Abstract
Background
It is known that gut microbiota can regulate cancer therapies. We hypothesized that gut microbiota may interact with androgen deprivation therapy (ADT) in the process of castration-resistant prostate cancer (CRPC). Here, the differences in gut microbiota between matched hormone-sensitive prostate cancer (HSPC) and CRPC were determined before and after ADT.
Methods
We profiled the fecal microbiota in matched HSPC and CRPC from 21 patients who received ADT at our urological center using 16S rRNA gene amplicon sequencing. Differences in microbiota were determined with α/β-diversity and LefSe analysis. Functional inference of microbiota was performed with PICRUSt.
Results
The results showed that the gut microbial community in CRPC was significantly altered with increased abundance of several bacterial flora including genus Phascolarctobacterium and Ruminococcus. For functional analyses, bacterial gene pathways involved in terpenoids/polyketides metabolism and ether lipid metabolism were significantly activated in CRPC.
Conclusions
Measurable differences in the gut microbiota were identified between HSPC and CRPC. Functional validations are further needed to ascertain the underlying mechanism of these differential microbiota in the process of CRPC, and their potential as new targets to enhance ADT responses.
Keywords: Prostate cancer (PCa), castration-resistant prostate cancer (CRPC), hormone-sensitive prostate cancer (HSPC), hormone therapy, gut microbiota
Introduction
The interaction between host and gastrointestinal microbial communities is involved in many aspects of carcinogenesis (1). Our commensal community can define the efficacy and toxicity of anticancer therapies through a large suite of chemical signaling pathways (2,3). Numerous studies have emerged providing evidence of the relationship between gut microbiota and cancer treatment. One research led by Viaud et al. using a murine sarcoma model proved that germ-free or bacteria-deprived mice showed resistance to cyclophosphamide treatment (4). The authors continuously found a specific set of bacteria e.g., Lactobacillus murinus and Enterococcus hirae that can stimulate type 17 T helper (TH17) cell and type1 T helper (TH1) cell responses, and restore the antitumor efficacy of cyclophosphamide. Bifidobacterium spp. is found to correlate with antitumor T-cell responses, Sivan et al. found with a murine melanoma model that oral administration of Bifidobacterium can strengthen the antitumor efficacy of anti-PD-L1 (5). Gut microbiota has also been demonstrated to associate with therapeutic efficacy of anti-CTLA-4, camptothecin, 5-fluorouracil, and cytosine-phosphate-guanosine oligodeoxynucleotides (CpG-ODN) (6-8). And its anticancer effects have been successfully corroborated with clinical studies. Transfer of chemotherapeutic response has been achieved by administrating human feces into murine, and the benefits of specific microbials in progression-free survival rates have been identified among patient cohorts (9-11).
The relationship between gut microbiota and prostate cancer (PCa) progression is largely unknown. Androgen deprivation therapy (ADT), also referred to hormone therapy is still the first-line therapeutic approach for PCa management, it lowers serum testosterone to castrate level and decreases the prostate-specific antigen (PSA) value to suppress cancer progression. ADT can be achieved by orchidectomy, luteinizing hormone-releasing hormone agonist (LHRHa) monotherapy, or combined LHRHa with anti-androgen agents (12). However, though ADT is initially successful, most patients may eventually experience biochemical relapse and progress to castration-resistant prostate cancer (CRPC). We hypothesize that gut microbiota may interact with ADT in the process of CRPC, particularly orally administered hormone therapies. ADT may promote the changes in microbiota, and microbiota in turn modulates the ADT efficacy. Here, we reported the first study of the compositional differences in gut microbiota that were examined from the same patients before and after ADT with eventual progression to CRPC, aiming to understand the potential role of gut microbiota in PCa progression and management. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tau-20-566).
Methods
Patient selection
All procedures performed in this study were in accordance with the Declaration of Helsinki (as revised in 2013) and approved by the institutional review board of Huashan hospital, Fudan University (reference number 2020-531). Informed consent was obtained from all individual participants included in the study. Clinical database of Department of Urology between Mar 2016 and Nov 2019 were searched with inclusion criteria as follows: (I) patients who were confirmed as PCa by histopathology evaluation; (II) patients who received ADT after initial diagnosis, with or without radical prostatectomy; (III) patients who were clinically diagnosed as CRPC. CRPC is defined as castrate serum testosterone (<50 ng/dL or 1.7 nmol/L) plus one of the following progressions: (I) biochemical progression: three consecutive rises in PSA 1 week apart, resulting in two 50% increases over the nadir, and PSA >2 ng/mL; (II) radiologic progression: the appearance of new lesions, either two or more new bone lesions or a soft tissue lesion (13). Men were excluded if they took antibiotic within 30 days of fecal collection, had a history of gastrointestinal diseases, or received other forms of hormonal or systemic therapy before enrollment. Eventually, 21 patients were enrolled, their clinical characteristics were collected. Paired feces were gathered at initial diagnosis before ADT, and at CRPC after ADT.
Sample collection
Fecal samples were collected by patients using a sterile “feces tube” with screw cap. The tube contains a spoon that is fixed to the cap and projects down the tube. Fresh feces were immediately stored frozen at –20 °C after production and then were transferred to –80 °C until used.
DNA extraction and 16S rRNA gene amplicon sequencing
Fecal DNA was extracted using the DNeasy PowerSoil Kit (QIAGEN, Inc., Netherlands) following the manufacturer’s instructions. Primers (forward: 5'-3'ACTCCTACGGGAGGCAGCA; reverse 5'-3'GGACTACHVGGGTWTCTAAT) with a 7-bp sample-specific barcode were used to amplify the regions V3–V4 of 16S rRNA gene. PCR reactions were run using the following program: 98 °C 10 min, 25 cycles of 98 °C 15 s, 55 °C 30 s, 72 °C 30 s, and 72 °C 5 min. After agarose gel electrophoresis, the PCR amplicons were purified using the Agencourt AMPure Kit (Beckman Coulter, Milan, Italy) and quantified using the PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA, USA). Equimolar amplicon pool was obtained and sequenced using the Illlumina MiSeq platform with MiSeq Reagent Kit v3 (Shanghai Personal Biotechnology Co., Ltd, Shanghai, China), 2×300 bp paired-end sequencing.
Sequence analysis
Sequencing data were processed using QIIME 1.8.0 package (14). Raw sequencing reads with exact matches to the barcodes were identified as valid sequences and assigned to respective samples. Sequences were excluded from analysis if they (I) were <150 nucleotides in length, (II) had average Phred scores of <20, (III) contained ambiguous bases or mononucleotide repeats of >8 bp. The sequences were assembled, and delineation of operational taxonomic units (OTUs) was conducted with UCLUST at a 97% cutoff (15). A representative sequence was selected from each OTU and subjected to BLAST to assign the taxonomic classification using the Greengenes database. Alpha and Beta diversity were calculated to indicate the richness and structure of microbial communities. LEfSe analysis was performed on http://huttenhower.sph.harvard.edu/galaxy to identify differentially abundant taxa with both statistical significance and biological relevance (16). Microbial functions were predicted by PICRUSt (17).
Results
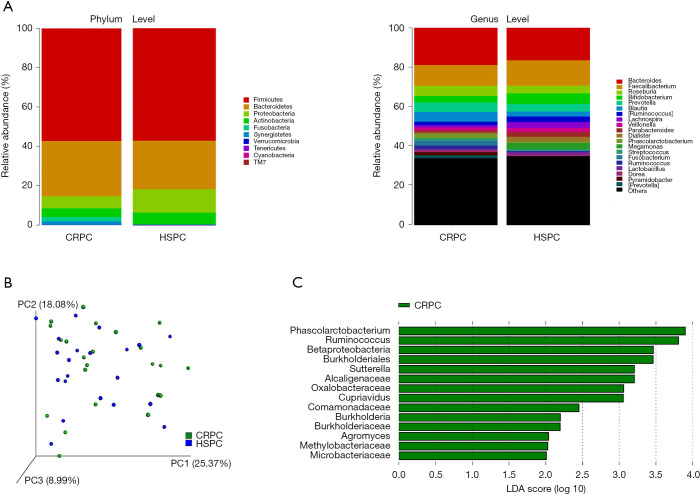
Clinical characteristics of the 21 patients was displayed (Table 1). We first determined the compositional differences in microbiome before [hormone-sensitive prostate cancer (HSPC)] and after ADT (CRPC). The community richness (Chao1) index of CRPC and HSPC were 913.80±359.36 and 842.39±360.29 (P=0.524), respectively. No significant differences were found. The average relative abundance of bacteria at phylum and genus levels were analyzed (Figure 1A). At the phylum level, CRPC had increased abundance of Bacteroidetes (28.04% vs. 24.59%), Fusobacteria (2.17% vs. 0.11%), Synergistetes (1.80% vs. 0.01%), Tenericutes (0.085% vs. 0.010%) and decreased abundance of Proteobacteria (6.12% vs. 11.76%), Actinobacteria (4.37% vs. 6.03%), and Cyanobacteria (0.006% vs. 0.013%). The Beta diversity, performed with principal co-ordinates analysis (PCoA) methods based on weighted UniFrac distance, showed no identifiable microbiota clusters between the two cohorts (Figure 1B). Though samples from the two groups did not cluster by their overall microbial composition, several biologically significant abundance differences were identified with LefSe analysis. As shown in Figure 1C, CRPC had increased abundance of 14 phylotypes, including class Betaproteobacteria, its order Betaproteobacteria, family Alcaligenaceae, Oxalobacteraceae, Comamonadaceae, Burkholderiaceae, genus Burkholderia, Cupriavidus, Sutterella; family Microbacteriaceae and its genus Agromyces; family Methylobacteriaceae; genus Phascolarctobacterium and Ruminococcus. The microbial functional inference analysis was performed with PICRUSt. We identified 11 KEGG pathways that were differentially abundant between the two cohorts. CRPC was enriched of pathways including terpenoids and polyketides metabolism, ether lipid metabolism, clavulanic acid biosynthesis, endocrine and excretory system, etc. (Table 2).
Table 1. Clinical characteristics of patients.
| Case | Age(y) | GS | PSA (ng/mL) | Methods of ADT | Time to CRPC (month) | Metastatic |
|---|---|---|---|---|---|---|
| 1 | 65 | 3+4 | 78.7 | LHRHa + antiandrogen | 36 | N |
| 2 | 71 | 4+5 | 15.8 | Orchiectomy + antiandrogen | 39 | Y |
| 3 | 68 | 4+5 | 113.18 | Orchiectomy + antiandrogen | 13 | Y |
| 4 | 73 | 5+4 | 5.54 | LHRHa + antiandrogen | 33 | N |
| 5 | 68 | 4+5 | 38.2 | LHRHa + antiandrogen | 12 | Y |
| 6 | 74 | 5+4 | 180.0 | LHRHa + antiandrogen | 6 | Y |
| 7 | 64 | 5+5 | 614.1 | Orchiectomy + antiandrogen | 22 | Y |
| 8 | 86 | 4+5 | 18.8 | LHRHa + antiandrogen | 38 | Y |
| 9 | 55 | 4+5 | 613 | LHRHa + antiandrogen | 28 | N |
| 10 | 61 | 4+4 | 85.15 | Orchiectomy + antiandrogen | 11 | N |
| 11 | 60 | 4+4 | 8.87 | LHRHa + antiandrogen | 25 | Y |
| 12 | 72 | 4+5 | 66.33 | LHRHa | 7 | Y |
| 13 | 61 | 4+5 | 100 | LHRHa + antiandrogen | 9 | Y |
| 14 | 55 | 5+4 | 593.5 | LHRHa + antiandrogen | 23 | Y |
| 15 | 67 | 4+5 | 420.6 | LHRHa + antiandrogen | 21 | Y |
| 16 | 70 | 4+5 | 22.79 | LHRHa + antiandrogen | 37 | Y |
| 17 | 65 | 5+4 | 35.7 | LHRHa + antiandrogen | 13 | Y |
| 18 | 63 | 4+5 | 11.20 | LHRHa + antiandrogen | 39 | N |
| 19 | 91 | 3+4 | 117.6 | LHRHa + antiandrogen | 8 | Y |
| 20 | 71 | 4+4 | 276.12 | LHRHa + antiandrogen | 22 | Y |
| 21 | 69 | 5+5 | 398.8 | LHRHa + antiandrogen | 14 | Y |
GS, Gleason score; PSA, prostate-specific antigen; ADT, androgen deprivation therapy; CRPC, castration-resistant prostate cancer; LHRHa, luteinizing hormone-releasing hormone agonist.
Figure 1.
Microbial community composition in CRPC and HSPC. (A) Bacterial average relative abundance between CRPC and HSPC at the phylum and genus level; (B) beta diversity based on weighted uniFrac principal coordinate analysis showed no identifiable microbiota clusters between the two cohorts; (C) LefSe analysis of phylotypes with biologically significant differential abundance in CRPC patients. CRPC, castration-resistant prostate cancer; HSPC, hormone-sensitive prostate cancer.
Table 2. Inference of the functional profile of microbial communities.
| KEGG level 3—differentiated pathways between CRPC and HSPC | Relative abundance | Ratio CRPC/HSPC | P value | |
|---|---|---|---|---|
| CRPC | HSPC | |||
| Metabolism; metabolism of terpenoids and polyketides; biosynthesis of 12-, 14- and 16-membered macrolides | 1.27E–07 | 1.31E–08 | 9.68 | 0.009 |
| Metabolism; metabolism of terpenoids and polyketides; biosynthesis of ansamycins | 1.36E–03 | 1.23E–03 | 1.11 | 0.018 |
| Metabolism; metabolism of terpenoids and polyketides; carotenoid biosynthesis | 2.67E–05 | 1.42E–05 | 1.87 | 0.029 |
| Metabolism; biosynthesis of other secondary metabolites; clavulanic acid biosynthesis | 6.20E–08 | 0E–12 | – | 0.002 |
| Metabolism; lipid metabolism; ether lipid metabolism | 2.42E–04 | 1.27E–04 | 1.90 | 0.022 |
| Human diseases; cardiovascular diseases; HCM | 1.23E–07 | 2.29E–09 | 53.7 | 0.027 |
| Organismal systems; endocrine system; renin-angiotensin system | 4.28E–07 | 2.67E–08 | 16.0 | 0.012 |
| Organismal systems; excretory system; vasopressin-regulated water reabsorption | 1.86E–08 | 0E–12 | – | 0.036 |
| Genetic information processing; replication, recombination and repair proteins | 7.86E–03 | 8.97E–03 | 0.88 | 0.039 |
| Genetic information processing; translation; mRNA surveillance pathway | 3.19E–08 | 2.53E–09 | 12.6 | 0.017 |
| Genetic information processing; replication and repair; NHEJ | 5.55E–05 | 3.26E–05 | 1.70 | 0.031 |
CRPC, castration-resistant prostate cancer; HSPC, hormone-sensitive prostate cancer; HCM, hypertrophic cardiomyopathy; NHEJ, non-homologous end-joining.
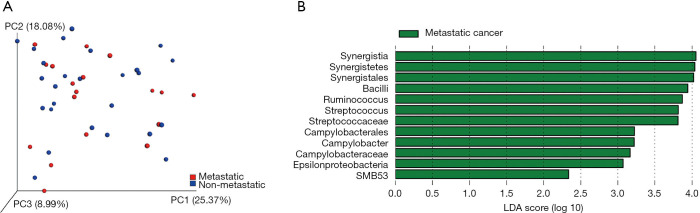
The microbial profiles of metastatic and non-metastatic PCa were also compared. The Chao1 index of metastatic and non-metastatic cancer were 913.80±359.36 and 842.39±360.29, respectively (P=0.524). For beta diversity analysis, no identified microbiota clusters were defined (Figure 2A). The LefSe analysis identified 12 microbial phylotypes that were increased in abundance in metastatic cancer: class Bacilli, its family Streptococcaceae, genus Streptococcus; pylum Synergistetes, its class Synergistia, order Synergistales; class Epsilonproteobacteria, its order Campylobacterales, family Campylobacteraceae, genus Campylobacter; genus Ruminococcus; and genus SMB53 (Figure 2B).
Figure 2.
Microbial community composition in metastatic and non-metastatic PCa. (A) Weighted uniFrac principal coordinate analysis showed no identifiable microbiota clusters between the two cohorts; (B) key phylotypes of gut microbiota in metastatic PCa patients. PCa, prostate cancer.
Discussion
In the present study, we have characterized for the first time the compositional differences in microbiota in matched HSPC and CRPC. CRPC harbored an altered composition of gut microbiome that was characterized by a significantly increased abundance of several microbial floras. We speculate that ADT may alter the patients’ gut microbiota, and the altered microbiota may conversely influence clinical responses to ADT and modulate the antitumor efficiency.
The microbial community structure altered from HSPC to CRPC, including increased abundance of phylum Bacteroidetes and Fusobacteria. Golombos et al. compared the gut microbiome of men with PCa or benign prostatic conditions, and found that Bacteroides massiliensis was significantly increased in cancer cases (18). Similar results were also confirmed in Liss’s study who performed 16S rRNA sequencing on rectal swab samples of 64 PCa and 41 non-cancer, and revealed that Bacteroides and Streptococcus species were enriched in cancer status (19). But in this study, we demonstrated that Bacteroidetes also has a role in PCa progression. Fusobacterium has been demonstrated its pro-tumorigenic role in colorectal cancer, it is persistently associated with distant metastases (20). Increased expression of Bacteroidetes and Fusobacteria were also found in patients with lung cancer (21).
Several key phylotypes that identified in CRPC have been proved to be associated with PCa progression. Genus Ruminococcus was over-expressed in CRPC, Ruminococcus is positively correlated with glutamate (22), which is extensively involved in tricarboxylic acid cycle and lipogenesis that are biologically relevant in prostate carcinogenesis and castrate-resistant progression (23). Koochekpour et al. found that glutamate deprivation could induce apoptosis in both androgen-stimulated and androgen-independent PCa cells. The serum glutamate levels were significantly higher in Gleason score (GS) ≥8 than in the GS ≤7, and in metastatic CRPC (mCRPC) patients than those with primary PCa (24). Phascolarctobacterium is known as acetate producer (25), it was also found to be elevated in CRPC. Acetate functions as a nutritional source for tumors, and has been proved to be implicated in the growth of hepatocellular carcinoma, breast cancer and PCa (26). In a recent study that examined the gut microbiome in epilepsy patients, Ruminococcus and Phascolarctobacterium also expressed higher relative abundance in patients with drug-resistant cases (27).
Among the 11 differentiated metabolic pathways, ether lipid metabolism was significantly enriched in CRPC. Ether lipid metabolism is often enhanced in cancer cells and tumors. Dramatic elevation in ether lipid contents has been correlated with the tumor growth and aggressiveness (28). Non-homologous end-joining (NHEJ) repair pathway was also activated in CRPC. NHEJ is the most efficient DNA double strand break (DSB) repair pathway that works through the entire cell cycle phase. Zhang et al. found that inhibition of androgen receptor in PCa cell lines could downregulate DNA-repair gene DNA-dependent protein kinase catalytic subunit (DNA-PKcs) expression and reduce NHEJ repair (29). Besides, NHEJ was also upregulated in hepatocellular and pancreatic carcinoma (30,31). Since PICRUSt only provides an indication of genetic potential, approaches such as metagenomics or metatranscriptomics are further required to ascertain the true functional activities of these gut microbial communities.
There are several limitations to our study including the relatively limited sample population, larger population is required to make sure the results were reproducible. Also, we are unable to account for changes in gut microbiome due to dietary or lifestyle factors. Finally, though we found altered gut microbiota in patients after ADT, the effects of these altered microbiota on ADT responses need further investigations.
Conclusions
In conclusion, our study provides preliminary evidence that gut microbiome is significantly altered between matched HSPC and CRPC. We found that ADT altered the composition of gut microbiota, and the microbiota in turn may influence the outcome of hormone therapy. Thus, functional validations are further needed to determine the underlying mechanism of these differentiated microbial communities in the process of CRPC, and their potential role to be treated as new targets to enhance ADT responses.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This study was supported by the National Natural Science Foundation of China, grant number: 81872102.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study were in accordance with the Declaration of Helsinki (as revised in 2013) and approved by the institutional review board of Huashan Hospital, Fudan University (reference number 2020-531). Informed consent was obtained from all individual participants included in the study.
Footnotes
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tau-20-566
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tau-20-566
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-20-566). The authors have no conflicts of interest to declare.
References
- 1.Roy S, Trinchieri G. Microbiota: a key orchestrator of cancer therapy. Nat Rev Cancer 2017;17:271-85. 10.1038/nrc.2017.13 [DOI] [PubMed] [Google Scholar]
- 2.Zhang J, Zhang J, Wang R. Gut microbiota modulates drug pharmacokinetics. Drug Metab Rev 2018;50:357-68. 10.1080/03602532.2018.1497647 [DOI] [PubMed] [Google Scholar]
- 3.Wilkinson EM, Ilhan ZE, Herbst-Kralovetz MM. Microbiota-drug interactions: Impact on metabolism and efficacy of therapeutics. Maturitas 2018;112:53-63. 10.1016/j.maturitas.2018.03.012 [DOI] [PubMed] [Google Scholar]
- 4.Viaud S, Saccheri F, Mignot G, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 2013;342:971-6. 10.1126/science.1240537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sivan A, Corrales L, Hubert N, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015;350:1084-9. 10.1126/science.aac4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vétizou M, Pitt JM, Daillère R, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015;350:1079-84. 10.1126/science.aad1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.García-González AP, Ritter AD, Shrestha S, et al. Bacterial metabolism affects the C. elegans response to cancer chemotherapeutics. Cell 2017;169:431-41.e8. 10.1016/j.cell.2017.03.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iida N, Dzutsev A, Stewart CA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 2013;342:967-70. 10.1126/science.1240527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matson V, Fessler J, Bao R, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 2018;359:104-8. 10.1126/science.aao3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Routy B, Le Chatelier E, Derosa L, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018;359:91-7. 10.1126/science.aan3706 [DOI] [PubMed] [Google Scholar]
- 11.Scott TA, Quintaneiro LM, Norvaisas P, et al. Host-microbe co-metabolism dictates cancer drug efficacy in C. elegans. Cell 2017;169:442-56.e18. 10.1016/j.cell.2017.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooperberg MR, Hinotsu S, Namiki M, et al. Trans-Pacific variation in outcomes for men treated with primary androgen-deprivation therapy (ADT) for prostate cancer. BJU Int 2016;117:102-9. 10.1111/bju.12937 [DOI] [PubMed] [Google Scholar]
- 13.Cornford P, Bellmunt J, Bolla M, et al. EAU-ESTRO-SIOG Guidelines on prostate cancer. Part II: treatment of relapsing, metastatic, and castration-resistant prostate cancer. Eur Urol 2017;71:630-42. 10.1016/j.eururo.2016.08.002 [DOI] [PubMed] [Google Scholar]
- 14.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7:335-6. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010;26:2460-1. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- 16.Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol 2011;12:R60. 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langille MG, Zaneveld J, Caporaso JG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 2013;31:814-21. 10.1038/nbt.2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Golombos DM, Ayangbesan A, O'Malley P, et al. The role of gut microbiome in the pathogenesis of prostate cancer: a prospective, pilot study. Urology 2018;111:122-8. 10.1016/j.urology.2017.08.039 [DOI] [PubMed] [Google Scholar]
- 19.Liss MA, White JR, Goros M, et al. Metabolic biosynthesis pathways identified from fecal microbiome associated with prostate cancer. Eur Urol 2018;74:575-82. 10.1016/j.eururo.2018.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bullman S, Pedamallu CS, Sicinska E, et al. Analysis of fusobacterium persistence and antibiotic response in colorectal cancer. Science 2017;358:1443-8. 10.1126/science.aal5240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang WQ, Zhao SK, Luo JW, et al. Alterations of fecal bacterial communities in patients with lung cancer. Am J Transl Res 2018;10:3171-85. [PMC free article] [PubMed] [Google Scholar]
- 22.Ottosson F, Brunkwall L, Ericson U, et al. Connection between bmi-related plasma metabolite profile and gut microbiota. J Clin Endocrinol Metab 2018;103:1491-501. 10.1210/jc.2017-02114 [DOI] [PubMed] [Google Scholar]
- 23.Koochekpour S. Glutamate, a metabolic biomarker of aggressiveness and a potential therapeutic target for prostate cancer. Asian J Androl 2013;15:212-3. 10.1038/aja.2012.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koochekpour S, Majumdar S, Azabdaftari G, et al. Serum glutamate levels correlate with Gleason score and glutamate blockade decreases proliferation, migration, and invasion and induces apoptosis in prostate cancer cells. Clin Cancer Res 2012;18:5888-901. 10.1158/1078-0432.CCR-12-1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu F, Guo X, Zhang J, et al. Phascolarctobacterium faecium abundant colonization in human gastrointestinal tract. Exp Ther Med 2017;14:3122-6. 10.3892/etm.2017.4878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schug ZT, Vande Voorde J, Gottlieb E. The metabolic fate of acetate in cancer. Nat Rev Cancer 2016;16:708-17. 10.1038/nrc.2016.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng A, Qiu X, Lai W, et al. Altered composition of the gut microbiome in patients with drug-resistant epilepsy. Epilepsy Res 2018;147:102-7. 10.1016/j.eplepsyres.2018.09.013 [DOI] [PubMed] [Google Scholar]
- 28.Benjamin DI, Cozzo A, Ji X, et al. Ether lipid generating enzyme AGPS alters the balance of structural and signaling lipids to fuel cancer pathogenicity. Proc Natl Acad Sci U S A 2013;110:14912-7. 10.1073/pnas.1310894110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang W, Liao CY, Chtatou H, et al. Apalutamide sensitizes prostate cancer to ionizing radiation via inhibition of non-homologous end-joining DNA repair. Cancers (Basel) 2019;11:1593. 10.3390/cancers11101593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evert M, Frau M, Tomasi ML, et al. Deregulation of DNA-dependent protein kinase catalytic subunit contributes to human hepatocarcinogenesis development and has a putative prognostic value. Br J Cancer 2013;109:2654-64. 10.1038/bjc.2013.606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li YH, Wang X, Pan Y, et al. Inhibition of non-homologous end joining repair impairs pancreatic cancer growth and enhances radiation response. PLoS One 2012;7:e39588. 10.1371/journal.pone.0039588 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as