Abstract
Daptomycin (DAP) is key in treating multidrug-resistant Staphylococcus infections. Diminished susceptibility to DAP is emerging among Staphylococcus epidermidis strains although mechanisms for non-susceptibility (NS) remain poorly understood. We report a case of persistent S. epidermidis bacteremia in which loss of DAP susceptibility arose during prolonged treatment. Whole genome sequencing identified two mutations, Q371del and P415L, in a single-affected gene, WalK, that coincided with the emergence of DAP-NS. Protein modeling of the mutations predicted a disruption of WalK protein configuration. The emergence of mutations in a single-gene during DAP exposure raises concerns in an era of increasingly treatment-resistant infections.
Lay summary: Daptomycin is an important antibiotic for fighting Staphylococcus infections. We identified variants in the WalK gene that were coincident with resistance in a clinical Staphylococcus epidermidis infection. Clinicians, hospital epidemiologists, and microbiology laboratories need to be aware of the potential for the evolution of drug resistance during prolonged daptomycin therapy.
Keywords: Staphylococcus epidermidis, daptomycin, antibiotic resistance
INTRODUCTION
Coagulase-negative Staphylococci (CoNS), including S. epidermidis, are capable of biofilm production, and are therefore one of the most common causes of endovascular device-related bloodstream infection (BSI) [1]. Staphylococcus epidermidis strains causing infections are increasingly multi-drug resistant (MDR) [2]. Guideline-based management of MDR-S. epidermidis endovascular device-related BSI includes parenteral antibiotics (primarily vancomycin) and removal of the device [3]. However, in some settings, such as patients with a left ventricular assist device (LVAD), device removal is often impractical. In these cases—absence of biofilm removal—prolonged courses of antimicrobial therapy may increase the potential for further drug-resistance development.
Daptomycin (DAP), a cyclic lipopeptide, demonstrates concentration-dependent bactericidal activity against Staphylococcus spp. [4]. Daptomycin is frequently used as an alternative to vancomycin for the treatment of VAD-related S. epidermidis BSI [5–7]. Although more than 98% of CoNS are reported to be susceptible to DAP [8], recent reports raise concern for emerging DAP resistance: (i) there are an increasing number of DAP non-susceptible (DAP-NS) CoNS isolates (minimum inhibitory concentrations [MICs] >2 μg/ml) and (ii) DAP-NS CoNS strains have been recovered from endovascular devices during DAP therapy [9].
Treatment-emergent DAP-NS S. aureus has been shown to occur through heterogenous pathways [10]; however, the mechanisms of clinically emergent DAP-NS S. epidermidis and other CoNS remain less well defined. One potentially overlapping mechanism across multiple Staphylococcus spp. is the environmental sensing apparatus, WalKR (syn: YycG/YycF). WalK is a sensor protein kinase and a member of the two-component regulatory system WalK/WalR (WalKR) that regulates genes involved in autolysis, biofilm formation, and cell-wall metabolism/degradation [11]. Increasing sub-inhibitory doses of DAP to a biofilm-producing laboratory S. epidermidis strain (ATCCRP62a) resulted in a DAP-NS phenotype via a single-nucleotide mutation in the WalK (V500F) [12, 13]. The functional relevance of V500F was demonstrated in an isogenic mutant RP62aWalKV500F with DAP-NS [13]. In a recent report, 6/17 of DAP-NS CoNS were found to have WalKR mutations [13]. However, whether WalKR mutations are sufficient to result in clinical-emergent DAP-NS CoNS and/or clinical failure has yet to be proven. Furthermore, given the potential for antimicrobial heteroresistance among bacterial sub-populations within a given biofilm, it is unclear to what extent antibiotic-resistant strains emerge through: (i) in vivo bacterial evolution during antibiotic pressure, (ii) antibiotic-mediated selection for a pre-existing intrinsically resistant subpopulation, and/or (iii) acquisition of an antibiotic resistant mutation from the external environment.
Here, we report on a case of persistent S. epidermidis bacteremia in a patient with an LVAD in which loss of susceptibility to both vancomycin and DAP arose during prolonged exposure to DAP. To the best of our knowledge, this is the first report to leverage whole genome sequencing (WGS) of isolates before and after antimicrobials to elucidate potential mechanisms of treatment-emergent DAP-NS within a patient host. We identified two mutations in the WalK gene from a single S. epidermidis strain (ST2) that became fixed after non-susceptible bloodstream isolates emerged, adding to the growing literature of the potential role of this gene in S. epidermidis DAP-NS.
CASE PRESENTATION
A 49-year-old woman with a history of non-ischemic cardiomyopathy and previous biventricular intra-cardiac defibrillator device and LVAD placement underwent LVAD exchange owing to device malfunction. Six weeks later, a percutaneous drain was placed to evacuate a persistent post-operative fluid collection near the LVAD. Ten days thereafter (52 days after LVAD exchange), she presented with fever. Multiple blood cultures grew MDR-S. epidermidis (resistant to oxacillin, clindamycin, gentamicin, and trimethoprim-sulfamethoxazole) that was susceptible to vancomycin and DAP by E-test (Supplementary Table 1; Supplementary Methods). Staphylococcus epidermidis was isolated from the draining fluid, confirming the putative source of infection. Vancomycin was initiated immediately for VAD-related S. epidermidis bloodstream infection, but despite achieving therapeutic vancomycin serum concentrations, bacteremia persisted. On day 5, vancomycin was changed to DAP (6 mg/kg IV), the drain was exchanged, and blood cultures resulted with no growth within 48 h. Daptomycin was continued for 6 weeks (46 total days) at which time the drain was removed. The patient was transitioned to oral doxycycline for suppression until heart transplant; however, on day 10 of doxycycline, fevers and MDR-S. epidermidis bacteremia recurred despite complete resolution of the fluid collection. Daptomycin was re-initiated, but S. epidermidis continued to grow in blood cultures, and the new isolate demonstrated intermediate susceptibility to vancomycin (MIC = 8.0 μg/ml) and loss of DAP susceptibility (MIC = 2.0 μg/ml).
CASE MANAGEMENT AND OUTCOME
Following the identification of a DAP-NS isolate, ceftaroline was initiated and bacteremia cleared within 48 h. However, eosinophilia and infusion-related chest pain after 20 days of ceftaroline prompted a change from ceftaroline to tigecycline. Surveillance blood cultures collected on day 6 of tigecycline again grew DAP-NS S. epidermidis (MIC = 4.0 μg/ml). Linezolid did not successfully clear bacteremia through 5-day monotherapy and another 4 days in combination with ceftaroline despite linezolid susceptibility (Supplementary Table 1). A combination of DAP–ceftaroline eventually led to sustained negative blood cultures and ultimately resolution of clinical symptoms. The patient was continued on DAP–ceftaroline without adverse events for more than 20 weeks until definitive VAD explant with heart transplantation was able to occur.
Microbiologic evaluation
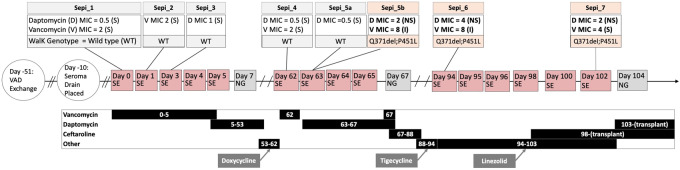
Vancomycin susceptibility testing (MIC) of the blood culture isolate from day 63 (Fig. 1) revealed both normal S. epidermidis growth around the E-test strip (Sepi_5a) and a smaller colony morphotype variant within the zone of inhibition of the E-test strip (Sepi_5b). To assess the colony heterogeneity, both colony morphotypes were sub-cultured, confirmed to be S. epidermidis, and vancomycin and DAP MICs were repeated on each morphotype separately and frozen separately (Supplementary Table 1). Given that distinct clones with different antimicrobial susceptibility patterns have recently been shown in a patient with VAD-related S. aureus endocarditis [14], we hypothesized that the presence of these two different morphotypes recovered from the same blood culture similarly indicated a mixed infection of two closely related strains.
Figure 1.
Timeline of clinical course: The clinical course of the patient (middle) with clinical events demarcated in circles while boxes indicate blood cultures (grey box: no growth; pink box: positive growth). The treatment regimen with days of antibiotic use is included (bottom) alongside the bloodstream isolate collection with susceptibility testing (top). VAD, ventricular assist device; SE, Staphylococcus epidermidis; NG, no growth; S, susceptible; NS, non-susceptible.
To determine if there was a genotypic basis for the differing phenotypic colony morphology and susceptibility patterns, we performed pulse-field gel electrophoresis (PFGE) and WGS on the patient’s isolates that were frozen and stored during each round of antimicrobial susceptibility testing, including both colony morphologies from day 63 (Sepi_5a and Sepi_5b).
Analysis of the PFGE patterns showed that the small colony morphotype, Sepi_5b, had only a slight shift in one band compared with Sepi_5a that was indistinguishable from the pre-antibiotic reference Sepi_1 (Supplementary Fig. 2). Furthermore, these three isolates were distinct from two archived VAD-related S. epidermidis bacteremia isolates at our institution, used for comparison.
Genomic analysis
DNA extraction was performed by picking multiple colonies from the pure subculture of each of the frozen bloodstream isolates to genetically characterize bloodstream isolates relevant to DAP exposure: seven time points, including both morphotypes from day 63 (Fig. 1). To capture as much genetic variation as possible, we performed short variant discovery and large structural variant discovery using a set of ‘raw’ (low stringency) and ‘filtered’ (high stringency) variants produced from reference guided mapping of paired-end WGS data (Supplementary Methods). In addition, we identified the species, multi-locus sequence type, resistance genes, and plasmids using a de novo assembly approach with the Centers for Genomic Epidemiology Bacterial Analysis Pipeline (Supplementary Fig. 1; Supplementary Methods) [15]. From these multiple approaches, we identified only two variants capable of completely segregating DAP-S isolates (Sepi_1, Sepi_2, Sepi_3, Sepi_4, and Sepi_5a) from DAP-NS isolates (Sepi_5b, Sepi_6, and Sepi_7; Supplementary Fig. 3). These sites encoded a three base-pair deletion (Q371del) and a missense mutation (P415L) in the WalK gene (NP_763574.1), respectively (Supplementary Fig. 4). Both variants were within the predicted kinase region (AA 384-602) of the protein (https://www.uniprot.org/uniprot/Q8CU87, July 2004, date last accessed) [13]. PROVEAN protein analysis indicated that the Q371del and P415L mutations had a deleterious predicted effect (Supplementary Table 2). Given the expected functional change from the mutation, we evaluated if structural changes in the protein could be predicted by homology modeling. The model indicates several disruptions in tertiary structures along the WalK protein when compared to wild-type protein, consistent with a non-neutral change in protein function (Supplementary Fig. 5).
DISCUSSION
We report that a deletion of Q371 together with a single substitution mutation in WalK (P415L) arose coinciding with the loss of DAP susceptibility in an MDR-S. epidermidis (ST2) blood stream infection after prolonged DAP exposure. To the best our knowledge, this is the first report of in vivo induction of the WalK mutation in a CoNS clinical isolate that is absent in other heterogenous mutations [9] including previously reported MprF S295L substitutions [2, 16]. We found no evidence of sub-populations of different strains using both traditional microbiological methods and whole genome analysis. As a result, we conclude that in vivo mutation resulting from antibiotic pressure was the most likely explanation for clinical failure owing to the emergence of a DAP-NS S. epidermidis strain. In addition, a previous study has shown that in vitro selection for DAP-NS resulted in a V500F WalK, further supporting the relevance of Q371del/P415L for phenotypic loss of DAP susceptibility [13]. Finally, the Q371del/P415L mutation persisted after discontinuation of DAP (day 67), suggesting that the mutated strain may not incur a fitness-cost relevant to this clinical setting. Thus, our findings add to an increasing recognition of the role of mutations in the WalK/R sensing system and DAP resistance among CoNS (Table 1).
Table 1.
Summary of WalK mutations and predicted effects
| Isolate | Case complication | Antibiotic exposures | WalK (yycG) variant | Provean score | Predicted functional effect | Reference |
|---|---|---|---|---|---|---|
|
UNC S. epidermidis (ST2) |
Ventricular-assist device | Vancomycin × 5 days | Q371del, P415L | −11.313, −9.022 | Deleterious, deleterious | This paper |
| Daptomycin × 46 days | ||||||
| Doxycycline × 10 days | ||||||
| S. capitis subsp. Ureolyticus | Ventricular-assist device | Vancomycin × 10 days | V220F | −1.505 | Neutral | PMC6310605 |
| Daptomycin × 6 days | ||||||
| S. capitis subsp. Ureolyticus | Aortic graft | Vancomycin × 14 days | N183I | −6.925 | Deleterious | PMC6310605 |
| S. epidermidis | in vitro | Daptomycin × 2 days | V500F | −3.058 | Deleterious | PMC6395924 |
| S. capitis | in vitro | Daptomycin × 11 days | PMC6395924 | |||
| S. epidermidis* | Clinical isolate | Not available | G312S, D472H | 2.28, 0.346 | Neutral, neutral | PMC6395924 |
| S. capitis*† | Clinical isolate | Not available | N48D | −1.05 | Neutral | PMC6395924 |
| S. capitis*† | Clinical isolate | Not available | N183I | −6.925 | Deleterious | PMC6395924 |
| S. capitis*† | Clinical isolate | Not available | G307D | −2.497 | Neutral | PMC6395924 |
| S. capitis*† | Clinical isolate | Not available | E573G | −6.526 | Deleterious | PMC6395924 |
| S. capitis*† | Clinical isolate | Not available | PMC6395924 | |||
| S. epidermidis | Pacemaker | Vancomycin × 1 day | M428T | −5.983 | Deleterious | PMC6408453 |
| S. epidermidis | Deep wound extract pocket site infection | Vancomycin × 1 day | M428T | −5.983 | Deleterious | PMC6408453 |
From a comprehensive literature review, we identified the previous reports of coagulase-negative Staphylococcus with daptomycin non-susceptible phenotypes and sequencing results. The isolate name, source (case complication), exposure to antibiotics, and the relevant mutations in the WalK gene are provided for each identified isolate. In addition, the PROVEAN score and predicted functional effect for each WalK variant are included. Given that PROVEAN estimates variant effects individually, the effects for the Q371del/P415L and G312S, D472H haplotypes cannot be estimated jointly. A subset of isolates underwent targeted Sanger sequencing and additional genome-wide effects are not available (*). Similarly, the co-occurrence of WalK and WalR mutations among a subset of S. capitis strains could not be determined from the manuscript (†).
How Q371del/P415L and V500F functional changes in WalK result in loss of DAP susceptibility is presently unknown. Similar to detergents, DAP is a bactericidal cell membrane-targeting lipopeptide. Perturbations in cell membrane events, including resistance to cell membrane depolarization and permeabilization, and reduced surface binding of DAP can lead to DAP resistance in S. aureus [17]. Interestingly, although overexpression of WalKR in S. aureus reduces susceptibility against vancomycin [18], the loss of WalKR expression resulting from mutations in regulatory genes leads to high resistance to cell lysis by Triton X-100 [11].
After prolonged exposure to a standard dose of DAP at 6 mg/kg IV, our patient developed a DAP-NS S. epidermidis infection with possible loss of vancomycin susceptibility. We cannot determine whether a higher initial dose of DAP may have averted DAP-NS evolution; however, increasing experience in the use of DAP for treatment of MRSA bloodstream infections suggests that the manufacturer-labeled dose may be inadequate, leading many experts to recommend doses of 8–10 mg/kg [19]. These considerations may be especially important for patients with an implanted cardiac device that cannot be removed, and when guideline-recommended adjuvant antibiotics such as aminoglycosides to promote bacterial clearance and/or rifampin to improve biofilm penetration [3] carry an unacceptably high risk of toxicity, as it is often the case of VAD-related bacteremia [1]. Further investigations are also needed to determine whether material-specific effects on biofilm production influence antibiotic resistance [20] in VAD-related infections, including regulation of WalKR [21].
Although we assumed that in our clinical setting, the Q371del/P415L mutation resulted in a low fitness cost, given its persistence even after the discontinuation of DAP treatment, it is possible that this persistence is not evidence of prosperity. In particular, the reversion of a deletion is highly unlikely and the Q371del/P415L mutation may represent an inescapable fitness in the absence of the DAP selective pressure. Future study with wild-type and isogenic knockout strains will be needed to elucidate this fitness landscape.
Increasing antimicrobial resistance among disease-associated S. epidermidis strains represents a global public health concern [2]. Given the current low incidence of DAP-R among S. epidermidis in the United States, DAP susceptibility is not universally reported by clinical microbiology laboratories and may often only be inferred. However, increasing reports of DAP-NS CoNS isolates raise the importance of vigilant monitoring and reporting of highly resistant isolates when they do occur. This may be of particular importance among patients with endovascular devices as a prior study found that only seven clones accounted for the majority of S. epidermidis infections in patients with VAD-related infection in the United States [22].
The utility of genomic and bioinformatic approaches for the surveillance, identification, and prediction of antibiotic resistance is being increasingly recognized as an aid or alternative to traditional clinical microbiology [23, 24]. In this manuscript, we combine these two disciplines together in a translational approach to elucidate the putative mechanism of antibiotic resistance observed in the clinic. In the future, similar translational approaches conducted in near real time would help to inform clinical practice by distinguishing the mechanism of resistance (e.g. in vivo evolution, selection of standing variation, and acquisition of resistance from the external environment), which thereby provides information for a tailored therapeutic response.
CONCLUSION
Clinicians, hospital epidemiologists, and microbiology laboratories need to be aware of the potential for drug-resistance development on therapy. Daptomycin dosing is undergoing scrutiny for MRSA and Enterococcal infections, and our data support the consideration of using higher than manufacturer label (6 mg/kg) when treating high-burden endovascular device-related S. epidermidis bacteremia. Our findings suggest that DAP promoted the Q371del/P415L WalK mutation, but given that other environmental triggers, like detergents and divalent metals, may also regulate WalKR; further investigations into the range of hospital chemical inducers of reduced susceptibility to vancomycin and DAP are warranted.
Supplementary Material
Acknowledgements
The authors thank the Research Computing group at the University of North Carolina at Chapel Hill, and in particular Sandeep Sarangi, for providing computational resources and support that have contributed to these research results. The authors also thank the University of North Carolina Office of Research for additional support.
FUNDING
N.F.B. was funded by the National Institute of Allergies and Infectious Diseases (F30AI143172). J.J.J. was funded by the Yang Biomedical Scholarship. L.A.B. was funded by the National Institute of Allergies and Infectious Diseases (K08AI108730).
AUTHOR CONTRIBUTIONS
N.F.B. developed software/pipelines, conducted analysis, and participated in manuscript preparation. K.J.L. performed microbiological testing, coordinated pulse-field gel electrophoresis and DNA library preparation, and participated in experimental design and manuscript preparation. A.S., I.H., P.I., C.C., A.B., D.D., A.L., and T.A. participated in clinical-decision making and participated in manuscript preparation. M.M. provided microbiology resources and participated in manuscript preparation. J.J.J. participated in experimental design, provided sequencing resources, developed software/pipelines, conducted analysis, and participated in manuscript preparation. L.A.B. served as primary investigator on IRB, experimental design, and participated in manuscript preparation.
ETHICAL APPROVALS
This study was approved by the Human Research Ethics/Institutional Review Board (OHRE/IRB) at the University of North Carolina at Chapel Hill (#17-3128).
REPRODUCIBILITY
All scripts and code used to generate these analyses are publicly available on Github (nickbrazeau/Sepi_Res_CaseStudy). Whole genome sequences are publicly available from the NCBI Short Read Archive (BioProject: PRJNA563309).
Conflict of interest: None declared.
Contributor Information
Nicholas F Brazeau, Department of Epidemiology, Gillings School of Global Public Health, University of North Carolina, Chapel Hill, NC 27599, USA; Medical Scientist Training Program, University of North Carolina School of Medicine, Chapel Hill, NC 27599, USA.
Kara J Levinson, Department of Pathology and Laboratory Medicine, School of Medicine, University of North Carolina, Chapel Hill, NC 27599, USA; Clinical Microbiology Laboratory, University of North Carolina Health Care, Chapel Hill, NC 27599, USA.
Asher Schranz, Division of Infectious Diseases, Department of Medicine, School of Medicine, University of North Carolina at Chapel Hill, Chapel Hill, NC, 27599, USA.
Kara A Moser, Department of Epidemiology, Gillings School of Global Public Health, University of North Carolina, Chapel Hill, NC 27599, USA.
Ian Hollis, University of North Carolina Health, UNC Eshelman School of Pharmacy, University of North Carolina at Chapel Hill, Chapel Hill, NC, 27599 USA.
Prashanth Iyer, University of North Carolina Health, UNC Eshelman School of Pharmacy, University of North Carolina at Chapel Hill, Chapel Hill, NC, 27599 USA.
Christopher Chien, Division of Cardiology, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA; Curriculum in Genetics and Molecular Biology, University of North Carolina, Chapel Hill, NC27599 USA.
Amanda Bowen, Division of Cardiology, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA; Curriculum in Genetics and Molecular Biology, University of North Carolina, Chapel Hill, NC27599 USA.
David van Duin, Division of Infectious Diseases, Department of Medicine, School of Medicine, University of North Carolina at Chapel Hill, Chapel Hill, NC, 27599, USA.
Anne Lachiewicz, Division of Infectious Diseases, Department of Medicine, School of Medicine, University of North Carolina at Chapel Hill, Chapel Hill, NC, 27599, USA.
Tessa Andermann, Division of Infectious Diseases, Department of Medicine, School of Medicine, University of North Carolina at Chapel Hill, Chapel Hill, NC, 27599, USA.
Melissa Jones, Clinical Microbiology Laboratory, University of North Carolina Health Care, Chapel Hill, NC 27599, USA.
Melissa Miller, Department of Pathology and Laboratory Medicine, School of Medicine, University of North Carolina, Chapel Hill, NC 27599, USA; Clinical Microbiology Laboratory, University of North Carolina Health Care, Chapel Hill, NC 27599, USA.
Jonathan J Juliano, Department of Epidemiology, Gillings School of Global Public Health, University of North Carolina, Chapel Hill, NC 27599, USA; Division of Infectious Diseases, Department of Medicine, School of Medicine, University of North Carolina at Chapel Hill, Chapel Hill, NC, 27599, USA.
Luther A Bartelt, Division of Infectious Diseases, Department of Medicine, School of Medicine, University of North Carolina at Chapel Hill, Chapel Hill, NC, 27599, USA.
REFERENCES
- 1. Koval CE, Stosor V, The AST ID Community of Practice. Ventricular assist device‐related infections and solid organ transplantation—Guidelines from the American Society of Transplantation Infectious Diseases Community of Practice. Clin Transplant 2019;33:e13552. [DOI] [PubMed] [Google Scholar]
- 2. Lee JYH, Monk IR, Gonçalves da Silva A. et al. Global spread of three multidrug-resistant lineages of Staphylococcus epidermidis. Nat Microbiol 2018;3:1175–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baddour LM, Wilson WR, Bayer AS, on behalf of the American Heart Association Committee on Rheumatic Fever, Endocarditis, and Kawasaki Disease of the Council on Cardiovascular Disease in the Young, Council on Clinical Cardiology, Council on Cardiovascular Surgery and Anesthesia, and Stroke Council et al. Infective endocarditis in adults: diagnosis, antimicrobial therapy, and management of complications: a scientific statement for healthcare professionals From the American Heart Association. Circulation 2015;132:1435–86. [DOI] [PubMed] [Google Scholar]
- 4. Steenbergen JN, Alder J, Thorne GM. et al. Daptomycin: a lipopeptide antibiotic for the treatment of serious Gram-positive infections. J Antimicrob Chemother 2005;55:283–8. [DOI] [PubMed] [Google Scholar]
- 5. Domínguez-Herrera J, Docobo-Pérez F, López-Rojas R. et al. Efficacy of daptomycin versus vancomycin in an experimental model of foreign-body and systemic infection caused by biofilm producers and methicillin-resistant Staphylococcus epidermidis. Antimicrob Agents Chemother 2012;56:613–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Duah M. Daptomycin for methicillin-resistant Staphylococcus epidermidis native-valve endocarditis: a case report. Ann Clin Microbiol Antimicrob 2010;9:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stewart PS, Davison WM, Steenbergen JN. Daptomycin rapidly penetrates a Staphylococcus epidermidis biofilm. Antimicrob Agents Chemother 2009;53:3505–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sader HS, Farrell DJ, Flamm RK. et al. Daptomycin activity tested against 164457 bacterial isolates from hospitalised patients: summary of 8 years of a Worldwide Surveillance Programme (2005–2012). Int J Antimicrob Agents 2014;43:465–9. [DOI] [PubMed] [Google Scholar]
- 9. Hagiya H, Sugawara Y, Kimura K. et al. Emergence of daptomycin non-susceptible coagulase-negative Staphylococci in patients with cardiovascular device infections: two cases report investigated by whole genome analysis. Medicine 2018;97:e13487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mangili A, Bica I, Snydman DR. et al. Daptomycin-resistant, methicillin-resistant Staphylococcus aureus bacteremia. Clin Infect Dis 2005;40:1058–60. [DOI] [PubMed] [Google Scholar]
- 11. Dubrac S, Boneca IG, Poupel O. et al. New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J Bacteriol 2007;189:8257–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Christensen GD, Simpson WA, Bisno AL. et al. Adherence of slime-producing strains of Staphylococcus epidermidis to smooth surfaces. Infect Immun 1982;37:318–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jiang J-H, Dexter C, Cameron DR. et al. Evolution of daptomycin resistance in coagulase-negative Staphylococci involves mutations of the essential two-component regulator WalKR. Antimicrob Agents Chemother 2019;63:e01926–18.DOI: 10.1128/AAC.01926-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harada S, Aoki K, Okamoto K. et al. Left ventricular assist device-associated endocarditis involving multiple clones of Staphylococcus aureus with distinct antimicrobial susceptibility patterns. Int J Infect Dis 2019;84:44–7. [DOI] [PubMed] [Google Scholar]
- 15. Thomsen MCF, Ahrenfeldt J, Cisneros JLB. et al. A bacterial analysis platform: an integrated system for analysing bacterial whole genome sequencing data for clinical diagnostics and surveillance. PLoS One 2016;11:e0157718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang S-J, Mishra NN, Rubio A. et al. Causal role of single nucleotide polymorphisms within the mprF gene of Staphylococcus aureus in daptomycin resistance. Antimicrob Agents Chemother 2013;57:5658–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bayer AS, Schneider T, Sahl H-G. Mechanisms of daptomycin resistance in Staphylococcus aureus: role of the cell membrane and cell wall. Ann N Y Acad Sci 2013;1277:139–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jansen A, Türck M, Szekat C. et al. Role of insertion elements and yycFG in the development of decreased susceptibility to vancomycin in Staphylococcus aureus. Int J Med Microbiol 2007;297:205–15. [DOI] [PubMed] [Google Scholar]
- 19. Liu C, Bayer A, Cosgrove SE. et al. Clinical Practice Guidelines by the Infectious Diseases Society of America for the treatment of methicillin-resistant Staphylococcus aureus infections in adults and children: executive summary. Clin Infect Dis 2011;52:285–92. [DOI] [PubMed] [Google Scholar]
- 20. Sakimura T, Kajiyama S, Adachi S. et al. Biofilm-forming Staphylococcus epidermidis expressing vancomycin resistance early after adhesion to a metal surface. Biomed Res Int 2015;2015:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Monk IR, Shaikh N, Begg SL. et al. Zinc-binding to the cytoplasmic PAS domain regulates the essential WalK histidine kinase of Staphylococcus aureus. Nat Commun 2019;10:3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gordon RJ, Miragaia M, Weinberg AD. et al. Staphylococcus epidermidis colonization is highly clonal across US cardiac centers. J Infect Dis 2012;205:1391–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Köser CU, Ellington MJ, Peacock SJ. Whole-genome sequencing to control antimicrobial resistance. Trends Genet 2014;30:401–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hendriksen RS, Bortolaia V, Tate H. et al. Using genomics to track global antimicrobial resistance. Front Public Health 2019;7:242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.