-7 in the absence of stroma did not generate significant amounts of NKp80+ cells, further establishing a requirement for both stromal cells and Notch in the generation of CD94+NKp80+ cells from stage 3 cells (Figure S2F). We saw similar results when culturing fresh tonsil-derived stage 4A NKDIs, comprising the stage directly preceding NKp80 acquisition during human NK cell development in SLTs (18). Stage 4A cells cultured in IL-7 alone maintained viability but did not acquire NKp80. The presence of OP9-DL1 stroma resulted in a significant increase in NKp80 acquisition from stage 4A cells compared to OP9 stroma following co-culture for 14 days (Figure 3D–3E, S2G). Of note, we confirmed using a time course experiment that 14 days was sufficient for NKp80 acquisition from stage 4A cells (and also for ILC3 differentiation from 4A cells, shown above) (Figure S2H). Additionally, we observed similar MFI patterns with stage 4A-derived cells as were seen with CD94+ cells derived from stage 3 cells. The stage 4A-derived NKp80+ cells generated in the presence of OP9-DL1 stroma had higher NKp80 MFIs compared to those generated in the presence of OP9 stroma, and treatment with DAPT decreased both the differentiation and MFIs of NKp80+ cells. Similar to what was seen in the experiments with stage 3 cells (Figure S1G above), we also observed that culture of fresh tonsil-derived stage 4A cells in the presence of plate-bound Notch ligands in the absence of any stromal cells was not sufficient to promote NKp80 acquisition (Figure S2I). Thus Notch signaling in the presence of stroma promoted the formation of Lin−CD94+NKp80+ cells.
Figure 3.
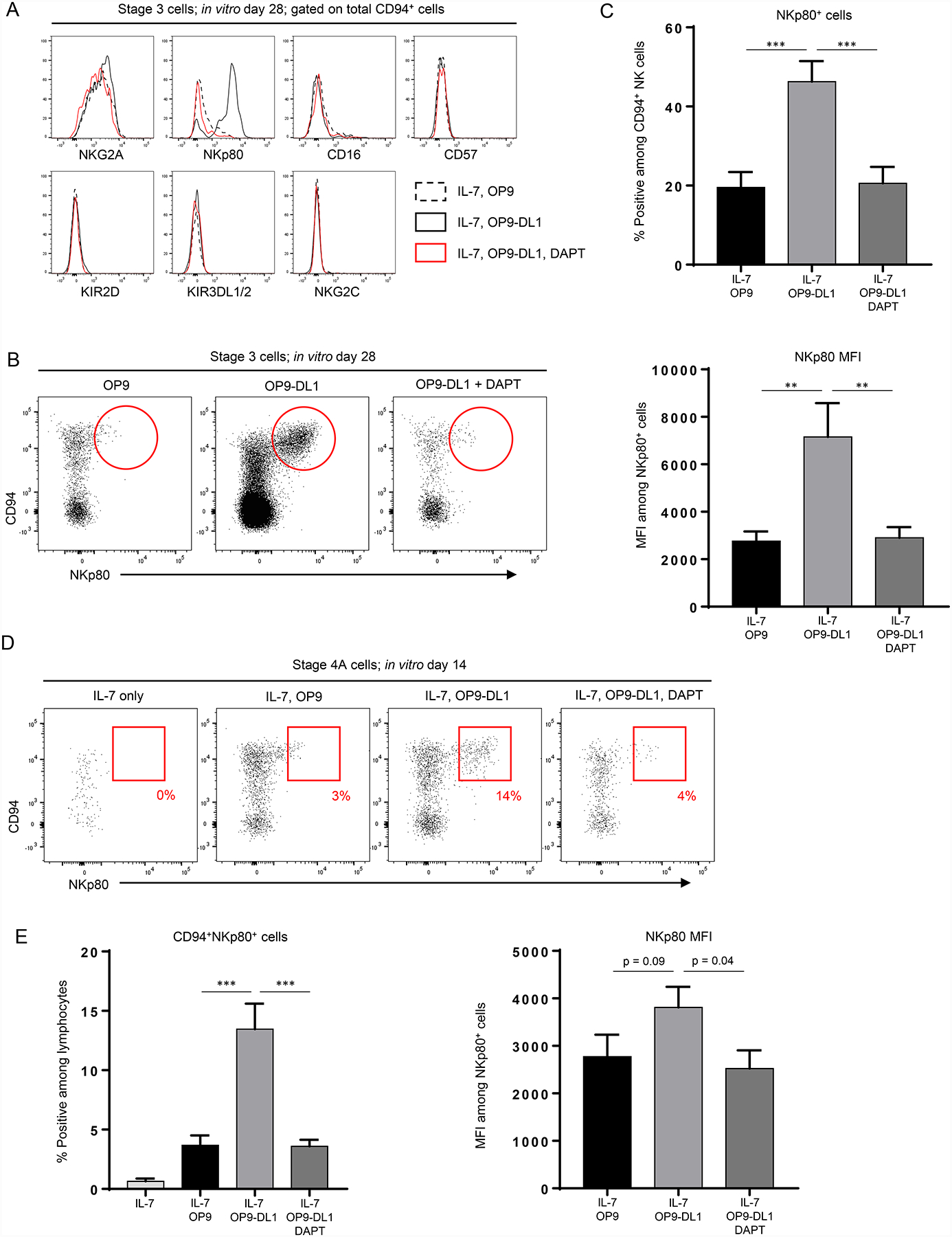
NKp80 expression is increased in vitro following co-culture with OP9-DL1 stroma. (A) Representative (n ≥ 4 for each marker) flow cytometry analyses of total CD94+ cells generated in vitro following 28 day culture of freshly purified tonsil-derived stage 3 cells with IL-7 + OP9 cells (dotted black histogram) or IL-7 + OP9-DL1 cells in the presence of either vehicle (solid black histogram) or DAPT (red histogram). (B) Representative (n = 16; 5 independent experiments) flow cytometry analyses of CD94+NKp80+/− populations generated in vitro following 28 day culture of freshly purified tonsil-derived stage 3 cells with IL-7 + OP9 or OP9-DL1 in the presence of either vehicle or DAPT. Red circles denote a qualitative increase of the CD94+NKp80+ population in the presence of Notch activation (middle panel). (C) Top, quantification of NKp80+ cells among in vitro-derived CD94+ cells generated from stage 3 cells cultured in the conditions described in (B). Bottom, mean fluorescence intensities (MFIs) of NKp80 among in vitro-derived NKp80+ cells from stage 3 cells cultured in the conditions described in (B). Data are represented as mean ± SEM. ** p < 0.01; *** p < 0.001. (D) Representative (n = 14; 4 independent experiments) surface flow cytometry analyses of ILCs generated in vitro following 14 day culture of freshly purified tonsil-derived stage 4A cells with IL-7 alone, IL-7 + OP9 cells, or IL-7 + OP9-DL1 cells and treated with either vehicle or DAPT. (E) Left, quantification of CD94+NKp80+ cells as a percentage of total in vitro-derived live CD45+ lymphocytes generated in vitro following 14 day culture of freshly purified tonsil-derived stage 4A cells in the conditions described in (D). Right, mean fluorescence intensities (MFIs) of NKp80 among in vitro-derived NKp80+ cells from stage 4A cells cultured in the conditions described in in (D). Data are represented as mean ± SEM. *** p < 0.001.
In vitro-derived NKp80+ cells show a tonsil stage 4B-like phenotype.
Next we phenotypically and functionally evaluated the Lin−CD94+NKp80+ stage 4B-like cells that were derived from fresh tonsil stage 4A NKDIs in the presence of IL-7 and OP9-DL1 stroma. In vitro-derived stage 3-like cells (most showing ILC3 features, Figures 2 and S2) and CD94+NKp80− cells that had retained a stage 4A-like surface phenotype were similarly evaluated in parallel. Of note, all of these populations expressed CD56. NKG2A and NKG2D were expressed by stage 4A-like and stage 4B-like cells but not by stage 3-like cells. CD16, CD57, NKG2C, KIR2D, and KIR3DL1/2 were low or absent in each of these populations, similar to what we observed when culturing tonsil stage 3 NKDIs (Figure 3A above). When comparing transcription factor expression, stage 4B-like cells and minor fractions of the stage 4A-like cells expressed EOMES. T-BET was not expressed by stage 3- or stage 4A-like cells, and only variably expressed in small proportions of stage 4B-like cells. RORγt expression was restricted to stage 3-like and some stage 4A-like cells but was not expressed by the stage 4B-like cells. Stage 4B-like cells variably expressed granzymes A and K, whereas perforin, granzyme B, and granzyme M were barely detected in any population (Figure 4).
Figure 4.
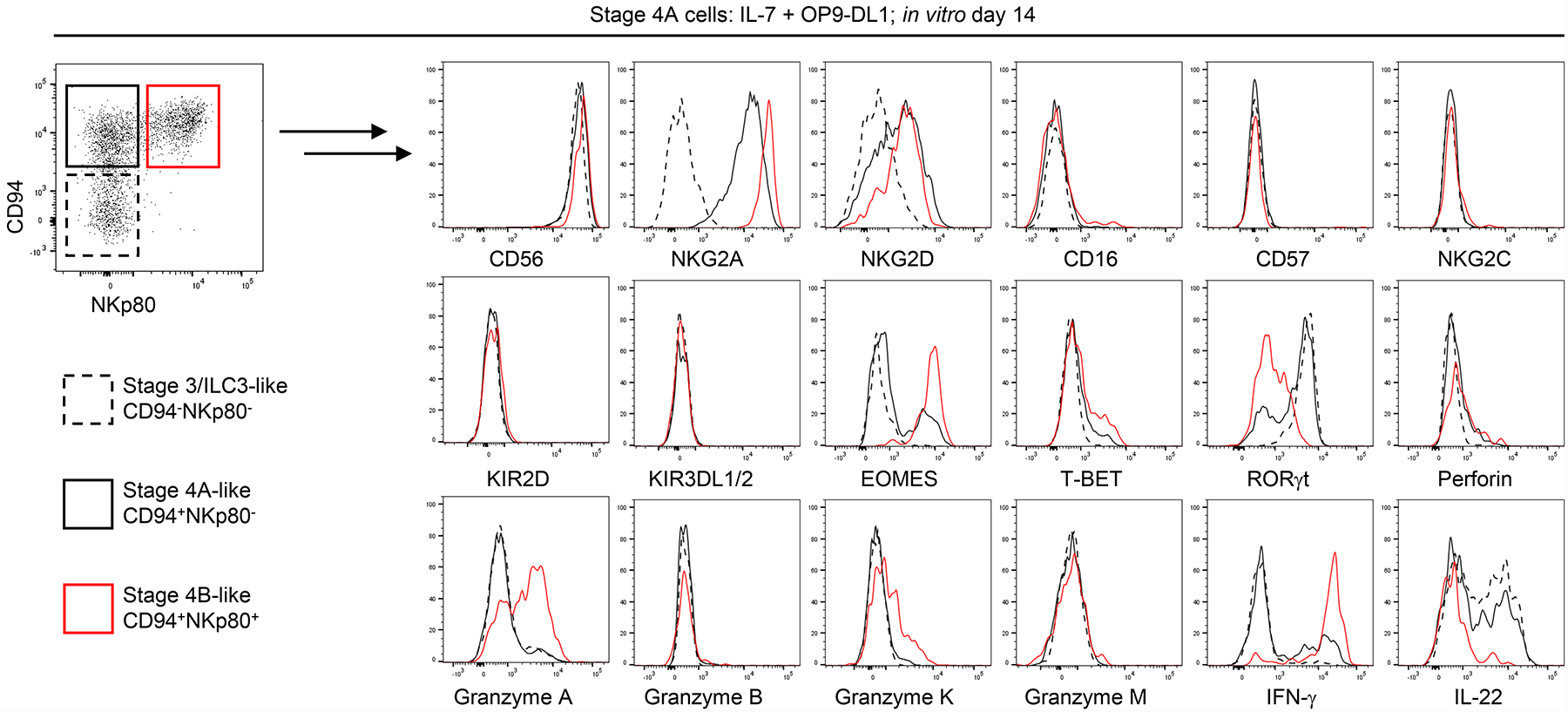
Co-culture of stage 4A cells with OP9-DL1 stroma promotes the transition to a stage 4B-like phenotype in vitro. Representative (n ≥ 4 for each marker) flow cytometry analyses depicting relative expression of selected surface and intracellular markers comparing ILC3/stage 3-like (CD94−NKp80−, dotted black histograms), stage 4A-like (CD94+NKp80−, solid black histograms) and stage 4B-like (CD94+NKp80+, red histograms) cells generated in vitro following 14 day culture of freshly purified tonsil-derived stage 4A cells with IL-7 and OP9-DL1 cells. Analysis of IFN-γ by flow cytometry was measured following post-culture incubation for 4 hr with IL-12, IL-15, and IL-18 (10 ng/ml each). Analysis of IL-22 by flow cytometry was measured following post-culture incubation for 4 hr with IL-2, IL-1β, and IL-23 (10 ng/ml each).
We also evaluated the functionalities of cells derived from stage 4A cells. Cytokine production profiles were similar to those of the corresponding tonsil-resident NKDIs. Stage 4B-like cells produced IFN-γ when stimulated following in vitro culture, whereas stage 3-like cells and most stage 4A-like cells did not. In contrast, IL-22 was produced by stage 3-like and stage 4A-like cells but not by stage 4B-like cells (Figure 4). OP9-DL1-derived stage 4B-like cells also expressed CD107a when co-cultured with K562 cells indicative of tumor cell induced degranulation (data not shown). Collectively, these data supported the conclusion that Notch activation in the presence of IL-7 and stroma promoted the differentiation of tonsil-derived stage 4A cells into CD94+NKp80+ stage 4B-like cells.
In vitro-derived stage 4A-like cells retain NK cell developmental potential.
Our findings thus far indicated that bulk cultures of stage 4A NKDIs with IL-7 and OP9-DL1 stroma produced distinct populations of CD94−NKp80− stage 3-like cells with ILC3-associated features, CD94+NKp80− stage 4A-like cells, and CD94+NKp80+ stage 4B-like cells that produced IFN-γ. In contrast, most if not all stage 4B cells maintained their surface CD94+NKp80+ NK cell phenotype and produced IFN-γ (Figure S3A). In order to determine the developmental potentials of individual tonsil-derived stage 4A and 4B NKDIs, we performed single cell clonal culture assays in which individually sorted stage 4A or stage 4B cells were cultured with IL-7, IL-15 (to promote proliferation), and OP9-DL1 stroma. As shown in Figures 5A and S3B, individually cultured stage 4A cells gave rise to variably mixed populations of stage 3/ILC3-like, 4A-like, and/or 4B-like populations, with approximately one-fourth of the clones containing cells of all three types. In contrast, all evaluated clones derived from tonsil stage 4B cells stably maintained their phenotype and did not differentiate into other populations. These data indicated that at least a subset of tonsil stage 4A NKDIs, despite expressing the NK cell-associated marker, CD94, were bona fide precursors with the potential to generate both ILC3s and NKp80+ NK cells.
Figure 5.
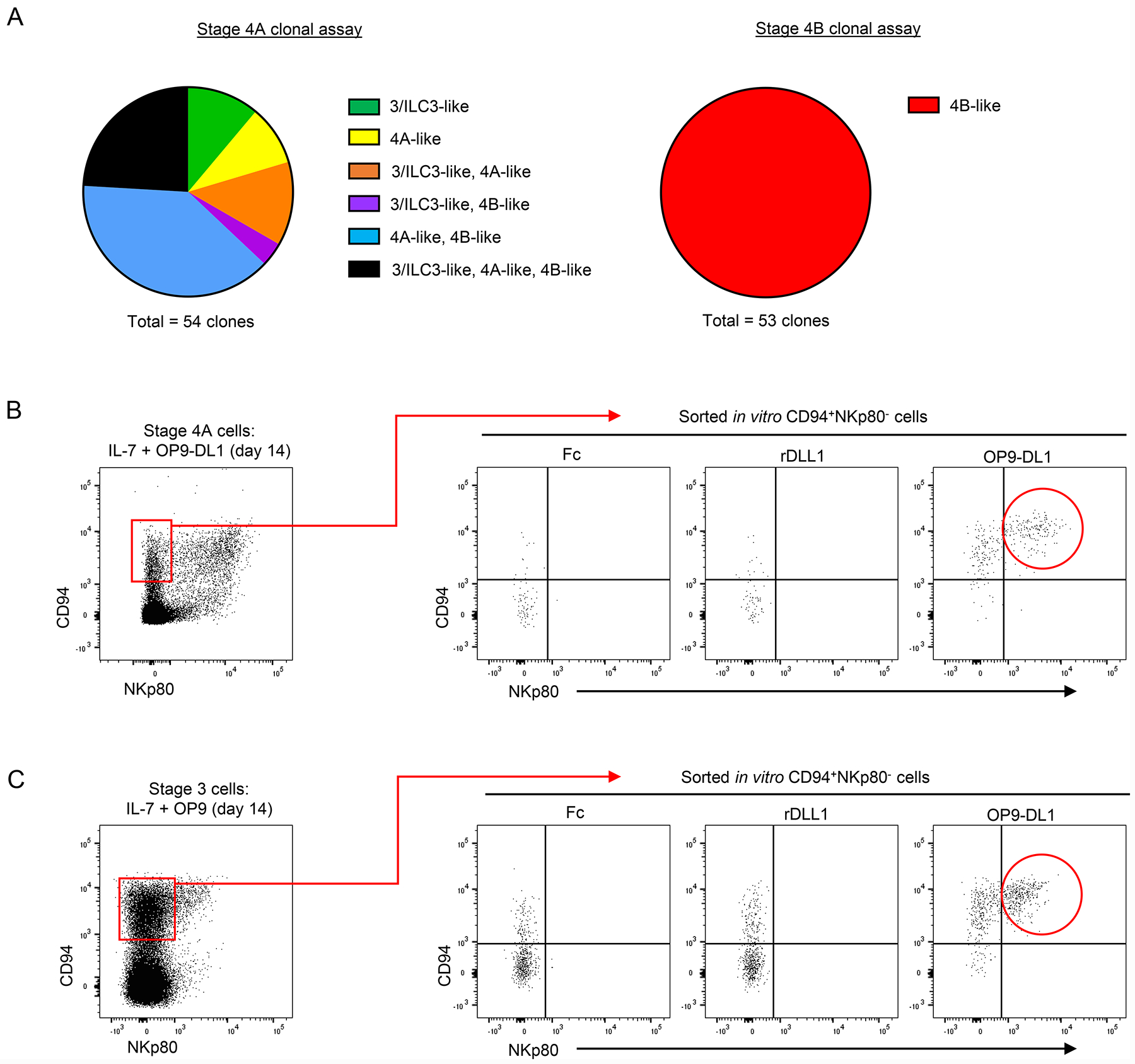
Stage 4A NKDIs are bipotent precursors to ILC3s and NKp80+ NK cells. (A) Clonal analyses of freshly purified tonsil-derived stage 4A and stage 4B cells individually sorted into wells containing OP9-DL1 cells supplemented with IL-7 and IL-15 (10 ng/ml) for 21 days. Clones were analyzed for stage 3/ILC3-like (CD94−NKp44+), stage 4A-like (CD94+NKp80−), and stage 4B-like (CD94+NKp80+) cells. (107 total clones evaluated from n = 6 donors; 3 independent experiments). Cloning efficiency was approximately 10% for stage 4A clones and 25% for stage 4B clones. (B-C) Representative (n = 3; 1 independent experiment) flow cytometry analyses depicting stage 4A-like (CD94+NKp80−, left panels, red boxes) cells that were sorted at in vitro culture day 14 and re-plated with IL-7 and Fc control, recombinant human Delta-like-1 (immobilized on plate at 4 μg/ml), or OP9-DL1 cells.
Given that some tonsil-derived stage 4A NKDIs retained their stage 4A-like phenotype post-culture (Figure 4A), we evaluated the capacity of these in vitro-derived stage 4A-like cells to differentiate into NKp80+ NK cells in secondary switch cultures. Fresh tonsil stage 4A cells were cultured for 14 days in the presence of IL-7 and OP9-DL1 stroma, and then the resultant CD94+NKp80− stage 4A-like cells were sorted and re-plated into culture wells containing IL-7 with either plate-bound Fc control, immobilized recombinant Delta-like-1 protein, or OP9-DL1 stroma. A significant proportion of the stage 4A-like cells acquired NKp80 when re-plated on OP9-DL1 stroma. However, re-plating stage 4A-like cells on recombinant Delta-like-1 protein (without stroma) was not sufficient to induce NKp80 expression and instead resulted in the majority of cells losing their CD94 expression (Figure 5B). In similar experiments, we also observed that stage 4A-like cells, which had been derived from stage 3 cells in the presence of IL-7 and OP9 stroma and then sorted and switched to IL-7 and OP9-DL1 stroma, upregulated NKp80 to attain a stage 4B-like phenotype. However, NKp80 was not acquired when the in vitro-derived stage 4A-like cells were re-cultured on recombinant Delta-like-1 protein without stroma (Figure 5C). These data demonstrated that continued interactions with the stroma were required for Notch-mediated NKp80 acquisition even by stage 4A-like cells that had been previously generated in the presence of IL-7 and either OP9 or OP9-DL1 stroma.
NOTCH1 and NOTCH2 are differentially expressed during human NK cell maturation.
Having established that Notch signaling was required for the differentiation of ILC2s and ILC3s, as well as for promoting the acquisition of NKp80 by CD94+ NK cells, we sought to determine which Notch receptors were involved in these processes. Using quantitative real-time RT-PCR, we measured the relative transcript amounts of the four human Notch receptor genes in stage 3, 4A, 4B, and 5 cells. While NOTCH1 and NOTCH2 were expressed by these populations, NOTCH3 and NOTCH4 were below the limit of detection. We observed opposite expression patterns of NOTCH1 and NOTCH2 at successive stages of maturation. NOTCH1 expression increased in stages 4B and 5 compared to the (less mature) stages 3 and 4A. Conversely, NOTCH2 expression was highest in stage 3 cells and decreased in stages 4B and 5. (Figure 6A). Next we looked for patterns of surface protein expression to see if they were comparable with those seen by transcript. Notch receptors were not detectable by surface flow cytometry among enriched ILCs when analyzed immediately ex vivo, perhaps due to exposure to Notch ligands in vivo (data not shown). However, following incubation of total ILCs for 3 days in vitro in media supplemented with IL-7 but in the absence of exogenous Notch ligands (24), NOTCH1 and NOTCH2 receptors could be detected at the surface (Figure 6B), whereas NOTCH3 and NOTCH4 were not detected (data not shown). Stage 3 cells expressed intermediate amounts of both NOTCH1 and NOTCH2; stage 4A cells had slightly higher expression of NOTCH1 but lower expression of NOTCH2 compared to stage 3 cells; and stage 4B and 5 cells were nearly uniformly positive for NOTCH1 and negative for NOTCH2 (Figure 6B). We also compared the relative expression patterns of NOTCH1 and NOTCH2 shown by total NK cells (expressing CD94, NKp80, and/or CD16) with those of other tonsil-resident ILC populations. In contrast to total NK cells which predominantly expressed NOTCH1 at the bulk population level, ILC2s and ILC3s showed intermediate expression of both NOTCH1 and NOTCH2, similar to stage 3 cells (Figure 6C). Taken together, these transcriptional and phenotypic data demonstrated that NOTCH1 and NOTCH2 were alternatively expressed in immature versus mature NK cell populations, and the combination of high NOTCH1 and low NOTCH2 expression was more specific to NK cells compared to ILC2s or ILC3s.
Figure 6.
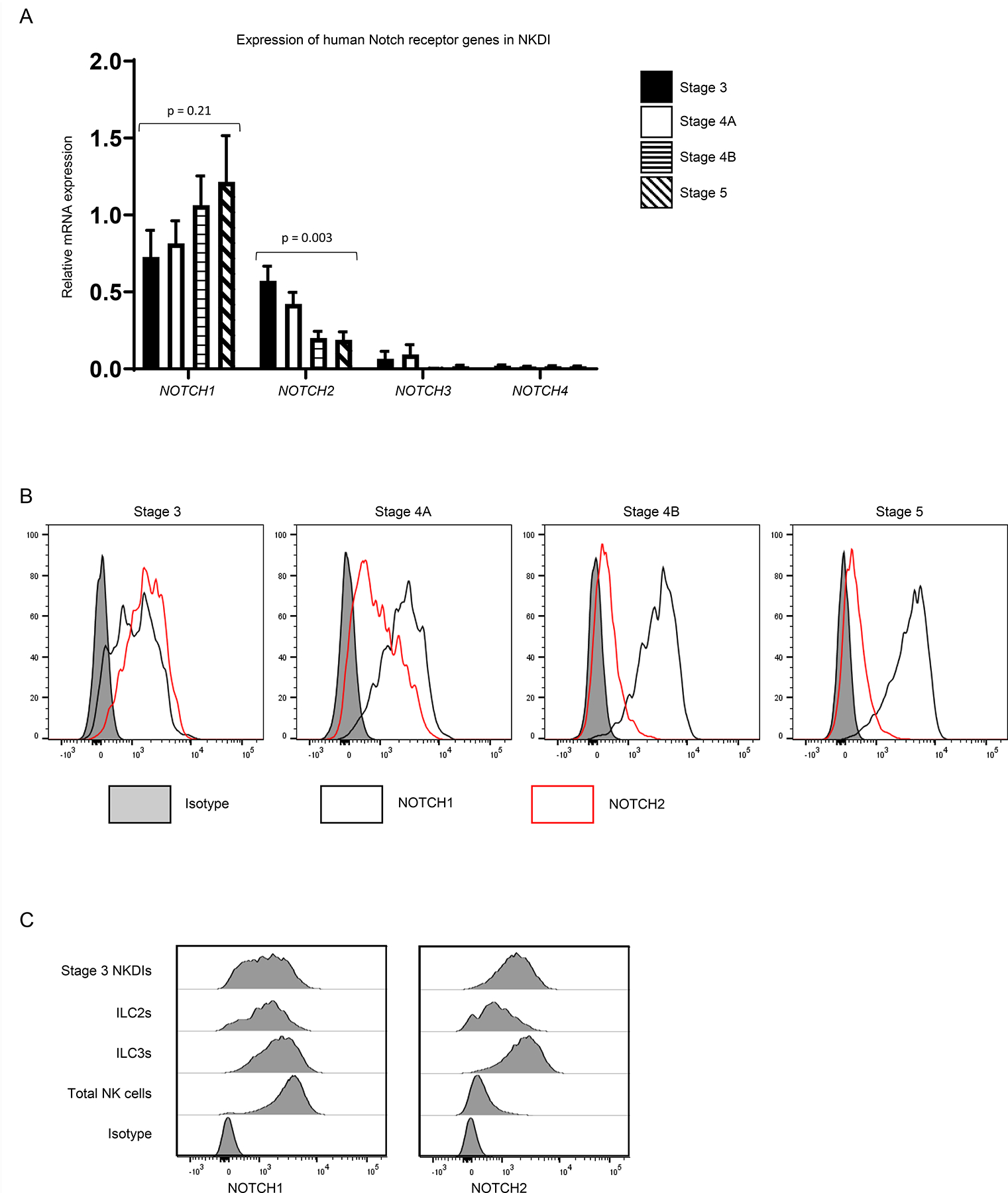
Notch receptors are expressed by human tonsil ILCs. (A) Quantification of relative transcript expression of NOTCH1, NOTCH2, NOTCH3, and NOTCH4 genes by quantitative real-time RT-PCR among freshly purified tonsil-derived stage 3, stage 4A, stage 4B, and stage 5 cells (n = 5; 5 independent experiments) as described in Methods. Gene expression was normalized to 18S internal control. (B) Representative (n = 4; 1 independent experiment) surface flow cytometry analysis of NOTCH1 and NOTCH2 in total tonsillar enriched Lin− lymphocytes following 3 days in vitro with IL-7 and gated on stage 3, stage 4A, stage 4B, and stage 5 cell populations. (C) Representative (n = 6; 2 independent experiments) surface flow cytometry analysis of NOTCH1 and NOTCH2 in total tonsillar enriched Lin− lymphocytes following 3 days in vitro with IL-7. Stage 3/ILCPs were gated as live Lin−CD34−CD117+CD94−NKp80−CD16−KIR2D−KIR3DL1/2−NKG2C−NKp44−CD294−KLRG1− lymphocytes. ILC2s were gated as live Lin−CD56−NKp44−CD94−NKp80−CD16−KIR2D−KIR3DL1/2−NKG2C−(CD294/KLRG1)+ lymphocytes. ILC3s were gated as live Lin−CD117+NKp44+CD94−NKp80−CD16−KIR2D−KIR3DL1/2−NKG2C−CD294−KLRG1− lymphocytes. NK cells were gated as any live Lin- lymphocytes expressing CD94, NKG2A, NKp80, CD16, KIR2D, KIR3DL1/2, and/or NKG2C as previously described (4).
NOTCH1 and NOTCH2 differentially regulate stage 3 and stage 4A cells.
Based on the above expression patterns, we hypothesized that inhibition of signaling through NOTCH1 and/or NOTCH2 would restrict the development of ILC2s, ILC3s, and NKp80+ NK cells. To test this hypothesis, we cultured stage 3 cells with IL-7 and OP9-DL1 stroma in the presence of monoclonal blocking antibodies against human NOTCH1 (anti-NRR1) and/or human NOTCH2 (anti-NRR2) (16). Individual blockade of either NOTCH1 or NOTCH2 resulted in decreased production of ILC2s from stage 3 cells, and the effects were additive when both NOTCH1 and NOTCH2 were blocked in combination (Figure 7A–7B, S3C). Indeed blockade of both receptors resulted in pronounced ILC2 inhibition to the same extent as was seen with DAPT. On the other hand, ILC3 production was not significantly affected by individual blockade of either NOTCH1 or NOTCH2, whereas treatment with both blocking antibodies significantly decreased the production of ILC3s to amounts detected in cultures with DAPT (Figure 7A–7B, S3C). Consistent with our earlier observations that Notch was not required for the differentiation of CD94+IFN-γ+/− cells from stage 3 cells (Figure 2), CD94+ cells were produced in the presence of either anti-NRR1 or anti-NRR2 blocking antibodies. Moreover, treatment with both antibodies in combination resulted in increased proportions of CD94+ cells, likely due to the concurrent inhibition of ILC2 and ILC3 differentiation as was seen with DAPT (Figure 7A–7B, S3C). Nonetheless, simultaneous blockade of both NOTCH1 and NOTCH2, but not with blockade of either receptor individually, inhibited NKp80 expression by the CD94+ NK cells that had been derived from stage 3 cells (Figure 7A–7B, S3C). In addition to changes in surface marker expression, these effects of Notch receptor inhibition on ILC differentiation from stage 3 cells were supported by further analysis of cytokine production. Blockade of NOTCH1 and NOTCH2 inhibited the production of IL-13+ cells (ILC2s) and IL-22+ cells (ILC3s), but IFN-γ+ cells (NK) were not inhibited (Figure S4A–S4C).
Figure 7.
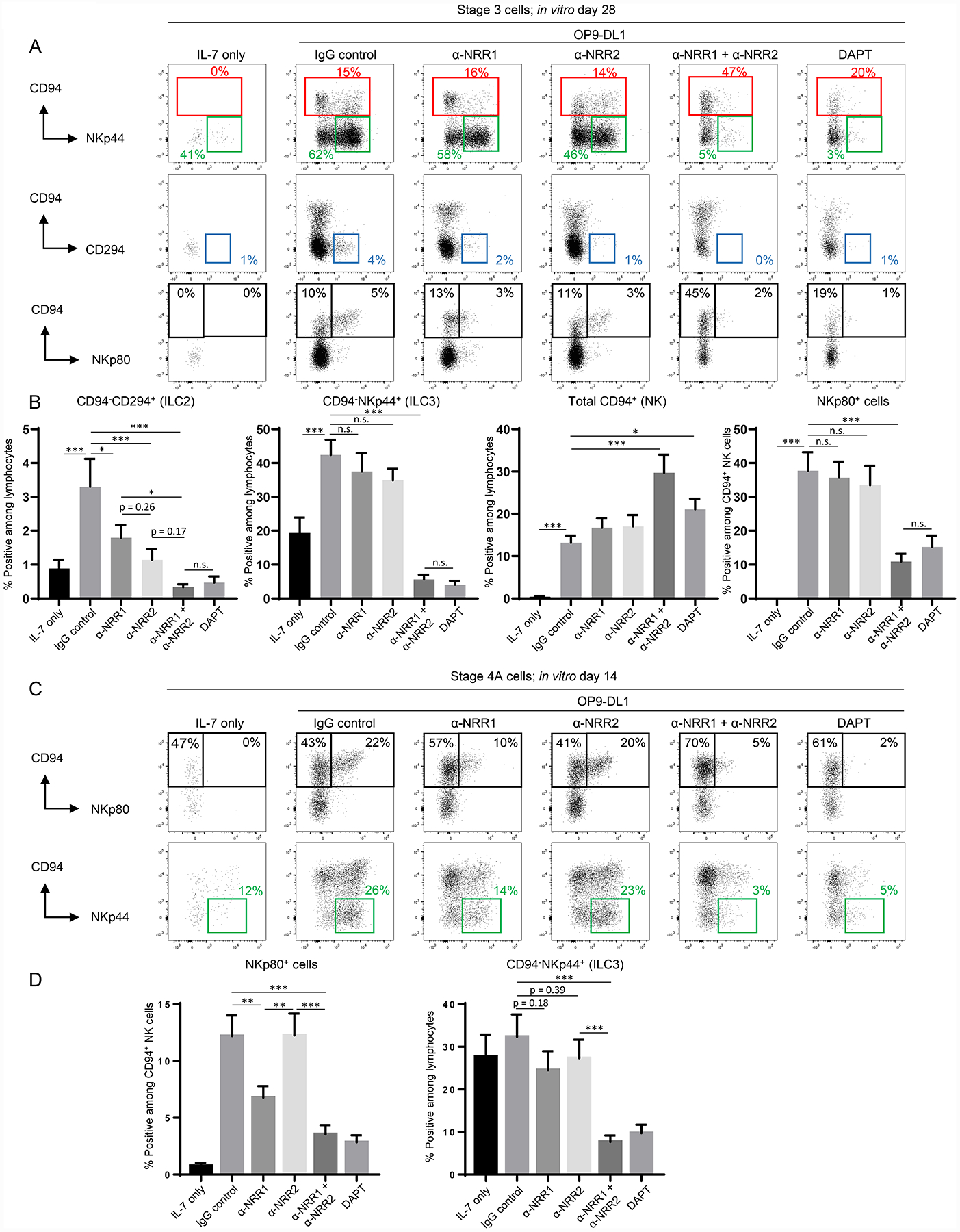
Stage-specific effects of NOTCH1 and NOTCH2 on ILC development. (A) Representative (n = 13; 4 independent experiments) surface flow cytometry analyses of ILCs generated in vitro following 28 day culture of freshly purified tonsil-derived stage 3 cells with IL-7 alone or IL-7 + OP9-DL1 cells with the addition of IgG control antibody, anti-NRR1 and/or anti-NRR2 (5 μg/ml each), or DAPT. (B) Quantification of CD94−CD294+ (ILC2s), CD94-NKp44+ (ILC3s), total CD94+ (NK), and NKp80+ cells generated in vitro following 28 day culture of freshly purified tonsil-derived stage 3 cells in the conditions described in (A). Data are represented as mean ± SEM. n.s. = not significant (p > 0.05); * p < 0.05; ** p < 0.01; *** p < 0.001. (C) Representative (n = 18; 6 independent experiments) surface flow cytometry of ILCs generated in vitro following 14 day culture of freshly purified tonsil-derived stage 4A cells with IL-7 alone or IL-7 + OP9-DL1 in the presence of IgG control, anti-NRR1 and/or anti-NRR2, or DAPT. (D) Quantification of NKp80+ and CD94−NKp44+ cells generated in vitro following 28 day culture of freshly purified tonsil-derived stage 4A cells in the same conditions described in (C). Data are represented as mean ± SEM. ** p < 0.01; *** p < 0.001.
We also determined the impact of Notch receptor blockade on differentiation starting with tonsil-derived stage 4A cells that expressed higher amounts of NOTCH1 than NOTCH2 (Figure 6B). Blockade of NOTCH1 resulted in significantly decreased proportions of NKp80+ cells compared to the control, whereas this effect was not seen when NOTCH2 alone was inhibited (Figure 7C–7D, S4D). Nonetheless, combined NOTCH1 and NOTCH2 blockade further decreased NKp80 expression compared to NOTCH1 blockade alone and at similar amounts to treatment with DAPT (Figure 7C–7D. S4D). When measuring ILC3 differentiation from stage 4A cells, a similar trend was seen, albeit to a lesser extent. Individual blockade of NOTCH1 marginally decreased the percentage of ILC3 differentiation, whereas NOTCH2 blockade had essentially no effect (neither result comparing the percentages of ILC3 differentiation was statistically significant; yet there was a significant decrease in the absolute number of ILC3s produced in the presence of NOTCH1 blockade, as shown in Figure S4D). However, blockade of both NOTCH1 and NOTCH2 significantly inhibited ILC3 differentiation to the same extent as was seen with DAPT (Figure 7C–7D). Collectively, these data indicate that the effects of NOTCH1 and NOTCH2 on ILC2 and ILC3 differentiation as well as on the acquisition of NKp80 expression were stage-specific and also distinct depending upon the measured outcome.
DISCUSSION
ILCs and NK cells comprise a heterogeneous family of effector lymphocytes with myriad roles in normal human physiology as well as in pathologic states including infection, autoimmunity, and cancer. Although it is not yet clear how long mature ILCs and NK cells survive in vivo, it is generally accepted that these cells are continuously produced in the body. Patients with gene mutations resulting in decreased or defective functional ILCs, particularly NK cells, are susceptible to fatal viral infections (25), and it is also well established that patient outcomes are impacted by ILC and NK cell reconstitution following allogeneic hematopoietic stem cell transplantation for acute myeloid leukemia (26, 27). For these and many other reasons, it is vital that we gain a thorough understanding of human innate lymphopoiesis.
In this study, we investigated mechanisms regulating intermediate steps in human NK cell development in tonsils where we previously demonstrated that NKDIs are naturally enriched and thus likely undergo differentiation commitment steps and complete their maturation. Here we have further refined our existing model of human NK cell development (1) by showing that the developmental potentials for ILC2 and then ILC3 differentiation were successively lost as NKDIs progressed from stage 3 to stage 4A to stage 4B, with the latter stage showing full NK cell commitment under the conditions tested. We also demonstrated that while the differentiation of IFN-γ-producing NK cells from stage 3 and 4A NKDIs in the presence of IL-7 required direct contact with OP9 stroma, simultaneous activation of Notch signaling was also required for ILC2 and ILC3 differentiation as well as for the acquisition of an NKp80+ stage 4B-like phenotype. Finally, we determined that the effects of Notch, specifically via NOTCH1 and NOTCH2 receptors, were stage-specific, stromal dependent, and also distinct depending upon the measured outcome. Collectively, our findings highlight a number of fundamental and clinically relevant mechanisms relating to the development of human ILCs.
Notch signaling has been previously shown in a number of murine and human studies to promote ILC2 and ILC3 differentiation from early hematopoietic progenitor cells (7, 28). However, to the best of our knowledge this is the first study to demonstrate the simultaneous requirement of Notch activation and direct contact with stroma for driving non-NK cell lineage plasticity from human tonsil NKDIs. Moreover, here we investigated the specific requirements of individual Notch receptors. We demonstrated that both NOTCH1 and NOTCH2 were expressed by stage 3 NKDIs as well as by ILC2s and ILC3s, whereas stage 4A, 4B, and 5 NKDIs primarily expressed NOTCH1. Consistent with these expression patterns ex vivo, our antibody-mediated Notch receptor blocking experiments showed that NOTCH1 and NOTCH2 were at least partially functionally redundant in promoting ILC2 and ILC3 differentiation from stage 3 NKDIs. Interestingly, we also observed that blockade of either NOTCH1 or NOTCH2 on stage 3 cells significantly reduced ILC2 differentiation, whereas ILC3 differentiation was only significantly impacted when both receptors were simultaneously blocked. One possible explanation to account for these findings is that the processes of ILC2 and ILC3 differentiation require distinct strengths of Notch signaling. This hypothesis is supported by published data from other investigators who used different concentrations of Notch ligands to support lymphocyte differentiation in vitro (29). Alternatively, NOTCH1 and NOTCH2 might provide qualitatively distinct effects on downstream gene expression in human stage 3 cells, and this might differentially influence ILC2 versus ILC3 lineage fate decisions. While it is clear that Notch regulates ILC2 and ILC3 developmental biology, there is still much unknown about the molecular mechanisms that drive the processes of differentiation. In addition to the kinetics of Notch activation during different stages of ILC development, other factors such as simultaneous activation of other signal pathways or the effects of soluble factors in the microenvironment may also impact which downstream pathways are modulated to promote ILC2 and/or ILC3 differentiation. Accordingly, further investigation is warranted to identify the target genes directly regulated by Notch in ILC2s and ILC3s, in addition to NK cells, in order to further understand how Notch regulates ILC plasticity during development.
Our results also revealed stage-specific effects of Notch activation on NK cell development in vitro that coincided with individual receptor expression patterns in tonsils. As mentioned earlier, NOTCH1 expression increased and NOTCH2 expression decreased as tonsil NKDIs proceeded through successive stages of maturation. We observed that while blockade of both NOTCH1 and NOTCH2 was required to significantly prevent NKp80 acquisition by stage 3-derived CD94+ NK cells, individual blockade of NOTCH1 but not of NOTCH2 was sufficient to significantly prevent NKp80 acquisition from tonsil stage 4A cells. This suggests that NOTCH1 is the primary receptor involved in regulating the maturation of stage 4A cells. Nonetheless, when considering the capacity of stage 4A cells to revert to ILC3s in the presence of Notch ligands and stroma, individual blockade of NOTCH1 resulted in a minor decrease of ILC3 differentiation, whereas combined blockade of NOTCH1 and NOTCH2 was required for the extent of inhibition seen with DAPT. One possible explanation for this is that when NOTCH1 was selectively inhibited, low amounts of NOTCH2 present on stage 4A cells compensated to promote reversion to ILC3s but not for NKp80 acquisition. Further investigation is needed to elucidate the molecular mechanisms by which NOTCH1 and NOTCH2 control differentiation at specific stages of human NK cell development.
Another notable finding was that while high NKp80 expression required Notch activation, the latter was not required for the differentiation of functional IFN-γ-producing CD94+ NK cells from tonsil stage 3 and 4A NKDIs, which do not produce IFN-γ ex vivo (18). Given that NKp80 expression closely correlates with the capacity for IFN-γ production during NK cell maturation in tonsils (18), the data shown here suggest that combined interactions with stroma and Notch ligands were required to promote synchronicity in NK cell function and surface phenotype that parallels NK cell development in vivo. Nonetheless, it is noted that while the stage 4B-like cells derived in vitro expressed CD94, NKp80, and EOMES, degranulated in response to co-culture with K562 target cells, and produced IFN-γ, they were still distinct from fresh tonsil-derived stage 4B cells that typically express much higher amounts of T-BET and perforin as well as variable amounts of granzyme B and KIRs ex vivo (18). Indeed even in the presence of OP9-DL1 stroma we did not observe significant expression of T-BET, KIRs, granzyme B, or CD16 (expressed by tonsil stage 5 NK cells) by the in vitro-derived NK cells, suggesting that additional factors were needed to promote the expression of these terminal NK cell differentiation-associated genes. Notably, these data contrast with those of a prior study in which Notch activation in vitro of cord blood CD34+ progenitor cell-derived NK cells was shown to promote higher amounts of KIR acquisition (12). We note that in addition to starting with a different progenitor population than was used in our studies here, in that study exogenous IL-15 was added to the cultures, whereas here we utilized IL-7. One possibility to account for these distinct experimental outcomes is that Notch may exert different biologic effects depending upon the specific developmental stage at which it becomes activated, and as discussed earlier we did observe distinct stage-specific effects of blocking either NOTCH1 or NOTCH2. Alternatively, KIR+ NK cells, which are generally enriched within the traditional CD56dim NK cell subset (that overlaps with stage 5 NKDIs), may originate from a progenitor population that is distinct from tonsil NKDIs giving rise to KIR− NK cells, which are largely CD56bright (and overlap with stage 4B NKDIs). Indeed the developmental origins and relationship between the traditionally described CD56bright and CD56dim human NK cell subsets is unclear and has been a topic of ongoing investigation and debate for many years (21, 30, 31).
Apart from the findings related to Notch signaling, another notable observation was that non-NK cell lineage differentiation plasticity was essentially retained until stage 4B of NK cell development in the tonsil. Stage 4B NKDIs were recently defined by the surface expression of NKp80, which during development in human tonsils accompanies the transition from a stage 4A NKDI that is non-cytotoxic and shows ILC3-associated features to a cytotoxic, IFN-γ+ NK cell (18). The data presented here now indicate that the stage 4A to 4B transition is also associated with the loss of ILC3 differentiation potential. These findings extend upon our observations from a previous study in which we demonstrated that NK cell commitment was associated with a CD117− surface phenotype (4). Given that CD117 is expressed (albeit weakly) on at least some tonsil stage 4B cells and that only NK cells were derived from cultured tonsil stage 4B cells in both our bulk and clonal differentiation assays, the findings here indicate that it is the acquisition of NKp80, rather than the loss of CD117, which most specifically identifies the commitment step during NK development in human tonsils. Nonetheless, as lineage differentiation plasticity is likely highly influenced by the microenvironment, we are cautious to conclude that stage 4B cells are necessarily lineage committed in all tissues or that in other settings phenotypically much less differentiated yet committed NK progenitor cells do not exist. Further study is warranted to determine if the same degree of developmental plasticity exists in other tissues as is seen in tonsils.
The results from our study also highlight the importance of coordinated signals from the microenvironment in driving innate lymphoid cell differentiation in human tonsils. We previously demonstrated through a series of ex vivo phenotypic, molecular, and functional studies that divergent pathways of ILC2, ILC3, and NK cell development likely occur physiologically within these tissues; however, to date it is not yet clear how these developmental events are regulated in vivo. The data we present here suggest that cell-cell interactions, likely with stromal cells (e.g. fibroblast reticular stromal cells in the extra-follicular regions) and/or dendritic cells, which can express Notch ligands (32), are critical, although very little is known about how the various cell types and intercellular signals are regulated and coordinated in the body. We previously reported that Delta-like-1 and Delta-like-4 are expressed in human tonsils, shown by immunohistochemical staining (33). These Notch ligands are expressed in the interfollicular regions of the tonsil, where we had also previously shown that stage 3 NKDIs reside (34). These observations support the premise that NK cell developmental intermediates can directly interact with Notch ligands in the lymphoid tissue microenvironment. As such, we believe that the OP9-DL1 stromal cell co-culture system is a relevant in vitro model for studying the role of Notch in regulating ILC differentiation in SLTs. In this study, we observed that simultaneous contact with stroma was required in order for the Notch-dependent effects to manifest. The stroma-dependent mechanism is not yet known, although based on the results of prior T cell differentiation studies, signals through vascular cell adhesion molecule 1 (VCAM-1), may be required for Notch activation in tonsil-derived NKDIs (35). OP9 cells express high amounts of VCAM-1, suggesting that expression of VCAM-1 may be similarly relevant in our system (36). Future studies are needed to determine which tonsil-resident cell types, Notch ligands, and stromal-derived factors are important to promote the synchronous patterns of ILC and NK cell development that are repeatedly detected ex vivo.
In summary, we have demonstrated that interactions with stromal cells were required for Notch-mediated ILC2 and ILC3 differentiation from stage 3 cells as well as for the promotion of functional and phenotypic NK cell maturation from tonsil-derived NKDIs. We also provide evidence that stage 4B likely represents the first step during development in human tonsils when cells become committed to the NK cell lineage. Moreover, we showed that NOTCH1 and NOTCH2 regulate human ILC differentiation in a stage-specific manner. This study raises interesting translational implications, such as the role of clinical grade anti-Notch receptor antibodies (16). The variability of Notch-mediated responses in different stages during NK cell development suggests that modulation of Notch in different populations may offer a more specific approach to novel therapies. Indeed while global antagonism of Notch (via pharmacologic inhibition) may have off-target effects, receptor blockade may provide greater specificity. NOTCH1 and NOTCH2 can exert different downstream effects, in part due to their respective regulation of specific target genes (37). Thus receptor blockade allows for greater specificity in modulation of the signal pathways downstream of Notch receptor activation. Further investigation to determine the in vivo effects on NK cells and other ILCs in the setting of Notch modulation will likely provide the basis for future translational studies involving Notch. Collectively, our data refine a development model in which combined interactions with stromal cells and Notch ligands support synchronous phenotypic and functional maturation of human NK cells.
Supplementary Material
KEY POINTS.
Stage 3 and 4A, but not 4B, NKDIs retain non-NK ILC developmental potential.
Notch activation in the presence of stroma promotes ILC2 and ILC3 differentiation.
NOTCH1 and NOTCH2 regulate ILC differentiation from stage 3 and 4A cells.
ACKNOWLEDGMENTS
The authors would like to thank Ms. Victoria Sellers for her outstanding assistance. We also thank the Cooperative Human Tissue Network of Nationwide Children’s Hospital (Columbus, OH) for providing us with human pediatric tonsil samples.
This work was supported by grants from the National Institutes of Health/National Cancer Institute (CA199447 and CA208353 to A.G.F., CA068458 to M.A.C., and CA236063 to A.P.N.), and funds from the Pelotonia Organization (OSU-CCC; A.P.N, B.L.M.-B., and A.G.F.). Research reported in this publication was supported by The Ohio State University Comprehensive Cancer Center and the National Institutes of Health (P30-CA016058).
Footnotes
DECLARATION OF INTERESTS
C.W.S. is an employee of Genentech, Inc., which has an interest in therapeutically targeting Notch signaling. Otherwise the authors declare no competing interests.
References
- 1.Scoville SD, Freud AG, and Caligiuri MA. 2017. Modeling Human Natural Killer Cell Development in the Era of Innate Lymphoid Cells. Front Immunol 8: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freud AG, Yu J, and Caligiuri MA. 2014. Human natural killer cell development in secondary lymphoid tissues. Semin Immunol 26: 132–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lim AI, Li Y, Lopez-Lastra S, Stadhouders R, Paul F, Casrouge A, Serafini N, Puel A, Bustamante J, Surace L, Masse-Ranson G, David E, Strick-Marchand H, Le Bourhis L, Cocchi R, Topazio D, Graziano P, Muscarella LA, Rogge L, Norel X, Sallenave J-M, Allez M, Graf T, Hendriks RW, Casanova J-L, Amit I, Yssel H, and Di Santo JP. 2017. Systemic Human ILC Precursors Provide a Substrate for Tissue ILC Differentiation. Cell 168: 1086–1100. [DOI] [PubMed] [Google Scholar]
- 4.Chen L, Youssef Y, Robinson C, Ernst GF, Carson MY, Young KA, Scoville SD, Zhang X, Sekhri P, Mansour AG, Chan WK, Nalin AP, Mao HC, Hughes T, Mace EM, Pan Y, Rustagi N, Chatterjee SS, Gunaratne PH, Behbehani GK, Mundy-Bosse BL, Caligiuri MA, and Freud AG. 2018. CD56 Expression Marks Human Group 2 Innate Lymphoid Cell Divergence from a Shared NK Cell and Group 3 Innate Lymphoid Cell Development Pathway. Immunity 49: 464–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freud AG, Yokohama A, Becknell B, Lee MT, Mao HC, Ferketich AK, and Caligiuri MA. 2006. Evidence for discrete stages of human natural killer cell differentiation in vivo. J Exp Med 203: 1033–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bachanova V, McCullar V, Lenvik T, Wangen R, Peterson KA, Ankarlo DE, Panoskaltsis-Mortari A, Wagner JE, and Miller JS. 2009. Activated notch supports development of cytokine producing NK cells which are hyporesponsive and fail to acquire NK cell effector functions. Biol Blood Marrow Transplant 15: 183–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kyoizumi S, Kubo Y, Kajimura J, Yoshida K, Hayashi T, Nakachi K, Moore MA, van den Brink MR, and Kusunoki Y. 2017. Fate Decision Between Group 3 Innate Lymphoid and Conventional NK Cell Lineages by Notch Signaling in Human Circulating Hematopoietic Progenitors. J Immunol 199: 2777–2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rolink AG, Balciunaite G, Demoliere C, and Ceredig R. 2006. The potential involvement of Notch signaling in NK cell development. Immunol Lett 107: 50–57. [DOI] [PubMed] [Google Scholar]
- 9.Andersson ER, Sandberg R, and Lendahl U. 2011. Notch signaling: simplicity in design, versatility in function. Development 138: 3593–3612. [DOI] [PubMed] [Google Scholar]
- 10.Haraguchi K, Suzuki T, Koyama N, Kumano K, Nakahara F, Matsumoto A, Yokoyama Y, Sakata-Yanagimoto M, Masuda S, Takahashi T, Kamijo A, Takahashi K, Takanashi M, Okuyama Y, Yasutomo K, Sakano S, Yagita H, Kurokawa M, Ogawa S, and Chiba S. 2009. Notch activation induces the generation of functional NK cells from human cord blood CD34-positive cells devoid of IL-15. J Immunol 182: 6168–6178. [DOI] [PubMed] [Google Scholar]
- 11.Beck RC, Padival M, Yeh D, Ralston J, Cooke KR, and Lowe JB. 2009. The Notch ligands Jagged2, Delta1, and Delta4 induce differentiation and expansion of functional human NK cells from CD34+ cord blood hematopoietic progenitor cells. Biol Blood Marrow Transplant 15: 1026–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felices M, Ankarlo DE, Lenvik TR, Nelson HH, Blazar BR, Verneris MR, and Miller JS. 2014. Notch Signaling at later stages of Natural Killer cell development enhances KIR expression and functional maturation. J Immunol 193: 3344–3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scoville SD, Keller KA, Cheng S, Zhang M, Zhang X, Caligiuri MA, and Freud AG. 2015. Rapid Column-Free Enrichment of Mononuclear Cells from Solid Tissues. Scientific Reports 5: 12490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scoville SD, Mundy-Bosse BL, Zhang MH, Chen L, Zhang X, Keller KA, Hughes T, Chen L, Cheng S, Bergin SM, Mao HC, McClory S, Yu J, Carson WE 3rd, Caligiuri MA, and Freud AG. 2016. A Progenitor Cell Expressing Transcription Factor RORgammat Generates All Human Innate Lymphoid Cell Subsets. Immunity 44: 1140–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmitt TM, and Zuniga-Pflucker JC. 2002. Induction of T Cell Development from Hematopoietic Progenitor Cells by Delta-like-1 In Vitro. Immunity 17: 749–756. [DOI] [PubMed] [Google Scholar]
- 16.Wu Y, Cain-Hom C, Choy L, Hagenbeek TJ, de Leon GP, Chen Y, Finkle D, Venook R, Wu X, Ridgway J, Schahin-Reed D, Dow GJ, Shelton A, Stawicki S, Watts RJ, Zhang J, Choy R, Howard P, Kadyk L, Yan M, Zha J, Callahan CA, Hymowitz SG, and Siebel CW. 2010. Therapeutic antibody targeting of individual Notch receptors. Nature 464: 1052–1057. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Wu S, Pu J, Huang X, and Zhang P. 2015. Dengue virus upregulates expression of notch ligands Dll1 and Dll4 through interferon-β signaling pathway. Immunology 144: 127–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freud AG, Keller KA, Scoville SD, Mundy-Bosse BL, Cheng S, Youssef Y, Hughes T, Zhang X, Mo X, Porcu P, Baiocchi RA, Yu J, Carson WE 3rd, and Caligiuri MA. 2016. NKp80 Defines a Critical Step during Human Natural Killer Cell Development. Cell Reports 16: 379–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Venkatesh K, Reddy LVK, Abbas S, Mullick M, Moghal ETB, Balakrishna JP, and Sen D. 2017. NOTCH Signaling Is Essential for Maturation, Self-Renewal, and Tri-Differentiation of In Vitro Derived Human Neural Stem Cells. Cellular Reprogramming 19: 372–383. [DOI] [PubMed] [Google Scholar]
- 20.Cupedo T, Crellin NK, Papazian N, Rombouts EJ, Weijer K, Grogan JL, Fibbe WE, Cornelissen JJ, and Spits H. 2009. Human fetal lymphoid tissue-inducer cells are interleukin 17-producing precursors to RORC+ CD127+ natural killer-like cells. Nat Immunol 10: 66–74. [DOI] [PubMed] [Google Scholar]
- 21.Freud AG, Mundy-Bosse BL, Yu J, and Caligiuri MA. 2017. The Broad Spectrum of Human Natural Killer Cell Diversity. Immunity 47: 820–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Smedt M, Taghon T, Van de Walle I, De Smet G, Leclercq G, and Plum J. 2007. Notch signaling induces cytoplasmic CD3e expression in human differentiating NK cells. Blood 110: 2696–2703. [DOI] [PubMed] [Google Scholar]
- 23.Crellin NK, Trifari S, Kaplan CD, Cupedo T, and Spits H. 2010. Human NKp44+IL-22+ cells and LTi-like cells constitute a stable RORC+ lineage distinct from conventional natural killer cells. J Exp Med 207: 281–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sauma D, Ramirez A, Alvarez K, Rosemblatt M, and Bono M. 2012. Notch Signalling Regulates Cytokine Production by CD8+ and CD4+ T Cells. Scand J Immunol 75: 389–400. [DOI] [PubMed] [Google Scholar]
- 25.Mace EM, and Orange JS. 2019. Emerging Insights into human health and NK cell biology from the study of NK cell deficiencies. Immunol Rev 287: 202–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mancusi A, Ruggeri L, and Velardi A. 2016. Haploidentical hematopoietic transplantation for the cure of leukemia: from its biology to clinical translation. Blood 128: 2616–2623. [DOI] [PubMed] [Google Scholar]
- 27.Ruggeri L, Capanni M, Urbani E, Perruccio K, Shlomchik WD, Tosti A, Posati S, Rogaia D, Frassoni F, Aversa F, Martelli MF, and Velardi A. 2002. Effectiveness of Donor Natural Killer Cell Alloreactivity in Mismatched Hematopoietic Transplants. Science 295: 2097–2100. [DOI] [PubMed] [Google Scholar]
- 28.Koga S, Hozumi K, Hirano K-I, Yazawa M, Terooatea T, Minoda A, Nagasawa T, Koyasu S, and Moro K. 2018. Peripheral PDGFRα(+)gp38(+) mesenchymal cells support the differentiation of fetal liver-derived ILC2. J Exp Med 215: 1609–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benne C, Lelievre JD, Balbo M, Henry A, Sakano S, and Levy Y. 2009. Notch increases T/NK protential of human hematopoietic progenitors and inhibits B cell differentiation at a pro-B stage. Stem Cells 27: 1676–1685. [DOI] [PubMed] [Google Scholar]
- 30.Cooper MA, Fehniger TA, and Caligiuri MA. 2001. The biology of human natural killer-cell subsets. Trends Immunol 22: 633–640. [DOI] [PubMed] [Google Scholar]
- 31.Wu C, Li B, Lu R, Koelle SJ, Yang Y, Jares A, Krouse AE, Metzger M, Liang F, Loré K, Wu CO, Donahue RE, Chen ISY, Weissman I, and Dunbar CE. 2014. Clonal tracking of rhesus macaque hematopoiesis highlights a distinct lineage origin for natural killer cells. Cell Stem Cell 14: 486–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoon S-O, Zhang X, Berner P, Blom B, and Choi YS. 2009. Notch Ligands Expressed by Follicular Dendritic Cells Protect Germinal Center B Cells from Apoptosis. J Immunol 183: 352–358. [DOI] [PubMed] [Google Scholar]
- 33.McClory S, Hughes T, Freud AG, Briercheck EL, Martin C, Trimboli AJ, Yu J, Zhang X, Leone G, Nuovo G, and Caligiuri MA. 2012. Evidence for a stepwise program of extrathymic T cell development within the human tonsil. J Clin Invest 122: 1403–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hughes T, Becknell B, Freud AG, McClory S, Briercheck E, Yu J, Mao C, Giovenzana C, Nuovo G, Wei L, Zhang X, Gavrilin M, Wewers MD, and Caligiuri MA. 2010. Interleukin-1beta Selectively Expands and Sustains Interleukin-22+ Immature Human Natural Killer Cells in Secondary Lymphoid Tissue. Immunity 32: 803–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shukla S, Langley MA, Singh J, Edgar JM, Mohtashami M, Zuniga-Pflucker JC, and Zandstra PW. 2017. Progenitor T-cell differentiation from hematopoietic stem cells using Delta-like-4 and VCAM-1. Nat Methods 14: 531–538. [DOI] [PubMed] [Google Scholar]
- 36.Supper E, Tahir S, Imai T, Inoue J, and Minato N. 2015. Modification of Gene Expression, Proliferation, and Function of OP9 Stroma Cells by Bcr-Abl-Expressing Leukemia Cells. PloS One 10: e0134026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baumgart A, Mazur PK, Anton M, Rudelius M, Schwamborn K, Feuchtinger A, Behnke K, Walch A, Braren R, Peschel C, Duyster J, Siveke JT, and Dechow T. 2014. Opposing role of Notch1 and Notch2 in a KrasG12D-driven murine non-small cell lung cancer model. Oncogene 34: 578. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


