Abstract
Xanthomonas oryzae is a serious pathogen causing bacterial leaf blight (BLB) disease in rice, markedly reducing its yield. In this study, the rice chorismate mutase (OsCM) gene was overexpressed in a bacterial leaf blight-susceptible rice line to investigate the functional role of OsCM in response to bacterial leaf blight stress. We reported that overexpression of OsCM altered the downstream pathway of aromatic amino acids, mitigating pathogen stress by altering stress-responsive genes and hormonal accumulation. Phenotypic evaluation showed that the lesion length in the transgenic line was significantly lesser than that in the wild-type, suggesting greater resistance in the transgenic line. Further analysis revealed that OsCM expression induced phenylalanine accumulation and suppressed tyrosine accumulation in response to bacterial leaf blight stress. Furthermore, bacterial leaf blight stress induced genes downstream of the phenylpropanoid pathway in conjunction with OsCM, suggesting that the phenylpropanoid pathway is dependent on OsCM gene expression. We reported high SA and low JA accumulation in response to bacterial leaf blight stress in the transgenic line. This higher SA accumulation suggested that SA induces immune responses by functioning as a promoter of nonexpresser pathogenesis-related genes 1 (NPR1) transcriptional regulation. Xa7 expression was induced with increase in nonexpresser pathogenesis-related genes 1, which is thought to be responsible for Xa7 expression, which is responsible for mitigating bacterial leaf blight stress.
Subject terms: Cell biology, Developmental biology, Genetics, Molecular biology, Plant sciences
Introduction
The yield of rice, a staple food for about 50% the world’s population, is decreased due to biotic and abiotic stresses. Among the biotic stresses, bacterial leaf blight (BLB) is one of the most severe and common rice diseases in most Asian countries; it is caused by Xanthomonas oryzae pv. oryza (Xoo) and results in considerable yield loss each year1. Xoo can be more successfully controlled by the development of transgenic resistant varieties of rice, usually through single-gene resistance2. Generally, the exposure of plants to pathogens enhances the induction of genes involved in the shikimate pathway and aromatic amino acids (AAAs), such as phenylalanine (Phe), tyrosine (Tyr), and tryptophan (Trp)3. Pathogenic attack stimulates the plant cell wall and releases oligogalacturonides, which in turn stimulate the expression of various genes encoding enzymes of the shikimate pathway and AAAs as well as those encoding secondary metabolites derived from Phe and Tyr4. Bacterial pathogens redirect normal host metabolism by delivering a constellation of type III effector proteins to enhance pathogen multiplication and nutrition5,6. However, genes associated with AAAs have been significantly modified to cope with bacterial challenges, including cell wall alteration to control the nutrients and water passage from plants to invading bacteria7,8. Phe, Tyr, and Trp are the main AAA molecules in plant metabolism which are responsible for synthesis of a number of hormones, such as salicylic acid and auxin, as well as for essential secondary metabolites with many biological functions9,10.
Chorismate along with the synthesis of salicylic acid (SA) through isochorismate synthase (ICS) activity11 is also the main precursor and initial branch point metabolite in the synthesis of all AAAs and their derivatives. The main route of Phe and Tyr biosynthesis is initiated from the same precursor, chorismite, catalyzed by the chorismate mutase (CM) enzyme to produce prephenate, which is further catalyzed to arogenate by the prephenate aminotransferase (PAT) enzyme. Phe and Tyr are synthesized in two ways. First, prephenate is converted to phenylpyruvate and hydroxyphenylpyruvate via the prephenate dehydratase (PDT) and prephenate dehydrogenase (PDH) enzymes, respectively. Further, phenylpyruvate and hydroxyphenylpyruvate are catalyzed into Phe and Tyr, respectively, through the aromatic amino acid aminotransferase (AAAAT) enzyme. Alternately, prephenate synthesizes arogenate through the prephenate aminotransferase (PAT) enzyme, which is further catalyzed into Phe and Tyr through the arogenate dehydratase (ADT) and arogenate dehydrogenase (TyrA) enzymes, respectively12–14. Previous reports support the observation that, in phenylpyruvate containing plants, it also act as a precursor of various secondary metabolites15. Phe and Tyr are catabolized to produce anabolic precursors of the most important secondary metabolites through the phenylpropanoid pathway. Phenylalanine ammonia lyase (PAL) is responsible for conversion of Phe into cinnamate; however, Tyr is the directly involved in the synthesis of coumarate in the phenylpropanoid pathway by the tyrosine ammonia lyase enzyme16. The gene responsible for the PAL enzyme is regulated during biotic stress and under conditions that increase the need for lignin as a cell wall component17. The metabolites of the phenylpropanoid pathway causes toughness to cells which act as pest deterrent and disease resistant18. The PAL enzyme is involved in the first step of the phenylpropanoid pathway, through which the synthesis of various secondary metabolites (flavonoid pigments, lignin, antimicrobial phenolics, UV protectants, and cell wall-associated phenolics) take place against biotic, mechanical, and UV stresses, which is a definitive defense reaction to pathogenic attack19. Expression of the CM gene induces PAL activity, providing precursors for the biosynthesis of lignin and SA, which change in response to pathogen infection. SA accumulation is essential for systemic acquired resistance (SAR), removing H2O2 and suppressing the oxidative burst necessary for the HR. A report has shown that SA plays an important role in Xoo resistance in rice and promotes basal as well as hypersensitive responses during Xoo infection in rice20. The accumulation of SA increases under stress conditions, which translocates NPR1 into the nucleus for SA-dependent transcriptomic changes21. NPR1 is hypothesized to be a transcription factor due to its lack of a DNA binding domain and responsibility for Xoo resistance in rice, given that it is responsible for PR gene expression22. Furthermore, the SA and JA signaling pathways intersect at various branches because they alter biotic stress antagonistically, which was reported for the first time in tomato23. SA inhibits JA accumulation by suppressing JA-responsive genes, which suggests that NPR1 is a key player in the antagonistic cross-talk of SA and JA24.
Despite of great importance of chorismate mutase in alteration of shikimate pathway and biosynthesis of AAAs, the role of chorismate mutase in the stress response has been mostly ignored. Although most of the researchers evaluated the function of chorismate mutase associated with the aromatic amino acids and secondary metabolites, but still it is not explored in response to biotic stress. Therefore, the present study focus on role of chorismate mutase in the alteration of the PAL pathway and biotic stress tolerance (BLB) in terms of aromatic amino acid and SA accumulation and transcriptional modulation of defense-related genes in Oryza sativa.
Results
Cloning and OsCM-overexpressing line development
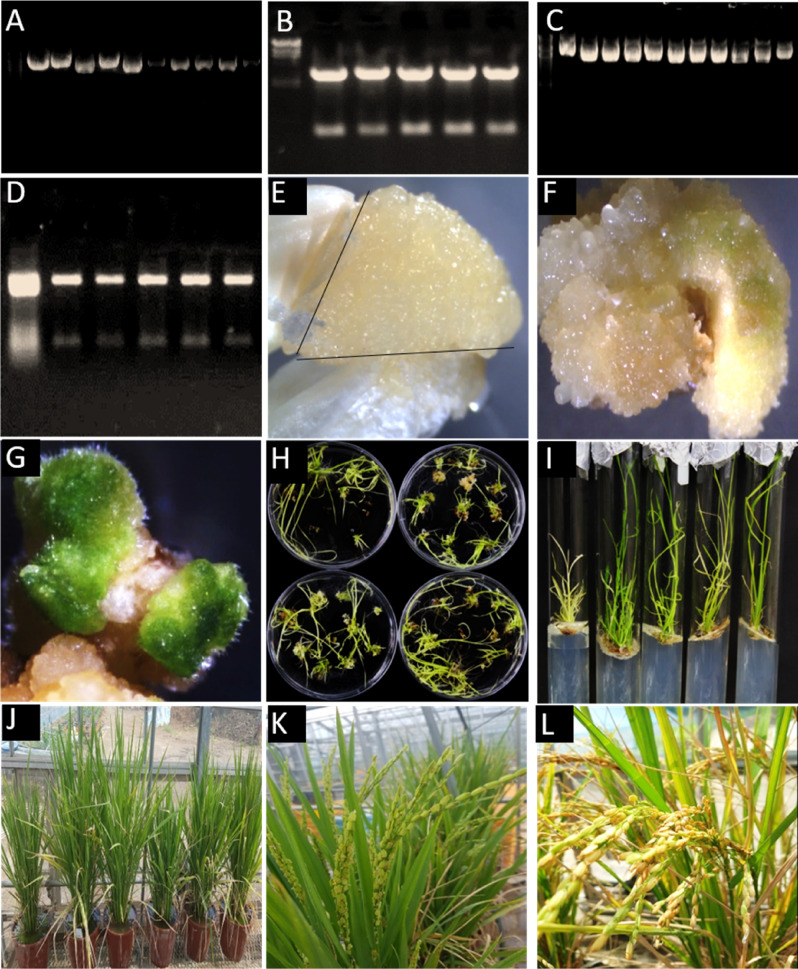
The complete ORF region of OsCM was successfully cloned into a cloning vector and transferred to E. coli cells for multiplication. The isolated plasmid and double digestion with the restriction enzymes Not1 and Asc1, are shown in Fig. 1A,B. The double-digested template was further cloned successfully into an expression vector and transferred into Agrobacterium cells. The plasmid isolated from Agrobacterium was double-digested with BamH1 and Xho1 restriction enzymes (Fig. 1C,D). The OsCM-overexpressing rice line was developed using callus culture; its various developmental stages are described in Fig. 1E–L. The efficiency of transgenic line development was about 18% using hygromycin as a selection marker. The seeds obtained were used for further analysis.
Figure 1.
Cloning and development of OsCM-overexpressing line. (A) Cloning into E. coli. (B) Double digestion of cloned plasmid isolated from E. coli. (C) Cloning into Agrobacterium. (D) Double digestion of cloned plasmid isolated from Agrobacterium. (E–L) Developmental stages of transgenic line through callus culture.
OsCM is functionally involved in BLB tolerance
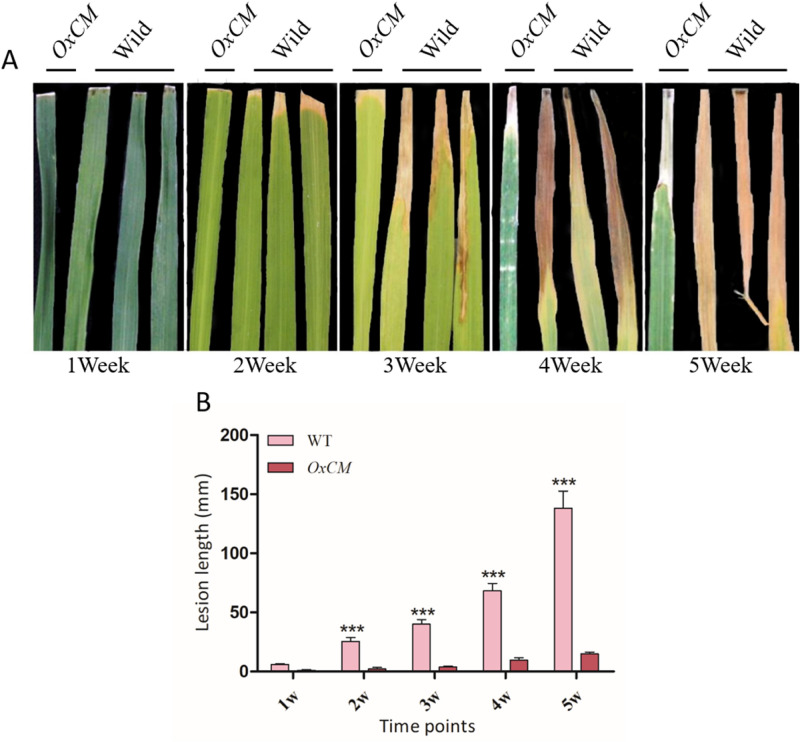
To evaluate the differences in morphological characteristics of OxCM and wild-type (WT) rice plants, using the leaf clipping method the plants were inoculated with K3a strain of Xoo. The lesion length upon BLB infection of WT as well as OxCM plants was measured from 1 week after inoculation of the K3a strain, for 5 consecutive weeks. The inoculation of K3a was confirmed through colony counting. The phenotypic evaluation showed that the lesion length increased markedly after each week, indicating that WT plants were highly susceptible to K3a infection (Fig. 2A). However, the lesion length of OxCM plants was much less throughout the five weeks, suggesting that OxCM expressing plant was highly tolerant of K3a.The infection of K3a in WT plants reached the severe stage and finally the leaves died after 5 weeks. The measurement of lesion length showed that the lesions increased from 6 to 138 mm from the first to the fifth week in WT plants (Fig. 2B). However, in OxCM plants, the lesions increased from 1.1 to 15 mm from the first to the fifth week. This shows that overexpression of OsCM in rice significantly (P < 0.001) enhanced tolerance to BLB.
Figure 2.
Phenotypic validation of pathogen infection. (A, B) Image and graphical representation of lesion length in response to pathogen infection, respectively. Bars represent mean ± standard deviation, while asterisks indicate a significant difference (p˂0.05, two-way ANOVA, Bonferroni posttest). 1w, 2w, 3w, 4w, and 5w represent time points for obtaining data (in weeks).
Pathogen infection enhances OsCM expression and phenylalanine accumulation and suppresses tyrosine
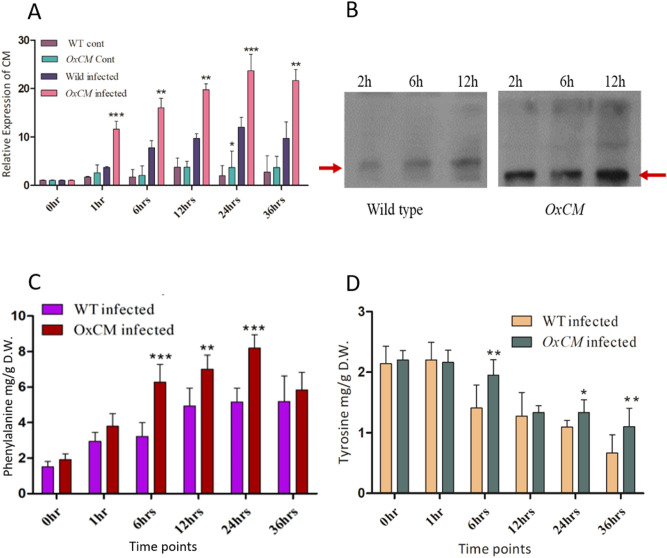
To further investigate the biological function of OsCM in Phe and Tyr accumulation in stressed as well as normal conditions, we infected WT plants and OxCM plants and compared them with uninfected WT and OxCM plants. The OsCM gene expression in the transgenic infected line was significantly different (P < 0.001) from that of WT infected plants (Fig. 3A). However, the expression of OsCM in uninfected plants was not significantly higher than that of WT uninfected plants. We also investigated the level of OsCM protein expression in transgenic and WT infected plants. The results showed that OsCM protein was significantly expressed in the OxCM line as compared to the WT plants (Fig. 3B). These results show that OsCM is positively involved in pathogen resistance. We further investigated Phe and Tyr accumulation in WT and OxCM plants under Xoo stress, as the CM activity enhances Phe and Tyr biosynthesis. Our results showed that Phe was significantly increased 83%, 62% and 52% after 6, 12 and 24 h respectively in OxCM infected plants compared with WT infected plants (Fig. 3C). On the other hand, Tyr accumulation was the same in both the WT and transgenic plants after 1 h post-infection, but the accumulation was more reduced in WT as compared to OxCM plants with the passage of time (Fig. 3D). These results suggested that, during BLB infection, OsCM expression upregulates Phe biosynthesis while down regulating Tyr biosynthesis.
Figure 3.
Functional expression of OsCM gene and Phe and Tyr accumulation. Bars represent mean ± standard deviation, while asterisks indicate a significant difference (p ˂ 0.05, two-way ANOVA, Bonferroni post-test). 0 h, 1 h, 6 h, 12 h, 24 h, and 36 h represent time points when data were obtained (in h). WT cont refers to the WT control, OxCM cont refers to the CM-overexpressing control line, wild infected is the WT infected with BLB, and OxCM infected is the CM-overexpressing line infected with BLB. (A) Relative expression of the OsCM gene in WT and overexpressing control and infected plants. (B) Protein expression in the WT and OxCM line at 2, 6, and 24 h after infection with BLB. The provided (B) is cropped due to the multiple samples blotting on same X-ray film due to time and resources limitation, the original non-processed picture is given in Supplementary Figure S1. (C, D) Phenylalanine and tyrosine quantification in WT and OxCM infected plants.
OsCM regulates aromatic amino acid biosynthesis genes under stressed conditions
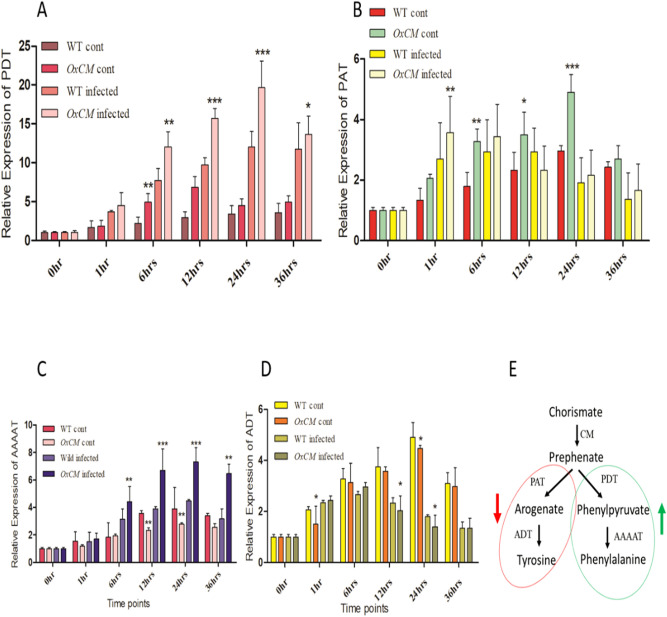
To shed light on the genes which are involved in AAAs in the WT and OxCM plants under Xoo stress, we compared the transcript abundance of prephenate dehydratase (PDT), prephenate aminotransferase (PAT), aromatic amino acid aminotransferase (AAAAT), and arogenate dehydratase (ADT) genes, which are involved in AAA biosynthesis, by quantitative PCR (Fig. 4). We observed different expression level of these genes in WT and OxCM plants in Xoo-infected as well as uninfected plants. The relative expression of the PDT gene was significantly increased in OxCM infected plants compared with that in WT infected plants until 24 h and was then reduced at 36 h (Fig. 4A). Meanwhile, the expression of PDT was also increased in OxCM uninfected plants compared with that in WT uninfected plants, which suggested that overexpression of the OsCM gene also activates the PDT gene involved in the catalysis of prephenate into phenylpyruvate. Unlike PDT, PAT was initially (1 h) upregulated but was gradually down regulated in both OxCM infected plants and WT infected plants; however, the expression level remained higher in OxCM plants than in WT throughout 36 h (Fig. 4B). However, PAT expression was significantly higher in OsCM-overexpressing uninfected plants than in WT uninfected ones. These results suggested that induction of the PAT gene decreased in response to Xoo infection in both WT and OxCM plants, while under normal conditions, OxCM plants exhibit increased PAT expression compared with WT plants. We assumed that both PDT and PAT act antagonistically during Xoo infection. Similarly, AAAAT, which is involved in the regulation of phenylpyruvate conversion into Phe, showed an expression pattern similar to PDT. AAAAT was significantly expressed in OxCM infected plants compared with that in WT infected plants (Fig. 4C). However, the expression was reduced in OxCM uninfected plants compared with that in WT uninfected plants. Unlike AAAAT, ADT was expressed in OxCM infected plants compared with that in WT infected plants until 6 h after the inoculation of Xoo, but was then significantly reduced (Fig. 4D). On the other hand, similar to PAT, ADT was also upregulated in WT and OxCM uninfected plants, but the expression of OxCM was lower than in WT plants. Figure 4E shows a graphical representation of the effects of OsCM-overexpression on all of the four genes (PDT, PAT, AAAAT, and ADT) under stress conditions. It was assumed that, under stress conditions, the overexpression of OsCM down regulates the genes involved in the Tyr biosynthesis pathway while up regulating the genes involved in the Phe biosynthesis pathway.
Figure 4.
Regulation of downstream genes by overexpression of chorismate mutase. Bars represent mean ± standard deviation, while asterisks indicate a significant difference (p ˂ 0 .05, two-way ANOVA, Bonferroni post-test). 0 h, 1 h, 6 h, 12 h, 24 h, and 36 h represent time points at which data were obtained (in hours). WT cont refers to the WT control, OxCM cont is the CM-overexpressing control line, wild infected is the WT infected with BLB, and OxCM infected is the CM-overexpressing line infected with BLB. (A) PDT is prephenate dehydratase, (B) PAT is prephenate aminotransferase, (C) AAAAT is the aromatic amino acid aminotransferase, and (D) is the arogenate dehydratase gene relative expression among the WT and OsCM-overexpressing infected and control plants. (E) A schematic representation of the related gene expression. A downward arrow indicates down regulation of PAT and ADT, while an upward arrow indicates up regulation of PDT and AAAAT genes at the same time.
JA and SA cross-talk in OxCM and WT plants under pathogen infection
To further verify the efficiency by which OxCM confers tolerance to Xoo infection with respect to that of WT plants, we evaluated the SA and JA accumulation in both types of plant under Xoo infection at six time points within 0–36 h. The results demonstrated that JA was significantly reduced in OxCM plants compared with that in WT plants (Fig. 5A). In the first hour of infection, the accumulation was increased in both types of plant, but later on it was consistently reduced. In the first hour of infection, about 70 ng/g D.W accumulated in WT plants, which was reduced to 30 ng/g D.W. which was 75%, after 36 h. Likewise, in the first hour of infection, 44 ng/g D.W. JA accumulated, which was reduced to 23 ng/g D.W. which was 47%, after 36 h. In contrast to JA, SA was significantly and continually enhanced in OxCM plants with increasing infection time, compared with that of WT plants (Fig. 5B). SA accumulated from 1 to 24 h in the range of 125–210 ng/g D.W. which was 68% however, from 36 h of infection, the SA level started to decrease. These results confirmed that JA and SA are regulated antagonistically.
Figure 5.
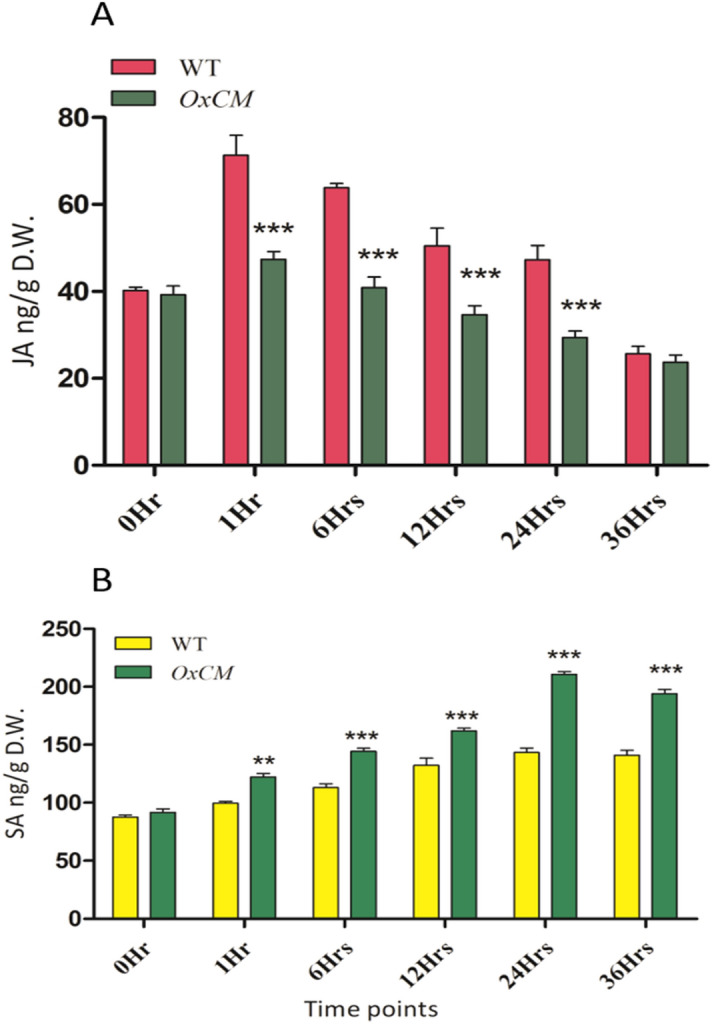
JA and SA accumulation in pathogen response. Bars represent mean ± standard deviation, while asterisks indicate a significant difference (p ˂ 0.05, two-way ANOVA, Bonferroni post-test). 0 h, 1 h, 6 h, 12 h, 24 h, and 36 h represent time points at which data were obtained (in h). WT refers to WT infected and OxCM is the CM-overexpressing line infected with BLB. (A) JA accumulation and (B) SA accumulation in WT and OxCM infected plants.
OsCM positively regulates the expression of disease resistance genes
It has been reported that NPR1 is involved in Xoo resistance via regulation of the defense-related gene Xa725,26. Thus, we examined the pattern of expression of NPR1 and Xa7 in the infected OxCM line with respect to infected WT plants. The results showed that NPR1 was significantly induced in the OxCM infected plants compared with the level in WT infected plants and the expression increased with increasing infection time until 24 h (Fig. 6A). After 24 h of infection, the NPR1 expression was reduced in infected and uninfected OxCM and WT plants. However, the NPR1 gene expression was also higher in OxCM uninfected plants than in WT uninfected plants. Likewise, the Xa7 gene also showed the same pattern of expression as NPR1. The Xa7 expression was also higher in OxCM infected plants than in WT infected ones (Fig. 6B). The expression gradually increased with increasing infection time until 24 h. The results suggest that overexpression of OsCM in rice plants increases BLB tolerance in terms of regulating the NPR1 gene, which in turn regulates the defense-related gene Xa7.
Figure 6.
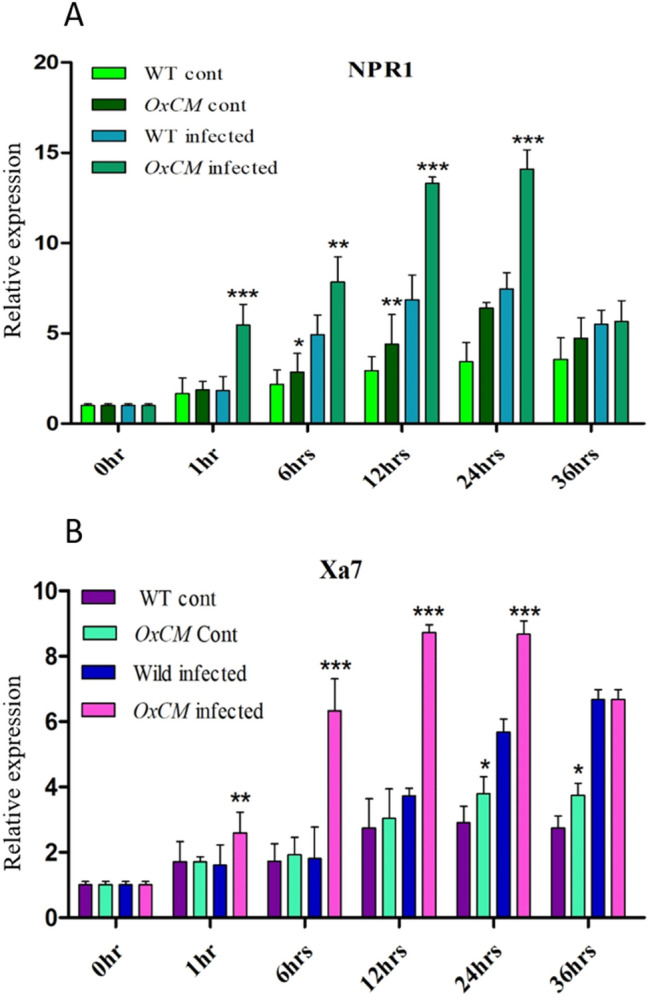
Expression of NPR1 and Xa7 under pathogen inoculation. Bars represent mean ± standard deviation, while asterisks indicate a significant difference (p ˂ 0.05, two-way ANOVA, Bonferroni post-test). 0 h, 1 h, 6 h, 12 h, 24 h, and 36 h represent time points at which data were obtained (in hours), WT refers to WT control, OxCM cont is CM-overexpressing control line, wild infected is the WT infected with BLB, and OxCM infected is the CM-overexpressing line infected with BLB. (A, B) Relative expression of NPR1 and Xa7 genes, respectively, among the WT and OsCM-overexpressing control and infected lines.
OsCM enhances lignin accumulation and amino acid content upon BLB stress
We studied lignin and total amino acid contents against Xoo stress in OxCM and WT plants, as lignin exhibits regulatory functions under biotic as well as abiotic stresses, while amino acids are involved in the biosynthesis of various secondary metabolites, which are directly involved in biotic stresses27,28. The results showed that more lignin and total amino acids were accumulated in the OxCM infected plants as compared to WT infected plants (Fig. 7A,B). Continuous increases of lignin and amino acids were found upon continued infection. Lignin was increased 115% from 13 mg/g D.W. to 28 mg/g D.W. from 0 to 36 h of infection in OxCM plants. Similarly, the concentration of amino acids in OxCM infected plants increased 21% from 107 to 130 mg/g D.W. over 36 h of infection. However, significance differences in both lignin and amino acids appeared after 6 h of infection. These results show that Xoo infection regulates lignin and total amino acids in rice plants.
Figure 7.
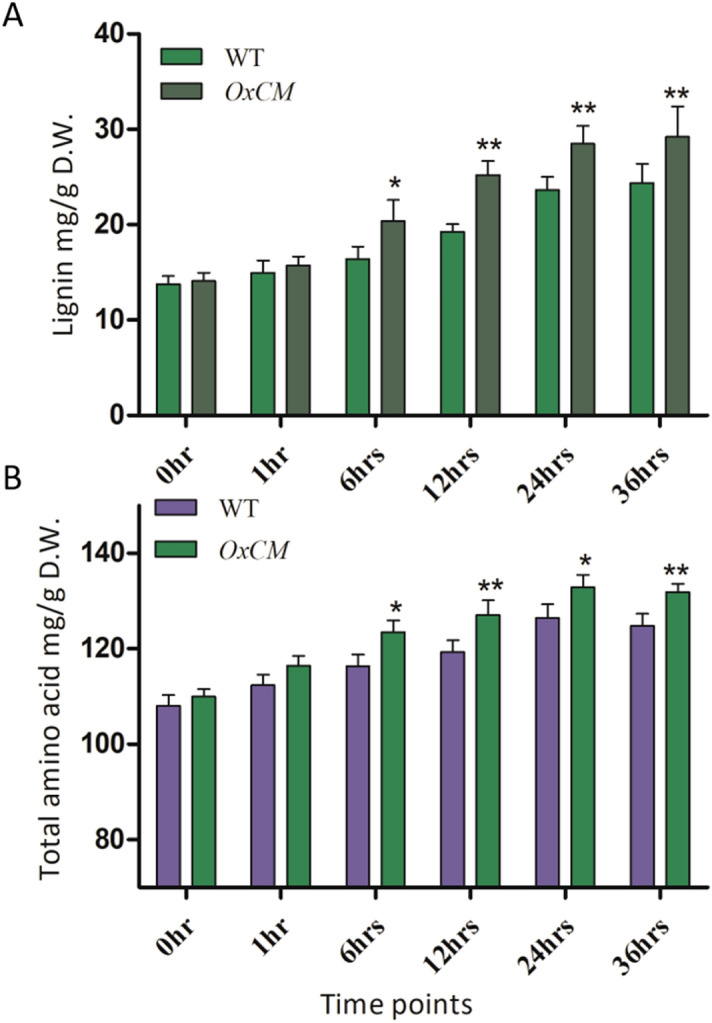
Lignin and amino acid accumulation upon pathogen stress. Bars represent mean ± standard deviation, while asterisks indicate a significant difference (p ˂ 0.05, two-way ANOVA, Bonferroni post-test). 0 h, 1 h, 6 h, 12 h, 24 h, and 36 h represent time points at which data were obtained (in h). WT refers to WT infected and OxCM is the CM-overexpressing line infected with BLB. (A) Lignin accumulation and (B) total amino acid accumulation in WT and OxCM infected plants.
Discussion
AAAs such as Phe and Tyr are central molecules in plant metabolism and synthesized in the shikimate pathway from the common precursor chorismate. The shikimate pathway connects primary metabolites to secondary metabolites via chorismate being the end product of shikimate, which initiates the synthesis of AAAs that in turn enhances secondary metabolites3. Phe and Tyr induces cinnamic acid which is the precursors of phenylpropanoid9. Plants have adopted several defense signaling pathways to mitigate severe environmental conditions and pathogenic attack, such as by altering secondary metabolites29. The modulation of secondary metabolites like phenylpropanoids against biotic and abiotic stresses is a clue to the involvement in stress tolerance30. A wide range of phenolic compounds are produced from phenylpropanoids such as flavonoids, isoflavonoids, anthocyanins, plant hormones, phytoalexins, and lignins31,32.
In current study, OsCM gene was functionally evaluated by generating an overexpressing line using Agrobacterium-mediated transformation in rice plants. The innate immunity of plants was altered upon infection of BLB and the suppression and induction of OsCM activity were confirmed by biological assays. The phenotypic evaluation of lesion length demonstrated that OxCM plants were highly tolerant of BLB stress. Evaluation of the two AAAs, Phe and Tyr, showed that they were adversely altered during BLB stress, which suggested that they are participants in pathogen tolerance. The results showed that Phe accumulation was increased in the OxCM line, while Tyr was decreased in Xoo after inoculation (Fig. 3C,D). We predicted that the alterations of Phe and Tyr were due to induction of the OsCM gene because the OsCM gene was significantly induced in the OxCM line at the transcriptomic and proteomic levels (Fig. 3A,B), which enhanced the accumulation of Phe in the transgenic line. Phe is considered to be the chief AAA accumulated in plants under normal conditions, being at a level higher than Tyr and Trp, and absorbing about 30% of photosynthetic carbon to produce phenylpropanoids33. We assumed that the induction of Phe under BLB stress in OxCM plants, enhanced the accumulation of downstream metabolites, which increased Xoo tolerance. A previous study showed that induction of the derivatives of Phe and Tyr (phenylpropanoids) under stress conditions was enhanced due to up regulation of the Phe catabolic enzyme, PAL, which uses Phe as a substrate34. Supported by the high accumulation of Phe in OxCM infected plants, the results showed that the genes downstream of OsCM such as PDT and AAAAT were positively regulated, whereas the genes responsible for Tyr biosynthesis such as PAT and ADT were negatively regulated (Fig. 4). The fold change of Phe was much greater than that of Tyr, which shows that Phe is highly involved in the biosynthesis of phenylpropanoids in response to pathogenic attack. A previous study confirmed that an increasing Phe level in petunia flowers increases the production of phenylpropanoids35. The signaling of a reduction in Tyr corresponding to Phe in BLB stress is not well known, but it was predicted that Tyr is not efficiently involved in stress mitigation compared with Phe.
Plant hormones are dynamic regulators of plant responses to environmental pressure by participating external stimuli with complex monitoring systems. SA is an important part of plant defense and is well known for its defensive role in plant defense system against pathogens36. SA has a vital role in rice defense system in response to Xoo, and enhances both basal defense and the hypersensitive response during Xoo infection in rice20. The SA signaling pathway might be involved in the regulation of glucosinolate, which is hydrolyzed by SA to various degradation products that play significant part in interactions with pathogens37. The current study investigated the regular increase of SA in rice plants in response to continued Xoo infection, and the SA accumulation was significantly higher in OxCM plants than in WT. This investigation proved that OsCM is significantly involved in the biosynthesis of SA. In most plants, SA can be synthesized by two key regulatory enzymes, PAL and ICS, whereas PAL uses phenylalanine and ICS uses chorismate as a substrate38. Although ICS is involved in SA accumulation in response to some plant stresses11,39, the pathogen and other stress-induced SA is mostly derived from the PAL-catalyzed pathway40. A previous study showed that Arabidopsis PAL mutant accumulated more SA than ICS mutant, which suggested that, in Arabidopsis, ICS is the key contributor to pathogen-induced SA biosynthesis11. However, these clarifications are based on independent analysis of either the PAL or the ICS-catalyzed pathway, and the comparative contributions of PAL vs. ICS to SA biosynthesis and pathogen defense have not been instantaneously assessed for any plant system. Although the PAL and ICS pathways are equally significant for SA biosynthesis, suppression of either pathway is enough to compromise SA biosynthesis in response to pathogenic attack. In the present study, we did not evaluate the relative expression of ICS, but the higher accumulation of Phe and SA suggested that PAL is sufficiently involved in the biosynthesis of SA in response to BLB stress. A previous study showed that SA regulation in response to stress may be feedback-regulated by the metabolites of biochemical pathways. This inference was supported by the feedback-regulation of CM by phenylalanine and feedback allosteric regulation assessed in amino acid biosynthesis41,42. Notably, the bacterial pathogen infection increased the expression level of the key leaf-expressed PAL isoform instead of ICS in soybean. Another previous study suggested that either PAL or ICS knockout eliminated pathogen-induced production of SA in soybean, which implied that the expression of ICS is not a requirement for SA biosynthesis38. It is possible that enhanced PAL induction as related to its role as a chief regulator of the phenylpropanoid pathway, which is well known to participate in defense43–45. Similar to SA, JA also has a main part in mitigating pathogen-related stress. However, SA and JA signaling pathways were reported to interact antagonistically against biotic stress46. Unlike SA, the JA accumulated in OxCM plants was efficiently reduced with increasing Xoo infection time, compared with that in the WT (Fig. 5A). Studies have shown that SA facilitates the down regulation of JA-responsive genes like lipoxygenase 2 (LOX2), vegetative storage protein, and PDF1.224. Pathogenic infection manipulates the defense-related regulatory network in plants in terms of phytohormone accumulation, which causes hormonal imbalance and activates untimely defense responses47. Previous studies showed that the synthesis of coronatine-1 JA-Ile mimic by Pseudomonas syringae pv. tomato (Pst) bacteria altered the stimulation of JA-dependent defense responses, leading to the inhibition of SA-dependent defense responses48,49. Plants usually resist a diverse range of attackers in the natural environment via regulating complex defense mechanisms to modulate effective defense responses against pathogens. However, the mechanism by which plants prioritize one response over another is unknown. This interpretation shows that not all pathogens activate JA signaling in plants, such as Bemisia tabaci enhancing SA and suppressing JA accumulation in Arabidopsis50, similar to our findings. This means that neither SA nor JA is more important, but both hormones are equally important for pathogen resistance. Positive or negative cross-talk between SA and JA may be induced depending on the pathogen51.
This study was extended to focus on the relative expression levels of NPR1 and Xa7 genes. NPR1 is one of the essential regulatory tools of SA signaling, which interacts with TGA transcription factors that are responsible for the activation of SA-responsive PR genes52. NPR1 in rice enhances tolerance to rice bacterial blight (Xanthomonas oryzae pv. oryzae) and rice blast fungus (Magnaporthe grisea), and also activates PR gene induction and regulates SAR22. Xa7 is a PR gene that enhances Xoo resistance in rice plants at high temperature, whereas resistance change due to other R genes is usually suppressed53. Xa7 regulates hypersensitivity and localized host cell death to control pathogenic infection in the plants54. It was previously shown that Xa7 expression suppressed the expression of ABA-related genes, which signal antagonistically with SA55. Our results suggested that SA and Xa7 interact with each other and are positively regulated in response to pathogens. Furthermore, our study revealed that the expression of NPR1 and Xa7 was highly induced in the OxCM line under Xoo stress, which was consistent with the higher SA accumulation (Fig. 6). A previous investigation evaluated that the enhancement of SA accumulation in pathogen-infected tissues resulted in enhancement of the PR gene expression that modulate the resistance of a broad range of pathogens46. NPR1 plays an essential part in SA–JA cross-talk because the NPR1 Arabidopsis plants were compromised in terms of the SA-mediated suppression of JA-responsive gene expression56. Various WRKY transcription factors downstream of NPR1 play key roles in altering SA-dependent defense responses in plants21. SA’s association with heme-containing enzymes, such as catalase, result in adequate redox stress to initiate the release of monomers of NPR1 and their entry into the nucleus57.
Lignin and amino acids are essential modulators of stress responses in plants produced via the shikimate pathway58, associated with the PAL pathway. Along with the accumulation of SA, PAL activity also offers a precursor (cinnamic acid) for lignin biosynthesis in response to pathogen infection59. We investigated that lignin quantification in response to Xoo infection was higher in OxCM rice plants than in the WT. It was assumed that overexpression of the OsCM gene was involved in the regulation of lignin via the alteration of PAL activity. In PAL-knockdown Brachypodium plants, lignin accumulation was decreased up to 40%, which enhanced pathogen susceptibility60. Lignin accumulation in Arabidopsis PAL1, PAL2, PAL3, and PAL4 mutants was decreased from 20 to 25% compared with that in WT plants, while the same plants also showed a reduced level of SA and enhanced pathogen susceptibility61. Although lignification enhances the toughness of the cell wall in response to pathogenic attack, the free radical-mediated polymerization of lignin precursors in intercellular spaces might also lignify pathogenic structures. Lignin hydrophobicity enables solute transformation in the vascular tissues and reduces water loss during evapo-transpirative processes. Similarly, total amino acid accumulation was enhanced in OxCM infected plants compared with that in WT plants because the higher accumulation of amino acids is essential for plants to respond to stress in term of ROS scavenging behavior, as well as potential regulatory and signaling molecules62,63. The higher accumulation of total amino acids in Xoo-infected OxCM plants suggested that the OsCM gene of the shikimate pathway is significantly involved in the accumulation of total amino acids in response to pathogenic attack.
Materials and methods
Generation of OsCM transgenic rice line
Rice plants (Cheongcheong) were selected for transformation and further study. The seeds were sterilized with 3% hypochlorite for 10 min and then incubated at 32 °C in water until they started to germinate, with their water changed every day. The germinated seeds were grown in a greenhouse and the samples were collected after 3 weeks for total RNA isolation. Total RNA was isolated using RNeasy Plant Mini Kit from Qiagen and the open reading frame (ORF) of OsCM (MH752192) was amplified using gene primers. The OsCM was cloned into the cloning vector pENTR/D-TOPO (pENTR Directional TOPO cloning kit; Invitrogen) and then into the PSB11 expression vector using the Gateway cloning system (Gateway LR Clonase enzyme mix kit; Invitrogen). The entry clone was first transferred into DH5α E. coli and then transferred into Agrobacterium cells LBA4404 (Takara) via the heat shock method and spread on hygromycin-containing LB medium. Both inserts were confirmed through double digestion using Not1 and Asc1 and BamH1 and Xho1, respectively. CM-overexpressing (OxCM) rice lines were developed through a callus culture technique. Seeds of good quality were dehulled, sterilized with 3% hypochlorite for 10 min, washed three times, sterilized again with 70% ethanol for 5 min, washed again with ddH2O, and dried in laminar flow. Dried seeds were inoculated in callus induction medium at 10–15 seeds per plate and placed in the dark for 12 days. The induced callus was further pre-cultured on callus induction medium for 3 days under dark conditions. At the same time, the OsCM-inserted Agrobacterium cells were grown on selection medium for transformation into callus. Agrobacterium cells were pelleted and resuspended in MS medium fortified with acetosyringone and the callus was immersed in the suspension for 30 min with continuous shaking. After incubation in Agrobacterium cells, the callus was dried for 30 min on sterilized filter paper and then inoculated into co-cultivation medium for 3 days in the dark. The excessive growth of Agrobacterium in the callus was controlled by washing three times with 500 mg/l carbenicillin, dried for 30 min, and again inoculated into the first selection medium containing 50 mg/l hygromycin. The callus was inoculated three times into selection medium under light conditions (16/8 h photoperiod). After three periods of selection, the callus was transferred into regeneration medium for 10 days in the dark. In the second phase, the callus was transferred to new identical medium and placed under light conditions until the plantlets developed; in the third phase, the plantlets were put in a test tube on the same medium to develop roots. After 20 days in a test tube, the plants developed appropriately and were transferred to soil in pots.
Inoculation of BLB and lesion length measurement
Wild-type and OxCM plants were grown in a greenhouse, following the described method. The K3a strain of Xanthomonas oryzae (Xoo) was inoculated into wild-type as well as OxCM plants using the leaf clipping method64. The experiment was conducted in two sets: one set of wild-type and OxCM plants were infected with Xoo, while the second set remained uninfected and was used as a control. The samples were collected in triplicate 0, 1, 6, 12, 24, and 36 h after the inoculation of Xoo for further analysis. However, lesion length was measured after each week until 5 weeks after Xoo infection.
RNA isolation and quantitative RT-PCR analysis
Total RNA was isolated from five leaves in triplicate and cDNA was synthesized using the qPCRBIO cDNA Synthesis Kit from PCRBIOSYSTEMS. Quantitative real-time RT-PCR (qRT-PCR) was performed using qPCRBIO SYBR Green Kit from PCRBIOSYSTEM, using cDNAs as templates and gene-specific primers. To normalize the level of relative expression of each gene, actin was used for each reaction and the expression level was calculated in wild-type plants relative to that in OxCM infected ones. The reaction was performed in a 20 µl volume containing 7 µl of ddH2O, 1 µl of primer, 10 µl of SYBR green, and 1 µl of cDNA, and was repeated in triplicate.
Western blot analysis
To check protein expression in the transgenic line as well as in the wild-type, western blotting was performed by a previously reported optimized method65 with slight modifications. Proteins of the OxCM line and the wild-type treated with BLB were collected at three time points: 2, 6, and 12 h after BLB inoculation. Total protein was isolated with 10 ml of TCA/acetone [10% trichloroacetic acid (TCA); 0.07% β-ME in acetone P.A.], by a previously reported method66. Equal amounts of protein were boiled for 5 min, separated by10% SDS-PAGE at 100 V for 150 min, and then transferred to an NC membrane (Whatman Japan) by a semi-dry method running for 90 min at 19 V using Trans-Blot DS semi-dry transfer cell (Bio-Rad). The membrane was blotted in TBST (0.1% Tween 20 in TBS) and 5% non-fat dry milk (w/v) for 2 h at room temperature. Proteins were further blotted with primary rabbit anti-CM synthase antibodies in 5% non-fat dry milk (w/v) in TBST overnight at 4 °C and subjected to three rinses for 10 min each in TBST solution. The membrane was then incubated with Gt anti-Ms IgG (H + L) secondary antibody (Invitrogen, USA) at a dilution of 1:1000 for 2 h at room temperature and rinsed three times for 10 min each in TBST solution. The blot was developed by Amersham ECL (GE Healthcare, UK) and protein bands were exposed on X-ray film.
Quantification of endogenous SA and JA hormones
To assess SA and JA accumulation in wild-type and OxCM plants in response to Xoo stress, we analyzed both of these hormones. Leaves of the plants were collected in liquid nitrogen after each time point and stored at − 80 °C. For SA analysis, frozen leaves were lyophilized and crushed into a fine powder in liquid nitrogen using a modified version of a previously reported protocol67. The lyophilized powder of each sample (0.3 g) was extracted with 90% ethanol and 100% methanol and centrifuged for 20 min at 1000 rpm. The supernatant was collected and methanol was dried in a vacuum centrifuge and again resuspended in 5% trichloroacetic acid (3 ml). The supernatant was further mixed with ethyl acetate/cyclopentane/isopropanol (49.5:49.5:1, v/v), and the uppermost organic layer was collected in a 4 ml vial and then dried with nitrogen gas. The extracted SA was analyzed by HPLC, with quantification through fluorescence detection. For JA analysis, freeze-dried leaves (0.2 g) were homogenized with liquid nitrogen and JA was extracted with acetone and 50 mM citric acid (70:30, v/v), following a previously reported protocol68,69. An internal standard, [9,10-2H2]-9,10-dihydro-JA (20 ng), was also added to the suspension. The extract was kept at low temperature overnight to evaporate highly volatile organic solvents and retain the less volatile fatty acids. The remaining solution was filtered and then extracted with 10 ml of diethyl ether three times. The extract was further loaded onto a solid-phase extraction cartridge (500 mg of sorbent, aminopropyl) and the cartridges were cleaned with 7.0 ml of 2-propanol and trichloromethane (1:2, v/v). The JA and related standard were eluted with 10 ml of diethyl ether and acetic acid (98:2, v/v). The residue of solvents after evaporation was esterified with diazomethane and analyzed by GS-MS (6890 N network GC system and the 5973 network mass-selective detector; Agilent Technologies, Palo Alto, CA, USA) in the selected ion mode. The ion fragment was monitored at m/z = 83 amu, consistent with the base peaks of JA and [9,10-2H2]-9,10-dihydro-JA; the JA was quantified from the peak areas corresponding to the respective standards.
Measurement of lignin content
Lignin content was assessed following a modified version of a previously reported method58. Samples weighing 0.6 g stored at − 80 °C were ground in liquid nitrogen and washed five times with 95% ethanol to eliminate soluble metabolites. The homogenate was further washed with acetone and dried in an oven. The dried sample was further disrupted in acetic acid using an ultrasonic machine and centrifuged at 3000 × g for 5 min. The remaining pellet was resuspended in 25% acetyl bromide and again centrifuged at 3000 × g for 5 min. The samples were mixed with a mixture of acetic acid and acetyl bromide (4:1, v/v) and heated at 70 °C for 2 h. The samples were cooled at room temperature and transferred to 50 ml of 2 M sodium hydroxide, 1.5 ml of acetic acid, and 7.5 M hydroxylamine hydrochloride. The volume of each sample was equalized with acetic acid. The supernatant’s absorbance was measured with a spectrophotometer at 280 nm (UV-2450; Shimadzu, Japan).
Statistical analysis
All experiments of each section were performed in triplicate, and the data collected from each replicate were pooled together. The data were analyzed using two-way ANOVA followed by Bonferroni post hoc test (significant difference: p ˂ 0.05). A completely randomized design was used to compare the mean values of different treatments. The data were graphically presented and the statistical analyses were performed using GraphPad Prism software (version 5.01; GraphPad, San Diego, CA, USA).
Conclusion
This study has provided evidence that the overexpression of OsCM potentially accelerated resistance to BLB stress in rice. The results revealed that the expression of OsCM significantly induced the accumulation of aromatic amino acids, as well as regulation of the PAL pathway and pathways downstream from AAAs. This induction led to the production of specialized metabolites such as lignin, the accumulation of hormones such as SA and JA, and alteration of some specialized amino acids that protect cells from oxidative damage. Here, we found that AAAs such as Phe and Tyr are not only essential for protein synthesis but also serve as precursors of the PAL pathway, which is responsible for the synthesis of a wide range of secondary metabolites. Increased PAL pathway function by the overexpression of OsCM has a positive impact on BLB resistance. Owing to the high accumulation of Phe and up regulation of the genes responsible for the synthesis of Phe compared with that for Tyr in the transgenic line, it was found that Phe is strongly associated with pathogen-related stress compared with Tyr. SA is an essential regulator of plant defense by inducing the expression of defense-related genes. The SA pathway was upregulated while JA was down regulated in infected transgenic plants, which suggested that SA accumulation enhances NPR1 and Xa7 expression in response to pathogen-related stress. The positive responses of lignin and total amino acid accumulation in response to pathogen-related stress in the OsCM-overexpressing line also indicated that lignin and amino acids are essential parts of plant defense. These results show that AAAs, the PAL pathway, hormones, and pathogen-resistant genes function cooperatively in response to BLB stress in rice.
Supplementary information
Acknowledgements
This work was supported by a Grant from the New breeding technologies development Program (Project No. PJ014793012020), Rural Development Administration, Republic of Korea.
Author contributions
R.J. made contributions as first author. R.J., J.S.B, K.M.K., and I.J.L. designed the study; R.J. performed experiments; M.A.K. and S.A. performed analyses; and R.J., J.S.B. and K.M.K. performed experiments and wrote the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jong-Sup Bae, Email: baejs@knu.ac.kr.
Kyung-Min Kim, Email: kkm@knu.ac.kr.
Supplementary information
is available for this paper at 10.1038/s41598-020-76675-1.
References
- 1.Reddy APK, MacKenzie DR, Rouse DI, Rao AV. Relationship of bacterial leaf blight severity to grain yield of rice. Phytopathology. 1979;69:967–969. doi: 10.1094/Phyto-69-967. [DOI] [Google Scholar]
- 2.Suh JP, et al. Development of breeding lines with three pyramided resistance genes that confer broad-spectrum bacterial blight resistance and their molecular analysis in rice. Rice (New York, N.Y.) 2013;6:5. doi: 10.1186/1939-8433-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tzin V, Galili G. New insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Mol. Plant. 2010;3:956–972. doi: 10.1093/mp/ssq048. [DOI] [PubMed] [Google Scholar]
- 4.Ferrari S, et al. Resistance to Botrytis cinerea induced in Arabidopsis by elicitors is independent of salicylic acid, ethylene, or jasmonate signaling but requires PHYTOALEXIN DEFICIENT3. Plant Physiol. 2007;144:367–379. doi: 10.1104/pp.107.095596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abramovitch RB, Martin GB. Strategies used by bacterial pathogens to suppress plant defenses. Curr. Opin. Plant Biol. 2004;7:356–364. doi: 10.1016/j.pbi.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 6.Alfano JR, Collmer A. Type III secretion system effector proteins: Double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 2004;42:385–414. doi: 10.1146/annurev.phyto.42.040103.110731. [DOI] [PubMed] [Google Scholar]
- 7.Truman W, de Zabala MT, Grant M. Type III effectors orchestrate a complex interplay between transcriptional networks to modify basal defence responses during pathogenesis and resistance. Plant J. Cell Mol. Biol. 2006;46:14–33. doi: 10.1111/j.1365-313X.2006.02672.x. [DOI] [PubMed] [Google Scholar]
- 8.Kocal N, Sonnewald U, Sonnewald S. Cell wall-bound invertase limits sucrose export and is involved in symptom development and inhibition of photosynthesis during compatible interaction between tomato and Xanthomonas campestris pv vesicatoria. Plant Physiol. 2008;148:1523–1536. doi: 10.1104/pp.108.127977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vogt T. Phenylpropanoid biosynthesis. Mol. plant. 2010;3:2–20. doi: 10.1093/mp/ssp106. [DOI] [PubMed] [Google Scholar]
- 10.Bartel B. Auxin biosynthesis. Annu. Rev. Plant Biol. 1997;48:51–66. doi: 10.1146/annurev.arplant.48.1.51. [DOI] [PubMed] [Google Scholar]
- 11.Garcion C, et al. Characterization and biological function of the ISOCHORISMATE SYNTHASE2 gene of Arabidopsis. Plant Physiol. 2008;147:1279–1287. doi: 10.1104/pp.108.119420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho MH, et al. Phenylalanine biosynthesis in Arabidopsis thaliana. Identification and characterization of arogenate dehydratases. J. Biol. Chem. 2007;282:30827–30835. doi: 10.1074/jbc.M702662200. [DOI] [PubMed] [Google Scholar]
- 13.Yamada T, et al. Mutation of a rice gene encoding a phenylalanine biosynthetic enzyme results in accumulation of phenylalanine and tryptophan. Plant Cell. 2008;20:1316–1329. doi: 10.1105/tpc.107.057455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maeda H, et al. RNAi suppression of AROGENATE DEHYDRATASE1 reveals that phenylalanine is synthesized predominantly via the arogenate pathway in petunia petals. Plant Cell. 2010;22:832–849. doi: 10.1105/tpc.109.073247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaminaga Y, et al. Plant phenylacetaldehyde synthase is a bifunctional homotetrameric enzyme that catalyzes phenylalanine decarboxylation and oxidation. J. Biol. Chem. 2006;281:23357–23366. doi: 10.1074/jbc.M602708200. [DOI] [PubMed] [Google Scholar]
- 16.MacDonald MJ, D’Cunha GB. A modern view of phenylalanine ammonia lyase. Biochem. Cell Biol. Biochimie et Biologie Cellulaire. 2007;85:273–282. doi: 10.1139/o07-018. [DOI] [PubMed] [Google Scholar]
- 17.Anterola AM, Lewis NG. Trends in lignin modification: A comprehensive analysis of the effects of genetic manipulations/mutations on lignification and vascular integrity. Phytochemistry. 2002;61:221–294. doi: 10.1016/s0031-9422(02)00211-x. [DOI] [PubMed] [Google Scholar]
- 18.Nair RB, Bastress KL, Ruegger MO, Denault JW, Chapple C. The Arabidopsis thaliana REDUCED EPIDERMAL FLUORESCENCE1 gene encodes an aldehyde dehydrogenase involved in ferulic acid and sinapic acid biosynthesis. Plant Cell. 2004;16:544–554. doi: 10.1105/tpc.017509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Somegowda M, Vigneshwaran V, Rajeshwar AN, Shivashankar S, Pramod SN. Biotic stress induced by Bacterocera cucurbitae (Melon Fly) triggers defense related phenylpropanoid pathway (PPP) and ROS detoxifying enzymes in cucurbits as adaptation. Asian J. Plant Sci. Res. 2017;7:18–29. [Google Scholar]
- 20.Le Thanh T, et al. Salicylic acid-induced accumulation of biochemical components associated with resistance against Xanthomonas oryzae pv. oryzae in rice. J. Plant Interact. 2017;12:108–120. doi: 10.1080/17429145.2017.1291859. [DOI] [Google Scholar]
- 21.Wang D, Amornsiripanitch N, Dong X. A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2006;2:e123. doi: 10.1371/journal.ppat.0020123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Withers J, Dong X. Posttranslational modifications of NPR1: A single protein playing multiple roles in plant immunity and physiology. PLoS Pathog. 2016;12:e1005707. doi: 10.1371/journal.ppat.1005707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verma V, Ravindran P, Kumar PP. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016;16:86. doi: 10.1186/s12870-016-0771-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spoel SH, et al. NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell. 2003;15:760–770. doi: 10.1105/tpc.009159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedrich L, et al. NIM1 overexpression in Arabidopsis potentiates plant disease resistance and results in enhanced effectiveness of fungicides. Mol. Plant Microbe Interact. 2001;14:1114–1124. doi: 10.1094/MPMI.2001.14.9.1114. [DOI] [PubMed] [Google Scholar]
- 26.Das B, Sengupta S, Prasad M, Ghose TK. Genetic diversity of the conserved motifs of six bacterial leaf blight resistance genes in a set of rice landraces. BMC Genet. 2014;15:82. doi: 10.1186/1471-2156-15-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moura JC, Bonine CA, de Oliveira Fernandes Viana J, Dornelas MC, Mazzafera P. Abiotic and biotic stresses and changes in the lignin content and composition in plants. J Integr Plant Biol. 2010;52:360–376. doi: 10.1111/j.1744-7909.2010.00892.x. [DOI] [PubMed] [Google Scholar]
- 28.Alcazar R, et al. Involvement of polyamines in plant response to abiotic stress. Biotech. Lett. 2006;28:1867–1876. doi: 10.1007/s10529-006-9179-3. [DOI] [PubMed] [Google Scholar]
- 29.Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 30.Payyavula RS, Navarre DA, Kuhl JC, Pantoja A, Pillai SS. Differential effects of environment on potato phenylpropanoid and carotenoid expression. BMC Plant Biol. 2012;12:39. doi: 10.1186/1471-2229-12-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dixon RA, Paiva NL. Stress-induced phenylpropanoid metabolism. Plant Cell. 1995;7:1085–1097. doi: 10.1105/tpc.7.7.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.La Camera S, et al. Metabolic reprogramming in plant innate immunity: the contributions of phenylpropanoid and oxylipin pathways. Immunol. Rev. 2004;198:267–284. doi: 10.1111/j.0105-2896.2004.0129.x. [DOI] [PubMed] [Google Scholar]
- 33.Rippert P, Matringe M. Purification and kinetic analysis of the two recombinant arogenate dehydrogenase isoforms of Arabidopsis thaliana. Eur. J. Biochem. 2002;269:4753–4761. doi: 10.1046/j.1432-1033.2002.03172.x. [DOI] [PubMed] [Google Scholar]
- 34.Manela N, et al. Phenylalanine and tyrosine levels are rate-limiting factors in production of health promoting metabolites in Vitis vinifera cv. Gamay Red cell suspension. Front. Plant Sci. 2015;6:538. doi: 10.3389/fpls.2015.00538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oliva M, et al. Enhanced formation of aromatic amino acids increases fragrance without affecting flower longevity or pigmentation in Petunia x hybrida. Plant Biotechnol J. 2015;13:125–136. doi: 10.1111/pbi.12253. [DOI] [PubMed] [Google Scholar]
- 36.Kachroo A, Kachroo P. Fatty acid-derived signals in plant defense. Annu. Rev. Phytopathol. 2009;47:153–176. doi: 10.1146/annurev-phyto-080508-081820. [DOI] [PubMed] [Google Scholar]
- 37.Yan X, Chen S. Regulation of plant glucosinolate metabolism. Planta. 2007;226:1343–1352. doi: 10.1007/s00425-007-0627-7. [DOI] [PubMed] [Google Scholar]
- 38.Shine MB, et al. Cooperative functioning between phenylalanine ammonia lyase and isochorismate synthase activities contributes to salicylic acid biosynthesis in soybean. New Phytol. 2016;212:627–636. doi: 10.1111/nph.14078. [DOI] [PubMed] [Google Scholar]
- 39.Catinot J, Buchala A, Abou-Mansour E, Metraux JP. Salicylic acid production in response to biotic and abiotic stress depends on isochorismate in Nicotiana benthamiana. FEBS Lett. 2008;582:473–478. doi: 10.1016/j.febslet.2007.12.039. [DOI] [PubMed] [Google Scholar]
- 40.Pallas JA, Paiva NL, Lamb C, Dixon RA. Tobacco plants epigenetically suppressed in phenylalanine ammonia-lyase expression do not develop systemic acquired resistance in response to infection by tobacco mosaic virus. Plant J. 1996;10:281–293. doi: 10.1046/j.1365-313X.1996.10020281.x. [DOI] [Google Scholar]
- 41.Yoo H, et al. An alternative pathway contributes to phenylalanine biosynthesis in plants via a cytosolic tyrosine:phenylpyruvate aminotransferase. Nat. Commun. 2013;4:2833. doi: 10.1038/ncomms3833. [DOI] [PubMed] [Google Scholar]
- 42.Less H, Galili G. Principal transcriptional programs regulating plant amino acid metabolism in response to abiotic stresses. Plant Physiol. 2008;147:316–330. doi: 10.1104/pp.108.115733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Subramanian S, Graham MY, Yu O, Graham TL. RNA interference of soybean isoflavone synthase genes leads to silencing in tissues distal to the transformation site and to enhanced susceptibility to Phytophthora sojae. Plant Physiol. 2005;137:1345–1353. doi: 10.1104/pp.104.057257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zabala G, et al. Transcriptome changes in the phenylpropanoid pathway of Glycine max in response to Pseudomonas syringae infection. BMC Plant Biol. 2006;6:26. doi: 10.1186/1471-2229-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Graham TL, Graham MY, Subramanian S, Yu O. RNAi silencing of genes for elicitation or biosynthesis of 5-deoxyisoflavonoids suppresses race-specific resistance and hypersensitive cell death in Phytophthora sojae infected tissues. Plant Physiol. 2007;144:728–740. doi: 10.1104/pp.107.097865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bari R, Jones JD. Role of plant hormones in plant defence responses. Plant Mol. Biol. 2009;69:473–488. doi: 10.1007/s11103-008-9435-0. [DOI] [PubMed] [Google Scholar]
- 47.Robert-Seilaniantz A, Navarro L, Bari R, Jones JD. Pathological hormone imbalances. Curr. Opin. Plant Biol. 2007;10:372–379. doi: 10.1016/j.pbi.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 48.Cui J, et al. Pseudomonas syringae manipulates systemic plant defenses against pathogens and herbivores. Proc. Natl. Acad. Sci. USA. 2005;102:1791–1796. doi: 10.1073/pnas.0409450102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laurie-Berry N, Joardar V, Street IH, Kunkel BN. The Arabidopsis thaliana JASMONATE INSENSITIVE 1 gene is required for suppression of salicylic acid-dependent defenses during infection by Pseudomonas syringae. Mol. Plant-Microbe Interact. MPMI. 2006;19:789–800. doi: 10.1094/mpmi-19-0789. [DOI] [PubMed] [Google Scholar]
- 50.Kempema LA, Cui X, Holzer FM, Walling LL. Arabidopsis transcriptome changes in response to phloem-feeding silverleaf whitefly nymphs. Similarities and distinctions in responses to aphids. Plant Physiol. 2007;143:849–865. doi: 10.1104/pp.106.090662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adie BA, et al. ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell. 2007;19:1665–1681. doi: 10.1105/tpc.106.048041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dong X. NPR1, all things considered. Curr. Opin. Plant Biol. 2004;7:547–552. doi: 10.1016/j.pbi.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 53.Webb KM, et al. A benefit of high temperature: Increased effectiveness of a rice bacterial blight disease resistance gene. New Phytol. 2010;185:568–576. doi: 10.1111/j.1469-8137.2009.03076.x. [DOI] [PubMed] [Google Scholar]
- 54.Hopkins CM, White FF, Choi SH, Guo A, Leach JE. (1992) Identification of a family of avirulence genes from Xanthomonas oryzae pv. oryzae. Mol. Plant-Microbe Interact. MPMI. 1992;5:451–459. doi: 10.1094/mpmi-5-451. [DOI] [PubMed] [Google Scholar]
- 55.Cohen SP, et al. RNA-Seq analysis reveals insight into enhanced rice Xa7-mediated bacterial blight resistance at high temperature. PLoS ONE. 2017;12:e0187625. doi: 10.1371/journal.pone.0187625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spoel SH, Johnson JS, Dong X. Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc. Natl. Acad. Sci. USA. 2007;104:18842–18847. doi: 10.1073/pnas.0708139104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Durner J, Klessig DF. Inhibition of ascorbate peroxidase by salicylic acid and 2,6-dichloroisonicotinic acid, two inducers of plant defense responses. Proc. Natl. Acad. Sci. USA. 1995;92:11312–11316. doi: 10.1073/pnas.92.24.11312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng M, et al. Manipulation of lignin metabolism by plant densities and its relationship with lodging resistance in wheat. Sci. Rep. 2017;7:41805. doi: 10.1038/srep41805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mauch-Mani B, Slusarenko AJ. Production of salicylic acid precursors is a major function of phenylalanine ammonia-lyase in the resistance of arabidopsis to Peronospora parasitica. Plant Cell. 1996;8:203–212. doi: 10.1105/tpc.8.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cass CL, et al. Effects of PHENYLALANINE AMMONIA LYASE (PAL) knockdown on cell wall composition, biomass digestibility, and biotic and abiotic stress responses in Brachypodium. J. Exp. Bot. 2015;66:4317–4335. doi: 10.1093/jxb/erv269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vanholme R, et al. A systems biology view of responses to lignin biosynthesis perturbations in Arabidopsis. Plant Cell. 2012;24:3506–3529. doi: 10.1105/tpc.112.102574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hildebrandt TM, Nesi AN, Araújo WL, Braun H-P. Amino acid catabolism in plants. Mol. Plant. 2015;8:1563–1579. doi: 10.1016/j.molp.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 63.Pratelli R, Pilot G. Regulation of amino acid metabolic enzymes and transporters in plants. J. Exp. Bot. 2014;65:5535–5556. doi: 10.1093/jxb/eru320. [DOI] [PubMed] [Google Scholar]
- 64.Chen H, Wang S, Zhang Q. New gene for bacterial blight resistance in rice located on chromosome 12 identified from minghui 63, an elite restorer line. Phytopathology. 2002;92:750–754. doi: 10.1094/phyto.2002.92.7.750. [DOI] [PubMed] [Google Scholar]
- 65.Hao P, et al. Roles of NlAKTIP in the growth and eclosion of the rice brown planthopper, Nilaparvata lugens Stål, as revealed by RNA interference. Int. J. Mol. Sci. 2015;16:22888–22903. doi: 10.3390/ijms160922888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu C, Xu Y, Huang B. Protein extraction for two-dimensional gel electrophoresis of proteomic profiling in turfgrass. Crop Sci. 2008;48:1608–1614. doi: 10.2135/cropsci2007.11.0624. [DOI] [Google Scholar]
- 67.Jan R, Khan MA, Asaf S, Lee I-J, Kim KM. Metal resistant endophytic bacteria reduces cadmium, nickel toxicity, and enhances expression of metal stress related genes with improved growth of Oryza sativa, via regulating its antioxidant machinery and endogenous hormones. Plants. 2019;8:363. doi: 10.3390/plants8100363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Khan MA, et al. Gibberellin application ameliorates the adverse impact of short-term flooding on Glycine max L. Biochem. J. 2018 doi: 10.1042/bcj20180534. [DOI] [PubMed] [Google Scholar]
- 69.McCloud ES, Baldwin IT. Herbivory and caterpillar regurgitants amplify the wound-induced increases in jasmonic acid but not nicotine in Nicotiana sylvestris. Planta. 1997;203:430–435. doi: 10.1007/s004250050210. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.