Abstract
The cytoplasmic fragile X mental retardation 1 (FMR1)-interacting protein 2 (CYFIP2) gene is associated with epilepsy, intellectual disability (ID), and developmental delay, suggesting its critical role in proper neuronal development and function. CYFIP2 is involved in regulating cellular actin dynamics and also interacts with RNA-binding proteins. However, the adult brain function of CYFIP2 remains unclear because investigations thus far are limited to Cyfip2 heterozygous (Cyfip2+/−) mice owing to the perinatal lethality of Cyfip2-null mice. Therefore, we generated Cyfip2 conditional knock-out (cKO) mice with reduced CYFIP2 expression in postnatal forebrain excitatory neurons (CaMKIIα-Cre). We found that in the medial prefrontal cortex (mPFC) of adult Cyfip2 cKO mice, CYFIP2 expression was decreased in both layer 2/3 (L2/3) and layer 5 (L5) neurons, unlike the L5-specific CYFIP2 reduction observed in adult Cyfip2+/− mice. Nevertheless, filamentous actin (F-actin) levels were increased only in L5 of Cyfip2 cKO mPFC possibly because of a compensatory increase in CYFIP1, the other member of CYFIP family, in L2/3 neurons. Abnormal dendritic spines on basal, but not on apical, dendrites were consistently observed in L5 neurons of Cyfip2 cKO mPFC. Meanwhile, neuronal excitability and activity were enhanced in both L2/3 and L5 neurons of Cyfip2 cKO mPFC, suggesting that CYFIP2 functions of regulating F-actin and excitability/activity may be mediated through independent mechanisms. Unexpectedly, adult Cyfip2 cKO mice did not display locomotor hyperactivity or reduced anxiety observed in Cyfip2+/− mice. Instead, both exhibited enhanced social dominance accessed by the tube test. Together, these results provide additional insights into the functions of CYFIP2 in the adult brain.
Keywords: CYFIP2, medial prefrontal cortex, neuronal activity, social dominance, excitability
Introduction
Two members of the cytoplasmic fragile X mental retardation 1 (FMR1)-interacting protein family, CYFIP1 and CYFIP2 (also referred to as SRA1 and PIR121, respectively), are evolutionarily highly conserved proteins which were originally identified as direct binding partners of FMR Protein (Schenck et al., 2001), an RNA-binding protein whose loss causes fragile X syndrome (Bagni and Zukin, 2019). Moreover, CYFIP1 and CYFIP2 are critical components of the Wiskott–Aldrich syndrome protein family verprolin-homologous protein (WAVE) regulatory complex (WRC; Chen et al., 2010), a heteropentameric protein complex consisting of CYFIP (either CYFIP1 or CYFIP2), WAVE, NAP1, ABI, and HSPC300. As a downstream effector of Rac1 GTPase, the WRC regulates Arp2/3 complex-mediated actin polymerization in various cellular compartments including neuronal synapses (Spence and Soderling, 2015; Chen et al., 2017).
Based on the above-mentioned protein interactions, CYFIP1 and CYFIP2 are known to be involved in the regulation of RNA-processing and actin dynamics, which are two critical molecular pathways for proper neuronal development and function (Bagni and Zukin, 2019; Zhang et al., 2019b). Notably, however, detailed in vivo studies of CYFIP1 and CYFIP2 have suggested their functions differ because they have distinct spatiotemporal expression patterns and different protein interactors in the brain (Zhang et al., 2019a; Lee Y. et al., 2020; Lee S. H. et al., 2020). Indeed, at least for survival, CYFIP1 and CYFIP2 cannot compensate for each other considering Cyfip1-null mice (Chung et al., 2015) and Cyfip2-null mice (Kumar et al., 2013; Han et al., 2015; Zhang et al., 2018) are nonviable owing to lethality at different developmental time points (early embryonic and perinatal lethality, respectively).
Both CYFIP1 and CYFIP2 genes are associated with various brain disorders (Abekhoukh and Bardoni, 2014; Zhang et al., 2019b). For example, deletions and duplications of CYFIP1 are associated with autism spectrum disorders, intellectual disability (ID), and schizophrenia (Yoon et al., 2014; Oguro-Ando et al., 2015). Large chromosomal deletions harboring CYFIP2 have been identified in patients with developmental delay, ID, and seizures (Spranger et al., 2000; Lee et al., 2016). Moreover, recent whole-exome and whole-genome sequencing studies have identified de novo CYFIP2 variants in patients with developmental delay, ID, and early-onset epileptic encephalopathy (Nakashima et al., 2018; Peng et al., 2018; Lee et al., 2019b; Zhong et al., 2019; Zweier et al., 2019).
To understand the neurobiological mechanisms underlying CYFIP-associated brain disorders, several rodent models have been generated and characterized for both CYFIP1 (Bozdagi et al., 2012; Pathania et al., 2014; Chung et al., 2015; Oguro-Ando et al., 2015; Hsiao et al., 2016; Bachmann et al., 2019; Davenport et al., 2019; Domínguez-Iturza et al., 2019; Silva et al., 2019) and CYFIP2 (Han et al., 2015; Lee S. H. et al., 2020). Nevertheless, in most of these studies, investigations on the adult brain functions of CYFIP1 and CYFIP2 were limited to the heterozygous mice (Cyfip1+/− or Cyfip2+/−) because of early developmental lethality of Cyfip1- and Cyfip2-null mice. In the case of CYFIP1, a conditional knock-out (cKO) strategy combining floxed-Cyfip1 mice and brain region-specific Cre-expressing mice was applied to reveal inhibitory synaptic functions of the protein (Davenport et al., 2019).
We recently reported prefrontal dysfunctions in adult Cyfip2+/− mice, including increased filamentous actin (F-actin), enlarged dendritic spines, and enhanced excitatory synaptic transmission and excitability (Lee S. H. et al., 2020). Notably, these changes were restricted to layer 5 (L5), but not in layer 2/3 (L2/3), neurons of the medial prefrontal cortex (mPFC) possibly because of a reduction in CYFIP2 protein exclusively in these L5 neurons of adult Cyfip2+/− mPFC. Importantly, it is not uncommon for protein levels to be unaffected when one copy of the gene is deficient. To overcome this, in this study, we crossed floxed-Cyfip2 mice with CaMKIIα-Cre mice to generate Cyfip2 cKO mice with reduced CYFIP2 expression specifically in postnatal forebrain excitatory neurons. Cyfip2 cKO mice displayed many of the common phenotypes that are observed in Cyfip2+/− mice, but with some distinct molecular, cellular, and behavioral abnormalities, providing additional insights into understanding the in vivo brain functions of CYFIP2.
Materials and Methods
Mice
The floxed-Cyfip2 and Cyfip2+/− mice used in this study have been previously described (Han et al., 2015; Zhang et al., 2018, 2019a; Lee S. H. et al., 2020). The CaMKIIα-Cre [B6.Cg-Tg(Camk2a-cre)T29-1Stl/J] and Thy1-YFP [B6.Cg-Tg(Thy1-YFP)HJrs/J] mice were obtained from the Jackson Laboratory. The mice were bred and maintained on a C57BL/6J background according to the Korea University College of Medicine Research Requirements. All procedures were approved by the Committee on Animal Research at Korea University College of Medicine (KOREA-2018-0174). The mice were fed ad libitum and housed under a 12 h light-dark cycle. All experiments were performed with an adult (8 to 10-week old) male Cyfip2 mice and their littermate controls.
RNA Purification and qRT-PCR
Real-time quantitative reverse transcription PCR (qRT-PCR) was performed as described previously (Choi et al., 2015; Jin et al., 2018; Lee et al., 2019a). Briefly, total RNA was extracted from the brain tissues using a miRNeasy Minikit (Qiagen) and two micrograms of total RNA were used for cDNA synthesis using the iScript™ cDNA Synthesis Kit (Bio-Rad). Target mRNAs were detected and quantified by a real-time PCR instrument (CFX96 Touch, Bio-Rad) using the SYBR Green master mix (Bio-Rad). The primer sequences for real-time PCR were described previously (Lee S. H. et al., 2020).
Biochemistry and Antibodies for Western Blotting
Whole and synaptosomal lysates of the mouse brains were prepared as described previously (Han et al., 2009; Jin et al., 2019a,b). The antibodies used for Western blot analysis were anti-Cofilin (1:1,000, Abcam, AB42824), anti-CYFIP1 (1:1,000, Millipore, AB6046), anti-CYFIP2 (1:3,000, Abcam, AB95969), anti-GABA-A-R-β2/3 (1:1,000, NeuroMab, 75-363), anti-GAPDH (1:3,000, Cell Signaling, #2118), anti-Gephyrin (1:500, Synaptic Systems, 147-011), anti-GluA1 (1:2,000, Millipore, 04-855), anti-GluA2 (1:1,500, Millipore, MAB397), anti-GluN1 (1:1,000, Millipore, MAB363), anti-Homer1b/c (1:1,000, Synaptic Systems, 160-002), anti-Neuroligin-3 (1:1,000, NeuroMab, 75-158), anti-PSD-95 (1:2,000, Thermo Fisher Scientific, MA1-046), and anti-WAVE1 (1:1,000, NeuroMab, 75-048). Western blot images were acquired using a ChemiDoc Touch Imaging System (Bio-Rad) and quantified using ImageJ software.
Immunohistochemistry
Mice were deeply anesthetized with isoflurane and transcardially perfused with heparinized (20 units/ml) phosphate-buffered saline (PBS) followed by 4% paraformaldehyde (PFA) in PBS. Brains were extracted and post-fixed in 4% PFA overnight. After post-fixation, the brains were washed with PBS and cryoprotected with 30% sucrose in PBS for 48 h. Brains were frozen in O.C.T compound (Sakura Tissue-Tek, 4583) and coronally sectioned (60 μm thickness) using a cryostat microtome (Leica, CM3050S). The sections were collected and stored in 50% glycerol in 2× PBS at −20°C until further processed. After being slightly rinsed in 1× PBS, sections were permeabilized by 0.5% Triton X-100 in 1× PBS three times for 10 min each. The sections were incubated with blocking solution (3% BSA in 0.5% Triton X-100 in 1× PBS) for 30 min at room temperature. Then, the sections were incubated with primary antibodies diluted in blocking solution overnight at 4°C. After washed three times for 15 min each with washing buffer (0.1% Triton X-100 in 1× PBS), the sections were incubated with Alexa Fluor-conjugated secondary antibodies diluted in blocking solution for 1 h at room temperature. The sections were washed with washing buffer three times for 15 min each. Finally, the sections were mounted on slide glasses with mounting media containing DAPI (Vector Laboratories, H-1400). The slide glasses were stored at 4°C until imaged. The list of primary and secondary antibodies used for immunohistochemistry is summarized in Supplementary Table 1. Confocal microscopy (Zeiss, LSM800) was used to acquire images (×20 objective and ×1 digital zoom) of the prelimbic mPFC (3.00–1.77 mm anterior from the Bregma) from coronal sections (Supplementary Figure 1). The prelimbic mPFC region was obtained by tile scanning (total of three tiles), and each frame was taken in Z-stacks of 5–10 slices (45–55 μm thickness in total). Using the anti-NeuN antibody, we examined antibody penetration to the mPFC section (Supplementary Figure 2). Tiled Z-projection images were aligned and converted into a single flattened image using ZEN software from Zeiss. From each tiled image, three to four regions of interest (ROI, 150 × 150 μm) were randomly selected from each cortical layer and were analyzed using ImageJ software. Specifically, CYFIP1 and cFos intensities in the cell body area of each ROI were measured and normalized by NeuN intensity for each cell body (Supplementary Figure 3A). For CYFIP2 and F-actin, the total intensity of each ROI was measured and normalized by total NeuN intensity for each ROI (Supplementary Figure 3B). All quantifications were performed by researchers who were blinded to the genotype and were involved in neither staining nor image acquisition processes.
Dendritic Spine Analysis
Mice were deeply anesthetized with isoflurane and transcardially perfused with heparinized (20 units/ml) PBS followed by 4% PFA in PBS. Brains were extracted and post-fixed in 4% PFA overnight. After post-fixation, coronal sections (100 μm thickness) of the mPFC region (3.00–1.77 mm anterior to Bregma) were obtained using a vibratome (VT1000S, Leica). The sections were collected and stored in 50% glycerol in 2 × PBS at −20°C until further processed. Blocking, permeabilization, and anti-GFP (1:1,000, Abcam, AB13970) primary and Alexa Fluor-conjugated (1:500, anti-chicken Alexa Fluor-488) secondary antibody incubation were performed as described above. Finally, the sections were mounted on slide glasses with mounting media (Biomeda, M02). Images of dendritic spines on the secondary or tertiary branches (apical or basal dendrites of YFP-positive L5 neurons in the prelimbic mPFC) were acquired by confocal microscopy (Zeiss, LSM800, ×63 objective, and ×3 digital zoom) and were analyzed using ImageJ software. All quantifications were performed by researchers who were blinded to the genotype and were involved in neither staining nor image acquisition processes.
Electrophysiology
Coronal slices of the mPFC were prepared (300 μm thickness) as described previously (Seo et al., 2012; Kang et al., 2017). After decapitation, brains were rapidly removed and placed in an ice-cold, oxygenated (95% O2 and 5% CO2), low-Ca2+/high-Mg2+ dissection buffer containing (in mM) 5 KCl, 1.23 NaH2PO4, 26 NaHCO3, 10 dextrose, 0.5 CaCl2, 10 MgCl2, and 212.7 sucrose. Slices were transferred to a storage chamber in an incubator containing oxygenated artificial cerebrospinal fluid (ACSF) containing (in mM) 124 NaCl, 2.5 KCl, 1.23 NaH2PO4, 2.5 CaCl2, 1.5 MgCl2, 26 NaHCO3, and 10-dextrose at 28–30°C for at least 30 min before recording. Slices were transferred to a recording chamber where they were perfused continuously with oxygenated ACSF (23–25°C) at a flow rate of 2 ml/min. Slices were equilibrated for 5 min before recordings. All recordings were performed in visually identified L2/3 or L5 pyramidal neurons in the prelimbic mPFC which were identified by their size and morphology. Patch pipettes (4–6 MΩ) were filled with (in mM) 145 K-gluconate, 5 NaCl, 10 HEPES, 0.2 EGTA, 1 MgCl2, 2 ATP-Mg, and 0.1 GTP-Na at a pH 7.4 and 280–290 mOsm. The extracellular recording solution consisted of ACSF supplemented with picrotoxin (100 μM). Data were acquired using an EPC-8 amplifier (HEKA), filtered at 3 kHz, digitized at 10 kHz with Digidata 1550B (Axon Instruments), and analyzed using pClamp 10 (Molecular Devices). Only cells with access resistance <20 MΩ and input resistance >100 MΩ were studied. Cells were discarded if the input or access resistance changed by more than 20%.
Behavioral Assays
Before each test, mice were habituated in the test room (400 lux, 60 dB white noise) for at least 30 min. The open-field test was performed as described previously (Han et al., 2013, 2015). Briefly, mice were placed in the center of a clear, open Plexiglas chamber (40 × 40 × 30 cm), and their activities were recorded for 30 min using a suspended digital camera and analyzed using the EthoVision video tracking software (Noldus). The center area was defined as a 20 × 20 cm zone in the middle of the chamber. The elevated plus-maze test was performed using a plus-maze raised 50 cm above the floor. Mice were placed at the intersection of the four arms and allowed to move freely for 10 min. The tube test was performed as described previously (Fan et al., 2019) using a transparent acryl tube with 30 cm length and 3 cm inside diameter, a size just sufficient to permit only one adult mouse to pass through without reversing direction. For the training session, each mouse was released at alternating ends of the tube and allowed to run through the tube. Each animal was given 10 training trials on each of two successive days. For the test session, a pair of mice were released at the two ends of a tube and allowed to stop at a guillotine door in the middle of the tube. During the test session (2 min maximum), the guillotine door was opened and the mouse that retreated out of the tube first was designated as the loser.
Gene Ontology and Pathway Analysis
Enrichment analysis was performed using the Enrichr web tool (Kuleshov et al., 2016) on Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) gene set libraries. Enrichr employs pre-defined gene set libraries to assist functional enrichment analysis of large gene lists. The enrichment of biological terms associated with the query list was assessed by Fischer’s exact test and the inferred P-values were further adjusted with Benjamini–Hochberg for multiple hypotheses testing. The enrichment terms with adjusted P-value less than 0.05 were considered significantly enriched.
Quantification and Statistical Analysis
Values from at least four independent experiments with biological replicates were used for quantification and statistical analysis. All quantifications were performed in a blinded manner. P values were calculated by two-tailed student’s t-test or analysis of variance (ANOVA) with Bonferroni’s post hoc test using the GraphPad Prism 6 software. All data are presented as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
Results and Discussion
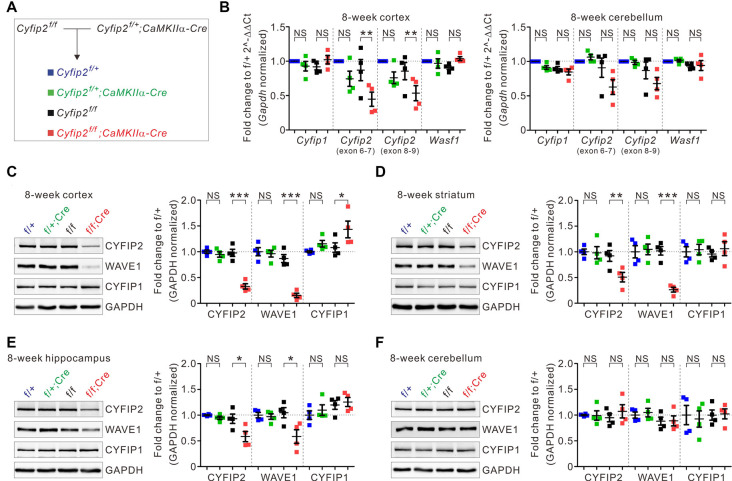
To generate postnatal forebrain excitatory neuron-specific Cyfip2 cKO mice, we crossed floxed-Cyfip2 (Cyfip2f/f) mice (Lee S. H. et al., 2020) with CaMKIIα-Cre mice, which start expressing Cre recombinase in the forebrain excitatory neurons during the third to fourth postnatal week (Kim et al., 2018; Figure 1A). As expected from the Cre expression pattern, qRT-PCR analyses showed reduced mRNA expression of Cyfip2 in the cortex, but not in the cerebellum, of adult (postnatal 8 weeks) Cyfip2 cKO (Cyfip2f/f;CaMKIIα-Cre) mice compared with those of Cre-negative control (Cyfip2f/f) mice (Figure 1B). However, cortical mRNA expression of Cyfip2 was not significantly decreased in Cyfip2 conditional heterozygous (Cyfip2f/+;CaMKIIα-Cre) mice compared with those of respective Cre-negative (Cyfip2f/+) control mice. Moreover, mRNA expression levels of Cyfip1 and Wasf1 (encoding WAVE1) were not changed in the cortex of Cyfip2 cKO mice. At the protein level, reduced expression of CYFIP2 and WAVE1 was observed in whole lysates of the cortex, striatum, and hippocampus but not the cerebellum of adult Cyfip2 cKO mice (Figures 1C–F), which is consistent with previous studies showing that WAVE protein in the WRC becomes less stable and is degraded by the proteasome without CYFIP (Kunda et al., 2003; Zhao et al., 2013; Han et al., 2015). Notably, CYFIP1 protein expression was increased in the cortex, but not in other brain regions, of Cyfip2 cKO mice (Figure 1C), suggesting a potential compensatory response of CYFIP1 in the cortex owing to a reduction in CYFIP2.
Figure 1.
Changes in the expression of cytoplasmic fragile X mental retardation 1 (FMR1)-interacting protein 1 (CYFIP1), CYFIP2, and Wiskott–Aldrich syndrome protein family verprolin-homologous protein 1 (WAVE1) in multiple brain regions of Cyfip2 conditional knock-out (cKO) mice. (A) The breeding scheme for the Cyfip2 cKO mice. (B) Real-time quantitative reverse transcription PCR (qRT-PCR) analyses of Cyfip1, Cyfip2, and Wasf1 (encoding WAVE1) mRNAs in the cortex (left panel) and cerebellum (right panel) of Cyfip2 cKO mice (n = 4 animals per genotype, one-way analysis of variance (ANOVA) with Bonferroni’s post hoc test). NS, not significant. (C–F) Western blot analysis of CYFIP1, CYFIP2, and WAVE1 proteins in the cortex, striatum, hippocampus, and cerebellum of Cyfip2 cKO mice (n = 4 animals per genotype, one-way ANOVA with Bonferroni’s post hoc test). All data are presented as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
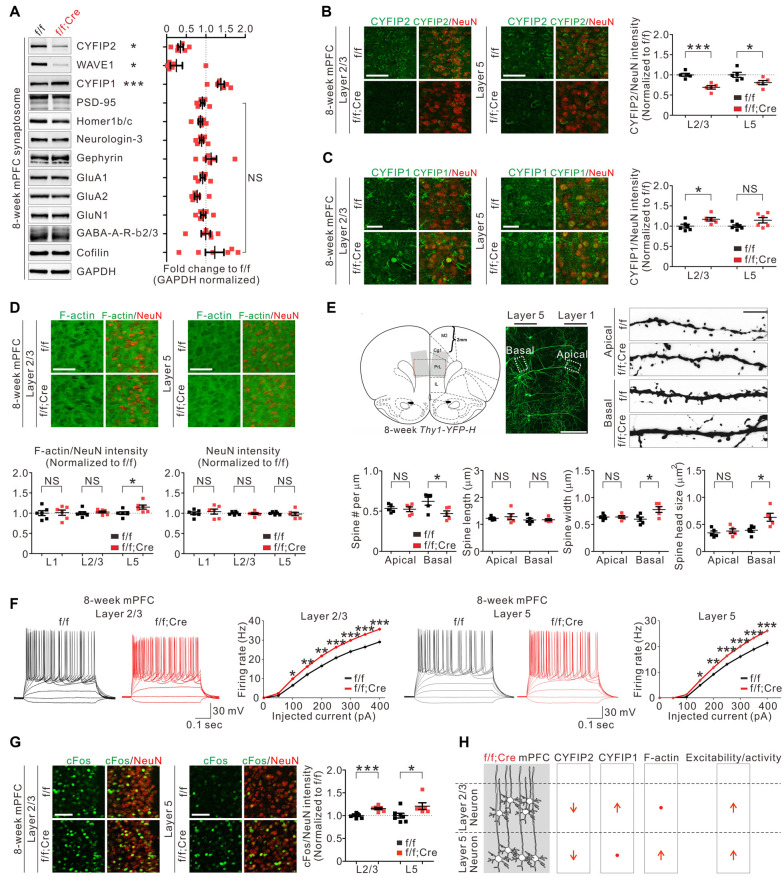
To further characterize Cyfip2 cKO mice, we focused on the mPFC because we recently identified several molecular, cellular, and functional changes in the mPFC excitatory neurons of adult Cyfip2+/− mice (Lee S. H. et al., 2020). First, we confirmed the decreased CYFIP2 and WAVE1, but increased CYFIP1, protein levels in synaptosomal lysates of adult Cyfip2 cKO mPFC (Figure 2A). Next, we assessed the levels of several other excitatory and inhibitory synaptic proteins, such as PSD-95 and Gephyrin, and found that they were normal (Figure 2A). We previously showed that in the mPFC of adult Cyfip2+/− mice, CYFIP2 protein expression is selectively reduced in L5 neurons but not in L2/3 neurons (Lee S. H. et al., 2020). However, in Cyfip2 cKO mice, we found that CYFIP2 levels were significantly reduced in both L2/3 and L5 neurons of the mPFC (Figure 2B and Supplementary Figure 4). We also observed CYFIP1 expression in both NeuN-positive or CaMKII-positive neurons and NeuN-negative non-neuronal cells in the mPFC of adult wild-type and Cyfip2 cKO mice (Supplementary Figure 5), which is similar to what we observed in the hippocampus of adult mice (Zhang et al., 2019a). When we selectively analyzed the CYFIP1 signals in NeuN-positive neurons of the mPFC, they were increased in L2/3 neurons, but not in L5 neurons, of Cyfip2 cKO mice compared with those in control mice (Figure 2C).
Figure 2.
Molecular, morphological, and functional changes in excitatory pyramidal neurons in the medial prefrontal cortex (mPFC) of Cyfip2 cKO mice. (A) Western blot analysis of various synaptic proteins in the mPFC synaptosome of Cyfip2 cKO mice (n = 6 animals per genotype, two-tailed student’s t-test). NS, not significant. (B) Fluorescence immunohistochemistry (IHC) analysis of CYFIP2 and NeuN in the mPFC of Cyfip2 cKO mice (n = 5 animals per genotype, two-tailed student’s t-test). Scale bar, 50 μm. L2/3, layer 2/3; L5, layer 5. (C) Fluorescence IHC analysis of CYFIP1 and NeuN in the mPFC of Cyfip2 cKO mice (n = 6 animals per genotype, two-tailed student’s t-test). Scale bar, 25 μm. (D) Fluorescence IHC analysis of F-actin and NeuN in the mPFC of Cyfip2 cKO mice (n = 6 animals per genotype, two-tailed student’s t-test). L1, layer 1. Scale bar, 50 μm. (E) Dendritic spine analysis of layer 5 pyramidal neurons in the mPFC of Cyfip2 cKO mice (n = 5 animals per genotype, two-tailed student’s t-test). Scale bars, 200 and 5 μm. (F) Intrinsic excitabilities of mPFC L2/3 (left panel) and L5 (right panel) neurons measured as firing rates against injected current (n = 4 animals per genotype, two-tailed student’s t-test). (G) Fluorescence IHC analysis of cFos and NeuN in the mPFC of Cyfip2 cKO mice (n = 7 animals for control and 6 for Cyfip2 cKO, two-tailed student’s t-test). Scale bar, 50 μm. (H) Summary of molecular and functional changes of pyramidal neurons in the mPFC of Cyfip2 cKO mice. All data are presented as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
In the basal state, CYFIP inhibits WAVE activity of the WRC (Chen et al., 2010) and its downstream actin polymerization (Zhao et al., 2013). As expected, the reduction in CYFIP2 protein observed in adult Cyfip2+/− mice coincided with an increase of F-actin levels in mPFC L5 neurons (Lee S. H. et al., 2020). Therefore, we also measured F-actin levels in the mPFC of Cyfip2 cKO mice and found that they were significantly increased in L5 but unchanged in layer 1 (L1) and L2/3 compared with those in control mPFC (Figure 2D). This L5-specific increase of F-actin levels was unexpected because CYFIP2 proteins were reduced in both L2/3 and L5 neurons (Figure 2B). One possible explanation is that the compensatory increase in CYFIP1 (Figure 2C) might normalize the net CYFIP levels in L2/3 neurons of Cyfip2 cKO mice, thereby maintaining their F-actin levels. NeuN intensities were normal in all layers of Cyfip2 cKO mPFC (Figure 2D).
Next, we analyzed dendritic spines of the mPFC L5 neurons by crossing Cyfip2 cKO mice with Thy1-YFP mice, which sparsely express yellow fluorescent proteins (YFPs) in L5 neurons visualizing their dendritic spines (Feng et al., 2000). In apical dendrites of L5 neurons, dendritic spine density, and morphology (length, width, and head size) were comparable between control and Cyfip2 cKO mice (Figure 2E). In contrast, in basal dendrites of L5 neurons, dendritic spine density was decreased, but spine width and head size were increased in Cyfip2 cKO mice compared with control mice.
We previously showed that, in the mPFC of adult Cyfip2+/− mice, neuronal excitability was increased in L5, but not in L2/3, neurons (Lee S. H. et al., 2020), suggesting that CYFIP2 negatively regulates neuronal excitability. Consistently, we found that neuronal excitability was increased in both L2/3 and L5 neurons of Cyfip2 cKO mPFC (Figure 2F). Furthermore, neuronal activity, as measured based on cFos expression levels, was also increased in both L2/3 and L5 neurons of Cyfip2 cKO mPFC (Figure 2G). It is intriguing that, unlike F-actin levels, which were increased only in L5, neuronal excitability and activity were enhanced in both L2/3 and L5 neurons of Cyfip2 cKO mPFC. These results suggest that the two functions of CYFIP2 (i.e., regulation of neuronal F-actin and excitability/activity) may be mediated by independent mechanisms and that CYFIP1 can only compensate for the F-actin-related function of CYFIP2 (see our “discussion” below; Figure 2H).
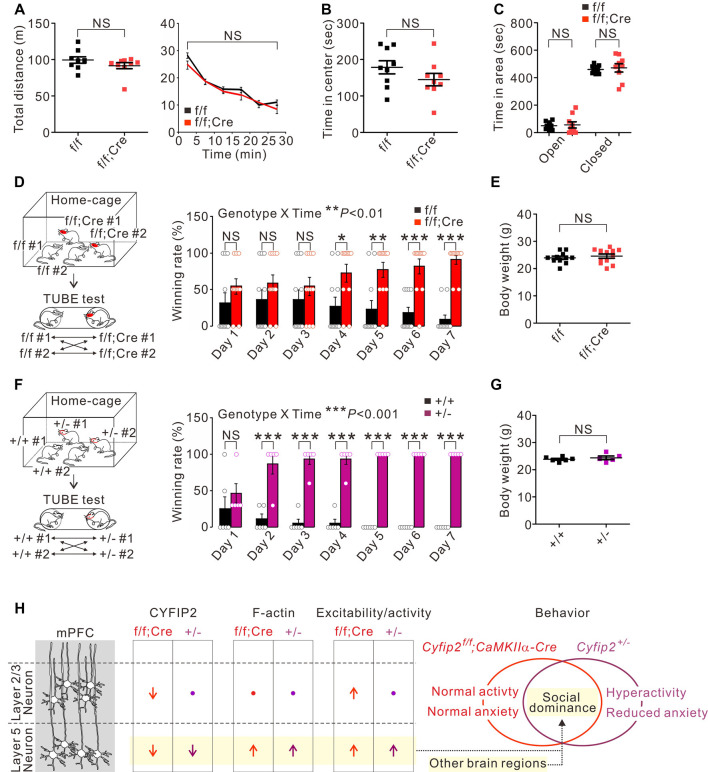
Next, we characterized behavioral phenotypes of adult Cyfip2 cKO mice. Based on the locomotor hyperactivity and reduced anxiety observed in adult Cyfip2+/− mice (Han et al., 2015; Lee S. H. et al., 2020), we performed the open field test and elevated plus maze test in adult Cyfip2 cKO mice. However, unlike Cyfip2+/− mice, Cyfip2 cKO mice displayed normal locomotor activity (Figure 3A), and anxiety (Figures 3B,C) levels. Changes in synaptic strength and neuronal activity in the mPFC are causally associated with social dominance behavior in mice (Wang et al., 2011, 2014). Therefore, we performed the tube test (Fan et al., 2019) to measure the social hierarchy among the group-housed Cyfip2 cKO mice and their control littermates (Figure 3D). Notably, we found that throughout the 7-day tube test, Cyfip2 cKO mice displayed a significantly higher winning rate over control mice from day 4 to day 7. There was no significant difference in body weights between the two groups (Figure 3E). These results prompted us to test the social dominance behavior in adult Cyfip2+/− mice (Figure 3F). Like Cyfip2 cKO mice, Cyfip2+/− mice displayed a significantly higher winning rate over their control littermates from day 2 to day 7 of the tube test. Body weights were similar between the two genotypes (Figure 3G). Altogether, these results suggest enhanced social dominance behavior in both adult Cyfip2+/− mice and Cyfip2 cKO mice.
Figure 3.
Enhanced social dominance behavior of Cyfip2 cKO and Cyfip2+/− mice. (A) Locomotor activity of control and Cyfip2 cKO mice in the open field test (n = 9 animals per genotype, two-tailed student’s t-test). NS, not significant. (B) Time spent in the center area during the open field test for control and Cyfip2 cKO mice. (C) Time spent in the open and closed arms during the elevated plus-maze test for control and Cyfip2 cKO mice (n = 9 animals per genotype, two-tailed student’s t-test). (D) Social dominance behavior of Cyfip2 cKO mice measured by the tube test (n = 11 animals per genotype, two-way ANOVA with Bonferroni’s post hoc test). (E) Body weights of control and Cyfip2 cKO mice were used for the tube test (two-tailed student’s t-test). (F) Social dominance behavior of Cyfip2+/− mice measured by the tube test (n = 6, 5 wild-type and Cyfip2+/− mice, respectively, two-way ANOVA with Bonferroni’s post hoc test). (G) Body weights of wild-type and Cyfip2+/− mice used for the tube test (two-tailed student’s t-test). (H) Summary of molecular and functional changes of pyramidal neurons in the mPFC of Cyfip2 cKO and Cyfip2+/− mice, and a hypothesis linking the changes in layer 5 neurons to increased social dominance behavior of the mice. Changes in other brain regions may also contribute to the phenotype. All data are presented as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
In this study, we generated Cyfip2 cKO mice with reduced CYFIP2 expression selectively in postnatal forebrain excitatory neurons. Specifically, in the mPFC, CYFIP2 protein levels were significantly reduced in L2/3 and L5 neurons of adult Cyfip2 cKO mice. Intriguingly, however, F-actin levels were increased only in L5, but not L2/3, neurons of Cyfip2 cKO mPFC, possibly because of a compensatory increase in CYFIP1 proteins in L2/3 neurons. This increase in CYFIP1 may substitute for CYFIP2 in the WRC of L2/3 neurons, thereby maintaining the basal levels of WRC activity and its downstream actin polymerization. However, unlike the L5-specific increase in F-actin levels, neuronal excitability and activity were enhanced in both L2/3 and L5 neurons of Cyfip2 cKO mPFC. Therefore, it is conceivable that the observed increase in CYFIP1 in L2/3 neurons is not sufficient to compensate for increased neuronal excitability and activity induced by a reduction in CYFIP2. This also suggests that the function of CYFIP2 regulating neuronal excitability and activity may be independent of the WRC activity and F-actin levels at least in L2/3 neurons.
Beyond the WRC, CYFIP1 and CYFIP2 can interact with a diverse range of proteins in the brain, such as RNA-binding proteins (RBPs) and postsynaptic scaffolding proteins (De Rubeis et al., 2013; Kumar et al., 2013; Li et al., 2017; Lee Y. et al., 2020). Notably, we recently showed that CYFIP1 and CYFIP2 can interact with distinct types of RBPs (Lee Y. et al., 2020). Specifically, among the 25 RBPs identified from the neonatal mouse forebrain CYFIP2 interactome, only two were found in common with the CYFIP1 brain interactome. Whether these differential interactions with RBPs contribute to the CYFIP2-specific roles in regulating neuronal excitability and activity is unclear. Future studies aimed at identifying target mRNAs of the CYFIP1-RBP and CYFIP2-RBP complexes and characterizing their functional implications in neurons are needed.
To better understand molecular mechanisms possibly contributing to the differential phenotypes of L2/3 and L5 neurons of Cyfip2 cKO mPFC, we investigated the relative mRNA expression levels between the forebrain L2/3 and L5 neurons for the 140 proteins of mouse forebrain CYFIP2 interactome (Lee Y. et al., 2020). Specifically, we used a recently established mouse brain single-cell gene expression database (Saunders et al., 2018) where we could get the relative mRNA expression ratios (L2/3 to L5) for 118 out of 140 proteins of the CYFIP2 interactome (Supplementary Table 2). We found that 39 out of 118 CYFIP2 interactors showed relatively higher mRNA expression in L2/3 neurons than in L5 neurons (L2/3-high), while 31 out of 118 CYFIP2 interactors showed the opposite relative expression ratio (L5-high). GO and pathway analyses on the L2/3-high 39 proteins revealed a significant enrichment of actin-related terms (Supplementary Table 3). Indeed, all key components of the WRC were included in the L2/3-high 39 proteins. However, no significant term was identified from the same analyses on the L5-high 31 proteins (Supplementary Table 4). These results suggest that the mPFC L2/3 neurons express relatively higher levels of mRNAs of actin-related CYFIP2 interactors than L5 neurons, and thus may be more resilient to CYFIP2 reduction in terms of maintaining normal F-actin levels (e.g., a compensatory increase in CYFIP1 proteins (Figure 2C) which were possibly translated from the abundant Cyfip1 mRNAs). Further experiments are necessary to validate this intriguing, yet premature, hypothesis.
Synaptic strength and neuronal activity in the mPFC are causally associated with social dominance status in mice (Wang et al., 2014). Specifically, an increase and decrease in excitatory synaptic efficacy in the mPFC cause an upward and downward change in social rank, respectively (Wang et al., 2011). Furthermore, direct optogenetic activation of the mPFC induces instantaneous winning in the tube test, while inhibition causes the opposite effect (Zhou et al., 2017; Tan et al., 2018). It has been proposed that, at the circuit level, the activation or inhibition of the mPFC modulates its downstream target regions including the striatum, basolateral amygdala, and dorsal raphe nucleus, thereby inducing changes in the social dominance behavior (Wang et al., 2014). Among the mPFC neurons, the deepest layer 5/6 neurons are the main output neurons targeting such subcortical nuclei (Riga et al., 2014). Considering the above-mentioned findings, it is conceivable that enhanced neuronal excitability/activity and/or increased F-actin levels of the mPFC L5 neurons may contribute to this enhanced social dominance behavior in adult Cyfip2+/− mice and Cyfip2 cKO mice (Figure 3H). The effect of F-actin increases on social dominance cannot be ignored because the involvement of the mPFC actin dynamics in social dominance behavior in mice has recently been reported (Tada et al., 2016). Importantly, neuronal changes in other brain regions, such as other PFC and subcortical areas, should also be carefully investigated to clearly understand critical brain regions and neuronal types involved in social dominance behavior of Cyfip2+/− mice and Cyfip2 cKO mice (Figure 3H).
It is somewhat counterintuitive that Cyfip2 cKO mice with lower CYFIP2 expressions in both L2/3 and L5 neurons did not display locomotor hyperactivity or reduced anxiety as observed in Cyfip2+/− mice considering that they also have lower CYFIP2 expressions but only in L5 neurons. One possible explanation is that the reduction in CYFIP2 in other neuronal cell types, such as inhibitory neurons, may contribute to such behavioral phenotypes in Cyfip2+/− mice. Indeed, we previously showed that β-galactosidase signals expressed from the LacZ cassette of Cyfip2+/− mice, which represent the promoter activity of the Cyfip2 gene, were detected in parvalbumin-positive inhibitory neurons of the mPFC (Lee S. H. et al., 2020). Therefore, CYFIP2 expression is also affected in inhibitory neurons of Cyfip2+/− mice, but not in Cyfip2 cKO mice that express Cre recombinase only in CaMKIIα-positive excitatory neurons. Another possibility is that this occurs because of the early developmental effects of CYFIP2 reduction in Cyfip2+/− mice. That is, brain CYFIP2 expression decreases in the early embryonic stages in Cyfip2+/− mice, whereas this decrease does not occur until the third to fourth postnatal week following the onset of Cre expression in Cyfip2 cKO mice. We previously showed that mRNA and protein levels of Cyfip2 were lowered by approximately 50% in the cortex of embryonic day 18.5 Cyfip2+/− mice (Zhang et al., 2018). Such a long-term reduction in CYFIP2 in Cyfip2+/− L5 neurons may have a larger effect on the associated brain circuits, thereby leading to more diverse behavioral phenotypes than those observed in Cyfip2 cKO mice. It is a prerequisite for validating the above-mentioned hypothesis to carefully characterize the spatiotemporal and cell-type-specific expression changes of CYFIP2 and its interactors, such as WAVE1, in both Cyfip2+/− mice and Cyfip2 cKO mice. Also, more comprehensive investigations on the cell-type- and age-specific brain functions of CYFIP2 are necessary, with a special emphasis on those factors implicated in the early-onset characteristics of CYFIP2-associated brain disorders (Zhong et al., 2019). To that end, our floxed-Cyfip2 mice represent a useful tool that can be combined with various available Cre-lines (Tsien, 2016) to more thoroughly assess other cell types of different developmental stages.
In conclusion, our results provide additional evidence that supports the critical roles CYFIP2 plays in regulating actin dynamics and neuronal excitability/activity in the mPFC principal neurons. Further investigations on the various mPFC target regions in Cyfip2 cKO mice and Cyfip2+/− mice may provide better insights into understanding the circuit-level mechanisms involved in the social dominance behavior of mice.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics Statement
The animal study was reviewed and approved and all procedures were approved by the Committee on Animal Research at Korea University College of Medicine (KOREA-2018-0174).
Author Contributions
YZ, HRK, S-HL, YKi, RM, CJ, J-EL, SK, YKa, SYK, S-KK, S-YC, and KH designed and performed the experiments. HRK, S-YC, and KH analyzed and interpreted the data. KH wrote the article. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government Ministry of Science and ICT (NRF-2018R1C1B6001235, NRF-2018M3C7A1024603, and NRF-2020R1A2C3011464), and by the Korea University grant K2004991.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2020.574947/full#supplementary-material.
References
- Abekhoukh S., Bardoni B. (2014). CYFIP family proteins between autism and intellectual disability: links with Fragile X syndrome. Front. Cell. Neurosci. 8:81. 10.3389/fncel.2014.00081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann S. O., Sledziowska M., Cross E., Kalbassi S., Waldron S., Chen F., et al. (2019). Behavioral training rescues motor deficits in Cyfip1 haploinsufficiency mouse model of autism spectrum disorders. Transl. Psychiatry 9:29. 10.1038/s41398-018-0338-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagni C., Zukin R. S. (2019). A synaptic perspective of fragile X syndrome and autism spectrum disorders. Neuron 101, 1070–1088. 10.1016/j.neuron.2019.02.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdagi O., Sakurai T., Dorr N., Pilorge M., Takahashi N., Buxbaum J. D. (2012). Haploinsufficiency of Cyfip1 produces fragile X-like phenotypes in mice. PLoS One 7:e42422. 10.1371/journal.pone.0042422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Borek D., Padrick S. B., Gomez T. S., Metlagel Z., Ismail A. M., et al. (2010). Structure and control of the actin regulatory WAVE complex. Nature 468, 533–538. 10.1038/nature09623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Chou H. T., Brautigam C. A., Xing W., Yang S., Henry L., et al. (2017). Rac1 GTPase activates the WAVE regulatory complex through two distinct binding sites. eLife 6:e29795. 10.7554/eLife.29795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S. Y., Pang K., Kim J. Y., Ryu J. R., Kang H., Liu Z., et al. (2015). Post-transcriptional regulation of SHANK3 expression by microRNAs related to multiple neuropsychiatric disorders. Mol. Brain 8:74. 10.1186/s13041-015-0165-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung L., Wang X., Zhu L., Towers A. J., Cao X., Kim I. H., et al. (2015). Parental origin impairment of synaptic functions and behaviors in cytoplasmic FMRP interacting protein 1 (Cyfip1) deficient mice. Brain Res. 1629, 340–350. 10.1016/j.brainres.2015.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport E. C., Szulc B. R., Drew J., Taylor J., Morgan T., Higgs N. F., et al. (2019). Autism and schizophrenia-associated CYFIP1 regulates the balance of synaptic excitation and inhibition. Cell Rep. 26, 2037.e6–2051.e6. 10.1016/j.celrep.2019.01.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S., Pasciuto E., Li K. W., Fernandez E., Di Marino D., Buzzi A., et al. (2013). CYFIP1 coordinates mRNA translation and cytoskeleton remodeling to ensure proper dendritic spine formation. Neuron 79, 1169–1182. 10.1016/j.neuron.2013.06.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez-Iturza N., Lo A. C., Shah D., Armendariz M., Vannelli A., Mercaldo V., et al. (2019). The autism- and schizophrenia-associated protein CYFIP1 regulates bilateral brain connectivity and behaviour. Nat. Commun. 10:3454. 10.1038/s41467-019-11203-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z., Zhu H., Zhou T., Wang S., Wu Y., Hu H. (2019). Using the tube test to measure social hierarchy in mice. Nat. Protoc. 14, 819–831. 10.1038/s41596-018-0116-4 [DOI] [PubMed] [Google Scholar]
- Feng G., Mellor R. H., Bernstein M., Keller-Peck C., Nguyen Q. T., Wallace M., et al. (2000). Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron 28, 41–51. 10.1016/s0896-6273(00)00084-2 [DOI] [PubMed] [Google Scholar]
- Han K., Chen H., Gennarino V. A., Richman R., Lu H. C., Zoghbi H. Y. (2015). Fragile X-like behaviors and abnormal cortical dendritic spines in cytoplasmic FMR1-interacting protein 2-mutant mice. Hum. Mol. Genet. 24, 1813–1823. 10.1093/hmg/ddu595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K., Holder J. L., Jr., Schaaf C. P., Lu H., Chen H., Kang H., et al. (2013). SHANK3 overexpression causes manic-like behaviour with unique pharmacogenetic properties. Nature 503, 72–77. 10.1038/nature12630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K., Kim M. H., Seeburg D., Seo J., Verpelli C., Han S., et al. (2009). Regulated RalBP1 binding to RalA and PSD-95 controls AMPA receptor endocytosis and LTD. PLoS Biol. 7:e1000187. 10.1371/journal.pbio.1000187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao K., Harony-Nicolas H., Buxbaum J. D., Bozdagi-Gunal O., Benson D. L. (2016). Cyfip1 regulates presynaptic activity during development. J. Neurosci. 36, 1564–1576. 10.1523/JNEUROSCI.0511-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C., Kang H. R., Kang H., Zhang Y., Lee Y., Kim Y., et al. (2019a). Unexpected compensatory increase in Shank3 transcripts in Shank3 knock-out mice having partial deletions of exons. Front. Mol. Neurosci. 12:228. 10.3389/fnmol.2019.00228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C., Kang H., Ryu J. R., Kim S., Zhang Y., Lee Y., et al. (2018). Integrative brain transcriptome analysis reveals region-specific and broad molecular changes in Shank3-overexpressing mice. Front. Mol. Neurosci. 11:250. 10.3389/fnmol.2018.00250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C., Kim S., Kang H., Yun K. N., Lee Y., Zhang Y., et al. (2019b). Shank3 regulates striatal synaptic abundance of Cyld, a deubiquitinase specific for Lys63-linked polyubiquitin chains. J. Neurochem. 150, 776–786. 10.1111/jnc.14796 [DOI] [PubMed] [Google Scholar]
- Kang M. S., Choi T. Y., Ryu H. G., Lee D., Lee S. H., Choi S. Y., et al. (2017). Autism-like behavior caused by deletion of vaccinia-related kinase 3 is improved by TrkB stimulation. J. Exp. Med. 214, 2947–2966. 10.1084/jem.20160974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim R., Kim J., Chung C., Ha S., Lee S., Lee E., et al. (2018). Cell-type-specific Shank2 deletion in mice leads to differential synaptic and behavioral phenotypes. J. Neurosci. 38, 4076–4092. 10.1523/JNEUROSCI.2684-17.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov M. V., Jones M. R., Rouillard A. D., Fernandez N. F., Duan Q., Wang Z., et al. (2016). Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids. Res. 44, W90–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V., Kim K., Joseph C., Kourrich S., Yoo S. H., Huang H. C., et al. (2013). C57BL/6N mutation in cytoplasmic FMRP interacting protein 2 regulates cocaine response. Science 342, 1508–1512. 10.1126/science.1245503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunda P., Craig G., Dominguez V., Baum B. (2003). Abi, Sra1 and Kette control the stability and localization of SCAR/WAVE to regulate the formation of actin-based protrusions. Curr. Biol. 13, 1867–1875. 10.1016/j.cub.2003.10.005 [DOI] [PubMed] [Google Scholar]
- Lee Y., Kang H., Jin C., Zhang Y., Kim Y., Han K. (2019a). Transcriptome analyses suggest minimal effects of Shank3 dosage on directional gene expression changes in the mouse striatum. Anim. Cells Syst. 23, 270–274. 10.1080/19768354.2019.1595142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Zhang Y., Ryu J. R., Kang H. R., Kim D., Jin C., et al. (2019b). Reduced CYFIP2 stability by Arg87 variants causing human neurological disorders. Ann. Neurol. 86, 803–805. 10.1002/ana.25598 [DOI] [PubMed] [Google Scholar]
- Lee J. H., Kim H. J., Yoon J. M., Cheon E. J., Lim J. W., Ko K. O., et al. (2016). Interstitial deletion of 5q33.3q35.1 in a boy with severe mental retardation. Korean J. Pediatr. 59, S19–S24. 10.3345/kjp.2016.59.11.S19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Zhang Y., Kang H., Bang G., Kim Y., Kang H. R., et al. (2020). Epilepsy-and intellectual disability-associated CYFIP2 interacts with both actin regulators and RNA-binding proteins in the neonatal mouse forebrain. Biochem. Biophys. Res. Commun. 529, 1–6. 10.1016/j.bbrc.2020.05.221 [DOI] [PubMed] [Google Scholar]
- Lee S. H., Zhang Y., Park J., Kim B., Kim Y., Lee S. H., et al. (2020). Haploinsufficiency of Cyfip2 causes lithium-responsive prefrontal dysfunction. Ann. Neurol. [Epub ahead of print]. 10.1002/ana.25827 [DOI] [PubMed] [Google Scholar]
- Li J., Zhang W., Yang H., Howrigan D. P., Wilkinson B., Souaiaia T., et al. (2017). Spatiotemporal profile of postsynaptic interactomes integrates components of complex brain disorders. Nat. Neurosci. 20, 1150–1161. 10.1038/nn.4594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima M., Kato M., Aoto K., Shiina M., Belal H., Mukaida S., et al. (2018). De novo hotspot variants in CYFIP2 cause early-onset epileptic encephalopathy. Ann. Neurol. 83, 794–806. 10.1002/ana.25208 [DOI] [PubMed] [Google Scholar]
- Oguro-Ando A., Rosensweig C., Herman E., Nishimura Y., Werling D., Bill B. R., et al. (2015). Increased CYFIP1 dosage alters cellular and dendritic morphology and dysregulates mTOR. Mol. Psychiatry 20, 1069–1078. 10.1038/mp.2014.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathania M., Davenport E. C., Muir J., Sheehan D. F., Lopez-Domenech G., Kittler J. T. (2014). The autism and schizophrenia associated gene CYFIP1 is critical for the maintenance of dendritic complexity and the stabilization of mature spines. Transl. Psychiatry 4:e374. 10.1038/tp.2014.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., Wang Y., He F., Chen C., Wu L. W., Yang L. F., et al. (2018). Novel west syndrome candidate genes in a chinese cohort. CNS Neurosci. Ther. 24, 1196–1206. 10.1111/cns.12860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riga D., Matos M. R., Glas A., Smit A. B., Spijker S., Van Den Oever M. C. (2014). Optogenetic dissection of medial prefrontal cortex circuitry. Front. Syst. Neurosci. 8:230. 10.3389/fnsys.2014.00230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A., Macosko E. Z., Wysoker A., Goldman M., Krienen F. M., De Rivera H., et al. (2018). Molecular diversity and specializations among the cells of the adult mouse brain. Cell 174, 1015.e16–1030.e16. 10.1016/j.cell.2018.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenck A., Bardoni B., Moro A., Bagni C., Mandel J. L. (2001). A highly conserved protein family interacting with the fragile X mental retardation protein (FMRP) and displaying selective interactions with FMRP-related proteins FXR1P and FXR2P. Proc. Natl. Acad. Sci. U S A 98, 8844–8849. 10.1073/pnas.151231598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J., Hong J., Lee S. J., Choi S. Y. (2012). c-Jun N-terminal phosphorylation is essential for hippocampal synaptic plasticity. Neurosci. Lett. 531, 14–19. 10.1016/j.neulet.2012.09.048 [DOI] [PubMed] [Google Scholar]
- Silva A. I., Haddon J. E., Ahmed Syed Y., Trent S., Lin T. E., Patel Y., et al. (2019). Cyfip1 haploinsufficient rats show white matter changes, myelin thinning, abnormal oligodendrocytes and behavioural inflexibility. Nat. Commun. 10:3455. 10.1038/s41467-019-11119-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence E. F., Soderling S. H. (2015). Actin out: regulation of the synaptic cytoskeleton. J. Biol. Chem. 290, 28613–28622. 10.1074/jbc.R115.655118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spranger S., Rommel B., Jauch A., Bodammer R., Mehl B., Bullerdiek J. (2000). Interstitial deletion of 5q33.3q35.1 in a girl with mild mental retardation. Am. J. Med. Genet. 93, 107–109. [DOI] [PubMed] [Google Scholar]
- Tada H., Miyazaki T., Takemoto K., Takase K., Jitsuki S., Nakajima W., et al. (2016). Neonatal isolation augments social dominance by altering actin dynamics in the medial prefrontal cortex. Proc. Natl. Acad. Sci. U S A 113, E7097–E7105. 10.1073/pnas.1606351113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S., Xiao Y., Yin H. H., Chen A. I., Soong T. W., Je H. S. (2018). Postnatal TrkB ablation in corticolimbic interneurons induces social dominance in male mice. Proc. Natl. Acad. Sci. U S A 115, E9909–E9915. 10.1073/pnas.1812083115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien J. Z. (2016). Cre-lox neurogenetics: 20 years of versatile applications in brain research and counting. Front. Genet. 7:19. 10.3389/fgene.2016.00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Kessels H. W., Hu H. (2014). The mouse that roared: neural mechanisms of social hierarchy. Trends Neurosci. 37, 674–682. 10.1016/j.tins.2014.07.005 [DOI] [PubMed] [Google Scholar]
- Wang F., Zhu J., Zhu H., Zhang Q., Lin Z., Hu H. (2011). Bidirectional control of social hierarchy by synaptic efficacy in medial prefrontal cortex. Science 334, 693–697. 10.1126/science.1209951 [DOI] [PubMed] [Google Scholar]
- Yoon K. J., Nguyen H. N., Ursini G., Zhang F., Kim N. S., Wen Z., et al. (2014). Modeling a genetic risk for schizophrenia in iPSCs and mice reveals neural stem cell deficits associated with adherens junctions and polarity. Cell Stem Cell 15, 79–91. 10.1016/j.stem.2014.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Kang H. R., Han K. (2019a). Differential cell-type-expression of CYFIP1 and CYFIP2 in the adult mouse hippocampus. Anim. Cells Syst. 23, 380–383. 10.1080/19768354.2019.1696406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Lee Y., Han K. (2019b). Neuronal function and dysfunction of CYFIP2: from actin dynamics to early infantile epileptic encephalopathy. BMB Rep. 52, 304–311. 10.5483/BMBRep.2019.52.5.097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Kang H., Lee Y., Kim Y., Lee B., Kim J. Y., et al. (2018). Smaller body size, early postnatal lethality and cortical extracellular matrix-related gene expression changes of Cyfip2-null embryonic mice. Front. Mol. Neurosci. 11:482. 10.3389/fnmol.2018.00482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Wang D., Wang Q., Rodal A. A., Zhang Y. Q. (2013). Drosophila cyfip regulates synaptic development and endocytosis by suppressing filamentous actin assembly. PLoS Genet. 9:e1003450. 10.1371/journal.pgen.1003450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong M., Liao S., Li T., Wu P., Wang Y., Wu F., et al. (2019). Early diagnosis improving the outcome of an infant with epileptic encephalopathy with cytoplasmic FMRP interacting protein 2 mutation: case report and literature review. Medicine 98:e17749. 10.1097/MD.0000000000017749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Zhu H., Fan Z., Wang F., Chen Y., Liang H., et al. (2017). History of winning remodels thalamo-PFC circuit to reinforce social dominance. Science 357, 162–168. 10.1126/science.aak9726 [DOI] [PubMed] [Google Scholar]
- Zweier M., Begemann A., Mcwalter K., Cho M. T., Abela L., Banka S., et al. (2019). Spatially clustering de novo variants in CYFIP2, encoding the cytoplasmic FMRP interacting protein 2, cause intellectual disability and seizures. Eur. J. Hum. Genet. 27, 747–759. 10.1038/s41431-018-0331-z [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.