Abstract
Background
Bladder urothelial carcinoma (BLCA) is still one of the most malignant diseases and has a dismal outcome. Angiogenesis has confirmed its critical role in the development of malignant neoplasms. In this study, we uncovered the prognostic implications of the angiogenesis-related gene panel in urothelial tumors.
Methods
The RNA-seq data and clinical records of 402 patients with BLCA were collected from the TCGA database. The panel, including 145 genes involved in angiogenesis, was retrieved from the Uniprot database and the published work. The patients with similar expressed profiles were clustered, and the differences in gene expression were compared. The correlation of gene expression and BLCA outcomes or clinical features were analyzed.
Results
There were two clusters of BLCA patients identified on the expressed basis of angiogenesis-related genes. A significant difference was detected in the tumor stages between the two clusters (P<0.001) and a striking advantaged prognosis shown in cluster_1 (86.83 vs. 27.06 months, P=0.001). According to statistics, 115 genes showed a discrepancy in expression between the two clusters, and 16 genes positively correlated to tumor stage progression. Separately analyzed the correlation of those stage-related genes and overall survivals (OS) revealed that high expression of 8 genes, including ECM1 (HR =1.72, P<0.001), FN1 (HR =1.564, P=0.004), FGF1 (HR =1.519, P=0.005), FAP (HR =1.449, P=0.020), JAM3 (HR =1.396, P=0.026), THBS1 (HR =1.402, P=0.028), MFGE8 (HR =1.394, P=0.028) and COL8A2 (HR =1.388, P=0.035), were showed worse prognosis of BLCA, respectively.
Conclusions
This study showed an integrated profile of angiogenesis-related genes and identified the different BLCA subgroups with favorable prognosis and poor prognosis depended on the expression pattern of angiogenesis-related genes. Furthermore, this work revealed the single gene expressions of ECM1, FN1, FGF1, FAP, JAM3, THBS1, MFGE8 and COL8A2 involved in angiogenesis associated the prognosis remarkably.
Keywords: Bladder urothelial carcinoma (BLCA), angiogenesis, gene expression profile, prognosis
Introduction
Bladder cancer has been identified as the most common genitourinary malignancy with high morbidity and mortality worldwide (1). Radical cystectomy with bilateral pelvic lymph node dissection is the gold-standard treatment for early bladder cancer (2). The cisplatin-based chemotherapy established from the 1970s as well as combined treatment of surgery, chemotherapy, and radiotherapy notably prolonged the bladder cancer survival in decades (3,4). However, the overall 5-year survival rate of bladder cancer remains dismal and less than 20% (5). To understand the molecular mechanisms of bladder carcinoma, previous research had identified multiple gene variations in TP53, MLL2, KRT14, KRT5, KRT6A, PPARG, E2F3, EGFR, FGFR3 and so on (6).Few molecularly targeted anticancer agents have been available to treat complex diseases (2). The major pathological types of bladder cancers are bladder urothelial carcinoma (BLAC), of which muscle-invasive type shows an aggressive manifestation and dismal prognosis compared to non-muscle invasive type (3). In muscle-invasive bladder cancer, it has been reported that KLK6, TNS1, TRIM56, IL1B, S100A8, S100A9 and EGFR were capable to predict the progression (7,8). BLCA is a highly heterogeneous disease caused by a genome harboring various genetic variations involving in somatic mutations and substantial chromosomal abnormalities or discrepancies in the functional gene expression (9,10). Hence, identification of effective subtypes with molecular features and developing available target drugs in BLCA is still a challenge.
Angiogenesis has known as clinicopathological factors as well as characteristics of biological aggressiveness in bladder cancer (11). It has well known that angiogenesis promoted the tumor extracellular milieu according to provided oxygen, nutrients and growth factors to the carcinoma cells (12). Molecules involving angiogenesis are commonly detected and the gene expression of VEGF, bFGF and TSP-1 and MVDCD31 were correlated to urothelial cancer prognosis (10). Previous work showed that the angiogenesis related genes expression tremendously varied in BLCA (13).In an earlier study, thrombospondin 1 and primary fibroblast growth factors were reported as an independent prognostic factor of BLCA (10). Multiple agents targeting the VEGF (vascular endothelial growth factor) pathway have been studied in several clinical trials in advanced urothelial cancer to suppress aggressiveness (14-16). The earliest evidence to indicate angiogenesis related genes as promising therapeutic targets approved from a randomized phase 2 trial revealing combination of ramucirumab and docetaxel could prolong PFS in patients with advanced disease (17). However, the clinical benefit has been limited to only some patients (14). Attributed to a complex biological process and multimolecular involved hallmark, the comprehensive observation of angiogenesis-related genes changing is crucial for BLCA management.
There have been many studies on how angiogenesis-related markers expression associated with prognosis in BLCA patients (10,11,18,19). A previous article studied the expression level of basic fibroblast growth factor (bFGF), vascular endothelial growth factor (VEGF) and interleukin-8 (IL-8), and indicated that bFGF and IL-8 expression is crucial in the regulation of angiogenesis at the early stage of tumor growth, progressing and subsequent metastases of bladder cancer (20). However, global analysis of angiogenesis-related gene profiles has not yet been performed for BLCA. Tumor angiogenesis plays a vital role in the multiple biological processes including formation of new blood vessel in the primary and distant metastatic tumor, promoting tumor growth according to supplying sufficient energy sources to cancer cells and facilitating tumor cells metastasize to distant sites (21,22). To systematically target tumor angiogenesis, genes coding particular proteins involved in angiogenesis, the sprouting or splitting of capillaries from pre-existing vasculature, should be taken into consideration (23). Keywords of angiogenesis on the Uniprot database provided a superior gene set to search the completed genes related to angiogenesis (24-26). Therefore, we focused on the expression profile of angiogenesis that involves genes in the RNA-seq data of BLCAs, of which hubs are potentially explored to develop agents targeting tumor angiogenesis as an efficient approach for BLCA therapy in combination with the conventional treatment .
In this study, transcriptome sequencing data in 402 BLCAs were assessed, and the association of expression of angiogenesis involved genes and clinical outcomes was investigated. We uncovered an expression profile of angiogenesis involving genes and the correlation of the gene set and the individuals to clinical outcomes in BLCA.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tau-20-1291).
Methods
Samples
The RNA sequencing data and the corresponding clinical record of 402 patients with BLCA from cbioportal were downloaded (cbioportal.org, a TCGA data platform) (27,28). We filter the data with whether the mRNA expression values histological diagnosis results and survival data are comprehensive. All those patients were pathologically diagnosed as muscle-invasive urothelial carcinoma. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Gene set construction
Angiogenesis related genes in the assessed panel were collected and arranged from the previous articles and Uniport-keyword database (KW0037, uniport.org/keywords), which has published work mentioned (24,29). One hundred forty-six genes were integrated into a gene set, and the corresponding expression levels were shown as mRNA z-score data. Then the genes were listed; among those data, transcriptome information of gene APELA was missing in the studied population. One hundred forty-five genes involved in angiogenesis were finally included. The interrelated gene expression and the single gene expression of distinct groups were compared to detect the correlation for clinical features of BLCA with TCGA data.
Bioinformatics
A cluster of the 145 genes expressional value in patients with muscle-invasive type BLCA was assessed to distinguish samples with gene expression patterns. The subjects with different gene expression patterns were identified from the entire data set. The transcriptional levels of contained genes were shown as mRNA z-scores and grouped using the hierarchical clustering algorithm by the Gene Cluster 3.0 program (30). The heat maps and cluster overview according to the genes variations were generated with the Java Treeview program (31).
Prognostic implication analysis
To assess the prognostic role of the cancer angiogenesis-related genes, we used GraphPad Prism version 7.0 (GraphPad Software, Inc., CA, USA) to compare the survival between the different clusters. The correlation of gene expression and outcomes were investigated to detect the difference in median survival between the subgroups with low and high gene expressional levels.
Statistical analysis
Survival curves were plotted according to the Kaplan-Meier method and compared using the log-rank test in GraphPad Prism version 7.0. The correlations of clinical characteristics and the variables to determine the clusters of patients were evaluated using Fisher’s exact test and the Pearson correlation analysis. The different gene expression between clusters were assessed using ANOVA. The regression analysis determines correlations between variables. All tests were performed with SPSS version 24.0 (IBM, Inc., New York, USA). A P value less than 0.05 was considered statistically significant.
Results
The expression profile of genes involved in angiogenesis dramatically heterogeneous in BLCA
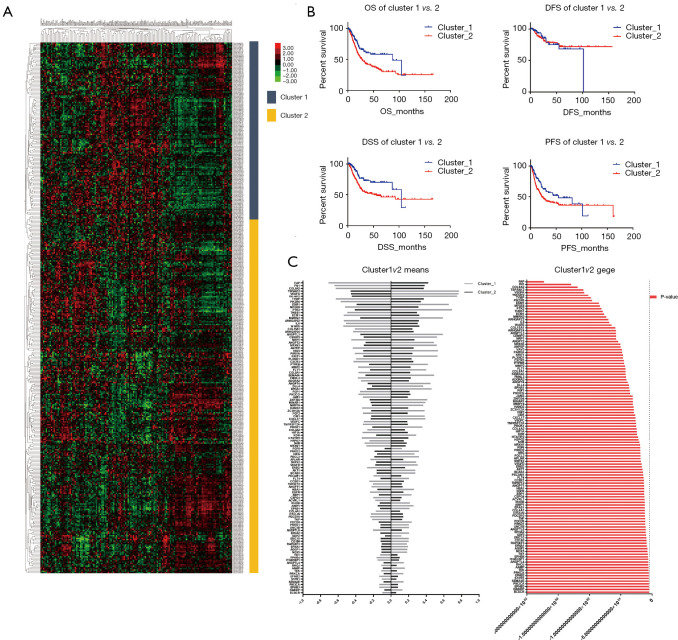
To investigate angiogenesis-related gene variation in BLCA, we grouped the patients with similar transcriptional angiogenesis genes. One hundred forty-five protein-coding genes involved in the sprouting or splitting of capillaries from pre-existing vasculature were included in the array in total online: https://cdn.amegroups.cn/static/application/791eea1171cf823cc20cd235ca04dc77/tau-20-1291-1.pdf. Differences in the gene expression sorted the patients with muscle invasion type BLCA according to the mRNA expression level. According to filtration, 402 patients with survival records were recruited. There were 150 patients in the first group defined as cluster_1 and the 252 patients in another group defined as cluster_2 primary divided from the entire population (Figure 1A). A comparison of the clinical characters regarding the two clusters revealed there was no difference in terms of age and sex (P>0.05, Table 1). However, a considerable discrepancy was shown in the AJCC stage (P<0.001). After analyzing the detailed pathological stage, we detected that cluster_1 tended to be with an earlier stage in M stage (P<0.001), N stage (P<0.05), and T stage (P<0.001) compared to cluster_2 (Table 1).
Figure 1.
There were two clusters identified in the bladder urothelial carcinoma (BLCA) according to angiogenesis related gene profile. (A) the entire cohort divided into cluster_1 and the cluster_2 basing on genes expression; (B) the patients in cluster_1 showed a favorable prognosis in the terms of overall survival (OS), progression-free survival (PFS), disease-free survival (DFS) and disease-specific survival (DSS); (C) angiogenesis related genes were differently expressed between the two clusters.
Table 1. The clinical features of each cluster.
| Clinical features | Cluster_1 (n=150) | Cluster_2 (n=251) | P value |
|---|---|---|---|
| Age | 67.16667 | 68.72112 | 0.208864 |
| Sex | 0.241233 | ||
| Male | 116 | 180 | |
| Female | 34 | 71 | |
| AJCC stage | <0.001*** | ||
| Stage I | 2 | 0 | |
| Stage II | 72 | 56 | |
| Stage III | 39 | 99 | |
| Stage IV | 35 | 96 | |
| NA& | 2 | 0 | |
| Path M stage | <0.001*** | ||
| M0 | 93 | 98 | |
| M1 | 2 | 9 | |
| MX | 54 | 143 | |
| NA& | 1 | 1 | |
| Path N stage | 0.045* | ||
| N0 | 97 | 136 | |
| N1 | 8 | 37 | |
| N2 | 24 | 50 | |
| N3 | 2 | 5 | |
| N4 | 16 | 20 | |
| NA& | 3 | 3 | |
| Path T stage | <0.001*** | ||
| T0 | 1 | 0 | |
| T1 | 2 | 1 | |
| T2 | 63 | 54 | |
| T3 | 51 | 140 | |
| T4 | 16 | 41 | |
| TX | 1 | 0 | |
| NA& | 16 | 15 |
&, represented data deficient; *, P<0.05; ***, P<0.001.
Angiogenesis related gene variation associated with prognosis in BLCA
To compare the survival between the two clusters, we assess overall survival (OS), progression-free survival (PFS), disease-free survival (DFS) and disease-specific survival (DSS) of each cluster (Table 2). The statistic showed cluster_1 was with a prolonged prognosis than the patients in cluster_2 regarding OS (86.83 vs. 27.06 months, P=0.001), DSS (104.6 vs. 51.16 months, P=0.001) and PFS (54.74 vs. 21.86 months, P=0.006) (Table 2, Figure 1B). Compared to cluster_2, patients with the expression pattern as cluster_1 tend to have a significantly better outcome (HR =0.5756, 0.5007, and 0.6304 in the OS, DSS, and PFS, respectively). We compared the expression of angiogenesis-related genes between the two clusters and detected the notable difference in the expression levels (P<0.05) of 115 genes (Figure 1C). Among those distinguishing genes, FAP, FN1, and COL8A2 showed the most significant differences (Table S1).
Table 2. The comparison of survival corresponds to each cluster.
| Prognosis | Cluster_1 | Cluster_2 | P value | Hazard ratio | 95% CI of the ratio |
|---|---|---|---|---|---|
| OS | 86.83 | 27.06 | 0.001** | 0.5756 | 0.4241 to 0.7812 |
| DFS | 101.5 | Undefined | 0.978 | 0.9899 | 0.4853 to 2.019 |
| DSS | 104.6 | 51.16 | 0.001** | 0.5007 | 0.3458 to 0.7249 |
| PFS | 54.74 | 21.86 | 0.006** | 0.6304 | 0.4636 to 0.8573 |
**, P<0.01.
Table S1. Differently expressed genes between the two clusters.
| Genes | P values | Mean expression value in Cluster_1 | Mean expression value in Cluster_2 |
|---|---|---|---|
| FAP | 1.22E-52 | −0.71219 | 0.42392 |
| FN1 | 1.47E-39 | −0.64355 | 0.383066 |
| COL8A2 | 1.86E-36 | −0.62956 | 0.374739 |
| TSPAN12 | 1.09E-33 | 0.774641 | −0.4611 |
| VEGFA | 2.49E-33 | 0.767452 | −0.45682 |
| TNFAIP2 | 2.64E-31 | 0.751268 | −0.44718 |
| TYMP | 4.53E-30 | −0.62808 | 0.373854 |
| PIK3R6 | 1.61E-29 | −0.57319 | 0.341186 |
| ESM1 | 3.09E-26 | 0.658365 | −0.39188 |
| MFGE8 | 1.02E-25 | −0.52427 | 0.312067 |
| PTK2 | 5.48E-25 | 0.69457 | −0.41343 |
| THBS1 | 2.35E-24 | −0.49913 | 0.297104 |
| ECM1 | 1.07E-23 | −0.53988 | 0.321357 |
| MMRN2 | 1.21E-22 | 0.590577 | −0.35153 |
| ARHGAP22 | 1.07E-21 | −0.53779 | 0.320115 |
| IL6 | 2.72E-21 | −0.51258 | 0.305104 |
| WARS | 3.85E-20 | −0.54455 | 0.324137 |
| COL15A1 | 2.52E-18 | −0.41217 | 0.245338 |
| ARHGAP24 | 4.2E-18 | −0.43441 | 0.258575 |
| ANGPTL3 | 4.82E-18 | 0.590072 | −0.35123 |
| CEMIP2 | 5.83E-18 | 0.524714 | −0.31233 |
| NRP1 | 6.06E-18 | −0.45278 | 0.269511 |
| ANGPT2 | 5.49E-17 | 0.489635 | −0.29145 |
| NRXN3 | 7.54E-16 | 0.52137 | −0.31034 |
| ANPEP | 8.2E-16 | −0.43563 | 0.259303 |
| VAV3 | 3.55E-15 | 0.534683 | −0.31826 |
| PARVA | 4.8E-15 | −0.41303 | 0.245851 |
| KRIT1 | 1.14E-14 | 0.530007 | −0.31548 |
| PLXND1 | 1.27E-14 | −0.39681 | 0.236198 |
| CXCR3 | 3.75E-14 | −0.42852 | 0.255069 |
| PTPRB | 4.31E-14 | 0.420305 | −0.25018 |
| MMP2 | 1.21E-13 | −0.37564 | 0.223598 |
| FLT1 | 1.81E-13 | 0.401674 | −0.23909 |
| COL8A1 | 2.02E-13 | −0.37136 | 0.22105 |
| SEMA4A | 2.2E-13 | 0.491908 | −0.2928 |
| FMNL3 | 3.48E-13 | −0.37989 | 0.226123 |
| ADGRA2 | 5.61E-13 | −0.34422 | 0.204892 |
| ANGPT4 | 2.62E-12 | 0.448416 | −0.26691 |
| DLL4 | 9.71E-12 | 0.366835 | −0.21835 |
| EPHA1 | 1.14E-11 | 0.463117 | −0.27567 |
| FGF1 | 2.43E-11 | −0.35539 | 0.211541 |
| PIK3CG | 3.08E-11 | −0.37258 | 0.221775 |
| JAM3 | 5.28E-10 | −0.32311 | 0.192326 |
| ZNF304 | 6.59E-10 | 0.413028 | −0.24585 |
| MINAR1 | 7.22E-10 | 0.392291 | −0.23351 |
| MMP19 | 7.84E-10 | 0.367841 | −0.21895 |
| HAND2 | 1.74E-09 | −0.33064 | 0.196807 |
| ZC3H12A | 1.99E-09 | 0.38305 | −0.22801 |
| FGF2 | 2.19E-09 | −0.32073 | 0.19091 |
| CIB1 | 9.77E-09 | 0.384205 | −0.22869 |
| CXCL17 | 1.22E-08 | 0.391253 | −0.23289 |
| VEGFC | 1.58E-08 | −0.31405 | 0.186933 |
| TNFRSF12A | 2.34E-08 | −0.32205 | 0.191699 |
| PROK1 | 2.72E-08 | 0.359058 | −0.21372 |
| COL4A2 | 4.02E-08 | −0.26925 | 0.160269 |
| HIF1A | 5.42E-08 | −0.32646 | 0.194324 |
| KDR | 6.94E-08 | 0.286836 | −0.17074 |
| HTATIP2 | 1.95E-07 | −0.33507 | 0.199448 |
| HSPG2 | 4.22E-07 | −0.24745 | 0.147291 |
| SHB | 9.83E-07 | −0.30112 | 0.179239 |
| NRXN1 | 1.98E-06 | −0.28212 | 0.167926 |
| FGF6 | 2.21E-06 | 0.126084 | −0.07505 |
| PRKD2 | 2.37E-06 | 0.31899 | −0.18987 |
| HRG | 2.76E-06 | 0.268654 | −0.15991 |
| PDCL3 | 3.42E-06 | 0.319955 | −0.19045 |
| APLNR | 3.52E-06 | −0.23848 | 0.141951 |
| SRPX2 | 4.98E-06 | 0.287026 | −0.17085 |
| VEGFD | 9.95E-06 | 0.305397 | −0.18178 |
| EREG | 1.37E-05 | −0.2746 | 0.163454 |
| AKT1 | 3.49E-05 | 0.278346 | −0.16568 |
| BCAS3 | 5.34E-05 | 0.27783 | −0.16538 |
| PECAM1 | 6.94E-05 | −0.1998 | 0.11893 |
| FLT4 | 0.000115 | 0.204357 | −0.12164 |
| CCBE1 | 0.000138 | −0.249 | 0.148212 |
| TNFSF12 | 0.000141 | −0.20855 | 0.124138 |
| ANGPT1 | 0.000177 | −0.22422 | 0.133465 |
| HBA1 | 0.000193 | 0.262918 | −0.1565 |
| EGFR | 0.000198 | −0.24009 | 0.142914 |
| XBP1 | 0.000208 | 0.235631 | −0.14026 |
| CSPG4 | 0.000265 | −0.20411 | 0.121496 |
| ACVRL1 | 0.000321 | −0.18018 | 0.107249 |
| RHOB | 0.000362 | 0.216142 | −0.12866 |
| AIMP1 | 0.000379 | 0.258381 | −0.1538 |
| EPAS1 | 0.000383 | 0.212453 | −0.12646 |
| COL4A1 | 0.000417 | −0.17544 | 0.104431 |
| OTULIN | 0.000423 | −0.23162 | 0.137869 |
| PDCD10 | 0.000471 | −0.23597 | 0.140461 |
| TNF | 0.000591 | −0.21641 | 0.128817 |
| PDCD6 | 0.000597 | 0.23889 | −0.1422 |
| PRKD1 | 0.000702 | 0.204308 | −0.12161 |
| UNC5B | 0.000942 | 0.206711 | −0.12304 |
| ANGPTL6 | 0.001225 | −0.20781 | 0.123699 |
| EPHA2 | 0.001262 | 0.207137 | −0.1233 |
| GDF2 | 0.001483 | 0.119507 | −0.07114 |
| EMC10 | 0.002135 | 0.203182 | −0.12094 |
| CD160 | 0.002441 | 0.201907 | −0.12018 |
| RAPGEF3 | 0.002685 | 0.182248 | −0.10848 |
| EFNA1 | 0.003462 | 0.195042 | −0.1161 |
| AGGF1 | 0.007101 | 0.186722 | −0.11114 |
| NOX5 | 0.007547 | 0.18537 | −0.11034 |
| TIE1 | 0.01076 | −0.12543 | 0.07466 |
| EPHB4 | 0.011818 | −0.16719 | 0.099516 |
| ITGB1BP1 | 0.018531 | −0.15539 | 0.092496 |
| ANGPTL4 | 0.019834 | 0.146825 | −0.0874 |
| RHOJ | 0.02072 | −0.11266 | 0.067058 |
| AAMP | 0.022248 | 0.159327 | −0.09484 |
| TEK | 0.02284 | −0.1134 | 0.067498 |
| PRKCA | 0.029006 | 0.141675 | −0.08433 |
| EFNB2 | 0.029184 | −0.13632 | 0.081143 |
| S1PR1 | 0.031866 | −0.10726 | 0.063843 |
| SEMA3E | 0.033715 | 0.145457 | −0.08658 |
| PIK3CA | 0.036101 | −0.13583 | 0.080854 |
| EPHB3 | 0.038304 | −0.13581 | 0.080837 |
| DAB2IP | 0.047238 | 0.130468 | −0.07766 |
| ECSCR | 0.049793 | −0.09987 | 0.059445 |
Variations in angiogenesis-related genes implicate tumor stage progression in BLCA
In the earlier data, we have detected that the clusters with a different distribution of angiogenesis are shown significantly in the tumor stage. To further investigate the implication of gene expression and tumor stage progression, we conducted regression analysis (Figure 2). There are 16 genes correlated to tumor stages in the BLCA with tremendously statistical significance (Table 3). Gene FN1 was with the strongest correlation to the BLCA tumor stage (r=0.34, P<0.001). Its expression was available to predict stage progression (AUC =0.720, 95% CI: 0.666–0.775, P<0.001) (Figure 3A). Sixteen genes contained a model set up to optimize the predictive performance, and a superior efficacy was shown in the stage’s prediction progression (AUC =0.834, 95% CI: 0.793–0.875, P<0.001) (Figure 3B).
Figure 2.
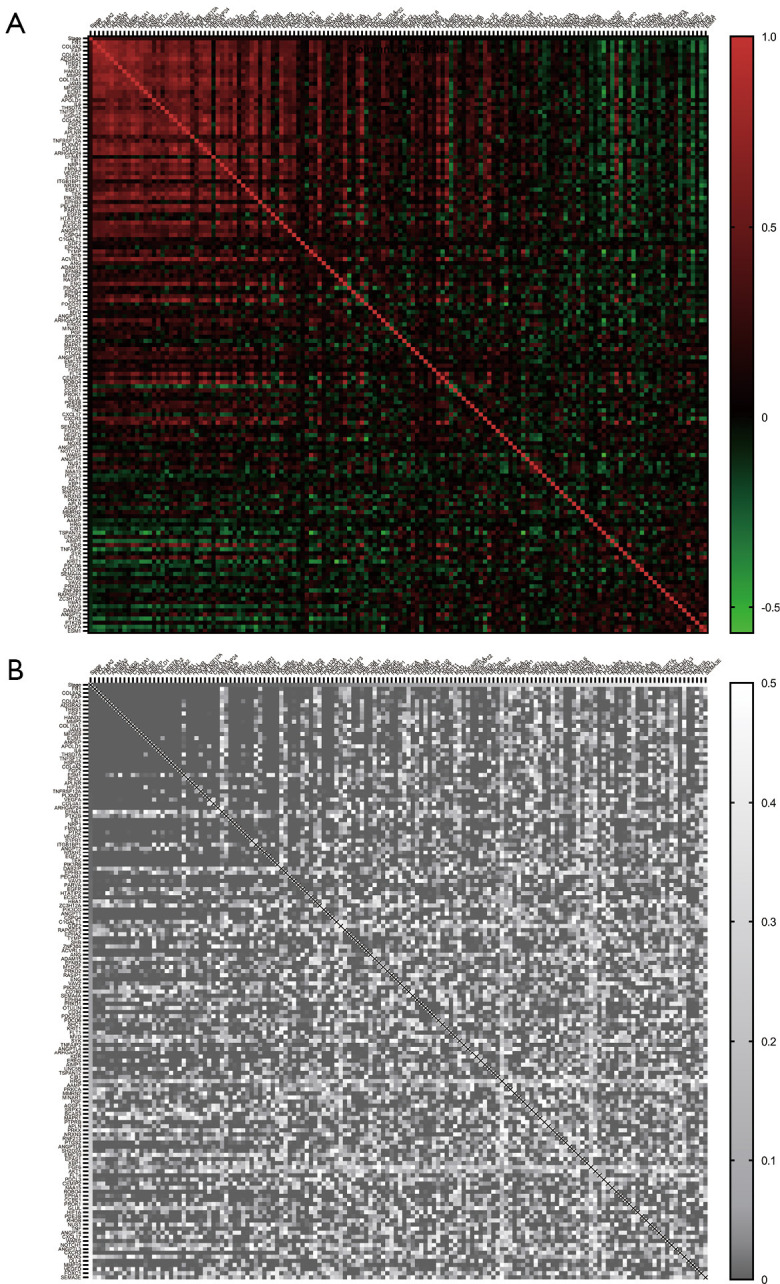
Regression analysis revealed the genes expression correlated with stage progression. (A) the Pearson correlation value (r) of stage and genes; (B) the statistical significance (P value) of corresponding correlation values.
Table 3. The angiogenesis-related gene correlated with tumor stages.
| Genes | Pearson correlation | P value |
|---|---|---|
| FN1 | 0.34 | 1.6E-12 |
| COL8A2 | 0.33 | 7.67E-12 |
| FAP | 0.33 | 7.71E-12 |
| COL8A1 | 0.29 | 9.26E-10 |
| ADGRA2 | 0.29 | 2.35E-09 |
| THBS1 | 0.27 | 2.14E-08 |
| FGF1 | 0.26 | 7.65E-08 |
| HAND2 | 0.26 | 1.11E-07 |
| MMP2 | 0.25 | 2.55E-07 |
| COL15A1 | 0.23 | 1.06E-06 |
| JAM3 | 0.23 | 1.28E-06 |
| MFGE8 | 0.23 | 2.5E-06 |
| ECM1 | 0.22 | 3.28E-06 |
| ANPEP | 0.21 | 7.83E-06 |
| APOLD1 | 0.21 | 8.36E-06 |
| IL6 | 0.21 | 1.02E-05 |
Figure 3.
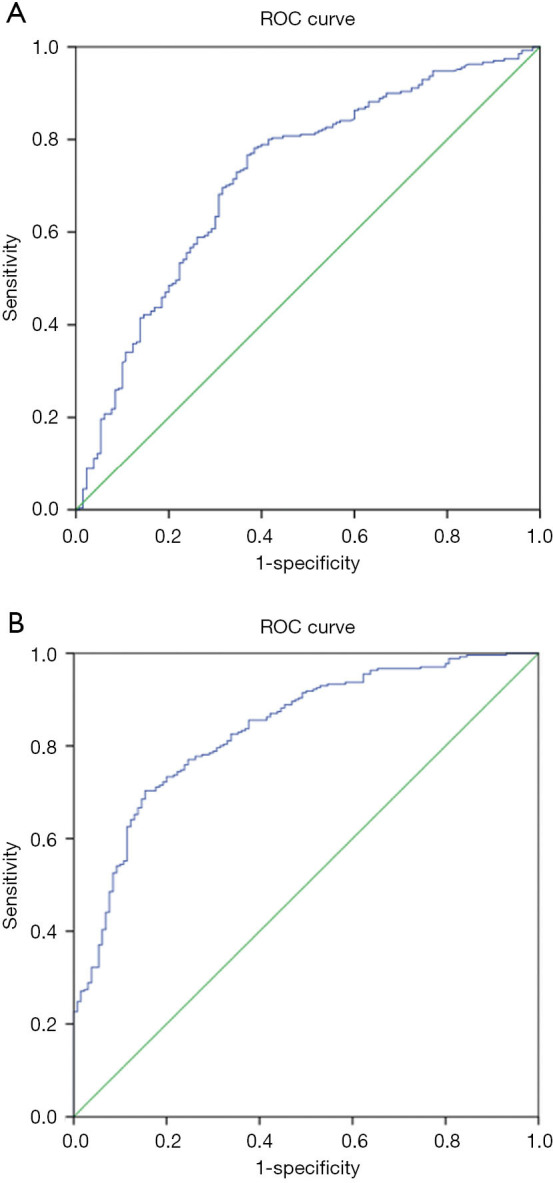
The angiogenesis related gene signatures were capable to predicate the stage in bladder urothelial carcinoma (BLCA). (A) the expression of single gene FN1 was available to predicted the stage progression; (B) the genes set combining 16 genes was with superior efficacy in the prediction of the stage progression.
The prognostic role of the angiogenesis-related gene in BLCA
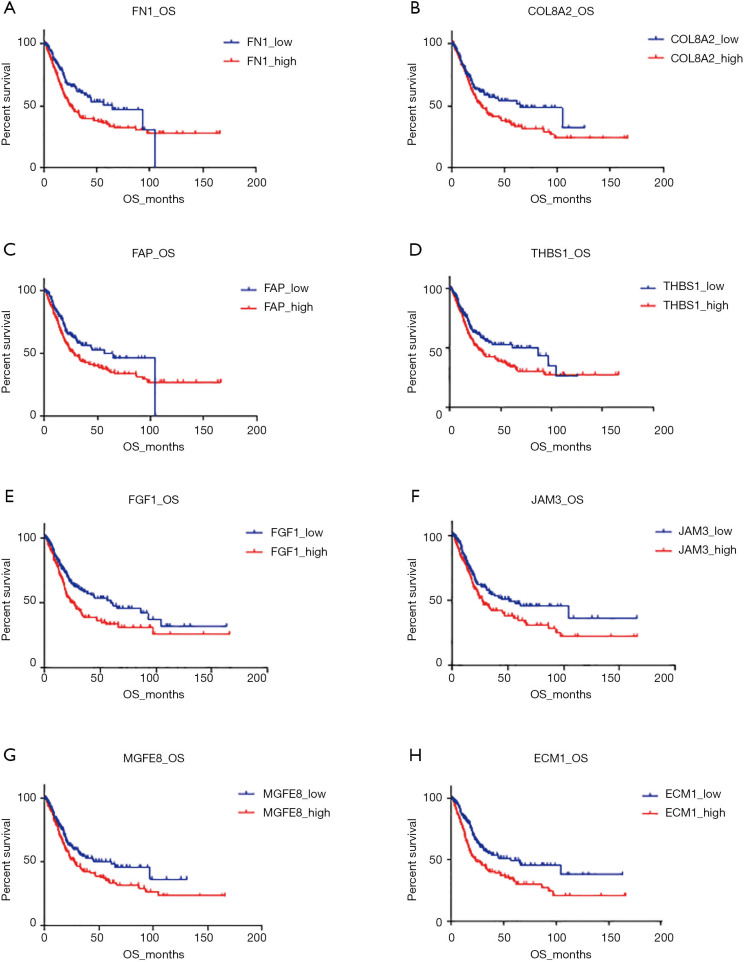
The comparison of the two clusters revealed the expression profile of angiogenesis-related genes was strictly correlated to the prognosis of BLCAs. To investigate the prognostic role of the individual gene, we divided the patients into two subgroups according to the different gene expression: high-expressed group and low-expressed group. Comparison of the OS in the paired groups showed that elevated expression of 8 genes, ECM1, FN1, FGF1, FAP, JAM3, THBS1, MFGE8, and COL8A2, were significantly shortened the OS in BLCA (P<0.05, Figure 4). Among those genes, the expression of ECM1 revealed the most detected negative correlation to OS (56.48 vs. 23.21 months, P<0.001; HR =1.72, 95% CI: 1.275–2.320) (Table 4).
Figure 4.
The elevated expression of 8 genes were significantly associated with worse prognosis in BLCA (bladder urothelial carcinoma). (A,B,C,D,E,F,G,H) The different OS (overall survivals) regarding to high expression population and low expression population of ECM1, FN1, FGF1, FAP, JAM3, THBS1, MFGE8 and COL8A2.
Table 4. The correlation of single gene expression with survival rate.
| Genes | Median survival in low expression (Month) | Median survival in high expression (Month) | P value | Hazard Ratio (log-rank) |
95% CI of the ratio |
|---|---|---|---|---|---|
| ECM1 | 56.48 | 23.21 | <0.001*** | 1.72 | 1.275 to 2.320 |
| FN1 | 64.8 | 26.14 | 0.004** | 1.564 | 1.162 to 2.105 |
| FGF1 | 59.31 | 23.64 | 0.005** | 1.519 | 1.129 to 2.044 |
| FAP | 56.48 | 27.45 | 0.020* | 1.449 | 1.073 to 1.959 |
| JAM3 | 51.16 | 27.06 | 0.026* | 1.396 | 1.037 to 1.879 |
| THBS1 | 86.83 | 27.45 | 0.028* | 1.402 | 1.043 to 1.885 |
| MFGE8 | 61.45 | 27.45 | 0.028* | 1.394 | 1.037 to 1.873 |
| COL8A2 | 64.8 | 28.24 | 0.035* | 1.388 | 1.031 to 1.868 |
| COL8A1 | 61.45 | 28.41 | 0.074 | 1.314 | 0.9739 to 1.773 |
| ADGRA2 | 44.32 | 28.24 | 0.124 | 1.263 | 0.9387 to 1.696 |
| APOLD1 | 41.75 | 28.41 | 0.132 | 1.255 | 0.9332 to 1.687 |
| HAND2 | 61.45 | 28.41 | 0.158 | 1.237 | 0.9204 to 1.662 |
| ANPEP | 41.75 | 33.01 | 0.216 | 1.205 | 0.8941 to 1.624 |
| COL15A1 | 56.48 | 32.02 | 0.323 | 1.162 | 0.8639 to 1.564 |
| MMP2 | 41.75 | 33.01 | 0.432 | 1.127 | 0.837 to 1.518 |
| IL6 | 31.2 | 34.98 | 0.872 | 1.025 | 0.7625 to 1.377 |
*, P<0.05; **, P<0.01; ***, P<0.001.
Discussion
Although multiple agents were developed to target specific proteins, bladder cancer still is an insensitive disease for clinical management. Platinum-based chemotherapy is an acceptable front-line regiment for patients with advanced or metastatic BLCA, while the outcomes are unsatisfactory (4). Prognosis in chemotherapy resistance patients remains poor, with a median overall survival with monotherapy of approximately seven months (32). Compared with chemotherapy in second-line treatment, a superior survival benefit has been showed in immune-checkpoint inhibitors therapy However, the survival benefit from immunotherapy was reported in only 20% of patients (33). Recently, an angiogenesis targeted study revealed ramucirumab combined docetaxel is the first regiment in a phase 3 study to show improved PFS in patients with platinum-insensitive advanced urothelial carcinoma (2). These data prompted that inhibition of angiogenesis-related signaling offered a potentially effective treatment possibility for patients with urothelial carcinoma. Therefore, a new targeted gene involved in the angiogenetic process must be investigated.
In the current study, we found that the profile of angiogenesis-related gene expression was significantly associated with the tumor stage and outcomes. The cluster_1, represented by depression of FAP, FN1, and COL8A2, shows a significant survival advantage. The elevated expression of those three genes is respectively correlated with the advanced tumor stage in the BLCA. The FN1 gene coding a fibronectin 1 protein was previously reported as a survival predictor and associated with the pathologic stage, which detection was inconsistent with our finding (34). Cytoplasmic immunostaining of FAP (fibroblast activation protein-coding gene) in cancer-associated fibroblasts showed worse DSS and were associated with tumor staging, which data supported our results (35). The COL8A2 gene coding a collagen alpha-2 (VIII) chain, a component of the endothelia of blood vessels, was rarely studied in the bladder cancer. However, the gene was consistently increased in the tumor content of prostate cancer (36). Our data suggested that high expression of COL8A2 was not only associated with advanced tumor stage but also showed a worse prognosis in BLCA. Hence, this gene deserves further study in BLCA.
The statistics showed that the different genes associated with the different prognosis were also correlated to the stage progression. This result showed that the prognostic role of angiogenesis-related genes might implicate by the effect to promote tumor progression. Therefore, we discovered the tumor stage-correlated genes and detected their performances to distinguish the early or advanced tumor stage, according to the ROC curve. We found no matter the single gene, FN1, or the combined 16 gene model that would capably show the tumor stage of the BLCA. To assess the prognostic effect of individual genes, we perform multiple survival comparisons. The data revealed that besides FAP, FN1, and COL8A2, the genes ECM1, FGF1, JAM3, THBS3, MFGE8, and COL8A2 also showed significant effects for indicating BLCA outcomes. Of which, the expression of ECM1 (Extracellular matrix protein 1 coding gene) was reported to display a significant increase compared with noncancerous counterparts and associated with a poor prognosis in bladder cancer. Depressing EMC1 affected not only cell proliferation and migration but also cell invasion ability and apoptosis potential (37). Downregulating the ECM1 in BLCA would significantly suppress the expression of GLUT1, LDHA and HIF-1α and reduced the lactate production in ECM1 suppressed cells, indicating that ECM1 potentially regulated the cancerous metabolism in BLCA (37). Consistent with our finding, ECM1 is with the potential to be developed a targeting for inhibiting BLCA.
In conclusion, we found that the expression of angiogenesis-related genes had significant differences in tumor stage, and the profile could predict the prognosis in BLCA. The results supply potential targets for the management of the BLCA.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tau-20-1291
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-20-1291). The authors have no conflicts of interest to declare.
(English Language Editor: J. Chapnick)
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Zhang W, Wang R, Ma W, et al. Systemic immune-inflammation index predicts prognosis of bladder cancer patients after radical cystectomy. Ann Transl Med 2019;7:431. 10.21037/atm.2019.09.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tachibana I, Bandali E, Calaway AC, et al. Urothelial carcinoma in situ response to cisplatin-based neoadjuvant chemotherapy, or lack thereof: Impact on patient selection for organ preservation in muscle-invasive disease? Urol Oncol 2020;38:850.e1-7. 10.1016/j.urolonc.2020.06.014 [DOI] [PubMed] [Google Scholar]
- 4.von der Maase H, Hansen SW, Roberts JT, et al. Gemcitabine and cisplatin versus methotrexate, vinblastine, doxorubicin, and cisplatin in advanced or metastatic bladder cancer: results of a large, randomized, multinational, multicenter, phase III study. J Clin Oncol 2000;18:3068-77. 10.1200/JCO.2000.18.17.3068 [DOI] [PubMed] [Google Scholar]
- 5.Lobo N, Mount C, Omar K, et al. Landmarks in the treatment of muscle-invasive bladder cancer. Nat Rev Urol 2017;14:565-74. 10.1038/nrurol.2017.82 [DOI] [PubMed] [Google Scholar]
- 6.Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014;507:315-22. 10.1038/nature12965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang PB, Huang ZL, Xu YH, et al. Systematic analysis of gene expression profiles reveals prognostic stratification and underlying mechanisms for muscle-invasive bladder cancer. Cancer Cell Int 2019;19:337. 10.1186/s12935-019-1056-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim WJ, Kim SK, Jeong P, et al. A four-gene signature predicts disease progression in muscle invasive bladder cancer. Mol Med 2011;17:478-85. 10.2119/molmed.2010.00274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan Y, Zhang T, Liang C. Circular RNA SMARCA5 is overexpressed and promotes cell proliferation, migration as well as invasion while inhibits cell apoptosis in bladder cancer. Transl Cancer Res 2019;8:1663-71. 10.21037/tcr.2019.08.08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shariat SF, Youssef RF, Gupta A, et al. Association of angiogenesis related markers with bladder cancer outcomes and other molecular markers. J Urol 2010;183:1744-50. 10.1016/j.juro.2010.01.018 [DOI] [PubMed] [Google Scholar]
- 11.Fus Ł P, Górnicka B. Role of angiogenesis in urothelial bladder carcinoma. Cent European J Urol 2016;69:258-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eelen G, Treps L, Li X, et al. Basic and Therapeutic Aspects of Angiogenesis Updated. Circ Res 2020;127:310-29. 10.1161/CIRCRESAHA.120.316851 [DOI] [PubMed] [Google Scholar]
- 13.Zhang PB, Huang ZL, Xu YH, et al. Systematic analysis of gene expression profiles reveals prognostic stratification and underlying mechanisms for muscle-invasive bladder cancer. Cancer Cell International 2019;19:337. 10.1186/s12935-019-1056-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Russi EG, Bensadoun RJ, Merlano MC, et al. Bio-radiation dermatitis: the need of a new grading: in regard to Bernier et al: Ann Oncol 2011;22:2191-200. Ann Oncol 2013;24:2463-5. 10.1093/annonc/mdt281 [DOI] [PubMed] [Google Scholar]
- 15.Choueiri TK, Ross RW, Jacobus S, et al. Double-blind, randomized trial of docetaxel plus vandetanib versus docetaxel plus placebo in platinum-pretreated metastatic urothelial cancer. J Clin Oncol 2012;30:507-12. 10.1200/JCO.2011.37.7002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harshman LC, Xie W, Bjarnason GA, et al. Conditional survival of patients with metastatic renal-cell carcinoma treated with VEGF-targeted therapy: a population-based study. Lancet Oncol 2012;13:927-35. 10.1016/S1470-2045(12)70285-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petrylak DP, Tagawa ST, Kohli M, et al. Docetaxel As Monotherapy or Combined With Ramucirumab or Icrucumab in Second-Line Treatment for Locally Advanced or Metastatic Urothelial Carcinoma: An Open-Label, Three-Arm, Randomized Controlled Phase II Trial. J Clin Oncol 2016;34:1500-9. 10.1200/JCO.2015.65.0218 [DOI] [PubMed] [Google Scholar]
- 18.Rahmani AH, Babiker AY, Alsahli MA, et al. Prognostic Significance of Vascular Endothelial Growth Factor (VEGF) and Her-2 Protein in the Genesis of Cervical Carcinoma. Open Access Maced J Med Sci 2018;6:263-8. 10.3889/oamjms.2018.089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ning X, Deng Y. Identification of key pathways and genes influencing prognosis in bladder urothelial carcinoma. Onco Targets Ther 2017;10:1673-86. 10.2147/OTT.S131386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chikazawa M, Inoue K, Fukata S, et al. Expression of angiogenesis-related genes regulates different steps in the process of tumor growth and metastasis in human urothelial cell carcinoma of the urinary bladder. Pathobiology 2008;75:335-45. 10.1159/000164218 [DOI] [PubMed] [Google Scholar]
- 21.Carmeliet P, Jain RK. Molecular mechanisms and clinical applications of angiogenesis. Nature 2011;473:298-307. 10.1038/nature10144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahmadi M, Rezaie J. Tumor cells derived-exosomes as angiogenenic agents: possible therapeutic implications. J Transl Med 2020;18:249. 10.1186/s12967-020-02426-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Potente M, Gerhardt H, Carmeliet P. Basic and therapeutic aspects of angiogenesis. Cell 2011;146:873-87. 10.1016/j.cell.2011.08.039 [DOI] [PubMed] [Google Scholar]
- 24.Ata SK, Fang Y, Wu M, et al. Disease Gene Classification with Metagraph Representations. Methods Mol Biol 2018;1807:211-24. 10.1007/978-1-4939-8561-6_16 [DOI] [PubMed] [Google Scholar]
- 25.Hinderer EW, 3rd, Moseley HNB. GOcats: A tool for categorizing Gene Ontology into subgraphs of user-defined concepts. PLoS One 2020;15:e0233311. 10.1371/journal.pone.0233311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.The UniProt C. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Research 2018;47:D506-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cerami E, Gao J, Dogrusoz U, et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discovery 2012;2:401. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masson P, Hulo C, de Castro E, et al. An integrated ontology resource to explore and study host-virus relationships. PLoS One 2014;9:e108075. 10.1371/journal.pone.0108075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Hoon MJ, Imoto S, Nolan J, et al. Open source clustering software. Bioinformatics 2004;20:1453-4. 10.1093/bioinformatics/bth078 [DOI] [PubMed] [Google Scholar]
- 31.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics 2004;20:3246-8. 10.1093/bioinformatics/bth349 [DOI] [PubMed] [Google Scholar]
- 32.Raggi D, Miceli R, Sonpavde G, et al. Second-line single-agent versus doublet chemotherapy as salvage therapy for metastatic urothelial cancer: a systematic review and meta-analysis. Ann Oncol 2016;27:49-61. 10.1093/annonc/mdv509 [DOI] [PubMed] [Google Scholar]
- 33.Bellmunt J, Bajorin DF. Pembrolizumab for Advanced Urothelial Carcinoma. N Engl J Med 2017;376:2304. 10.1056/NEJMoa1613683 [DOI] [PubMed] [Google Scholar]
- 34.Riester M, Taylor JM, Feifer A, et al. Combination of a novel gene expression signature with a clinical nomogram improves the prediction of survival in high-risk bladder cancer. Clin Cancer Res 2012;18:1323-33. 10.1158/1078-0432.CCR-11-2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calvete J, Larrinaga G, Errarte P, et al. The coexpression of fibroblast activation protein (FAP) and basal-type markers (CK 5/6 and CD44) predicts prognosis in high-grade invasive urothelial carcinoma of the bladder. Hum Pathol 2019;91:61-8. 10.1016/j.humpath.2019.07.002 [DOI] [PubMed] [Google Scholar]
- 36.Angel PM, Spruill L, Jefferson M, et al. Zonal regulation of collagen-type proteins and posttranslational modifications in prostatic benign and cancer tissues by imaging mass spectrometry. Prostate 2020;80:1071-86. 10.1002/pros.24031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Z, Zhou Q, Li A, et al. Extracellular matrix protein 1 (ECM1) is associated with carcinogenesis potential of human bladder cancer. Onco Targets Ther 2019;12:1423-32. 10.2147/OTT.S191321 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as