Abstract
Corona viruses (CoV) are known to cause extreme pandemics in the globe. The year 2020 will be a pandemic with the spread of the novel coronavirus (SARS-CoV-2) across the globe. Coronavirus 2019 (COVID-19) has been a part of our scary life for more than a quarter of a year in 2020. The Wuhan market and China have been the most commonly used terms in the world for at least a quarter of 2020. A zoonotic coronavirus has entered organisms to affect organisms for the third season in several centuries. CoV is a global pandemic prompted a drastic and rapid reconfiguration of society. CoV have extraordinary broad genomes of about 30 kilobases of RNA. There is no genetic relationship between the SARS-CoV, MERS and SARS-CoV-2. For health care strategies and for anticipating and preventing potential outbreaks, adequate description of the international spread of COVID-19 virus is imperative. The WHO has declared COVID-19 as endemic to pandemic in the first trimester of 2020. The biggest issues for diagnosis COVID-19 is not established apart from Real-time reverse transcriptase polymerase chain reaction (RT-PCR). In order to monitor the COVID-19 pandemic, testing of active SARS-CoV-2 infections is a fundamental public health method. The vast use of SARS-CoV-2 RT-PCR tests around the world has led to increased availability of test kits, which is also a major bottleneck. The technique RT-PCR was generally agreed in the present scenario to detect SARS-CoV-2 in the human body. This review discusses about the importance of molecular technique for diagnosing the pandemic disease of 2019. In conclusion, RT-PCR was found to be an apt technique for identification of SARS-CoV-2.
Keywords: Coronavirus, COVID-19, SARS-CoV-2, Pandemic and from Real-time reverse transcriptase polymerase chain reaction
1. Introduction
It is known that infectious diseases impact in human development is through a complex interaction between infection and the host. Intrinsically, contagious infections are triggered by a solitary infectious agent. However, even before the present era of molecular biology and heritability training, the predominant indication was that part of the inter-individual variations was attributed to the genetic makeup of the host (Di Maria et al., 2020). We completed the year 2019 with a tangible feeling of hope. Many patients have obtained medication for contagious diseases such as HIV, tuberculosis and malaria that have taken life-saving treatment than ever before. About 18 years after the occurrence of severe acute respiratory syndrome (SARS) in China and eight years after the occurrence of Middle East Respiratory Syndrome (MERS) in Saudi Arabia, a novel Coronavirus (CoV) infection has recently been described by the World Health Organization (WHO) as a pandemic that threatens the global human population. The epidemic is confirmed as coronavirus 2019 (COVID-19) caused by the new human CoV, initially referred to as coronavirus 2019 (2019-nCoV) and subsequently updated as coronavirus SARS 2 (SARS-CoV-2) by the Coronavirus Study Community of the International Committee on Virus Taxonomy (Decaro and Lorusso, 2020) (see Fig. 1).
Fig. 1.
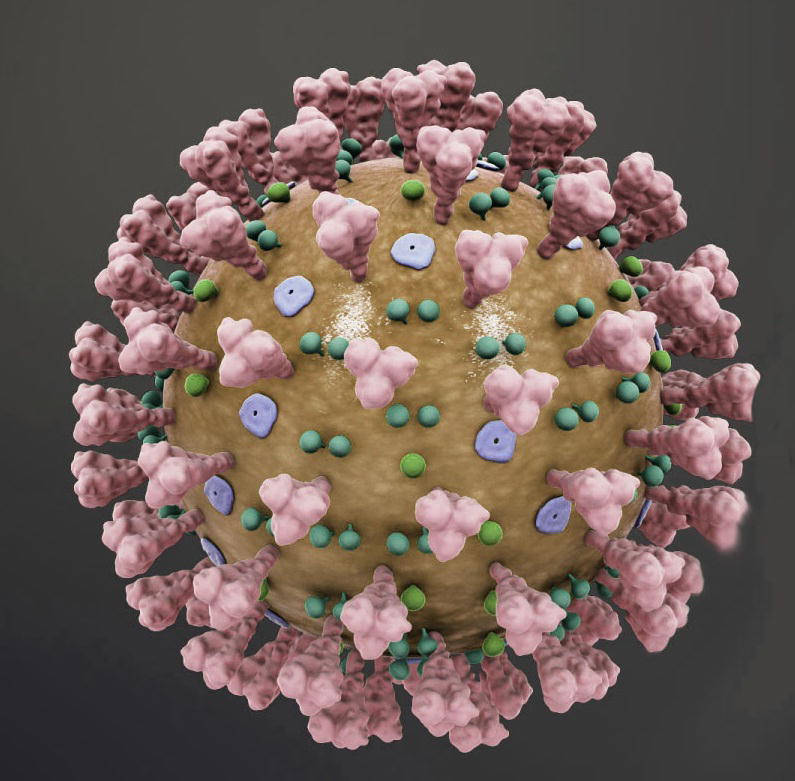
Describes the structure of CoV.
2. SARS-CoV-2
CoV is known as SARS-CoV-2 affected by a significant respiratory disorder called infection with COVID-19 (Lou et al., 2020). However, WHO officially named COVID-19 as SARS-CoV-2 and WHO announced a public health emergency of international importance on 30 January 2020 and a pandemic on 11 March 2020 (Lv et al., 2020, Wang et al., 2019). The outbreak of viral pneumonia originated in the Chinese town of Wuhan in December 2019, identified with a new coronavirus called Wuhan virus or novel coronavirus in 2019 (Zhu et al., 2020). In the closing months of 2019, the virus emerged from an animal source that has not yet been detected and has subsequently spread across the globe (Wijsman et al., 2020). This laboratory-confirmed cases have been identified worldwide and WHO has reported this virus to be COVID-19 (Zhang et al., 2020). The 2019-nCoV or SARS-CoV-2, which has now been identified, has spread rapidly from the origin of Wuhan to the rest of the world from the City of Hubei (Singhal, 2020). The early detection of COVID-19 was mysterious pneumonia, with signs of pneumonia in the first clinical instances. It was subsequently diagnosed as a SARS-CoV-2 infection consistent with severe pneumonia. It was initially known as a novel coronavirus or nCOVID19 (Xu et al., 2020). The COVID-19 clinical characteristics include dry cough, fever, diarrhea, myalgia and vomiting. People with many co-morbidities are vulnerable to severe infections (Sohrabi et al., 2020).
3. Human CoV and its family
COVs appear in a variety of species as well as in humans. Human coronaviruses (HCoVs) comprise HCoV-229E and HCoV-NL63 in the Alphacoronavirus family and HCoV-OC43 and HCoV-HKU1 in the Betacoronavirus lineage family (Embecovirus subgenus). CoVs belong to the Coronavirus Alphacoronavirus (αCoV), betacoronavirus (βCoV), deltacoronavirus (ΔCoV) and gammacoronavirus (γCoV) genus of Coronaviridae, containing four genera, as well as several subgenera and species (Loeffelholz and Tang, 2020). CoVs have been reported as well-known sources of serious respiratory, enteric and systemic infections in humans and numerous animal hosts such as goats, pigs, camels, rodents, horses, cats / dogs / bats / palm cats / ferrets / mink / rabbits / snakes and numerous other wild animals (Malik et al., 2020). SARS-CoVs is an enveloped virus with a diameter ~50–200 nm and a single strand of positive RNA genome varying from 26 to 32 kb (Dhama et al., 2020). SARS-CoV-2 is a β-coronavirus carrying a single positive RNA strand within it. Its envelope, which is about 60–140 nm in diameter, gives it a squared, elliptical morphology. There are special elements in its genome that have been derived from nine Wuhan patients and consisting of a tightly linked 29,903 base pairs of single-stranded RNA (88%) of a couple of β-coronavirus-isolated bats (Lu et al., 2020). The SARS-CoV-2 genome consists of 5′untranslated area (UTR) comprising 5′leader sequence, free read frame 1a / b replicase genes, spike protein, envelope protein, membrane / matrix protein and accessory protein, nucleoprotein and 3′UTR in sequence (Han et al., 2020). Four CoVs, such as HKU1, NL63, 229E and OC43, are mainly responsible for mild respiratory disorders in human circulation (Cao et al., 2020).
4. COVID-19 in China
In the initial report of Huang et al (Huang et al., 2020) confirmed the clinical characteristics of 41 COVID-19 patients, 32% of whom had cardiovascular disorders, diabetes, and chronic pneumonic obstructive diseases, and hypertension (Yang et al., 2020). The mortality rate of COVID-19 is relatively high and moderately contagious; but the literature awareness is increasing rapidly (Harapan et al., 2020). A cluster of affected family and medical personnel reported a person-to-person transmission. Furthermore, the transmission from person to person between close contacts is expected to occur mainly through respiratory droplets that develop after cough and sneezing. Fomite could be a major transmission source, as SARS-CoV has been generated for up to 96 h and CoVs for up to 9 days (Harapan et al., 2020). Fomite could have been a major transmission source. In contrast to Wuhan's counterparts, Xu et al. (2020) find that out of Wuhan patients had milder illness and less serious laboratory anomalies (Lou et al., 2020, Fu et al., 2020).
5. Protein structure of COVID-19
For the alignment and virulence factors of SARS-CoV-2 the current genome defines four structural proteins: “Surface glycoprotein S, E enveline, membrane protéin M, N protein nucleocapsid” and several others that interfere with the immune response. Glycoprotein S is situated on the surface of the envelope and forms athree-dimensional structure within the binding domain of the host cell which promotes virus anchoring. It consists of two subunits: the tropism specifier S1 for the same receiver and the cell and viral membrane fusion phase S2 (Zou et al., 2020). The SARS-CoV-2 genome review demonstrated that 79.5 and 96% of SARS-CoV-RaTG’s total SARS-CoV and bat SARS coronavirus sequence detection rates were respectively (Chen et al., 2020).
6. Genomic RNA and COVID-19
The SARS-CoV-2 / CoV's genetic structure is RNA viruses with a diameter of 80–120 nm on a single strand consisting of a subtype of CoV (α, β, Δ and γ). Before SARS-CoV-2, 6 CoV was believed to cause human disease. This is a β-CoV of SARS-CoV, MERS-CoV and SARS-CoV-2. SARS and SARS-CoV-2 have ~79% of the genome sequence (Wang et al., 2019, Yao et al., 2020). The genomic RNAs can be the strongest RNA molecules in the cell cytoplasm. CoVs therefore evolved a relatively complex multiplication mechanism to facilitate virus reproduction (Chen and Guo, 2016). In their viral life cycle CoVs propagate genomes and subgenomic RNAs only from RNA templates, and do not need a step of DNA. In CoV specifically, the 3′-5′ exonuclease activity of the Nonstructural Protein 14 (NSP14) provides revision and enhanced replication reliability. CoVs use exonuclease NSP14, which is the first known RNA virus-encoded proofreading enzyme, and which, in comparison with other error-prone RNA viruses, could be adapted to handle CoVs' large RNA genomes (Feng et al., 2020).
7. Global wide COVID-19 cases
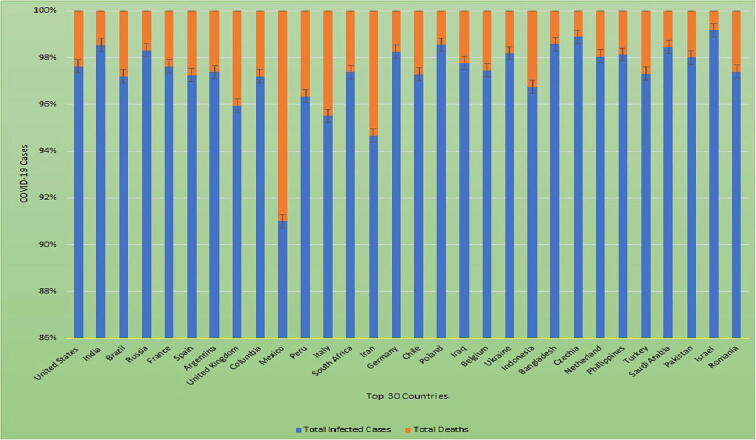
Currently, as of 6th November 2020, there were 49,527,230 cases of COVID-19 worldwide, with a death rate of 2.5% (Di Maria et al., 2020) (https://www.worldometers.info/coronavirus/). Fig. 2 lists the top 30 countries with total and death cases of COVID-19.
Fig. 2.
Top 30 countries infected with COVID-19 deaths.
8. Sequencing analysis
Symptoms may appear 2–14 days after exposure and period incubation ranges from 4 to 7 days, during which any infected patient may be asymptomatic and contagious (Kaye et al., 2020). While several COVID-19 patients are asymptomatic and/or healthy, with neutralization and cell mediated tolerance of the anti-proliferative antibodies and an independent immune action. In hospitalized patients, standard laboratory abnormalités contain almost 83% lymphopenia and abnormal inflammatory markers, such as the rate of erythrocytes, C-reactive protein, ferritin, factor α tumor necrosis, interleukin 1 and 6, and uncommon parameters of coagulation. But ~10% of COVID-19 events are extreme in dyspnoea, lymphopenia and significant chest X-ray anomaly and most of these are severely compromised by respiratory and multi-organ failure. There seems to be a connection between COVID-19′s clinical and immunological character, as its magnitude is related to certain immunological markers (Sohn et al., 2020, Wiersinga et al., 2020). Many diagnostic tests were tried to suit with the reliable COVID-19 diagnostic marker, and none was effective other than the Real Time Chain Reaction Polymerase (RT-PCR). The diagnosis was nevertheless rendered of COVID19 by the RT-PCR and Next Generation sequencing (NGS) studies in patients with contaminated pneumonia (Jiang et al., 2019).
9. COVID-19 and RT-PCR
There are different techniques to detect the COVID-19 in the human body. Real-time reverse-transcriptase polymerase chain reaction (RT-PCR) assay has been widely recommended and used for detection of SARS-CoV-2 (Li et al., 2020, Pan et al., 2020). The purpose of performing RT-PCR for COVID-19 patients is to identify the novel virus precisely. However, RT-PCR result was also taken as an indication of isolation, discharge or transfer by the Chinese government for COVID-19 diagnosed patients. Notably, patient's isolation can be revoked after two consecutive negative RT-PCR tests and patients discharged (China, 2020). An online viral genome sequence (GenBank Accession number MN908947), followed by four other genomes deposited on 12 January 2020 in the Viral Sequence Curated Database by the Global Influenza Data Sharing Initiative, has been published through group online tools virological.org on 10 January 2020 (Wuhan-Hu-1). The genome sequences indicate that a virus is near linked to members of a viral community called the SARS, a species identified by the 2002/03 human SARS disease agent as a serious respiratory syndrome (Corman et al., 2020, de Groot et al., 2012, Peiris et al., 2003).
10. Amplification of RT-PCR analysis in COVID-19 patients
RT-PCR assay is based on automated fluorescent capillary electrophoresis. Infections in the lower respiratory tract account for 50–90% of viruses Reverse transcription-PCR research provides the ability to improve sensitivity and diagnose respiratory infections in more timely fashion. RT-PCR showed rapid results with equal or greater sensitivity to detect respiratory viruses than direct antigen detection or virus isolation. While numerous RT-PCR assays for each respiratory virus have been established, the conditions under which they are extracted, amplified and detected differ widely. A recent trend has been the development of RT-PCR tests which, in separate reactions with standard amplification requirements, simultaneously detect multiple respiratory pathogens with a combination of primer pairs to individual respiratory viruses, with widely reactive primers to highly preserved genes, covering different genes of a virus (Erdman et al., 2003). WHO has recommended the technique RT-PCR to detect the COVI-19 virus in the humans and initially China has implemented this technique. The test was performed by collecting the swab sample either from nose or throat (Saliva, mucous or fluid from a patient’s lungs can also be used). Initially, the swab of an effected or suspected person with COVID-19 will contain a mixture of human cells, virus particles and other microbes. The second step is to extract the viral RNA from the collected sample. RNA and DNA are the genetic material which are unique to an organism. The purpose of using RNA to detect the limited viruses such as COVID-19, HIV and others have RNA as their genetic material. However, chemically RNA is similar to DNA, but has only a single strand. The virus RNA is surrounded by a nucleocapsid protein within the virus envelope. The other proteins are embedded in the envelope itself. The SARS-CoV-2 genome contains genes that carry the directions from making these and other proteins that are required to replicate the virus inside the human cell. In general, N gene which carries directions for making the nucleocapsid protein. Limited viral RNA will be present when we collect the samples and using PCR technique, we will amplify the small quantity of cDNA into the large copies of a segment of the N gene. Short single stranded sequences (1–30 bp) are known as primers recognize unique RNA sequences within the viral genome that bracket the target region of the N gene. After the first primer binds, an enzyme called reverse transcriptase extends (synthesizes) a single-stranded DNA copy of the viral RNA, known as cDNA. After the RNA is removed, the second primer binds to the other side of the single-stranded cDNA. Then, a second enzyme, Taq DNA polymerase, extends a second strand to produce a double-stranded DNA copy of the target region of the viral RNA (Fig. 3).
Fig. 3.
Amplification of RT-PCR by extending the 2nd strand to yields double-stranded DNA of the patient RNA.
This DNA copy then undergo successive rounds (cycles) of amplification during which the DNA denatures into single-strands, both primers bind to their target sequences and Taq polymerase synthesizes a new DNA strand. The number of copies of the target region of the viral genome doubles with each cycle. Up to a billion DNA copies of viral RNA are produced in PCR after 30 cycles. In practice, 30–45 PCR cycles are usually used to identify the virus. The inclusion of a fluorescent sample facilitates a quantification of the target DNA after each PCR loop in real time. (Fig. 4). The extracted RNA is converted into complimentary DNA (cDNA) through the enzyme reverse transcriptase. The third step involves the replicating cDNA into multiple copies through RT-PCR. The patient cDNA will be matching with SARS-CoV-2 using the specific probes to detect the variations in the human sequence either the patient is positive or negative towards the COVID-19.
Fig. 4.
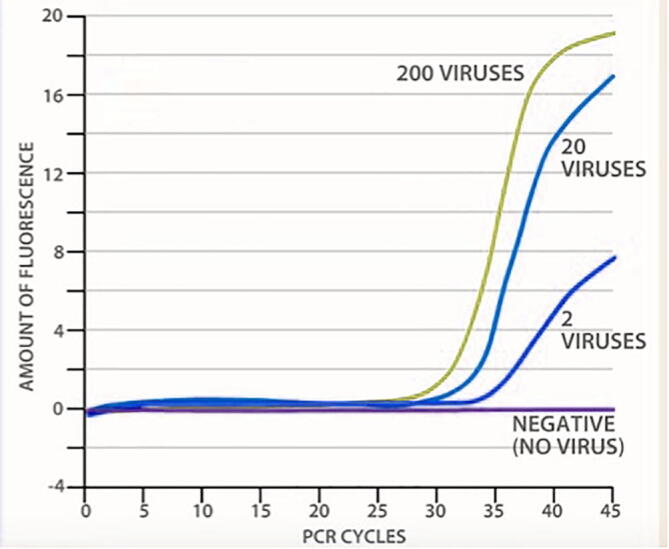
The curves represent the analysis of COV ID-19 test using RT-PCR.
11. Diagnosis of COVID-19 testing through Real Time-Polymerase chain reaction
RT-PCR is a robust in vitro technique that plays a central role in the fields of medical and laboratory sciences. RT-PCR is one of the key techniques used for accurate analysis of COVID-19 detection. This technique has been developed in 1977 mostly during the study of genetic material viral replication which led to the formation of the RT-PCR. In the field of molecular biology, RT-PCR methodology is widely used to detect gene expression using cDNA. A nuclear-derived technique for perceiving the presence of unique genetic material in any pathogen, including a virus such as Ebola, Zika and COVID-19 is through real time RT–PCR. Presently, one of the most commonly used laboratory methods for detecting the COVID-19 virus is real time RT-PCR for accurate analysis. The basic concept of RT-PCR is used isotope markers of radioactive for detecting the targeted genetic material (DNA or RNA) and subsequently refining the replacement of isotopic labelling with special markers, most frequently fluorescent dyes (Dwivedi et al., 2017). The most commonly used approach for direct diagnosis of COVID-19 is nucleic acid amplification using RT-PCR, which can be conducted in either single or double step strategies. The single step technique both RT and DNA polymerase are quicker, whereas, double step strategy encompasses RT and RNA in one tube and ensuing DNA polymerization in a different reaction tube. Based on the specific format of assay, a single RT-PCR apparatus can test maximum of 96 samples in a single PCR plate (Cheng et al., 2020). The possibility of getting false-negative and false-positive results is an essential problem with the RT-PCR test (Wang et al., 2020).
12. Screening of human viruses with PCR and RT-PCR
Numerous methods have been employed for detecting viruses, such as serology, viral culture, PCR and RT-PCR. However, PCR and RT-PCR are currently widely used because of their greater detection ability compared to normal viral culture and serology. Viruses are essential etiological agents and differ from region to region in their prevalence study (Mohan et al., 2010). Viruses are regarded as the leading causes of foodborne and waterborne diseases and several methods for their identification in food and water have been established. The Norwalk-like viruses (NLVs) of gastroenteritis and hepatitis A (HAV) are the two most important foodborne and waterborne viral pathogens. There are no lab hosts for NLVs has yet been identified and cell cultures have found it difficult or impossible to reproduce HAV strains causing outbreaks. Thus, these agents usually were identified using PCR techniques, which followed them by a reverse transcription, since they contain RNA instead of DNA. Special problems when applied to specified examples are found when RT-PCR detection even when combined with antibody capture or immunomagnetic capture (Nuanualsuwan and Cliver, 2002). The PCR method is known as the “gold standard” for detecting certain viruses and is characterized as quick, high sensitivity and specialty testing among nucleic acid tests. In view of its benefit as a basic qualitative test, RT-PCR is nowadays especially involved in the identification of SARS-CoV-2 (Shen et al., 2020, Wan et al., 2016, Noh et al., 2017). In addition, RT-PCR is susceptible to detection at an early stage of infection. The RT-PCR measure, which has been cited by criteria, may therefore be treated as a principal tool for the identification of COVID-19′s trigger agent, SARS-CoV-2 (Tahamtan and Ardebili, 2020). Payungporn et al. (2004) studies confirmed RT-PCR is one of the best techniques for identification of virus with a single step multiplex RT-PCR for simultaneous detection of 3 particular target genes. Respiratory viral identification can be enhanced with the use of molecular biology approaches. Numerous researches on PCR or RT-PCR methods for detection and typing of respiratory viruses have been developed and assessed (Bellau-Pujol et al., 2005). Reliable, simple and economical procedures are necessary for routine diagnosis, so the possibility of PCR techniques is given. Multiplex experiments could further minimize costs and the number of samples. An internal monitoring would be ideal to ensure reliability and make it easier to perceive negative outcomes. There have been numerous publications of various internal plant management primers. But none differ between RNA and DNA templates so complete removal of DNA is necessary before RT-PCR is performed. One alternative is to design primers that amplify various fragments because of the presence of introns in the target gene, but each additional templates raises the risk that the viral templates are out compressed on the RT-PCR multiplex, and that other fragments on the gel are complicated to recognize (Menzel et al., 2002).
Despite there are enormous strengths and limitations in diagnostic tests, RT-PCR was confirmed for detecting the novel SARS-CoV-2 in the human swabs. In conclusion, RT-PCR was used to validate the COVID-19 tests in humans as a diagnostic tool. The exact diagnostic test for COVID-19 could not be verified by other serological tests, CT-Scan and X-ray.
Footnotes
Peer review under responsibility of King Saud University.
References
- Bellau-Pujol S., Vabret A., Legrand L., Dina J., Gouarin S., Petitjean-Lecherbonnier J. Development of three multiplex RT-PCR assays for the detection of 12 respiratory RNA viruses. J. Virol. Methods. 2005;126(1–2):53–63. doi: 10.1016/j.jviromet.2005.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Li L., Feng Z., Wan S., Huang P., Sun X. Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discovery. 2020;6(1):1–4. doi: 10.1038/s41421-020-0147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Guo D. Molecular mechanisms of coronavirus RNA capping and methylation. Virologica Sinica. 2016;31(1):3–11. doi: 10.1007/s12250-016-3726-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. The Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M.P., Papenburg J., Desjardins M., Kanjilal S., Quach C., Libman M. Diagnostic testing for severe acute respiratory syndrome–related coronavirus-2: a narrative review. Ann. Intern. Med. 2020 doi: 10.7326/M20-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- China N. National Health Commission of the People's Republic of China; Beijing: 2020. Diagnosis and treatment protocols of pneumonia caused by a novel coronavirus (trial version 5) [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25(3):2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R.J., Baker S., Baric R., Enjuanes L., Gorbalenya A., Holmes K. Family coronaviridae. Virus Taxon. 2012:806–828. [Google Scholar]
- Decaro N., Lorusso A. Novel human coronavirus (SARS-CoV-2): a lesson from animal coronaviruses. Vet. Microbiol. 2020;108693 doi: 10.1016/j.vetmic.2020.108693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhama K., Patel S.K., Sharun K., Pathak M., Tiwari R., Yatoo M.I. SARS-CoV-2 jumping the species barrier: zoonotic lessons from SARS, MERS and recent advances to combat this pandemic virus. Travel Med. Infect. Dis. 2020;101830 doi: 10.1016/j.tmaid.2020.101830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Maria E., Latini A., Borgiani P., Novelli G. Genetic variants of the human host influencing the coronavirus-associated phenotypes (SARS, MERS and COVID-19): rapid systematic review and field synopsis. Human Genom. 2020;14(1):1–19. doi: 10.1186/s40246-020-00280-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi S., Purohit P., Misra R., Pareek P., Goel A., Khattri S. Diseases and molecular diagnostics: a step closer to precision medicine. Indian J. Clin. Biochem. 2017;32(4):374–398. doi: 10.1007/s12291-017-0688-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdman D.D., Weinberg G.A., Edwards K.M., Walker F.J., Anderson B.C., Winter J. GeneScan reverse transcription-PCR assay for detection of six common respiratory viruses in young children hospitalized with acute respiratory illness. J. Clin. Microbiol. 2003;41(9):4298–4303. doi: 10.1128/JCM.41.9.4298-4303.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W., Newbigging A.M., Le C., Pang B., Peng H., Cao Y. Molecular diagnosis of COVID-19: challenges and research needs. Anal. Chem. 2020;92(15):10196–10209. doi: 10.1021/acs.analchem.0c02060. [DOI] [PubMed] [Google Scholar]
- Fu L., Wang B., Yuan T., Chen X., Ao Y., Fitzpatrick T. Clinical characteristics of coronavirus disease 2019 (COVID-19) in China: a systematic review and meta-analysis. J. Infect. 2020 doi: 10.1016/j.jinf.2020.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Q., Lin Q., Jin S., You L. Coronavirus 2019-nCoV: a brief perspective from the front line. J. Infect. 2020;80(4):373–377. doi: 10.1016/j.jinf.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harapan H., Itoh N., Yufika A., Winardi W., Keam S., Te H. Coronavirus disease 2019 (COVID-19): a literature review. J. Infect. Public Health. 2020 doi: 10.1016/j.jiph.2020.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F., Deng L., Zhang L., Cai Y., Cheung C.W., Xia Z. Review of the clinical characteristics of coronavirus disease 2019 (COVID-19) J. Gen. Intern. Med. 2020:1–5. doi: 10.1007/s11606-020-05762-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye K., Paprottka F., Escudero R., Casabona G., Montes J., Fakin R. Elective, non-urgent procedures and aesthetic surgery in the wake of SARS–COVID-19: considerations regarding safety, feasibility and impact on clinical management. Aesthetic Plast. Surg. 2020;1 doi: 10.1007/s00266-020-01752-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Yao L., Li J., Chen L., Song Y., Cai Z. Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19. J. Med. Virol. 2020 doi: 10.1002/jmv.25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeffelholz M.J., Tang Y.-W. Laboratory diagnosis of emerging human coronavirus infections–the state of the art. Emerging Microbes Infect. 2020;9(1):747–756. doi: 10.1080/22221751.2020.1745095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou J., Tian S.-J., Niu S.-M., Kang X.-Q., Lian H.-X., Zhang L.-X. Coronavirus disease 2019: a bibliometric analysis and review. Eur. Rev. Med. Pharmacol. Sci. 2020;24(6):3411–3421. doi: 10.26355/eurrev_202003_20712. [DOI] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. The Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv M., Luo X., Estill J., Liu Y., Ren M., Wang J. Coronavirus disease (COVID-19): a scoping review. Eurosurveillance. 2020;25(15):2000125. doi: 10.2807/1560-7917.ES.2020.25.15.2000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik Y.S., Sircar S., Bhat S., Sharun K., Dhama K., Dadar M. Emerging novel coronavirus (2019-nCoV)—current scenario, evolutionary perspective based on genome analysis and recent developments. Vet. Quart. 2020;40(1):68–76. doi: 10.1080/01652176.2020.1727993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel W., Jelkmann W., Maiss E. Detection of four apple viruses by multiplex RT-PCR assays with coamplification of plant mRNA as internal control. J. Virol. Methods. 2002;99(1–2):81–92. doi: 10.1016/s0166-0934(01)00381-0. [DOI] [PubMed] [Google Scholar]
- Mohan A., Chandra S., Agarwal D., Guleria R., Broor S., Gaur B. Prevalence of viral infection detected by PCR and RT-PCR in patients with acute exacerbation of COPD: a systematic review. Respirology. 2010;15(3):536–542. doi: 10.1111/j.1440-1843.2010.01722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh J.Y., Yoon S.-W., Kim D.-J., Lee M.-S., Kim J.-H., Na W. Simultaneous detection of severe acute respiratory syndrome, Middle East respiratory syndrome, and related bat coronaviruses by real-time reverse transcription PCR. Arch. Virol. 2017;162(6):1617–1623. doi: 10.1007/s00705-017-3281-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuanualsuwan S., Cliver D.O. Pretreatment to avoid positive RT-PCR results with inactivated viruses. J. Virol. Methods. 2002;104(2):217–225. doi: 10.1016/s0166-0934(02)00089-7. [DOI] [PubMed] [Google Scholar]
- Pan Y., Zhang D., Yang P., Poon L.L., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet. Infect. Dis. 2020;20(4):411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payungporn S., Phakdeewirot P., Chutinimitkul S., Theamboonlers A., Keawcharoen J., Oraveerakul K. Single-step multiplex reverse transcription–polymerase chain reaction (RT-PCR) for influenza A virus subtype H5N1 detection. Viral Immunol. 2004;17(4):588–593. doi: 10.1089/vim.2004.17.588. [DOI] [PubMed] [Google Scholar]
- Peiris J.S., Yuen K.Y., Osterhaus A.D., Stöhr K. The severe acute respiratory syndrome. N. Engl. J. Med. 2003;349(25):2431–2441. doi: 10.1056/NEJMra032498. [DOI] [PubMed] [Google Scholar]
- Shen M., Zhou Y., Ye J., Al-Maskri A.A.A., Kang Y., Zeng S. Recent advances and perspectives of nucleic acid detection for coronavirus. J. Pharm. Anal. 2020 doi: 10.1016/j.jpha.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal T. A review of coronavirus disease-2019 (COVID-19) Indian J. Pediatr. 2020;1–6 doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn K.M., Lee S.-G., Kim H.J., Cheon S., Jeong H., Lee J. COVID-19 patients upregulate toll-like receptor 4-mediated inflammatory signaling that mimics bacterial sepsis. bioRxiv. 2020 doi: 10.3346/jkms.2020.35.e343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrabi C., Alsafi Z., O’Neill N., Khan M., Kerwan A., Al-Jabir A. World Health Organization declares global emergency: a review of the 2019 novel coronavirus (COVID-19) Int. J. Surg. 2020 doi: 10.1016/j.ijsu.2020.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahamtan A., Ardebili A. Taylor & Francis; 2020. Real-Time RT-PCR in COVID-19 Detection: Issues Affecting the Results. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Z., Zhang Y., He Z., Liu J., Lan K., Hu Y. A melting curve-based multiplex RT-qPCR assay for simultaneous detection of four human coronaviruses. Int. J. Mol. Sci. 2016;17(11):1880. doi: 10.3390/ijms17111880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Kang H., Liu X., Tong Z. Combination of RT-qPCR testing and clinical features for diagnosis of COVID-19 facilitates management of SARS-CoV-2 outbreak. J. Med. Virol. 2020;92(6):538–539. doi: 10.1002/jmv.25721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wang Y., Ye D. Review of the 2019 novel coronavirus (SARS-CoV-2) based on current. Int. J. Antimicrob. Agents. 2019 doi: 10.1016/j.ijantimicag.2020.105948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiersinga W.J., Rhodes A., Cheng A.C., Peacock S.J., Prescott H.C. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;324(8):782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- Wijsman L., Molenkamp R., Reusken C., Meijer A. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. J. Clin. Virol.: Off. Publ. Pan Am. Soc. Clin. Virol. 2020;128 doi: 10.1016/j.jcv.2020.104412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y.-H., Dong J.-H., An W.-M., Lv X.-Y., Yin X.-P., Zhang J.-Z. Clinical and computed tomographic imaging features of novel coronavirus pneumonia caused by SARS-CoV-2. J. Infect. 2020 doi: 10.1016/j.jinf.2020.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X.-W., Wu X.-X., Jiang X.-G., Xu K.-J., Ying L.-J., Ma C.-L. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS-Cov-2) outside of Wuhan, China: retrospective case series. BMJ. 2020;368 doi: 10.1136/bmj.m606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Zheng Y., Gou X., Pu K., Chen Z., Guo Q. Prevalence of comorbidities and its effects in patients infected with SARS-CoV-2: a systematic review and meta-analysis. Int. J. Infect. Dis. 2020;94:91–95. doi: 10.1016/j.ijid.2020.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T.T., Qian J.D., Zhu W.Y., Wang Y., Wang G.Q. A systematic review of lopinavir therapy for SARS coronavirus and MERS coronavirus—A possible reference for coronavirus disease-19 treatment option. J. Med. Virol. 2020;92(6):556–563. doi: 10.1002/jmv.25729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Zhang L., Zha D., Hu C., Wu X. Clinical characteristics and risks of Chinàs 2019 novel coronavirus patients with AKI: a systematic review and meta-analysis. Ren. Fail. 2020;42(1):926–931. doi: 10.1080/0886022X.2020.1812401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X., Chen K., Zou J., Han P., Hao J., Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020,:1–8. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]