Abstract
The severe acute respiratory syndrome coronavirus, identified as SARS-CoV-2, initially established in Wuhan, China at the end of 2019, affects respiratory infections known as COVID-19. In an extraordinary manner, COVID-19 is affecting human life and has transformed a global public health issue into a crisis. Natural products are already recognized owing to the massive advantageous window and efficient antioxidant, antiviral immunomodulatory, and anti-inflammatory belongings. Additionally, the object of the present study was to demonstrate the inhibitory potential of the natural products coumarins and its analogues alongside SARS coronavirus. The present work, focuses on the synthesis of new coumarin analogues and characterized by FT-IR, 1H and 13C NMR, elemental analyses, and mass spectra. The recently synthesised compounds were projected conceptual association for COVID-19 protease and also to explore in anticipation if this protein will help target protease inhibitor drugs such as Calanolide A, Cardatolide A, Collinin, Inophyllum A, Mesuol, Isomesuol, Pteryxin, Rutamarin, Seselin and Suksdorin. The natural coumarin analogues docking scores were compared to standard Hydroxychloroquine. While the 3D module of SARS coronavirus main protease was predicted with the SWISS MODEL web server, as well as biochemical interaction tests were performed with the AutoDock Vina tool between the target protein with ligands. This research further showed that all the protease inhibitors accessed the target protein with negative dock energy. Molecular docking studies found that the natural coumarin analogue Inophyllum A showed an exceptional potential for inhibition with a binding energy of −8.4 kcal/mol. The synthetic coumarin analogues 1m and 1p both demonstrated a similar binding energy, inhibition potential of −7.9 kcal / mol as opposed to hydroxychloroquine and co-crystallized ligand alpha-ketoamide with binding energy values of −5.8 and −6.6 kcal / mol. All compounds evaluated were known as drug-like in nature, passing Lipinski's “Law of 5” with 0 violations except for alpha-ketoamide, passing Lipinski's “Rule of 5” with 1 violation (MW > 500). The inhibitor binding in silico research thus offers a structural understanding of COVID-19 and molecular interactions across the known protease inhibitors centred on the findings of the multiple sequence alliance.
Keywords: Coumarin, Molecular docking, ADME prediction, SARS-CoV-2, COVID-19
1. Introduction
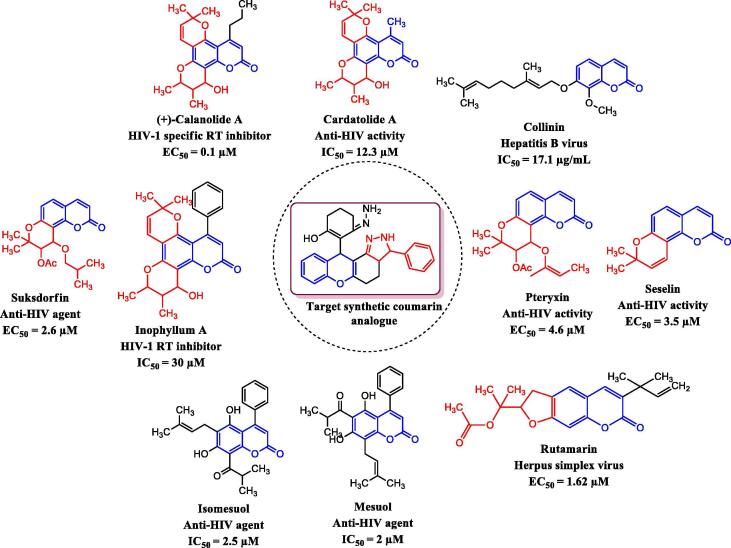
In humans and animals, coronaviruses are negotiators of diseases that may trigger illness in the respiratory region and even in the gastrointestinal area. Sooner CoV inspections revealed that CoVs, and also reptiles, avian species and mammals, may influence some forms of animals (Malik et al., 2020). At the end of 2019, in Wuhan, China (Lee and Hsueh 2020), the latest CoV species were recognized and initially named 2019-nCoV, and occurred by an eruption. WHO acknowledged an eruption in China on January 30, 2020, which had been meant as a public health crisis of international importance (Rodríguez-Morales et al., 2019). The WHO officially identified this COVID-19 (coronavirus disease 2019) on February 11, 2020. There are now no specific treatments available for COVID-19 and findings on the management of COVID-19 are incomplete (Rodríguez-Morales et al., 2019). However, the activities initiated appear to be confined to preventive and supportive treatments aimed at avoiding future complications and organ damage (Rodríguez-Morales et al., 2019). Antiviral like α-ketoamides have also been described in the literature as inhibitors of the coronavirus main protease (Kim et al., 2012). Environmental characteristics will greatly stimulate the discharge from tropical plants of secondary metabolites such as phytochemicals. As a result, secondary metabolites hidden by plants are prodigiously regarded in tropical areas and can be advanced as therapies (Yang et al., 2018, Guerriero et al., 2018). Several therapeutic plant phytochemicals have testified to antiviral function (Thayil and Thyagarajan, 2016, Jo et al., 2020, Zakaryan et al., 2017). In comparison to both AZTresistant G-9106 and pyridinone-resistant A17 with an EC50 value of 0.1 μM, a coumarin analogue named Calanolide A from the steamy rainforest tree, Calophyllum lanigerum was active in HIV-1 specific RT inhibitors (Kashman et al., 1992). Novel HIV-1 exact RT inhibitor Inophyllum A was extracted from Calophyllum inophyllum Linn fractions (Malaysian indigenous) with the IC50 value of 30 µM (Patil et al., 1993). Cordatolide A was extracted from the medicinal plant Calophyllum cordato-oblongum (a Srilankan indigenous) and having anti-HIV activity with the IC50 value of 12.3 µM (Dharmaratne et al., 1998). A new natural geranyloxycoumarin analogue Collinin isolated from Zanthoxylum schinifolium (Rutaceae) root bark extract and displayed the effect of antihepatitis B virus (HBV) with (IC50 = 17.1 µg/mL) related with DNA-synthesis inhibition (Chang et al., 1997). Marquez et al. stated two natural 4-phenylcoumarin derivatives, namely, Mesuol and Isomesuol acquired from Marila pluricostata tree as a significant anti-HIV agents with the IC50 value of 2 and 2.5 µM (Marquez et al., 2005). A new class of anti-HIV tricyclic coumarin termed as Suksdofin, which is a pyranocoumarin attained from the medicinal plant of Lomatium suksdorfii (Umbelliferae) and displayed significant anti-HIV with the EC50 value of 2.6 µM (Huang et al., 1994). The natural pyranocoumarin analogue Pteryxin having anti-HIV activity with the EC50 value of 4.6 µM (Willette and Soine 1962). The unsubstituted natural pyranocoumarin analogue seselin having significant anti-HIV property with the EC50 value of 3.5 µM (Hassan et al., 2016). Natural furanocoumarin Rutamarin achieved from Ruta graveolens L (common rue) plant having a significant effect against herpus simplex virus with the EC50 value of 1.62 µM (Xu et al., 2014). The natural antiviral coumarin analogues were represented in Fig. 1. Keep it in mind, natural coumarin analogues Calanolide A, Cardatolide A, Collinin, Inophyllum A, Mesuol, Isomesuol, Pteryxin, Rutamarin, Seselin, Suksdorin have been investigated as possible candidates for SARS coronavirus major protease complex with inhibitor alpha-ketoamide (PDB ID: 5N5O) and contrasted with Hydroxychloroquine and synthesized coumarin analogues (1m-1z). The findings of this analysis will have prospects for other researchers to identify the precise drug to combat COVID-19.
Fig. 1.
Natural antiviral coumarin analogues.
2. Material and methods
Both chemicals and reagents were obtained from Sigma-Aldrich. Chemical characterization was utilized by follow equipment such as IR spectra recording at 4000–400 cm−1 for used KBr pellets on Shimadzu 8201 pc, 1H & 13C NMR spectra was recorded on Bruker DRX instrument at 300 MHz. Elemental Analyzer (Varian EL III) was utilized to analyse, elements C, H, N present in the synthetic compounds. Thin layer chromatography (TLC) was used to analyzed the purity of the compounds.
2.1. General method for preparation of synthetic coumarin analogue
The compounds 9-(2-hydroxy-6-oxocyclohex-1-en-1-yl)-2,3,4,9-tetrahydro-1H-xanthen-1-one (0.001 mol, 0.3 g), hydrazine hydrate (0.1 mol, 0.5 mL), aldehyde (0.005 mol) and catalyst CuCl2·2H2O were mixed together in a mortar. To this few drops of con.HCl was added and grinded well. The reaction solution was poured into water then filtered and dried in a vacuum. The final solid was recrystallized from hot alcohol for further purification.
2.1.1. 3-hydrazono-2-(3-phenyl-2,3,3a,4,5,11-hexahydrochromeno[2,3-g]indazol-11-yl) cy clohex-1-enol (1m)
Light yellow solid; Yield: 92%; mp 261–264 °C; IR (KBr): 3172.28, 3072.84, 3032.46, 2594.68 cm−1; 1H NMR (300 MHz): δ 10.78 (s, 1H, OH), 9.84 (s, 1H), 8.62 (s, 2H, NH2), 6.80–7.80 (m, 9H, Phenyl), 4.56 (s, 1H), 2.23–2.80 (m, 2H), 1.34–2.00 (m, 10H); 13C NMR (75 MHz): 176.9 (1C), 160.9 (1C), 153.9 (1C), 155.6 (1C), 146.1 (1C), 101.8 (1C), 95.0 (1C), 129.7, 126.4, 123.9, 122.8, 117.1 (5C, Ar ring), 140.5, 128.5, 128.1, 125.9 (6C, Phenyl ring), 33.7 (1C), 53.8 (1C), 46.2 (1C), 29.7 (1C), 26.7 (1C), 22.8 (1C), 23.1 (1C), 16.2 (1C); EI-MS: 427.21 (M+, 28.5%); elemental analysis (C26H26N4O2): calculated: C, 73.22; H, 6.14; N, 13.14%; found: C, 73.23; H, 6.12; N, 13.15%.
2.1.2. 2-(3-(4-chlorophenyl)-2,3,3a,4,5,11-hexahydrochromeno[2,3-g]indazol-11-yl)-3-hydrazonocyclohex-1-enol (1n)
Yellow solid; Yield: 90%; mp 192–194 °C; IR (KBr): 3174.32, 3070.48, 3034.24, 2596.25 cm−1; 1H NMR (300 MHz): δ 10.76 (s, 1H, OH), 9.82 (s, 1H), 8.60 (s, 2H, NH2), 6.78–7.84 (m, 8H, Phenyl), 4.52 (s, 1H), 2.20–2.82 (m, 2H), 1.36–2.02 (m, 10H); 13C NMR (75 MHz): 176.8 (1C), 160.7 (1C), 153.8 (1C), 155.4 (1C), 146.3 (1C), 101.6 (1C), 95.2 (1C), 129.5, 126.2, 123.7, 122.6, 117.3 (5C, Ar ring), 140.3, 128.7, 128.4, 125.6 (6C, Phenyl ring), 33.5 (1C), 53.6 (1C), 46.4 (1C), 29.5 (1C), 26.4 (1C), 22.6 (1C), 23.3 (1C), 16.4 (1C); EI-MS: 461.17 (M+, 28.5%); elemental analysis (C26H25ClN4O2): calculated: C, 67.75; H, 5.47; N, 12.15%; found: C, 67.72; H, 5.49; N, 12.16%.
2.1.3. 3-hydrazono-2-(3-(3-nitrophenyl)-2,3,3a,4,5,11-hexahydrochromeno[2,3-g]indazol-11-yl)cyclohex-1-enol (1o)
Light yellow solid; Yield: 86%; mp 190–192 °C; IR (KBr): 3173.26, 3068.40, 3032.26, 2598.28 cm−1; 1H NMR (300 MHz): δ 10.74 (s, 1H, OH), 9.86 (s, 1H), 8.64 (s, 2H, NH2), 6.76–7.82 (m, 8H, Phenyl), 4.50 (s, 1H), 2.24–2.86 (m, 2H), 1.32–2.04 (m, 10H); 13C NMR (75 MHz): 176.4 (1C), 160.5 (1C), 153.2 (1C), 155.2 (1C), 146.4 (1C), 101.3 (1C), 95.4 (1C), 129.6, 126.4, 123.6, 122.4, 117.1 (5C, Ar ring), 140.5, 128.6, 128.2, 125.3 (6C, Phenyl ring), 33.6 (1C), 53.8 (1C), 46.2 (1C), 29.3 (1C), 26.5 (1C), 22.7 (1C), 23.5 (1C), 16.6 (1C); EI-MS: 472.19 (M+, 30.1%); elemental analysis (C26H25N5O4): calculated: C, 66.23; H, 5.34; N, 14.85%; found: C, 66.25; H, 5.32; N, 14.86%.
2.1.4. 4-(11-(6-hydrazono-2-hydroxycyclohex-1-en-1-yl)-2,3,3a,4,5,11-hexahy drochro meno[2,3-g]indazol-3-yl)phenol (1p)
Green solid; Yield: 88%; mp 210–212 °C; IR (KBr): 3170.26, 3062.24, 3036.26, 2592.28 cm−1; 1H NMR (300 MHz): δ 10.74 (s, 1H, OH), 10.40 (s, 1H, OH), 9.82 (s, 1H), 8.60 (s, 2H, NH2), 6.78–7.84 (m, 8H, Phenyl), 4.53 (s, 1H), 2.20–2.90 (m, 2H), 1.36–2.06 (m, 10H); 13C NMR (75 MHz): 178.4 (1C), 161.5 (1C), 154.2 (1C), 156.2 (1C), 145.4 (1C), 102.3 (1C), 96.4 (1C), 129.4, 126.8, 123.4, 122.6, 117.3 (5C, Ar ring), 140.7, 128.8, 128.6, 125.5 (6C, Phenyl ring), 34.6 (1C), 52.8 (1C), 47.2 (1C), 29.5 (1C), 26.3 (1C), 22.9 (1C), 23.7 (1C), 16.8 (1C); EI-MS: 443.20 (M+, 29.7%); elemental analysis (C26H26N4O3): calculated: C, 70.57; H, 5.92; N, 12.86%; found: C, 70.58; H, 5.93; N, 12.84%.
2.1.5. 4-(11-(6-hydrazono-2-hydroxycyclohex-1-en-1-yl)-2,3,3a,4,5,11-hexahydro chrom eno[2,3-g]indazol-3-yl)-2-methoxyphenol (1q)
Light yellow solid; Yield: 90%; mp 182–184 °C; IR (KBr): 3170.54, 3062.36, 3036.42, 2592.65 cm−1; 1H NMR (300 MHz): δ 10.72 (s, 1H, OH), 10.46 (s, 1H, OH), 9.80 (s, 1H), 8.64 (s, 2H, NH2), 6.74–7.86 (m, 7H, Phenyl), 4.50 (s, 1H), 3.83 (s, 3H), 2.24–2.94 (m, 2H), 1.38–2.08 (m, 10H); 13C NMR (75 MHz): 178.2 (1C), 161.7 (1C), 154.3 (1C), 156.4 (1C), 145.6 (1C), 102.5 (1C), 96.2 (1C), 129.6, 126.5, 123.7, 122.3, 117.6 (5C, Ar ring), 140.5, 128.2, 128.0, 125.3 (6C, Phenyl ring), 56.1 (1C), 52.5 (1C), 34.2 (1C), 47.5 (1C), 29.3 (1C), 26.0 (1C), 22.6 (1C), 23.5 (1C), 16.4 (1C); EI-MS: 473.21 (M+, 30.7%); elemental analysis (C27H28N4O4): calculated: C, 68.63; H, 5.97; N, 11.86%; found: C, 68.65; H, 5.95; N, 11.84%.
2.1.6. 2-(3-(4-(dimethylamino)phenyl)-2,3,3a,4,5,11-hexahydrochromeno[2,3-g]indazol-11-yl)-3-hydrazonocyclohex-1-enol (1r)
Red solid; Yield: 92%; mp 154–156 °C; IR (KBr): 3172.24, 3072.80, 3032.42, 2594.64 cm−1; 1H NMR (300 MHz): δ 10.78 (s, 1H, OH), 9.84 (s, 1H), 8.62 (s, 2H, NH2), 6.80–7.80 (m, 8H, Phenyl), 4.56 (s, 1H, —CH), 3.04 (s, 6H), 2.23–2.80 (m, 2H, —CH), 1.34–2.02 (m, 10H); 13C NMR (75 MHz): 176.9 (1C), 160.9 (1C), 153.9 (1C), 155.6 (1C), 146.1 (1C), 101.8 (1C), 95.0 (1C), 129.7, 126.4, 123.9, 122.8, 117.1 (5C, Ar ring), 140.5, 128.5, 128.1, 125.9 (6C, Phenyl ring), 33.7 (1C), 53.8 (1C), 46.2 (1C), 41.3 (2C), 29.7 (1C), 26.7 (1C), 22.8 (1C), 23.1 (1C), 16.2 (1C); EI-MS: 470.25 (M+, 30.7%); elemental analysis (C28H31N5O2): calculated: C, 71.62; H, 6.65; N, 14.91%; found: C, 71.60; H, 6.66; N, 14.89%.
2.1.7. 3-hydrazono-2-(3-(4-methoxyphenyl)-2,3,3a,4,5,11-hexahydrochromeno[2,3-g]inda zol-11-yl)cyclohex-1-enol (1s)
Light yellow solid; Yield: 84%; mp 150–152 °C; IR (KBr): 3172.28, 3072.76, 3032.68, 2594.60 cm−1; 1H NMR (300 MHz): δ 10.72 (s, 1H, OH), 9.82 (s, 1H), 8.60 (s, 2H, NH2), 6.76–7.74 (m, 8H, Phenyl), 4.52 (s, 1H), 3.06 (s, 3H), 2.26–2.82 (m, 2H, —CH), 1.30–2.04 (m, 10H); 13C NMR (75 MHz): 176.6 (1C), 160.3 (1C), 153.4 (1C), 155.3 (1C), 146.5 (1C), 101.6 (1C), 95.2 (1C), 129.6, 126.6, 123.8, 122.6, 117.3 (5C, Ar ring), 140.3, 128.6, 128.3, 125.6 (6C, Phenyl ring), 33.5 (1C), 53.2 (1C), 46.5 (1C), 41.6 (1C), 29.8 (1C), 26.5 (1C), 22.2 (1C), 23.3 (1C), 16.5 (1C); EI-MS: 457.22 (M+, 29.6%); elemental analysis (C27H28N4O3): calculated: C, 71.03; H, 6.18; N, 12.27%; found: C, 71.06; H, 6.19; N, 12.29%.
2.1.8. 2-(3-(furan-2-yl)-2,3,3a,4,5,11-hexahydrochromeno[2,3-g]indazol-11-yl)-3-hydrazo nocyclohex-1-enol (1t)
Brown solid; Yield: 82%; mp 147–149 °C; IR (KBr): 3171.23, 3073.46, 3034.70, 2596.30 cm−1; 1H NMR (300 MHz): δ 10.68 (s, 1H, OH), 9.86 (s, 1H), 8.62 (s, 2H, NH2), 6.78–7.80 (m, 7H, Phenyl), 4.54 (s, 1H), 2.30–2.86 (m, 2H), 1.36–2.08 (m, 10H); 13C NMR (75 MHz): 176.8 (1C), 160.5 (1C), 153.6 (1C), 155.8 (1C), 146.4 (1C), 101.3 (1C), 95.1 (1C), 129.3, 126.2, 123.5, 122.2, 117.6 (5C, Ar ring), 151.0, 141.5, 110.0, 109.2 (4C, Furan ring), 33.3 (1C), 53.5 (1C), 46.2 (1C), 29.3 (1C), 26.2 (1C), 22.4 (1C), 23.6 (1C), 16.3 (1C); EI-MS: 417.19 (M+, 26.3%); elemental analysis (C24H24N4O3): calculated: C, 69.21; H, 5.81; N, 13.45%; found: C, 69.20; H, 5.83; N, 13.46%.
2.1.9. 2-(11-(6-hydrazono-2-hydroxycyclohex-1-en-1-yl)-2,3,3a,4,5,11-hexahydro chrom eno[2,3-g]indazol-3-yl)phenol (1u)
Green solid; Yield: 88%; mp 128–130 °C; IR (KBr): 3170.23, 3072.46, 3033.70, 2595.30 cm−1; 1H NMR (300 MHz): δ 10.68 (s, 1H, OH), 10.40 (s, 1H, OH), 9.78 (s, 1H), 8.58 (s, 2H, NH2), 6.82–7.86 (m, 8H, Phenyl), 4.52 (s, 1H), 2.34–2.90 (m, 2H), 1.32–2.04 (m, 10H); 13C NMR (75 MHz): 176.6 (1C), 160.3 (1C), 153.3 (1C), 155.5 (1C), 146.1 (1C), 101.0 (1C), 94.8 (1C), 129.0, 125.9, 123.2, 121.9, 117.3 (5C, Ar ring), 140.0, 128.3, 128.0, 125.3 (6C, Phenyl ring), 33.0 (1C), 53.2 (1C), 45.9 (1C), 29.0 (1C), 25.9 (1C), 22.1 (1C), 23.3 (1C), 16.0 (1C); EI-MS: 443.20 (M+, 29.7%); elemental analysis (C26H26N4O3): calculated: C, 70.57; H, 5.92; N, 12.66%; found: C, 70.55; H, 5.93; N, 12.65%.
2.1.10. 2-(2,3,3a,4,5,11-hexahydrochromeno[2,3-g]indazol-11-yl)-3-hydrazonocyclohex-1-enol (1v)
Green solid; Yield: 90%; mp 216–218 °C; IR (KBr): 3172.23, 3074.46, 3035.70, 2597.30 cm−1; 1H NMR (300 MHz): δ 10.70 (s, 1H, OH), 9.80 (s, 1H), 8.60 (s, 2H, NH2), 6.84–7.88 (m, 4H, Phenyl), 4.54 (s, 2H), 2.36–2.92 (m, 2H, —CH), 1.34–2.06 (m, 10H); 13C NMR (75 MHz): 176.2 (1C), 160.3 (1C), 153.3 (1C), 155.5 (1C), 146.1 (1C), 101.0 (1C), 94.8 (1C), 129.0, 125.9, 123.6, 121.5, 117.7 (5C, Ar ring), 33.4 (1C), 53.6 (1C), 45.5 (1C), 29.4 (1C), 25.5 (1C), 22.5 (1C), 23.7 (1C), 16.4 (1C); EI-MS: 351.18 (M+, 22.0%); elemental analysis (C20H22N4O2): calculated: C, 68.55; H, 6.33; N, 15.99%; found: C, 68.52; H, 6.30; N, 15.95%.
2.1.11. 3-hydrazono-2-(3-methyl-2,3,3a,4,5,11-hexahydrochromeno[2,3-g]indazol-11-yl)cyclohex-1-enol (1w)
Green solid; Yield: 86%; mp 254–256 °C; IR (KBr): 3170.23, 3072.46, 3033.70, 2595.30 cm−1; 1H NMR (300 MHz): δ 10.76 (s, 1H, OH), 9.86 (s, 1H), 8.66 (s, 2H, NH2), 7.00–7.94 (m, 4H, Phenyl), 4.50 (s, 1H), 2.42–2.96 (m, 2H, —CH), 1.40–2.12 (m, 10H), 1.12 (s, 3H); 13C NMR (75 MHz): 176.8 (1C), 160.5 (1C), 153.5 (1C), 155.7 (1C), 146.3 (1C), 101.2 (1C), 95.0 (1C), 129.2, 126.1, 123.9, 121.7, 117.9 (5C, Ar ring), 33.6 (1C), 53.8 (1C), 45.7 (1C), 29.6 (1C), 25.7 (1C), 22.7 (1C), 23.9 (1C), 18.0 (1C), 16.6 (1C); EI-MS: 365.19 (M+, 24.3%); elemental analysis (C21H24N4O2): calculated: C, 69.21; H, 6.64; N, 15.37%; found: C, 69.25; H, 6.62; N, 15.35%.
2.1.12. 3-hydrazono-2-(3-((E)-prop-1-en-1-yl)-2,3,3a,4,5,11-hexahydrochromeno[2,3-g]in dazol-11-yl)cyclohex-1-enol (1x)
Green solid; Yield: 82%; mp 222–224 °C; IR (KBr): 3169.42, 3071.86, 3032.60, 2594.26 cm−1; 1H NMR (300 MHz): δ 10.70 (s, 1H, OH), 9.82 (s, 1H), 8.60 (s, 2H), 7.04–7.92 (m, 4H, Phenyl), 5.61 (d, 2H, J = 6.80 Hz, CH), 4.46 (s, 1H), 2.40–2.92 (m, 2H), 1.38–2.10 (m, 10H), 1.12 (s, 3H); 13C NMR (75 MHz): 175.8 (1C), 161.5 (1C), 152.5 (1C), 154.7 (1C), 145.3 (1C), 141.5 (1C), 125.2 (1C), 100.2 (1C), 94.0 (1C), 128.2, 125.1, 122.9, 120.7, 116.9 (5C, Ar ring), 32.6 (1C), 52.8 (1C), 44.7 (1C), 28.6 (1C), 24.7 (1C), 21.7 (1C), 22.9 (1C), 17.0 (1C), 15.6 (1C); EI-MS: 391.21 (M+, 25.3%); elemental analysis (C23H26N4O2): calculated: C, 70.75; H, 6.71; N, 14.35%; found: C, 70.72; H, 6.70; N, 14.32%.
2.1.13. 3-hydrazono-2-(3-((E)-styryl)-2,3,3a,4,5,11-hexahydrochromeno[2,3-g]indazol-11-yl)cyclohex-1-enol (1y)
Brown solid; Yield: 84%; mp 162–164 °C; IR (KBr): 3172.42, 3073.86, 3034.60, 2596.26 cm−1; 1H NMR (300 MHz): δ 10.76 (s, 1H, OH), 9.88 (s, 1H), 8.66 (s, 2H), 7.98–7.50 (m, 9H, Phenyl), 5.61 (d, 2H, J = 6.80 Hz, CH), 4.48 (s, 1H), 2.42–2.90 (m, 2H), 1.40–2.14 (m, 10H); 13C NMR (75 MHz): 176.8 (1C), 162.5 (1C), 153.2 (1C), 155.2 (1C), 146.3 (1C), 142.5 (1C), 124.2 (1C), 101.2 (1C), 95.0 (1C), 129.2, 126.1, 123.9, 121.7, 117.9 (5C, Ar ring), 140.0, 128.3, 128.0, 125.3 (6C, Phenyl ring), 33.6 (1C), 53.8 (1C), 45.7 (1C), 29.6 (1C), 25.7 (1C), 22.7 (1C), 23.9 (1C), 16.6 (1C); EI-MS: 453.22 (M+, 31.8%); elemental analysis (C28H28N4O2): calculated: C, 74.31; H, 6.24; N, 12.38%; found: C, 74.30; H, 6.25; N, 12.40%.
2.1.14. 2-(3-(2,6-dimethylhepta-1,5-dien-1-yl)-2,3,3a,4,5,11-hexahydrochromeno[2,3-g]indazol-11-yl)-3-hydrazonocyclohex-1-enol (1z)
Brown solid; Yield: 86%; mp 95–97 °C; IR (KBr): 3169.40, 3071.82, 3032.40, 2594.30 cm−1; 1H NMR (300 MHz): δ 10.70 (s, 1H, OH), 9.82 (s, 1H), 8.60 (s, 2H, NH2), 7.04–7.92 (m, 4H, Phenyl), 5.61 (d, 2H, J = 6.80 Hz, CH), 4.46 (s, 1H), 2.40–2.92 (m, 2H), 2.00 (m, 4H), 1.38–2.10 (m, 10H), 1.82 (s, 9H, 3CH3); 13C NMR (75 MHz): 175.8 (1C), 161.5 (1C), 152.5 (1C), 154.7 (1C), 145.3 (1C), 141.5 (1C), 125.2 (1C), 132.0 (1C), 123.5 (1C), 100.2 (1C), 94.0 (1C), 128.2, 125.1, 122.9, 120.7, 116.9 (5C, Ar ring), 33.7 (1C), 32.6 (1C), 52.8 (1C), 44.7 (1C), 28.6 (1C), 26.4 (1C), 24.7 (2C), 22.4 (1C), 21.7 (1C), 22.9 (1C), 18.6 (1C), 15.6 (1C); EI-MS: 473.29 (M+, 31.9%); elemental analysis (C29H36N4O2): calculated: C, 73.70; H, 7.68; N, 11.85%; found: C, 73.68; H, 7.70; N, 11.87%.
2.2. Molecular docking
2.2.1. Preparation of receptor
The SARS corona virus main protease crystal structure (PDB ID: 5N5O) was accessed from protein data bank. The water molecules and co-crystallized ligand were removed from the protein via Discovery Studio 2019 program. Finally, the protein was energy minimized by using Swiss PDB viewer.
2.2.2. Preparation of ligand
The 3D structures of ligand molecules (1a-1z) were sketched by ChemDraw Ultra 12.0 and energy minimized via MM2 force field in Chem3D Pro 12.0 softwares.
2.2.3. Identification of binding pocket
The identification of binding pocket was utilized by discovery studio program via co-crystallized ligand α-ketoamide and the residues of aminoacids Thr26, His41, Met49, Phe140, Leu141, Asn142, Gly143, Ser144, Cys145, His163, His164, Met165, Glu166, His172, Asp187, Gln189 and Thr190 were positioned on the binding pocket. The docking grid box selection was based upon the binding pocket.
2.2.4. Docking
Molecular interaction of docking were studied by following mode, between compounds Calanolide A, Cardatolide A, Collinin, Inophyllum A, Mesuol, Isomesuol, Pteryxin, Rutamarin, Seselin, Suksdorin, Hydroxychloroquine, (1m-1z) and co-crystallized ligand α-ketoamide with protein 5N5O using Autodock vina 1.1.2 (Trott and Olson 2010). AutoDock Tools 1.5.6 program package was used to build the input files for Autodock Vina. The docking grid box of SARS main protease was renowned as center_x: –23.002, center_y: −3.023, and center_z: 4.681 with size dimensions x,y,z: 24 with 1.0 Å spacing. The exhaustively value was set at 8. For Autodock Vina, the more constrictions were set to default and not specified. The best-scoring compounds results were visually investigated through Discovery studio 2019 program.
2.3. ADME and molecular property prediction
ADME analysis, Lipinski’s “Rule of five” was used to predict theoretical in silico ADME of compounds Calanolide A, Cardatolide A, Collinin, Inophyllum A, Mesuol, Isomesuol, Pteryxin, Rutamarin, Seselin, Suksdorin, Hydroxychloroquine, (1m-1z) and α-ketoamide (Lipinski et al., 2001). To predict Lipinski 's parameters, a Swiss ADME web tool was used (Swiss ADME 2016). To predict bioavailability and transport through the blood-brain barrier, the topological polar surface area (tPSA) was used (Ertl et al., 2000). Bioavailability is strongly multifactorial, mainly concerned with gastrointestinal absorption (Daina and Zoete 2016).The absorption percent was calculated from the formula, % ABS = 109 – (0.345 × TPSA).
3. Results
3.1. Chemistry
The synthetic coumarin analogues (1m-1z) were synthesized according to the synthetic sequence illustrated in scheme 1. Compounds (1m-1z) were synthesized by employing CuCl2·2H2O as a catalyst via grindstone chemistry technique.
Scheme 1.
Synthesis of coumarin analogues (1m-1z).
3.2. Docking studies
Docking simulations were conducted in order to advance the understanding of the possible process of biological activities. The compounds Calanolide A, Cardatolide A, Collinin, Inophyllum A, Mesuol, Isomesuol, Pteryxin, Rutamarin, Seselin, Suksdorfin, Hydroxychloroquine, (1m-z) were assessed for 5N5O protein with Autodock Vina tool. The tested inhibitors Inophyllum A (−8.4 kcal/mol), Calanolide A (−6.8 kcal/mol), Cardatolide A (−7.5 kcal/mol), Collinin (−6.1 kcal/mol), Mesuol (−7.6 kcal/mol) Isomesuol (−7.2 kcal/mol), Pteryxin (−7.3 kcal/mol), Rutamarin (−7.0 kcal/mol), Seselin (−6.6 kcal/mol), Suksdorfin (−7.0 kcal/mol), hydroxychloroquine (−5.8 kcal/mol), 1m (−7.9 kcal/mol), 1n (−7.4 kcal/mol), 10 (−7.1 kcal/mol), 1p (−7.9 kcal/mol), 1q (−7.6 kcal/mol), 1r (−7.1 kcal/mol), 1s (−7.1 kcal/mol), 1t (−7.4 kcal/mol), 1u (−7.8 kcal/mol), 1v (−6.5 kcal/mol), 1w (−6.8 kcal/mol), 1x (−7.1 kcal/mol), 1y (−7.3 kcal/mol), 1z (−7.5 kcal/mol), and α-ketoamide (−6.6 kcal/mol) in 5N5O protein correspondingly. The compounds 1m and 1p demonstrated substantial inhibition capacity with the binding energy of −7.9 kcal/mol. In table 1, the results have been abridged.
Table 1.
Molecular docking values of natural and synthesis coumarin analogues (1a-z).
| Main protease of SARS coronavirus (PDB ID: 5N5O) |
|||
|---|---|---|---|
| Binding affinity (kcal/mol) | No. of H-bonds | H-bonding residues | |
| Calanolide A (1a) | −6.8 | 1 | His164 |
| Cardatolide A (1b) | −7.5 | 1 | His164 |
| Collinin (1c) | −6.1 | 5 | Gly143, Ser144, Cys145, Gly166 |
| Inophyllum A (1d) | −8.4 | 1 | Cys145 |
| Mesuol (1e) | −7.6 | 2 | His41, Gly143 |
| Isomesuol (1f) | −7.2 | 0 | – |
| Pteryxin (1 g) | −7.3 | 1 | Gly143 |
| Rutamarin (1 h) | −7.0 | 4 | Ala46, Gly143, Ser144, Cys145 |
| Seselin (1i) | −6.6 | 0 | – |
| Suksdorin (1j) | −7.0 | 1 | Gly143 |
| α-ketoamide (1 k) | −6.6 | 0 | – |
| Hydroxychloroquine (1l) | −5.8 | 2 | Ser144, Cys145 |
| 1m | −7.9 | 0 | – |
| 1n | −7.4 | 2 | Asn142, Gln189 |
| 1o | −7.1 | 2 | Thr26, Gln189 |
| 1p | −7.9 | 1 | His163 |
| 1q | −7.6 | 1 | His163 |
| 1r | −7.1 | 1 | Asn142 |
| 1s | −7.1 | 3 | Asn142, His164 |
| 1t | −7.4 | 0 | – |
| 1u | −7.8 | 1 | Asn142 |
| 1v | −6.5 | 2 | His163, Glu166 |
| 1w | −6.8 | 2 | His163, Glu166 |
| 1x | −7.1 | 2 | Thr26, Leu141 |
| 1y | −7.3 | 2 | His163, Glu166 |
| 1z | −7.5 | 1 | Thr26 |
3.3. ADME and molecular property prediction
Bioavailability plays a key part in the progress of bioactive compounds as healing agents (Newby et al., 2015). The primary forecasters of this study were represented such as low polar surface area, hydrogen-bonding capacity, intestinal absorption, and decreased molecular flexibility (Azam et al., 2012). “All compounds tested pass Lipinski's” Rule of 5 “with 0 violations, except that alpha-ketoamide passes Lipinski's” Rule of 5 “with 1 MW > 500 violations (Table 2). Tested compounds with the exception of alpha-ketoamide (17 rotatable bonds) were 10 rotatable bonds. The tPSA value of testing compounds was less than 140 Å2. The absorption percent of tested compounds were devising under 50%. The prediction of solubility nature of testing compounds based on LogS value. Compounds Seselin, α-ketoamide and Hydroxychloroquine shows better solubility in water.
Table 2.
ADME value of natural and synthesis coumarin analogues (1a-1z).
| Comp. | tPSAa | %Absb | MWc | RoBd | HBDe | HBAf | MRg | IlogPh (MlogP) | LogSi | CYP2D6 Inhibitor |
|---|---|---|---|---|---|---|---|---|---|---|
| Rule | ≤140 ′Å2 | >50 | ≤500 | ≤10 | ≤5 | ≤10 | 40–130 | <5 | >−4 | – |
| Calanolide A (1a) | 68.90 | 85.22 | 370.44 | 2 | 1 | 5 | 106.11 | 3.83 (2.85) | −4.70 | Yes |
| Cardatolide A (1b) | 68.90 | 85.22 | 342.39 | 0 | 1 | 5 | 96.50 | 3.40 (2.40) | −4.07 | Yes |
| Collinin (1c) | 48.67 | 92.20 | 342.43 | 8 | 0 | 4 | 102.59 | 3.95 (3.38) | −5.35 | No |
| Inophyllum A (1d) | 68.90 | 85.22 | 404.46 | 1 | 1 | 5 | 116.97 | 3.73 (3.25) | −5.28 | No |
| Mesuol (1e) | 87.74 | 78.72 | 392.44 | 5 | 2 | 5 | 115.49 | 3.22 (2.98) | −5.87 | No |
| Isomesuol (1f) | 87.74 | 78.72 | 392.44 | 5 | 2 | 5 | 115.49 | 2.90 (2.98) | −5.87 | No |
| Pteryxin (1g) | 74.97 | 83.13 | 358.39 | 4 | 0 | 6 | 97.03 | 3.22 (2.06) | −4.14 | No |
| Rutamarin (1h) | 65.74 | 86.31 | 356.41 | 5 | 0 | 5 | 100.79 | 3.67 (3.09) | −4.80 | No |
| Seselin (1i) | 39.44 | 95.39 | 228.24 | 0 | 0 | 3 | 66.61 | 2.66 (2.37) | −3.47 | No |
| Suksdorin (1j) | 74.97 | 83.13 | 360.40 | 5 | 0 | 6 | 97.50 | 3.23 (2.13) | −4.07 | No |
| α-ketoamide (1k) | 136.63 | 61.86 | 534.65 | 17 | 5 | 5 | 150.47 | 3.58 (1.11) | −3.72 | No |
| Hydroxychloroquine (1l) | 48.39 | 92.30 | 335.87 | 9 | 2 | 3 | 98.57 | 3.58 (2.35) | −3.91 | Yes |
| 1m | 92.23 | 77.18 | 426.51 | 2 | 3 | 4 | 132.36 | 3.22 (2.96) | −4.90 | No |
| 1n | 92.23 | 77.14 | 460.96 | 2 | 3 | 4 | 137.37 | 3.47 (3.43) | −5.51 | No |
| 1o | 138.05 | 61.37 | 471.51 | 3 | 3 | 6 | 141.18 | 2.80 (2.07) | −4.99 | No |
| 1p | 112.46 | 70.20 | 442.51 | 2 | 4 | 5 | 134.38 | 2.81 (2.43) | −4.77 | No |
| 1q | 121.69 | 67.01 | 472.54 | 3 | 4 | 6 | 140.87 | 3.42 (2.11) | −4.86 | No |
| 1r | 95.47 | 76.06 | 469.58 | 3 | 3 | 4 | 146.57 | 3.65 (2.83) | −5.16 | No |
| 1s | 101.46 | 73.99 | 456.54 | 3 | 3 | 5 | 138.85 | 3.68 (2.63) | −4.99 | No |
| 1t | 105.37 | 72.64 | 416.47 | 2 | 3 | 5 | 124.62 | 2.90 (1.78) | −4.26 | No |
| 1u | 112.46 | 70.20 | 442.51 | 2 | 4 | 5 | 134.38 | 2.95 (2.43) | −4.77 | No |
| 1v | 92.23 | 77.18 | 350.41 | 1 | 3 | 4 | 107.87 | 2.60 (1.91) | −3.43 | No |
| 1w | 92.23 | 77.18 | 366.44 | 1 | 3 | 4 | 112.68 | 2.96 (2.13) | −3.79 | No |
| 1x | 92.23 | 77.18 | 390.48 | 2 | 3 | 4 | 121.82 | 3.24 (2.48) | −4.17 | No |
| 1y | 92.23 | 77.18 | 452.55 | 3 | 3 | 4 | 142.29 | 3.54 (3.29) | −5.39 | No |
| 1z | 92.23 | 77.18 | 472.62 | 5 | 3 | 4 | 150.19 | 4.21 (3.63) | −6.01 | No |
4. Discussion
4.1. Chemistry
The key projections of the synthesized compounds (1m-1z) by FT-IR spectra embrace peaks at 3169.42–3174.32, 3062.36–3074.46, and 2592.65–2598.26 cm−1, conforming the —NH, Aromatic —CHstr, and —C N functional groups. The 1H NMR spectra was shows signals at δ 10.70–10.80, 9.78–9.88 and 8.58–8.66 ppm, conforming the OH, NH, and NH2 protons individually. The 13C NMR spectra was exhibited peaks at δ 176.2–178.4, 155.2–156.8 and 15.6–16.8 ppm, imitates —C—OH, —C N and CH2 carbon atoms. The compounds were additionally confirmed via elemental analysis and mass spectroscopy techniques.
4.2. Docking studies
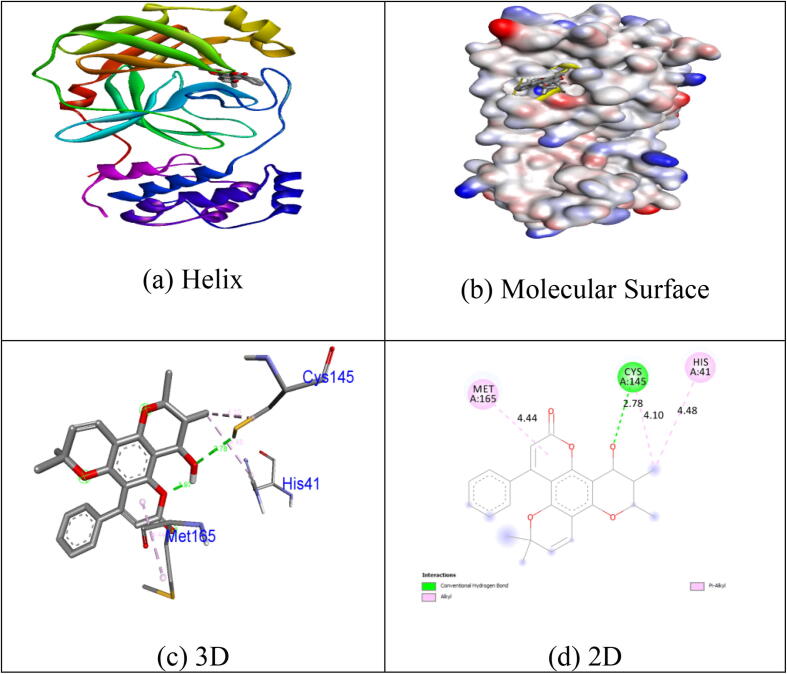
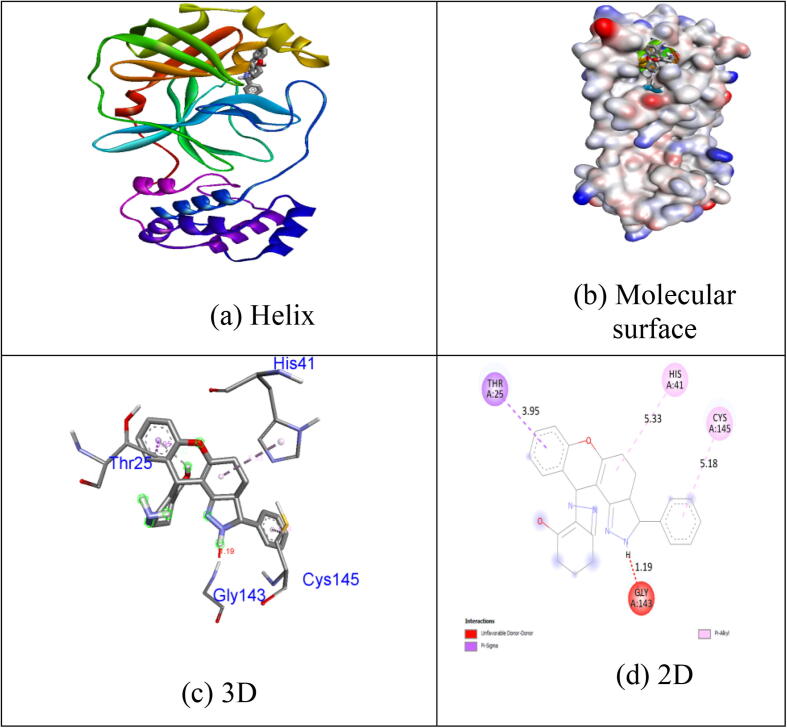
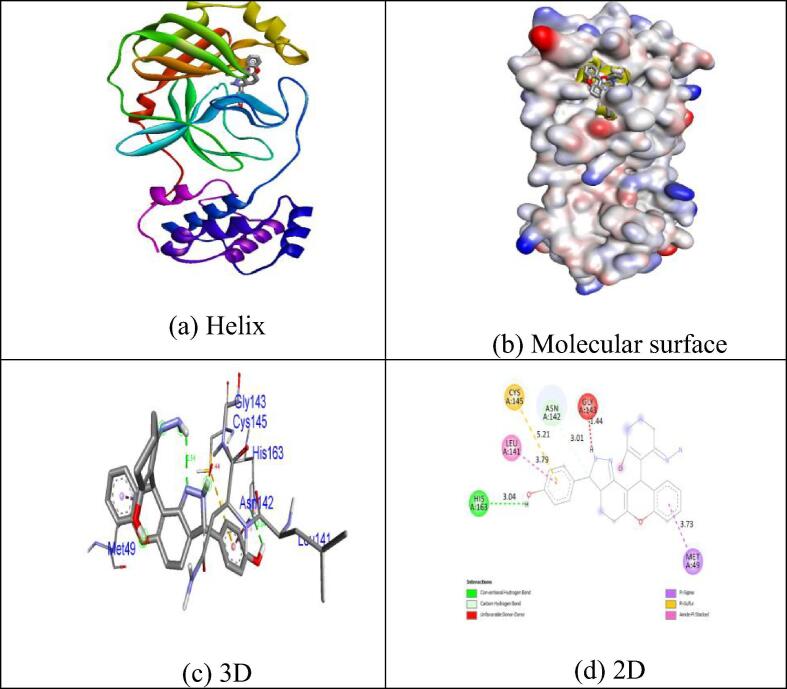
Many of the measured ligands demonstrate substantial inhibition potential by a binding energy of −6.6 kcal / mol excluding Collinin (−6.1 kcal / mol) relative to in-build co-crystallized ligand alpha-ketoamide. Unlike other chemicals, the natural coumarin equivalent Inophyllum A displays exceptional binding affinity (−8.4 kcal / mol). H-donor and H-acceptor atoms was approved bond gap is less than 3.5 Å (Taha et al., 2015). The compounds in the target protein 5N5O were less than 3.5 Å, suggesting the resilient hydrogen bonding between protein and ligands. Compound Inophyllum A shows one H-bond interaction with 5N5O receptor. In hydrogen bonding activity with the bond lengths of 2.78 Å, the residues of Cys145 amino acids became intertwined. The His41, Cys145 and Met165 amino acid residues became tangled in hydrophobic contacts. Hydrogen bonding and hydrophobic interactions of 5N5O protein amino acid residues with the Inophyllum A compound have been seen in Fig. 2. No association with the 5N5O receptor is seen in the synthesised coumarin analogue 1m. Hydrophobic contacts were tangled in the residues Thr25, His41, Gly143 and Cys145. Hydrogen bonding and hydrophobic connections of 5N5O protein amino acid residues with compound 1m is seen in Fig. 3. One hydrogen bond associated with the receptor 5N5O as seen by the synthesised coumarin analogue 1p. In hydrogen bonding contact with the bond lengths of 3,04 Å., the residues of His163 became intertwined. In the hydrophobic encounters, the residues Met49, Leu141, Asn142, Gly143 and Cys145 were intertwined. Hydrogen bonding and hydrophobic connections of 5N5O protein with compound 1p is seen in Fig. 4. The fallouts revealed that the compound Inophyllum A had the extraordinary capacity to suppress the respective target protein than other compounds.
Fig. 2.
Interactions between binding site of 5N5O protein with Inophyllum A.
Fig. 3.
Interactions between binding site of 5N5O protein with 1m.
Fig. 4.
Interactions between binding site of 5N5O protein with 1p.
4.3. ADME and molecular property prediction
The amount of rotatable bonds and also the capacity to attach to receptors were represented by the molecules. With the exception of alpha-ketoamide (17 rotatable bonds) devised under 10 rotatable bonds and without a chirality core, the checked compounds pass one of the oral bioavailability conditions, exhibiting low versatility of conformation. In addition to the blood–brain barrier, passive molecular transport across membranes has been associated with the Topological Polar Surface Area (tPSA) property (Ertl et al., 2000). Checked substances with a tPSA value <140 Å2 pass the gastrointestinal absorption requirements and the oral administration criterion later. In divergence, all verified compounds excepting α-ketoamide (tPSA = 136.63 Å2) and 1m (tPSA = 92.23 Å2), 1n (tPSA = 92.23 Å2), 1o (tPSA = 138.05 Å2), 1p (tPSA = 112.46 Å2), 1q (tPSA = 121.69 Å2), 1r (tPSA = 95.47 Å2), 1s (tPSA = 101.46 Å2), 1t (tPSA = 105.37 Å2), 1u (tPSA = 112.46 Å2), 1v (tPSA = 92.23 Å2), 1w (tPSA = 92.23 Å2), 1x (tPSA = 92.23 Å2), 1y (tPSA = 92.23 Å2), 1z (tPSA = 92.23 Å2), that suggests that in compound alpha-ketoamide and compounds (1m-1z), but not in other compounds, the side effects of the central nervous system are compact or inattentive. Absorption percent (percent Abs = > 50) was demonstrated by the studied substances, suggesting strong bioavailability. The appropriate oral bioavailability was (>50 percent). The seasoned compounds Seselin, α-ketoamide Hydroxychloroquine, 1v, and 1w showed tremendous water solubility (−logS value of >−4) excepting compounds Calanolide A (−logS value of −4.70), Cardatolide A (−logS value of −4.07), Collinin (−logS value of −5.35), Inophyllum A (−logS value of −5.28), Mesuol (−logS value of −5.87), Isomesuol (−logS value of −5.87), Pteryxin (−logS value of −4.14), Rutamarin (−logS value of −4.80), Suksdorin (−logS value of −4.07) 1m (−logS value of −4.90), 1n (−logS value of −5.51), 1o (−logS value of −4.99), 1p (−logS value of −4.77), 1q (−logS value of −4.86), 1r (−logS value of −5.16), 1s (−logS value of −4.99), 1t (−logS value of −4.26), 1u (−logS value of −4.77), 1x (−logS value of −4.17), 1y (−logS value of −5.39), and 1z (−logS value of −6.01) shows moderate water solubility. Side effects of liver dysfunction were not expected for Collinin, Inophyllum A, Mesuol, Isomesuol, Pteryxin, Rutamarin, Seselin, Suksdorfin, compounds (1m-1z) and co-crystallized alpha-ketoamide ligands since they were projected to be non-inhibitors of CYP2D6 and were anticipated for Calanolide A, Cardatolide A and hydroxychloroquine owing to the receptor properties of CYP2D6. The P-glycoprotein (P-gp) part of the ATP-binding transporter family includes brain penetration, intestinal absorption, and drug metabolism, and its reticence may significantly alter the safety of bioavailability (Fromm 2000). Dose-convinced phospholipidosis is a condition known to have promoted toxicity through the additional accretion of phospholipids in tissues and linked to the drug (Nonoyama and Fukuda 2008). The findings indicate that the compounds evaluated for P-gp were not components of Collinin, Mesuol, Isomesuol, Pteryxin, Rutamarin, Seselin and Suksdorin and did not promote phospholipidosis. The compounds 1m, 1p, alpha-ketoamide and Inophyllum A indicate respectable pharmacokinetic properties in preserving the overhead effects of ADME and toxicity. All compounds evaluated were known as drug-like in nature, passing Lipinski's “Law of 5” with 0 violations except for alpha-ketoamide, passing Lipinski's ”Rule of 5” with 1 MW > 500 violations. In the spectrum of accepted ideals, the predicted restrictions are.
5. Conclusion
COVID-19 existed in the general community at this period, which is a prospective danger to all health worldwide. Nevertheless, there is no scientifically agreed drug to treat the condition. Currently available pharmaceutical drugs for COVID-19 that interact with key proteases. Any therapeutic plant-derived coumarin analogues that could be cast off to fight COVID-19 were inspected in the report. The most prescribed compounds derived from therapeutic plants were Calanolide A, Collinin, Inophyllum A, Mesuol, Isomesuol, Pteryxin, Rutamarin, Seselin and Suksdorfin, which may serve as major inhibitors of the main protease COVID-19 (PDB ID: 5N5O). Molecular docking tests have shown that the natural coumarin analogue Inophyllum A has an exceptional binding energy inhibition potential of −8.4 kcal / mol relative to other compounds. The synthetic coumarin analogues 1m and 1p both demonstrated comparable binding energy inhibition potential of −7.9 kcal / mol. However, in order to analyse the likely applications of the medicinal substance and further research currently ongoing for compounds 1m and 1p, advance investigation is necessary.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Authors are thankful to the Researchers Supporting Project number (RSP-2020/182), King Saud University, Riyadh, Saudi Arabia.
Footnotes
Peer review under responsibility of King Saud University.
References
- Azam F., Madi A.M., Ali H.I. Molecular Docking and Prediction of Pharmacokinetic Properties of Dual Mechanism Drugs that Block MAO-B and Adenosine A2A Receptors for the Treatment of Parkinson’s Disease. J. Young Pharm. 2012;4:184–192. doi: 10.4103/0975-1483.100027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.T., Doong S.L., Tsai I.L., Chen I.S. Coumarins and anti-HBV constituents from Zanthoxylum schinifolium. Phytochem. 1997;45:1419–1422. [Google Scholar]
- Daina A., Zoete V. A BOILED-Egg To Predict Gastrointestinal Absorption and Brain Penetration of Small Molecules. Chem. Med. Chem. 2016;11:1117–1121. doi: 10.1002/cmdc.201600182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmaratne H.R.W., Wanigasekera W.M.A.P., Mata-Greenwood E., Pezzuto J.M. Inhibition of human immunodeficiency virus type 1 reverse transcriptase activity by cordatolides isolated from Calophyllum cordatooblongum. Planta. Med. 1998;64:460–461. doi: 10.1055/s-2006-957483. [DOI] [PubMed] [Google Scholar]
- Ertl P., Rohde B., Selzer P. Fast calculation of molecular polar surface area as a sum of fragment-based contributions and its application to the prediction of drug transport properties. J. Med. Chem. 2000;43:3714–3717. doi: 10.1021/jm000942e. [DOI] [PubMed] [Google Scholar]
- Fromm M.F. P-glycoprotein: A defense mechanism limiting oral bioavailability and CNS accumulation of drugs. Int. J. Clin. Pharmacol. Ther. 2000;38:69–74. doi: 10.5414/cpp38069. [DOI] [PubMed] [Google Scholar]
- Guerriero G., Berni R., Muñoz-Sanchez J.A., Apone F., Abdel-Salam E.M., Qahtan A.A., Alatar A.A., Cantini C., Cai G., Hausman J.F., Siddiqui K.S., Hernández- Sotomayor S.M.T., Faisal M. Production of plant secondary metabolites: Examples, tips and suggestions for biotechnologists. Genes (Basel). 2018;9:34–46. doi: 10.3390/genes9060309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan M.Z., Osman H., Ali M.A., Ahsan M.J. Therapeutic potential of coumarins as antiviral agents. Eur. J. Med. Chem. 2016;123:236–255. doi: 10.1016/j.ejmech.2016.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Kashiwada Y., Cosentino L.M., Fan S., Lee K.H. 3’,4’-Di-o-(-)- camphanoyl- (+)-ciskhellactone and related compounds: a. new class of potent anti- HIV agents. Bioorg. Med. Chem. Lett. 1994;4:593–598. [Google Scholar]
- Jo S., Kim S., Shin D.H., Kim M.S. Inhibition of SARS-CoV 3CL protease by flavonoids. J. Enzyme Inhib. Med. Chem. 2020;35:145–151. doi: 10.1080/14756366.2019.1690480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashman Y., Gustafson K.R., Fuller R.W., Cardellina J.H., McMahon J.B., Currens M.J., Buckheit R.W., Hughes S.H., Cragg G.M., Boyd M.R. The calanolides, a novel HIV-inhibitory class of coumarin derivatives from the tropical rainforest tree, Calophyllum lanigerum. J. Med. Chem. 1992;35:2735–2743. doi: 10.1021/jm00093a004. [DOI] [PubMed] [Google Scholar]
- Kim Y., Lovell S., Tiew K.C., Mandadapu S.R., Alliston K.R., Battaile K.P., Groutas W.C., Chang K.O. Broad-spectrum antivirals against 3C or 3C-like proteases of picornaviruses, noroviruses, and coronaviruses. J. Virol. 2012;86:11754–11762. doi: 10.1128/JVI.01348-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P.I., Hsueh P.R. Emerging threats from zoonotic coronaviruses-from SARS and MERS to 2019-nCoV. J. Microbiol. Immunol. Infect. 2020;1–3 doi: 10.1016/j.jmii.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- Malik Y.S., Sircar S., Bhat S., Sharun K., Dhama K., Dadar M., Tiwari R., Chaicumpa W. Emerging novel Coronavirus (2019-nCoV) - Current scenario, evolutionary perspective based on genome analysis and recent developments. Vet. Q. 2020;40:1–12. doi: 10.1080/01652176.2020.1727993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez N., Sancho R., Bedoya L.M., Alcami J., Lopez-Perez J.L., Feliciano A.S., Fiebich B.L., Munoz E. Mesuol, a natural occurring 4-phenylcoumarin, inhibits HIV-1 replication by targeting the NF-kappa B pathway. Antivir. Res. 2005;66:137–145. doi: 10.1016/j.antiviral.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Newby D., Freitas A.A., Ghafourian T. Decision trees to characterise the roles of permeability and solubility on the prediction of oral absorption. Eur. J. Med. Chem. 2015;90:751–765. doi: 10.1016/j.ejmech.2014.12.006. [DOI] [PubMed] [Google Scholar]
- Nonoyama T., Fukuda R. Drug induced phospholipidosis pathological aspects and its prediction. J. Toxicol. Pathol. 2008;21:9–24. [Google Scholar]
- Patil A.D., Freyer A.J., Eggleston D.S., Haltiwanger R.C., Bean M.F., Taylor P.B., Caranfa M.J., Breen A.L., Bartus H.R., Johnson R.K., Hertzberg R.P., Westley J.W. The inophyllums, novel inhibitors of HIV-1 reverse transcriptase isolated from the malaysian tree, Calophyllum inophyllum Linn. J. Med. Chem. 1993;36:4131–4138. doi: 10.1021/jm00078a001. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Morales A.J., MacGregor K., Kanagarajah S., Patel D., Schlagenhauf P. Going global – Travel and the 2019 novel coronavirus. Travel Med. Infect. Dis. 2019;33:1015178. doi: 10.1016/j.tmaid.2020.101578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiss ADME. Available online: http://www.swissadme.ch (accessed on 03 November (2020).
- Taha M., Ismail N.H., Khan A., Shah S.A.A., Anwar A., Halim S.A., Fatmi M.Q., Imran S., Rahim F., Kha K.M. Synthesis of novel derivatives of oxindole, their urease inhibition and molecular docking studies. Bioorg. Med. Chem. Let. 2015;25:3285–3289. doi: 10.1016/j.bmcl.2015.05.069. [DOI] [PubMed] [Google Scholar]
- Thayil S.M., Thyagarajan S.P. Pa-9: A flavonoid extracted from plectranthus amboinicus inhibits HIV-1 protease. Int. J. Pharmacogn. Phytochem. Res. 2016;8:1020–1024. [Google Scholar]
- Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willette R.E., Soine T.O. Coumarins I: isolation, purification, and structure determination of pteryxin and suksdorfin. J. Pharm. S. C. 1962;51:149–156. doi: 10.1002/jps.2600510215. [DOI] [PubMed] [Google Scholar]
- World Health Organization (WHO), 2020. Novel Coronavirus (2019-nCoV. WHO Bull. January, 1-7.
- Xu, B., Wang, L., Gonz_alez-Molleda, L., Wang, Y., Xu, J., Yuan, Y., 2014. Antiviral activity of (+)-rutamarin against Kaposi's sarcoma-associated herpesvirus by inhibition of the catalytic activity of human topoisomerase II. Antimicrob. Agents. Chemother. 58, 563- 573. [DOI] [PMC free article] [PubMed]
- Yang L., Wen K.S., Ruan X., Zhao Y.X., Wei F., Wang Q. Response of plant secondary metabolites to environmental factors. Molecules. 2018;23(4):1–26. doi: 10.3390/molecules23040762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakaryan H., Arabyan E., Oo A., Zandi K. Flavonoids: promising natural compounds against viral infections. Arch. Virol. 2017;162(9):2539–2551. doi: 10.1007/s00705-017-3417-y. [DOI] [PMC free article] [PubMed] [Google Scholar]