Abstract
Biomaterial scaffolds are increasingly being used to drive tissue regeneration. The limited success so far in human tissues rebuilding and therapy application may be due to inadequacy of the functionality biomaterial scaffold. We developed a new decellularized method to obtain complete anatomical skin biomatrix scaffold in situ with extracellular matrix (ECM) architecture preserved, in this study. We described a skin scaffold map by integrated proteomics and systematically analyzed the interaction between ECM proteins and epidermal cells in skin microenvironment on this basis. They were used to quantify structure and function of the skin’s Matrisome, comprised of core ECM components and ECM-associated soluble signals that are key regulators of epidermal development. We especially revealed that ECM played a role in determining the fate of epidermal stem cells through hemidesmosome components. These concepts not only bring us a new understanding of the role of the skin ECM niche, they also provide an attractive combinational strategy based on tissue engineering principles with skin biomatrix scaffold materials for the acceleration and enhancement of tissue regeneration.
Keywords: Skin, extracellular matrix, biomatrix scaffold, epidermal stem cell, hemidesmosome
Introduction
Skin is a typical stratified tissue with distinct layers of epidermis and dermis, where the epidermis is a stratified squamous epithelium. Keratinocytes within the basal layer occupy a unique niche distinguished by their contact with the basement membrane (BM), composed of extracellular matrix (ECM) components and growth factors that contribute both to the epidermis and the underlying dermis. Adhesion of keratinocytes to the BM provides not only a physical supporting tissue mechanics for myosin-dependent contractions, but is also a highly specialized ECM microenvironment, which plays a crucial role in skin development and maintaining skin function.1 If they lose contact with the BM, the innermost keratinocytes, also called epithelial stem cells (EpSCs), lose stemness, and undergo terminal differentiation. This process has been found to be associated with migration to more superficial skin layers.2 The EpSCs cease to proliferate and move upward to produce the skin’s barrier, which is the feature that maintains their proliferative status.3 Therefore, skin ECM is of fundamental importance in regulating EpSCs maintenance, proper mobilization, and differentiation.4
ECM contains numerous components, primarily proteins, proteoglycans (PGs), and carbohydrates, which constitute a complex, three-dimensional microenvironment. This microenvironment organizes the tissue macroscopically and also provides many chemical and biophysical signals required for cell functions. The exact ECM composition, the interplay between the skin ECM and microenvironment, and how they cooperate to regulate the biology of EpSCs in skin, are largely unknown despite several reports on cell fate determination through cell-matrix interactions in stem-cell niches.4–6 It is therefore important to acquire more knowledge of ECM composition and functions.
There was no effective method to fully analyze the ECM composition of any tissue, given its complexity until recently, with the rapid development of proteomics tools. Martin et al. coined the term “Matrisome” in 1984, to define the components of ECM.7 Naba et al. proposed to extend this term to define structural ECM components and ECM interactions or remodeling ECM components using bioinformatics tools and experimental approaches.8 They established the Matrisome database in silico and with experimental data on ECM genes and components of 14 different tissues and tumor types.9
Matrisome components are distinguished as the “core Matrisome components,” comprising collagens, PGs, and glycoproteins that serve as adhesion molecules and play other roles, versus “Matrisome-associated components,” based on the structural and functional features of ECM. They comprise ECM-bound proteins and carbohydrates (e.g. glycosaminoglycan (GAG) chains not linked to a core protein but bound to proteins by anionic charge binding), which include regulatory signals and secreted factors that are bound to one or more core Matrisome components. The Matrisome composition of multiple organs has been identified at least in part and including that for aorta, bone, colon, lung, liver, heart, and mammary gland.9–11 The Matrisome components in skin tissue have not yet been identified or characterized. This is because of the lack of an effective method to obtain the skin matrix, while retaining as much as possible of the Matrisome components, even though several functions of some ECM components on different cell types of skin have been reported.12,13 However, most prior methods of decellularization are quite harsh and preserve only cross-linked ECM primarily (or entirely), thus eliminating many biologically potent ECM components, especially ones that are newly synthesized and not yet cross-linked. For example, it was found that the most widely used surfactant (sodium dodecyl sulfate, SDS) damages structural and signaling proteins and components.14,15 Another surfactant—t-octyl phenoxy polyethoxy ethanol, triton X-100—can decrease native tissue GAG content.15
We have identified gentler decellularization strategies16 based on protocols using known solubility properties of collagens.17 Conditions are used that keep all of the collagens insoluble and preserve also factors bound to them. The method has been demonstrated to retrieve a tissue extract, referred to as “biomatrix scaffolds,” containing more than 98% of the collagens of the tissue and associated proteins and PGs and essentially all of the known signals (growth factors, cytokines) that bind to these core components. We recently reported our successful application of liver biomatrix scaffolds (LBSs) to reconstitute a stable and reproducible engineered whole liver to model the nonalcoholic fatty liver disease (NAFLD) development process and a hepatic organoids culture system constructed with cryopulverized LBSs to identify hepatotoxic compounds.18,19 More and more investigations have shown that the decellularized biomatrix represents the secreted products of resident cells of tissues and organs and provides cues affecting cell functions and fate. Understanding the exact Matrisome composition and potential beneficial effects of biologic scaffolds on tissue homeostasis can accelerate the development of the field of tissue engineering and regenerative medicine.
We modified the established protocol for LBSs, tailoring it for porcine skin, and obtained skin biomatrix scaffolds (SBSs) that support EpSCs growth during wound healing in this study. We revealed an in-depth whole profile of skin ECM, the modulatory properties of ECM components, the bound signals and regulators-all collectively influencing skin tissue maintenance and epithelization for the first time. These discoveries provide us with enhanced knowledge of skin ECM composition and provide attractive combinational strategies of tissue engineering principles using skin ECM components for accelerating and enhancing tissue regeneration or for establishing bioengineered skin constructs.
Materials and methods
Human tissue resource
We obtained human foreskin tissues from patients undergoing circumcision from the first and fifth medical center of Chinese PLA General Hospital, after obtaining ethical permission and informed consent. This study was approved by the Medical Ethics Committee of Chinese PLA General Hospital (No. S2018-123-02).
Animal models
All animal experiments were conducted according to the approved standards established by the institutional Animal Care and Use Committee at Beijing Institute of Health Service and Transfusion Medicine at the Laboratory Animal Center of the Academy of Military Medical Sciences.
Full-thickness skin was harvested from 3-month-old male piglets, which were derived from the SiPeiFu Biotechnology Company. Six male mice were anesthetized with Ketamine and hair was removed from their backs using a depilatory cream before the surgeries. Two 6-mm full-thickness wounds were excised from the dorsum of mice on each side of midline using a biopsy punch. A rubber ring was placed in each wound to prevent them from shrinking. The acellular skin bioscaffolds (SBSs) were broken into powder before dissolving in them phosphate buffered saline (PBS) to form a solution of 10 µg/µl protein concentration. About 150 µl of SBSs powder solution was applied on the wounds of the experimental group every 3 days. Mice were sacrificed after 3, 7, and 14 days of surgery and the wounds were fixed in 4% formaldehyde for staining analysis.
Preparation of acellular, porcine SBSs
A 3 to 6-mm thick segment of porcine skin tissue was cut with an electric skin picker. The skin tissues were rinsed with cold PBS, followed by delipidation with phospholipase A2 combined with sodium deoxycholate for 2.5 to 3.5 h in a shaker at 37°C until the tissue segments became oyster white. Then, the surface of the skin was scraped gently and carefully with the back of a knife to remove the epidermis. The samples were rinsed with high salt washes (3.4 M NaCl) for 1 h and then washed for the last time with PBS containing nucleases (DNase 10 μg/ml, RNase 5 μg/ml) at 37°C for 1 h. Collagen, glycosaminoglycan (GAG), and nuclear acid content in SBSs was evaluated to confirm the efficiency of the decellularization. The amount of collagen was quantified with Sircol dye by reading the absorbance at 555 nm, and GAG content was determined with a Blyscan assay and normalized with a heparin sodium standard curve. Total residual DNA was assessed by using a Fluorescent DNA Quantitation Kit. Finally, the acellularized scaffolds were flash frozen prior to Western blot and proteomic analyses.
Protein and peptide extraction
The acellularized, biomatrix scaffolds of porcine skin were quickly frozen on dry ice and pulverized using a freezer mill filled with liquid nitrogen. About 1 ml Protein Extraction Reagent Type 4 was added per 125 mg of pulverized tissue, and the sample was placed in a homogenizer for ECM protein extraction. The supernatants were collected for decellularization quality control analysis and to extract protein peptide segments after centrifugation at 14,000 × g for 30 min. Next, for peptide extraction, 60 μg samples (the supernatants mentioned above) were solubilized in 1M DTT at 37°C for 1 h. Samples were added with 1M IAA and kept in the dark for 1 h. All samples were collected, and the suspension was removed after centrifugation at 12,000 × g. Then, 100 μl UA was added, and the suspension was removed after centrifugation twice. NH4HCO3 (50 mM) was added, and the suspension was removed after centrifugation three times. Final digestion was conducted using trypsin at 37°C overnight, at a ratio of 1:50 enzyme: substrate. The peptide mixtures were analyzed using an Orbitrap Q-Exactive Mass Spectrometer equipped with an Easy-nLC nanoflow LC system.
Mass spectrometry and data analysis
All MS/MS raw files were processed with the MaxQuant software20 (version 1.5.8.3), searched against the UniProt (https://www.uniprot.org/) database for porcine samples (downloaded on August of 2017). Peptide and fragment ion mass tolerances were set to 4.5 ppm and 20 ppm, respectively. Fixed cysteine carbamido methylation (C + 57.022), variable methionine oxidation (M + 15.995), and Acetyl (Protein N-term) were specified. Up to two missed cleavages were allowed, and trypsin was set as the enzyme specificity. Automatic target and reverse database searches were used, and a false discovery rate of 1% at both the peptide and the protein level were allowed. Protein was quantified using the intensity-based absolute quantification (iBAQ) method21 embedded in MaxQuant.
We used DAVID22 to annotate identified proteins according to biological processes, cellular components, and molecular functions of Gene Ontology,23 as well as the pathways involved based on KEGG.24 A global protein interactomic network was built with the Cytoscape (version 3.6.1) software.25 Protein-protein interactions were retrieved from the major public online tool STRING database.26 We acquired a protease list for annotation analysis by using several protease-related keywords when searching the UniProt database, followed by a manual check.
Cell culture
The human foreskin biopsies were cut into squares measuring approximately 2 mm2, using sterile scalpels. The epidermal sheets were separated from the dermis and were allowed to continue digesting with 37°C pre-warmed 0.25% trypsin/EDTA for 15 min following digestion with 2 mg/ml Dispase II at 37°C for 1 h. Digestion was stopped with fetal bovine serum, and the epidermal cells were washed twice with cold PBS to remove trypsin before filtering with a 40-mm strainer and centrifuged at 1000 rpm for 3 min. In the case of 2D (monolayer) cultures, cells were seeded onto plates pre-coated with Matrigel. Advanced DMED/F-12 medium supplemented with 1% NEAA, 2% B-27, 1% Glutamax, 1% HEPES, 1.25 mM N-ace, 50 ng/ml hEGF, 50 ng/ml Wnt3a, 1 µM A83, 10 µM Fosklin, and 10 µl/ml of 1× penicillin/streptomycin was added. For 3D cultures (organoids), the isolated epithelial cells of human foreskins were cultured on ultralow-attachment culture dishes, and in the aforementioned medium with 0.2% sodium hyaluronate, with or without pulverized skin biomatrix scaffold material. All culture medium was removed from the wells and replaced with fresh medium after 24 h. The 3D organoids were harvested for the next experiment after 8 days of culture.27
Recellularized artificial skin equivalent construction with the cells directly in contact or without contact with the BM layer
An artificial skin equivalent was constructed through the recellularization of human EpSCs seeded onto the acellularized SBSs. The primary EpSCs were enriched in organoids, then digested into a single cell suspension, and seeded onto the SBSs at 50,000 cells/cm2 in six-well plates. For DC, EpSCs were seeded in SBSs with the cells directly in contract with the BM layer. For non-contact (NC), EpSCs were seeded in back-to-BM, which was not connected to the BM layer. The artificial skins were maintained in the air/liquid interphase for 20 days, as indicated in the text, in all experiments.
TEM
The samples were washed in PBS and fixed overnight in 0.1% formalin and 2 h in 2.5% glutaraldehyde. The samples were then washed in PBS again, dehydrated in ascending concentrations of ethanol, and air-dried. The ultrastructure of the artificial skins was gold spattered and mounted for imaging on a transmission electron microscope.
Hematoxylin/eosin staining, histology, and immunofluorescence
Samples were fixed in 4% formaldehyde overnight at 4°C and then processed with a dehydration gradient. The sections were cut to a thickness of 4 μm for H&E staining or immunohistochemistry and immunofluorescence analysis after the tissues were embedded in paraffin.
The sections were deparaffinized, heated with a microwave until they boiled for at least 12 min with antigen retrieval buffer, and then removed with endogenous catalase in 0.3% H2O2 for 30 min after cooling for immunohistochemical staining. Then, the sections were blocked with normal horse serum in Tris-buffered saline solution for 1 h, blocked with Avidin/Biotin Blocking Kit, and stained with antibodies overnight at 4°C. The sections were colored after staining with a secondary antibody.
The cells were fixed in 4% formaldehyde for 20 min and washed in PBS for immunofluorescence staining. Then, the cells were treated with 0.25% Triton X-100 for 20 min to eliminate the plasma membranes, blocked in 10% serum for 1 h at room temperature (RT), and incubated with primary antibodies overnight at 4°C. The sections were sealed with Fluoro-Gel for Photography after being incubated for 1 h at RT with secondary antibodies and counterstained with DAPI. Negative control samples were incubated with secondary antibody alone. The photographs were taken at 20×/40× magnification and analyzed with Volocity Demo (× 64).
Confocal microscopy
Immunofluorescence imaging was used to observe polarity, cell junctions and functions of artificial skin equivalents, and 3D cell spheres. Materials were fixed in 4% formaldehyde for 20 min and washed in PBS. Samples were then subjected to 0.5% Triton X-100 for 15 min and subsequently blocked for 1 h with 10% serum for 1 h at RT and incubated with primary antibodies overnight at 4°C. Images were taken using an LSM 700-point scanning confocal microscope equipped with 5× and 10× objectives and adjusted linearly for presentation using Photoshop (Adobe Inc., Seattle, WA) after being incubated for 1 h at RT with secondary antibodies and counterstained with DAPI.
Co-immunoprecipitation and Western blot analysis
EpSCs were solubilized in 1 ml cold lysis buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1% Triton X-100) containing complete protease inhibitors and shaken by ultrasonic cell disruptor at 4°C until the buffer solution was clear. The cell lysates were incubated with 2 µg anti-plectin for 2 h at 4°C, followed by incubation with 30 µl Protein A/G PLUS agarose for 12 h at 4°C. Protein A/G PLUS agarose was then washed with cold lysis buffer at 4°C three times. The proteins in the immunocomplexes were extracted in SDS sample buffer and used for immunoblotting to identify interacting partner proteins.
For Western blot analysis, the Integrin β4 antibodies were incubated for 2 h at 37°C. After washing three times, blots were incubated with secondary antibodies for 50 min at 37°C. The signals were detected using Pierce™ ECL Western Blotting Substrate and Millipore Immobilon™ Western Chemiluminescent HRP Substrate at RT after washing three times.
Statistical analysis
The statistical data are shown as means ± SEM for all results. Student’s t test was used to compare data between groups. ANOVA was used to compare three or more groups. Replicates used were biological replicates. Results were considered significant at p ⩽ 0.05. Statistical tests were carried out using GraphPad Prism 6 (La Jolla, CA, USA) software for Apple Mac.
Data and materials availability
All mass spectrometry data have been deposited to the ProteomeXchange Consortium via the iProX partner repository28 with the dataset identifier PXD014454.
Results
SBSs support EpSCs growth during wound healing
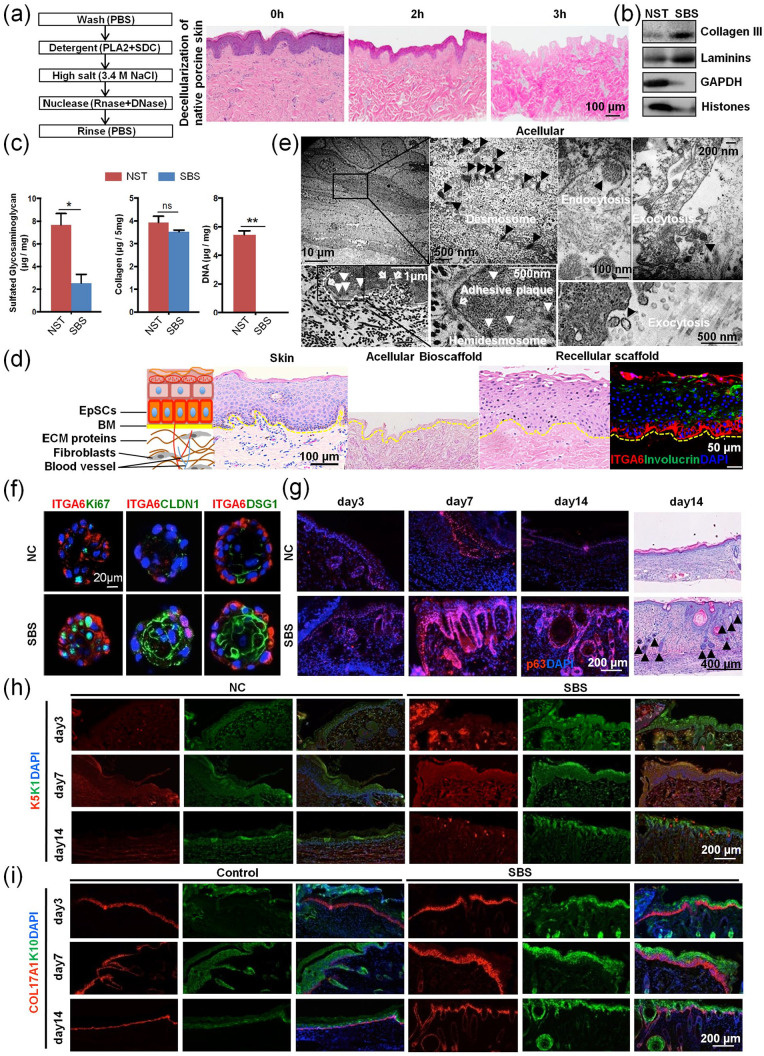
We obtained SBSs from freshly harvested porcine skin, and then used it for recellularization (Figure 1(a)), by using modifications of our established strategy of preparing biomatrix scaffolds from liver tissue.16 All cells, their cytosolic content (e.g. cytoskeleton, ribosomes, GAPDH, and lipids), and all nucleic acids (DNA and RNA) were completely removed from the native skin tissue (NSTs), while preserving the skin ECM. It is comprised of multiple collagen types, adhesion molecules (e.g. laminins) and PGs and factors bound to them and that were preserved in SBSs (Figure 1(a)–(c)).
Figure 1.
Acellular skin biomatrix scaffolds supports epidermal stem cells (EpSCs) growth during wound healing. (a) Three-step decellularization method for generating biomatrix scaffolds and hematoxylin/eosin (HE) staining analysis on native porcine dermal tissue versus skin biomatrix scaffolds, prepared for 2 h and 3 h, respectively (scale bar = 100 μm). (b) Immunoblotting analysis of GAPDH (cytosol) and histones (nucleus) in NST and SBS after three-step decellularization. (c) Quantitative analysis of free glycosaminoglycans (GAGs), independent of proteoglycans; collagens and DNA in NST and SBS. (a) Overview of stratified skin structure with representative hematoxylin/eosin staining (scale bar: 100 μm) and immunofluorescence (scale bar: 50 μm) on normal skin, on acellularized biomatrix scaffolds, and on scaffolds that have been recellularized artificial skin, respectively. (e) TEM observations on recellularized artificial skin. The black arrowheads point to the desmosome, endocytosis, and exocytosis (scale bars: 10 μm, 100, 200, and 500 nm). The white arrowheads point to the hemidesmosome, and the hollow arrowheads point to the adhesive plaque on the junction of cells on the skin biomatrix scaffolds along the newly formed BM (scale bar: 1 μm, 500 nm). (f) Immunofluorescence analysis of ITGA6, Ki67, CLDN1, and DSG1 in human EpSCs in organoid cultures with pulverized SBS for 7 days (scale bars: 20 μm). (g) Immunofluorescence analysis of p63 during the process of mouse wound healing (day 3, 7, and 14) (scale bar: 200 μm) and hematoxylin/eosin staining of mouse repaired skin in day 14 (scale bar: 400 μm). Black arrowheads point to the epidermal appendage. (h, i) Immunofluorescence analysis of K5, K1, COL17A1, and K10 during the process of mouse wound healing (day 3, 7, and 14) (scale bar: 200 μm).
An artificial skin equivalent can be obtained by seeding hEpSCs onto the SBSs with stratification and polarity preserved. The skin equivalent showed a specific hierarchical structure that retained the morphological characteristics of normal, native skin, with the Integrin subunit alpha 6 (ITGA6)+ EpSCs located on the basal layers and involucrin (IVL)+ terminal differentiated epithelia cells on the superficial layers that equate to the spinous and granular layers in real skin tissue (Figure 1(d)), after 20 days of recellularization cultures. The ultrastructure of the skin equivalent with well-stratified epithelial layers on top of the SBSs, with multiple desmosomes on the intracellular membranes of the keratinocytes and with hemidesmosome complexes and adhesive plaques on the junctions of basal layer cells to the surface of the SBSs (Figure 1(e)) was observed with transmission electron microscopy (TEM). By maintaining the cells with the support of SBSs, the layers of epidermal cells showed active exocytosis; many extracellular vesicles could be found around the interface between cells and matrix.
The SBSs were pulverized into powders and mixed with EpSCs to facilitate the generation of cultures of floating 3D spheres. Results indicated that in the presence of the pulverized SBSs, the EpSCs formed organoids demonstrating a maturational gradient, with the outer layer, that in contact with the pulverized powder, showing expression of the stem-cell marker ITGA6, while cells on several inner layers showing expression of the proliferation marker Ki67, and the differentiated epidermal cell junctional markers Claudin 1 (CLDN1) and desmosomal glycoprotein 1 (DSG1). Conversely, the organoids that formed without the pulverized SBSs did not form a typical epithelial cell differentiation gradient (Figure 1(f)). These results indicated that the SBSs are functionally supportive for both formation of an artificial skin equivalent and for EpSCs’ differentiation and maturation. The wound repair of skin needs the dynamic process of EpSCs’ proliferation and differentiation to complete reepithelialization.29 A mouse wound healing model was established (Supplemental Figure S1A) in order to further study whether SBSs promote wound healing by regulating the function of EpSCs. The results indicated that the function of EpSCs was enhanced in the presence of the pulverized SBSs, and expression of EpSCs transcription factor p63, stem-cell marker K5 and K14, epidermal maturation marker K1 and K10, and hemidesmosome COL17A1 increased significantly during the wound healing process on days 3, 7, and 14 (Figure 1(g)–(i) and Supplemental Figure S1B). EGFR and integrins are important EpSCs receptors in skin.30,31 The results indicate that Integrin Subunit Beta 1(ITGB1) and EGFR of EpSCs were activated more strongly at the wound site with SBSs treatment (Supplemental Figure S1B). Furthermore, we were surprised to find that SBSs can promote the formation of epidermal appendages in the dermis after the final stage of wound healing on day 14 (Supplemental Figure S1C and Figure 1(g)). In contrast, most of the injured parts did not form epidermal appendages after reepithelialization without SBSs treatment. These results indicated that the SBSs are functionally supportive for wound healing through regulating EpSCs’ reepithelialization and epidermal appendages formation. The ability of SBSs to fully support EpSCs motivated us to explore possible mechanisms through further study of the components and the assembly of SBSs. The schematics of a label-free proteomic Matrisome analysis strategy and construction of an artificial skin equivalent with hEpSCs and SBSs are shown in Supplemental Figure S2.
Unbiased analysis on overall skin Matrisome with SBSs
The integrity of the skin Matrisome was analyzed specifically in SBSs compared with that in NSTs. The quantitative proteomics analysis was employed to characterize ECM components by using liquid chromatography in combination with tandem mass spectrometry (LC-MS/MS). Parallel analyses independently performed on two groups of samples showed a large overlap of the identified proteins (Supplemental Figure S3), which indicated the reproducibility of our experimental pipeline and a good correlation between the two groups (Supplemental Table S1). More bioactive proteins with low abundance were identified as supplements by using a cytokine array to display an integrated skin ECM map (Supplemental Table S2).
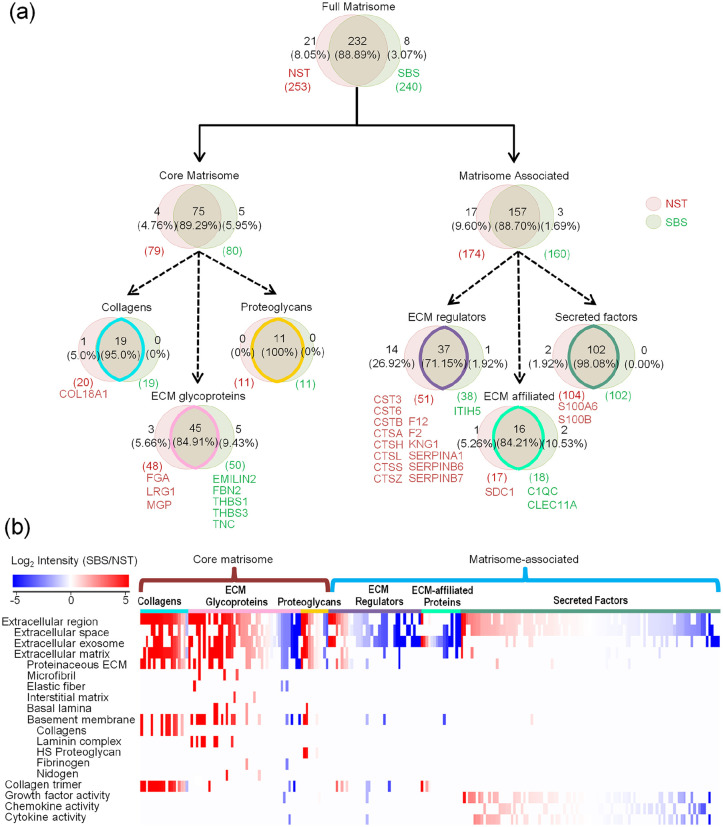
Briefly, a total of 261 ECM proteins constitute the Matrisome components annotated in a published Matrisome database,9 with 240 of them identified from SBSs, while 253 of which are from NSTs (Figure 2(a), Supplemental Tables S3). Interestingly, 232 out of 261 Matrisome components were identified in both, which means that SBSs have an 88.89% identity to that in the NSTs. The overlap of core Matrisome components and that of Matrisome-associated was more than 89.29% and 88.70%, respectively. The heatmap analysis showed an enrichment of core Matrisome components. The top components were enriched in the annotated terms of extracellular vesicles, collagens, laminin complexes, various fibers, heparan sulfate (HS) PGs, fibrinogens (FG), and nidogens (NID) from the composition ratio (Figure 2(b)).
Figure 2.
Unbiased analysis on overall skin Matrisome with SBSs. (a) Pie charts represent the proportion of the six categories (collagens, ECM glycoproteins, proteoglycans, ECM regulators, ECM-affiliated proteins, and secreted factors) that make up the Matrisome in both NST and SBS. The red and green circulars represented NST and SBS, respectively. (b) Functional term analysis of the six ECM categories identified according to log2 fold changes in SBS relative to NST. Red and blue boxes indicate proteins with increased and decreased abundance in SBS, respectively. Clusters of proteins associated with a similar set of functions were grouped as indicated at left side. Cyan, pink, yellow, purple, green, and atrovirens lines correspond to the six components of the skin Matrisome.
The core Matrisome components are known to support overall scaffold structures and mechanical physical properties.32 Almost all of the collagens and PGs and most of the glycoproteins have been preserved in the SBSs. Glycoproteins are major components of the non-collagenous core of Matrisome components in skin ECM and are known to regulate cell proliferation, migration, differentiation, and wound healing.13 Laminins, elastin fibers, elastin-associated microfibrils, elastin fiber interfacers, fibrillins (FBN), NID, FG, fibronectins (FN), and fibulins (FBLN)—all found highly enriched in SBSs—are included in this group (Figure 3(a)).
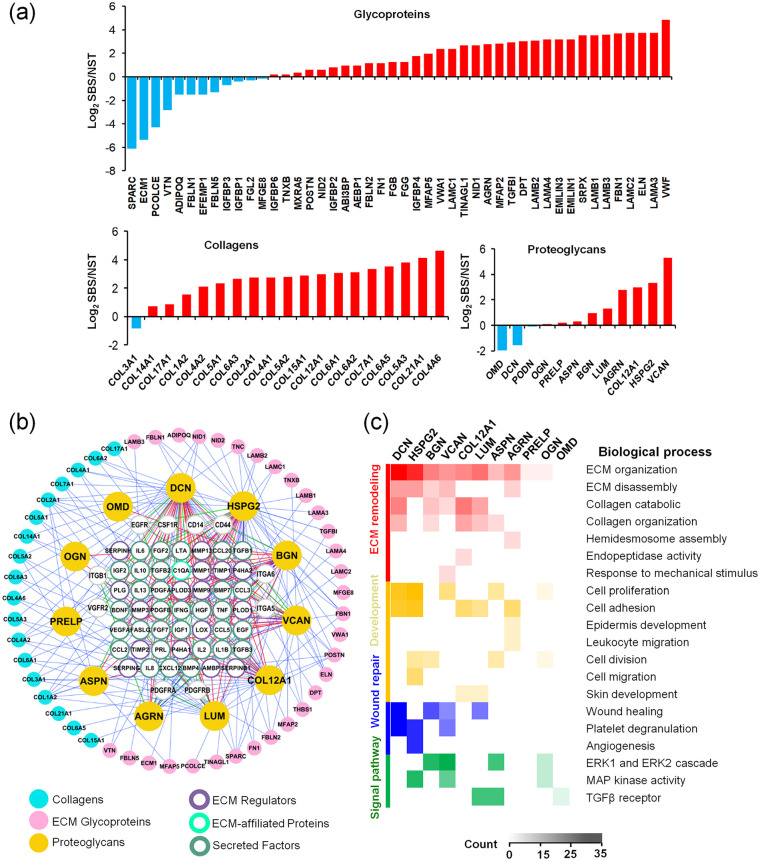
Figure 3.
Soluble regulators (Matrisome-associated components) can be highly preserved in SBS through interactions with scaffold proteins or PGs. (a) Quantitative proteomic analysis of NST and SBS. Types of collagens, proteoglycans, and ECM glycoproteins identified in NST and SBS. Y-axis represented the value of log2 SBS/NST. (b) Overview of the integrative analysis of the direct interactions between soluble Matrisome-associated proteins and PGs, collagens, glycoproteins, and receptors. Cyan, pink, and yellow circles correspond to collagens, ECM glycoproteins, and PGs; purple, green, and atrovirens circles correspond to ECM regulators, ECM-affiliated proteins, and secreted factors. Gray circles correspond to receptors. Red, green, and blue lines represent the direct interactions between PGs and Matrisome-associated proteins, receptors, and collagens and glycoproteins. (c) Biological process analyses of PGs (DCN, HSPG2, BGN, VCAN, COL12A1, LUM, ASPN, AGRN, PRELP, OGN, OMD) found in SBS and the proteins with which they interact. The columns with different colors on the left of the heatmap represent different functional categories.
Collagens are the principle structural component of ECM cross-linked networks. Among the known genetically distinct collagens, we identified 12 types of them in SBSs, which were highly enriched compared with those in NSTs (Figure 3(a)). Collagen IV is an important structural protein in BM. We found that it was the most retained collagen in SBSs, and three protein isoforms were identified. Other collagens including COL15A1, COL2A1, COL5A1, as well as COL7A1 and COL17A1 (especial for skin) in BM were also found enriched. These anchor fibrillar collagens are important structures connecting basal layer EpSCs to the BM, and play critical roles in the proliferation and migration of keratinocytes and fibroblasts, which are important events of reepithelialization.13 COL1A2, COL5A2, and COL5A3 are fibrillar-forming collagen, which are widely found in skin tissue. COL6A1, COL6A2, and COL6A5 are major structural components of microfibrils in skin. COL12A1 and COL14A1 are also fibril-associated collagens, which are involved in tissue remodeling, wound healing, and regulation of fibrillogenesis, respectively.
COL12A1 is one of the collagen PGs found enriched in SBSs and to which specific GAG chains and/or oligosaccharides are attached. Collagen PGs are major components of the ECM and form an intricate lattice in the spaces between cells by binding to a variety of other ECM proteins. Furthermore, cell surface PG, Biglycan (BGN), pericellular and BM PGs, Agrin (AGRN), Heparan Sulfate PG 2 (Perlecan, HSPG2) and Proline-Arginine-Rich, End Leucine-Rich Repeat Protein (PRELP), extracellular PGs and small leucine-rich PGs (SLRPs) lumican (LUM), decorin (DCN), osteoglycin (OGN), asporin (ASPN), and podocan (PODN) were the PGs identified. Most of them have been reported enriched in skin and involved in the skin hydration, viscoelasticity,33 ECM assembly, vasculogenesis, fibrosis,34 and immunity35 functions. Among them, versican (VCAN), HSPG2, LUM, and BGN were four of the top six PGs specifically enriched in SBSs, which were important for skin function and were well preserved (Figure 3(a)). In contrast, some bone-specific PGs such as osteomodulin (OMD), OGN, PRELP, ASPN, and smooth-muscle-specific PG PODN, were not preserved well in SBSs, compared to the NSTs. Their relevance to skin structural and functional maintenance must await future studies.
Soluble regulators (Matrisome-associated components) can be highly preserved in SBSs through interactions with Core Matrisome components
Most of Matrisome-associated components are growth factors, chemokines, and cytokines that act as regulators to facilitate cell attachment and growth by interacting with core ECM components. We found that there were 141 Matrisome-associated proteins that were well retained in the SBSs, with minor loss of some of them (fold change >1/8 compared to NSTs)10 (Supplemental Figure S4, Tables S1 and S2). The rest of the Matrisome-associated proteins included PLA2 inhibitors, S100 calcium-binding proteins, serpin peptidase inhibitors, and transglutaminases. They were more likely to appear in multiple tumors, skin diseases, and wound healing, than in normal skin tissue. Theoretically, those hydrophilic, secretory factors are extremely easy to lose during the decellularization processes unless attached by strong intermolecular bonds. The fact that they were preserved enables them to be considered as constituents of ECM. The Venn diagram analysis shows that those soluble regulators may be bound to the scaffold through interaction with multiple types of core Matrisome components, and some of them can be loaded and transported with exosomes (Supplemental Figure S5).
PGs can store and release biologically active soluble signals through their binding to the sulfated carbohydrate chains of GAGs.36 Epitopes of the GAG chains form a complex with the soluble signals and work synergistically through specific receptors to elicit intracellular signal transduction processes. The PGs of SBS were found relevant to several important receptors in our study, including EGFR, Integrin Subunit Beta 1(ITGB1), CD44 (hyaluronan receptors), CD14, Platelet-Derived Growth Factor Receptor Alpha, Platelet-Derived Growth Factor Receptor Beta, Kinase Insert Domain Receptor, Colony Stimulating Factor 1 Receptor (CSF1R), and Fas Cell Surface Death Receptor (FAS) (Figure 3(b)). A map of interaction networks showed 48 Matrisome-associated proteins (31 out of 48 were secreted factors) interacted with the 11 PGs, which could be further bound to the scaffold with interactions with other core Matrisome components (Figure 3(b)). For example, DCN ranked first, were relevant to Vascular Endothelial Growth Factors (VEGFs), Insulin Like Growth Factor, Hepatocyte Growth Factors (HGF), and inflammatory factors such as Interleukin 10 (IL10), Lymphotoxin Alpha (LTA), and Tumor Necrosis Factor (TNF) (Figure 3(b) and Table 1). These have been found abundant throughout the human dermal layers, especially in the papillary dermis, and interacting with collagen fibrils and aggregated skin ECM.37 Results showed that perlecan, the major PG of BM, is relevant with several soluble ECMs in skin including Epidermal Growth Factor (EGFs), HGFs, Fibroblast Growth Factor (FGFs), Platelet-Derived Growth Factor (PDGFs), VEGFs and Prolactin (PRL), inflammatory factors Interferon (IFN), Interleukin 2 (IL2), IL10 through the GAG chains of HSs and chondroitin sulfates.
Table 1.
Information on the proteoglycans identified in SBS and their interacted cytokines.
| Number | Gene name | Abbreviation | GAGPGs | GAG chains | Skin ECM location | Interacted factors |
|---|---|---|---|---|---|---|
| 1 | Decorin | DCN | DSPG2 | CS/DS | Ubiquitous collagen fibril associated | TGFB1, MMP3, TGFB2, TGFB3, IGF1, MMP9, EGF, VEGFA, CXCL12, FGF2, IL10, IL1B, TNF, C1QA, IGF2, TIMP2, TIMP1, MMP1, LOX, PLG, LTA, HGF, MMP13, BDNF, CCL2, CCL3, FGF7, IFNG, IL8 |
| 2 | Perlecan | PLC | HSPG2 | HS/CS/DS | Basal epidermis/dermis, basement | FGF2, PDGFB, PDGFA, PLG, MMP3, VEGFA, MMP13, PRL, FGF7, HGF, IL2, IFNG, EGF, MMP9, BDNF, CCL2, CCL3, CCL5, CXCL12, IL1B, IL6, IL8, TGFB1, TNF |
| 3 | Biglycan | BGN | DSPG1 | CS/DS | Dennoepidermal border, BM and connective tissue sheath of the hair follicle | TGFB1, TGFB3, TGFB2, BMP4, FGF2, VEGFA, IL6, BMP7, TNF, IL13, TIMP1, MMP9, MMP3, LOX, BDNF, CCL2, CCL3, EGF, FGF7, HGF, IFNG, IL8 |
| 4 | Versican | VCAN | CSPG2 | CS/DS | Widespread distribution in epidermis, dermis and vasculature and elastic networks in skin | CCL2, CXCL12, MMP9, CCL5, PLG, EGF, TIMP1, TGFB1, MMP3, FGF2, MMP1, VEGFA, CCL20, BDNF, CCL3, FGF7, HGF, IFNG, IL8 |
| 5 | Collagen type XII alpha 1 | COL12A1 | First widely distributed and becomes restricted in the upper, papillary dermis after 6 months of gestation | SERPINH1, PLOD1, P4HA1, P4HA2, PLOD3, LOX, MMP9, SERPING1, SERPINB1, MMP3 | ||
| 6 | Lumican | LUM | KSPG | KS | Dermis | TGFB1, TGFB3, TGFB2, FASLG, AMBP, FGF2 |
| 7 | Asporin | ASPN | Dermis | FGF2, FGF7, CCL3, CCL5, CXCL12, IL1B, IL6, TNF | ||
| 8 | Agrin | AGRN | HS | Basement membrane | TGFB1, TGFB3, TGFB2 | |
| 9 | Prolargin | PRELP | KS | Basement membrane | FGF2 | |
| 10 | Osteoglycin | OGN | KS | In normal vessels | FGF2, AMBP | |
| 11 | Osteomodulin | OMD | KSPG | KS | N/A | TGFB1 |
Further biological process analyses on the above PGs showed that almost all of the PGs were responsible for ECM remodeling such as ECM organization and disassembly or collagen organization and catabolic processes (Figure 3(c)). They were involved in several signaling pathways, such as the Extracellular Signal-Regulated Kinase (ERK1/2) cascade, Mitogen-Activated Protein Kinase cascade, lipopolysaccharide and TGFβ receptor pathways. Altogether, core Matrisome PGs act as major reservoirs for soluble and biologically active factors in the skin microenvironment and play critical roles facilitating their mechanisms of action in tissues.
PGs also interact actively with matrix metalloproteinases (MMPs), which can not only catalyze the normal turnover of ECM macromolecules such as the interstitial and BM collagens, but also interact with PGs to release secreted factors in addition to the binding and interaction with secreted factors.38,39 Our results identified MMP1 and MMP13, collagenases with a wide substrate specificity and capable of degrading HSPG2 and VCAN.40 MMP9 is a gelatinase and capable of releasing cytokines, chemokines and HSPG2.41 MMP3, a stromelysin, can degrade many ECM proteins such as fibronectin, denatured collagens, laminin, and PGs such as HSPG2.42 The interaction map indicated that MMPs and its inhibitors were likely to bind more PGs as substrates in skin (Figure 3(b) and Table 1). Given that dozens of MMPs have been identified to trigger angiogenesis, turnover of the ECM, and tumor cell motility action and up regulated only in the context of skin diseases or wound healing processes, it is understandable that only a few MMPs with low abundance expression and many inhibitors of metalloproteinases (TIMPs) can be found in SBSs in this study focused on normal tissue under homeostasis conditions.
Together, the results from the above analyses suggest that preservation of soluble Matrisome-associated components is from their interactions with scaffold proteins and especially their binding to the GAG chains of PGs. The fully, biologically active scaffolds enabled us to further explore how these Matrisome components support skin-specific functions.
Interactions between soluble factors and core Matrisome components are necessary for wound healing
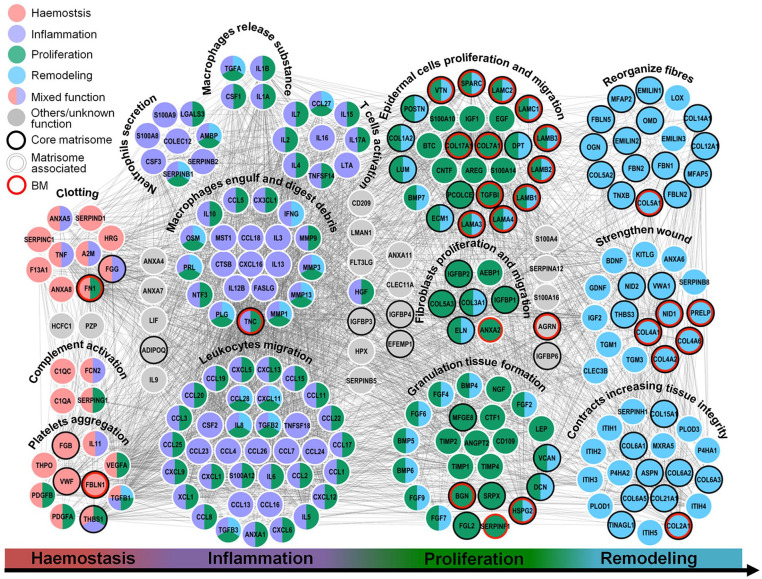
The process of wound healing is a continuous, seamless process and can be divided into hemostasis, inflammation, proliferation, and remodeling phases. Some soluble factors such as growth factors and inflammatory factors are captured by interactions with core Matrisome components and assist with rapid repair throughout the different phases of the healing process. We studied further how ECM molecules are involved in the biological and pathological process as an extracellular microenvironment comprised of interactome networks of core Matrisome proteins, PGs and soluble factors. From the landscape, we found that some Matrisome components occurred in a specific stage, while most of them were involved in more than one phase (Figure 4). Most of the soluble Matrisome-associated components (chemokines, interleukins, and growth factors) demonstrated effects at the phase of hemostasis or inflammation to initiate cell reactions and/or support cell proliferation through endocrine, paracrine, autocrine, or intracrine pathways in the target cells. As an example, TNF, PDGF, and VEGF were responsible for hemostasis and Colony Stimulating Factor 1 (CSF1), Transforming Growth Factor Alpha, Interleukin 1 Alpha (IL1A/B), and Interferon (IFN) were responsible for inflammation, according to our results.43 Some components demonstrated biological activity in more phases. TGFB1, for example, was of particular importance for the entire processes by exerting pleiotropic effects on wound healing by modulating the immune response, regulating cell proliferation, differentiation, and ECM production.44
Figure 4.
Interactions between soluble factors and core Matrisome components necessary for wound healing. The primary biological processes and functional classes analyses show the map of the Matrisome interactome networks that are involved with the four major phases of wound healing: hemostasis (red), inflammation (purple), proliferation (green), and remodeling (cyan), respectively. Black, white shadow, and red hollow circles represent core Matrisome components, Matrisome-associated factors, and Matrisome components of the BM. Interactions of Matrisome components are indicated by gray lines.
Both core Matrisome components and soluble factors are involved in the proliferation and migration of epidermal cells and fibroblasts, and the granulation tissue formation, the most important stage of completing reepithelialization. Granulation tissue is a vascularized and cell-rich tissue replacing the fibrin clot in the wound zone. At least four MMPs (MMP1, MMP3, MMP9, and MMP13) have been identified in SBSs, acting on angiogenesis and vascular remodeling through release of several soluble factors. In addition, those MMPs’ involvement continues into the last remodeling phase, modulating matrix function, and degradation of ECM. For example, MMP13 is capable of cleavage of type II collagen and fibrinogen,45 and MMP1 is capable of proteolytic cleavage of fibrillar collagen leading to unwinding of the triple-helical structure that leaves collagen (or gelatin) highly susceptible to degradation by other proteases to reduce hypertrophic scars.46 Although MMP3 is incapable of degrading triple-helical collagens, it can degrade FN, denatured collagens (gelatin) and laminin.46
ECM organization and disassembly requires a battery of proteases working together to efficiently degrade collagens and elastins to regulate the state of macromolecules within the ECM to provide outside-in signals for cells through ECM receptors. We identified 10 and eight proteases produced for collagen and elastin organization respectively; 10 proteases for ECM organization; and 18 proteases or peptidases for post-translational modifications (Supplemental Figure S6 and Table S4). ECM degradation results in a multitude of cytokines and growth factors secreted by immune and tissue-resident cells released by the action of proteases during wound healing. The biologically active form of VEGF is released by MMP9 and together they are capable of angiogenesis Altogether, skin Matrisome components play an important role throughout the whole process of wound healing by synergistic effects of core Matrisome components and soluble factors.
Interaction network analyses revealed Matrisome components of BM are necessary for establishment of skin structure and functions
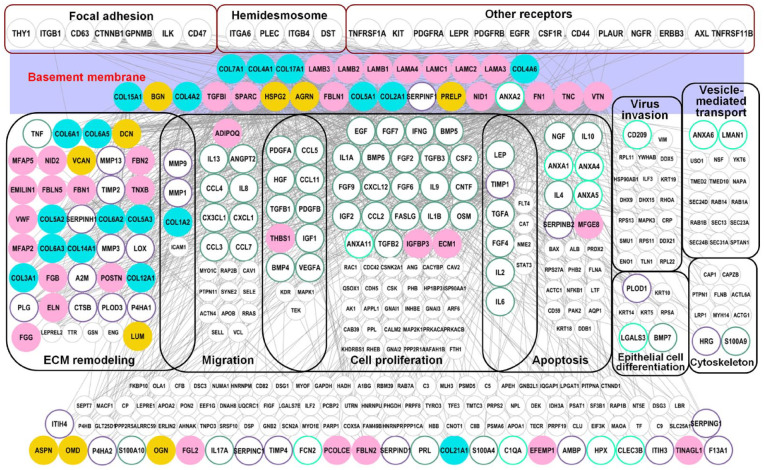
As the major component of the natural stem-cell niche for the basal EpSCs, ECM polymers, and growth factors of BM, and their mechano-physical properties, along with those of the underlying dermis provide necessary complex stimuli for the behavior of basal cells.47 Our results indicated that most Matrisome components in BM were responsible for various biological processes especially for the reepithelialization stage (Figure 4). There were 28 Matrisome components including eight collagens, 14 ECM glycoproteins, four PGs, and two Matrisome-associated proteins identified in SBSs. An interactome network was constructed between Matrisome components in BM and other factors in SBSs (Figure 5) in order to further study the effect of Matrisome components of BM on EpSCs biology. Firstly, our results indicated that Matrisome proteins of BM were closely related to anchoring complexes including the hemidesmosome, focal adhesion, and other receptors, which are important in mediating cell-matrix interactions. Hemidesmosomes, which provide mechanical strength to the cytoplasm below the nucleus of the columnar basal cells, have a major role in promoting epithelial stromal attachment in stratified and complex epithelia. Furthermore, in addition to linking the actin cytoskeleton indirectly to BM, α3β1-rich focal adhesions have a major role in basal layer growth and migration through their ligands such as laminin 5. Other receptors, such as hyaluronan receptors CD44, EGFR, PDGFR, and other all have important roles in intracellular signal transduction of extracellular important ligands. Secondly, we also found that Matrisome components in BM are involved in many biological processes such as cell proliferation, differentiation, migration, apoptosis, viral invasion, skeleton rearrangement, and vesicle transport. The above biological processes are important functions for skin homeostasis maintenance under normal conditions and facilitating responses appropriate to injuries.
Figure 5.
Interactome network analysis revealed Matrisome components in BM are necessary for skin functions and maintenance. Interactome network between Matrisome components of BM and identified in SBS is highly associated with the main biological processes and functional classes of skin. Gray circles correspond to non-Matrisome components. Purple rectangles represent BM structure. Black and red solid frame represent different skin functional areas and anchoring complexes of the sites between basal cells and biomatrix scaffolds, respectively.
In addition, proteases are involved in the supporting of BM to basal EpSCs to provide space for the vertically differentiation and expansion and formed layered distribution of the skin tissue. We identified collagenases (MMP1, MMP3, and MMP9) secreted by epidermal cells in SBSs that are supposed to break down collagens of BM and dermis, and plasminogen activator, which stimulates the production of plasmin. These results indicate that Matrisome components in BM are necessary for epidermal cells to function through anchoring complexes of epidermal cells and the interactome networks with other proteins in SBSs.
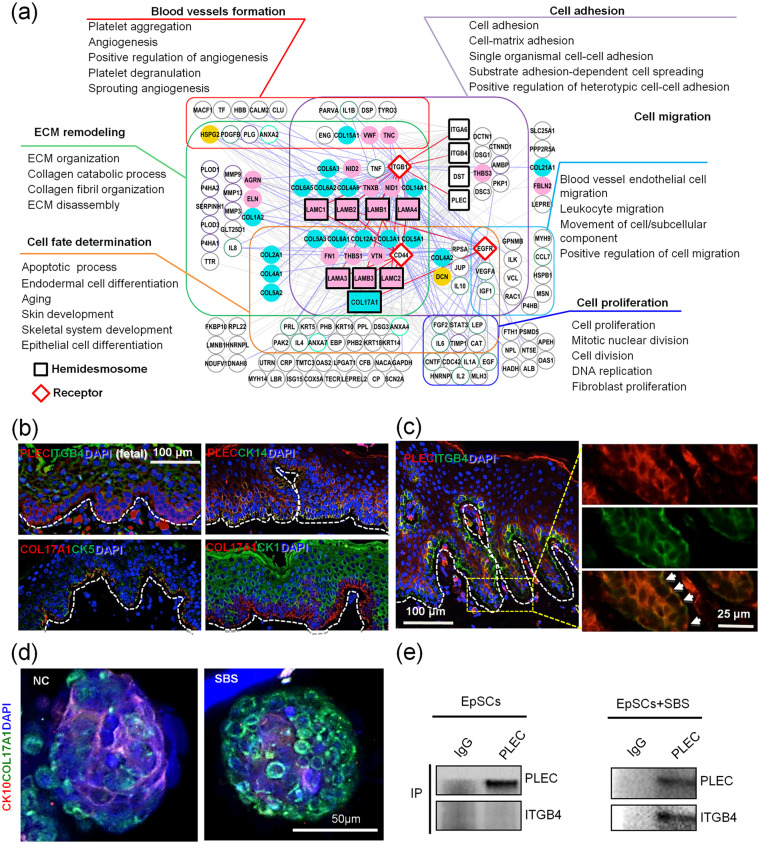
Functional hemidesmosomes are necessary for EpSCs’ fate decisions and bioengineered skin construction in BM
The hemidesmosomes are the specialized anchoring complexes of skin, which connects basal EpSCs to the BM and underlying ECM proteins, and affects the migration and differentiation of epidermal cells through adhesion.48 Our maps of the interactome between hemidesmosome components and other proteins in SBSs suggest that the most enriched biological processes are those involved in cell adhesion, migration, and proliferation, which is consistent with findings in previous reports.48 Most importantly, we found that blood vessel formation, ECM remodeling and cell fate determination also had close relationships with hemidesmosome assembly and important epidermal cell receptors (Figure 6(a)). Here we found that not only COL17A1, but also hemidesmosome component, Plectin (PLEC), are co-expressed with EpSCs markers Keratin 14 (CK14) (Figure 6(b)). Most importantly, we found that when mixed with pulverized SBSs powders, EpSC organoids formed complete hemidesmosome complexes more quickly compared to the controls (Figure 6(c)). The interaction between PLEC and ITGB4 in human skin tissue proved that the hemidesmosome is functional49 (Figure 6(b) and (c)). In order to identify whether Matrisome components influence hemidesmosome activity, we tested the assemble ability of hemidesmosome complexes in primary cultured EpSCs. Co-IP data showed that only with the support of SBSs powder, interactions between PLEC and ITGB4 could be found, which suggest that functional hemidesmosomes organization needs functional SBS support (Figure 6(e)).
Figure 6.
Functional analysis on hemidesmosome-associated factors found in SBS. (a) Map of the interactome networks between hemidesmosome components and other identified proteins in SBS are highly involved with the main biological processes and functional classes of skin. Squares and diamonds represent hemidesmosome components and receptors, respectively. Purple, cyan, blue, orange, green, and red frames and solid lines represent different biological processes. (b and c) Immunofluorescence analyses of PLEC, ITGB4 and other proteins expressed in fetal and adult skin (scale bar: 100 and 25 μm). Arrowheads point to the hemidesmosome complexes. (d) Immunofluorescence analyses of CK10 and COL17A1 expression in skin organoids cultured with pulverized SBS (scale bar: 50 μm). (e) Immuno-co-precipitation analysis of PLEC and ITGB4 on EpSCs and recellularized scaffolds.
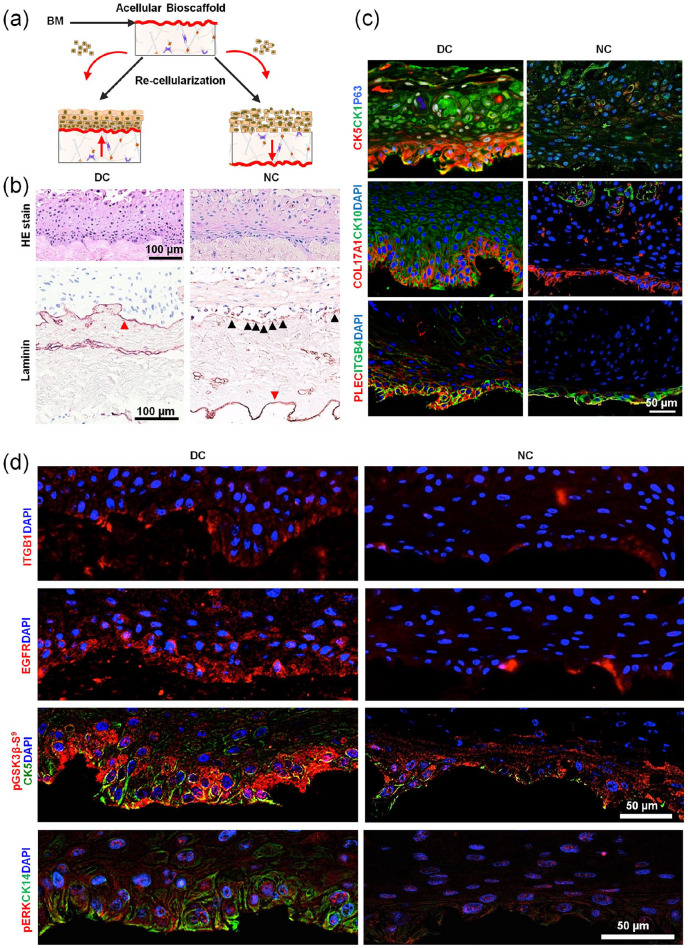
Finally, EpSCs have been recellularized either upright or inverted onto the decellularized scaffolds to represent direct contact (DC) and NC cultures, respectively (Figure 7(a)) in order to further identify whether hemidesmosomes influence functional skin equivalent construction. Results indicated that the recellularized artificial skin equivalents in the DC group were well stratified and polarized, whereas the one in the NC cultures was disordered (Figure 7(b) and (c)). Laminins, the important interacting molecules of hemidesmosomes in BM, expressed continuously in the DC cultures, while EpSCs in the NC one only secreted a little laminins distributed along the BM discontinuously (Figure 7(b)). In addition, the hemidesmosome complexes of the former expressed well in the basal layer, which was similar to normal human skin, with PLEC and ITGB4 co-expression in the bottom of the EpSC layer. This suggested that the functional hemidesmosome structure was established through the interactions between PLEC and ITGB4. The expressions of them on the NC cultures were weak and disordered (Figure 7(c)).
Figure 7.
Hemidesmosomes are necessary for EpSCs’ fate determination. (a) Schematic illustration of recellularization strategies by connecting EpSCs directly to biomatrix scaffolds directly versus indirectly to scaffolds. (b) Hematoxylin/eosin (HE) staining and immunohistochemistry analysis of laminin on the recellularized skin-like tissue (scale bar: 100 μm). (c) Immunofluorescence analysis of CK5, CK1, P63, COL17A1, CK10, PLEC, and ITGB4 in the recellularized scaffold (scale bar: 50 μm). (d) Immunofluorescence analysis of CK5, CK1, P63, on the recellularized skin-like tissue (scale bar: 50 μm).
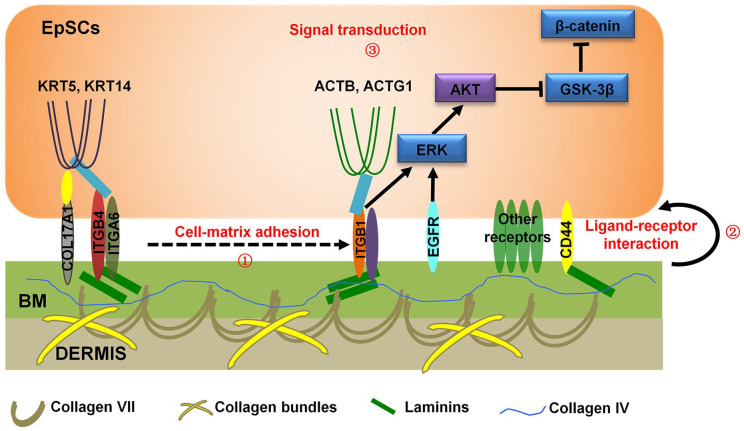
Wnt and ERK pathways are important for supporting the development of EpSCs.50,51 Results indicated that ITGB1 and EGFR of EpSCs were activated more strongly in cells bound directly to the SBSs than those without direct contact (Figure 7(d)). In addition, Phospho-Glycogen Synthase Kinase 3 Beta (p-GSK3β) protein of the Wnt pathway was activated only in cells directly bound to SBSs, while p-ERK protein of the ERK pathway was activated in both.50,51 Briefly, first, our results demonstrated that cell-matrix adhesion related components including hemidesmosomes (COL17A1, ITGB4, ITGA6), BM components (COL4A1, Laminin, COL7A1), and other collagen bundles, which were identified by proteomic on SBSs, physically support the interaction between EpSCs and BM (Figure 8). Second, we identified several receptors that mediate signal transduction from ECM of BM to EpSCs. Third, key proteins of signal pathways could be activated to affect EpSCs growth. Together, these results indicate that BM determines EpSCs fates in the presence of hemidesmosome complexes expressing correct polarity, performed by EGFR/ITGB1-Wnt pathways.
Figure 8.
Schematic mechanism of the regulation of cell fate determination by hemidesmosomes. Schematic representation of the proposed molecular functions of anchoring complexes on BM.
Discussion
ECM is a tissue-specific, insoluble, chemical complex outside cells and binding the cells into a community to form a tissue; it is composed of multiple proteins, PGs, and carbohydrates.52 We used a mild method to make a new kind of skin bioscaffold, which contains most ECM components and improves the efficiency of engineering than previously reported (Figure 1). The scaffolds not only effectively support the growth of 3D organs of EpSCs, but also enhance the functional repair of skin tissues during the process of wound healing. ECM-based biomaterials elicit a variety of favorable cellular responses such as angiogenesis, stem-cell recruitment, modulation of innate immune responses, and functional tissue reconstruction.53 It is the stiffness and the chemical signals that are responsible for their bioactivity with respect to various cell types. Studies on matrix extracts have focused on phenomenological effects of Matrigel or of matrix scaffolds prepared with distilled-water-perfused tissues (resulting in isolation of only cross-linked matrix components).54,55 There have not been systematic studies on the native matrix of a tissue, its composition and its regulatory functions. For the first time, we have analyzed comprehensively the ECM composition and functions in a normal skin tissue. We used quantitative proteomics to reveal an in-depth, whole profile of skin ECM components and their modulatory properties and regulatory effects on skin tissue maintenance, with our developed in situ decellularization method for skin to preserve both the cross-linked and nascent, uncross-linked ECM and the many factors that bind to the core components of the matrix.
The simple three step protocol enabled us to succeed at preserving far more of the matrix components than has been done previously including preservation of the bioactive regulators hidden in the core Matrisome components. The method, which focuses on collagen chemistry, can be used to prepare substrate of various tissue extracts. Until now, we have applied it on liver, pancreas, and skin. Remarkably, although the reagents used are the same, the processes (such as whether or not using perfusion methods, velocity, and enzyme concentration) of the method used are slightly different for different species of animals. The factors of SBSs are easily lost during the process of decellularization because of their low abundance and high solubility. The factors (e.g. TGF-β, BMPs, PDGFs, FGFs, Wnts, Hedgehogs, S100 proteins, chemokines, etc.) are always at a low level under homeostatic conditions and rapidly up-regulate once injury or trauma occurs. Cytokine array has been used to help us identify the low abundance Matrisome-associated components, which cannot be identified with mass spectrometry methods. The complete ECM map was obtained by integration of it with the results of mass spectrometry. Our results indicate that the regulators are retained in SBSs based on the interactions with core Matrisome components. Among them, skin-specific PGs play critical roles as storage reservoirs for signals that can be released as needed for ECM remodeling, wound healing, cell migration, and proliferation. We also found that PGs in skin contribute more soluble regulators, unexpected ones, that target cells, including members of the Serpin family (SERPINH, SERPING, and SERPINB1), Dioxygenases (PLOD1 and 2), Proly 4-Hydroxylase Subunits (P4HA1 and 2), Fas Ligand (FASLG), Lysyl Oxidase (LOX), and Growth-Inhibiting Protein (AMBP).
The ECM scaffold not only shapes the cells into the tissue and provides the microenvironment on which cells depend for survival and preservation of homeostasis, and also ensures fast and efficient responses to alterations or injuries. We sought to study the interaction between ECM microenvironment and cells. Therefore, we systematically demonstrated that ECM components in the basal stem-cell niches, the BM layer, mediate the structural function of skin through anchorage complexes and important receptors on the cell-matrix boundary. We also presented the interactions between core Matrisome components and regulators working together to participate in stages of wound healing. Recently, it has been reported that scaffolds can directly promote tissue regeneration through inflammatory factors.53 ECM dynamics are tightly regulated to ensure normal development, physiology, and robustness of organ systems. This is achieved by the mechanisms of modulating functions through ECM modifying enzymes secreted by different types of cells at multiple levels.45,46 We also identified phosphatases, aldehyde dehydrogenases, carbohydrate modification proteases, isomerases, deubiquitinases, methyltransferases, caspases, cathepsins, and several types of proteins involved in the organization and disassembly of important ECM proteins, in addition to some common enzymes such as MMPs. They play an important role in stem-cell maintenance of skin homeostasis, and cell debris removal, reepithelialization, and ECM remodeling of wound healing. These results indicated that ECM is modulated by cells to maintain the balance of the matrix microenvironment while creating specific functional microenvironments for cells.
Among the many functions of ECM on skin tissue development and wound healing, what is especially interesting to us is how ECM components regulate EpSCs’ fate. Understanding that should help to develop new approaches for skin tissue regeneration and in vitro bionic construction of artificial skin equivalents. We found that hemidesmosomes, an important structure in the BM of the EpSCs’ niche, are closely involved in EpSC’s fate determination. EpSCs were unable to polarize and differentiate into fully mature epidermal cells without hemidesmosome support. When skin is injured, the strong adhesion of keratinocytes to the BM is disrupted due to the disrupted hemidesmosomes, resulting in the basal EpSCs arrangement without polarity. When epithelialization is complete, basal keratinocytes revert to a stationary phenotype in which they have apical polarity and are firmly anchored to BM through reconstituted hemidesmosomes. Recently, it has been reported that COL17A1, an important component of the hemidesmosome, is closely related to the EpSCs’ fate determination.56 Therefore, our results indicated that the formation of functional hemidesmosomes is necessary to support the scaffolds with intact active ingredients.
We performed experiments to construct recellularized artificial skin equivalents in order to verify whether the hemidesmosomes themselves play a role in intracellular signaling from the ECM to the interior of the cell. To our surprise, we achieved very different results when the EpSCs recellularized on BM directly in contrast with when were not in direct contact. Only the EpSCs in direct contact with the BM layer provided the stem cells more opportunities to communicate with adhesion complexes on the BM, especially forming more functional hemidesmosomes. The ECM-receptor signals in BM can be efficiently transmitted to EpSCs and could polarize normally and maintain the characteristics of an epidermal stem cell under those conditions. They lose direct contact with BM when the EpSCs mature vertically, and with those adhesion complexes it results in a weakened interaction and leads to the hierarchical structure in the artificial skin equivalents. These results suggest that the interaction between epidermal stem cells and their receptors is a prerequisite for the existence of hemidesmosomes to help epidermal stem cells establish polarity.
Continued basic research into the human skin Matrisome, especially skin structural- and functional-specific intracellular machineries responsible for tissue development and regeneration may eventually give rise to novel approaches through which particular ECM or regulator proteins or peptides can be applied for future artificial skin preparations or dermatological treatments. For example, a recent study reported a microarchitechtural analysis of decellularized unscarred and scarred dermis for future dermal substitute scaffold design.57 Our research provided the functional skin microenvironment information of healthy and damaged status, which can provide a standardized nomenclature for Matrisome across laboratories would also benefit the field.
Conclusion
In conclusion, we have developed a new decellularized method to obtain complete anatomical skin biomatrix scaffold in situ with ECM architecture preserved. The acellularized scaffolds with complete Matrisome components not only support the construction of an artificial skin equivalent with stratification and polarity preserved but also promote functional repair during wound healing. On this basis, we described a skin scaffold map by integrated proteomics. We systematically analyzed the important role of the interaction between extracellular matrix (ECM) proteins and epidermal cells in skin microenvironment. Especially, we revealed that ECM was closely related with epidermal stem cells fate determination through hemidesmosome components. These concepts not only bring us the new understanding on the role of the skin ECM niche, but also provide attractive combinational strategy of tissue engineering principles with skin biomatrix scaffold materials accelerating and enhancement of tissue regeneration.
Supplemental Material
Supplemental material, SupplementaryFiles for Comprehensive proteomic atlas of skin biomatrix scaffolds reveals a supportive microenvironment for epidermal development by Ling Leng, Jie Ma, Xuer Sun, Baolin Guo, Fanlu Li, Wei Zhang, Mingyang Chang, Jinmei Diao, Yi Wang, Wenjuan Wang, Shuyong Wang, Yunping Zhu, Fuchu He, Lola M Reid and Yunfang Wang in Journal of Tissue Engineering
Acknowledgments
We thank Drs Ping Xu, Xin Wang, and Jian Wang for critical discussions and technical support.
Footnotes
Author contributions: Author contributions: Y.W. and L.L. conceived the overall study and designed experiments. L.L. and J.M. performed proteomics experiments and bioinformatics analysis. X.S., B.G., F.L., and W.Z. performed most of biological and functional experiments and analyzed the data. M.C., J.D., Y.W., S.W., and W.W. participated in cell and tissue culture. L.L., J.M., L.M.R., and Y.W. wrote and edited the manuscript. Y.Z. and F.H. conceived and supervised the proteomics experiments. All co-authors made important comments to the manuscript.
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Natural Science Foundations of China (No. 81730052), the Interdisciplinary Cooperation Project of Beijing Nova Program (Z1811100006218127), the National Major Scientific and Technological Special Project for “Significant New Drugs Development” (2018ZX09711003-001-002), and the National Key Research and Development Program of China (No. 2016YFC1101305).
ORCID iD: Yunfang Wang  https://orcid.org/0000-0003-2522-3182
https://orcid.org/0000-0003-2522-3182
Supplemental material: Supplemental material for this article is available online.
References
- 1. Chen CS, Mrksich M, Huang S, et al. Geometric control of cell life and death. Science 1997; 276(5317): 1425–1428. [DOI] [PubMed] [Google Scholar]
- 2. Jones PH, Simons BD, Watt FM. Sic transit gloria: farewell to the epidermal transit amplifying cell? Cell Stem Cell 2007; 1(4): 371–381. [DOI] [PubMed] [Google Scholar]
- 3. Gonzales KAU, Fuchs E. Skin and its regenerative powers: an alliance between stem cells and their niche. Dev Cell 2017; 43(4): 387–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chermnykh E, Kalabusheva E, Vorotelyak E. Extracellular matrix as a regulator of epidermal stem cell fate. Int J Mol Sci 2018; 19(4): 1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hsu YC, Li L, Fuchs E. Emerging interactions between skin stem cells and their niches. Nat Med 2014; 20(8): 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lane SW, Williams DA, Watt FM. Modulating the stem cell niche for tissue regeneration. Nat Biotechnol 2014; 32(8): 795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Martin GR, Kleinman HK, Tekranova VP, et al. The regulation of basement membrane formation and cell-matrix interactions by defined supramolecular complexes. Ciba Found Symp 1984; 108: 197–212. [DOI] [PubMed] [Google Scholar]
- 8. Naba A, Clauser KR, Hoersch S, et al. The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Mol Cell Proteomics 2012; 11(4): M111.014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Naba A, Clauser KR, Ding H, et al. The extracellular matrix: tools and insights for the “omics” era. Matrix Biol 2016; 49: 10–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li Q, Uygun BE, Geerts S, et al. Proteomic analysis of naturally-sourced biological scaffolds. Biomaterials 2016; 75: 37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mayorca-Guiliani AE, Madsen CD, Cox TR, et al. ISDoT: in situ decellularization of tissues for high-resolution imaging and proteomic analysis of native extracellular matrix. Nat Med 2017; 23(7): 890–898. [DOI] [PubMed] [Google Scholar]
- 12. Wells A, Nuschke A, Yates CC. Skin tissue repair: matrix microenvironmental influences. Matrix Biol 2016; 49: 25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rousselle P, Montmasson M, Garnier C. Extracellular matrix contribution to skin wound re-epithelialization. Matrix Biol 2019; 75–76: 12–26. [DOI] [PubMed] [Google Scholar]
- 14. Kawasaki T, Kirita Y, Kami D, et al. Novel detergent for whole organ tissue engineering. J Biomed Mater Res A 2015; 103(10): 3364–3373. [DOI] [PubMed] [Google Scholar]
- 15. García-Gareta E, Abduldaiem Y, Sawadkar P, et al. Decellularised scaffolds: just a framework? Current knowledge and future directions. J Tissue Eng 2020; 11: 2041731420942903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang Y, Cui CB, Yamauchi M, et al. Lineage restriction of human hepatic stem cells to mature fates is made efficient by tissue-specific biomatrix scaffolds. Hepatology 2011; 53(1): 293–305. [DOI] [PubMed] [Google Scholar]
- 17. Bornstein P, Sage EH. Matricellular proteins: extracellular modulators of cell function. Curr Opin Cell Biol 2002; 14(5): 608–616. [DOI] [PubMed] [Google Scholar]
- 18. Wu Q, Liu J, Liu L, et al. Establishment of an ex vivo model of nonalcoholic fatty liver disease using a tissue-engineered liver. ACS Biomater Sci Eng 2018; 4(8): 3016–3026. [DOI] [PubMed] [Google Scholar]
- 19. Liu J, Li R, Xue R, et al. Liver extracellular matrices bioactivated hepatic spheroids as a model system for drug hepatotoxicity evaluations. Adv Biosyst 2018; 2(10): 1800110. [Google Scholar]
- 20. Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 2008; 26(12): 1367–1372. [DOI] [PubMed] [Google Scholar]
- 21. Schwanhausser B, Busse D, Li N, et al. Global quantification of mammalian gene expression control. Nature 2011; 473(7347): 337–342. [DOI] [PubMed] [Google Scholar]
- 22. Huang da W, Sherman BT, Lempicki RA, et al. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009; 37(1): 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25(1): 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ogata H, Goto S, Sato K, et al. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 1999; 27(1): 29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003; 13(11): 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Szklarczyk D, Morris JH, Cook H. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 2017; 45(D1): D362–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hisha H, Tanaka T, Kanno S, et al. Establishment of a novel lingual organoid culture system: generation of organoids having mature keratinized epithelium from adult epithelial stem cells. Sci Rep 2013; 3: 3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ma J, Chen T, Wu S, et al. iProX: an integrated proteome resource. Nucleic Acids Res 2019; 47(D1): D1211–D1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Aragona M, Dekoninck S, Rulands S, et al. Defining stem cell dynamics and migration during wound healing in mouse skin epidermis. Nat Commun 2017; 8: 14684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Barrandon Y, Green H. Cell migration is essential for sustained growth of keratinocyte colonies: the roles of transforming growth factor-alpha and epidermal growth factor. Cell 1987; 50(7): 1131–1137. [DOI] [PubMed] [Google Scholar]
- 31. Jones PH, Harper S, Watt FM. Stem cell patterning and fate in human epidermis. Cell 1995; 80(1): 83–93. [DOI] [PubMed] [Google Scholar]
- 32. Hynes RO, Naba A. Overview of the matrisome–an inventory of extracellular matrix constituents and functions. Cold Spring Harb Perspect Biol 2012; 4(1): a004903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hasegawa K, Yoneda M, Kuwabara H, et al. Versican, a major hyaluronan-binding component in the dermis, loses its hyaluronan-binding ability in solar elastosis. J Invest Dermatol 2007; 127(7): 1657–1663. [DOI] [PubMed] [Google Scholar]
- 34. Iozzo RV, Goldoni S, Berendsen AD, et al. Small leucine-rich proteoglycans. In: RP Mecham. (ed.) The extracellular matrix: an overview. Biology of extracellular matrix. Berlin/Heidelberg: Springer-Verlag, 2011, pp.197–231. [Google Scholar]
- 35. Iozzo RV, Schaefer L. Proteoglycans in health and disease: novel regulatory signaling mechanisms evoked by the small leucine-rich proteoglycans. FEBS J 2010; 277(19): 3864–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Farach-Carson MC, Warren CR, Harrington DA, et al. Border patrol: insights into the unique role of perlecan/heparan sulfate proteoglycan 2 at cell and tissue borders. Matrix Biol 2014; 34: 64–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Danielson KG, Baribault H, Holmes DF, et al. Targeted disruption of decorin leads to abnormal collagen fibril morphology and skin fragility. J Cell Biol 1997; 136(3): 729–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 2003; 113(6): 685–700. [DOI] [PubMed] [Google Scholar]
- 39. Gubbiotti MA, Neill T, Iozzo RV. A current view of perlecan in physiology and pathology: a mosaic of functions. Matrix Biol 2017; 57–58: 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McCawley LJ, Matrisian LM. Matrix metalloproteinases: they’re not just for matrix anymore! Curr Opin Cell Biol 2001; 13(5): 534–540. [DOI] [PubMed] [Google Scholar]
- 41. Van den Steen PE, Dubois B, Nelissen I, et al. Biochemistry and molecular biology of gelatinase B or matrix metalloproteinase-9 (MMP-9). Crit Rev Biochem Mol Biol 2002; 37(6): 375–536. [DOI] [PubMed] [Google Scholar]
- 42. Cauwe B, Opdenakker G. Intracellular substrate cleavage: a novel dimension in the biochemistry, biology and pathology of matrix metalloproteinases. Crit Rev Biochem Mol Biol 2010; 45(5): 351–423. [DOI] [PubMed] [Google Scholar]
- 43. Zomer HD, Trentin AG. Skin wound healing in humans and mice: challenges in translational research. J Dermatol Sci 2018; 90(1): 3–12. [DOI] [PubMed] [Google Scholar]
- 44. Kiritsi D, Nystrom A. The role of TGFbeta in wound healing pathologies. Mech Ageing Dev 2018; 172: 51–58. [DOI] [PubMed] [Google Scholar]
- 45. Malemud CJ. Matrix metalloproteinases (MMPs) in health and disease: an overview. Front Biosci 2006; 11: 1696–1701. [DOI] [PubMed] [Google Scholar]
- 46. Klein T, Bischoff R. Physiology and pathophysiology of matrix metalloproteases. Amino Acids 2011; 41(2): 271–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fuchs E, Nowak JA. Building epithelial tissues from skin stem cells. Cold Spring Harb Symp Quant Biol 2008; 73: 333–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nievers MG, Schaapveld RQ, Sonnenberg A. Biology and function of hemidesmosomes. Matrix Biol 1999; 18(1): 5–17. [DOI] [PubMed] [Google Scholar]
- 49. Osmani N, Labouesse M. Remodeling of keratin-coupled cell adhesion complexes. Curr Opin Cell Biol 2015; 32: 30–38. [DOI] [PubMed] [Google Scholar]
- 50. Lim X, Tan SH, Koh WLC, et al. Interfollicular epidermal stem cells self-renew via autocrine Wnt signaling. Science 2013; 342(6163): 1226–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Scholl FA, Dumesic PA, Barragan DI, et al. Mek1/2 MAPK kinases are essential for Mammalian development, homeostasis, and Raf-induced hyperplasia. Dev Cell 2007; 12(4): 615–629. [DOI] [PubMed] [Google Scholar]
- 52. Hynes RO. The extracellular matrix: not just pretty fibrils. Science 2009; 326(5957): 1216–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sadtler K, Estrellas K, Allen BW, et al. Developing a pro-regenerative biomaterial scaffold microenvironment requires T helper 2 cells. Science 2016; 352(6283): 366–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Benton G, Kleinman HK, George J, et al. Multiple uses of basement membrane-like matrix (BME/Matrigel) in vitro and in vivo with cancer cells. Int J Cancer 2011; 128(8): 1751–1757. [DOI] [PubMed] [Google Scholar]
- 55. Arenas-Herrera JE, Ko IK, Atala A, et al. Decellularization for whole organ bioengineering. Biomed Mater 2013; 8(1): 014106. [DOI] [PubMed] [Google Scholar]
- 56. Liu N, Matsumura H, Kato T, et al. Stem cell competition orchestrates skin homeostasis and ageing. Nature 2019; 568(7752): 344–350. [DOI] [PubMed] [Google Scholar]
- 57. Khan U, Bayat A. Microarchitectural analysis of decellularised unscarred and scarred dermis provides insight into the organisation and ultrastructure of the human skin with implications for future dermal substitute scaffold design. J Tissue Eng 2019; 10: 2041731419843710. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, SupplementaryFiles for Comprehensive proteomic atlas of skin biomatrix scaffolds reveals a supportive microenvironment for epidermal development by Ling Leng, Jie Ma, Xuer Sun, Baolin Guo, Fanlu Li, Wei Zhang, Mingyang Chang, Jinmei Diao, Yi Wang, Wenjuan Wang, Shuyong Wang, Yunping Zhu, Fuchu He, Lola M Reid and Yunfang Wang in Journal of Tissue Engineering