Abstract
Knee osteoarthritis (KOA) is a highly prevalent disabling joint disease in aged people. Progressive cartilage degradation is the hallmark of KOA, but its deeper mechanism remains unclear. Substantial evidence indicates the importance of the synovium for joint homeostasis. The present study aimed to determine whether the synovium regulates cartilage metabolism through chondrogenesis-related microRNAs (miRNAs) in the KOA microenvironment. Clinical sample testing and in vitro cell experiments screened out miR-455 and miR-210 as effective miRNAs. The levels of both were significantly reduced in KOA cartilage but increased in KOA synovial fluid compared with controls. We further revealed that transforming growth factor β1 (TGF-β1) can significantly upregulate miR-455 and miR-210 expression in synoviocytes. The upregulated miRNAs can be secreted into the extracellular environment and prevent cartilage degeneration. Through bioinformatics and in vitro experiments, we found that Runx1 can bind to the promoter regions of miR-455 and miR-210 and enhance their transcription in TGF-β1-treated synoviocytes. Collectively, our findings demonstrate a protective effect of the synovium against cartilage degeneration mediated by chondrogenesis-related miRNAs, which suggests that Runx1 is a potential target for KOA therapy.
Keywords: synovium, cartilage degeneration, Smad2/3-Runx1 cascade, microRNA, knee osteoarthritis
Graphical Abstract
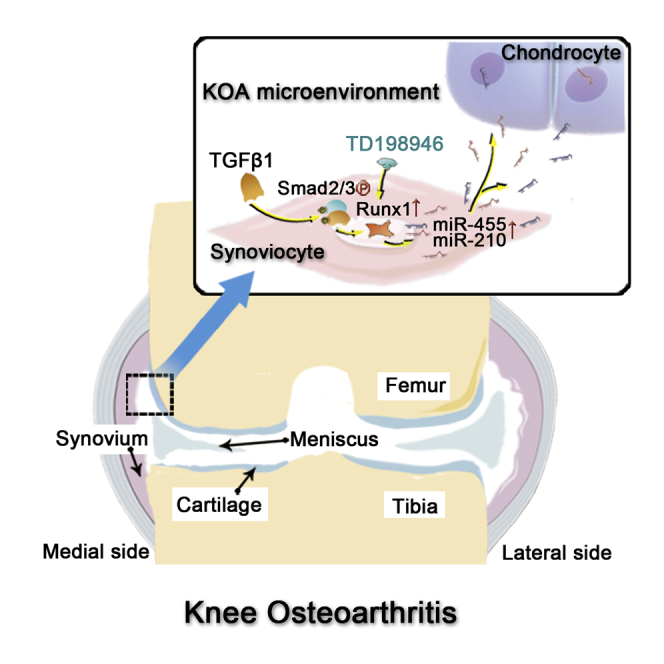
Aged people suffer severely from knee osteoarthritis. Zhang and colleagues demonstrate that the release of miR-455 and miR-210 from TGF-β1-stimulated synoviocytes in knee osteoarthritis synovial fluid could prevent cartilage degeneration. The whole process was regulated by the Smad2/3-Runx1 cascade, which revealed the potential role of Runx1 in KOA therapy.
Introduction
Knee osteoarthritis (KOA) is a complex, multifactorial disease that causes chronic joint pain, stiffness, and swelling in aged people.1,2 It is a highly prevalent, disabling joint disease that accounts for more than 80% of the total osteoarthritic burden.3 Articular cartilage degeneration caused by mechanical loading and inflammation is central to KOA pathology. Due to low blood supply, articular cartilage has poor self-healing ability once damaged. Progenitor cell-based cartilage engineering is a promising strategy for repair of injured articular cartilage.4 However, chondrocyte hypertrophy after implantation is of grave concern.5,6 The synovial fluid microenvironment in which cartilage resides plays an important role in regulating cartilage metabolism.7 Recent studies have demonstrated that the levels of not only catabolic factors but also anabolic factors are elevated in KOA synovial fluid.8,9 A key question is how to modify the microenvironment to favor cartilage regeneration; to answer this question, it is necessary to understand the influencing factors of the synovial fluid microenvironment and their effects on metabolic balance in chondrocytes.
Synovitis is frequently observed in KOA and is associated with the severity of joint symptoms and cartilage degeneration.10,11 Interleukin-1β (IL-1β) and transforming growth factor β1 (TGF-β1) have been reported to play important roles in the continuation of synovial inflammation and hyperplasia during KOA.12,13 The levels of TGF-β1, which has historically been considered to be a reparative mediator, are elevated and TGF-β1 is activated in synovial fluid from KOA patients.14 However, the role of TGF-β1 in KOA is controversial.15 IL-1β is one of the major proinflammatory cytokines involved in the pathophysiology of KOA and induces the expression of inflammatory cytokines and degradative enzymes in chondrocytes.13 We previously identified eight chondrogenesis-related microRNAs (miRNAs) (miR-193b, miR-199a, miR-455, miR-210, miR-381, miR-92a, miR-320c, and miR-136) that are significantly upregulated during chondrogenesis in human adipose-derived stem cells.16 Some of these miRNAs were found to exert protective effects on chondrocyte metabolism in an IL-1β-mediated chondrocyte degenerative model.17, 18, 19 miRNAs are a class of small (22-∼24-nt) noncoding RNAs that can directly bind to mRNAs to regulate their expression at the posttranscriptional level. Accumulating evidence suggests that miRNAs not only function in the intracellular space but also can be released from cells and disseminated through biological fluid to remotely alter the transcriptomes of recipient cells.20,21
Given the roles of miRNAs in intercellular communication and cartilage metabolism, we hypothesized that increased IL-1β and TGF-β1 in the joint microenvironment may target the synovium and regulate KOA development through chondrogenesis-related miRNAs.22,23 To confirm this hypothesis and explore potential therapeutic targets in KOA, we designed and performed a series of experiments.
Results
Expression Patterns of Chondrogenesis-Related miRNAs in Articular Cartilage, Synovial Fluid, and Plasma during KOA
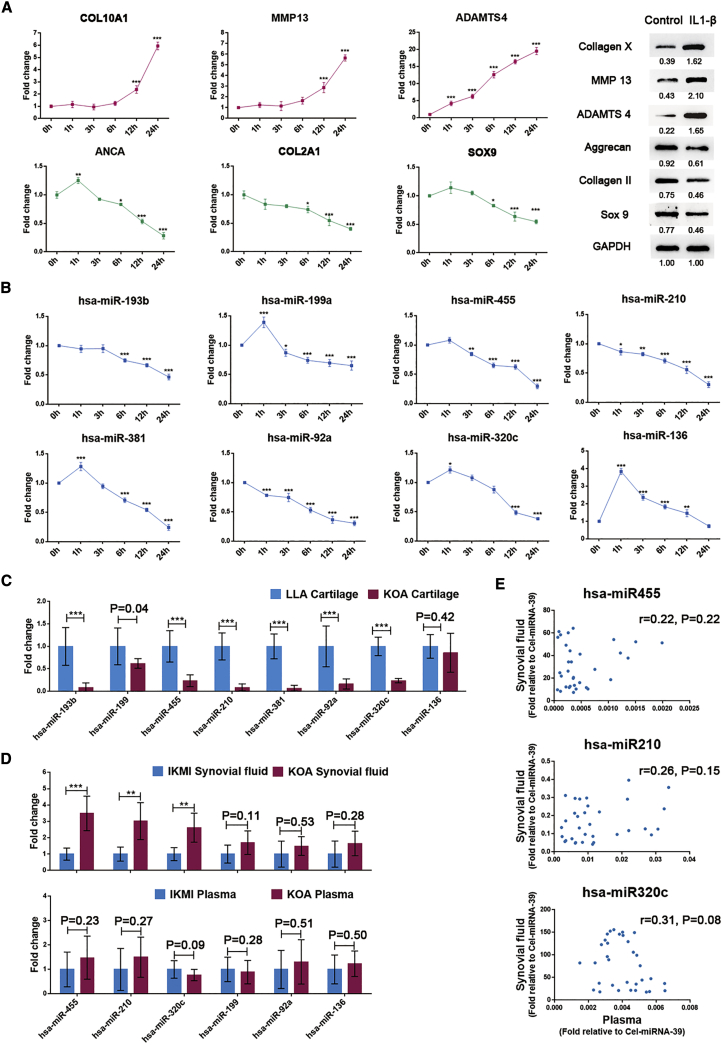
To determine the expression patterns of the eight chondrogenesis-related miRNAs during the degeneration of articular cartilage, we detected their expression levels in vitro and in vivo. IL-1β-induced chondrocyte degeneration was verified by the upregulation of hypertrophic genes (COL10A1, ADAMTS4, and MMP13) and the downregulation of chondrogenic genes (ACAN, COL2A1, and SOX9), as shown in Figure 1A. The expression of the eight chondrogenesis-related miRNAs showed an inverse correlation with the degree of chondrocyte degeneration in a time-dependent manner (Figure 1B). We further examined the expression of these miRNAs in clinical samples, specifically in healthy articular cartilage (n = 10, from lower-limb amputation [LLA] patients) and osteoarthritic articular cartilage (n = 12, from KOA patients), and observed similar expression trends (Figure 1C).
Figure 1.
Analysis of the Relative Expression Levels of the Eight Chondrogenesis-Related miRNAs in Articular Cartilage, Synovial Fluid, and Plasma during KOA
(A) The expression patterns of chondrogenic genes and hypertrophic genes in chondrocytes treated with IL-1β (5 ng/mL) as determined by qRT-PCR and western blot analysis. (B) Time-dependent changes in the eight chondrogenesis-related miRNAs in IL-1β (5 ng/mL)-treated chondrocytes. Healthy chondrocytes from LLA patients were used for the above experiments. (C) The expression patterns of the eight chondrogenesis-related miRNAs in cartilage samples from LLA (n = 10) and KOA (n = 12) patients. (D) The expression patterns of the eight chondrogenesis-related miRNAs in synovial fluid and plasma samples from KOA patients (n = 18 for synovial fluid and n = 26 for plasma) and IKMI patients (n = 15 for synovial fluid and n = 25 for plasma). (E) The correlations of miR-455, miR-210, and miR320c between synovial fluid and plasma samples were analyzed (n = 33 KOA and IKMI patients with both synovial fluid and plasma samples). The qRT-PCR data are presented as the mean and SD; GAPDH and U6 snRNA were detected as endogenous controls for mRNA and miRNA, respectively. Exogenous spike-in Cel-miRNA-39 was used to normalize miRNA levels in synovial fluid and plasma. A representative image of western blot data is shown. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Six chondrogenesis-related miRNAs (mir-199a, mir-455, mir-210, mir-92a, mir-320c, and mir-136) were detectable in synovial fluid. Interestingly, in contrast with their expression trends in cartilage, mir-455, mir-210, and mir-320c were significantly upregulated in KOA synovial fluid (n = 18) compared with isolated knee meniscus injury (IKMI) synovial fluid (n = 15) (Figure 1D, upper). However, their expression levels in plasma were similar between KOA patients (n = 26) and IKMI patients (n = 25) (Figure 1D, lower). Our data also suggested no correlations between plasma miRNA and synovial fluid miRNA expression (Figure 1E), although the concentrations of these miRNAs were significantly higher in synovial fluid than in plasma. These results imply that the overexpressed miR-455, miR-210, and miR-320c in KOA synovial fluid are generated by tissues in the joint cavity rather than by penetration of plasma miRNAs into the synovial fluid.
TGF-β1 Induces the Overexpression of miR-455 and miR-210 in KOA Synoviocytes
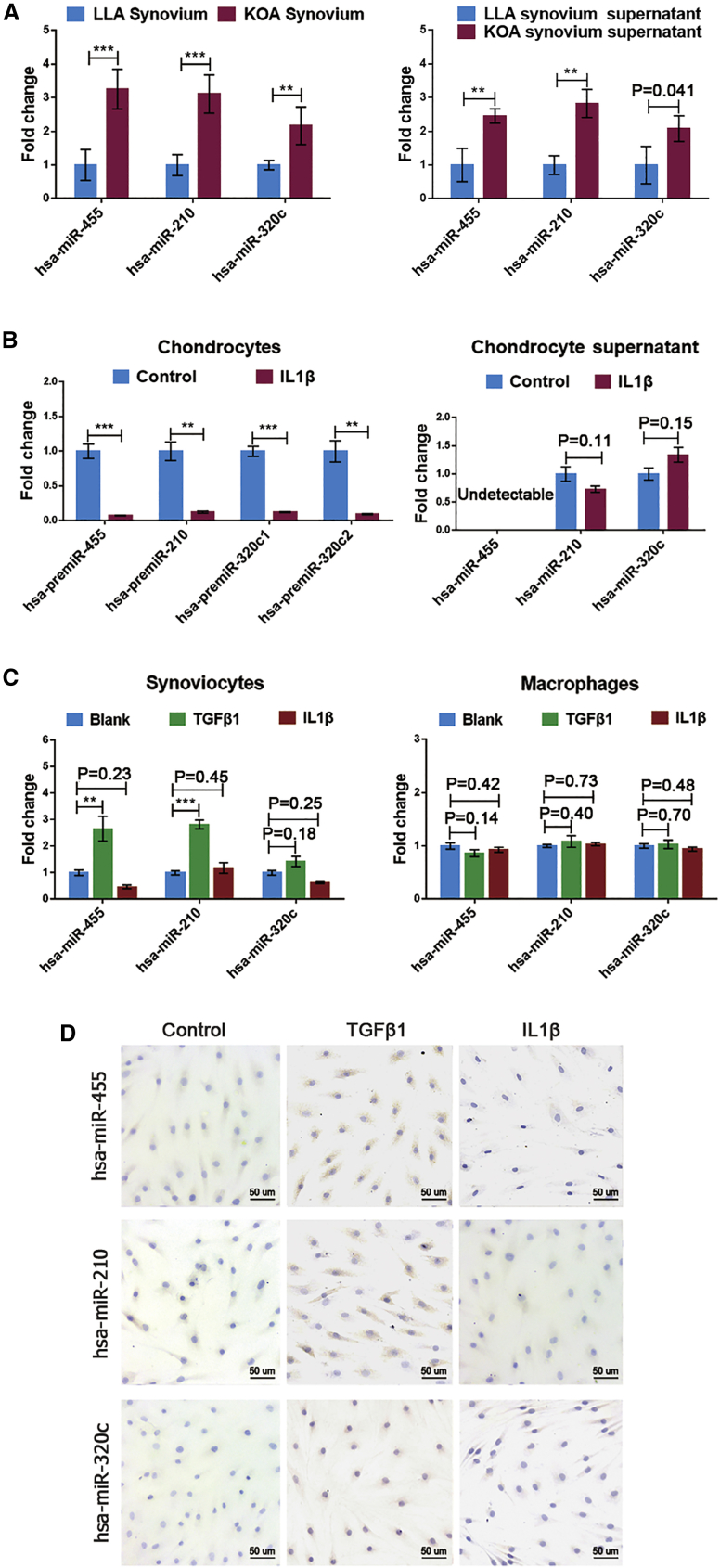
To determine the origin of the overexpressed miRNAs in KOA synovial fluid, we obtained and incubated hyperplastic synovium from KOA patients (n = 10) and nonhyperplastic synovium from LLA patients (n = 6) in complement medium for 24 h. The qRT-PCR results showed that hyperplastic synovia, in the contexts of both tissues and culture supernatants, had significantly higher miR-455, miR-210, and miR-320c levels than nonhyperplastic synovium (Figure 2A). Notably, IL-1β treatment significantly decreased the expression of these miRNA precursors in chondrocytes but did not significantly increase the levels of the mature miRNAs in culture supernatants (Figure 2B). Mature miR-455 was undetectable in degenerated chondrocyte culture supernatants, with CT values over 35. It seems that the hyperplastic synovia, rather than degenerated chondrocytes, is the source of these miRNAs in synovial fluid.
Figure 2.
Identification of the Source of the Differentially Expressed miRNAs (DEMs) in Synovial Fluid
(A) The relative expression levels of miR-455, miR-210, and miR320c in synovial tissue and culture supernatant (n = 10 for KOA and n = 6 for LLA patients). (B) Under IL-1β treatment (5 ng/mL), the precursor levels of the three DEMs in chondrocytes and their mature levels in culture supernatant were determined by qRT-PCR. Healthy chondrocytes from LLA patients were used for the above experiments. (C and D) The regulatory effects of IL-1β (5 ng/mL) and TGF-β1 (10 ng/mL) on the expression of the three DEMs in synoviocytes and macrophages were estimated by qRT-PCR (C) and in situ hybridization (D). Synoviocytes and monocyte-derived macrophages from KOA patients were used for the above experiments. qRT-PCR data are presented as the mean and standard deviation (SD); U6 snRNA was detected as an endogenous control for miRNA in synovial tissue and cells. A representative image of in situ hybridization data is shown. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
The causes of the overexpression of these miRNAs in KOA synovium were further investigated in synoviocytes and macrophages, the two main cell types within hyperplastic synovia. Upon TGF-β1 stimulation, we observed significant increases in miR-455 and miR-210 expression in synoviocytes (Figures 2C and 2D). Neither IL-1β nor TGF-β1 changed the expression of these miRNAs in macrophages (Figure 2C). This finding suggests that TGF-β1-activated synoviocytes may be major contributors to the overexpression of miR-455 and miR-210 in hyperplastic synovia.
Synoviocyte-Derived miR-455 and miR-210 Attenuate Cartilage Degeneration
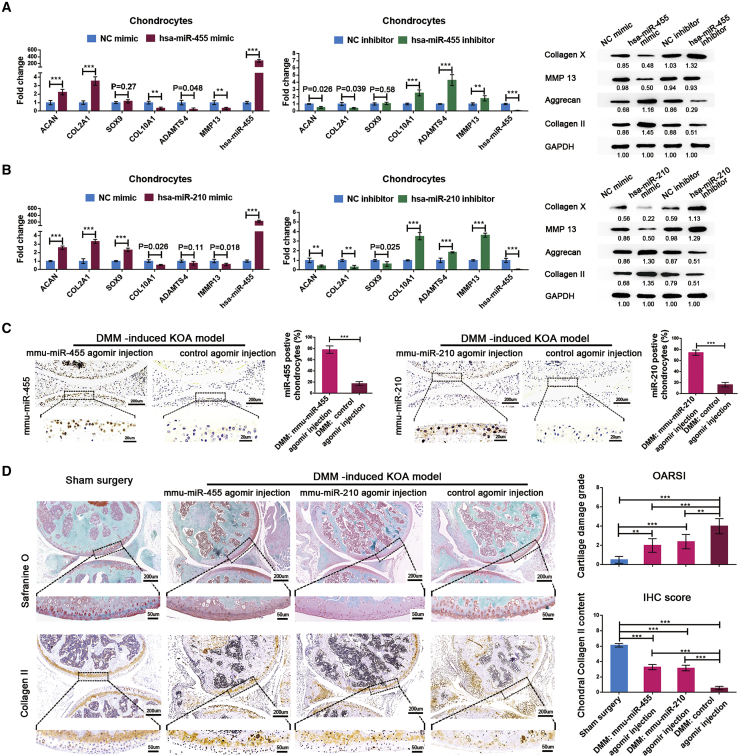
The functions of miR-455 and miR-210 in articular cartilage degeneration were determined in degenerated chondrocytes using mimics and inhibitors of these identified miRNAs and corresponding NCs. Overexpression of miR-455 or miR-210 in chondrocytes not only significantly reduced the expression of hypertrophic genes but also increased the expression of chondrogenic genes. Downregulation of the expression of these miRNAs in chondrocytes produced the opposite results (Figures 3A and 3B; Figure S2). Next, we constructed destabilization of the medial meniscus (DMM) mouse models to validate the protective roles of miR-455 and miR-210 in vivo. The expression level of each miRNA in cartilage was significantly increased after intra-articular injection of the corresponding mmu-miRNA agomir (Figure 3C). The mmu-miR-455 agomir group and the mmu-miR-210 agomir group presented significantly higher cartilage OARSI grade scores and collagen II content than the control agomir group (Figure 3D). These data indicate that miR-455 and miR-210 exert protective effects against cartilage degeneration during KOA.
Figure 3.
miR-455 and miR-210 Attenuate Cartilage Degeneration In Vitro and In Vivo
(A and B) The expression patterns of chondrogenic genes and hypertrophic genes in chondrocytes after transfection with a mimic NC, miRNA mimic, inhibitor NC, or miRNA inhibitor for miR-455 (A) and miR-210 (B), as determined by qRT-PCR and western blot analysis. Degenerated chondrocytes from KOA patients were used for NC-mimic and miR-mimic transfection, and healthy chondrocytes from LLA patients were used for NC-inhibitor and miR-inhibitor transfection. (C) The levels of mmu-miR-455 and mmu-miR-210 in mouse cartilage after intra-articular injection of the corresponding mmu-miRNA agomir or control agomir were measured by in situ hybridization and quantified as the average percentage of brown cells in five randomly selected high-power fields. (D) Left: Safranin O staining and immunohistochemical (IHC) staining of collagen II in sham surgery mice, DMM mice with mmu-miR-455 agomir injection, DMM mice with mmu-miR-210 agomir injection, and DMM mice with control agomir injection. Right: the OARSI scoring system was used to grade mouse cartilage degeneration, and IHC staining scores were used to quantify collagen II content in articular cartilage. The qRT-PCR data are presented as the mean and SD; GAPDH and U6 snRNA were detected as endogenous controls for mRNA and miRNA, respectively. Representative images of western blot, in situ hybridization, Safranin O staining, and immunohistochemistry results are shown. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
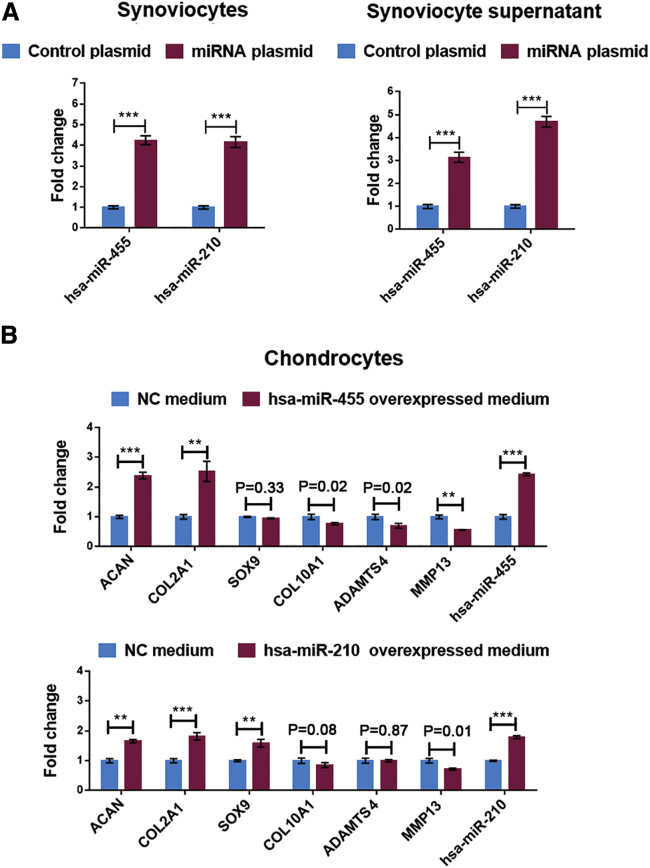
To further validate the direct effects of synoviocyte-released miRNAs on cartilage homeostasis, we treated degenerated chondrocytes with synoviocyte-derived conditioned medium. As shown in Figure 4A, transfection of synoviocytes with the miRNA overexpression plasmids significantly upregulated the miRNA levels in both cells and culture supernatant. After treatment with the above-mentioned conditioned medium, chondrocytes showed significantly reduced hypertrophic gene expression and increased chondrogenic gene expression, indicating that synoviocyte-derived miR-455 and miR-210 can attenuate chondrocyte degeneration (Figure 4B).
Figure 4.
Effects of Synoviocyte-Derived miR-455 and miR-210 on Cartilage Homeostasis
miRNA overexpression plasmid vectors and a control plasmid were designed and transfected into SW982 cells. (A) miRNA expression in cells and supernatant was assessed by qRT-PCR. (B) The expression patterns of chondrogenic genes and hypertrophic genes in degenerated chondrocytes treated with conditioned medium as determined by qRT-PCR. Degenerated chondrocytes from KOA patients were used for the above experiments. The qRT-PCR data are presented as the mean and SD; GAPDH and U6 snRNA were detected as endogenous controls for mRNA and miRNA, respectively. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
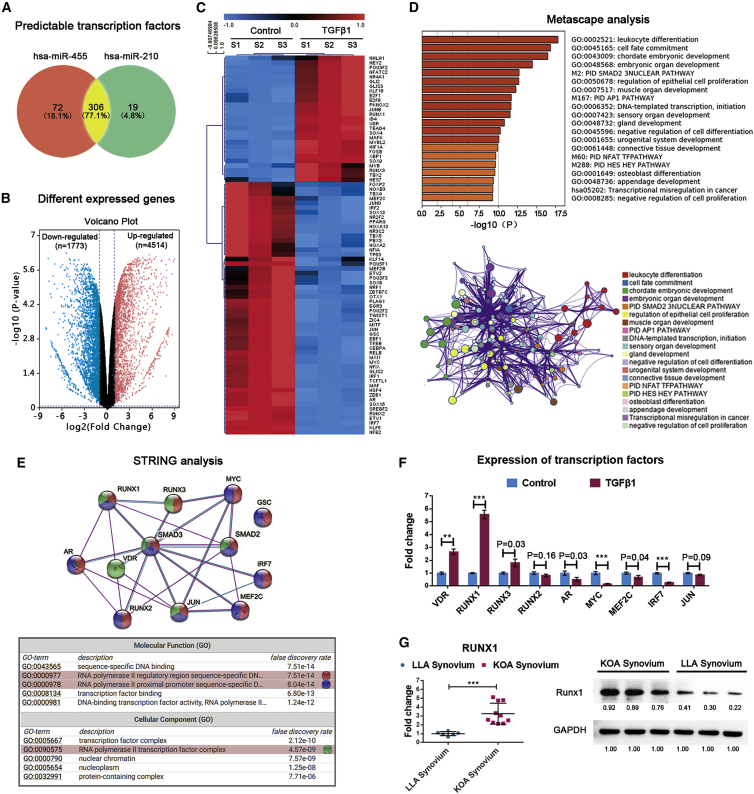
Smad2/3-Runx1 Signaling Is Responsible for TGF-β1-Induced miR-455 and miR-210 Upregulation in Synoviocytes
To predict the transcription factors (TFs) of miR-455 and miR-210, we applied the MEME tool Find Individual Motif Occurrences (FIMO; v.4.9.1) to search all known TF motifs from the JASPAR database in sequences 10 kb upstream and downstream of each precursor miRNA.24 In total, 378 TFs for miR-455 and 325 TFs for miR-210 were predicted (Figure 5A). Next, we compared the gene expression between TGF-β1-activated synoviocytes and control synoviocytes. A large set of DEGs was identified, including 1,773 upregulated genes and 4,514 downregulated genes (Figure 5B). Of these DEGs, 81 were identified as potential TFs that might regulate miR-455 and miR-210 transcription (Figure 5C). The candidate TFs were mapped to Metascape database entries (including Gene Ontology [GO] biological processes, Kyoto Encyclopedia of Genes and Genomes [KEGG] pathways, canonical pathways, and Reactome gene sets) to analyze their enriched pathways. Notably, the enriched processes were highly connected and clustered around the Smad2/3 pathway, which is an important downstream signaling pathway of TGF-β125 (Figure 5D). The close relationships among the above TFs and Smad2/3 were assessed using the STRING experimentally determined and curated databases (Figure 5E). The changes in these TFs in synoviocytes after TGF-β1 stimulation were verified by qRT-PCR (Figure 5F). According to the GO annotation, most of the TFs are components of the RNA polymerase II complex and participate in binding of the complex to the proximal promoters or regulatory regions of transcribed genes. In clinical samples, Runx1 expression was also higher in hyperplastic synovium (from KOA patients, n = 10) than in nonhyperplastic synovium (from LLA patients, n = 6) (Figure 5G). Based on its expression patterns in synovial cells and tissues and its positive role in KOA development, Runx1 shows the most promise as a TF that affects the transcription of primary miRNA-455 and primary miRNA-210.4,26,27
Figure 5.
Prediction of TFs for miR-455 and miR-210
(A) Venn diagrams showing the numbers of predicted TFs for miR-455 and miR-210. Transcriptome sequencing was conducted on synoviocytes from three KOA patients. Volcano plots of DEGs in synoviocytes treated with or without TGF-β1 (10 ng/mL). (B) Red indicates significantly upregulated genes, blue indicates significantly downregulated genes, and gray indicates genes with no differential expression based on the criteria of p < 0.05 and fold change > 2.0. (C) Heatmap of the predicted TFs that were significantly changed in TGF-β1-activated synoviocytes. Pathways significantly associated with these potential TFs are shown. (D) The enriched terms are depicted in a heatmap and an interaction network according to their p values. (E) A protein-protein interaction (PPI) network of the TFs involved in the Smad2/3 pathway was constructed, and the corresponding GO terms were mapped. (F) The expression patterns of Smad2/3 pathway-related TFs in synoviocytes treated with TGF-β1 (10 ng/mL) as determined by qRT-PCR. Synoviocytes from KOA patients were used for the above cell experiments. (G) Left: Runx1 expression changes in synovial tissue from KOA patients (n = 10) and LLA patients (n = 6) as detected by qRT-PCR. Right: the Runx1 levels in three random samples from each group were validated by western blot analysis. The qRT-PCR data are presented as the mean and SD, and representative images of western blots are shown. GAPDH was detected as an endogenous control. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
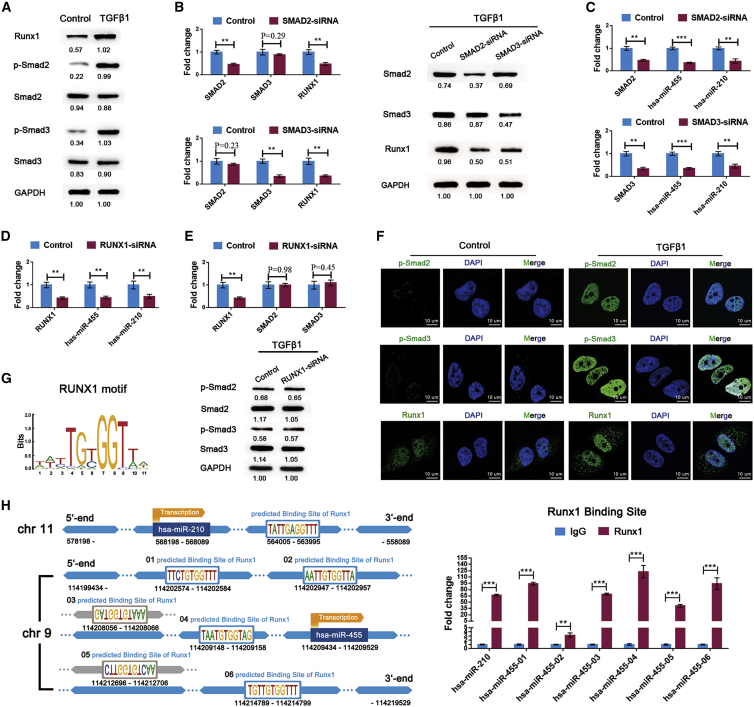
Subsequently, we investigated the downstream molecular events mediated by TGF-β1 stimulation in synoviocytes. TGF-β1 significantly increased the phosphorylation of Smad2 and Smad3 and the expression of Runx1 (Figure 6A). Following transfection with SMAD2-siRNA (small interfering RNA) or SMAD3-siRNA, Runx1 expression was significantly decreased (Figure 6B). Furthermore, Smad2, Smad3, or Runx1 knockdown significantly abrogated TGF-β1-induced miR-455 and miR-210 overexpression (Figures 6C and 6D). However, the levels of Smad2, Smad3, phospho-Smad2, and phospho-Smad3 were not affected by Runx1 knockdown (Figure 6E). These results suggest that TGF-β1-induced Smad2/3 phosphorylation increases Runx1 expression and then activates the transcription of miR-455 and miR-210 in synoviocytes. This relationship was also validated by immunofluorescence staining, which showed that the increase in Runx1 was associated with nuclear import of phospho-Smad2 and phospho-Smad3 under TGF-β1 treatment (Figure 6F). The binding of Runx1 to the promoter regions of miR-455 and miR-210 was also confirmed by CUT&Tag assays in TGF-β1-treated SW982 cells (Figure 6H).
Figure 6.
Smad2/3-Runx1 Signaling Promotes the Transcription of miR-455 and miR-210 in Synoviocytes
(A) The protein levels of Smad2, Smad3, phospho-Smad2, phospho-Smad3, and Runx1 were measured in synoviocytes treated with TGF-β1 (10 ng/mL). (B) Under TGF-β1 (10 ng/mL) treatment, the expression pattern of Runx1 was assessed after SMAD2 siRNA and SMAD3 siRNA transfection into synoviocytes. (C) miR-455 and miR-210 expression in the above groups were analyzed using qRT-PCR. (D) The regulatory effects of Runx1 on miR-455 and miR-210 expression were determined in TGF-β1 (10 ng/mL)-treated synoviocytes following transfection with RUNX1 siRNA. (E) The influence of Runx1 knockdown on Smad2/3 phosphorylation was evaluated by qRT-PCR and western blot analysis. (F) The changes in Runx1, phospho-Smad2, and phospho-Smad3 in synoviocytes after TGF-β1 (10 ng/mL) treatment were detected by immunofluorescence staining. Synoviocytes from KOA patients were used for the above cell experiments. (G) The Runx1 binding motif is shown. (H) Left: schematic diagram of the putative Runx1 binding sites in the promoter regions of miR-455 and miR-210. Right: the putative Runx1 binding sites were confirmed by CUT&Tag assays in SW982 cells . The qRT-PCR data are presented as the mean and SD; GAPDH and U6 snRNA were detected as endogenous controls for mRNA and miRNA, respectively. Representative images of western blots and immunofluorescence are shown. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
TD-198946, a Strong Inducer of Runx1, Upregulates miR-455 and miR-210 in Synoviocytes
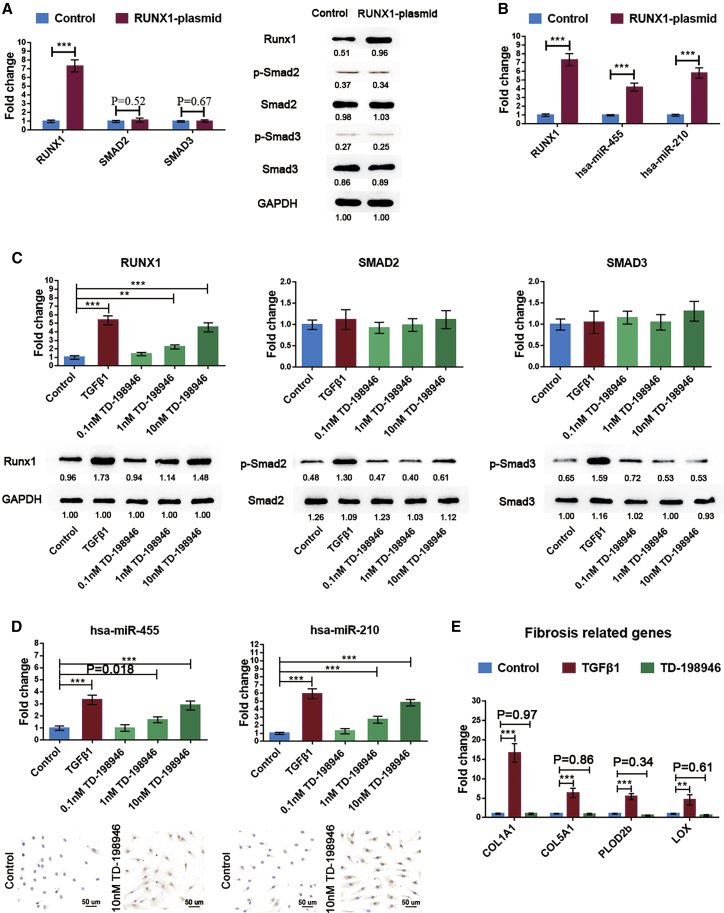
To verify the role of Runx1 in the regulation of miR-455 and miR-210 transcription, we transfected synoviocytes with a plasmid overexpressing Runx1. As expected, Runx1 overexpression significantly upregulated miR-455 and miR-210 in the absence of TGF-β1 stimulation (Figures 7A and 7B).
Figure 7.
Efficiency of Runx1 in Promoting miR-455 and miR-210 Transcription
Synoviocytes were transfected with a Runx1 overexpression plasmid. (A and B) The activation of Smad2/3 signaling (A) and the levels of miR-455 and miR-210 (B) were evaluated by qRT-PCR and western blot analysis. (C) The dose-dependent changes in Runx1, Smad2, Smad3, phospho-Smad2, and phospho-Smad3 expression in synoviocytes treated with TD-198946 were measured by qRT-PCR (top) and western blot analysis (bottom). (D) TD-198946 dose-response effects on miR-455 and miR-210 transcription in synoviocytes as determined by qRT-PCR and in situ hybridization. The expression patterns of synovial fibrosis-related genes in synoviocytes incubated with TD-198946 (10 nM) or TGF-β1 (10 ng/mL) were assessed. Synoviocytes from KOA patients were used for cell experiments. The qRT-PCR data are presented as the mean and SD; GAPDH and U6 snRNA were detected as endogenous controls for mRNA and miRNA, respectively. Representative images of western blots and in situ hybridization are shown. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
TD-198946 is a thienoindazole derivative recently described by Yano et al.28 that can increase the expression of Runx1 in chondrocytes. Our data showed that TD-198946 exerted the same effect on Runx1 expression in synoviocytes in a dose-dependent manner, and that this activation appeared to be independent of Smad2/3 signaling activation (Figure 7C). Importantly, TD-198946, similar to the Runx1 overexpression plasmid, significantly enhanced the transcription of miR-455 and miR-210 (Figure 7D) but did not increase the expression of TGF-β1-associated synovial fibrosis-related genes,29 as shown in Figure 7E.
Discussion
The knee is a typical synovial joint. The articular cartilage is normally avascular; therefore, it relies upon adjacent tissues to provide nutrients and to remove metabolic waste through the synovial fluid. The synovium, as the source of synovial fluid, plays an important role in modifying the joint microenvironment during KOA pathology.30 Low-grade synovitis presents early in and persists throughout the disease.31 Given this information, we can speculate that synovitis, the critical feature of OA, greatly influences the balance of cartilage homeostasis.32
In this study, IL-1β was found to induce the degeneration of chondrocytes and correspondingly decrease the expression of chondrogenesis-related miRNAs. Similar results were observed in cartilage samples, whereas an opposite trend was observed in synovial fluid, because miR-455 and miR-210 were significantly upregulated in KOA patients compared with IKMI patients. Further investigation suggested that miR-455 and miR-210 in synovial fluid were secreted by the hyperplastic synovium in KOA.
Moreover, evidence from the present and previously published studies has suggested a protective role of these miRNAs, which can directly bind to their target genes and regulate chondrocyte metabolism during KOA.33, 34, 35 These findings imply that the synovium not only contributes to the inflammatory responses in KOA but also plays a role in joint tissue homeostasis and repair. Although promising results were observed in DMM mice, the roles of miR-455 and miR-210 in other joint tissues or cells besides chondrocytes remained uncertain and needed to be further investigated.
Indeed, both proinflammatory and anti-inflammatory mediators are elevated in OA synovial tissue and fluid.36,37 TGF-β1 is a pluripotent factor that is crucial in maintaining joint homeostasis. The protective effect of TGF-β1 on healthy cartilage signals mainly via ALK5 activation and subsequently via Smad2/Smad3 phosphorylation. Knockout of SMAD3 or TGFBR2 in mice results in aberrant chondrocyte hypertrophy and progressive joint OA.38,39 Genetic linkage analyses have also demonstrated that patients with SMAD3 mutations are more susceptible to joint OA than those without such mutations.40 However, the role of TGF-β1 in KOA joints is very different from its role in healthy joints. In KOA joints, permanent high levels of active TGF-β1 and increasing ALK1:ALK5 ratios in cartilage together result in Smad1/Smad5/Smad8 pathway activation, leading to chondrocyte hypertrophy and cartilage degeneration.15 In addition, injecting TGF-β1 directly into naive mouse knee joints leads to synovial fibrosis and osteophyte formation similar to those observed in experimental KOA.41,42 Thus, it seems that TGF-β1 itself is not an ideal KOA therapeutic target.
Interestingly, 30 days after intra-articular injection of an adenovirus overexpressing active TGF-β1, fibrotic synovium have been found to undergo chondrometaplasia, as indicated by a change to a rounded, chondrocyte-like morphology and dramatic increases in collagen II and other cartilage link proteins in the matrix.43 In the present study, treating human synoviocytes with TGF-β1 significantly increased the transcription and secretion of miR-455 and miR-210 via the Smad2/Smad3 pathway activation. These findings imply that the elevation in TGF-β1 in the hyperplastic synovium in KOA represents an attempt to repair the tissue. However, KOA is a joint destruction that results from a combination of risk factors. Not only low-grade inflammation but also genetics, aging, obesity, injury, and knee malalignment contribute to the development of KOA. Considering the deleterious effects of TGF-β1 in OA joints, targeting TGF-β1 is apparently inadequate for reversal of joint failure and may ultimately impair joint function. To enable more predictable targeting of TGF-β1-dependent pathways, we further studied the mechanisms of miR-455 and miR-210 upregulation in TGF-β1-treated synoviocytes.
Runx1 is an essential TF required for chondrogenesis and the suppression of subsequent hypertrophy. In a mouse model, deletion of RUNX1 in chondrocytes significantly enhances KOA development,26 whereas Runx1 overexpression significantly attenuates OA-related pathologies.27 However, the role of this molecule in synoviocyte has rarely been reported. A recent study reported that TD-198946 can synergistically enhance synoviocyte chondrogenesis.44 Similar results were observed in our study, in which Runx1 overexpression independently promoted miR-455 and miR-210 transcription in synoviocytes. Through bioinformatics analysis and verification experiments, we demonstrated that Runx1 can bind to the promoter regions of miR-455 and miR-210 and enhance their transcription in TGF-β1-treated synoviocytes. These findings suggest that Runx1 is a potential KOA therapeutic target that does not trigger TGF-β1-associated adverse side effects, such as synovial fibrosis, cartilage degeneration, and osteophyte formation.
Although KOA is the result of joint failure, we cannot completely exclude the existence of protective mechanisms in joints. Taken together, our results reveal that Runx1, downstream of the TGF-β1 signal, can independently promote miR-455 and miR-210 transcription in synoviocytes. Upregulated miR-455 and miR-210 can be extracellularly secreted to form a prochondrogenic environment and in turn attenuate chondrocyte degeneration. Compared with previous studies, the present study provides a more nuanced view of the synovium in KOA and supports TD-198946 as a disease-modifying drug against KOA. Further studies on the mechanisms by which the synovium participates in the regeneration and repair of injured joints are needed.
Materials and Methods
Study Subjects
This study was approved by the Institutional Review Board (IRB) of the First Affiliated Hospital of Sun Yat-sen University (IRB no. 2013C-110) and was conducted in accordance with the Declaration of Helsinki. Convenience samples were used. Between February 2015 and December 2018, 26 KOA patients (14 female and 12 male; age 62.34 ± 12.17 years) who underwent total knee arthroplasty (TKA), 25 patients presenting for IKMI without cartilage defect (10 female and 15 male; age 32.03 ± 9.63 years) verified by arthroscopy, and 10 patients without knee injury or chronic pain history who had undergone LLA for serious trauma (4 female and 6 male; age 33.30 ± 10.63 years) were recruited. Patients with other forms of knee arthritis or exposure to any disease or treatment known to influence cartilage metabolism were not included in the study. Written informed consent was obtained from all patients prior to their participation in this study. Demographics, comorbidities, medications, and the histories of the affected knees were prospectively collected.
Sample Collection
Venous blood samples were collected in ethylenediamine tetraacetic acid (EDTA)-containing Vacutainer tubes prior to surgery. Plasma was separated by centrifugation at 1,000 × g for 15 min and stored at −80°C until use. Synovial fluid aspiration was performed before skin incision during surgeries, including TKA and arthroscopy. Samples with obvious blood contamination were excluded. Synovial fluid from the affected knees was centrifuged at 3,000 × g for 10 min to remove debris and stored at −80°C until use. Both plasma and synovial fluid were available for 18 KOA patients and 15 IKMI patients; only plasma samples were available for the remaining 8 KOA patients and 10 IKMI patients. Synovium (10 mm × 10 mm) and hyaline cartilage (10 mm × 5 mm × 2 mm) samples were washed in phosphate-buffered saline (PBS), soaked in RNAlater stabilization solution (Thermo Fisher Scientific, USA), and stored in liquid nitrogen until utilization. Hyaline cartilage of the medial femoral condyle was collected from 10 LLA patients and 12 KOA patients, and the synovium of the suprapatellar bursa was obtained from 6 LLA patients and 10 KOA patients.
Human Primary Cells and Tissue Culture
Primary human chondrocytes were isolated as previously described.33 The chondrocytes were grown in DMEM/F-12 (GIBCO Life Technologies, USA) with 5% fetal bovine serum (FBS; GIBCO Life Technologies, USA), 1% penicillin/streptomycin (PS; GIBCO Life Technologies, USA), and ITS premix (BD Biosciences, USA). Primary human synoviocytes were isolated following Xu et al.’s45 description and then seeded into flasks containing DMEM/F-12, 10% FBS, and 1% PS. Circulating monocytes were isolated from fresh whole blood with an EasySep Human Monocyte Isolation Kit (STEMCELL Technologies, Canada) and then matured in RPMI 1640 medium (GIBCO Life Technologies, USA) containing 10% FBS, 1% PS, and macrophage colony-stimulating factor (M-CSF; 40 ng/mL; Sino Biological, PR China) for 7 days. SW982 human synovial sarcoma cells (ATCC, USA) were cultured in DMEM/F-12, 10% FBS, and 1% PS. All cells and tissues were incubated in a humidified atmosphere at 37°C under 5% CO2; the medium was changed every 2–3 days. Chondrocytes at passages 1–2 and synoviocytes at passages 4–846 were used in experiments. Synovial tissue (10 mm × 10 mm) was washed in PBS three times, cultured in DMEM/F-12 with 10% exosome-depleted FBS (System Biosciences, USA) and 1% PS for 24 h, and then harvested for miRNA analysis.
Animals and Establishment of a DMM-Induced KOA Model
Twelve-week-old male C57BL/6J mice were purchased from the Model Animal Research Center (Nanjing, PR China). The mice were randomized into four groups: the sham surgery group (n = 5), the DMM group with mmu-miR-455 agomir injection (n = 5), the DMM group with mmu-miR-210 agomir injection (n = 5), and the DMM group with control agomir injection (n = 5). Experimental KOA was induced in mice by transection of the medial anterior meniscotibial ligament in the right knee as previously described;47 sham surgery was used as a control. In KOA mice, intra-articular injections of mmu-miRNA agomir or control agomir (5 nmol; RiboBio, PR China) were administered through the patellar tendon at 7, 21, 35, and 49 days after surgery. At the 10th week after surgery, the mice were sacrificed, and the knee joints were harvested. The degree of cartilage degeneration was assessed by Safranin O staining and immunohistochemistry.
The sections were scored by two observers in a blinded fashion. When discrepancies were noted, a score was assigned after consensus had been reached. The Osteoarthritis Research Society International (OARSI) scoring system was used to grade mouse cartilage degeneration. A 0–6 subjective scoring system was applied to four quadrants (normal = 0; loss of Safranin O without structural changes = 0.5; superficial fibrillation without loss of cartilage = 1; loss of surface lamina = 2; erosion to the calcified layer lesion for 1%–25% = 3; erosion to the calcified layer lesion for 25%–50% = 4; erosion to the calcified layer lesion for 50%–75% = 5; erosion to the calcified layer lesion for 75%–100% = 6).48 The severity of cartilage destruction is presented as an average score of the three highest scores in all slides of the joint. The collagen II content in articular cartilage was histochemically quantified by multiplying the staining score (negative = 0; weak = 1; moderate = 2; strong = 3) and proportion score (<5% = 0; 6%–25% = 1; 26%–50% = 2; 51%–75% = 3; >76% = 4). The average score of five high-power fields was recorded.
Transfection
For RNA interference/overexpression and miRNA mimic/inhibitor analyses, primary human chondrocytes or synoviocytes were seeded in six-well plates and allowed to grow to 80% confluence. The cells were then transfected with an homo sapiens (hsa) miRNA mimic/inhibitor (RiboBio, PR China), siRNA (Gene Pharma, PR China), or overexpression plasmid (Vigene Bioscience, PR China) using Lipofectamine 3000 (Invitrogen, USA) according to the manufacturer’s instructions. The properties of the plasmid vectors are listed in Figure S1. Nonspecific hsa-miRNAs (mimic negative control [NC] and inhibitor NC), NC siRNA oligos, and a plasmid with a scrambled sequence were used as NCs. Cells were harvested after 24 h for qRT-PCR or after 48 h for western blot analysis. The specific siRNA sequences used in the study are listed in Table S1.
Conditioned Medium Exchange
SW982 cells were transfected with a miRNA overexpression or control plasmid and then incubated for 18 h in complete DMEM/F-12 followed by 24 h in serum-free DMEM/F-12 supplemented with 1% PS. The conditioned medium was concentrated with an Amicon Ultra-15 (3 kDa) and used to treat chondrocytes immediately. Chondrocytes were harvested after 24 h for qRT-PCR.
CUT&Tag Assays
SW982 cells were grown to confluence in 100-mm dishes, treated for 24 h with 10 ng/mL TGF-β1, and used for CUT&Tag assay analysis. A NovoNGS ChiTag Transposon Kit (Novoprotein, PR China) was used following the manufacturer’s recommended protocol.49 An anti-Runx1 antibody (ab23980; Abcam, USA) was used to pull down DNA-protein complexes, and rabbit IgG was used as an NC. Purified DNA was quantified by qRT-PCR, and GAPDH was used as a reference. The primer sequences used to amplify the predicted binding sites of Runx1 are provided in Table S2.
RNA Extraction, Reverse Transcription, and qRT-PCR
Total RNA from cells and tissue samples was extracted using an RNeasy Mini Kit (QIAGEN, USA) according to the manufacturer’s instructions. Samples of tissues such as synovium and hyaline cartilage were weighed and ground in liquid nitrogen prior to RNA isolation. A miRNeasy Serum/Plasma Kit (QIAGEN, USA) was used to purify miRNA from biofluid samples, including plasma and synovial fluid samples. The RNA samples were qualified and quantified using a NanoDrop spectrophotometer (NanoDrop Technologies, USA).
Next, cDNA was synthesized using a miScript II RT Kit (QIAGEN, USA). qRT-PCR for the target genes was performed in a CFX96 qRT-PCR machine (Bio-Rad, USA) using a miScript SYBR Green PCR Kit (QIAGEN, USA). Specific primer sequences for hsa-precursor-miRNAs were purchased from RiboBio, and the others used are listed in Tables S3 and S4. The transcript levels were normalized to those of the reference gene GAPDH for mRNA and U6 small nuclear (snRNA) for miRNA in cells, culture supernatants, and tissue samples. To normalize the miRNA in biofluid samples, we added a fixed quantity (5.6 × 108 copies) of exogenous spike-in Cel-miRNA-39 to each sample (200 μL) following the manufacturer’s instructions. Relative gene expression was calculated using the comparative cycle threshold (CT) method (ΔΔCt).
Transcriptome Analysis
Total RNA was extracted from synoviocytes treated with or without TGF-β1. Synoviocytes from three KOA patients were used for transcriptome sequencing. The sequencing libraries were prepared using a TruSeq RNA Sample Preparation Kit (Illumina) according to the manufacturer’s instructions. Sequencing was performed using an Illumina HiSeq 4000 System with 2 × 150 bp paired-end reads (Majorbio, PR China). The raw sequence data were filtered to remove adaptor sequences, low-quality reads, sequences with a high content of N, and reads <50 bp long by using SeqPrep and Sickle. The filtered data were aligned against the human reference genome using HISAT2 (v.2.1) with the default parameters and then input into StringTie (v.1.3.6) software. The gene expression levels were calculated as transcripts per million (TPM) reads values by using RNA-Seq by Expectation-Maximization (RSEM) software. The fold changes and p values of the differentially expressed genes (DEGs) were derived with DEG sequencing (DEG-seq).
Western Blot Analysis
Western blot analysis of whole-cell extracts was performed as previously described.33 The primary antibodies were anti-Aggrecan (1:1,000, MABT83; Millipore, USA), anti-Collagen II (1:1,000, ab188570; Abcam, USA), anti-SOX9 (1:1,000, AB5535; Millipore, USA), anti-Collagen X (1:1,000, ab182563; Abcam, USA), anti-ADAMTS4 (1:800, ab185722; Abcam, USA), anti-MMP13 (1:2,000, ab39012; Abcam, USA), anti-Smad2 (1:1,000, #5339; Cell Signaling Technology [CST], USA), anti-Smad3 (1:1,000, #9523; CST, USA), anti-phospho-Smad2 (1:1,000, ab53100; Abcam, USA), anti-phospho-Smad3 (1:1,000, ab52903; Abcam, USA), anti-Runx1 (1:800, ab23980; Abcam, USA), and anti-GAPDH (1:2,000, #97166; CST, USA). GAPDH was utilized as an internal control. The grayscale values of the blots were quantitated with ImageJ software by following the instructions. The grayscale value of each target protein was normalized to that of GAPDH.
In Situ Hybridization, Immunohistochemistry, and Immunofluorescence
Human and mouse tissues were fixed in 4.0% paraformaldehyde at 4°C overnight. Human tissues were subsequently dehydrated with a graded series of ethanol, embedded in paraffin, and cut into 5-μm-thick sections. Mouse joints were submitted to this process after decalcification in 10% EDTA for 4 weeks. Cells on coverslips were fixed at room temperature for 45 min. To detect miRNA expression, we subjected sections and cells on coverslips to in situ hybridization analysis using miRNA-specific probes (Servicebio, PR China) (Table S5). In brief, paraffin sections were deparaffinized and incubated for 30 min at 37°C with 20 μg/mL Proteinase K. Then endogenous peroxidases were blocked in 1% hydrogen peroxide. The sections were prehybridized in hybridization buffer for 1 h at 50°C and then subjected to hybridization with a miRNA probe (8 ng/μL) overnight at 37°C. Then the sections were washed for subsequent Digoxigeni (DIG) detection methods. The nuclei were stained with hematoxylin. For cells on coverslips, a similar procedure was performed after the cells were treated with 20 μg/mL Proteinase K for 5 min at 37°C.
For immunohistochemistry analysis, sections were treated as previously described.19 Primary antibodies against collagen II (GB12021; Servicebio, PR China) were used at a 1:100 dilution. For immunofluorescence analysis, the indicated cells were incubated with primary antibodies (1:100) against phospho-Smad2, phospho-Smad3, and Runx1, incubated with secondary antibodies, and finally counterstained with DAPI.
Statistical Analysis
Statistical analysis was performed using SPSS v.13.0 statistical software (SPSS, USA). Continuous variables are expressed as the mean plus the standard deviation (SD). Unpaired t tests and Mann-Whitney U tests were used to identify differences between groups. One-way analysis of variance (ANOVA) followed by the Bonferroni test was carried out for multiple-group comparisons. Correlations between two quantitative variables were analyzed using Pearson’s correlation coefficient. A p value <0.05 and a fold change >2 were considered to indicate statistical significance. For all cell experiments, the analyses were performed on biological triplicates.
Author Contributions
Study conception and design: Z.Z. and Y.K. Acquisition, analysis, and interpretation of the data: X.Z., H.H., R.P., X.W., and S.H. Patient recruitment and sample collection: F.M. and Z.Y. Manuscript drafting and editing: X.Z. and F.M. Critical revision of the article for important intellectual content: Z.Z. and Y.K. All authors approved the final version of the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (grants 81601921, 81672145, 81874016, 81702142, 81972051, and 81972049); the Natural Science Foundation of Guangdong Province, China (grants 2016A030310156, 2017A030313804, and 2019A1515011946); the Science and Technology Planning Project of Guangdong Province, China (grant 2019A030317007); the Science and Technology Project of Guangzhou City, China (grant 201710010164); and the Basic Research Funding of Sun Yat-sen University (grant 19ykpy63).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.10.004.
Contributor Information
Yan Kang, Email: neokang@163.com.
Zhiqi Zhang, Email: zhzhiqi@mail.sysu.edu.cn.
Supplemental Information
References
- 1.Hunter D.J., Bierma-Zeinstra S. Osteoarthritis. Lancet. 2019;393:1745–1759. doi: 10.1016/S0140-6736(19)30417-9. [DOI] [PubMed] [Google Scholar]
- 2.Loeser R.F., Collins J.A., Diekman B.O. Ageing and the pathogenesis of osteoarthritis. Nat. Rev. Rheumatol. 2016;12:412–420. doi: 10.1038/nrrheum.2016.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wallace I.J., Worthington S., Felson D.T., Jurmain R.D., Wren K.T., Maijanen H., Woods R.J., Lieberman D.E. Knee osteoarthritis has doubled in prevalence since the mid-20th century. Proc. Natl. Acad. Sci. USA. 2017;114:9332–9336. doi: 10.1073/pnas.1703856114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nukavarapu S.P., Dorcemus D.L. Osteochondral tissue engineering: current strategies and challenges. Biotechnol. Adv. 2013;31:706–721. doi: 10.1016/j.biotechadv.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Armiento A.R., Alini M., Stoddart M.J. Articular fibrocartilage—Why does hyaline cartilage fail to repair? Adv. Drug Deliv. Rev. 2019;146:289–305. doi: 10.1016/j.addr.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 6.Chen S., Fu P., Cong R., Wu H., Pei M. Strategies to minimize hypertrophy in cartilage engineering and regeneration. Genes Dis. 2015;2:76–95. doi: 10.1016/j.gendis.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mobasheri A., Rayman M.P., Gualillo O., Sellam J., van der Kraan P., Fearon U. The role of metabolism in the pathogenesis of osteoarthritis. Nat. Rev. Rheumatol. 2017;13:302–311. doi: 10.1038/nrrheum.2017.50. [DOI] [PubMed] [Google Scholar]
- 8.Wojdasiewicz P., Poniatowski Ł.A., Szukiewicz D. The role of inflammatory and anti-inflammatory cytokines in the pathogenesis of osteoarthritis. Mediators Inflamm. 2014;2014:561459. doi: 10.1155/2014/561459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balakrishnan L., Nirujogi R.S., Ahmad S., Bhattacharjee M., Manda S.S., Renuse S., Kelkar D.S., Subbannayya Y., Raju R., Goel R. Proteomic analysis of human osteoarthritis synovial fluid. Clin. Proteomics. 2014;11:6. doi: 10.1186/1559-0275-11-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robinson W.H., Lepus C.M., Wang Q., Raghu H., Mao R., Lindstrom T.M., Sokolove J. Low-grade inflammation as a key mediator of the pathogenesis of osteoarthritis. Nat. Rev. Rheumatol. 2016;12:580–592. doi: 10.1038/nrrheum.2016.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X., Hunter D.J., Jin X., Ding C. The importance of synovial inflammation in osteoarthritis: current evidence from imaging assessments and clinical trials. Osteoarthritis Cartilage. 2018;26:165–174. doi: 10.1016/j.joca.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 12.Remst D.F., Blaney Davidson E.N., van der Kraan P.M. Unravelling osteoarthritis-related synovial fibrosis: a step closer to solving joint stiffness. Rheumatology (Oxford) 2015;54:1954–1963. doi: 10.1093/rheumatology/kev228. [DOI] [PubMed] [Google Scholar]
- 13.Kapoor M., Martel-Pelletier J., Lajeunesse D., Pelletier J.-P., Fahmi H. Role of proinflammatory cytokines in the pathophysiology of osteoarthritis. Nat. Rev. Rheumatol. 2011;7:33–42. doi: 10.1038/nrrheum.2010.196. [DOI] [PubMed] [Google Scholar]
- 14.Albro M.B., Cigan A.D., Nims R.J., Yeroushalmi K.J., Oungoulian S.R., Hung C.T., Ateshian G.A. Shearing of synovial fluid activates latent TGF-β. Osteoarthritis Cartilage. 2012;20:1374–1382. doi: 10.1016/j.joca.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van der Kraan P.M. The changing role of TGFβ in healthy, ageing and osteoarthritic joints. Nat. Rev. Rheumatol. 2017;13:155–163. doi: 10.1038/nrrheum.2016.219. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Z., Kang Y., Zhang Z., Zhang H., Duan X., Liu J., Li X., Liao W. Expression of microRNAs during chondrogenesis of human adipose-derived stem cells. Osteoarthritis Cartilage. 2012;20:1638–1646. doi: 10.1016/j.joca.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 17.Mao G., Wu P., Zhang Z., Zhang Z., Liao W., Li Y., Kang Y. MicroRNA-92a-3p regulates aggrecanase-1 and aggrecanase-2 expression in chondrogenesis and IL-1β-induced catabolism in human articular chondrocytes. Cell. Physiol. Biochem. 2017;44:38–52. doi: 10.1159/000484579. [DOI] [PubMed] [Google Scholar]
- 18.Chang Z.K., Meng F.G., Zhang Z.Q., Mao G.P., Huang Z.Y., Liao W.M., He A.S. MicroRNA-193b-3p regulates matrix metalloproteinase 19 expression in interleukin-1β-induced human chondrocytes. J. Cell. Biochem. 2018;119:4775–4782. doi: 10.1002/jcb.26669. [DOI] [PubMed] [Google Scholar]
- 19.Meng F., Zhang Z., Chen W., Huang G., He A., Hou C., Long Y., Yang Z., Zhang Z., Liao W. MicroRNA-320 regulates matrix metalloproteinase-13 expression in chondrogenesis and interleukin-1β-induced chondrocyte responses. Osteoarthritis Cartilage. 2016;24:932–941. doi: 10.1016/j.joca.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 20.Raab-Traub N., Dittmer D.P. Viral effects on the content and function of extracellular vesicles. Nat. Rev. Microbiol. 2017;15:559–572. doi: 10.1038/nrmicro.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anfossi S., Babayan A., Pantel K., Calin G.A. Clinical utility of circulating non-coding RNAs—an update. Nat. Rev. Clin. Oncol. 2018;15:541–563. doi: 10.1038/s41571-018-0035-x. [DOI] [PubMed] [Google Scholar]
- 22.Malemud C.J. MicroRNAs and Osteoarthritis. Cells. 2018;7:92. doi: 10.3390/cells7080092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malda J., Boere J., van de Lest C.H.A., van Weeren P.R., Wauben M.H.M. Extracellular vesicles—new tool for joint repair and regeneration. Nat. Rev. Rheumatol. 2016;12:243–249. doi: 10.1038/nrrheum.2015.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bandyopadhyay S., Bhattacharyya M. PuTmiR: a database for extracting neighboring transcription factors of human microRNAs. BMC Bioinformatics. 2010;11:190. doi: 10.1186/1471-2105-11-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Derynck R., Budi E.H. Specificity, versatility, and control of TGF-β family signaling. Sci. Signal. 2019;12:eaav5183. doi: 10.1126/scisignal.aav5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yano F., Ohba S., Murahashi Y., Tanaka S., Saito T., Chung U.I. Runx1 contributes to articular cartilage maintenance by enhancement of cartilage matrix production and suppression of hypertrophic differentiation. Sci. Rep. 2019;9:7666. doi: 10.1038/s41598-019-43948-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aini H., Itaka K., Fujisawa A., Uchida H., Uchida S., Fukushima S., Kataoka K., Saito T., Chung U.I., Ohba S. Messenger RNA delivery of a cartilage-anabolic transcription factor as a disease-modifying strategy for osteoarthritis treatment. Sci. Rep. 2016;6:18743. doi: 10.1038/srep18743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yano F., Hojo H., Ohba S., Fukai A., Hosaka Y., Ikeda T., Saito T., Hirata M., Chikuda H., Takato T. A novel disease-modifying osteoarthritis drug candidate targeting Runx1. Ann. Rheum. Dis. 2013;72:748–753. doi: 10.1136/annrheumdis-2012-201745. [DOI] [PubMed] [Google Scholar]
- 29.Remst D.F., Blom A.B., Vitters E.L., Bank R.A., van den Berg W.B., Blaney Davidson E.N., van der Kraan P.M. Gene expression analysis of murine and human osteoarthritis synovium reveals elevation of transforming growth factor β-responsive genes in osteoarthritis-related fibrosis. Arthritis Rheumatol. 2014;66:647–656. doi: 10.1002/art.38266. [DOI] [PubMed] [Google Scholar]
- 30.Scanzello C.R., Goldring S.R. The role of synovitis in osteoarthritis pathogenesis. Bone. 2012;51:249–257. doi: 10.1016/j.bone.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Atukorala I., Kwoh C.K., Guermazi A., Roemer F.W., Boudreau R.M., Hannon M.J., Hunter D.J. Synovitis in knee osteoarthritis: a precursor of disease? Ann. Rheum. Dis. 2016;75:390–395. doi: 10.1136/annrheumdis-2014-205894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Asghar S., Litherland G.J., Lockhart J.C., Goodyear C.S., Crilly A. Exosomes in intercellular communication and implications for osteoarthritis. Rheumatology. 2020;59:57–68. doi: 10.1093/rheumatology/kez462. [DOI] [PubMed] [Google Scholar]
- 33.Sun H., Zhao X., Zhang C., Zhang Z., Lun J., Liao W., Zhang Z. MiR-455-3p inhibits the degenerate process of chondrogenic differentiation through modification of DNA methylation. Cell Death Dis. 2018;9:537. doi: 10.1038/s41419-018-0565-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang D., Cao X., Li J., Zhao G. MiR-210 inhibits NF-κB signaling pathway by targeting DR6 in osteoarthritis. Sci. Rep. 2015;5:12775. doi: 10.1038/srep12775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Z., Meng D., Li G., Xu J., Tian K., Li Y. Overexpression of microRNA-210 promotes chondrocyte proliferation and extracellular matrix deposition by targeting HIF-3α in osteoarthritis. Mol. Med. Rep. 2016;13:2769–2776. doi: 10.3892/mmr.2016.4878. [DOI] [PubMed] [Google Scholar]
- 36.Daghestani H.N., Kraus V.B. Inflammatory biomarkers in osteoarthritis. Osteoarthritis Cartilage. 2015;23:1890–1896. doi: 10.1016/j.joca.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X., Hunter D., Xu J., Ding C. Metabolic triggered inflammation in osteoarthritis. Osteoarthritis Cartilage. 2015;23:22–30. doi: 10.1016/j.joca.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 38.Li T.F., Gao L., Sheu T.J., Sampson E.R., Flick L.M., Konttinen Y.T., Chen D., Schwarz E.M., Zuscik M.J., Jonason J.H., O’Keefe R.J. Aberrant hypertrophy in Smad3-deficient murine chondrocytes is rescued by restoring transforming growth factor β-activated kinase 1/activating transcription factor 2 signaling: a potential clinical implication for osteoarthritis. Arthritis Rheum. 2010;62:2359–2369. doi: 10.1002/art.27537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen J., Li J., Wang B., Jin H., Wang M., Zhang Y., Yang Y., Im H.J., O’Keefe R., Chen D. Deletion of the transforming growth factor β receptor type II gene in articular chondrocytes leads to a progressive osteoarthritis-like phenotype in mice. Arthritis Rheum. 2013;65:3107–3119. doi: 10.1002/art.38122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van de Laar I.M., Oldenburg R.A., Pals G., Roos-Hesselink J.W., de Graaf B.M., Verhagen J.M., Hoedemaekers Y.M., Willemsen R., Severijnen L.A., Venselaar H. Mutations in SMAD3 cause a syndromic form of aortic aneurysms and dissections with early-onset osteoarthritis. Nat. Genet. 2011;43:121–126. doi: 10.1038/ng.744. [DOI] [PubMed] [Google Scholar]
- 41.Schelbergen R.F., de Munter W., van den Bosch M.H., Lafeber F.P., Sloetjes A., Vogl T., Roth J., van den Berg W.B., van der Kraan P.M., Blom A.B., van Lent P.L. Alarmins S100A8/S100A9 aggravate osteophyte formation in experimental osteoarthritis and predict osteophyte progression in early human symptomatic osteoarthritis. Ann. Rheum. Dis. 2016;75:218–225. doi: 10.1136/annrheumdis-2014-205480. [DOI] [PubMed] [Google Scholar]
- 42.van Beuningen H.M., Glansbeek H.L., van der Kraan P.M., van den Berg W.B. Osteoarthritis-like changes in the murine knee joint resulting from intra-articular transforming growth factor-β injections. Osteoarthritis Cartilage. 2000;8:25–33. doi: 10.1053/joca.1999.0267. [DOI] [PubMed] [Google Scholar]
- 43.Watson R.S., Gouze E., Levings P.P., Bush M.L., Kay J.D., Jorgensen M.S., Dacanay E.A., Reith J.W., Wright T.W., Ghivizzani S.C. Gene delivery of TGF-β1 induces arthrofibrosis and chondrometaplasia of synovium in vivo. Lab. Invest. 2010;90:1615–1627. doi: 10.1038/labinvest.2010.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yano F., Hojo H., Ohba S., Saito T., Honnami M., Mochizuki M., Takato T., Kawaguchi H., Chung U.I. Cell-sheet technology combined with a thienoindazole derivative small compound TD-198946 for cartilage regeneration. Biomaterials. 2013;34:5581–5587. doi: 10.1016/j.biomaterials.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 45.Xu S., Xiao Y., Zeng S., Zou Y., Qiu Q., Huang M., Zhan Z., Liang L., Yang X., Xu H. Piperlongumine inhibits the proliferation, migration and invasion of fibroblast-like synoviocytes from patients with rheumatoid arthritis. Inflamm. Res. 2018;67:233–243. doi: 10.1007/s00011-017-1112-9. [DOI] [PubMed] [Google Scholar]
- 46.Manferdini C., Paolella F., Gabusi E., Silvestri Y., Gambari L., Cattini L., Filardo G., Fleury-Cappellesso S., Lisignoli G. From osteoarthritic synovium to synovial-derived cells characterization: synovial macrophages are key effector cells. Arthritis Res. Ther. 2016;18:83. doi: 10.1186/s13075-016-0983-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Glasson S.S., Blanchet T.J., Morris E.A. The surgical destabilization of the medial meniscus (DMM) model of osteoarthritis in the 129/SvEv mouse. Osteoarthritis Cartilage. 2007;15:1061–1069. doi: 10.1016/j.joca.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 48.Zirlik A., Maier C., Gerdes N., MacFarlane L., Soosairajah J., Bavendiek U., Ahrens I., Ernst S., Bassler N., Missiou A. CD40 ligand mediates inflammation independently of CD40 by interaction with Mac-1. Circulation. 2007;115:1571–1580. doi: 10.1161/CIRCULATIONAHA.106.683201. [DOI] [PubMed] [Google Scholar]
- 49.Kaya-Okur H.S., Wu S.J., Codomo C.A., Pledger E.S., Bryson T.D., Henikoff J.G., Ahmad K., Henikoff S. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 2019;10:1930. doi: 10.1038/s41467-019-09982-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.