Abstract
Cells rely on protein degradation by AAA+ proteases. A well-known example is the hexameric ClpX unfoldase, which captures ATP hydrolysis to feed substrates into the oligomeric ClpP peptidase. Recent studies show that an asymmetric ClpX spiral cycles protein translocation upon ATP hydrolysis. However, how this cycle affects peptide products is less explored in part because ClpP cleavage is thought to be solely defined by sequence constraints. Here, we comprehensively characterize peptides from Caulobacter crescentus ClpXP degradation of three different substrates using high-resolution mass spectrometry and find that cleavage of translocated substrates is driven by factors other than sequence. We report that defined locations in a translocated protein are especially sensitive to cleavage spaced on average every 10–13 residues. These sites are not exclusively controlled by sequence and are independent of bulk changes in catalytic peptidase sites, ATP hydrolysis, or efficiency of initial recognition. These results fit a model where processive translocation through ClpX starts at a specific location in a polypeptide and pauses during reset of the ClpX hexamer after a cycle of translocation. Our work suggests that defined peptides, which could be used as signaling molecules, can be generated from a given substrate by a nonspecific peptidase.
Graphical Abstract
Introduction:
AAA+ proteases are required in all kingdoms of life during both normal growth and stress responses. In bacteria, the highly conserved ClpXP system is responsible for degradation of hundreds of substrates,1,2 including products of stalled translation through the tmRNA-mediated trans-translation system.1,2 ClpX consumes ATP to power recognition and unfolding of a target substrate, gripping proteins with a series of pore-loops, and translocating the unfolded polypeptides through a central pore to the chambered ClpP peptidase, where the protein is hydrolyzed to small peptides.1,2 ClpXP is able to degrade proteins of diverse sequence and structural composition, without particular selectivity once substrate degradation is initiated.1,2 Unfolded substrates are degraded more rapidly than folded substrates, but the relationship between target structure and unfolding is principally dependent on unraveling of local kinetically stable elements rather than global thermodynamic stability.3 Importantly, how these parameters translate into the peptide products of degradation is less clear.
Initial studies have shown that the ClpP family of proteases produces peptides that range from 8–12 residues in length.4–6 Structural and biochemical studies7 reveal dynamic pores that circumscribe the ClpP barrel and prevent larger peptides from exiting the chamber. Peptide product identity has been far less explored than other parameters of the ClpXP processing cycle as ClpP is relatively nonspecific based on preferences established with short peptide reporters.5 Interestingly, during proteotoxic conditions, peptides released from the mitochondria ClpP trigger activation of the mitochondria unfolded protein response8 suggesting that release of specific peptides could be a ‘canary in the coalmine’ signal for cells to respond to stress. Therefore, understanding what constraints drive peptide cleavage in the ClpP-family of proteases may aid in our discovery of these signals.
Here, we use a series of structurally distinct substrates and high-resolution mass spectrometry to determine the specific sites and patterns of substrate cleavage. We find that translocation speed, recognition efficiency, or number of active catalytic sites have negligible effects on product peptide identities. Rather, local structural constraints and product length superimposed on intrinsic peptide site preferences are the major drivers for where cleavage ultimately occurs. Interestingly, we also find a substrate-independent enrichment of peptides with a 10–13 residue average periodic cleavage pattern. In light of recent structural and single molecule studies, we interpret this preference as the existence of a pause during ClpXP degradation of the Caulobacter crescentus species, driven by resetting of the ClpX ring and cleavage of the polypeptide extended into the ClpP chamber at sites constrained by primary sequence preference.
Results
Cleavage preference is unaffected by substrate extrinsic reaction parameters
Three proteins, GFP, RcdA, and β2m, that differ in size and secondary structure were expressed with an ssrA tag on the C-terminus and subjected to in vitro degradation by ClpXP. The peptides that are produced upon degradation of GFPSsrA, RcdAssrA, and disulfide-reduced β2mssrA were identified by LC/MS/MS (Figure 1A). Sequence coverages ranging from 92–100%, with an average of 96% ± 3% (n=14), were obtained under standard degradation conditions. This high sequence coverage provides unprecedented insight into how substrates are digested by ClpXP. From these experiments, we find that cleavage is preferred when Leu, Met, Ser, or Ala residues are directly adjacent to the cleave site in the n-terminal direction, or the P1 position, while other positions show no obvious cleavage specificity (Figure 1B). When each protein substrate is considered separately, the preference for the hydrophobic residues, Leu and Met, at the P1 position persists (Figure 1C, D, E).
Figure 1:
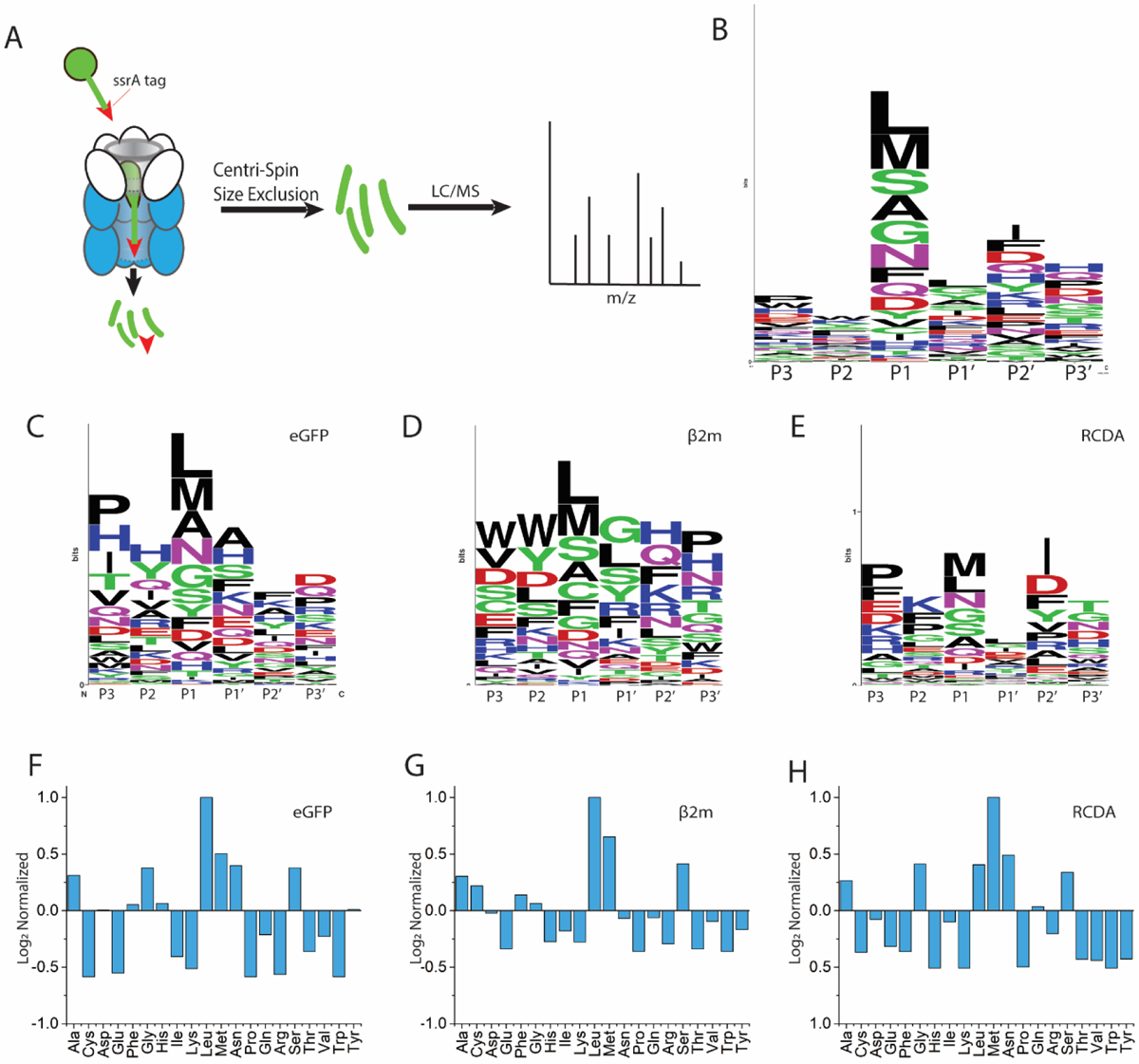
Primary structure cleavage specificity of ClpXP. (A) Schematic depiction of tagged protein digestion, separation, and analysis. Residues directly adjacent to the cleavage site in the N-terminal direction are indicated as P1, P2 and P3, and residues directly adjacent to the cleavage site in the c-terminal direction are indicated as P1’, P2’ and P3’, numerically increasing as they are further from the cleavage site. Weblogo representation of cleavage preferences for all substrates (B), GFP (C), reduced β2m (D), and RcdA (E). Amino acid preferences at the P1 position normalized to presence in the given protein for GFP (F), reduced β2m (G), and RcdA (H).
While this method of data analysis indicates preferred residues at cleavage sites, it does not clearly represent nonpreferred residues nor does it account for how often each residue appears in the sequence of the protein. To address these issues, the frequency with which a residue appears at the C-terminus of a peptide fragment was normalized to the frequency with which it is found in the protein sequence. Figures 1F, G, and H depict this normalized data, where values greater than zero indicate a preference for cleavage at the P1 position, values less than zero indicate that cleavage at a given residue is disfavored, and a value of zero indicates that cleavage at this residue is as likely as random chance. From this analysis, we find a preference for cleavage at Leu and Met residues but also at Ala, Gly, Ser, and Asn residues. These cleavage preferences persist across the studied substrates, with 81% agreement (Pearson correlation) between reduced β2m and GFP and 77% agreement between RcdA and GFP for cleavage preference at the P1 position. The intrinsic preference of ClpXP for hydrophobic residues at the P1 position is consistent with previous work in which Gersch and coworkers found general cleavage preferences at Met, Ser, Leu, and Ala at the P1 position for human ClpP, Escherichia coli ClpP, and Staphylococcus aureus ClpP upon digestion of both endogenous substrates and peptide libraries.5 Our data with a fourth species, Caulobacter crescentus ClpP, demonstrates that the preference is conserved across all these species. It is possible that differences observed in peptide products from ClpXP originating in different species could be a result of differences in dynamics or sequence of each unique species.
We next considered the length of produced peptides and found average peptide lengths of 13 ± 5, 13 ± 5, and 15 ± 6 residues for GFP, β2m, and RcdA, respectively, based on at least three replicate experiments for each protein. Peptide distributions are skewed towards higher molecular weights with peptides up to 28 amino acids observed in some cases (Figure S1). When the distribution of peptides is weighted by their mass spectral intensities, we observe that the average lengths for GFP, reduced β2m, and RcdA are 11 ± 3, 12 ± 3, and 12 ± 4 residues, respectively. A similar MS-based analysis from Sieber and co-workers showed peptide product length distributions ranging from 8–12 residues derived from protein degradation by ClpXP,5 largely consistent with what we find.
Having determined that cleavage specificity or length is not substrate dependent, we investigated if degradation rate influences these parameters. For example, slowing proteolysis may increase the dwell time a substrate spends within the barrel of ClpXP which may affect cleavage specificity or product length. Using GFPSsrA, we first investigated changes in ATP concentration as ATP hydrolysis affects degradation rate. By using sufficiently low ATP concentrations ranging from 7 to 31 μM, we could adjust the degradation rates substantially as monitored by loss of fluorescence (Figure 2A). It is worth noting that at 7 μM ATP virtually no loss of full-length protein is observed by the fluorescence assay, but peptides are still generated and detected by MS, highlighting the sensitivity of our measurements. Overall, there is no significant difference in peptide length or cleavage specificity as compared to saturating ATP concentrations (Figure S3). An analysis of the cleavage specificity of protease under optimal cleavage conditions compared with low ATP levels indicates greater than 97% agreement (Pearson correlation) for the preferred residues in the P1 position (Table 1). The fact that peptide distributions are the same regardless of ATP concentration further supports our understanding that ClpXP is a highly processive protease and once engaged with a substrate will fully degrade that target. We note that this effect could also be contributed by ClpP itself given prior work suggesting that peptide bond hydrolysis alone may be sufficient to power processive degradation.9
Figure 2:
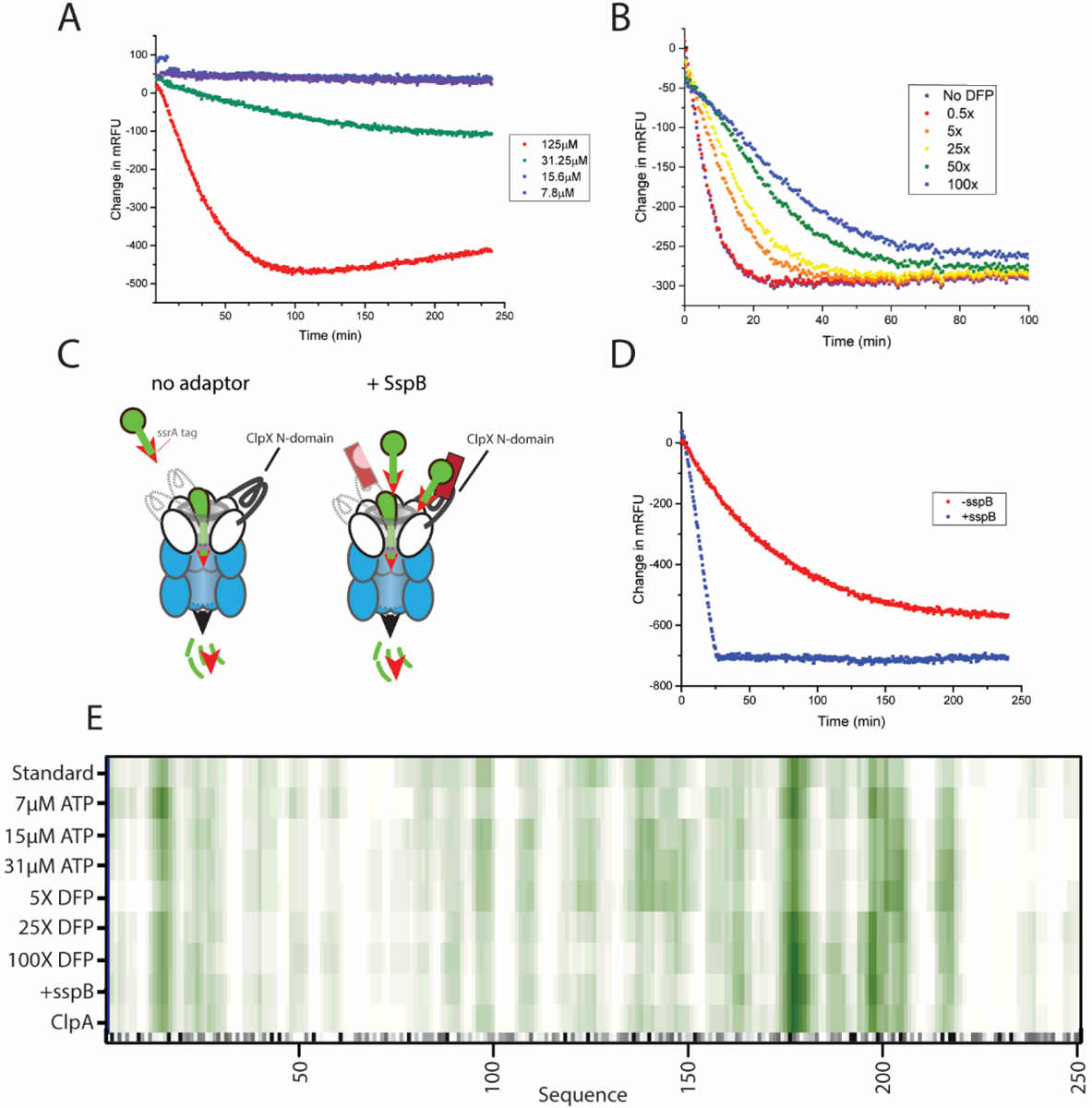
Altered reaction conditions and rates do not change ClpXP cleavage specificities. (A) GFP degradation rate at varying ATP concentrations; 125 μM ATP (red) 31.25 μM (green), 15.6 μM (blue), and 7.8 μM (purple). (B) GFP degradation rate at 0.5, 5, 25, 50, and 100-fold molar excesses of the inhibitor DFP. (C) Cartoon depiction of the sspB adapter facilitating substrate orientation. (D) GFP degradation rate in the presence(blue) and absence(red) of the sspB adapter. (E) Heat map that summarizes the GFP cleavage locations under various reaction conditions. A darker shade of green indicates a higher incidence of cleavage, while white indicates a low incidence of cleavage. At the bottom of the plot, the gray scale indicates the cleavage specificity derived from Figure 1F with darker colors corresponding to amino acids that are more preferred in the P1 position.
Table 1:
A summary of the Pearson correlations of primary sequence cleavage specificity and peptide length distribution as compared to native GFPSsrA degradation under standard conditions
| Primary sequence cleavage in P1 position | Peptide length distribution | Cleavage location on sequence | |
|---|---|---|---|
| RcdA | 77% | 86% | |
| Reduced β2m | 81% | 93% | |
| β2m | 75% | 92% | |
| GFP +7μM ATP | 98% | 95% | 97% |
| GFP +15μM ATP | 98% | 95% | 96% |
| GFP +30μM ATP | 97% | 97% | 97% |
| GFP +5X DFP | 93% | 93% | 82% |
| GFP +25X DFP | 97% | 95% | 86% |
| GFP +100XDFP | 97% | 93% | 87% |
| GFP +sspB | 92% | 89% | 91% |
ClpP degradation rate was also reduced by inhibiting active sites with the protease inhibitor diisopropyl fluorophosphate (DFP). DFP inhibits ClpP activity in a dose-dependent manner (Figure 2B) by binding covalently and irreversibly to the active site serine. Previous studies have shown that degradation of substrates by DFP-treated ClpP results in release of partially processed intermediates.10 We speculated that by inhibiting some active sites, longer peptides with different cleavage specificities might be produced as there are fewer active sites available to cleave the protein. However, digestion of GFP in the presence of 5, 25, and 100-fold molar excesses of DFP resulted in both peptide length and cleavage specificity that were very similar to control experiments without DFP (i.e. >93% similar; see Table 1). These results indicate that inhibiting active sites does not change cleavage preference or distribution of final peptide products, even though overall proteolysis is slowed.
We also increased degradation rates by using the SspB adaptor, which delivers ssrA-tagged substrates more effectively to ClpXP (Figure 2C). Similar to slowing the reaction, accelerating the reaction has little effect on cleavage specificity or the resulting peptide length distribution (Figure S1 and Table 1). The primary sequence cleavage specificity and the peptide length distributions were >92% and >89% correlated, respectively, to GFP degradation under standard conditions (Table 1). Together, our data show that the fundamental distribution of cleavage sites within a substrate degraded by ClpXP is mostly unaffected by energy consumption, number of peptide hydrolysis sites, or efficiency of substrate recognition, as summarized in Figure 2E and Table 1
Cleavage specificity is constrained by position
Interestingly, the preferences shown in the heatmaps of Figure 2E suggest that there is positional specificity in the cleavage products of ClpXP. When mass spectral weighted cleavage sites are plotted as a function of substrate length for all substrates (Figure 3), it becomes clear that there are “hot spots” that are not solely correlated to sites most likely to be cleaved based exclusively on primary sequence preference (Figures 3 and 1F). The average amino acid spacing between each peak in Figure 3 is 12 ± 3 (GFP), 12 ± 3 (β2m), and 13 ± 6 (RcdA), consistent with the peptide length distributions shown previously. Moreover, a power spectral analysis of the plots in Figure 3, from a Fast Fourier Transform, reveals defined frequency components for the cleavage of each substrate, while no such defined frequency components are observed when the same analysis is done on the sequence preference (Figure S7). These defined frequency components presumably correspond to discrete cleavage step sizes. Cleavage patterns also deviate when comparing the C-terminus (which would be the first to enter the ClpP chamber). In particular, near the initiation site at the C-terminus, cleavage site seems mostly driven by amino acid identity until a structured region is encountered, then cleavage appears to occur at regular intervals thereafter (Figure S4). This observation suggests that upon initiation, cleavage of the pioneering sequence entering the ClpP chamber is random (resulting in no significant accumulation of a particular site), but once structured regions are being translocated, there are deliberate pauses in the processive degradation which result in enrichment of cleavage sites spaced 10–13 residues apart (Figure 3).
Figure 3:
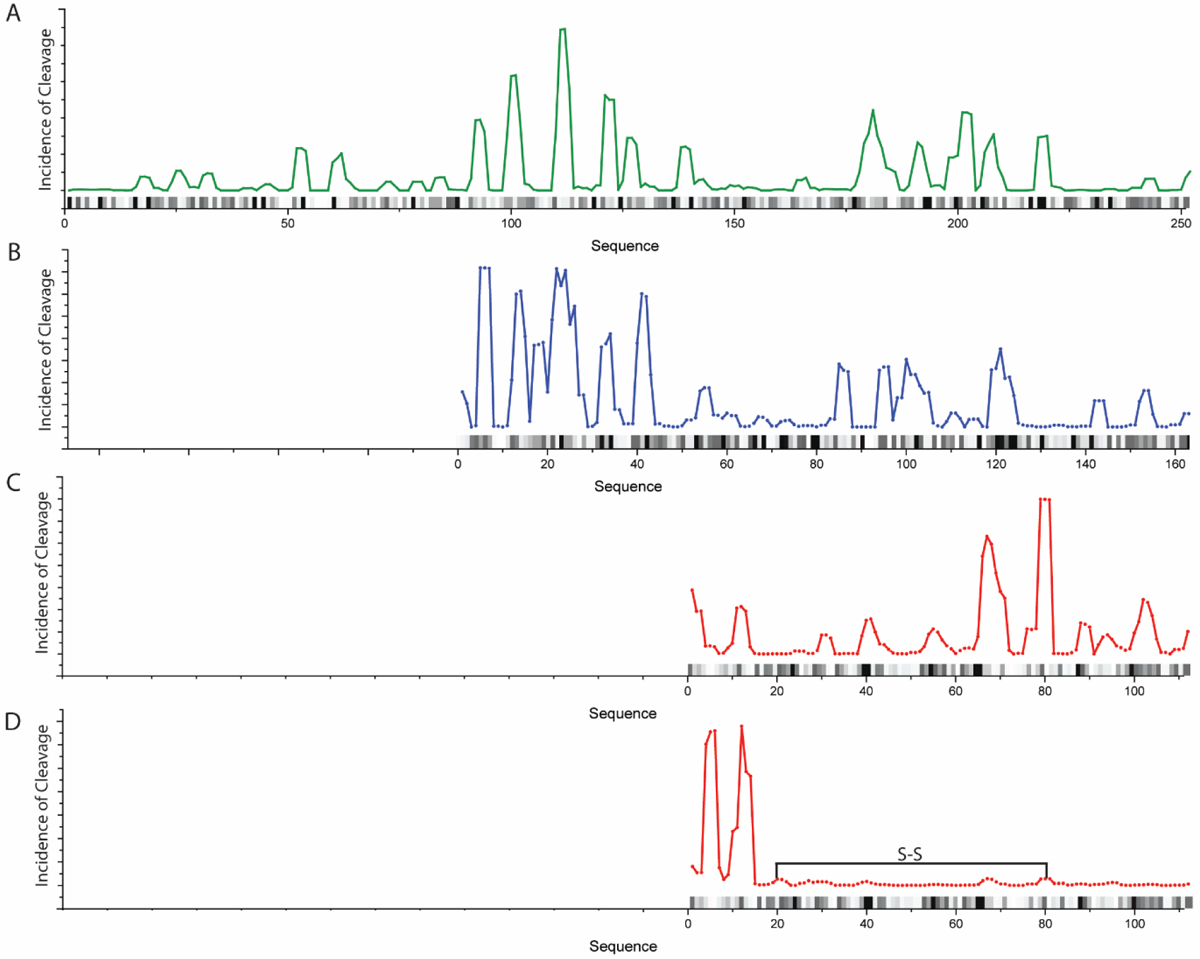
Cleavage incidence locations on the studied substrates weighted for mass spectral intensity. (A) GFP, (B) RcdA, (C) reduced β2m and (D) oxidized β2m. The location of the disulfide bond is depicted as a black line. Data were subject to a moving average smoothing over 3 data points. The black and gray bars on each graph represent primary sequence cleavage specificity, where darker colors indicate higher likelihood of cleavage.
Disulfide effects on positional cleavage specificity
To further investigate the effect of protein higher order structure on ClpXP cleavage patterns, we examined the peptide products of the ssrA-tagged disulfide-bonded form of β2m. Peptides resulting from digestion of β2m with an intact disulfide bond are notably different than peptides produced from the reduced form of the protein (Figures 3C and 3D), with only a 55% correlation in the cleavage locations. This difference is even more apparent when the observed cleavage sites are weighted for mass spectral intensity (Figure 3D). Considered in this way, there is a preference for cleavage at residues 66 and 78–80 in the reduced form but a preference at residues 4 and 12 for the oxidized form. We do not observe a significant difference in the degradation efficiency of between oxidized and reduced β2m, nor do we observe any aggregation of either form of this substrate. Because ClpX is known to accommodate a disulfide-bond linked polypeptide,11 this suggests that the translocation of this more structured region results in a reset of cleavage site preference.
Discussion
Rather than a simple two-step process of recognition and degradation, our data indicate additional steps occur after the initial translocation during the degradation process of Caulobacter crescentus ClpXP that are influenced by the tertiary structure of the substrate (Figure 3D). We propose that initiation of degradation is followed by several cycles of translocation and a defined pause such that unfolded substrates dwell in the peptidase chamber and those preferred sites most accessible to the ClpP active sites can be cleaved, while others remain out of reach (Figure 4). Indeed, prior work shows that while ClpP is highly specific for certain residues when degrading small peptides, larger polypeptides show less discrimination in their products.5 Given the high measured sequence coverage we obtain in our study, we predict that this difference in stringent selectivity stems from the position dependent cleavages we describe here.
Figure 4:
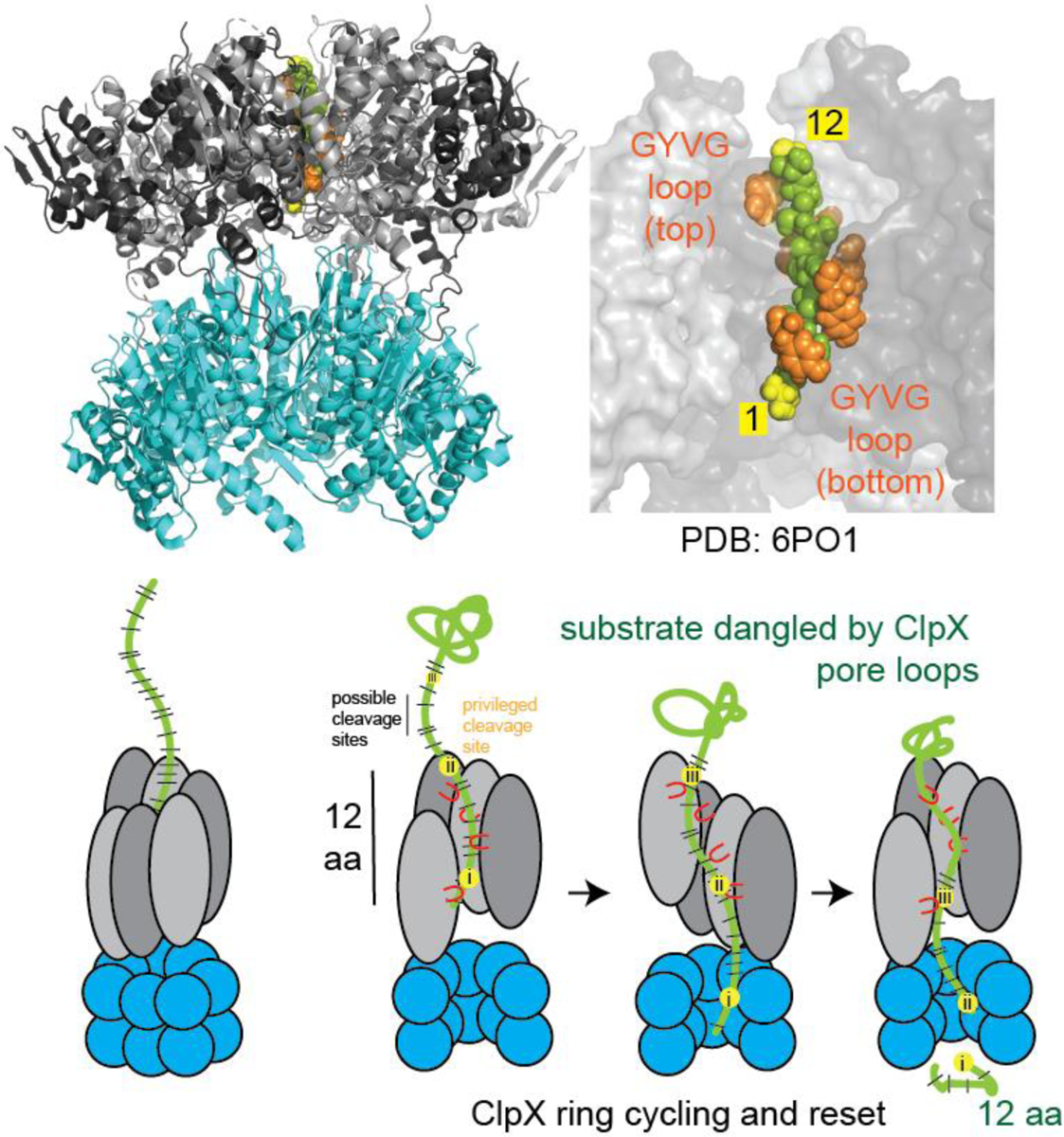
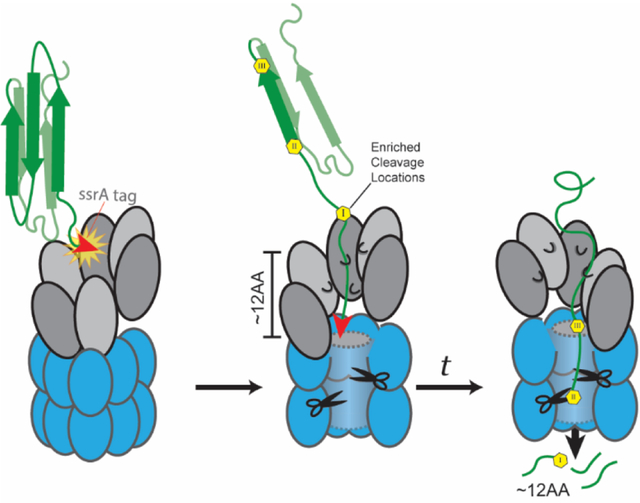
Depiction of ClpXP degradation mechanism. Upper: the ClpXP structure (PDB 6PO1) can accommodate a 12-residue span of peptide in its grip. Lower: After initial engagement of the substrate, ClpX enters the processive state of degradation where two residues are translocated for every ATP hydrolyzed. After a complete cycle, ClpX pauses to reset. During this time, the dangled substrate is cleaved by the ClpP peptidase, at the sites most accessible to the catalytic active sites.
We consider a possible model which explains our results based on recent structural and single molecule experiments. Recent cryo-electron microscopy studies of ClpXP bound to substrate show that subunits of ClpX form a shallow right-handed spiral, resulting in a grip that easily spans 10–13 residues (Figure 4).7,12 In the continuous spiral portion of these static structures, the pore loops of each subunit grip the substrate two residues away from the comparable position of the neighboring subunit. Given the hexameric nature of ClpX, our 12-residue average spacing between cleavage sites is consistent with a pause that occurs after all subunits in the ring has hydrolyzed ATP (6 ATP x 2 residues). During this pause, the unfolded polypeptide dangles into the ClpP chamber for a sufficient time to rapidly degrade at cleavage sites most favored by the intrinsic specificity of ClpP but is constrained by the hold of ClpX on the upstream sequence. After the ring is reset, hydrolysis begins again to start another cycle. This model is also consistent with several recent optical trapping based single molecule experiments that demonstrate bursts of translocation that generally report 1–2 nm (or 6–12 aa) runs of translocation, followed by a brief pause.13–15 Therefore, both static structural and dynamic solution experiments are consistent with our proposal that cycling of translocation and pausing results in dangling of substrates to yield defined substrate cleavage positions and spacing.
Interestingly, structures of ClpA, another hexameric unfoldase, have revealed similar processive translocation steps16 and we observe similar peptide distributions with that enzyme (Supporting Figures 5 and 6). Indeed, prior biochemical studies with ClpAP using low resolution HPLC data forecasted a translocation/proteolysis pausing mechanism4 similar to what we describe here with ClpXP. This is also in line with ClpAP ‘step sizes’ of 1–2 nm seen with single molecule studies using optical traps17 or 5–14 residues seen with single-turnover population measurements.18,19 Collectively, our data suggest that AAA+ proteases capture a series of ATP hydrolysis events collectively across all subunits to facilitate a run of translocation, followed by a pause for degradation. This pause would allow for more complete degradation of substrates to prevent the release of improperly sized products.
Regardless of the specific source of the pausing, our primary result is that we observe enrichment of specific periodically spaced peptides, which supports our hypothesis that ClpXP degradation has a defined start and fixed spacing in cleavage afterwards. Our main point (as illustrated in Figure 3) is that not all highly preferred cleavage sites are used (e.g., region 60–80 in RcdA), but rather there is a defined spacing consistent with processive translocation of substrates into the cleavage chamber. Moreover, residues that are not the most preferred seem to still result in cleavage (e.g., region 90–110 in GFP). The specific pattern of peptides therefore seems to be a combination of inherent primary chemical preference and a restriction on spacing that we propose is driven by translocation. This observation implies that peptides arising from ClpXP processing are not randomly distributed across a given protein and reasons that that specific peptides can be deliberately generated. This leads to a tempting hypothesis that peptides generated during a particular response, such as toxic stress, could be used as signaling molecules for the cell to respond to this damage.
Materials and Methods:
Protein expression/purification
ClpX, ClpP-his, eGFPSsrA, and SspB were purified as before.20,21 ClpX and ClpP are from the C. Crescentus species. HisSUMO tagged RcdA and β2m were appended with a C-terminal ssrA tag (AANDNFAEEFAVAA) using appropriate oligonucleotides, expressed and purified as other similar constructs.20 For β2mssrA, purified protein was also reduced with 2 mM tris(2-carboxyethyl)phosphine (TCEP) and the free cystines were then alkylated with 4 mM iodoacetamide (IAM) unless otherwise specified.
Fluorescence eGFPSsrA experiments
Adaptor, ATP and DFP (di-isopropyl fluorophosphate) concentration-based changes in ClpXP activity were experimentally determined by monitoring the fluorescence loss of model substrate eGFPSsrA over time in 20 μL reactions using a Spectramax fluorescence plate reader. All reactions used 1 μM ClpX6, 2 μM ClpP14 and 10 μM of substrate in 20 mM MOPS pH 7.5, 100 mM KCl, 10 mM MgCl2, 10 % glycerol and an ATP regeneration mixture (4 mM ATP, 16 mM creatine phosphate, 0.75 μg/ml creatine kinase) unless otherwise described. ATP limiting conditions: ATP provided in the regeneration mixture was titrated as shown in Figure 2A. Adaptor based reactions: Initially a titration of adaptor SspB was performed to determine maximal adaptor activity, adaptor:unfoldase molar ratio. No significant inhibition was observed at the highest concentrations used here. This molar ratio was preserved for all other adaptor-substrate experiments. DFP analysis: 0–100 fold molar excess of DFP was preincubated with ClpP alone in H-buffer for 1 h at 30 °C prior to substrate digestion.
Degradation Preparation and Peptide Recovery
Degradation of substrates for peptide recovery took place in a 50 μL reaction volume with 20 mM MOPS buffer, 100 mM KCl, 10 mM MgCl2, and 5 mM ATP. Concentrations of substrate, ClpX, and ClpP were 10 μM, 1 μM, and 2 μM, respectively. These ratios were used to guarantee a proper ClpX to ClpP ratio, as well as to ensure an excess of substrate to facilitate degradation and thus peptide production. ATP was the last constituent added, and once added, the reaction was incubated at 30 °C for 1 h. SDS-PAGE separation and Coomassie staining was used to validate substrate depletion. After 1 h, the remaining proteins were separated from peptides by a Centri-Spin-10 size exclusion column. Columns were preequilibrated in water. Two spin downs were performed, the first to remove substances over 5 kDa, such as intact substrate or ClpXP, while retaining peptide fragments. The remaining peptides were eluted from the spin column with 50 μL of 50/50 ACN/H2O. The 5 kDa cut off mass was determined to be acceptable as peptides above 30 mers were rarely detected. The peptide mixture was dried down with a speedVac for 1 h at 45 °C to remove the organic solvent and subsequently resuspend in HPLC grade H2O. Peptides produced from di-sulfide intact β2m were reduced with 2 mM TCEP and the free cystines were then alkylated with 4 mM IAM.
Liquid Chromatography and Mass Spectrometry
LC/MS/MS analyses were carried out with a Thermo Scientific EASY-nLC 1200 System (Thermo Fisher Scientific) coupled to a Thermo Fisher Orbitrap Fusion mass spectrometer. A 20 mm by 75 μm Thermo Acclaim Pepmap trap column was used preceding a 150 mm by 75 μm Thermo Acclaim Pepmap RSCL analytical column packed with 2 μm particles. Solvent A and B were HPLC grade water with 0.1% formic acid and HPLC grade ACN with 0.1% formic acid, respectively. Separation was achieved with gradients 0 to 60% B in 60 minutes at 300 nL/min, with a 30-minute hold at 95% B as a post separation wash. A 30-minute blank was run between each injection to ensure no peptide carry over. 2 μL of sample were injected for all methods. The electrospray voltage was set to 1900–2100 V with the ion transfer tube temperature at 300 °C. Full mass range scans were performed with a m/z range of 200 to 5000 with a resolution of 60,000. Collision-induced dissociation was performed on ions with an intensity of 5000 counts or higher with a collision energy of 35 eV and a 10 ms activation time.
Data Analysis
Peptide detection data processing was performed with Proteome Discoverer. Because nonspecific cleavage events were included in the search, a larger false positive discovery rate is expected. Consequently, only peptides detected with high confidence under strict validation criteria were considered for subsequent analysis. Peptides were analyzed for cleavage location, structural cleavage preference, and length. All structural results are normalized to the occurrence of that structure in the protein.
Supplementary Material
Acknowledgements
This work was supported by grants from the National Institutes of Health (NIH CBI training grant T32GM008515 to RHV and CT, NIGMS R35GM130320 to PC, and R01 GM075092 to RWV). The data were acquired on a Thermo Fisher Orbitrap Fusion Tribrid mass spectrometer funded by NIH grant 1S10OD010645- 01A1. The authors wish to thank the Institute of Applied Life Sciences Mass spectrometry center and Prof. Stephen J. Eyles for his help with the operation of the mass spectrometer.
Abbreviations
- β2m
β−2-microglobulin
- GFP
green fluorescent protein
- RcdA
Regulator of CtrA Degradation
- MS
Mass spectrometry
- DFP
diisopropyl fluorophosphate
- HPLC
High Performance Liquid Chromatography
- PDB
Protein Data Bank
- AAA+
ATPases Associated with diverse cellular Activities
- TCEP
tris(2-carboxyethyl)phosphine
- IAM
iodoacetamide
- ACN
acetonitrile
Footnotes
Supporting information available.
Histogram plots of peptide length distributions both unweighted and weighted for mass spectral intensity for standard degradation conditions, degradation facilitated by sspB, limited by DFP inhibition, and limited by ATP concentration. Cleavage incidence location plots with secondary structure. Comparison of peptide length distribution and cleavage location of GFP digested with ClpXP and ClpAP.
Accession IDs:
Caulobacter crescentus ClpX YP_002517412.1
Caulobacter crescentus ClpP YP_002517414.1
Caulobacter crescentus RcdA YP_002518777.1
Homo sapiens β2Microglobulin XP_005254606.1
References
- (1).Mahmoud SA; Chien P Regulated Proteolysis in Bacteria. Annu. Rev. Biochem 2018, 87, 677–696. 10.1146/annurev-biochem-062917-012848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Sauer RT; Baker TA AAA + Proteases : ATP-Fueled Machines of Protein Destruction. Annu. Rev. Biochem 2011, 80, 587–612. 10.1146/annurev-biochem-060408-172623. [DOI] [PubMed] [Google Scholar]
- (3).Lee C; Schwartz MP; Prakash S; Iwakura M; Matouschek A ATP-Dependent Proteases Degrade Their Substrates by Processively Unraveling Them from the Degradation Signal. Mol. Cell 2001, 7 (3), 627–637. 10.1016/S1097-2765(01)00209-X. [DOI] [PubMed] [Google Scholar]
- (4).Choi K; Licht S Control of Peptide Product Sizes by the Energy-Dependent Protease ClpAP †. Biochemistry 2005, 44 (42), 13921–13931. 10.1021/bi0505060. [DOI] [PubMed] [Google Scholar]
- (5).Gersch M; Stahl M; Poreba M; Dahmen M; Dziedzic A; Drag M; Sieber SA Barrel-Shaped ClpP Proteases Display Attenuated Cleavage Specificities. ACS Chem. Biol 2016, 11 (2), 389–399. 10.1021/acschembio.5b00757. [DOI] [PubMed] [Google Scholar]
- (6).Sprangers R; Gribun A; Hwang PM; Houry WA; Kay LE Quantitative NMR Spectroscopy of Supramolecular Complexes: Dynamic Side Pores in ClpP Are Important for Product Release. Proc. Natl. Acad. Sci. U. S. A 2005, 102 (46), 16678–16683. 10.1073/pnas.0507370102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Ripstein ZA; Vahidi S; Houry WA; Rubinstein JL; Kay LE A Processive Rotary Mechanism Couples Substrate Unfolding and Proteolysis in the ClpXP Degradation Machinery. Elife 2020, 9, 1–25. 10.7554/eLife.52158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Haynes CM; Yang Y; Blais SP; Neubert TA; Ron D The Matrix Peptide Exporter HAF-1 Signals a Mitochondrial Unfolded Protein Response by Activating the Transcription Factor ZC376.7 in C. Elegans. Mol. Cell 2011, 37 (4), 529–540. 10.1016/j.molcel.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Jennings LD; Lun DS; Médard M; Licht S ClpP Hydrolyzes a Protein Substrate Processively in the Absence of the ClpA ATPase: Mechanistic Studies of ATP-Independent Proteolysis. Biochemistry 2008, 47 (44), 11536–11546. 10.1021/bi801101p. [DOI] [PubMed] [Google Scholar]
- (10).Thompson MW; Singh SK; Maurizi MR Processive Degradation of Proteins by the ATP-Dependent Clp Protease from Escherichia Coli. Requirement for the Multiple Array of Active Sites in ClpP but Not ATP Hydrolysis. J. Biol. Chem 1994, 269 (27), 18209–18215. [PubMed] [Google Scholar]
- (11).Bolon DN; Grant RA; Baker TA; Sauer RT Nucleotide-Dependent Substrate Handoff from the SspB Adaptor to the AAA ClpXP Protease. Mol. Cell 2004, 16, 343–350. [DOI] [PubMed] [Google Scholar]
- (12).Fei X; Bell TA; Jenni S; Stinson BM; Baker TA; Harrison SC; Sauer RT Structures of the ATP-Fueled ClpXP Proteolytic Machine Bound to Protein Substrate. Elife 2020, 9, 1–22. 10.7554/eLife.52774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Maillard RA; Chistol G; Sen M; Righini M; Tan J; Kaiser CM; Hodges C; Martin A; Bustamante C ClpX(P) Generates Mechanical Force to Unfold and Translocate Its Protein Substrates. Cell 2011, 145 (3), 459–469. 10.1016/j.cell.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Aubin-Tam M-E; Olivares AO; Sauer RT; Baker TA; Lang MJ Single-Molecule Protein Unfolding and Translocation by an ATP- Fueled Proteolytic Machine. Cell 2011, 145 (2), 257–267. 10.1038/jid.2014.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Rodriguez-Aliaga P; Ramirez L; Kim F; Bustamante C; Martin A Substrate-Translocating Loops Regulate the Mechanochemical Coupling and Power Production in a AAA+ Protease. Nat Struct Mol Biol 2016, 23 (11), 974–981. 10.1038/nsmb.3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Lopez KE; Rizo AN; Tse E; Lin JB; Scull NW; Thwin AC; Lucius AL; Shorter J; Southworth DR Conformational Plasticity of the ClpAP AAA+ Protease Couples Protein Unfolding and Proteolysis. Nat. Struct. Mol. Biol 2020, 27 (5), 406–416. 10.1038/s41594-020-0409-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Olivares AO; Nager AR; Iosefson O; Sauer RT; Baker TA Mechanochemical Basis of Protein Degradation by a Double-Ring AAA+ Machine Adrian. Nat Struct Mol Biol 2014, 21 (10), 871–875. 10.1038/nsmb.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Miller JM; Lin J; Li T; Lucius ALE Coli ClpA Catalyzed Polypeptide Translocation Is Allosterically Controlled by the Protease ClpP Justin. J. Mol. Biol 2013, 425 (15), 2795–2812. 10.1038/jid.2014.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Rajendar B; Lucius AL Molecular Mechanism of Polypeptide Translocation Catalyzed by the Escherichia Coli ClpA Protein Translocase. J. Mol. Biol 2010, 399 (5), 665–679. 10.1016/j.jmb.2010.03.061. [DOI] [PubMed] [Google Scholar]
- (20).Bhat NH; Vass RH; Stoddard PR; Shin DK; Chien P Identification of ClpP Substrates in Caulobacter Crescentus Reveals a Role for Regulated Proteolysis in Bacterial Development. Mol. Microbiol 2013, 88 (6), 1083–1092. 10.1111/mmi.12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Chien P; Perchuk BS; Laub MT; Sauer RT; Baker TA Direct and Adaptor-Mediated Substrate Recognition by an Essential AAA+ Protease. Proc. Natl. Acad. Sci. U. S. A 2007, 104 (16), 6590–6595. 10.1073/pnas.0701776104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


