Abstract
Objectives
The Network for Genomic Surveillance in South Africa (NGS-SA) was formed to investigate the introduction and understand the early transmission dynamics of the SARS-CoV-2 epidemic in South-Africa.
Design
This paper presents the first results from this group, which is a molecular epidemiological study of the first 21 SARS-CoV-2 whole genomes sampled in the first port of entry – KwaZulu-Natal (KZN) – during the first month of the epidemic. By combining this with calculations of the effective reproduction number (R), it aimed to shed light on the patterns of infections in South Africa.
Results
Two of the largest provinces – Gauteng and KZN – had a slow growth rate for the number of detected cases, while the epidemic spread faster in the Western Cape and Eastern Cape. The estimates of transmission potential suggested a decrease towards R = 1 since the first cases and deaths, but a subsequent estimated R average of 1.39 between 6–18 May 2020. It was also demonstrated that early transmission in KZN was associated with multiple international introductions and dominated by lineages B1 and B. Evidence for locally acquired infections in a hospital in Durban within the first month of the epidemic was also provided.
Conclusion
The COVID-19 pandemic in South Africa was very heterogeneous in its spatial dimension, with many distinct introductions of SARS-CoV2 in KZN and evidence of nosocomial transmission, which inflated early mortality in KZN. The epidemic at the local level was still developing and NGS-SA aimed to clarify the dynamics in South Africa and devise the most effective measures as the outbreak evolved.
Keywords: SARS-CoV-2, South Africa, NGS-SA, Next Generation Sequencing, Epidemiology, Phylogenetic, Transmission genomics
Evidence before this study
PubMed, BioRxiv and MedRxiv were searched for reports on epidemiology and phylogenetic analysis using whole genome sequencing (WGS) of SARS-CoV-2. The following keywords were used: SARS-CoV-2, COVID-19, 2019-nCoV or novel coronavirus and transmission genomics, epidemiology, and phylogenetic or reproduction number. The search identified an important lack of molecular epidemiology studies in the southern hemisphere, with a few reports from Latin America and one from Africa. In other early transmission reports on SARS-CoV-2 infections in Africa, authors focused on transmission dynamics, but molecular and phylogenetic methods were missing.
Added value of this study
With a growing sampling bias in the study of transmission genomics of the SARS-CoV-2 pandemic, it is important for high-quality whole genome sequencing (WGS) of local SARS-CoV-2 samples to be reported and in-depth phylogenetic analyses to be conducted of the first month of infection in South Africa. This molecular epidemiological investigation identified the early transmission routes of the infection in KZN and reported 13 distinct introductions from many locations and a cluster of localised transmissions linked to a healthcare setting that caused most of the initial deaths in South Africa. Furthermore, a national consortium was formed in South Africa, funded by the Department of Science and Innovation and the South African Medical Research Council, to capacitate 10 local laboratories to produce and analyse SARS-CoV-2 data in near real-time.
Implications of all the available evidence
The COVID-19 pandemic is progressing around the world and in Africa. Early transmission genomics and dynamics of SARS-CoV-2 throw light on the early stages of the epidemic in a given region. This facilitates the investigation of localised outbreaks and serves to inform public health responses in South Africa.
Introduction
The novel coronavirus disease 2019 (COVID-19) was detected in China in late December 2019. On 30 January 2020, it was declared a Public Health Emergency of International Concern by the World Health Organization (WHO) (Sohrabi et al. 2020). By 15 May 2020, there were 4,621,410 COVID-19 cases and 308,542 related deaths (Worldometer, 2020) worldwide, involving almost every country in the world. Within five months, the virus had spread to Europe, America and eventually to Africa. The first case in Africa was reported in Nigeria on 28 February 2020 (Adepoju, 2020). At the time of writing, the pandemic had spread to almost all countries on the African continent. South Africa had the highest number of COVID-19 cases to date, with a total of 13,524 people infected and 247 deaths (on 15 May 2020) (COVID-19 WEEKLY EPIDEMIOLOGY BRIEF PROVINCES AT A GLANCE, n.d.).
The first confirmed case of COVID-19 in South Africa was reported on 05 March 2020. Decisive early action was taken by the government: a national state of disaster was declared on 15 March 2020 and a nationwide lockdown was enforced on 27 March 2020 to avoid the first wave overwhelming the health system. While initially only people who had travelled to at-risk countries and their contacts received PCR tests for severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2), the recommendation broadened to include all people with an acute respiratory illness. Furthermore, a program of community-based screening and testing was rolled out across the country (NICD, 2020). Testing increased rapidly and by the middle of May 2020, over 600,000 tests had been carried out in South Africa (approximately 10,000 per million population) (Roser et al., 2020).
As the global pandemic has expanded, whole genome sequencing and genomic epidemiology (Grubaugh et al., 2019) have been consistently used to investigate COVID-19 transmission and outbreaks (Deng et al., 2020, Eden et al., 2020, Gonzalez-Reiche et al., 2020, Grubaugh, 2020, Leung et al., 2020, Lu et al., 2020, Munnink et al., 2020). In response to the COVID-19 pandemic, the South African Network for Genomics Surveillance of COVID (NGS-SA) was formed (Msomi et al., 2020), which is a network of five large government laboratories and five public universities funded by the Department of Science and Innovation and the South African Medical Research Council. This paper focused on a detailed analysis of the epidemic in South Africa and preliminary genomic analysis of some of the first introductions of SARS-CoV-2 in KwaZulu-Natal (KZN). It shows that although the South African epidemic started in KZN, which had the first cases and deaths, other provinces in the country – namely the Western Cape (WC), Gauteng (GP) and the Eastern Cape (EC) – overtook KZN in the number of confirmed cases. Evidence of many distinct introductions of SARS-CoV-2 in KZN and early evidence suggesting nosocomial transmission is also presented.
Methods
Data sources
Publicly released data up to 11 May 2020 from the National Department of Health (NDoH) and the NICD in South Africa, which were collected in the repository of the Data Science for Social Impact Research Group at the University of Pretoria (Marivate et al., 2020), as well as global data on confirmed cases from the Johns Hopkins Coronavirus Resource Centre were used (Dong et al., 2020). The NDoH releases daily updates on the number of newly confirmed cases, with a breakdown by province. In the early stages of the epidemic, individual-level information on sex, age and travel history was released, but detailed reporting was discontinued on 23 March. In addition, the National Institute of Communicable Diseases (NICD) releases daily updates on the number of reverse-transcriptase polymerase chain reaction (RT-PCR) tests performed across all public and private sector laboratories, as well as the number of cases testing positive for SARS-CoV-2. Information from government press releases and speech transcripts were also included to chart a timeline of the government response to the epidemic. To understand the epidemic trajectory, the cumulative number of confirmed cases by province since the report of the hundredth case in the country by province was plotted.
Epidemiological analysis and reproductive number estimation
The effective reproduction number (R) was estimated by taking into account the observed epidemic growth rate r and two theoretical relationships (i, ii) of R, with r previously described in the literature. (i) The current study used the relationship R = (1+r/b)a as described in Imperial College London’s COVID-19 report 13 (Flaxman et al., 2020), where a = m2/s2 and b = m/s2, m the serial interval (SID) mean and the SID standard deviation. The SID distribution used was the one estimated by Nishiura et al. (Nishiura et al., 2020), with m = 4.7 and s = 2.9. This approach was termed the Flaxman et al. approach (Flaxman et al., 2020). (ii) The relationship R = (1+r/sigma)(1+r/delta), with 1/sigma being the infectious period and 1/delta the incubation period, was used as described by Wallinga and Lipsitch (Wallinga and Lipsitch, 2007), which is based on an SEIR modelling framework and expects both periods to be exponentially distributed. Exponential distributions were used with a mean 5.1 days for incubation (Kucharski et al., 2020, Linton et al., 2020) and 4 days for the infection (Kucharski et al., 2020, Linton et al., 2020); this approach was termed the Wallinga et al. approach. To obtain the epidemic growth rate r, maximum likelihood estimation in R (function optim) was used, by fitting the exponential growth model A0ert to the reported time series of cases and deaths (independently), where t is time, A0 is the number of reports at t = 0, and r the growth rate. Daily reported deaths and cases were used. The time periods for which there were data for deaths was 27 March to 11 May, and for cases was 5 March to 12 May. This approach is similar to that implemented by Xavier et al. (2020).
Ethics statement
The project was approved by University of KwaZulu-Natal Biomedical Research Ethics Committee. Protocol reference number: BREC/00,001,195/2020. Project title: COVID-19 transmission and natural history in KwaZulu-Natal, South Africa: Epidemiological Investigation to Guide Prevention and Clinical Care.
SARS-CoV-2 sample collection and preparation
Remnant samples from nasopharyngeal and oropharyngeal swabs collected from symptomatic patients were used for SARS-CoV-2 whole genome sequencing. These samples comprised either the primary swab sample or extracted RNA. The swab samples were heat inactivated in a water bath at 60 °C for 30 min, in a biosafety level 3 laboratory, prior to RNA extraction. RNA was extracted using the Viral NA/gDNA Kit on the Chemagic 360 system (Perkin Elmer, Hamburg, Germany) using the automated Chemagic 360 instrument (Perkin Elmer, Hamburg, Germany) or manually using the Qiagen Viral RNA Mini Kit (QIAGEN, California, USA).
Real-time RT-PCR
In order to detect the SARS-CoV-2 virus by PCR, the TaqPath COVID-19 CE-IVD RT-PCR Kit (Life Technologies, Carlsbad, CA) was used according to the manufacturer’s instructions. The assays target genomic regions (ORF1ab, S protein and N protein) of the SARS-CoV-2 genome. RT-PCR was performed on a QuantStudio 7 Flex Real-Time PCR instrument (Life Technologies, Carlsbad, CA). Cycle thresholds (Ct) were analysed using auto-analysis settings with the threshold lines falling within the exponential phase of the fluorescence curves and above any background signal. To accept the results, a Ct value for RNAse P (i.e. an endogenous internal amplification control) and/or the target gene in each reaction was confirmed, with undetermined Ct values in the no template control. The Ct values were reported for each target gene.
Tiling polymerase chain reaction
cDNA synthesis was performed on the RNA using random primers followed by gene-specific multiplex PCR using the ARTIC protocol (Quick, 2020) (Supplementary Table S6). Briefly, extracted RNA was converted to cDNA using the Protoscript II First Strand cDNA synthesis Kit (New England Biolabs, Hitchin, UK) and random hexamer primers. SARS-CoV-2 whole genome amplification by multiplex PCR was carried out using primers designed on Primal Scheme (http://primal.zibraproject.org/) to generate 400 bp amplicons with an overlap of 70 bp that covered the 30 Kb SARS-CoV-2 genome. PCR products were cleaned up using AmpureXP purification beads (Beckman Coulter, High Wycombe, UK) and quantified using the Qubit dsDNA High Sensitivity assay on the Qubit 4.0 instrument (Life Technologies Carlsbad, CA, USA).
Illumina MiSeq sequencing
PCR products for samples yielding sufficient material were included in this sequencing platform. Illumina® TruSeq® Nano DNA Library Prep kits were used according to the manufacturer’s protocol to prepare uniquely indexed paired end libraries of genomic DNA. The libraries were quantified using the Qubit dsDNA high-sensitivity assay on the Qubit 4.0 instrument (Life Technologies) and the fragments were analysed using the LabChip GX Touch (Perkin Elmer, Hamburg, Germany). Sequencing libraries were normalised to 4 nM, pooled and denatured with 0.2 N sodium acetate. The 12 pM sample library was spiked with 1% PhiX (PhiX Control v3 adapter-ligated library used as a control). Libraries consisting of 12 samples each were loaded onto a 500-cycle MiSeq Nano Reagent Kit v2 nano v2 Miseq reagent kit and run on the Illumina MiSeq instrument (Illumina, San Diego, CA, USA).
Bioinformatics assembly of genomes
Raw reads coming from both Nanopore and Illumina sequencing were assembled using Genome Detective 1.126 (https://www.genomedetective.com/) and the Coronavirus Typing Tool (Cleemput et al., 2020, Vilsker et al., 2019). The initial assembly obtained from Genome Detective was polished by aligning mapped reads to the references and filtering out low-quality mutations using bcftools 1.7–2 mpileup method. All mutations were confirmed visually with bam files using Geneious software (Biomatters Ltd, New Zealand). All of the sequences were deposited in GISAID (https://www.gisaid.org/) (Shu and McCauley, 2017).
Reference dataset
All sequences and associated metadata were downloaded from the GISAID sequence database (https://www.gisaid.org/) (Shu and McCauley, 2017) as of 01 May 2020 (n = 15,793). Due to the low variability of SARS-CoV-2, it was decided to only include high-quality sequences in the downstream analyses. To this end, sequences that were <25 kbp in length as well as sequences with a high proportion of ambiguous sites (>5%) were filtered out. Additionally, sequences that lacked any geographic and/or sampling date information were also removed. The resulting 10,959 sequences were analysed along with 20 sequences that were generated by the KwaZulu-Natal Research Innovation and Sequencing Platform (KRISP) laboratory. The dataset also contained one additional KZN sequence (EPI_ISL_417186) that was generated by the National Institute for Communicable Diseases (NICD) and represented a distant contact of the first diagnosed case in South Africa.
Lineage classification
No established nomenclature system currently exists for SARS-CoV-2. A dynamic lineage classification method proposed by Rambault et al. was used in this study (Rambaut et al., 2020) via the Phylogenetic Assignment of named Global Outbreak LINeages (PANGOLIN) software suite (https://github.com/hCoV-2019/pangolin). This was aimed at identifying the most epidemiologically important lineages of SARS-CoV-2 at the time of analysis, allowing researchers to more effectively monitor the epidemic in a particular geographical region. Two main SARS-CoV-2 lineages are currently recognised: lineage A, defined by Wuhan/WH04/2020 and lineage B, defined by Wuhan-Hu-1 strain. Although Wuhan-Hu-1 was the first published genome from SARS-CoV-2, it was classified as lineage B. Phylogenetic analyses of SARS-CoV-2 identified sequences from lineage A to be more closely related to a bat corona virus (Rambaut et al., 2020), which suggests that this is the first lineage (hence A). Lineage A genomes are characterised by two unique mutations (8782C > T and 28,144 T > C), relative to lineage B. Lineage B, on the other hand, shares no common mutations since this lineage contains the global SARS-CoV-2 genome reference (Wuhan-Hu-1). From these lineages, sub-lineages (e.g. A.1, A.2, A.3, and so forth) are then designated, each defined by an additional set of unique mutations. For example, for sub-lineage A.1, these mutations would be 11747C > T, 1785A > G and 18060C > T. Sub-lineages can further diversify into sub sub-lineages (e.g. A.1.1). Please refer to the schema provided in Supplementary Figure 5 for more information.
Phylogenetic analysis
A total of 10,959 GISAID reference genomes and 20 KRISP sequences were aligned in Mafft v7·313 (FF-NS-2) followed by manual inspection and editing in the Geneious Prime software suite (Biomatters Ltd, New Zealand). A maximum likelihood (ML) tree topology was constructed in IQ-TREE (GTR + G+I, no support) (Nguyen et al., 2015, Tavaré and Miura, 1986). Due to the large size of the alignment and low variability, it was opted to not infer support for splits in this tree topology. In any tree topology of SARS-CoV-2 the majority splits will be poorly supported, with only the major splits separating the major lineages having good support. The resulting ML tree topology was transformed into a time-scaled phylogeny using TreeTime (Sagulenko et al., 2018) with a clock rate of 8 × 10−4 and rooted along the branch of Wuhan-WH04 (GISAID: hCoV19/Wuhan/WH04/2020) and Wuhan-Hu1 (Genbank: MN908947). The resulting phylogeny was viewed and annotated in FigTree and ggtree.
Based on this large phylogeny of SARS-CoV-2, the GISAID reference sequences that passed initial sequence quality checks were randomly down-sampled to ∼10% of the original size. All African sequences in the GISAID subset, the 20 genotypes that were generated in this study, as well as a select few external references (e.g. Wuhan-Hu-1) were included. The resulting dataset of 1848 sequences was used in a custom build on the NextStrain analysis platform (Hadfield et al., 2018). To infer support for the splits in this tree topology, an additional 100 bootstrap trees in IQ-Tree under the same model parameters as NextStrain were inferred. These trees were then used to infer transfer support for splits in the phylogeny (Lemoine et al., 2018).
Bayesian tree
Bayesian coalescent analyses were performed on major lineages of the NextStrain build in which KZN sequences fell. The purpose of these analyses were to: (i) confirm the estimated date of origin for SARS-CoV-2, as proposed in recent literature (Andersen et al., 2020, Li et al., 2020), (ii) infer the estimated date of the most recent common ancestor (MRCA) for major lineages and (iii) infer the estimated dates of viral introductions into South Africa.
Due to the dynamic lineage assignment system of pangolins, many sub sub-lineages (e.g. A.1.1 or A.1.1.1) have emerged since the start of the outbreak. In order to keep things neat and tidy B lineages were organised into B, B.1 and B.2. Due to the large number of B and B.1 lineages, these were randomly down-sampled while all South African genotypes were retained. This resulted in three datasets for Bayesian coalescent inference: (B = 128, B.1 = 178 and B.2 = 69). Since none of the KZN sequences were classified as lineage A, A genotypes were exclude from the Bayesian analyses.
In short, sequences were aligned in mafft v7.313 and visualised and manually edited in Geneious software (Biomatters Ltd, New Zealand) as previously described. ML-tree topologies were inferred from each alignment in IQ-TREE v1.6.9 (GTR + G+I, with transfer support values) (Nguyen et al., 2015, Tavaré and Miura, 1986). Resulting tree topologies were analysed in TempEst software suite for temporal clock signal (Supplementary Figure S4). Coalescent molecular clock analyses were performed in BEAST v1.8. Analyses were run under a strict molecular clock assumption at a constant evolutionary rate of 8 × 10−4 nucleotide substitutions per site per year and an exponential growth coalescent tree prior. The Markov Chains were run in duplicate for a total length of 100 million steps, sampling every 10,000 iterations in the chains. Runs were assets in Tracer for good convergence (ESS > 200) and TreeAnnotator after discarding 10% of runs as burn-in.
Data availability
The SARS-CoV-2 genome sequences generated in this study were deposited in the GISAID database (https://www.gisaid.org/) under the following accession IDs: EPI_ISL_421572, EPI_ISL_421573, EPI_ISL_421574, EPI_ISL_421575 EPI_ISL_421576 EPI_ISL_436684 EPI_ISL_436685 EPI_ISL_436686 EPI_ISL_436687. In addition, raw short and long reads were submitted to the Short Read Archive (SRA) and can be accessed under BioProject Accession: PRJNA636748 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA636748).
Results
Epidemiology of COVID-19 in KZN and South Africa
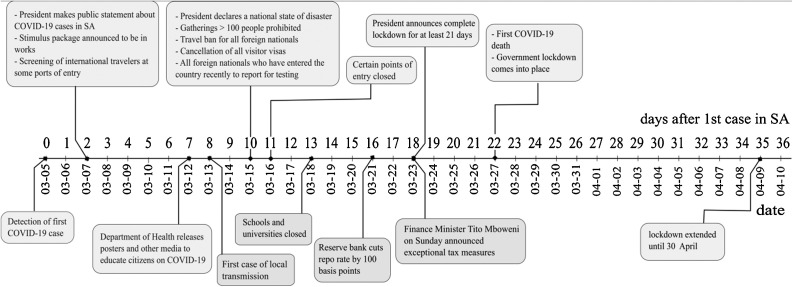
The first confirmed case of COVID-19 in South Africa was reported on 05 March 2020 in KZN. He was a South African citizen returning home from a skiing holiday in Italy. A steady increase in the number of confirmed cases in South Africa (all imported cases) followed over the next week, with the first suspected case of local transmission reported on 13 March 2020 in Durban, KZN. The early cases were predominantly located in the three provinces with the main urban populations and international travel hubs, namely: GP (main cities Pretoria and Johannesburg), the WC (Cape Town) and KZN (Durban). In these three provinces, the doubling time for confirmed cases was approximately three days prior to the lockdown (Figure 1 ). However, since the lockdown on 27 March 2020, the epidemic seems to have grown at different rates in South Africa.
Figure 1.
Timeline of measures implemented in South Africa from the first detected COVID-19 case on 05 March to the expansion of the lockdown in April 2020.
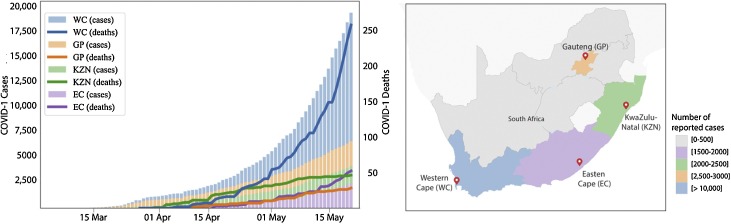
The South African epidemic has been very heterogeneous. For example, the first cases and deaths happened in KZN and GP. This was more pronounced in KZN, as a large nosocomial outbreak in a private hospital in Durban caused KZN to lead the country in number of deaths until the WC overtook it on 21 April 2020. In addition, GP, home of the largest metropolitan area of Johannesburg, had an unusual epidemic, as the majority of initial cases were in middle-age and wealthy individuals who travelled overseas for holidays. This translated in a very small number of deaths over time and infections were concentrated in the wealthy suburb of Sandton in Johannesburg. However, the epidemic expanded the fastest in the WC province, especially in Cape Town, which is the capital and most populated city of the WC. At the time of writing this report, this province had >60% of all of the cases and deaths in South Africa (Figure 2 ). There is mounting evidence that the WC is seeding the growing epidemic in the Eastern Cape, as the funerals from some of the deaths in the WC are taking place in the Eastern Cape.
Figure 2.
Summary of the COVID-19 epidemic in South Africa. A) Numbers of COVID-19 cases and deaths in the Western Cape (WC), Gauteng (GP), KwaZulu-Natal (KZN) and the Eastern Cape (EC). B) Geographic map showing the location of South African provinces.
This dynamic and heterogeneous epidemic complicates the estimation of effective reproductive number (R) over time and space. For example, deaths, which is normally one of the gold standard data for estimation of R0, for South Africa in May 2020 were stable at 1.12 (1.0–1.2) (Supplementary Fig. 1, Supplementary Table S1, Supp). KZN, the first province affected by COVID-19, initially had the highest death rate but in the last analysed period had three deaths. This study therefore attempted to estimate R from two data sources: aggregated reported cases and deaths at the country level (See Methods). Similarly, to that observed in other regions of the world, the estimates of R for South Africa suggest a decreasing transmission potential towards R = 1 since the first cases and deaths were reported, independent of the data source used. By the last analysed period between 6–18 May, using the Wallinga et al. approach (Wallinga and Lipsitch, 2007), it was found that R was still 1.39 (95% CI 1.04–2.15), suggesting the potential of sustained transmission for the near future.
SARS-CoV-2 genomes from KZN
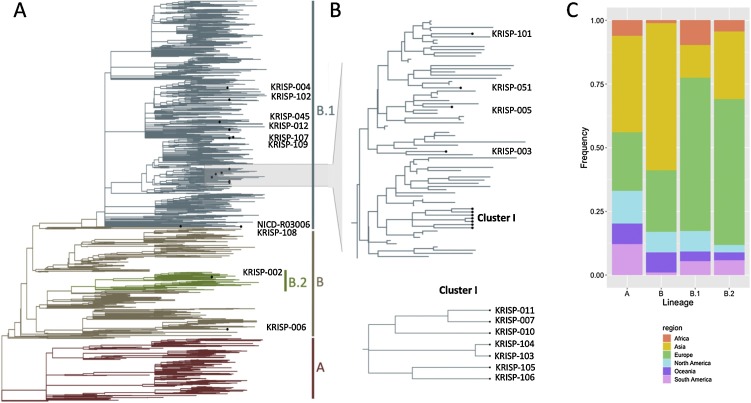
In order to determine the route of introduction of SARS-CoV-2 in KZN, 27 of some of the first confirmed cases in the province were assessed. Samples obtained from nasopharyngeal swabs represented 14 females and 10 males between the ages of 23–74 years. It managed to produce 20 near-whole genome sequences (>90% coverage) from these samples, and six partial genomes (Supplementary Table S2, Table S3). An extra genome was added to this dataset from the NICD, which was sampled in KZN (a close contact of the first reported case) on 07 March 2020. The 21 KZN whole genomes (20 KRISP and one NICD) were assigned to SARS-CoV-2 sub-lineages according to the nomenclature proposed and lineage classification obtained from >5000 genomes analysed by Rambaut et al. (Rambaut et al., 2020) (Supplementary Table S5). Given the uncertainties pertaining to the low diversity of this virus (Moreno et al., 2020), lineage assignment was restricted to the four most prominent subgroups: A, B, B.1 and B.2. Of the 21 KZN isolates being investigated in the present study, one was assigned to lineage B (KRISP-006) and one to sub-lineage B.2 (KRISP-002) (Figure 3 ). The remaining 19 KZN sequences were all assigned to lineage B.1. The B.1 lineage primarily consists of cases originating in Europe (Figure 3C), suggesting that introductions from Europe accounted for many of the early cases in KZN.
Figure 3.
Phylogenetic analysis. (A) A time-scaled maximum likelihood tree of 1849 sequences, including 21 genotypes from KwaZulu-Natal, South Africa. Major lineages of SARS-CoV-2 are labelled. B) Monophyletic cluster of KZN sequences. (C) Stacked bar plot showing the lineage breakdown of the dataset by region.
Although this investigation contained only a small number of samples from the first month of the epidemic in South Africa, it identified at least 13 distinct introductions (Figure 3, Figure 4 ) and one monophyletic cluster involving seven sequences. Three of the sequences (KRISP-007, KRISP-010 and KRISP-011) were identical and contained five mutations (241C > T, 3037C > T, 14408C > T, 16376C > T and 23403A > G). After investigation, it was found that these samples were from healthcare workers at a private hospital in Durban, KZN, with no history of travel outside the country. A detailed investigation is currently being conducted in this hospital, but preliminary findings suggest a point-source nosocomial outbreak (Lessells et al. manuscript in preparation). The other four sequences in the cluster contained two pairs (KRISP-103; KRISP-104 and KRISP-105; KRISP-106). Samples 103 and 104 are identical to one another and characterised by three additional mutations (5672C > A, 10592A > G and 26,063 G > T) on top of the ones reported above (Supplementary Table S4). Sample 106 acquired one additional mutation (24034C > T) on top of the five mutations common to the hospital outbreak, while sample 105 acquired another two mutations (13766A > T and 18,411 T > C) on top of the mutations found in 106. These two pairs were derived from random sampling within the Durban metropolitan area, suggesting early evidence of localised transmission in Durban (Figure 3B).
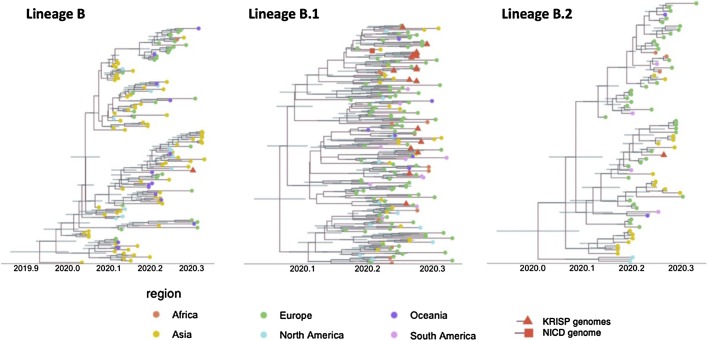
Figure 4.
Time-stamped phylogenetic trees of the three lineages of SARS-CoV-2 found in KwaZulu-Natal (KZN). The genomes produced in this study are marked with a red triangle, and the NICD genome by a red square. The geographic region of the other sequences is marked with coloured circles.
Time-resolved analysis of three main lineages circulating in KZN
To determine the evolutionary relationship of the KZN sequences to the world-wide SARS-CoV-2 pandemic, a Bayesian molecular clock analysis was conducted for each of the lineages found in KZN (Figure 4). Coalescent molecular clock analyses of lineage B place KRISp-006 at the base of a subclade along with sequences from Canada with high posterior support (p = 1.0). The remainder of the subclade contains sequences from a large number of Asian countries (Singapore, Philippines and Malaysia), Australia, the United States and the United Kingdom. The B.1 Bayesian analysis, which contained 19 KZN sequences, suggests multiple introductions into KZN from European countries. Due to low diversity in this lineage, the posterior support for splits in the tree were very low. Furthermore, due to the small number of nucleotide differences between isolates, the monophyletic clade of seven KZN sequences previously observed now only contain five sequences. Samples 103 and 104, though clustering together with one another, were separated from the rest of the clade by other European reference sequences. The time to the most recent common ancestor (tMRCA) for the monophyletic KZN clade of five sequences was inferred around 23 March 2020, with the 95% highest posterior density (HPD) between 10–31 March 2020, which is consistent with the dates of the nosocomial outbreak in Durban (Lessells et al. manuscript in preparation).
This coalescent analyses in BEAST placed the origin of B and B.1 around the first week of December 2019 [95% HPD], with the 95% HPD ranging between mid-October and the last week of December 2019. Coalescent analyses placed the tMRCA for sub-lineage B.2 around late December 2019 (95% HPD mid-November 2019 – first week of January 2020). These temporal estimates are consistent with the SARS-CoV-2 date of origin.
Discussion and conclusion
The spread of SARS-CoV-2 across the globe has given rise to one of the largest evolving pandemics in modern times. South Africa currently has the highest number of infections in Africa. South Africa seems to be moving to the next stage of the COVID-19 pandemic, with increasing community transmission even during the stringent lockdown and the epidemic growing at different rates in different regions of the country. At the time of writing this report, Cape Town, the main city in the WC, has the fastest increase in new infections and deaths in South Africa. Recent data indicate that >62% of the new infections and deaths are happening in this province, although only 17% of the South African population lives in this region. The fast spread of COVID-19 in the WC is not fully explained by the higher testing rates, as this province has performed between 20–22% of the tests in South Africa, but the positivity rate has been around 9%, whereas in the other provinces the positivity rate is around 1–2%. The estimates of transmission potential for South Africa suggest a decreasing transmission potential towards R = 1 since the first cases and deaths were reported, which is similar to that observed in other regions of the world. By the last period analysed between 6–18 May, when using the Wallinga et al. estimation approach applied to time series of reported cases, it was estimated that R was on average 1.39 (95% CI 1.04–2.15). Overall, these results are suggestive of an epidemic still in expansion at that time, despite a very early lockdown.
Sequencing of viral isolates from early COVID-19 cases in KZN, which is the province of South Africa with the first infections and early deaths, provided useful insights into the origins and transmission of SARS-CoV-2. From the first 21 analysed genomes, 13 independent introductions were found in KZN. These introductions were related to lineages B, B.1 and B.2, which have spread widely in Europe and North America. A cluster of cases was also found in healthcare workers in Durban, highlighting the potential importance of nosocomial transmission in this pandemic and potentially two other transmission pairs. The production of genomes from the WC will be crucial to understand the drivers of transmission during the lockdown period, and particularly whether healthcare facilities, prisons, workplaces and other institutions were acting as amplifiers of transmission. This is one of the main activities that the NGS-SA is currently working on.
Genomic analysis of SARS-CoV-2 in Africa has proven challenging on many fronts. First, sequencing of high-quality SARS-CoV-2 genomes is not a straightforward task. For example, a survey of thousands of sequences deposited in public databases has revealed a number of putative sequencing issues that appear to be the result of contamination, recurrent sequencing errors or hypermutability (Virological, 2020). These might arise from laboratory-specific techniques of sample preparation, sequencing technology or consensus calling. Furthermore, the low diversity of this virus and the small number of mutations that define lineages have prompted caution in the interpretation of early phylogenetic analysis worldwide (Lu et al., 2020). Often apparent local transmission clusters can in fact be the result of multiple introductions from under-sampled regions from non-uniform sequencing efforts (Grubaugh et al., 2019, Kraemer et al., 2019). To mitigate this, this study confirmed phylogenetic results by manual inspection of mutations relative to the reference of SARS-CoV-2 (Supplementary Table S4). Second, the pandemic is still evolving and grouping of SARS-CoV-2 into lineages and sub-clades is likely to be dynamic at this stage and it is influenced by proportionally larger numbers of sequences produced in the northern hemisphere (Rambaut et al., 2020). Third, the travel histories of apparent community transmissions need to be thoroughly investigated in order to elucidate the true dynamics of transmission in a particular area. In the current case, a subsequent investigation into the samples comprising the monophyletic cluster revealed an association with a big hospital outbreak of SARS-CoV-2 infections in Durban, KZN (Lessells et al. manuscript in preparation 2020).
This study had some important limitations. The first was related to estimation of R from a limited number of deaths in a highly heterogeneous epidemic, both in time and space - for which R was only able to be estimated at the aggregated country level. The second was a lack of well-set-up genomics laboratories that can sequence the virus in Africa. This is also amplified by the difficulty in acquiring reagents that are in high demand, coupled with the disruption of air freight. It is therefore a high priority for NGS-SA to evaluate and share protocols among national laboratories in South Africa that could generate high-quality sequences and capacitate laboratories with the protocols and bioinformatics pipelines to properly investigate virus introduction and to validate the call of variants with a detailed and reliable bioinformatics system. NGS-SA is also working with the Africa Center for Disease Control (CDC) and the World Health Organization (WHO) to strengthen genomics surveillance in the African continent.
This paper provides an early analysis of the COVID-19 pandemic in South Africa, showing very heterogeneous epidemics in the different provinces. It also estimated SARS-CoV-2 genetic diversity in KZN using the first 21 genomes from some of the first cases in the country. It found that KZN had many distinct introductions of SARS-CoV2, but also had early evidence of nosocomial transmission. The pandemic at a local level is still developing and the objective of NGS-SA is to clarify the dynamics of the epidemic in South Africa and devise the most effective measures as the outbreak evolves.
Funding statement
This work was based upon research supported by the UKZN Flagship Program entitled: Afrocentric Precision Approach to Control Health Epidemic; by the Saouth African Medical Research Council Self-Initiated Research grant (MRC SIR HIVDR-POC); by a research Flagship grant from the South African Medical Research Council (MRC-RFA-UFSP-01-2013/UKZN HIVEPI); by the Technology Innovation Agency and the Department of Science and Innovation; and by the National Human Genome Research Institute of the National Institutes of Health under Award Number U24HG006941. H3ABioNet is an initiative of the Human Health and Heredity in Africa Consortium (H3Africa). The content is solely the responsibility of the authors and does not necessarily represent the official views of any of the funders.
Conflict of interest
None to declare.
Acknowledgments
We wish to extend our thanks to all laboratory personnel that have worked hard to genotype SARS-CoV-2 samples and who have generously made them public via the GISAID database. Without this free data-sharing environment, this research would not have been possible. A full list of acknowledgments to contributing laboratories can be found in Supplementary Table S7.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.ijid.2020.11.128.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Adepoju P. Nigeria responds to COVID-19; first case detected in sub-Saharan Africa. Nat Med. 2020;26:444–448. doi: 10.1038/d41591-020-00004-2. [DOI] [PubMed] [Google Scholar]
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleemput S., Dumon W., Fonseca V., Abdool Karim W., Giovanetti M., Alcantara L.C., et al. Genome Detective Coronavirus Typing Tool for rapid identification and characterization of novel coronavirus genomes. Bioinformatics. 2020;36:3552–3555. doi: 10.1093/bioinformatics/btaa145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Gu W., Federman S., Du Plessis L., Pybus O., Faria N., et al. A Genomic Survey of SARS-CoV-2 Reveals Multiple Introductions into Northern California without a Predominant Lineage. MedRxiv. 2020 doi: 10.1101/2020.03.27.20044925. 2020.03.27.20044925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden J.-S., Rockett R., Carter I., Rahman H., de Ligt J., Hadfield J., et al. An emergent clade of SARS-CoV-2 linked to returned travellers from Iran. Virus Evol. 2020;6 doi: 10.1093/VE/VEAA027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaxman S., Mishra S., Gandy A., Unwin H.J.T., Coupland H., Mellan T., et al. 2020. Estimating the number of infections and the impact of non-pharmaceutical interventions on COVID-19 in 11 European countries. Imperial College COVID-19 response team. March. [Google Scholar]
- Gonzalez-Reiche A.S., Hernandez M.M., Sullivan M., Ciferri B., Alshammary H., Obla A., et al. Introductions and early spread of SARS-CoV-2 in the New York City area. MedRxiv. 2020 doi: 10.1101/2020.04.08.20056929. 2020.04.08.20056929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubaugh . Coast-to-coast spread of SARS-CoV-2 during the early epidemic in the United States. Cell Press. 2020. JRFEPEBHKSHYEAGWCBFVAFBTAAMJRRDNRCALWCCKI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubaugh N.D., Ladner J.T., Lemey P., Pybus O.G., Rambaut A., Holmes E.C., et al. Tracking virus outbreaks in the twenty-first century. Nat Microbiol. 2019;4:10–19. doi: 10.1038/s41564-018-0296-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer M.U.G., Cummings D.A.T., Funk S., Reiner R.C., Faria N.R., Pybus O.G., et al. Reconstruction and prediction of viral disease epidemics. Epidemiol Infect. 2019;147 doi: 10.1017/S0950268818002881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucharski A.J., Russell T.W., Diamond C., Liu Y., Edmunds J., Funk S., et al. Early dynamics of transmission and control of COVID-19: a mathematical modelling study. Lancet Infect Dis. 2020 doi: 10.1016/S1473-3099(20)30144-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine F., Entfellner J.-B.D., Wilkinson E., Correia D., Felipe M.D., De Oliveira T., et al. Renewing Felsenstein’s phylogenetic bootstrap in the era of big data. Nature. 2018;556:452–456. doi: 10.1038/s41586-018-0043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung K.S.-S., Ng T.T.-L., Wu A.K.-L., Yau M.C.-Y., Lao H.-Y., Choi M.-P., et al. A territory-wide study of early COVID-19 outbreak in Hong Kong community: A clinical, epidemiological and phylogenomic investigation. MedRxiv. 2020 doi: 10.1101/2020.03.30.20045740. 2020.03.30.20045740. [DOI] [Google Scholar]
- Li X., Zai J., Zhao Q., Nie Q., Li Y., Foley B.T., et al. Evolutionary history, potential intermediate animal host, and cross‐species analyses of SARS‐CoV‐2. J Med Virol. 2020;92:602–611. doi: 10.1002/jmv.25731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linton N.M., Kobayashi T., Yang Y., Hayashi K., Akhmetzhanov A.R., Jung S., et al. Incubation period and other epidemiological characteristics of 2019 novel coronavirus infections with right truncation: a statistical analysis of publicly available case data. J Clin Med. 2020;9:538. doi: 10.3390/jcm9020538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., du Plessis L., Liu Z., Hill V., Kang M., Lin H., et al. Genomic Epidemiology of SARS-CoV-2 in Guangdong Province, China. Cell. 2020 doi: 10.1016/j.cell.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marivate V., Arbi R., Combrink H., de Waal A., Dryza H., Egersdorfer D., et al. 2020. Coronavirus disease (COVID-19) case data - South Africa. [DOI] [Google Scholar]
- Roser M., Ritchie H., Esteban Ortiz-Ospina J.H. Coronavirus Pandemic (COVID-19) Publ Online. 2020 OurWorldInDataOrg. [Google Scholar]
- Moreno G.K., Braun K.M., Halfmann P.J., Prall T.M., Riemersma K.K., Haj A.K., et al. Limited SARS-CoV-2 diversity within hosts and following passage in cell culture. BioRxiv. 2020 doi: 10.1101/2020.04.20.051011. 2020.04.20.051011. [DOI] [Google Scholar]
- Msomi N., Mlisana K., de Oliveira T., Msomi N., Mlisana K., Willianson C., et al. A genomics network established to respond rapidly to public health threats in South Africa. Lancet Microbe. 2020;1:e229–30. doi: 10.1016/S2666-5247(20)30116-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnink B.B.O., Nieuwenhuijse D.F., Stein M., O’Toole A., Haverkarte M., Mollers M., et al. Rapid SARS-CoV-2 whole genome sequencing for informed public health decision making in the Netherlands. BioRxiv. 2020 doi: 10.1101/2020.04.21.050633. 2020.04.21.050633. [DOI] [PubMed] [Google Scholar]
- Nguyen L.-T., Schmidt H.A., Von Haeseler A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiura H., Linton N.M., Akhmetzhanov A.R. Serial interval of novel coronavirus (COVID-19) infections. Int J Infect Dis. 2020 doi: 10.1016/j.ijid.2020.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick J. 2020. Forked from Ebola virus sequencing protocol. [DOI] [Google Scholar]
- Rambaut A., Holmes E.C., Hill V., OToole A., McCrone J., Ruis C., et al. A dynamic nomenclature proposal for SARS-CoV-2 to assist genomic epidemiology. BioRxiv. 2020 doi: 10.1101/2020.04.17.046086. 2020.04.17.046086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagulenko P., Puller V., Neher R.A. TreeTime: Maximum-likelihood phylodynamic analysis. Virus Evol. 2018;4 doi: 10.1093/ve/vex042. vex042–vex042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y., McCauley J. GISAID: Global initiative on sharing all influenza data–from vision to reality. Eurosurveillance. 2017;22:30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrabi C., Alsafi Z., O’Neill N., Khan M., Kerwan A., Al-Jabir A., et al. World Health Organization declares global emergency: A review of the 2019 novel coronavirus (COVID-19) Int J Surg. 2020 doi: 10.1016/j.ijsu.2020.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavaré S., Miura R.M. Lectures on Mathematics in the Life Sciences; 1986. Some Mathematical Questions in Biology: DNA Sequence Analysis. [Google Scholar]
- Vilsker M., Moosa Y., Nooij S., Fonseca V., Ghysens Y., Dumon K., et al. Genome Detective: an automated system for virus identification from high-throughput sequencing data. Bioinformatics. 2019;35:871–873. doi: 10.1093/bioinformatics/bty695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallinga J., Lipsitch M. How generation intervals shape the relationship between growth rates and reproductive numbers. Proc R Soc B Biol Sci. 2007;274:599–604. doi: 10.1098/rspb.2006.3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier J., Giovanetti M., Adelino T., Fonseca V., da Costa A.V.B., Ribeiro A.A., et al. The ongoing COVID-19 epidemic in Minas Gerais, Brazil: insights from epidemiological data and SARS-CoV-2 whole genome sequencing. MedRxiv. 2020 doi: 10.1080/22221751.2020.1803146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The SARS-CoV-2 genome sequences generated in this study were deposited in the GISAID database (https://www.gisaid.org/) under the following accession IDs: EPI_ISL_421572, EPI_ISL_421573, EPI_ISL_421574, EPI_ISL_421575 EPI_ISL_421576 EPI_ISL_436684 EPI_ISL_436685 EPI_ISL_436686 EPI_ISL_436687. In addition, raw short and long reads were submitted to the Short Read Archive (SRA) and can be accessed under BioProject Accession: PRJNA636748 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA636748).