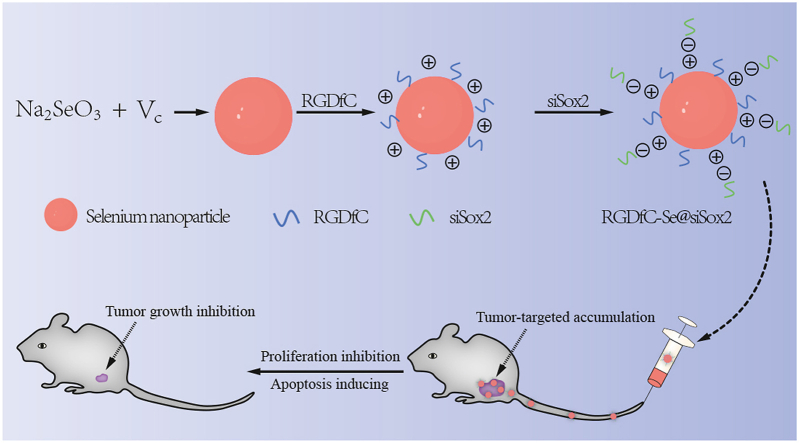
Abstract
RNA interference (RNAi) is one of the most promising methods for the treatment of malignant tumors. However, developing an efficient biocompatible delivery vector for small interfering RNA (siRNA) remains a challenging issue. This study aimed to prepare a non-viral tumor-targeted carrier, named RGDfC-modified functionalized selenium nanoparticles (RGDfC-SeNPs). RGDfC-SeNPs were used to selectively deliver siSox2 to HepG2 liver cancer cells and tissues for the treatment of hepatocellular carcinoma (HCC). In the current study, RGDfC-SeNPs were successfully synthesized and characterized. It was shown that RGDfC-SeNPs could effectively load siSox2 to prepare an antitumor prodrug RGDfC-Se@siSox2. RGDfC-Se@siSox2 exhibited selective uptake in HepG2 liver cancer cells and LO2 normal liver cells, indicating RGDfC-SeNPs could effectively deliver siSox2 to HepG2 liver cancer cells. RGDfC-Se@siSox2 entered HepG2 cells via clathrin-mediated endocytosis by firstly encircling the cytoplasm and then releasing siSox2 in the lysosomes. RGDfC-Se@siSox2 could effectively silence Sox2 and inhibit the proliferation, migration and invasion of HepG2 cells. RGDfC-Se@siSox2 induced HepG2 cells apoptosis most likely via overproduction of reactive oxygen species and disruption of the mitochondrial membrane potentials. Most importantly, RGDfC-Se@siSox2 significantly inhibited the tumor growth in HepG2 tumor-bearing mice without obvious toxic side effects. These studies indicated that RGDfC-SeNPs may be an ideal gene carrier for delivering siSox2 to HepG2 cells and that RGDfC-Se@siSox2 may be a novel and highly specific gene-targeted prodrug therapy for HCC.
Keywords: Nanoparticle, Cancer, Gene therapy, siRNA delivery, Sox2
Graphical abstract
Highlights
-
•
The biocompatible selenium nanoparticle (RGDfC-SeNPs) selectively deliver siRNA to HepG2 cells.
-
•
RGDfC-Se@siSox2 induced HepG2 cells apoptosis most likely via ROS overproduction and disruption of MMP.
-
•
RGDfC-Se@siSox2 exhibited excellent in vivo antitumor efficacy without obvious toxic side effect.
1. Introduction
Hepatocellular carcinoma (HCC) is one of prevalent cancers with increasing incidence and high mortality rates globally over the past decade [[1], [2], [3]]. Conventional treatments for HCC are limited and largely ineffective necessitating an urgent need for new treatment options [4,5]. RNA interference (RNAi) is a very efficient gene-silencing technology that is developing as a novel promising cancer therapeutic option [6]. Small interfering RNA (siRNA) refers to effector molecules of RNAi that can selectively silence expression of target genes in a sequence‐specific manner [7,8]. The effective silencing of functionalized targeted genes by siRNA is a powerful tool to inhibit cancer cell survival, migration, invasion and tumorigenicity. However, the safe and efficient delivery of siRNA into cancer cells remains challenging [9,10]. Traditional viral gene carriers have been widely used in clinical studies but the toxicity and immunogenicity of viral gene carriers have hindered their widespread clinical application [11,12]. Non-viral gene vectors are an attractive approach due to their bio-safety and easy modification and are attracting significant attention from researchers in the field of gene delivery systems [13,14].
Selenium nanoparticles (SeNPs) are superior non-viral gene carriers that have been widely investigated due to attractive properties including biocompatibility, biodegradability, low toxicity and permeability enhancing features [[15], [16], [17]]. In this study, SeNPs were used as a gene vector to deliver specific gene-targeted siRNA to cancer cells for the treatment of HCC. However, the tumor-targeted gene delivery ability of siRNA is still a crucial issue in the design of gene carriers [18,19]. Therefore, to enhance the tumor-targeted capability of SeNPs, a positively charged peptide RGDfC (Arg-Gly-Asp-DPhe-Cys) was used to modify the surface of SeNPs to fabricate functionalized selenium nanoparticles RGDfC-SeNPs. RGDfC is specifically capable of binding with αvβ3 integrin that is overexpressed in cancer cells [20]. In addition, positively charged RGDfC can contribute to the link between nanoparticles and siRNA via their electrostatic interactions [21].
Sox2 as a critical transcriptional regulator is overexpressed in the majority of solid cancers, such as HCC, lung cancer, colorectal and breast cancer [22]. Overexpression of Sox2 promotes the proliferation, migration, invasion and tumorigenicity of cancer cells [23]. Thus, knockdown of Sox2 expression can effectively inhibit these events. Previous studies have reported that the level of Sox2 in normal liver is much lower compared to HCC tissues [24]. Therefore, the Sox2 gene has emerged as a crucial target for HCC therapy. In the current paper, we fabricated RGDfC-modified functionalized selenium nanoparticles (RGDfC-SeNPs) which were used to load siSox2 and fabricate the nanoscale anticancer prodrug RGDfC-Se@siSox2, aiming at silencing the gene expression of Sox2 in liver cancer cells and tumor tissues. The anti-hepatocellular carcinoma efficacy and mechanisms of RGDfC-Se@siSox2 were systematically elucidated by a HepG2 cell model and a HepG2 tumor model.
2. Materials and methods
2.1. materials
Lyso-tracker red, ascorbic acid (vitamin C, Vc), propidium iodide (PI), sodium selenite and DAPI were purchased from Sigma (MO, USA). All antibodies were purchased from CST (MA, USA). Fetal bovine serum (FBS) was purchased from Gibco. siRNA was provided by RIBRIO Co., Ltd (Guangzhou, China) directly against the sequence of siSox2: 5′-CCCGCAUGUACAACAUGAUUU-3’.
2.2. Fabrication and characterizations of RGDfC-SeNPs
RGDfC-modified SeNPs (RGDfC-SeNPs) were synthesized according to previous methods with a minor revision [25]. In brief, 0.25 mL of 20 mM Na2SeO3 and 80 mM Vc stock solutions were freshly prepared. 5 mL of Na2SeO3 solution was dripped into an equal volume of Vc solution and the mixed solutions were stirred for 30 min. 5 mL of RGDfC solution (1.5 mg/mL) was dripped into the mixed solutions that were further stirred for 1 h. The excess Na2SeO3, Vc and RGDfC were removed by dialysis for 12 h. The concentration of RGDfC-SeNPs was determined by ICP-MS analysis. Elemental composition analysis of R-SeNPs was carried out using energy dispersive X-Ray (EDX) [26]. The morphology of RGDfC-SeNPs was visualized by transmission electron microscope (TEM). The zeta potential and size distributions of nanoparticles were examined using a Zetasizer Nano ZS particle size analyzer [27].
2.3. Gel electrophoresis assay
The affinity of RGDfC-SeNPs to siSox2 was determined by a gel mobility shift assay [28]. The RGDfC-Se@siSox2 complexes were prepared at RGDfC-SeNPs/siSox2 weight ratios at the range of 1:1 to 20:1 and incubated for 30 min. The samples were loaded on an agarose gel (2%) and run for electrophoretic mobility at 100 V for 20 min siSox2 retardation was visualized using a gel documentation system. To study the binding stability, degradation of siSox2 under physiology condition, the naked siSox2 and RGDfC-Se@siSox2 in 50% Fetal Bovine Serum (FBS) was investigated using the similar method mentioned above.
2.4. Cellular uptake and lysosome escape of RGDfC-Se@siSox2
Selective cellular uptakes of RGDfC-Se@siSox2 in both of LO2 cells and HepG2 cells were investigated using fluorescence microscopy. Briefly, 5 × 105 LO2 cells and 2 × 105 HepG2 cells in a 6-well plate were incubated for 24 h to adhere the bottom of cell plate. The cells were co-cultured with RGDfC-Se@siSox2 (200 nM equivalent concentration of FAM-labeled siSox2) for different times and the nuclei were stained with DAPI for 15 min. To further verify whether RGDfC-mediated uptake occurred in HepG2 cell, a competitive inhibition experiment has been performed. In brief, HepG2 cells were pretreated with free RGDfC (0.5 μg/mL) for 2 h. Subsequently, the cells were washed with cold PBS and exposed to RGDfC-Se@siSox2 for 1 h, 2 h and 4 h. Finally, cells were washed and the imaging of cells was captured under a fluorescence microscope.
The uptake of RGDfC-Se@siSox2 in HepG2 cells was further demonstrated by transmission electron microscope (TEM). Briefly, HepG2 cells were exposed to RGDfC-Se@siSox2 (200 nM equivalent dose of siSox2) for 24 h and washed with PBS. Cells were then digested with trypsin, fixed with 3% glutaraldehyde overnight and post-fixed in 1% osmium tetroxide before being dehydrated using ethanol. Samples were then processed and embedded in Spurr's resin. Sections were loaded onto the copper grid and contrasted with lead citrate and uranyl acetate. The samples were analyzed using a TEM [29].
The lysosome escape of RGDfC-Se@siSox2 in HepG2 cells was investigated using LysoTracker® Red that indicates endosome site. HepG2 cells were processed according to the description above and then stained with LysoTracker Red (80 nM) for 30 min. Cells were washed and visualized by fluorescence microscopy.
2.5. Endocytosis pathways of RGDfC-Se@siSox2
For the energy-dependent effect on uptake, HepG2 cells were pre-incubated for 60 min at either 4 °C or 37 °C with NaN3/DOG (2-deoxy-d-glucose). To further study the endocytosis pathways by which RGDfC-Se@siSox2 entered HepG2 cells, three inhibitors, amiloride (macropinocytosis), nystatin (caveolae-mediated endocytosis) and chlorpromazine (clathrin-associated endocytosis) were used to pretreat HepG2 cells for 60 min. Cells were treated with RGDfC-Se@siSox2 for 4 h and then washed with PBS. Finally, the uptake of RGDfC-Se@siSox2 in HepG2 cells was detected using inductively coupled plasma mass spectrometry (ICP-MS) [30].
2.6. Release of siSox2 from RGDfC-SeNPs
The release of si Sox2 was tested by RGDfC-SeNPs containing FAM-siSox2. To analyze the pH-sensitive release profile, RGDfC-Se@FAM-siSox2 containing 1 μg of FAM-siSox2 was dissolved in PBS at pH 7.4 or 5.4, and dialyzed with a dialysis bag (50 kDa). The dialysis bag was placed in PBS and stirred at 100 rpm. A small volume of the media was removed at intervals and the equal fresh PBS was supplemented into the media. The concentrations of released siSox2 were determined by measuring fluorescence intensities [31].
2.7. Quantitative real-time
PCR Sox2 mRNA levels were tested via quantitative real-time PCR. HepG2 cells were exposed to RGDfC-Se@siSox2 or RGDfC-Se@siNC for 4 h and then previous medium was exchanged with fresh one. The treated cells were incubated for 48 h at 37 °C and collected to analyze mRNA level of Sox2 gene. Trizol reagent (Takara Biotechnology, China) was employed to extract the total RNA and the synthesis of cDNA was conducted using the Prime Script™ RT reagent kit in accordance with the manufacturer's instruction. The relative mRNA expression analysis of Sox2 gene was carried out via the 2-ΔΔCT method [32].
2.8. Cytotoxicity assays
The cytotoxicity of RGDfC-Se@siNC and RGDfC-Se@siSox2 was identified by MTT assay [33]. 5 × 103 HepG2 cells per well in 96-well plate were incubated for 12 h, and then exposed to RGDfC-Se@siNC or RGDfC-Se@siSox2 with different concentrations of siRNA. The untreated cells were used as controls. After 48 h of incubation, cells were treated with 20 μL of MTT (0.5 mg/mL) for another 4 h. Then, the formazan product was solubilized using 150 μL of dimethyl sulfoxide. The absorbance was set at 570 nm. The cytotoxicity of RGDfC-Se@siSox2 against LO2 cells was detected using similar method. Cell viabilities were determined according to the formula: Cell viabilities = Asample-Ablank/Acontrol-Ablank
2.9. Wound scratch and cell invasion assays
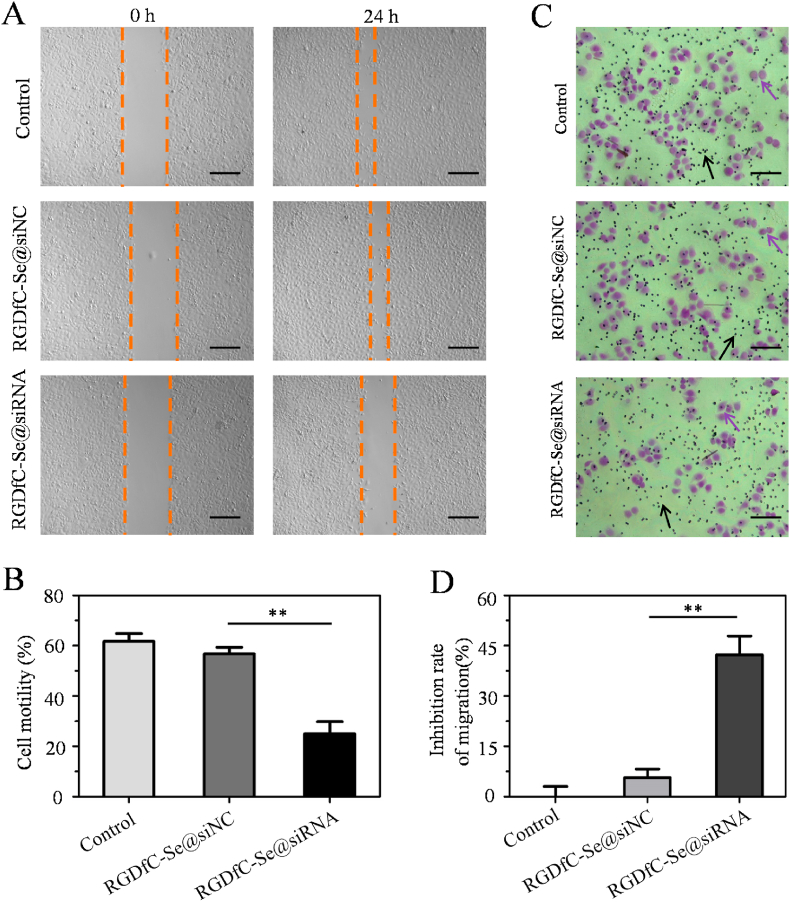
The wound scratch assay was applied to demonstrate the migration of HepG2 cells [34]. Briefly, the mono-layer cells were scratched using a 200 μL pipette tip to create a scratch in a line and the detached cells were cleared away using PBS. The HepG2 cells were then supplemented with fresh medium and exposed to RGDfC-Se@siSox2 or RGDfC-Se@siNC at a concentration of 100 nM siRNA for 24 h. Representative photographs were taken under a microscope. The migration rate was quantified as: cell motility (%) = (the migrated cell surface width during 24 h/the wound width at 0 h) × 100.
For invasion assays, HepG2 cells in medium containing 2% FBS were added to the upper chambers. The bottom chambers were added with 0.4 mL of complete medium. After 24 h of incubation with RGDfC-Se@siSox2 or RGDfC-Se@siNC (100 nM siRNA), cells passed through the membrane. A cotton swab was used to gently remove the upper cells. Cells on the lower surface were visualized by staining with crystal violet and counted under a microscope. The cell migration inhibition rate was quantified as follows: Inhibition of migration (%) = (Migration cellscontrol - Migration cellstreated)/Migration cellscontrol × 100. Each assay was carried out at least in triplicate.
2.10. Cell cycle and apoptosis were analyzed by flow cytometer
To examine the effect of Sox2-silencing on cell cycle, 2 × 105 HepG2 cells in 6-well plates were incubated overnight and co-cultured with RGDfC-Se@siSox2 and RGDfC-Se@siNC (100 nM siRNA). After 24 h, cells were fixed with 70% ice-cold ethanol and stained with PI solution. Cell cycle distribution analysis was carried out by a FACS flow cytometer. For cellular apoptosis examination, cells were treated using a similar approach mentioned above. Collected cells were examined by FACS flow cytometer after staining with FITC-Annexin V/PI [35].
2.11. Intracellular reactive oxygen species
(ROS) and mitochondrial membrane potential (MMP) measurementsThe ROS level of HepG2 cells was tested as previously described [36]. In brief, treated cells were co-incubated with 10 μM of DCFH-DA for 25 min, rinsed in PBS and photographed using a fluorescence micrograph. For mitochondrial membrane potential (MMP) examination, the treated HepG2 cells were treated with JC-1 (10 μg/mL) for 20 min and tested via a FACS flow cytometer.
2.12. Living images
All of the animal experiments were approved by the Animal Experimental Ethical Committee of Guangzhou Medical University. HepG2 tumor-bearing nude mice models were used to assess in vivo biodistributions of RGDfC-Se@siSox2 using cy5.5 labeling. BALB/c nude mice (6-week-old, n = 3) were subcutaneously inoculated with 5 × 106 HepG2 cells in the left hind leg. 3 mg/kg of RGDfC-Se@cy5.5-siSox2 was intravenously injected into mice after the tumor reached a volume of 200–300 mm3. Mice without intravenous injection were set as controls. Fluorescence images of the mice after injection for 3 and 6 h were acquired using an IVIS imaging system (Xenogen, USA) [37].
2.13. In vivo therapeutic efficacy in HepG2 tumor-bearing mice
A HepG2 tumor xenograft model was established to assess the therapeutic efficacy of RGDfC-Se@siSox2. 5 × 106 HepG2 cells were subcutaneously (s.c.) injected into the left hind legs of mice (n = 6). Tumor-bearing mice were intravenously injected with saline (control), RGDfC-Se@siSox2 (3 mg/kg) or RGDfC-Se@siNC (3 mg/kg) after the tumors reached a volume of 100 mm3. Mice were weighted and the volumes of tumor were recorded once every three days during 21 days of treatment. The volumes of tumor were calculated using the following formula (length × width2)/2. Major organs and tumors were excised from the sacrificed mice and the images of tumors were photographed. Major organs and tumors were fixed in 3.7% paraformaldehyde for more than three days and then sectioned into 5-μm slice. Tissue sections of major organs were prepared by hematoxylin and eosin staining. The expressions of caspase-3, CD31, ki67 and phosphorylated p53 (pp53) in tumors were measured by immuno-histochemical staining [38].
2.14. Statistical analysis
All experiments are repeated at least three times. Data are presented as the means ± SD. A One-way ANOVA test was used to determine significance among groups. *p < 0.05 and **p < 0.01 were considered significant and highly significant, respectively.
3. Results and discussion
3.1. Characterization of nanoparticles
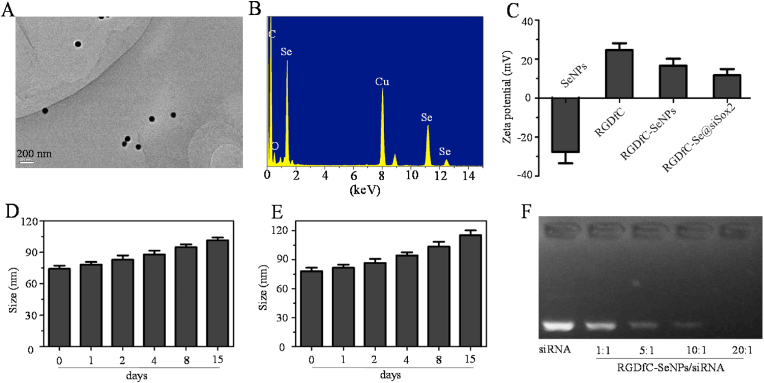
The morphology and size of the prepared functionalized selenium nanoparticles RGDfC-SeNPs was visualized by transmission electron microscopy (TEM). From Fig. 1A, RGDfC-SeNPs exhibited uniform spherical morphology with a size of ~75 nm. The elemental compositions of RGDfC-SeNPs are shown in Fig. 1B. The signals of C and O from RGDfC appeared in the EDS spectra, indicating successful link between RGDfC and SeNPs. The signal of Cu originated from the copper mesh. The zeta potential of RGDfC-SeNPs was around +16 mV after RGDfC modification, which was prone to the load negative charge of siSox2 (Fig. 1C), and the zeta potential of RGDfC-Se@siSox2 was approximately +12 mV. As shown in Fig. 1D&E, the average sizes of RGDfC-SeNPs in water and PBS were observed for 15 days. The results showed that the size of RGDfC-SeNPs gradually increased from ~75 to ~115 nm.
Fig. 1.
Characteristics of nanoparticle carrier RGDfC-SeNPs. (A) The morphology and size characterization of RGDfC-SeNPs by TEM. (B) Elemental composition examination of RGDfC-SeNPs using energy dispersive X-ray spectroscopy (EDX). (C) Zeta potential distributions of SeNPs, RGDfC, RGDfC-SeNPs and RGDfC-Se@siSox2. In vitro stability of RGDfC-SeNPs in water solution (D) and phosphate buffer saline solution (E). (F) Agarose gel electrophoresis image of RGDfC-Se@siSox2 nanoparticles at different RGDfC-SeNPs/siSox2 weight ratios.
3.2. Gene binding ability of RGDfC-SeNPs
An agarose gel assay was adopted to investigate the complex efficiencies of the positively charged nanoparticles RGDfC-SeNPs for siSox2. RGDfC-Se@siSox2 was prepared at different RGDfC-SeNPs/siSox2 weight ratios (w.t.) and the movement of siSox2 was observed by agarose gel electrophoresis. As seen in Fig. 1F, free siSox2 could easily migrate to the opposite end. However, siSox2 binding with the RGDfC-SeNPs was gradually retarded when the weight ratio increased from 1:1 to 20:1. The siSox2 migration was completely retarded when the weight ratio of RGDfC-SeNPs/siSox2 was 20:1, indicating the effective loading of siSox2 in RGDfC-SeNPs. In order to ensure stable and efficient delivery of siSox2, the RGDfC-SeNPs/siSox2 weight ratio of 20:1 was chosen to prepare RGDfC-Se@siSox2.
The instability of siRNA in the physiological environment due to its susceptibility to serum-nuclease catalyzed degradation, is a major limitation in RNA interference (RNAi)-based gene therapy. Therefore, we studied the siSox2 protection efficiency of RGDfC-SeNPs and its ability to prevent siSox2 degradation in the presence of serum. As shown in Fig. S1, Gel retardation assay showed that, the naked siSox2 was partially degraded after 1 h of incubation in 50% serum and almost totally degraded after 2 h of incubation. However, the siSox2 loaded onto the RGDfC-SeNPs was efficiently protected when incubated in the presence of 50% FBS during 2 h. This finding suggests that the RGDfC-SeNPs can prevent the siSox2 from degradation by serum nucleases.
3.3. Selective delivery of siSox2 and lysosomes escape in HepG2 cells
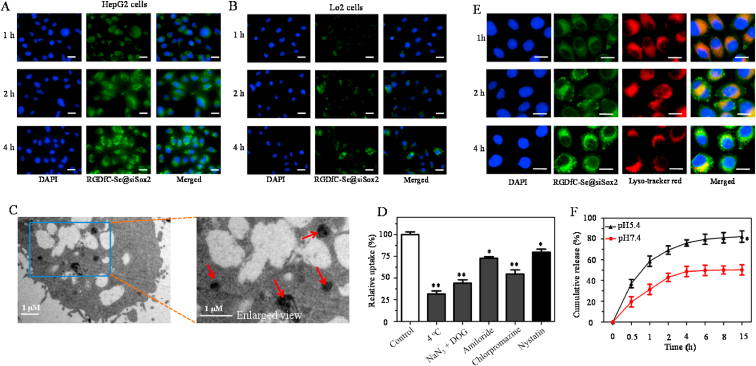
The selective delivery of siRNA to cancer cells is a very important issue for the anticancer therapeutic application of siRNA [39]. The uptake levels of the FAM-labeled RGDfC-Se@siSox2 in HepG2 cells and normal liver LO2 cells were determined by a fluorescence microscope. From Fig. 2A&B, uptakes of RGDfC-Se@siSox2 in both cells increased with time, indicating RGDfC-Se@siSox2 entered cells in a time-dependence manner. HepG2 cells exhibited significantly stronger fluorescence signals than LO2 cells, suggesting RGDfC-modified functionalized selenium nanoparticles could selectively deliver siSox2 to cancer cells. A competitive inhibition experiment has been performed to further verify whether RGDfC-mediated uptake occurred in HepG2 cell. Free RGDfC was added to HepG2 cells first to block the interaction between RGDfC and its receptor αvβ3, and then RGDfC-Se@siSox2 was added to the medium to study whether less RGDfC-Se@siSox2 were taken by HepG2 cells. As shown in figS2,Fig. S2 the pretreatment with free RGDfC in HepG2 cells resulted in less uptake of RGDfC-Se@siSox2 compared to pretreated HepG2 cells, suggesting RGDfC-mediated targeting played an important role in the uptake of RGDfC-Se@siSox2 in HepG2 cells. In addition, TEM imaging was further used to demonstrate the internalization of RGDfC-Se@siSox2 into HepG2 cells. From Fig. 2C, RGDfC-Se@siSox2 mainly gathered in the cytoplasm of HepG2 cells after 2 h of co-incubation. The above results consistently verified that RGDfC-Se@siSox2 had significant cellular uptake in HepG2 cells.
Fig. 2.
(A) Cellular uptakes of RGDfC-Se@siSox2FAM (100 nM) were analyzed by a fluorescence microscope after 1 h, 2 h and 4 h of incubation in HepG2 cells (A) and LO2 cells (B). Scale bars = 20 μm. (C) TEM image of HepG2 cells after exposure to RGDfC-Se@siSox2 for 24 h. Red arrows indicate the areas where the RGDfC-Se@siSox2 is present. (D) Effects of low temperature and endocytic inhibitors on cellular uptake of RGDfC-Se@siSox2. *p < 0.05, **p < 0.01 vs control. (E) The observation of the escape of RGDfC-Se@siSox2FAM from lysosomes after incubation for different times. Scale bars = 10 μm. (F) In vitro siSox2 release from RGDfC-Se@siSox2 in PBS at different pH values. *p < 0.05 vs pH 7.4.
3.4. RGDfC-Se@siSox2 enters HepG2 cells by clathrin-mediated endocytosis
Endocytosis is a key cellular process that leads to the internalization of nanoparticles in cancer cells [40]. Many previous studies have shown that selenium nanoparticles enter cancer cells via an energy-dependent endocytic manner. As shown in Fig. 2D, the uptake of RGDfC-Se@siSox2 by cells incubated at 4 °C or co-incubated with NaN3/DOG (a cell energy metabolism inhibitor) at 37 °C was dramatically decreased to approximately 25% compared to the untreated group (control), showing an active and energy-dependent internalization of RGDfC-Se@siSox2 in HepG2 cells. To further explore the manner of endocytosis by which RGDfC-Se@siSox2 entered the cells, three chemical substances including amiloride, nystatin and chlorpromazine, were used as endocytosis inhibitors for the macropinocytosis, caveolae-mediated endocytosis and clathrin-associated endocytosis, respectively. Fig. 2D shows that the uptakes of RGDfC-Se@siSox2 in the amiloride and nystatin-treated groups were reduced by 27.3% and 20.3%, respectively. However, chlorpromazine treatment resulted in maximum inhibition (45.4%) of intracellular uptake of RGDfC-Se@siSox2, indicating RGDfC-Se@siSox2 entered HepG2 cells via endocytosis way and clathrin-mediated endocytosis plays a major role in the internalization of RGDfC-Se@siSox2.
3.5. Lysosomes escape of RGDfC-Se@siSox2
The selective delivery of siRNA to cancer cells by nanoparticles is essential to guarantee effective gene silencing [41]. Nanoparticles are always trafficked into lysosomes in cancer cells and the endocytosed siRNA must escape from lysosomes to reveal gene-silencing efficacy. Therefore, the escape of siSox2 from lysosomes was assessed by observing the co-localization of lysosomes (lysotracker red) and RGDfC-Se@siSox2. The RGDfC-Se@siSox2 (FAM-labeled siSox2) was visualized as green fluorescence. As shown in Fig. 2E, at 1 h, the yellow fluorescence produced by the merging of the lysosomes (red) and RGDfC-Se@siSox2 (green) was observed, indicating that the RGDfC-Se@siSox2 accumulated in the lysosomes. However, from 2 to 4 h of incubation, some portions of the green fluorescence started to dissociate from the red areas, suggesting that siSox2 could successfully escape from lysosomes. The endosome escape of siSox2 could reduce siSox2 degradation in the lysosome and improve the efficacy of gene-silencing.
3.6. Release of siRNA
The in vitro release of siSox2 from RGDfC-Se@siSox2 was carried out in PBS under pH 5.4 and pH 7.4, mimicking the lysosome environment in tumors and normal physiological environment, respectively. From Fig. 2F, RGDfC-Se@siSox2 exhibited a burst release of siSox2 at both pH 5.4 (58.6%) and pH 7.4 (31.3%) within 1 h. The siSox2 release under pH 7.4 achieved a plateau at 4 h and the release rate was approximately 48.7%. The release rate of siSox2 at pH 5.4 reached 76.1% within 4 h and maintained a slight upward trend. RGDfC-Se@siSox2 indicated an obviously higher release rate of Sox2 at pH 5.4 (82.4%) compared to that at pH 7.4 (50.3%) during 15 h. The above results showed that RGDfC-Se@siSox2 exhibited a rapid release of siSox2 in an acidic environment allowing for efficient delivery to the targeted cancer cells.
3.7. Gene silencing efficacy and toxicity of RGDfC-Se@siSox2
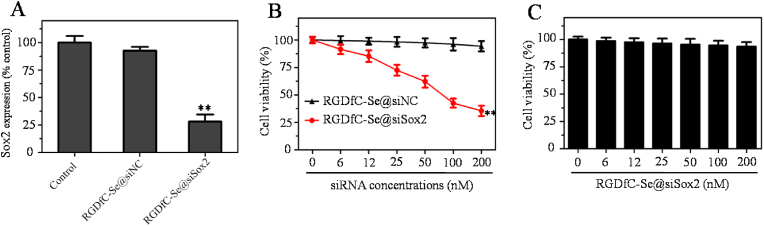
The silencing efficiency of Sox2 gene was assessed by real-time qPCR [42]. RGDfC-Se@siSox2 or RGDfC-Se@siNC with 100 nM equivalent siRNA was incubated with HepG2 cells for 48 h and mRNA expression levels of the cells were tested. As shown in Fig. 3A, the RGDfC-Se@siSox2 significantly decreased the expression of mRNA compared to the control (non-treated cells) and RGDfC-Se@siNC groups. These data confirmed the sequence specificity of siSox2. The results of qPCR showed that RGDfC-Se@siSox2 could efficiently silence the Sox2 gene in HepG2 cells.
Fig. 3.
(A) Gene silencing ability of RGDfC-Se@siSox2 or RGDfC-Se@siNC in HepG2 cells. The concentration of siRNA was 200 nM **p < 0.01 vs control. (B) Viabilities of HepG2 cells treated with RGDfC-Se@siSox2 or RGDfC-Se@siNC for 48 h **p < 0.01 vs RGDfC-Se@siNC. (C) Cell viability of LO2 cells incubated with RGDfC-Se@siSox2 for 48 h.
The cytotoxicity of RGDfC-Se@siSox2 was evaluated by MTT assay. From Fig. 3B, the proliferation of HepG2 cells was inhibited by RGDfC-Se@siSox2 at equivalent siSox2 concentrations in the range of 6–200 nM for 48 h. However, there was no significant cytotoxicity for RGDfC-Se@siNC under the same conditions, further confirming the sequence specificity of siSox2. To further investigate the biocompatibility of RGDfC-Se@siSox2, the cytotoxicity of RGDfC-Se@siSox2 against LO2 normal liver cells was also considered (Fig. 3C). The results showed that RGDfC-Se@siSox2 exhibited no significant cytotoxicity against LO2 cells. Taking together, RGDfC-Se@siSox2 had the capability of inhibiting the proliferation of HepG2 cells with good biocompatibility.
3.8. RGDfC-Se@siSox2 inhibits migration and invasion of HepG2 cells
Metastatic and invasive abilities are significant characteristics of malignant tumors [43]. Thus, the wound scratch and trans-well assays were performed to determine whether knockdown of Sox2 expression by RGDfC-Se@siSox2 could inhibit the migration and invasion behaviors of HepG2 liver cancer cells. From Fig. 4A&B, the wound repair ability in RGDfC-Se@siSox2-treated cells was delayed in comparison to the untreated control and RGDfC-Se@siNC-treated cells, indicating RGDfC-Se@siSox2 has an inhibitory effect on cell migration. However, RGDfC-Se@siNC (negative control group), had almost no effect on cell motility. In addition, cell invasion inhibition was evaluated using a Transwell assay after the cells had been exposed to RGDfC-Se@siSox2 or RGDfC-Se@siNC. As shown in Fig. 4C&D, we observed a distinct decrease in the number of penetrated cells in RGDfC-Se@siSox2-treatment group compared to untreated control group, indicating that RGDfC-Se@siSox2 could suppress HepG2 cell invasion. As expected, RGDfC-Se@siNC did not obviously inhibit cell invasion. Taken together, the aforementioned results indicated that RGDfC-Se@siSox2 was able to inhibit the migration and invasion behaviors of HepG2 cells by silencing Sox2 expression.
Fig. 4.
(A) RGDfC-Se@siSox2 suppressed the migration of HepG2 cells for 24 h by wound healing assay. Scale bar is 400 mm. (B) Quantitative assessment of migration rate. **p < 0.01 vs RGDfC-Se@siNC. (C) RGDfC-Se@siSox2 suppressed the invasion of HepG2 cells for 24 h by transwell invasion assay. Purple arrows indicate the cells which passed through the membrane. Black arrows indicate the hole on the membrane. Scale bar is 200 mm. (D) Quantitative assessment of the invasion inhibition rate. **p < 0.01 vs RGDfC-Se@siNC.
3.9. RGDfC-Se@siSox2 induces apoptosis in HepG2 cells
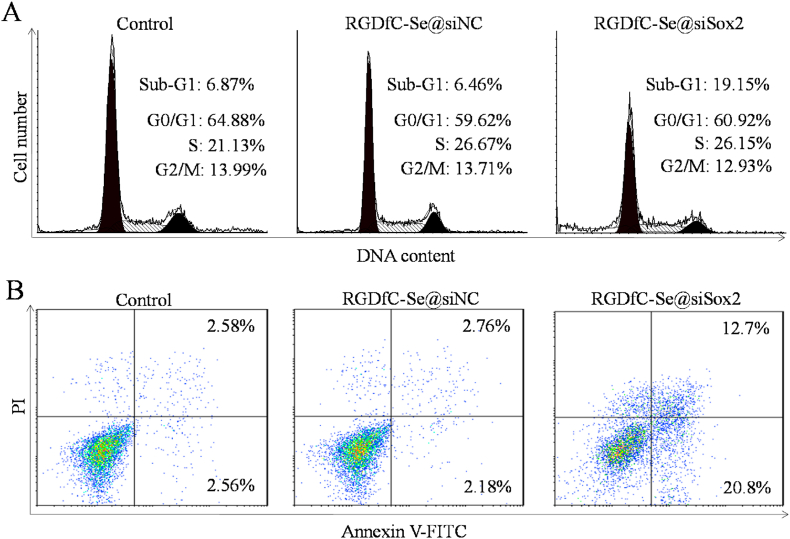
Apoptosis is a process of programmed cell death that is considered the preferred way to eliminate tumor cells [44]. Apoptosis was measured by flow cytometer using propidium iodide (PI) staining. As shown in Fig. 5A, a significant increase in the percentage of apoptosis presented as Sub-G1 peak was found after treatment with RGDfC-Se@siSox2 (19.15%), compared to the control (untreated cells, 6.87%) and RGDfC-Se@siNC groups (6.46%). However, no obvious differences in cell cycle were observed between the RGDfC-Se@siSox2-treated and control groups. In addition, an annexin V/PI-based staining assay was used to precisely examine the proportions of early and late apoptotic HepG2 cells. As shown in Fig. 5B, the proportions of both the early and late apoptotic cells were increased in RGDfC-Se@siSox2-treated group (early: 12.7%; late: 20.8%) compared to the untreated (early: 2.58%; late: 2.56%) and RGDfC-Se@siNC-treated groups (early: 2.76%; late: 2.18%). The apoptosis assay further proved that RGDfC-Se@siSox2 had the potential to exhibit anticancer effects by inducing the apoptosis of cancer cells.
Fig. 5.
(A) Cell cycle phase distribution of nuclear DNA in the treated HepG2 cells was determined by flow cytometry. (B) The treated HepG2 cells were detected using Annexin-V/PI staining by flow cytometry.
3.10. Overproduction of ROS in the RGDfC-Se@siSox2-treated HepG2 cells
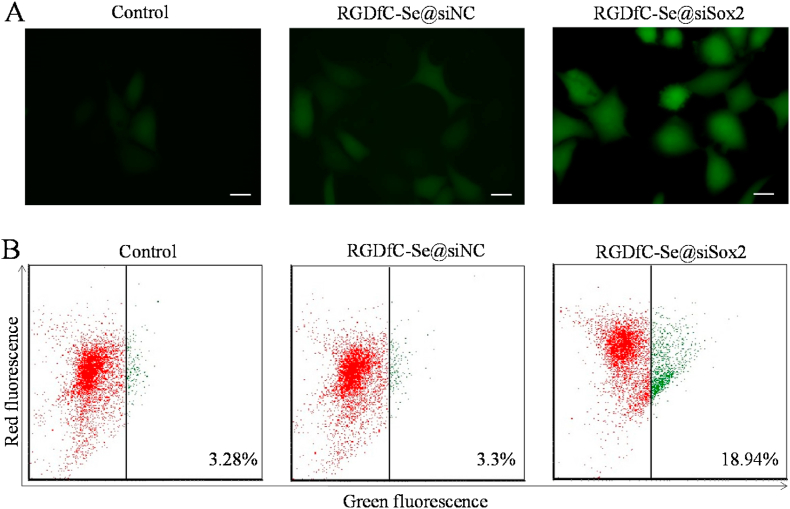
Overproduction of reactive oxygen species (ROS) caused by antitumor drugs play an essential role in the apoptosis and death of cancer cells [45]. Many studies have shown that ROS overproduction can efficiently damage tumor cells. The ROS of HepG2 cells was visualized using a fluorescence microscope. From Fig. 6A, HepG2 cells exhibited significant ROS generation after RGDfC-Se@siSox2-treatment compared to untreated and RGDfC-Se@siNC-treated negative groups, indicating RGDfC-Se@siSox2 triggered HepG2 cells apoptosis by increasing the ROS overproduction.
Fig. 6.
(A) RGDfC-Se@siSox2 increased the ROS level of HepG2 cells. The treated HepG2 cells were co-incubated with DCFDA for 25 min and then observed by a fluorescence microscope to measure intracellular ROS levels. (B) RGDfC-Se@siSox2 induced changes of MMP in HepG2 cells. The MMP values of treated cells were examined using JC-1 staining by flow cytometry.
3.11. RGDfC-Se@siSox2 disrupts the MMP of HepG2 cells
Previous studies have shown that the loss of MMP can initiate apoptosis of tumor cells [46]. In healthy cells, the JC-1 dye enters the mitochondrial matrix to form the JC-1 aggregates due to the negative charge established by the intact MMP; In apoptotic cells, the JC-1 dye accumulates in the cytoplasm in monomeric form due to the collapse of the MMP [47]. Thus, we used flow cytometry to examine whether RGDfC-Se@siSox2 triggered HepG2 cells apoptosis by damaging the MMP. Changes in MMP were examined via a fluorescent dye JC-1 in which the red and green fluorescence signals indicate normal and apoptotic cells with mitochondrial dysfunction, respectively. As shown in Fig. 6B, a high red fluorescence signal was observed in the control group. However, the red signal in RGDfC-Se@siSox2-treated cells was partially shifted to a green fluorescence signal indicating RGDfC-Se@siSox2 induced the HepG2 cell apoptosis by activating the dysfunction of MMP.
3.12. In vivo biodistribution
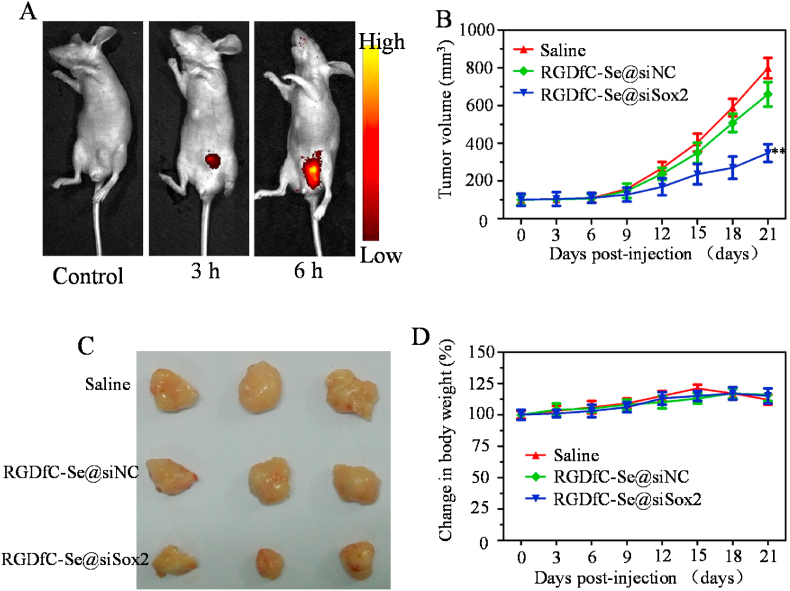
The tumor -targeted biodistribution of cy5.5-labeled RGDfC-Se@siSox2 was assessed in tumor-bearing mice model. Mice were subcutaneously inoculated with 5 × 106 HepG2 cells and administered with PBS (control) or cy5.5-labeled RGDfC-Se@siSox2 via tail vein injection after tumors reached ~100 mm3. From Fig. 7A, the obvious fluorescence signal was observed in the tumors at 3 h post-injection of cy5.5-labeled RGDfC-Se@siSox2. We also observed that the fluorescence signal at 6 h post-injection with cy5.5-labeled RGDfC-Se@siSox2 was much stronger than that at 3 h, suggesting that RGDfC-Se@siSox2 had significantly accumulated in tumors at 6 h post-injections. These results indicated that RGDfC-SeNPs can effectively deliver siSox2 to tumors for HCC therapy.
Fig. 7.
(A) In vivo biodistributions of RGDfC-Se@siSox2 in HepG2 tumor-bearing mice were observed at 3 h and 6 h after injection with Cy5.5-labeled RGDfC-Se@siSox2. The mice without injection were used as controls. (B) Tumor volume change curves. **p < 0.01 vs saline group. (C) The photographs of tumors in each group removed from the sacrificed mice at the end of study. (D) Body weight observation of mice during treatment time.
3.13. In vivo antitumor efficacy
The anti-tumor potentia l of RGDfC-Se@siSox2 was measured by an animal xenograft model. Mice were subcutaneously inoculated with the HepG2 cells (5 × 106). After the tumors reached 100 mm3, mice were administered with 100 μL of saline, RGDfC-Se@siSox2 and RGDfC-Se@siNC (100 μg/kg siRNA) via the tail vein once every three days. From Fig. 7B, the average volume of tumors in the saline group dramatically increased after 21 days of treatment. However, tumor growth was inhibited by RGDfC-Se@siSox2 during the 21 days of treatment. The images of tumors obtained from mice on day 21 further confirmed the significant antitumor efficacy of RGDfC-Se@siSox2 (Fig. 7C). The body weight of mice was also observed. As shown in Fig. 7D, no obvious weight change was found in all groups during the treatment period, indicating low toxicity of RGDfC-Se@siSox2.
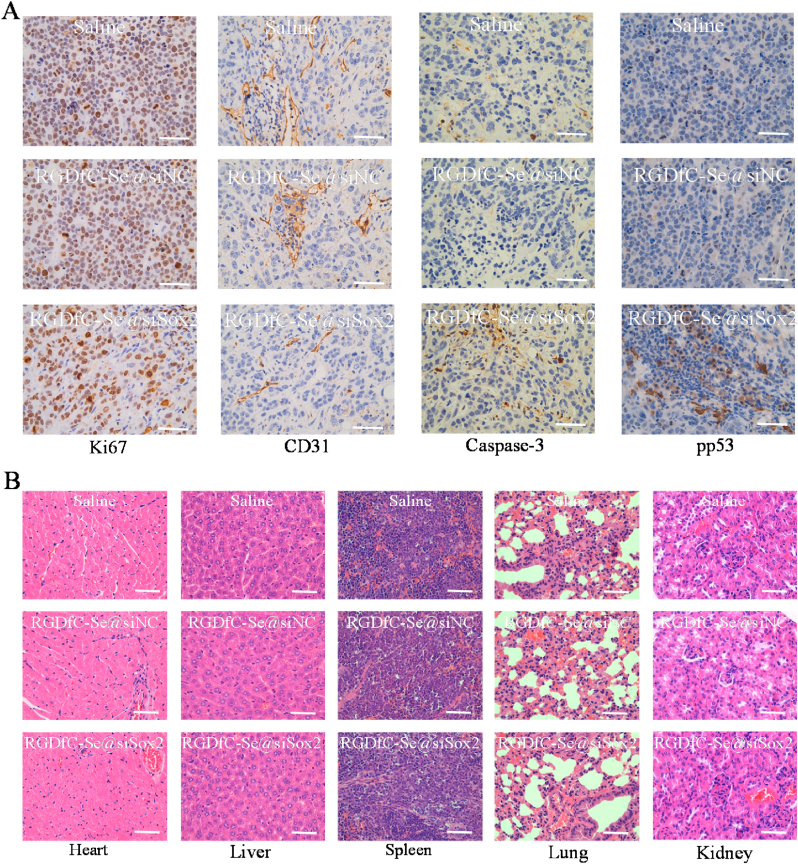
Immuno-histochemical examination was conducted to elucidate the antitumor mechanism of RGDfC-Se@siSox2 by analyzing the expression of the cell proliferation-related and apoptosis-related proteins in tumors. As shown in Fig. 8A, RGDfC-Se@siSox2 significantly down-regulated the protein expression of ki67 (cell proliferation-related protein) compared to saline (control) and RGDfC-Se@siNC (negative control) groups because of the siSox2-silencing effect [48]. RGDfC-Se@siSox2 also inhibited the expression of the CD31 (a tumor angiogenesis-related protein) and up-regulated the expression of apoptosis-promoting proteins caspase-3 and phosphorylated p53, compared to saline and RGDfC-Se@siNC groups. These results indicated that RGDfC-Se@siSox2 effectively delayed tumor growth by inhibiting the proliferation of tumor cells as well as triggering apoptosis of tumor cells. H&E staining was used to analyze the toxicity of RGDfC-Se@siSox2 on the major organs of mice. From Fig. 8B, no obvious histological changes were found among different groups, which further suggest that RGDfC-Se@siSox2 is a promising prodrug for the treatment of HCC.
Fig. 8.
(A) Ki67, CD31, caspase-3 and pp53 immunohistochemistry analysis of tumors in the treated mice. (B) H&E analysis of heart, kidney, liver, lung and spleen of the treated mice. Scale bars = 40 μm.
4. Conclusions
In summary, we developed biocompatible RGDfC-modified functionalized selenium nanoparticle RGDfC-SeNPs and an easy approach to fabricate an RGDfC-Se@siSox2 complex. RGDfC-Se@siSox2 could selectively enter the HepG2 liver cancer cells via clathrin-mediated endocytosis way and release siSox2 in the lysosomal region. RGDfC-Se@siSox2 can specifically silence the Sox2 gene and inhibit HepG2 cell proliferation, migration and invasion. In addition, RGDfC-Se@siSox2 triggers HepG2 cell apoptosis probably by the combined effects of ROS overproduction and dysfunction of MMP. Importantly, RGDfC-Se@siSox2 can selectively accumulate in tumors and effectively inhibit the growth of tumors with low toxicity in a tumor model of HepG2 cells. Taken together, these studies provide a potential strategy for HCC-targeted gene therapy.
CRediT authorship contribution statement
Yu Xia: Methodology, Formal analysis, Data curation, Conceptualization, Writing - original draft, Writing - review & editing. Guoyi Tang: Methodology, Software, Formal analysis, Data curation, Conceptualization. Yi Chen: Formal analysis, Conceptualization, Data curation. Changbing Wang: Conceptualization, Formal analysis, Data curation. Min Guo: Methodology, Software, Validation, Formal analysis, Data curation, Visualization. Tiantian Xu: Writing - original draft, Writing - review & editing. Mingqi Zhao: Methodology, Software, Validation, Formal analysis, Data curation, Visualization. Yongjian Zhou: Methodology, Investigation, Resources, Validation, Conceptualization, Supervision, Writing - review & editing.
Declaration of competing interest
The authors declared no conflict of interest.
Acknowledgement
his work was supported by National Natural Science Foundation of China (81970507), Natural Science Foundation of Guangdong Province (2020A1515010100), Technology Planning Project of Guangzhou City (201904010132), Innovative Clinical Technique of Guangzhou (2019GX05), China Postdoctoral Science Foundation (2017M612632), Yu Xia thanks to Yuyan Huang and all the staff of Guangzhou First People's Hospital for supporting this research.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.bioactmat.2020.10.019.
Contributor Information
Yu Xia, Email: eyyuxia@scut.edu.cn.
Yongjian Zhou, Email: eyzhouyongjian@scut.edu.cn.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.He Y., Guo S., Wu L., Chen P., Wang L., Liu Y., Ju H. Near-infrared boosted ROS responsive siRNA delivery and cancer therapy with sequentially peeled upconversion nano-onions. Biomaterials. 2019;225:119501. doi: 10.1016/j.biomaterials.2019.119501. [DOI] [PubMed] [Google Scholar]
- 2.Yu Z., Guo J., Hu M., Gao Y., Huang L. Icaritin exacerbates mitophagy and synergizes with doxorubicin to induce immunogenic cell death in hepatocellular carcinoma. ACS Nano. 2020;14:4816–4828. doi: 10.1021/acsnano.0c00708. [DOI] [PubMed] [Google Scholar]
- 3.Hajiasgharzadeh K., Somi M.H., Shanehbandi D., Mokhtarzadeh A., Baradaran B. Small interfering RNA-mediated gene suppression as a therapeutic intervention in hepatocellular carcinoma. J. Cell. Physiol. 2019;234:3263–3276. doi: 10.1002/jcp.27015. [DOI] [PubMed] [Google Scholar]
- 4.Zatsepin T.S., Kotelevtsev Y.V., Koteliansky V. Lipid nanoparticles for targeted siRNA delivery - going from bench to bedside. Int. J. Nanomed. 2016;11:3077–3086. doi: 10.2147/IJN.S106625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anilmis N.M., Kara G., Kilicay E., Hazer B., Denkbas E.B. Designing siRNA-conjugated plant oil-based nanoparticles for gene silencing and cancer therapy. J. Microencapsul. 2019;36:635–648. doi: 10.1080/02652048.2019.1665117. [DOI] [PubMed] [Google Scholar]
- 6.Yalcin E., Kara G., Celik E., Pinarli F.A., Saylam G., Sucularli C., Ozturk S., Yilmaz E., Bayir O., Korkmaz M.H., Denkbas E.B. Preparation and characterization of novel albumin-sericin nanoparticles as siRNA delivery vehicle for laryngeal cancer treatment. Prep. Biochem. Biotechnol. 2019;49:659–670. doi: 10.1080/10826068.2019.1599395. [DOI] [PubMed] [Google Scholar]
- 7.Xia Y., Tang G., Guo M., Xu T., Chen H., Lin Z., Li Y., Chen Y., Zhu B., Liu H., Cao J. Silencing KLK12 expression via RGDfC-decorated selenium nanoparticles for the treatment of colorectal cancer in vitro and in vivo. Mater. Sci. Eng. C-Mater. 2020;110:110594. doi: 10.1016/j.msec.2019.110594. [DOI] [PubMed] [Google Scholar]
- 8.Kozielski K.L., Ruiz-Valls A., Tzeng S.Y., Guerrero-Cazares H., Rui Y., Li Y., Vaughan H.J., Gionet-Gonzales M., Vantucci C., Kim J., Schiapparelli P., Al-Kharboosh R., Quinones-Hinojosa A., Green J.J. Cancer-selective nanoparticles for combinatorial siRNA delivery to primary human GBM in vitro and in vivo. Biomaterials. 2019;209:79–87. doi: 10.1016/j.biomaterials.2019.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin A., Wang Y., Lin K., Jiang L. Nanoparticles modified by polydopamine: working as "drug" carriers. Bioactive Mater. 2020;5:522–541. doi: 10.1016/j.bioactmat.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hao Y., Chen Y., He X., Yang F., Han R., Yang C., Li W., Qian Z. Near-infrared responsive 5-fluorouracil and indocyanine green loaded MPEG-PCL nanoparticle integrated with dissolvable microneedle for skin cancer therapy. Bioactive Mater. 2020;5:542–552. doi: 10.1016/j.bioactmat.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia Y., Xu T., Zhao M., Hua L., Chen Y., Wang C., Tang Y., Zhu B. Delivery of doxorubicin for human cervical carcinoma targeting therapy by folic acid-modified selenium nanoparticles. Int. J. Mol. Sci. 2018;19:3582. doi: 10.3390/ijms19113582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xia Y., Chen Y., Hua L., Zhao M., Xu T., Wang C., Li Y., Zhu B. Functionalized selenium nanoparticles for targeted delivery of doxorubicin to improve non-small-cell lung cancer therapy. Int. J. Nanomed. 2018;13:6929–6939. doi: 10.2147/IJN.S174909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J., Li X., Zhao X., Wu Q., Zhu H., Mao Z., Gao C. Doxorubicin-conjugated pH-responsive gold nanorods for combined photothermal therapy and chemotherapy of cancer. Bioactive Mater. 2018;3:347–354. doi: 10.1016/j.bioactmat.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parmar M.B., Meenakshi Sundaram D.N., K C.R., Maranchuk R., Montazeri Aliabadi H., Hugh J.C., Lobenberg R., Uludag H. Combinational siRNA delivery using hyaluronic acid modified amphiphilic polyplexes against cell cycle and phosphatase proteins to inhibit growth and migration of triple-negative breast cancer cells. Acta Biomater. 2018;66:294–309. doi: 10.1016/j.actbio.2017.11.036. [DOI] [PubMed] [Google Scholar]
- 15.Zhao S., Yu Q., Pan J., Zhou Y., Cao C., Ouyang J.M., Liu J. Redox-responsive mesoporous selenium delivery of doxorubicin targets MCF-7 cells and synergistically enhances its anti-tumor activity. Acta Biomater. 2017;54:294–306. doi: 10.1016/j.actbio.2017.02.042. [DOI] [PubMed] [Google Scholar]
- 16.Shan D., Ma C., Yang J. Enabling biodegradable functional biomaterials for the management of neurological disorders. Adv. Drug Deliv. Rev. 2019;148:219–238. doi: 10.1016/j.addr.2019.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y., Lin Z., Gong G., Guo M., Xu T., Wang C., Zhao M., Xia Y., Tang Y., Zhong J., Chen Y., Hua L., Huang Y., Zeng F., Zhu B. Inhibition of H1N1 influenza virus-induced apoptosis by selenium nanoparticles functionalized with arbidol through ROS-mediated signaling pathways. J. Mater. Chem. B. 2019;7:4252–4262. [Google Scholar]
- 18.Ye J., Yang Y., Jin J., Ji M., Gao Y., Feng Y., Wang H., Chen X., Liu Y. Targeted delivery of chlorogenic acid by mannosylated liposomes to effectively promote the polarization of TAMs for the treatment of glioblastoma. Bioactive Mater. 2020;5:694–708. doi: 10.1016/j.bioactmat.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zou J., Zhu B., Li Y. Functionalization silver nanoparticles loaded with paclitaxel-induced A549 cells apoptosis through ROS-mediated signaling pathways. Curr. Top. Med. Chem. 2020;20:89–98. doi: 10.2174/1568026619666191019102219. [DOI] [PubMed] [Google Scholar]
- 20.Xia Y., Lin Z., Li Y., Zhao M., Wang C., Guo M., Zhang B., Zhu B. Targeted delivery of siRNA using RGDfC-conjugated functionalized selenium nanoparticles for anticancer therapy. J. Mater. Chem. B. 2017;5:6941–6952. doi: 10.1039/c7tb01315a. [DOI] [PubMed] [Google Scholar]
- 21.Trac N.T., Chung E.J. Peptide-based targeting of immunosuppressive cells in cancer. Bioactive Mater. 2020;5:92–101. doi: 10.1016/j.bioactmat.2020.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Menendez S.T., Rey V., Martinez-Cruzado L., Gonzalez M.V., Morales-Molina A., Santos L., Blanco V., Alvarez C., Estupinan O., Allonca E., Rodrigo J.P., Garcia-Castro J., Garcia-Pedrero J.M., Rodriguez R. SOX2 expression and transcriptional activity identifies a subpopulation of cancer stem cells in sarcoma with prognostic implications. Cancers. 2020;12:964. doi: 10.3390/cancers12040964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H., Wang L., Li Z., Geng X., Li M., Tang Q., Wu C., Lu Z. SOX2 has dual functions as a regulator in the progression of neuroendocrine prostate cancer. Lab. Invest. 2020;100:570–582. doi: 10.1038/s41374-019-0343-5. [DOI] [PubMed] [Google Scholar]
- 24.Xiao W., Zheng S., Xie X., Li X., Zhang L., Yang A., Wang J., Tang H., Xie X. SOX2 promotes brain metastasis of breast cancer by upregulating the expression of FSCN1 and HBEGF. Mol. Ther-Oncolytics. 2020;17:118–129. doi: 10.1016/j.omto.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia Y., Xu T., Wang C., Li Y., Lin Z., Zhao M., Zhu B. Novel functionalized nanoparticles for tumor-targeting co-delivery of doxorubicin and siRNA to enhance cancer therapy. Int. J. Nanomed. 2018;13:143–159. doi: 10.2147/IJN.S148960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su Y., Yang H., Gao J., Qin Y.X., Zheng Y., Zhu D. Interfacial zinc phosphate is the key to controlling biocompatibility of metallic zinc implants. Adv. Sci. 2019:1900112. doi: 10.1002/advs.201900112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qu X., Yang H., Yu Z., Jia B., Qiao H., Zheng Y., Dai K. Serum zinc levels and multiple health outcomes: implications for zinc-based biomaterials. Bioactive Mater. 2020;5:410–422. doi: 10.1016/j.bioactmat.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cristofolini T., Dalmina M., Sierra J.A., Silva A.H., Pasa A.A., Pittella F., Creczynski-Pasa T.B. Multifunctional hybrid nanoparticles as magnetic delivery systems for siRNA targeting the HER2 gene in breast cancer cells. Mater. Sci. Eng. C-Mater. 2020;109:110555. doi: 10.1016/j.msec.2019.110555. [DOI] [PubMed] [Google Scholar]
- 29.Li K., Lu X., Zhang Y., Liu K., Huang Y., Liu H. Bi3TaO7/Ti3C2 heterojunctions for enhanced photocatalytic removal of water-borne contaminants. Environ. Res. 2020;185:109409. doi: 10.1016/j.envres.2020.109409. [DOI] [PubMed] [Google Scholar]
- 30.Huang Y., Long B., Tang M., Rui Z., Balogun M.-S., Tong Y., Ji H. Bifunctional catalytic material: an ultrastable and high-performance surface defect CeO2 nanosheets for formaldehyde thermal oxidation and photocatalytic oxidation. Appl. Catal. B Environ. 2016;181:779–787. [Google Scholar]
- 31.Sun X., Dong S., Li X., Yu K., Sun F., Lee R.J., Li Y., Teng L. Delivery of siRNA using folate receptor-targeted pH-sensitive polymeric nanoparticles for rheumatoid arthritis therapy. Nanomed. Nanotechnol. 2019;20:102017. doi: 10.1016/j.nano.2019.102017. [DOI] [PubMed] [Google Scholar]
- 32.Nikkhoo A., Rostami N., Farhadi S., Esmaily M., Moghadaszadeh Ardebili S., Atyabi F., Baghaei M., Haghnavaz N., Yousefi M., Aliparasti M.R., Ghalamfarsa G., Mohammadi H., Sojoodi M., Jadidi-Niaragh F. Codelivery of STAT3 siRNA and BV6 by carboxymethyl dextran trimethyl chitosan nanoparticles suppresses cancer cell progression. Int. J. Pharm. (Amst.) 2020;581:119236. doi: 10.1016/j.ijpharm.2020.119236. [DOI] [PubMed] [Google Scholar]
- 33.Xia Y., Xiao M., Zhao M., Xu T., Guo M., Wang C., Li Y., Zhu B., Liu H. Doxorubicin-loaded functionalized selenium nanoparticles for enhanced antitumor efficacy in cervical carcinoma therapy. Mater. Sci. Eng. C-Mater. 2020;106:110100. doi: 10.1016/j.msec.2019.110100. [DOI] [PubMed] [Google Scholar]
- 34.Li J., Zhang J., Wang Y., Liang X., Wusiman Z., Yin Y., Shen Q. Synergistic inhibition of migration and invasion of breast cancer cells by dual docetaxel/quercetin-loaded nanoparticles via Akt/MMP-9 pathway. Int. J. Pharm. (Amst.) 2017;523:300–309. doi: 10.1016/j.ijpharm.2017.03.040. [DOI] [PubMed] [Google Scholar]
- 35.Li Y., Xia Y., Liu K., Ye K., Wang Q., Zhang S., Huang Y., Liu H. Constructing Fe-MOF-Derived Z-scheme photocatalysts with enhanced charge transport: nanointerface and carbon sheath synergistic effect. ACS Appl. Mater. Interfaces. 2020;12:25494–25502. doi: 10.1021/acsami.0c06601. [DOI] [PubMed] [Google Scholar]
- 36.Xia Y., Tang G., Wang C., Zhong J., Chen Y., Hua L., Li Y., Liu H., Zhu B. Functionalized selenium nanoparticles for targeted siRNA delivery silence Derlin1 and promote antitumor efficacy against cervical cancer. Drug Deliv. 2020;27:15–25. doi: 10.1080/10717544.2019.1667452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shuai Q., Zhao G., Zhang X., Yu B., Lee R.J., Su W.K. Selection of fluorescent dye for tracking biodistribution of paclitaxel in live imaging. Colloids Surf., B. 2019;181:872–878. doi: 10.1016/j.colsurfb.2019.06.035. [DOI] [PubMed] [Google Scholar]
- 38.Jiang Y., Yang N., Zhang H., Sun B., Hou C., Ji C., Zheng J., Liu Y., Zuo P. Enhanced in vivo antitumor efficacy of dual-functional peptide-modified docetaxel nanoparticles through tumor targeting and Hsp90 inhibition. J. Contr. Release. 2016;221:26–36. doi: 10.1016/j.jconrel.2015.11.029. [DOI] [PubMed] [Google Scholar]
- 39.Xia Y., Guo M., Xu T., Li Y., Wang C., Lin Z., Zhao M., Zhu B. siRNA-loaded selenium nanoparticle modified with hyaluronic acid for enhanced hepatocellular carcinoma therapy. Int. J. Nanomed. 2018;13:1539–1552. doi: 10.2147/IJN.S157519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xia Y., Zhong J., Zhao M., Tang Y., Han N., Hua L., Xu T., Wang C., Zhu B. Galactose-modified selenium nanoparticles for targeted delivery of doxorubicin to hepatocellular carcinoma. Drug Deliv. 2019;26:1–11. doi: 10.1080/10717544.2018.1556359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jhala D., Rather H., Kedaria D., Shah J., Singh S., Vasita R. Biomimetic polycaprolactone-chitosan nanofibrous substrate influenced cell cycle and ECM secretion affect cellular uptake of nanoclusters. Bioactive Mater. 2019;4:79–86. doi: 10.1016/j.bioactmat.2018.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meng Y., Xu Q., Chen L., Wang L., Hu X. The function of SOX2 in breast cancer and relevant signaling pathway. Pathol. Res. Pract. 2020:153023. doi: 10.1016/j.prp.2020.153023. [DOI] [PubMed] [Google Scholar]
- 43.Yang X., Zhang Y., Malichewe C., Shi Z., Wang L., Lu Z., Guo X. Chitosan nanoparticle mediated upregulation of microRNA34a expression to suppress the proliferation, migration, invasion of MDA-MB-231 cells. J. Drug Deliv. Sci. Technol. 2019;52:1061–1069. [Google Scholar]
- 44.Patil S., Kuman M.M., Palvai S., Sengupta P., Basu S. Impairing powerhouse in colon cancer cells by hydrazide-hydrazone-based small molecule. ACS Omega. 2018;3:1470–1481. doi: 10.1021/acsomega.7b01512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Faustova M., Nikolskaya E., Sokol M., Zabolotsky A., Mollaev M., Zhunina O., Fomicheva M., Lobanov A., Severin E., Yabbarov N. High-effective reactive oxygen species inducer based on Mn-tetraphenylporphyrin loaded PLGA nanoparticles in binary catalyst therapy. Free Radical Bio. Med. 2019;143:522–533. doi: 10.1016/j.freeradbiomed.2019.09.008. [DOI] [PubMed] [Google Scholar]
- 46.Gupta P.K., Pappuru S., Gupta S., Patra B., Chakraborty D., Verma R.S. Self-assembled dual-drug loaded core-shell nanoparticles based on metal-free fully alternating polyester for cancer theranostics. Mater. Sci. Eng. C-Mater. 2019;101:448–463. doi: 10.1016/j.msec.2019.03.041. [DOI] [PubMed] [Google Scholar]
- 47.Marino J.E., Yeisley D.J., Chen H., Levin M., Kaplan D.L., Hahn M.S. Interferon-gamma stimulated murine macrophages in vitro: impact of ionic composition and osmolarity and therapeutic implications. Bioelectricity. 2020;2:48–58. doi: 10.1089/bioe.2019.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim G.B., Aragon-Sanabria V., Randolph L., Jiang H., Reynolds J.A., Webb B.S., Madhankumar A., Lian X., Connor J.R., Yang J., Dong C. High-affinity mutant Interleukin-13 targeted CAR T cells enhance delivery of clickable biodegradable fluorescent nanoparticles to glioblastoma. Bioactive Mater. 2020;5:624–635. doi: 10.1016/j.bioactmat.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.