Abstract
Background:
In a recent trial of milk oral immunotherapy (MOIT) with or without omalizumab in 55 patients with milk allergy treated for 28 months, 44 of 55 subjects passed a 10-g desensitization milk protein challenge; 23 of 55 subjects passed the 10-g sustained unresponsiveness (SU) challenge 8 weeks after discontinuing MOIT.
Objective:
We sought to determine whether IgE and IgG4 antibody binding to allergenic milk protein epitopes changes with MOIT and whether this could predict the development of SU.
Methods:
By using a novel high-throughput Luminex-based assay to quantitate IgE and IgG4 antibody binding to 66 sequential epitopes on 5 milk proteins, serum samples from 47 subjects were evaluated before and after MOIT. Machine learning strategies were used to predict whether a subject would have SU after 8 weeks of MOIT discontinuation.
Results:
MOIT profoundly altered IgE and IgG4 binding to epitopes, regardless of treatment outcome. At the initiation of MOIT, subjects achieving SU exhibited significantly less antibody binding to 40 allergenic epitopes than subjects who were desensitized only (false discovery rate ≤ 0.05 and fold change > 1.5). Based on baseline epitope-specific antibody binding, we developed predictive models of SU. Using simulations, we show that, on average, IgE-binding epitopes alone perform significantly better than models using standard serum component proteins (average area under the curve, >97% vs 80%). The optimum model using 6 IgE-binding epitopes achieved a 95% area under the curve and 87% accuracy.
Conclusion:
Despite the relatively small sample size, we have shown that by measuring the epitope repertoire, we can build reliable models to predict the probability of SU after MOIT. Baseline epitope profiles appear more predictive of MOIT response than those based on serum component proteins.
Keywords: Cow’s milk allergy, oral immunotherapy, omalizumab, desensitization, sustained unresponsiveness, allergenic epitopes, epitope-specific antibodies, machine learning, elastic net algorithm, bootstrap aggregating strategy
Graphical Abstract
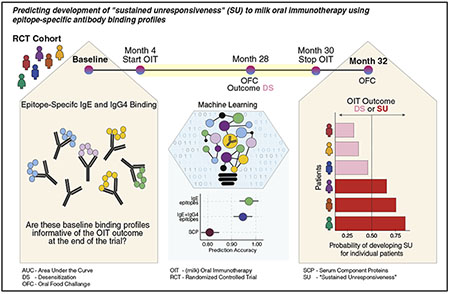
IgE-mediated cow’s milk allergy typically presents in the first year of life and is one of the most common food allergies worldwide, affecting 2% to 3% of infants in developed countries.1,2 Fortunately, the majority of children outgrow their milk allergy in the first decade of life, and only 10% to 15% remain allergic in their late teenage years.3,4 However, those who retain their allergy often experience severe anaphylactic reactions after accidental consumption of milk. In surveys of severe and fatal food-induced anaphylaxis, milk ranks second to peanut in provoking fatal anaphylactic reactions in the United States and is the leading cause in the United Kingdom and Israel.5,6
In the past decade, there have been a number of studies evaluating the efficacy of oral immunotherapy (OIT) for the treatment of persistent food allergies. However, milk oral immunotherapy (MOIT) has been associated with significant adverse reactions, with some experiencing anaphylaxis, and 15% to 20% of patients were forced to discontinue therapy because of the adverse reactions.7–11 In addition to the high rate of adverse reactions, the response to MOIT is typically not sustained once therapy is discontinued (ie, patients are temporarily desensitized but do not achieve long-term tolerance).10,12–16
In an attempt to reduce adverse reactions and possibly enhance the sustainability of MOIT, we previously undertook a placebo-controlled trial of omalizumab.17 Omalizumab has been shown to reduce adverse effects in small pilot studies of milk and peanut OIT.18–21 The trial randomized 57 subjects to either omalizumab or placebo in combination with MOIT, with a primary end point of sustained unresponsiveness (SU) defined as persistence of desensitization for 8 weeks after discontinuation of treatment. Overall, 55 of the 57 patients with milk allergy were treated for 28 months; 44 (80%) subjects (24 in the omalizumab group and 20 in the placebo group) tolerated 10 g of milk protein during an oral food challenge (OFC) at the end of therapy, but only 23 (42%) subjects (13 in the omalizumab group and 10 in the placebo group) tolerated 10 g of milk protein 8 weeks after discontinuing MOIT (ie, SU). In this study we sought to determine whether IgE and IgG4 antibody binding to allergenic epitopes changed after MOIT and whether baseline profiles could be used as predictive biomarkers to identify patients who would achieve SU 8 weeks after discontinuing MOIT.
METHODS
Study design
The MOIT plus omalizumab trial design has been described previously.17 In brief, 57 subjects (age, 7-35 years) with IgE-mediated cow’s milk allergy were randomized 1:1 to receive omalizumab (n = 28) or placebo (n = 29) in a double-blind, placebo-controlled trial to evaluate whether the addition of omalizumab to MOIT would reduce treatment-related adverse reactions and increase the frequency of SU.1 MOIT dosing was administered for 12 months, after which treatment assignment was unblinded (at month 16). Omalizumab was continued for an additional 12 months in the active group, and injections were discontinued in placebo-treated participants (see Fig E1 in this article’s Online Repository at www.jacionline.org). At month 28, omalizumab was discontinued, and all subjects underwent a 10-g OFC. Subjects passing this challenge received 8 additional weeks of MOIT and then discontinued treatment for 8 weeks, followed by a final 10-g OFC. SU was defined as the persistence of desensitization for 8 weeks after discontinuation of MOIT.
Library of milk peptides
A meta-analysis of data from previously published peptide microarray studies22–26 was performed and identified 66 informative and clinically relevant peptides belonging to αS1-casein (n = 18), αS2-casein (n = 13), β-casein (n = 14), β-lactoglobulin (n = 8), and κ-casein (n = 13). Because the peptides have been preselected to represent milk proteins, the terms peptide and epitope are used interchangeably.
Luminex-based peptide assay
Milk peptides (CS Bio, Menlo Park, Calif) were coupled to LumAvidin beads (Luminex, Austin, Tex) and stored in 1× PBS plus 0.02% Tween 20 plus 0.1% BSA (PBS-TBN) buffer. A master mix of peptide-coupled beads was made in PBS-TBN buffer, and 100 μL of the master mix was added to filter plates. After washing the beads, 100 μL of the subject’s plasma at 1:50 dilution was added to the wells in triplicates. The plates were incubated on a shaker for 2 hours at 300 rpm at room temperature. Excess plasma was then removed, and the plates were washed. Mouse anti-human IgE-phycoerythrin (50 μL/well; Clone BE5, lot SA2333415, diluted 1:50 in PBS-TBN; Thermo Fisher Scientific, Rockford, Ill) or mouse anti-human IgG4 Fc-phycoerythrin (clone HP6025, lot B3317-PN67, diluted 1:400 in PBS-TBN; SouthernBiotech, Birmingham, Ala) secondary antibody was added, and plates were incubated for 30 minutes. After a final wash, 100 μL of PBS-TBN was added to each well to resuspend the beads, which were then transferred to fixed-bottom 96-well reading plates and quantified on the Luminex 200 instrument (Luminex 100/200 System; Luminex).
Preprocessing of Luminex signal
All samples were processed in triplicates. PBS-TBN buffer was also processed in triplicates in each plate to eliminate background intensity. The median fluorescence intensity (MFI) for each epitope and sample was obtained directly from Luminex xPONENT software. For each sample i and epitope j, the binding measurements Bij was defined as follows:
where ns represents the nonspecific binding (PBS-TBN) samples.
This quantitative outcome denotes epitope-specific antibody binding (ESAB) and was found to have essentially a normal distribution. The plate effect was evaluated by using principal variance component analysis,27 indicating the presence of a plate effect responsible for 25% and 13% of the variance for IgE and IgG4, respectively. This effect was corrected by using the ComBat algorithm (sva R package).28,29 Agreement among replicates was evaluated by using the intraclass correlation coefficient (ICC).30,31 High reproducibility among replicates was observed (ICC > 0.8 for 87% of IgE epitopes and ICC > 0.9 for all but 1 IgG4 epitope, see Fig E2 in this article’s Online Repository at www.jacionline.org), and triplicates were then averaged for further analysis.
Epitope diversity was estimated as the proportion of epitopes present in each sample. An epitope was considered present if its median fluorescence intensity was greater than the upper 95% CI limit of the nonspecific binding samples.
Statistical analysis
Descriptive analyses were presented for demographics, clinical characteristics, and serum component proteins (SCPs) in the cohort by using summary statistics. SCPs were analyzed on a log10 scale; for the ratio analysis, IgE (in kilounits of antigen per liter) and IgG4 (in milligrams per milliliter) values were converted to nanograms per milliliter as follows:
Changes induced by MOIT on the ESAB were assessed by using linear mixed-effect models, with time, treatment, and their interaction as fixed-effect factors and a random intercept for each patient. A model including desensitized and SU outcomes as a fixed effect (and interactions) was also considered to evaluate changes in ESAB by using the MOIT outcome. Model estimation used an empiric Bayes approach that allows estimation of the variance parameters acquiring information across all peptides and is widely used in high-throughput analyses.32,33 Hypotheses of interest were tested by using moderated t tests (or F-tests), and P values were adjusted for multiple hypotheses by using the Benjamini-Hochberg approach, controlling the false discovery rate (FDR). Differential ESAB was defined as an FDR of 0.05 or less and fold change of greater than 1.5. Linear mixed-effect models were also used for analysis of SCPs, epitope diversity, and ESAB z scores.
Machine learning approach
To identify the baseline epitope signature that would predict SU with MOIT, we used elastic net, a machine learning algorithm suitable for classification problems with more features (epitopes) than instances (patients). Elastic net is a regularized logistic regression method that linearly combines the L1 and L2 penalties (for the selection of covariates) of the lasso and ridge methods.34 The regularization parameter is estimated through k-fold cross-validation, maximizing the accuracy statistics. To compensate for the small sample size available for prediction modeling, we conducted a bootstrapping simulation experiment to obtain CIs for performance statistics. This allowed us to better evaluate the potential of epitope profiling as a base for building predictive models and to compare models built with different sets of variables without relying on a point estimate that might be susceptible to outliers. Additionally, we coupled the elastic net with a bootstrapping aggregate procedure to obtain more robust epitope signatures. In each simulation (n = 300) a balanced random sample with replacement is selected from the original cohort and is used for model estimation, which includes selection of the predictive epitopes and estimation of the predictive model. The bagging frequency (BF) is then computed as the number of times that an epitope is identified in the predictive signature. Robust epitopes are then defined as those that have a BF of greater than 60% to 80%, and those are used for the final model (see Fig E3 in this article’s Online Repository at www.jacionline.org). Model performance was evaluated by using performance statistics, including accuracy, specificity, sensitivity, and area under the curve (AUC).
RESULTS
Plasma samples were available at baseline and after 2 years of MOIT in 47 of 55 subjects. Forty-four (94%) of these 47 subjects completing 2 years of MOIT passed the 10-g milk protein OFC and were considered desensitized. After 8 weeks off MOIT, only 50% (23/44) of the desensitized patients achieved SU at the final 10-g OFC, with rates of SU being similar between the 2 treatment arms (see Fig E1).17
MOIT induces profound changes in ESAB profiles
MOIT induced profound changes in ESAB profiles regardless of the adjuvant treatment, with significant changes in 48 (73%) IgE-specific and 60 (91%) IgG4-specific peptides after MOIT (Fig 1, A, and see Tables E1 and E2 in this article’s Online Repository at www.jacionline.org). There was an overall decrease in IgE ESAB (especially for αS1-casein epitopes) and IgG4 ESAB (Fig 1, A and B). Interestingly, subjects treated with omalizumab experienced a less pronounced reduction in epitope-specific IgE binding than those treated with placebo, whereas for IgG4 peptides, increased binding was seen in subjects receiving omalizumab compared with those receiving placebo (Fig 1, B). Overall, the decreases observed in IgE binding after 30 months of MOIT are attenuated when omalizumab is used in combination with MOIT, with 19 peptides exhibiting significant differences in MOIT-induced changes compared with placebo (FDR ≤ 0.05 and fold change > 1.5). In contrast, the increases in IgG4 binding to allergenic epitopes are accentuated by the use of omalizumab, with 34 peptides showing greater changes than in the placebo group (see Tables E1 and E3 in this article’s Online Repository at www.jacionline.org). This IgE attenuation with IgG4 accentuation effect of omalizumab was also mirrored in the SCPs of milk- and casein-specific IgE and casein-specific IgG4 (Fig 1, C, and see Fig E4, A, in this article’s Online Repository at www.jacionline.org).
FIG 1.
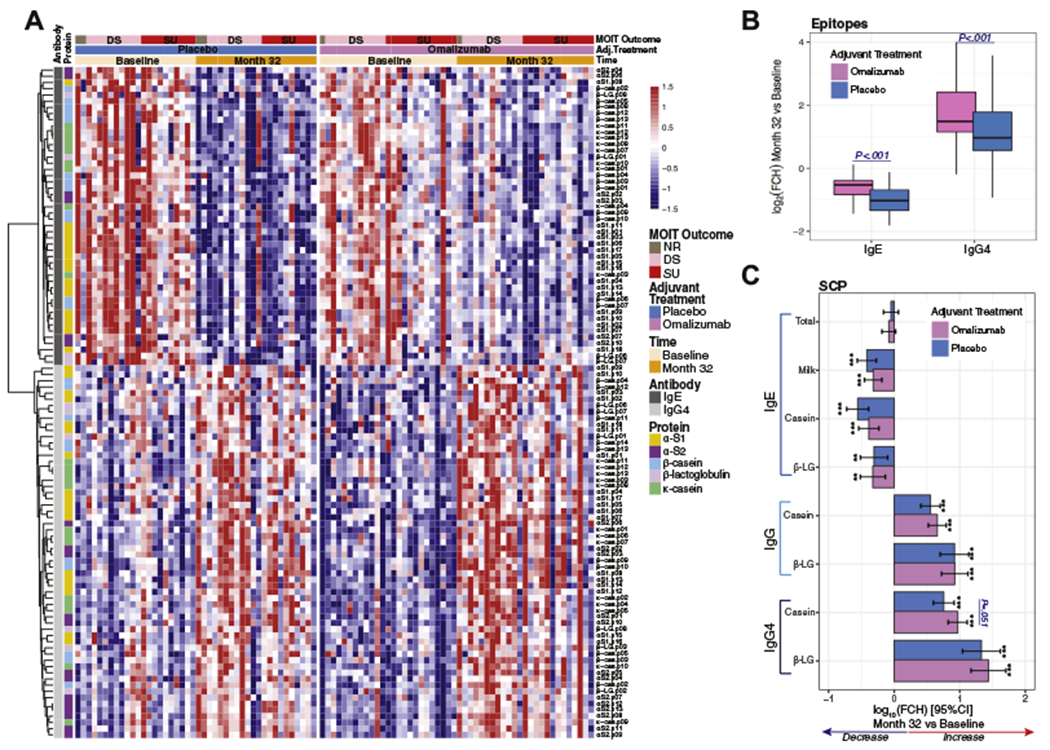
Changes induced by MOIT in epitope-specific IgE or IgG4 binding by adjuvant treatment arm. A, Heat map representing (standardized) binding of 108 epitopes (48 IgE and 60 IgG4), with significant changes (fold change [FCH] > 1.5 and FDR ≤ 0.05) at month 32 from baseline. B, Box plot of the MOIT-induced log2 FCHs in epitope binding by adjuvant treatment. C, Bar plots of MOIT-induced changes and 95% CIs in log10 levels of SCPs. Asterisks represent a significant change at month 32 from baseline: *P < .05, **P < .01, and ***P < .001. DS, Desensitization; β-LG, β-lactoglobulin.
Overall, changes in ESAB and diversity after MOIT were very similar among desensitized subjects and those who achieved SU after 8 weeks off MOIT (Fig 2 and see Figs E5 and E6, A and B, in this article’s Online Repository at www.jacionline.org). IgE epitope diversity (defined as the percentage of epitopes present) was significantly reduced with MOIT, whereas more than 90% of the IgG4 epitopes were present at baseline and after MOIT (see Fig E6, A and B). Only a few peptides had greater MOIT-induced changes in binding in desensitized patients than in those achieving SU (Fig 2 and see Table E3). Lastly, no significant association between MOIT-induced changes and outcomes were found in milk SCP IgE and IgG4 levels (see Fig E4, B).
FIG 2.
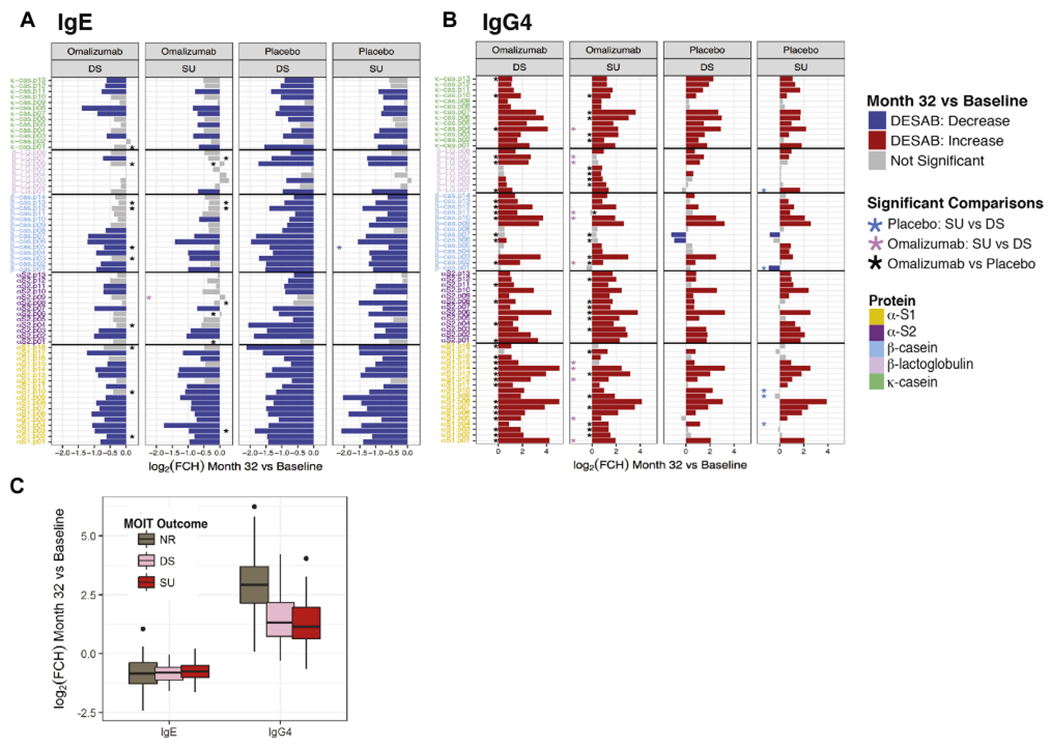
Changes in ESAB by adjuvant treatment and MOIT outcome. A and B, Bar plots representing changes induced by MOIT in patients achieving only desensitization (DS) or SU by adjuvant treatment arm for each IgE and IgG4 epitope. The height of the bars represents the log2 fold change (FCH) in binding from baseline to month 32. Colors represent the direction of change (red, increase; blue, decrease) for the differential epitope-specific antibody binding (DESAB; FCH > 1.5 and FDR < 0.05). Colored asterisks indicate significance in changes between SU and DS outcomes for each adjuvant treatment (light blue, placebo; light pink, omalizumab); black asterisks represent DESAB, with significant differences between omalizumab and placebo for each outcome. C, Box plot of MOIT-induced changes in ESAB by MOIT outcome. NR, Nonresponder.
SU to MOIT is associated with lower levels of baseline epitope-specific IgE
Although no substantial association between MOIT-induced changes in ESAB and development of SU were observed, there were striking differences in baseline levels of epitope-specific IgE binding (Fig 3, B, and see Fig E7 in this article’s Online Repository at www.jacionline.org). At a global level, lower levels of IgE binding to peptides were seen in subjects who achieved SU (Fig 3, A, and see Fig E6, C), with significant differences in IgE binding to 36 peptides (FDR ≤ 0.05 and fold change > 1.5; Fig 3, B) and only one IgG4 peptide (α-casein.p08; see Fig E7). Subjects achieving SU also exhibited less baseline epitope-specific IgE diversity than the desensitized group (76% vs 92%, P < .01; see Fig E6, A and B). No outcome differences were observed at baseline in demographic variables (Table I),17,35 but subjects achieving SU exhibited significantly lower levels of IgE specific to milk, casein, and β-lactoglobulin before MOIT (Fig 3, C, and see Fig E4, C).
FIG 3.
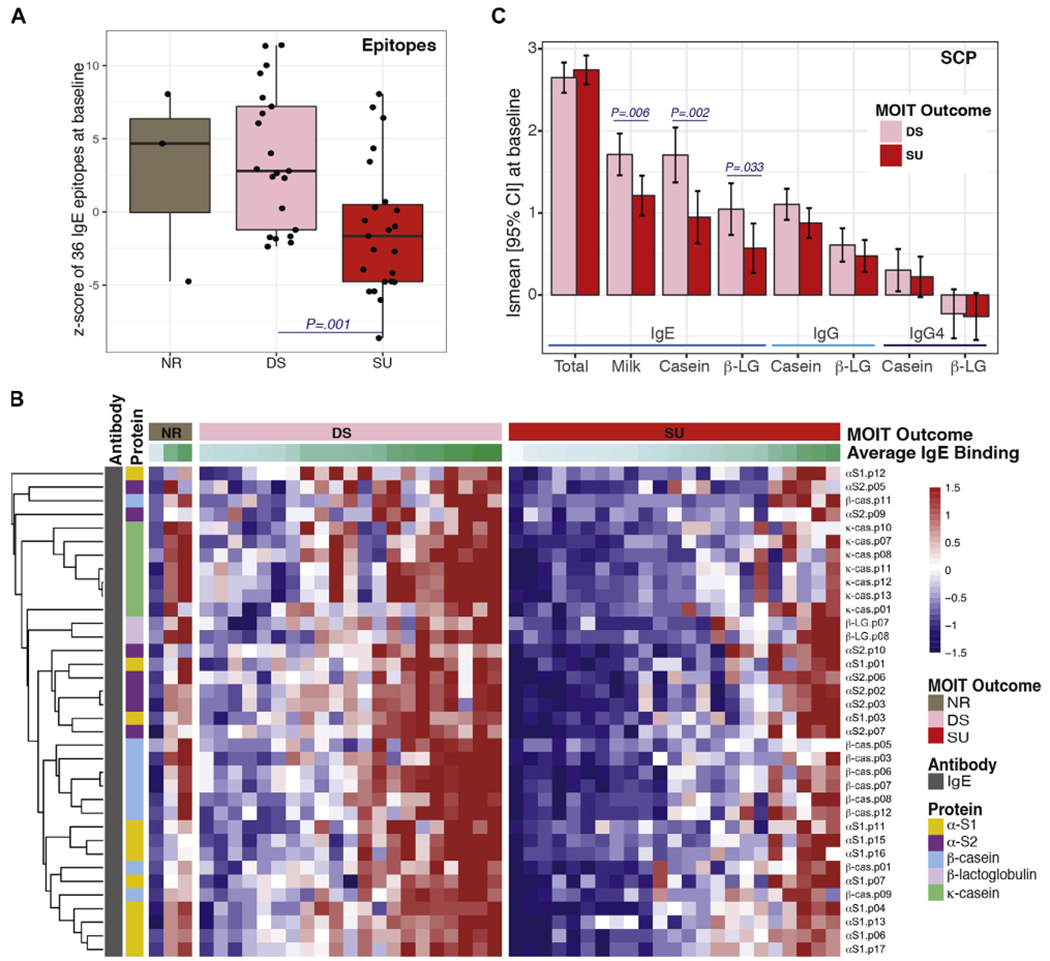
Differences in baseline epitope and SCP levels between groups achieving desensitization (DS) or SU 8 weeks after MOIT discontinuation. A, Box plots of the baseline overall (z score) epitope-specific IgE binding by MOIT outcome, with significantly different z scores between the SU and DS groups. NR, Nonresponders. B, Heat map representing baseline epitope-specific IgE binding for each patient for the set of epitopes with significant differences (fold change [FCH] > 1.5 and FDR < 0.05) between DS and SU outcomes at baseline. β-LG, β-Lactoglobulin. C, Bar plots of the least square mean (Ismean) with the 95% CI of log10 SCP levels at baseline by MOIT outcome.
TABLE I.
Baseline characteristics of the mechanistic cohort by adjuvant treatment arm and MOIT outcome at 32 months
| Adjuvant treatment | MOIT outcome* | ||||
|---|---|---|---|---|---|
| Placebo (n = 22) | Omalizumab (n = 25) | DS (n = 21) | SU (n = 23) | P value | |
| Demographics | |||||
| Age | 11.6 (6.2) | 12.5 (4.3) | 12.3 (4.5) | 12.2 (6.0) | .97 |
| Female sex | 6 (27.3%) | 7 (28.0%) | 6 (28.6%) | 6 (26.1%) | .99 |
| Hispanic or Latino | 2 (9.1%) | 0 (0.0%) | 2 (9.5%) | 0 (0.0%) | .22 |
| Height | 139.2 (21.1) | 145.4 (17.7) | 144.5 (21.6) | 142.2 (17.8) | .70 |
| Weight | 35.4 (16.9) | 37.7 (14.0) | 37.9 (17.2) | 36.3 (14.3) | .74 |
| Medical history | |||||
| Asthma | 17 (77.3%) | 18 (72.0%) | 17 (81.0%) | 15 (65.2%) | .32 |
| Rhinitis | 18 (81.8%) | 18 (72.0%) | 17 (81.0%) | 16 (69.6%) | .50 |
| AD | 7 (31.8%) | 9 (36.0%) | 7 (33.3%) | 7 (30.4%) | .99 |
| Additional food allergy | 14 (63.6%) | 18 (72.0%) | 11 (52.4%) | 18 (78.3%) | .14 |
| SCPs | |||||
| Total IgE (kU/L) | 604.5 (348.0-818.8) | 568.0 (233.0-918.0) | 556.0 (215.0-750.0) | 578.0 (343.0-823.5) | .43 |
| Milk IgE (kUA/L) | 35.8 (12.9-99.1) | 39.4 (9.9-80.3) | 55.6 (25.8-137.0) | 23.7 (5.1-38.9) | .01 |
| Casein IgE (kUA/L) | 39.4 (6.5-129.0) | 19.8 (7.5-88.4) | 68.1 (23.0-163.0) | 14.6 (1.9-38.3) | .003 |
| β-Lactoglobulin IgE (kUA/L) | 4.7 (1.7-35.4) | 4.4 (2.1-20.6) | 15.9 (3.0-40.3) | 3.2 (1.4-13.8) | .02 |
| Casein IgG (mg/mL) | 14.9 (7.0-17.8) | 9.5 (5.9-13.8) | 12.5 (7.8-21.5) | 8.3 (4.8-14.9) | .08 |
| β-Lactoglobulin IgG (mg/mL) | 3.8 (2.0-5.7) | 4.0 (2.4-5.4) | 4.6 (2.6-5.8) | 3.3 (2.0-4.8) | .14 |
| Casein IgG4 (mg/mL) | 2.2 (0.8-5.5) | 2.0 (0.4-2.7) | 2.2 (1.2-3.4) | 1.9 (0.6-4.8) | .75 |
| β-Lactoslobulin IgG4 (mg/mL) | 0.8 (0.2-1.1) | 0.6 (0.3-1.4) | 0.7 (0.3-1.4) | 0.5 (0.3-1.2) | .59 |
| Skin prick test | 8.9 (3.5) | 8.8 (3.1) | 10.2 (3.4) | 7.5 (2.4) | .004 |
Continuous variables are presented as either means and SD or medians and first and third quartiles; categorical variables are reported as frequencies and percentages. MOIT outcome comparison was tested by using either the 2-sample t test or Wilcoxon-Mann-Whitney test when the normality assumption was not met; categorical variables were compared by using either the χ2 test or Fisher exact test for variables with low cell frequencies.
DS, Desensitization.
The nonresponder group (n = 3) is not presented. Because patients were randomized to receive either omalizumab or placebo, baseline comparisons between adjuvant treatment arms were presented as descriptive statistics, and no tests of statistical significance were performed.35
Baseline ESAB profiling can predict development of SU and has better predictive ability than use of SCPs
Using baseline ESAB profiles from 23 subjects achieving SU and 21 desensitized subjects (averaged across triplicates), machine learning algorithms were used to build a model that predicted which patients would have SU 8 weeks after discontinuing MOIT. When building predictive models, it is common practice to divide the data set in a 3:1 ratio, where 75% of the observations are used for model development and 25% are retained for testing the performance of the model. However, because of the limited number of patients in the clinical trial, such division would greatly reduce the number of samples available for model building, and evaluating model performance on the remaining small test set would not reflect its true predictive ability. Therefore we opted to use a machine learning approach that combined regularization algorithms (which are more suitable for cases in which the number of potential predictors is larger than the number of subjects) and simulation techniques that allowed us to evaluate the performance range of the model and to measure the robustness/importance of each epitope (see Fig E3).
To evaluate the predictive power of ESAB profiles compared with milk component protein antibody measurements currently available, (ImmunoCAP assay; Thermo Fisher, Uppsala, Sweden), we developed models that considered serum epitope-specific IgE or IgG4 binding and serum total IgE, milk-specific IgE, and casein- and β-lactoglobulin–specific IgE, IgG, and IgG4, as well as various combinations of these biomarkers. Models built using the epitope-specific IgE profiles alone greatly outperformed those using component proteins alone based on various performance metrics, including AUC, accuracy, sensitivity, specificity, and positive and negative predictive values (Fig 4, A, and see Fig E6, A). Additionally, the combination of SCPs and ESAB profiles did not improve the model’s predictive ability; the IgE-specific epitope repertoire alone provided maximal model performance (Fig 4, A, and see Fig E8 in this article’s Online Repository at www.jacionline.org).
FIG 4.
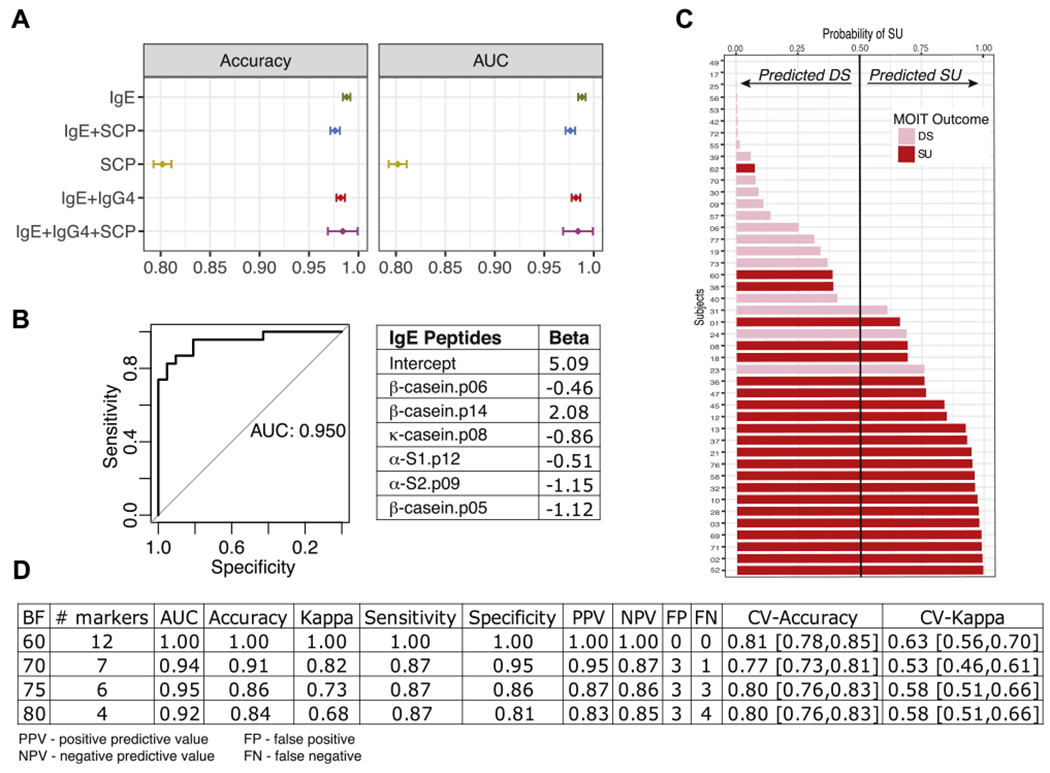
Prediction of desensitization (DS) versus SU outcome at month 32 (8 weeks after MOIT discontinuation) by using baseline levels with machine learning algorithms. A, Performance metrics of models fitted with different features: epitope-specific IgE or IgG4 binding only, SCPs (total IgE; milk-specific IgE; casein-specific IgE, IgG, and IgG4; and β-lactoglobulin–specific IgE and IgG), or the combination of SCPs and IgE and IgG4 epitopes in 300 bootstrapping simulations. Means and 95% CIs are presented. IgE epitopes are sufficient to achieve maximal predictive performance. Accuracy and receiver operating characteristic (ROC) curve are presented here; other metrics can be found in Fig E8. B, ROC curve and coefficients for the best model with the 6 most robust IgE epitopes identified as important predictors in at least 75% of the bootstrapping simulations (BF = 75%). C, Predicted probabilities (x-axis) versus actual MOIT outcomes of all subjects (colored bars) as predicted by using an IgE-based epitope model with a BF of 75%. If the predicted probability is greater than 0.5, the subject is classified as SU and DS otherwise. Pink and red bars represent actual MOIT outcomes: DS (pink) and SU (red). D, Performance metrics of 4 models, including epitopes with BFs of 60% to 80%. Estimates with 95% CIs are presented for cross-validation (CV) statistics.
When models were fitted using only informative peptides (those that appeared in the majority of the bootstrapping runs with BF 60% to 80%), IgE models also performed best in terms of cross-validation accuracy, a measure of how well the model might perform in future data sets (see Fig E8, B and C). For the model built using informative IgE epitopes, the performance statistics improved (see Fig E8, B). Using the 6 most informative (BF > 75) IgE epitopes (Fig 4, B), we can predict the probability of SU for each patient (Fig 4, C) with 86% accuracy, 87% sensitivity, and an AUC of 0.95 (Fig 4, B–D). A model using 12 peptides was 100% predictive, but the cross-validation accuracy was no better than models with only 4 or 6 IgE epitopes (Fig 4, D).
We ran a sensitivity analysis by developing models using one of the triplicates and testing them on the remaining 2 triplicates. Performance statistics were very similar to those based on triplicate averages (data not shown). This analysis should not be seen as a validation of the predictive performance, which requires a true validation set, but supports the internal consistency of our approach.
DISCUSSION
In the clinical trial of MOIT plus omalizumab, about 80% of patients with milk allergy achieved desensitization, whereas only about 40% achieved SU.17 This indicates that about 60% of patients treated with MOIT would be at risk for an allergic reaction if they stopped therapy for any prolonged period of time (eg, 4–8 weeks). Consequently, it is recommended that patients continue daily MOIT administration indefinitely, and if therapy is disrupted for any sustained period of time, the daily MOIT dose must be decreased and then re-escalated to the maintenance level under a physician’s supervision. In counseling patients about the risks and benefits of MOIT before initiating therapy, knowing the likelihood that a patient would achieve SU might influence a patient’s willingness to undergo such therapy. Patients achieving this level of protection would have much more flexibility in MOIT dosing and would be at less risk of experiencing an anaphylactic reaction if they missed several MOIT doses.36 In this study we demonstrated that analyzing IgE binding to specific informative milk protein epitopes can accurately predict whether patients with milk allergy will achieve SU 8 weeks after discontinuing MOIT.
In previous studies 66 peptides were identified representing sequential epitopes on the 5 major milk allergenic proteins: αS1-, αS2-, β-, and κ- caseins and β-lactoglobulin22,24,37. As depicted in Figs 1 and 2, decreases in IgE binding to milk protein epitopes appeared less pronounced in subjects after 30 months of MOIT plus omalizumab compared with those receiving MOIT plus placebo, whereas IgG4 binding to these epitopes appeared increased in those receiving omalizumab. Overall, IgG4 binding was greater in about one half of the allergenic epitopes (34 epitopes) after 30 months of MOIT plus omalizumab compared with MOIT plus placebo, but interestingly, IgG4 binding was greater (11 epitopes) in subjects only achieving desensitization compared with those achieving SU.
Our results show that MOIT reduces the amount of epitope-specific IgE and increases the amount of epitope-specific IgG4 (Fig 1), but we did not find this effect to be associated with the treatment response (Fig 2) either for epitope-specific IgE or IgG4 binding. These findings were mirrored in a reanalysis of the original serum milk component protein-specific data (Fig 1, C, and see Fig E4, A). Our analyses suggest that patients achieving desensitization start with greater quantities and diversity of epitope-specific IgE (Fig 3 and see Figs E6 and E7) than patients who achieved SU, and this difference appears sufficient to predict the response after 8 weeks of MOIT discontinuation. We did not find baseline differences in the quantity or diversity of epitope-specific IgG4 (see Figs E6 and E7), with similar observations in SCP IgG4 levels (Fig 3, C, and see Fig E4, B).
As depicted in Figs 4, E3, and E8, machine learning algorithms were developed from available data to predict the likelihood of achieving SU based on the baseline profiles of serum IgE and IgG4 epitope-specific and SCP-specific antibodies. Predictive models using informative epitopes identified subjects who would have SU 8 weeks after discontinuing MOIT, with an average accuracy of 92% to 95%. Additional models combining baseline levels of IgE and IgG4 epitope-specific antibodies did not improve the performance of models using IgE epitopes alone (Fig 4). Of note, algorithm-generated models selected IgE epitope-specific antibodies over IgG4 antibodies; among the 15 most informative epitopes, only 3 were IgG4 specific, and none were selected for the final model (data not shown). Collectively, our findings suggest that having both lower binding and lower diversity of IgE specific to allergenic epitopes at baseline is the strongest predictor of SU.
Milk component proteins have been previously shown to have some utility in identifying patients with persistent or more severe cow’s milk allergy,38 and baseline levels of IgE specific to milk, caseins, and β-lactoglobulin had significant association with SU outcome of MOIT when adjusted for the adjuvant treatment group.17 However, the epitope-specific IgE model outperformed the best model using only standard laboratory data (ie, SCP-specific IgE, IgG, and IgG4; AUC = 0.80 and accuracy = 80%). If validated in future studies, these epitope-specific IgE algorithms might enable allergists to predict which patients undergoing MOIT will achieve SU and which patients might require other forms of immunotherapy. Although nonresponder ESAB profiles were much closer to desensitization than those in patients achieving SU, conclusions regarding the ability of our algorithm to predict nonresponders are unwarranted (n = 3).
The ultimate goal of developing personalized medicine algorithms is to apply them to medical practice. To achieve this, a sizeable data set must be available to train the models and validate their performance in an independent data set. The small size of this available cohort, the small number of nonresponders, and the lack of an independent validation cohort are clear limitations of this study, which cannot be overcome at this point. In this study we set out to use the available cohort as a pilot proof of concept (ie, to determine whether baseline epitope profiles could be predictive biomarkers of the development of SU with OIT treatment). In the classical approach a predictive model would be fit by using a multivariable logistic regression starting with a small set of informative epitope-specific antibodies identified through a univariate analysis and/or by using a stepwise selection procedure. Such approaches have major limitations when the sample size is smaller than the number of (possibly correlated) features: collinearity among variables leads to biased model estimators, and feature selection steps can lead to overfitting and thus result in overoptimistic predictions, which can also be affected by the effect of outliers in a small cohort.39 To avoid such pitfalls, we used an analytic pipeline that allowed us to take full advantage of the available data by using regularized algorithms and combining computational techniques to ascertain the robustness of the predictive performance. This modeling strategy, which was previously used by our group to develop predictive models of treatment outcomes in psoriasis,40 uses bootstrapping simulations to (1) obtain not only point estimates for the performance but also CIs of ESAB’s predictive performance and (2) allow us to identify a robust set of epitopes that lead to the most accurate predictions. With this strategy, we found that epitope-based predictions are significantly superior to SCPs across all performance statistics and that assessing IgE binding to epitopes alone is sufficient to predict the clinical outcome. We used a well-known machine learning method, the elastic net, to build our predictive models. Although more sophisticated machine learning algorithms could render a greater predictive performance in this particular data set, such efforts can lead to overoptimistic estimations and should be saved for an adequately powered study with an independent validation cohort. Despite the limitations discussed above, our conservative analytic pipeline leads to a simple model with a highly predictive performance, showing the predictive capacity of IgE epitope profiling as a biomarker of sustained clinical response to OIT in patients with cow’s milk allergy.
To our knowledge, this is the first attempt to use epitope profiling and machine learning methods to predict OIT outcomes. The results presented in this proof-of concept study will hopefully encourage new and greater efforts to incorporate precision medicine in food allergy treatment strategies, with studies specifically designed for testing predictive performance with a more diverse population in terms of demographics, environmental exposures, and OIT outcomes.
Supplementary Material
Key messages.
OIT in patients with cow’s milk allergy induces significant changes in IgE and IgG4 antibody binding to allergenic milk protein epitopes.
Epitope profiles have a better predictive accuracy than models based on SCPs alone, and adding the latter will not improve predictive performance.
IgE-based epitope profiles taken at the beginning of treatment can predict SU after MOIT with an AUC of 95%.
Acknowledgments
We thank the clinical trial team (Jennifer S. Kim, MD; Robert Lindblad, MD; Alice K. Henning; Peter Dawson, PhD: and Marshall Plaut, MD) for their collaboration on data generation, data management, and expertise.
M.S.-F. received research grants to Mount Sinai by AllerGenis. This research was funded in part by the David H. and Julia Koch Research Program in Food Allergy Therapeutics and U19 AI44236, a grant from the National Institutes of Health (NIH)/National Institute of Allergy and Infectious Diseases (NIAID).
Disclosure of potential conflict of interest: M. Suárez-Fariñas reports grants from AllerGenis during the conduct of the study and personal fees from DBV. R. Getts has a patent pending (PCT/US2015/02171). K. Nadeau reports grants from the NIH and Food Allergy Research & Education during the conduct of the study and grants from Novartis, Genentech, Before Brand, Alladapt, Regeneron, Astellas, and Sanofi outside of the submitted work. R. A. Wood reports grants from the National Institute of Allergy and Infectious Diseases (NIAID), DBV, Aimmune, Astellas, HAL Allergy, and Regeneron and royalties from UpToDate outside the submitted work. H. A. Sampson reports grants from the NIAID, Immune Tolerance Network, and NIH/NIAID during the conduct of the study and personal fees from Hycor, UCB, N-Fold, DBV Technologies, UpToDate, and Elsevier outside of the submitted work.
Abbreviations used
- AUC
Area under the curve
- BF
Bagging frequency
- ESAB
Epitope-specific antibody binding
- FDR
False discovery rate
- ICC
Intraclass correlation coefficient
- MOIT
Milk oral immunotherapy
- OFC
Oral food challenge
- OIT
Oral immunotherapy
- PBS-TBN
1× PBS plus 0.02% Tween 20 plus 0.1% BSA
- SCP
Serum component protein
- SU
Sustained unresponsiveness
REFERENCES
- 1.Boyce JA, Assa’ad A, Burks AW, Jones SM, Sampson HA, Wood RA, et al. Guidelines for the diagnosis and management of food allergy in the United States: summary of the NIAID-sponsored Expert Panel Report. J Allergy Clin Immunol 2010;126:1105–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sicherer SH. Epidemiology of food allergy. J Allergy Clin Immunol 2011;127: 594–602. [DOI] [PubMed] [Google Scholar]
- 3.Skripak JM, Matsui EC, Mudd K, Wood RA. The natural history of IgE-mediated cow’s milk allergy. J Allergy Clin Immunol 2007;120:1172–7. [DOI] [PubMed] [Google Scholar]
- 4.Wood RA, Sicherer SH, Vickery BP, Jones SM, Liu AH, Fleischer DM, et al. The natural history of milk allergy in an observational cohort. J Allergy Clin Immunol 2013;131:805–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cianferoni A, Muraro A. Food-induced anaphylaxis. Immunol Allergy Clin North Am 2012;32:165–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grabenhenrich LB, Dolle S, Moneret-Vautrin A, Kohli A, Lange L, Spindler T, et al. Anaphylaxis in children and adolescents: the European Anaphylaxis Registry. J Allergy Clin Immunol 2016;137:1128–37, e1. [DOI] [PubMed] [Google Scholar]
- 7.Skripak JM, Nash SD, Rowley H, Brereton NH, Oh S, Hamilton RG, et al. A randomized, double-blind, placebo-controlled study of milk oral immunotherapy for cow’s milk allergy. J Allergy Clin Immunol 2008;122:1154–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Longo G, Barbi E, Berti I, Meneghetti R, Pittalis A, Ronfani L, et al. Specific oral tolerance induction in children with very severe cow’s milk-induced reactions. J Allergy Clin Immunol 2008;121:343–7. [DOI] [PubMed] [Google Scholar]
- 9.Meglio P, Giampietro PG, Gianni S, Galli E. Oral desensitization in children with immunoglobulin E-mediated cow’s milk allergy—follow-up at 4 yr and 8 months. Pediatr Allergy Immunol 2008;19:412–9. [DOI] [PubMed] [Google Scholar]
- 10.Keet CA, Frischmeyer-Guerrerio PA, Thyagarajan A, Schroeder JT, Hamilton RG, Boden S, et al. The safety and efficacy of sublingual and oral immunotherapy for milk allergy. J Allergy Clin Immunol 2012;129:448–55, e1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Staden U, Rolinck-Werninghaus C, Brewe F, Wahn U, Niggemann B, Beyer K. Specific oral tolerance induction in food allergy in children: efficacy and clinical patterns of reaction. Allergy 2007;62:1261–9. [DOI] [PubMed] [Google Scholar]
- 12.Keet CA, Seopaul S, Knorr S, Narisety S, Skripak J, Wood RA. Long-term follow-up of oral immunotherapy for cow’s milk allergy. J Allergy Clin Immunol 2013;132:737–9, e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burks AW, Jones SM, Wood RA, Fleischer DM, Sicherer SH, Lindblad RW, et al. Oral immunotherapy for treatment of egg allergy in children. N Engl J Med 2012; 367:233–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Syed A, Garcia MA, Lyu SC, Bucayu R, Kohli A, Ishida S, et al. Peanut oral immunotherapy results in increased antigen-induced regulatory T-cell function and hypomethylation of forkhead box protein 3 (FOXP3). J Allergy Clin Immunol 2014;133:500–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Narisety SD, Frischmeyer-Guerrerio PA, Keet CA, Gorelik M, Schroeder J, Hamilton RG, et al. A randomized, double-blind, placebo-controlled pilot study of sublingual versus oral immunotherapy for the treatment of peanut allergy. J Allergy Clin Immunol 2015;135:1275–82, e1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorelik M, Narisety SD, Guerrerio AL, Chichester KL, Keet CA, Bieneman AP, et al. Suppression of the immunologic response to peanut during immunotherapy is often transient. J Allergy Clin Immunol 2015;135:1283–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wood RA, Kim JS, Lindblad R, Nadeau K, Henning AK, Dawson P, et al. A randomized, double-blind, placebo-controlled study of omalizumab combined with oral immunotherapy for the treatment of cow’s milk allergy. J Allergy Clin Immunol 2016;137:1103–10, e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nadeau KC, Schneider LC, Hoyte L, Borras I, Umetsu DT. Rapid oral desensitization in combination with omalizumab therapy in patients with cow’s milk allergy. J Allergy Clin Immunol 2011;127:1622–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneider LC, Rachid R, LeBovidge J, Blood E, Mittal M, Umetsu DT. A pilot study of omalizumab to facilitate rapid oral desensitization in high-risk peanut-allergic patients. J Allergy Clin Immunol 2013;132:1368–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belliveau PP. Omalizumab: a monoclonal anti-IgE antibody. MedGenMed 2005;7:27. [PMC free article] [PubMed] [Google Scholar]
- 21.Pennington LF, Tarchevskaya S, Brigger D, Sathiyamoorthy K, Graham MT, Nadeau KC, et al. Structural basis of omalizumab therapy and omalizumab-mediated IgE exchange. Nat Commun 2016;7:11610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beyer K, Jarvinen KM, Bardina L, Mishoe M, Turjanmaa K, Niggemann B, et al. IgE-binding peptides coupled to a commercial matrix as a diagnostic instrument for persistent cow’s milk allergy. J Allergy Clin Immunol 2005;116:704–5. [DOI] [PubMed] [Google Scholar]
- 23.Cerecedo I, Zamora J, Shreffler WG, Lin J, Bardina L, Dieguez MC, et al. Mapping of the IgE and IgG4 sequential epitopes of milk allergens with a peptide microarray-based immunoassay. J Allergy Clin Immunol 2008;122:589–94. [DOI] [PubMed] [Google Scholar]
- 24.Han N, Jarvinen KM, Cocco RR, Busse PJ, Sampson HA, Beyer K. Identification of amino acids critical for IgE-binding to sequential epitopes of bovine kappa-casein and the similarity of these epitopes to the corresponding human kappa-casein sequence. Allergy 2008;63:198–204. [DOI] [PubMed] [Google Scholar]
- 25.Wang J, Lin J, Bardina L, Goldis M, Nowak-Wegrzyn A, Shreffler WG, et al. Correlation of IgE/IgG4 milk epitopes and affinity of milk-specific IgE antibodies with different phenotypes of clinical milk allergy. J Allergy Clin Immunol 2010; 125:695–702, e1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Savilahti EM, Kuitunen M, Valori M, Rantanen V, Bardina L, Gimenez G, et al. Use of IgE and IgG4 epitope binding to predict the outcome of oral immunotherapy in cow’s milk allergy. Pediatr Allergy Immunol 2014;25:227–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Bushel PR, Chu TM, Wolfinger RD. Principal variance components analysis: estimating batch effects in microarray gene expression data In: Scherer A, editor. Batch effects and noise in microarray experiments: sources and solutions. John Wiley & Sons; 2009. pp. 141–54. [Google Scholar]
- 28.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007;8:118–27. [DOI] [PubMed] [Google Scholar]
- 29.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012;28:882–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartko JJ. The intraclass correlation coefficient as a measure of reliability. Psychol Rep 1966;19:3–11. [DOI] [PubMed] [Google Scholar]
- 31.Gamer M, Lemon J, Fellows I, Singh P. irr: various coefficients of interrater reliability and agreement. R package. 0.84 ed. CRAN2012. Available at:http://CRAN.R-project.org/package=irr. [Google Scholar]
- 32.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 2004;3:Article 3. [DOI] [PubMed] [Google Scholar]
- 33.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 35.Palesch YY. Some common misperceptions about P values. Stroke 2014;45:e244–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dantzer JA, Wood RA. The use of omalizumab in allergen immunotherapy. Clin Exp Allergy 2018;48:232–40. [DOI] [PubMed] [Google Scholar]
- 37.Lin J, Bardina L, Shreffler WG, Andreae DA, Ge Y, Wang J, et al. Development of a novel peptide microarray for large-scale epitope mapping of food allergens. J Allergy Clin Immunol 2009;124:315–22, e1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petersen TH, Mortz CG, Bindslev-Jensen C, Eller E. Cow’s milk allergic children—can component-resolved diagnostics predict duration and severity? Pediatr Allergy Immunol 2018;29:194–9. [DOI] [PubMed] [Google Scholar]
- 39.Costello JC, Heiser LM, Georgii E, Gonen M, Menden MP, Wang NJ, et al. A community effort to assess and improve drug sensitivity prediction algorithms. Nat Biotechnol 2014;32:1202–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Correa da Rosa J, Kim J, Tian S, Tomalin LE, Krueger JG, Suarez-Farinas M. Shrinking the psoriasis assessment gap: early gene-expression profiling accurately predicts response to long-term treatment. J Invest Dermatol 2017;137:305–12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


