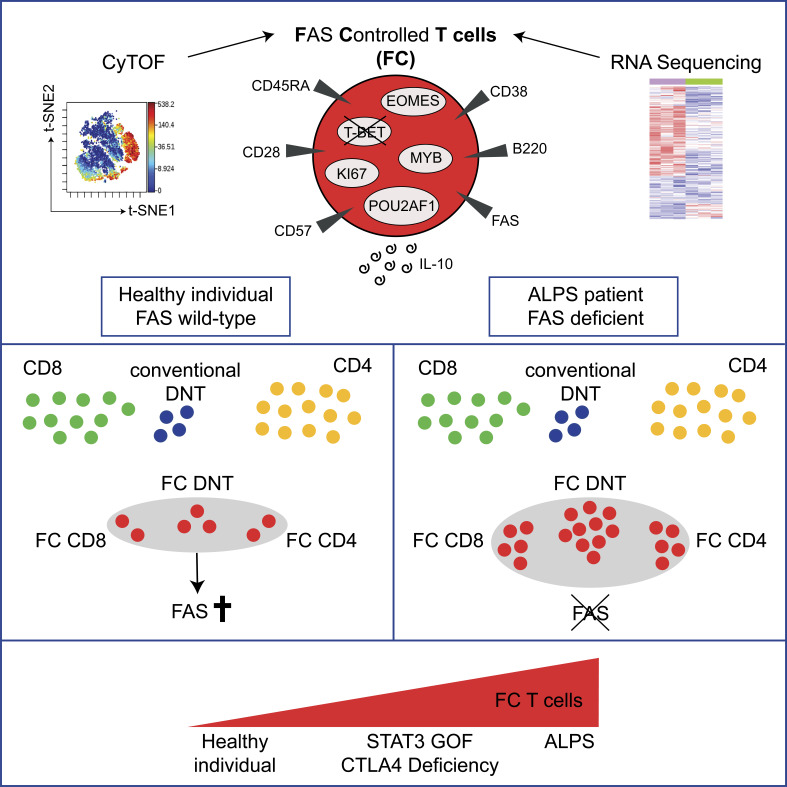
Maccari et al. identify a physiological population of highly proliferative CD38+CD45RA+, IL-10–producing TCRαβ+ T cells. They can be CD4+, CD8+, or double-negative and are controlled by FAS and CTLA4, while their survival is enhanced by mTOR and STAT3 signals.
Abstract
The identification and characterization of rare immune cell populations in humans can be facilitated by their growth advantage in the context of specific genetic diseases. Here, we use autoimmune lymphoproliferative syndrome to identify a population of FAS-controlled TCRαβ+ T cells. They include CD4+, CD8+, and double-negative T cells and can be defined by a CD38+CD45RA+T-BET− expression pattern. These unconventional T cells are present in healthy individuals, are generated before birth, are enriched in lymphoid tissue, and do not expand during acute viral infection. They are characterized by a unique molecular signature that is unambiguously different from other known T cell differentiation subsets and independent of CD4 or CD8 expression. Functionally, FAS-controlled T cells represent highly proliferative, noncytotoxic T cells with an IL-10 cytokine bias. Mechanistically, regulation of this physiological population is mediated by FAS and CTLA4 signaling, and its survival is enhanced by mTOR and STAT3 signals. Genetic alterations in these pathways result in expansion of FAS-controlled T cells, which can cause significant lymphoproliferative disease.
Graphical Abstract
Introduction
Inborn errors of immunity can be instructive for understanding physiological and abnormal human immune cell differentiation and regulation. In autoimmune lymphoproliferative syndrome (ALPS), unusually differentiated T and B cells determine the disease phenotype, which is characterized by chronic lymphadenopathy, splenomegaly, and autoimmunity. The disease is caused by germline and/or somatic FAS (sFAS) mutations (Fisher et al., 1995; Holzelova et al., 2004; Rieux-Laucat et al., 1995). FAS is a transmembrane receptor expressed on all activated lymphocytes. It induces apoptotic cell death but can also mediate nonapoptotic functions (Klebanoff et al., 2016; Le Gallo et al., 2017; Peter et al., 2007).
Lymphoproliferation in ALPS is mainly caused by accumulation of polyclonal CD3+TCRαβ+CD4−CD8− double-negative T cells (DNTs). ALPS DNTs are predominantly CCR7−CD45RA+CD57+, resembling terminally differentiated T cells reexpressing CD45RA (Bleesing et al., 2002; Bristeau-Leprince et al., 2008; Rensing-Ehl et al., 2014), but they express CD27 and CD28, similar to naive or early differentiated T cells, and lack expression of T-BET and KLRG1 (Rensing-Ehl et al., 2014). Moreover, they express CD45R B220, are highly proliferative, and excessively produce IL-10 (Bleesing et al., 2001; Ohga et al., 2002). A small population of TCRαβ+ DNTs can also be found in healthy individuals. These DNTs are nonproliferative with a predominant CCR7−CD45RA− effector memory phenotype and do not produce IL-10 (Fischer et al., 2005). Physiological DNTs are also present in gut and kidney and in some reports have been shown to express B220 and/or to secrete IL-10 (Martina et al., 2016; Mohamood et al., 2008). Increased numbers of DNTs have been observed in patients with STAT3-activating mutations and CTL-associated protein 4 (CTLA4) deficiency (Besnard et al., 2018; Milner et al., 2015; Nabhani et al., 2017). Whether these “conventional” and “disease-related” DNT populations represent separate lineages or different states of the same lineage has not been defined.
Models of ALPS DNT ontogeny must explain both their accumulation and their unusual differentiation. We previously observed that small populations of CD57+CD4+ and CD8+ T cells of ALPS patients were enriched for TCR sequences that were also prevalent among DNTs (Rensing-Ehl et al., 2014). While these data support that peripheral single-positive T cells (SPTs) can transition to ALPS DNTs by loss of coreceptor expression, they cannot easily account for the significant phenotypic differences between conventional T cell subsets and ALPS DNTs. It is possible that the unusual profile results from defective FAS signaling, driving conventional T cells into the differentiation state of ALPS DNTs. Alternatively, the unusual differentiation program could evolve independently of FAS. In this case, T cells with this program should be detectable in healthy individuals.
The presence of T cells with this unusual differentiation profile has not been documented in healthy humans so far. In this study, we combined mass cytometry and RNA sequencing analyses to further explore the relationship of ALPS DNTs to other T cell subsets at high resolution and identified their distinct molecular signature, which was not restricted to DNTs but also present in subsets of CD4+ and CD8+ T cells. The detailed characterization allowed us to identify these cells in healthy individuals. The cellular signature of these T cells characterizes a highly proliferative, unconventional CD38+CD45RA+T-BET− IL-10–producing polyclonal T cell population tightly controlled by FAS and CTLA4 and maintained by mammalian target of rapamycin (mTOR) and STAT3 signals.
Results
DNTs in ALPS patients show a unique transcriptional profile
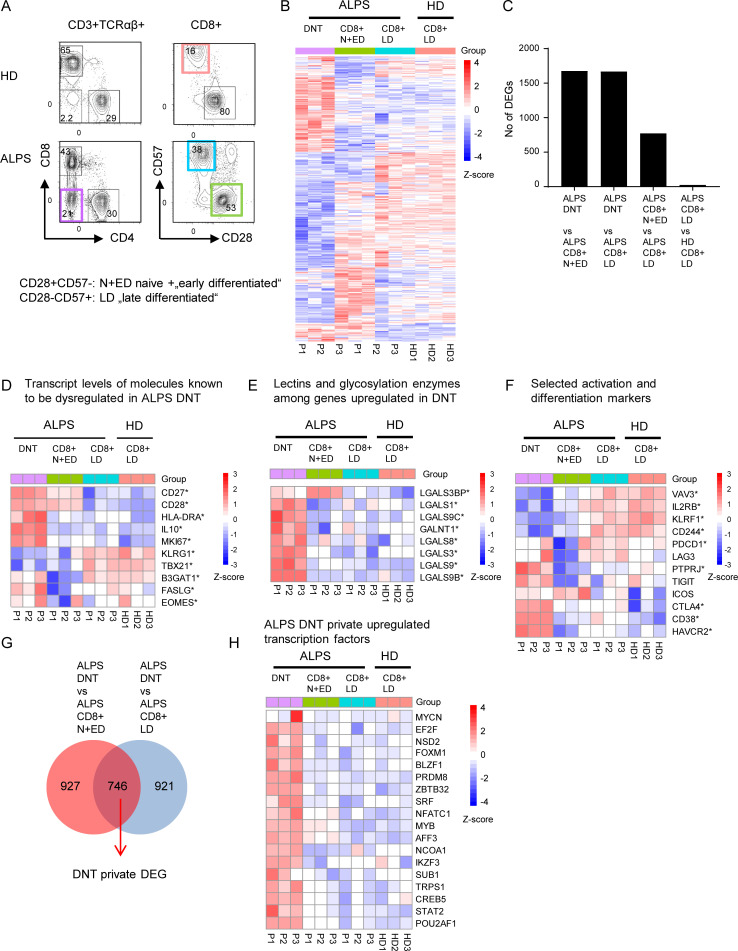
When initiating this study, we considered DNTs in ALPS patients a distinct, disease-specific, pathological T cell population. We studied the transcriptional basis of their unusual differentiation pattern and possible relation to other defined T cell subsets by RNA sequencing analysis. We sorted DNTs, CD8+CD57−CD28+ naive and early differentiated T cells (N+EDs) and CD8+CD57+CD28− late differentiated effector memory T cells (LDs) from three untreated ALPS-FAS patients (P1–P3; Table S1). We also included CD8+ LDs of three healthy donors (HDs) to analyze whether the FAS mutation had an impact on gene expression in this cell compartment (Fig. 1 A). The transcriptome of ALPS DNTs clearly differed from CD8+ N+EDs and LDs of patients and HDs (Figs. 1 B and S1 A). While transcriptional profiles of ALPS versus HD CD8+ LDs showed only 25 differentially expressed genes (DEGs; adjusted P < 0.05), 1,675 and 1,667 genes were differentially expressed between ALPS DNTs versus CD8+ N+EDs and ALPS DNTs versus CD8+ LDs, respectively (Fig. 1, B and C). As expected, these included molecules known to be dysregulated, such as IL-10, FAS ligand (FASLG), KLRG1, T-BET (TBX21), and beta-glucuronosyltransferase 1 (B3GAT1), responsible for the generation of the CD57 epitope (Fig. 1 D). Expression of the CD45R B220 glycoform on ALPS DNTs results from high glycosylation activity (Imai et al., 1988). Interestingly, GALNT1, encoding a glycosylation enzyme, and seven galectin-encoding genes were among the top up-regulated genes (Fig. 1 E). We also found strong up-regulation of CD38, HAVCR2 (TIM3), TIGIT, and CTLA4, typically expressed on activated, exhausted, and/or regulatory T cells (T reg cells; Fig. 1 F). We then focused on the 746 DNT-private DEGs, dysregulated compared with both CD8+ N+EDs and CD8+ LDs (Fig. 1 G). We identified 19 private up-regulated transcription factors, including MYB and OBF1 (POU2AF1), previously found up-regulated in DNTs of Fas-deficient lpr mice (Mountz et al., 1984; Zuo et al., 2007; Mountz and Steinberg, 1989; Figs. 1 H and S1 B). 26 transcription factors were down-regulated (Fig. S1 C) in ALPS DNTs. Forkhead box protein P3 (FOXP3) expression was low (data not shown). Moreover, many of the top up- or down-regulated genes in ALPS DNTs were not dysregulated in T reg cells when we analyzed published RNA sequencing data (Schmiedel et al., 2018; Fig. S1 D). Thus, although ALPS DNTs shared features with N+EDs, LDs, exhausted T cells, and T reg cells, their transcriptional signature unambiguously differed from these conventional T cell populations and was not suggestive of any close relationship.
Figure 1.
DNTs in ALPS-FAS patients show a unique transcriptional profile. (A) Gating strategy for DNTs, CD8+CD28+CD57− N+EDs, and CD8+CD28-CD57+ LDs of three ALPS-FAS patients (P1–P3) and CD8+ LDs of three adult HDs (HD1–HD3) sorted for RNA sequencing analysis. (B) Heatmap showing all DEGs significantly dysregulated (adjusted P < 0.05) in any comparison of ALPS DNTs versus ALPS CD8+ N+EDs, ALPS DNTs versus ALPS CD8+ LDs, ALPS CD8+ N+EDs versus ALPS CD8+ LDs, or ALPS CD8+ LDs versus HD CD8+ LDs. (C) Number of DEGs (adjusted P < 0.05) for each comparison. (D) Heatmap showing transcripts of molecules known to be dysregulated in ALPS DNTs. (E) Heatmap of selected genes coding for lectins or enzymes involved in glycosylation. (F) Heatmap of selected genes involved in T cell activation or differentiation. * in D–F indicates that a gene was significantly (adjusted P < 0.05) dysregulated in at least one comparison. (G) Venn diagram showing the intersection of shared DEGs in the comparisons of ALPS DNTs versus ALPS CD8+ N+EDs and ALPS DNTs versus ALPS CD8+ LDs (ALPS DNT “private” genes). (H) Heatmap of up-regulated transcription factors among ALPS DNT private DEGs. For all heatmaps, the relative expression (Z-score) of genes is shown and is color coded according to the legend. Rows are scaled to have a mean value of 0 and an SD of 1.
Figure S1.
ALPS DNTs show a unique transcriptional profile. (A) Principal component (PC) analysis of RNA sequencing data on DNTs, CD8+ N+EDs, and CD8+ LDs of three ALPS-FAS patients (P1–P3) and CD8+ LDs of three adult HDs (HD1–HD3). (B) Heatmap depicting up-regulated transcription factors among DNT private DEGs (all significantly up-regulated in both comparisons of DNTs versus CD8+ N+EDs and DNTs versus CD8+ LDs). Log2 fold changes (log2 FCs) are shown for the comparisons of DNTs versus CD8+ N+EDs and DNTs versus CD8+ LDs to compare the degree of dysregulation. (C) Heatmap of down-regulated transcription factors among DNT private DEGs (all significantly down-regulated in both comparisons of DNTs versus CD8+ N+EDs and DNTs versus CD8+ LDs). Relative expression (Z-score) of genes is shown and is color coded according to the legend. Rows are scaled to have a mean value of 0 and an SD of 1. (D) Heatmaps showing expression levels (color code = log2 FC) of the top 30 up- or down-regulated genes identified in ALPS DNTs versus CD8+ N+EDs and CD8+ LDs by RNA sequencing analysis (RNAseq) or selected up- or down-regulated molecules on ALPS DNTs by CyTOF/FACS in human naive T reg cells (T reg_naive = CD3+CD4+CD25hiCD127lowCD45RA+) and memory T reg cells (T reg_mem = CD3+CD4+CD25hiCD127lowCD45RA−) compared with naive CD4+ T cells (Tn = CD3+CD4+CD45RA+CCR7+). Tn, T naive.
ALPS patients have two phenotypically distinct DNT populations
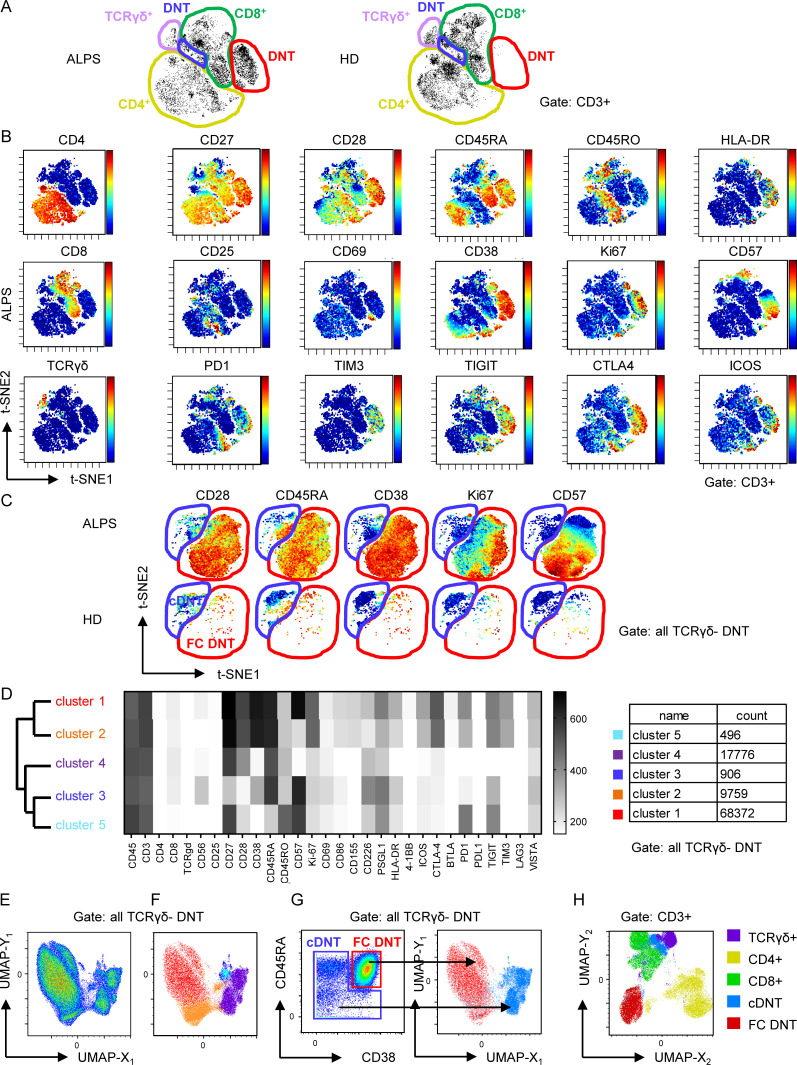
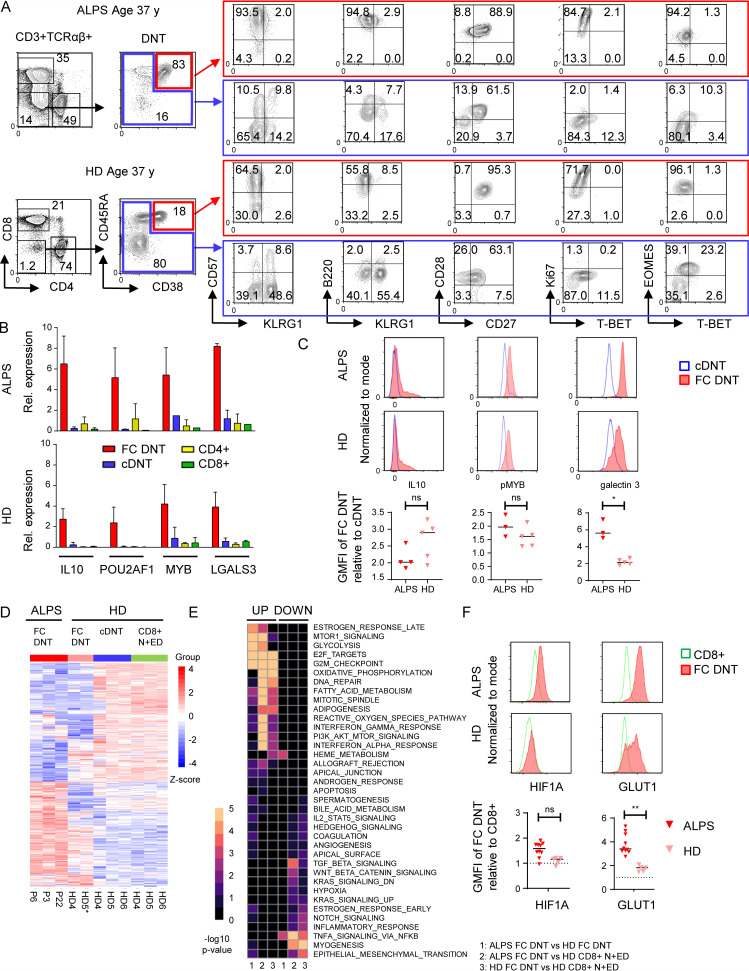
To further define the ALPS DNT profile, we performed mass cytometry by time of flight (CyTOF), analyzing 34 markers on peripheral blood mononuclear cells (PBMCs) of seven untreated ALPS-FAS patients (P4–P10; Table S1) and seven HDs. Interestingly, t-stochastic neighbor embedding (t-SNE)–based dimensionality reduction delineated two major DNT subpopulations. One population was specifically prominent in ALPS patients (Fig. 2 A, encircled in red) and showed a distinct profile characterized by high CD38, CD45RA, CD27, and CD28 expression. The majority of these DNTs expressed Ki67, CD57, TIM3, and TIGIT, and a subset expressed HLA-DR, CD69, PD1, and ICOS (Fig. 2 B). We termed this unique profile a “FAS-controlled (FC) T cell signature” because cells with this signature are expanded in FAS-deficient patients. Subsequent t-SNE–based visualization restricted to TCRγδ− DNTs again separated two major populations: FC DNT, expanded in ALPS, and a second, small DNT population with a resting naive to effector memory phenotype (CD45RA+/−CD28+/−CD38−Ki67−CD57−/+), which was present in both HDs and patients with ALPS and therefore termed “conventional DNTs” (cDNTs; Fig. 2 C). To further validate the two DNT clusters, we analyzed mass cytometry data using a self-organizing map (FlowSOM) for unsupervised clustering of all TCRγδ− DNTs in the 14 datasets. Again, two main clusters were generated containing subclusters with expression profiles of FC DNTs (clusters 1 and 2) or cDNTs (clusters 3–5; Fig. 2 D). To visualize relationships between clusters, we used Uniform Manifold Approximation and Projection (UMAP), which also generated two main DNT clusters (Fig. 2 E). When the five subclusters identified by FlowSOM were projected onto the two-dimensional UMAP plot, they localized with the two main DNT populations (Fig. 2 F). Because FC DNTs most homogeneously expressed CD38 and CD45RA, we evaluated whether this combination could separate FC DNTs and cDNTs. Indeed, projection of DNT populations manually gated for these two markers matched the two major populations in the UMAP plot (Fig. 2 G). Of note, when projected to a UMAP plot depicting all T cells, FC DNTs formed a distinct population, while cDNTs aligned with CD8+ and TCRγδ+ T cells, indicating their phenotypic relationship (Figs. 2 H and S2 A).
Figure 2.
FC DNTs show a distinct protein expression profile differing from that of cDNTs. (A) t-SNE plots of CD3+ T cells of one ALPS-FAS patient and one HD with manual gating of T cell subsets based on CD4, CD8, and TCRγδ expression. Major DNT populations: cDNT (blue gate) and FC DNTs (red gate). (B) t-SNE plots of one ALPS-FAS patient showing expression levels of the indicated markers. Color codes represent expression levels of respective markers, scaled relative to the maximum (red) and minimum (blue) expression for each individual marker across all samples. (C) t-SNE plots restricted to DNT (CD3+TCRγδ−CD4−CD8−) shown for one ALPS-FAS patient and one HD. Red, FC DNT; blue, cDNT. Comparable results were obtained for six additional ALPS-FAS patients and six HDs. (D) Unsupervised clustering using FlowSOM on manually gated DNTs (CD3+TCRγδ−). Dendrogram for cluster relationships and expression levels of all included markers. (E) UMAP plots for manually gated DNTs (CD3+TCRγδ−) including all samples. (F) Distribution of the five DNT subclusters identified by FlowSOM among the two main DNT populations in the UMAP visualization. (G) Density plot of all DNTs (CD3+TCRγδ−) showing CD38 and CD45RA expression with manual gates on FC DNTs (red) and cDNTs (blue). Manually gated populations were projected onto the UMAP plot. Red, FC DNT; blue, cDNT. (H) Distribution of TCRγδ+, CD4+, CD8+, FC DNT, and cDNT populations among total CD3+ T cells using UMAP.
Figure S2.
ALPS patients and HDs have two main DNT clusters. (A) UMAP projection of indicated T cell populations in seven ALPS-FAS patients and seven HDs. (B) Heatmap comparing transcript levels of genes known to be dysregulated in ALPS DNTs in sorted ALPS FC DNT, HD FC DNT, HD cDNT, and HD N+ED CD8+ T cells. Cells were sorted from ALPS patients P3, P6, and P22 and from HD4, HD5, and HD6. For one HD FC DNT sample, RNA was pooled from four HDs to achieve sufficient RNA (HD5*). * indicates that a gene was significantly (adjusted P < 0.05) dysregulated compared with the reference HD CD8+ N+ED population. (C) Heatmap of selected genes coding for lectins or enzymes involved in glycosylation. (D) Heatmap of selected genes involved in T cell activation or differentiation. (E) Heatmap of up-regulated transcription factors found among ALPS DNT private DEGs in the first RNA sequencing analysis. For all heatmaps, the relative expression (Z-score) of genes is shown and is color coded according to the legend. Rows are scaled to have a mean value of 0 and an SD of 1. (F) Top 100 up- and down-regulated genes based on log2 fold changes in ALPS FC DNTs versus HD CD8+ N+EDs. Color code indicates log2 fold changes for the indicated comparisons (1, 2, or 3).
Unconventional, highly proliferative FC DNTs are present in HDs
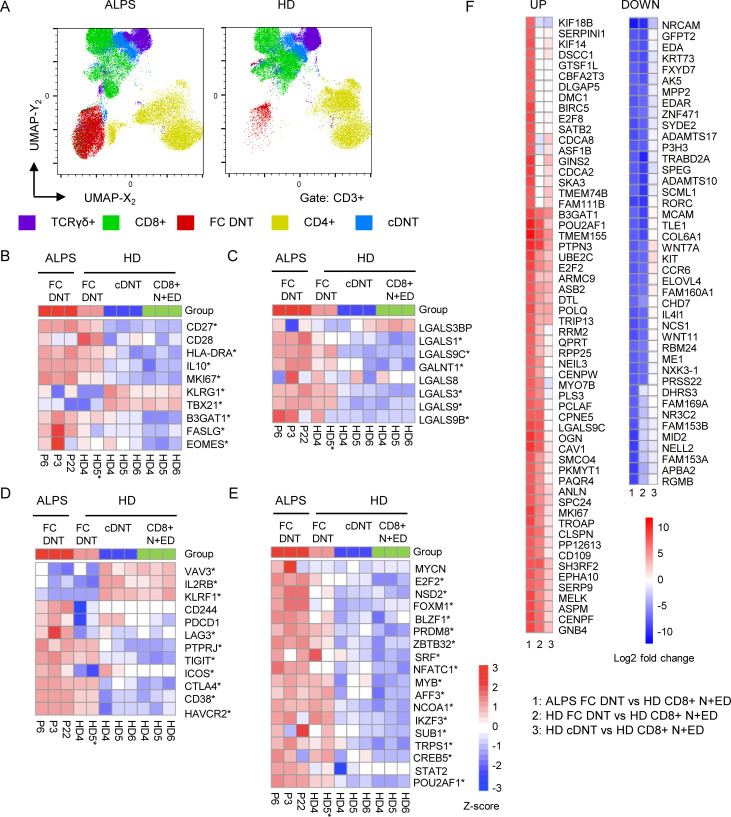
Unexpectedly, CyTOF analysis identified not only cDNTs but also FC DNTs in HDs (Fig. 2, A and C; and Fig. S2 A). Subsequent flow cytometric analyses confirmed a population of CD38+CD45RA+ cells among HD DNTs with the same CD27+CD28+Ki67+/−CD57+/− profile as FC DNTs in ALPS patients. These T cells also highly expressed B220 and eomesodermin (EOMES) while lacking T-BET and KLRG1 (Fig. 3 A). Using RT-PCR, we confirmed that the characteristic FC T cell signature genes IL10, POU2AF1, MYB, and LGALS3 were highly up-regulated in sorted CD38+CD45RA+ FC DNTs of HDs (Fig. 3 B). Transcript levels of these genes in HD FC DNTs were not as high as in FC DNTs of ALPS patients, but differences were not statistically significant. Protein expression levels of IL-10 and constitutively phosphorylated MYB were also similar, but galectin 3 expression was lower in FC DNTs of HDs than in ALPS patients (Fig. 3 C). To corroborate cellular identity of FC DNTs in HDs and ALPS patients and further explore their relationship to cDNTs, we performed additional transcriptomic analyses. We used HD N+EDs (CD28+CD57−) CD8+ T cells as the reference population analogous to the initial RNA sequencing analysis. Notably, because FC DNTs represent <1% of TCRαβ+ T cells in HDs, we had to sort cells from buffy coats, which may have introduced a relevant variable when comparing HD FC DNTs with those from fresh blood of ALPS patients. HD cDNTs differed from CD8+ N+EDs by only few DEGs and could be clearly separated from both ALPS and HD FC DNTs when we analyzed overall transcriptional patterns (Fig. 3 D) as well as transcript levels of selected genes encoding activation/differentiation markers, lectins, and transcription factors (Fig. S2, B–E). When comparing ALPS with HD FC DNTs, we found that approximately one-third of DEGs were similarly up-regulated, one-third of DEGs were similarly down-regulated, and one-third of DEGs were discordantly regulated (Fig. 3 D) relative to the reference population. The similarly regulated DEGs included most characteristic signature genes analyzed in Fig. 1 (Fig. S2, B–E). Moreover, >80% of the top 100 DEGs between ALPS FC DNTs and CD8+ N+EDs were concordantly dysregulated in ALPS and HD FC DNTs (Fig. S2 F). To better understand the transcriptional differences, we performed gene set enrichment analysis, which showed an overall concordance of pathways predicted to be up- or down-regulated in ALPS and FC DNTs compared with CD8+ N+EDs. Significant differences in gene set enrichment mainly affected pathways involved in cell cycle progression, glucose metabolism, and mTOR signaling (Fig. 3 E). Comparative analysis of protein expression of the transcription factor HIF1A and the glucose transporter GLUT1, which are both regulated by mTOR and promote glycolysis, confirmed higher upregulation in ALPS FC DNTs than in HD FC DNTs (Fig. 3 F). In summary, FC DNTs are a physiological T cell population representing a small fraction of total TCRαβ+ DNTs in healthy individuals that shares key features of the distinct signature seen in FC DNTs expanded in ALPS patients.
Figure 3.
FC DNTs are present in HDs. (A) Example flow cytometric data showing selected surface marker and transcription factor expression in FC DNTs and cDNTs in one ALPS-FAS patient compared with one age-matched HD. Numbers in gates indicate percentages. Another 5 ALPS-FAS patients and 10 HDs (ages 0–73 yr) were analyzed with the same panel, yielding similar results (data not shown). (B) Relative (Rel.) expression of four genes selected from the top up-regulated ALPS DNT private DEGs identified by RNA sequencing analysis was quantified in sorted FC DNTs and cDNTs (based on CD38 and CD45RA expression) as well as total CD4+ and CD8+ T cells from three ALPS-FAS patients and four HDs by quantitative RT-PCR. Expression was normalized to HPRT, and mean and SD values are shown. (C) Representative histograms showing IL-10, pMYB, and galectin 3 expression in FC DNTs and cDNTs of one ALPS patient and one HD. Summary plots showing geometric mean fluorescence intensity (GMFI) of IL-10, pMYB, and galectin 3 in FC DNTs relative to cDNTs in three or four ALPS patients and 5 HD. Lines indicate median values. Statistical comparisons were performed using the Mann-Whitney U test. (D) Heatmap showing all DEGs significantly dysregulated (adjusted P < 0.05) in any comparison between ALPS FC DNT, HD FC DNT, HD cDNT, and HD N+ED CD8+ T cells. Cells were sorted from P3, P6, and P22 and from HD4, HD5, and HD6. For the FC DNT HD5 sample, RNA was pooled from four HDs to achieve sufficient RNA (HD5*). Relative expression (Z-score) of genes is shown and is color coded according to the legend. Rows are scaled to have a mean value of 0 and an SD of 1. (E) Gene set enrichment analysis heatmap showing the top 10 up- and down-regulated hallmark pathways in ALPS FC DNTs versus HD FC DNTs (1), ALPS FC DNTs versus HD + N+EDs (2), or HD FC DNTs versus HD CD8+ N+EDs (3). Color code represents the −log10 P value. (F) Representative histograms showing hypoxia-inducible factor-1α (HIF1A) and GLUT1 expression in FC DNTs compared with CD8+ T cells of one ALPS patient and one HD. Summary plots showing GMFI of FC DNTs relative to CD8+ T cells in 10 ALPS patients and 5 HDs. Lines indicate median values. Statistical comparisons were performed using the Mann-Whitney U test. *, P < 0.05; **, P < 0.01. ns, not significant.
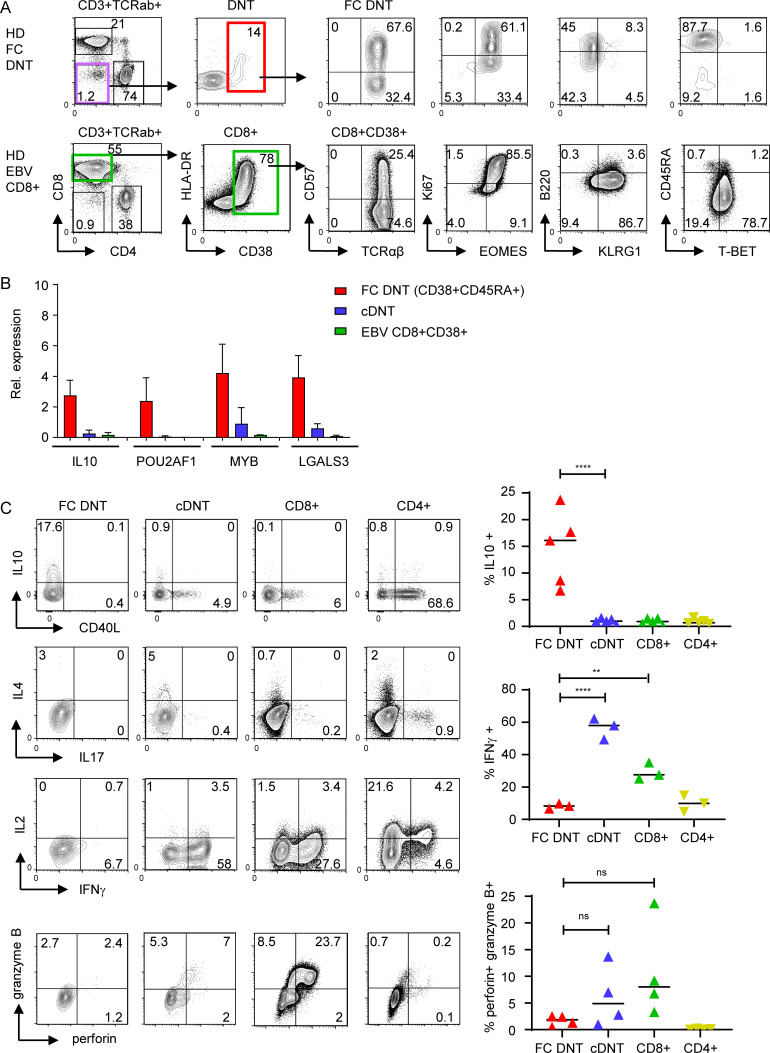
An IL-10–biased cytokine expression profile and lack of cytolytic molecules delineates CD38+HLA-DR+Ki67+ FC from virus-activated T cells
FC DNTs show several key features of activated and proliferating T cells, such as CD38, HLA-DR, and Ki67 expression. To determine the degree of phenotypic and functional overlap with in vivo activated T cells, we compared FC DNTs of HDs with activated CD8+ T cells from individuals with acute EBV infection. In these subjects, CD8+ T cells were massively expanded and showed raised CD38, HLA-DR, and Ki67 expression, similar to FC DNTs. However, CD8+ T cells activated in the context of acute EBV infection were predominantly CD45RA− and B220−, highly expressed KLRG1 and T-BET, and had not up-regulated IL10, POU2AF1, MYB, and LGALS3 (Fig. 4, A and B). For functional analysis, we compared FC DNTs with other T cell populations in the same HDs. FC DNTs expressed high levels of IL-10 but lacked IFNγ, IL-2, IL-4, IL-17, and CD40 ligand, while IFNγ was the key cytokine produced by CD8+ T cells. Moreover, FC DNTs did not express perforin and granzyme B, indicating a lack of cytolytic capacity (Fig. 4 C). Notably, cDNTs expressed IFNγ and cytolytic molecules, further supporting their relationship to CD8+ T cells. Taken together, FC DNTs showed clear transcriptional, phenotypic, and functional differences from virus-activated T cells and cDNTs, supporting their classification as an independent T cell subset.
Figure 4.
FC DNTs differ from virus-activated T cells. (A) Flow cytometric analyses showing the differentiation pattern of human highly activated CD38+CD8+ effector T cells of one individual with acute EBV infection compared with HD FC DNTs. Similar results were acquired in 10 additional patients with acute EBV infection (data not shown). (B) Transcriptional levels of four up-regulated FC genes analyzed by quantitative RT-PCR in sorted CD38+CD8+ T cells from three donors with acute EBV infection compared with those of HD FC DNTs and HD cDNTs (n = 4). Expression levels normalized to HPRT with mean and SD values. (C) Representative plots showing the cytokine profile and expression of perforin and granzyme B in FC DNTs, cDNTs, and CD4+ and CD8+ of one HD. Summary plots showing percentages of IL-10+, IFNγ+, and perforin+/granzyme B+ cells in all subjects tested. Lines indicate median values. Statistical comparisons were performed using Ordinary one-way ANOVA with Tukey’s multiple comparisons test. **, P < 0.01; ****, P < 0.0001. ns, not significant.
The FC T cell signature characterizes T cells across CD4/CD8 identity
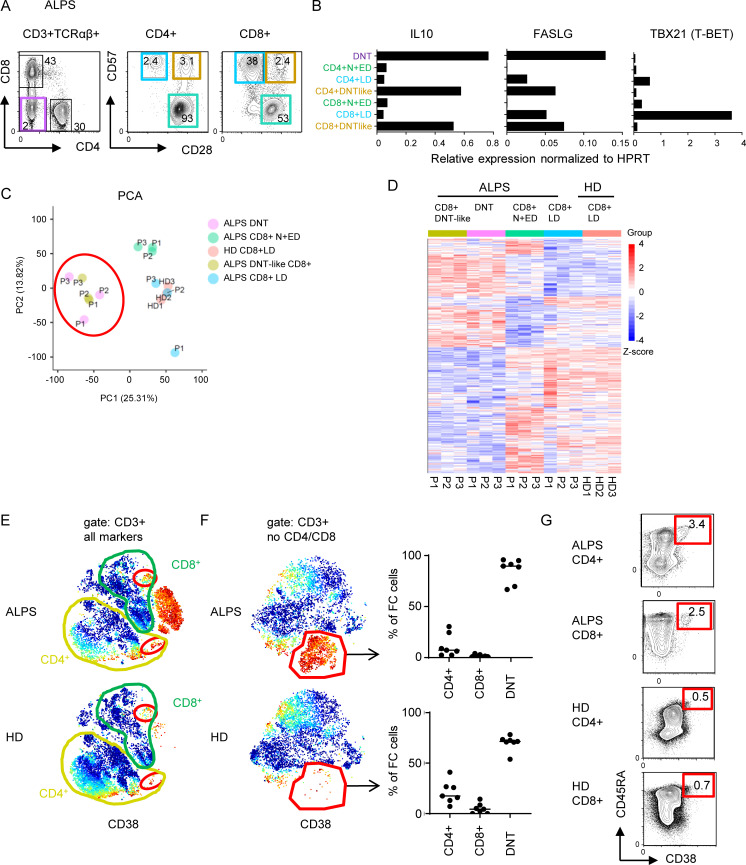
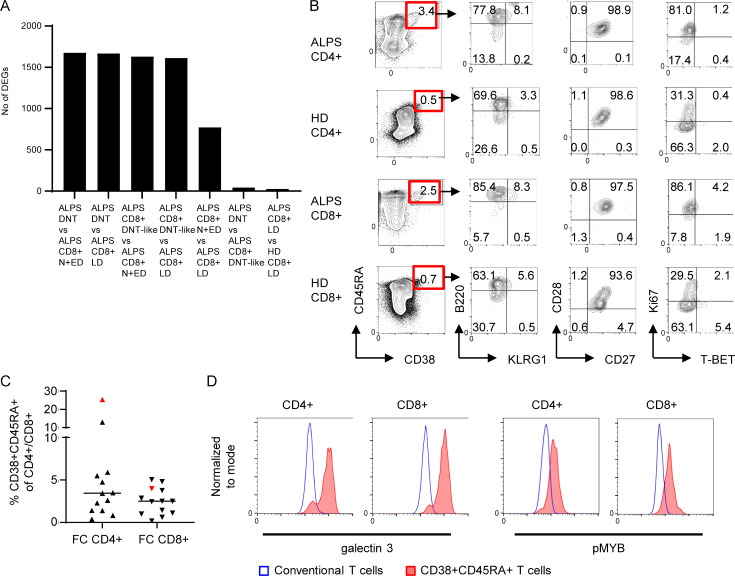
We have previously identified subsets of CD4+ or CD8+ T cells in ALPS patients with a CD28+CD57+ phenotype resembling ALPS DNTs (Rensing-Ehl et al., 2014; Fig. 5 A). Sorting and transcriptomic analysis of these DNT-like CD4+ and CD8+ T cells revealed high IL10 and FASLG and low TBX21 (T-BET) expression characteristic of FC DNTs (Fig. 5 B). Moreover, in our RNA sequencing analysis, the CD8+CD28+CD57+ “DNT-like” population clustered with ALPS DNTs upon principal component analysis with a transcriptional profile separated from DNTs by only 43 DEGs (Fig. 5, C and D; and Fig. S3 A). Small CD4+ and CD8+ T cell populations resembling FC DNTs were also detectable in our CyTOF t-SNE analysis (Fig. 5 E). To define the FC signature independent of coreceptor expression, t-SNE was performed excluding CD4 and CD8. This allowed separation of the FC T cell population into FC DNTs, FC CD4+ T cells, and FC CD8+ T cells (Fig. 5 F). The presence of CD38+CD45RA+ cells among CD4+ and CD8+ T cells with the expected B220+KLRG1−CD28+Ki67+T-BET− phenotype and overexpression of MYB and galectin 3 was confirmed by flow cytometry with frequencies up to 25% of CD4+/CD8+ T cells in ALPS patients and <1% in HDs (Fig. 5 G; Fig. S3, B–D; and data not shown). Thus, the FC T cell signature is not restricted to DNTs, and FC SPTs exist also in HDs.
Figure 5.
The FC T cell signature can be found among CD4+ and CD8+ T cells. (A) Sorting strategy for ALPS DNTs, CD4+CD28+CD57− N+EDs, CD4+CD28−CD57+ LDs, CD4+CD28+CD57+ (ALPS DNT-like CD4+), and the respective CD8+ T cell subsets for quantitative RT-PCR and/or RNA sequencing. (B) Expression levels of IL10, FASLG, and TBX21 (T-BET) in indicated T cell subsets of one ALPS-FAS patient (P1). Expression was normalized to HPRT. (C) Principal component (PC) analysis (PCA) of RNA sequencing data on DNTs, CD8+ N+EDs, CD8+ LDs, and DNT-like CD8+ of three ALPS-FAS patients (P1–P3) and CD8+ LDs of three HDs (HD1–HD3). (D) Heatmap of all genes differentially expressed (adjusted P < 0.05) in any comparison are shown for all analyzed subsets. The relative expression (Z-score) of genes is shown and color coded according to the legend. Rows are scaled to have a mean value of 0 and an SD of 1. (E) Example t-SNE plots showing all T cells and CD38 expression for one ALPS-FAS patient and one HD. Small populations among CD4+ and CD8+ T cells resembling FC DNTs are highlighted with red gates. Similar results were seen in all seven ALPS patients and all seven HDs. (F) t-SNE visualization of all T cells without CD4 and CD8 lineage markers. Example plots for one ALPS-FAS patient and one HD are shown with CD38 expression. The FC population was manually gated. Percentages of CD4+, CD8+, and DNTs within the FC cluster shown for all ALPS patients (n = 7) and all HDs (n = 7). Lines indicate medians. (G) Example plots with gating and percentages of FC CD4+ of total CD4+ and FC CD8+ of total CD8+ T cells determined by flow cytometry. Similar results were obtained in another 13 ALPS-FAS patients and 54 controls (Fig. S3 and data not shown).
Figure S3.
Small populations of CD4+ and CD8+ T cells share the FC T cell signature. (A) Number of significantly dysregulated genes (DEGs; adjusted P < 0.05) between the indicated comparisons by RNA sequencing analysis. Patient DNTs, CD8+ N+EDs, CD8+ LDs, and DNT-like CD8+ (CD8+CD28+CD57+) as well as HD CD8+ LDs were included. (B) Example FACS plots showing percentages and phenotypes of FC CD4+ and CD8+ T cells in one ALPS patient and one HD. Comparable results were generated in 9 ALPS patients and 20 HDs (data not shown). (C) Percentages of CD38+CD45RA+ cells of CD4+ T cells and of CD8+ T cells in peripheral blood (black symbols) and spleen (red symbol) of 13 ALPS patients. Lines indicate medians. (D) Representative histograms showing galectin 3 and pMYB expression in conventional and CD38+CD45RA+ T cells of one ALPS-FAS patient. Comparable results were generated in three ALPS patients.
FC T cells are present in cord blood and do not expand in the context of acute infection
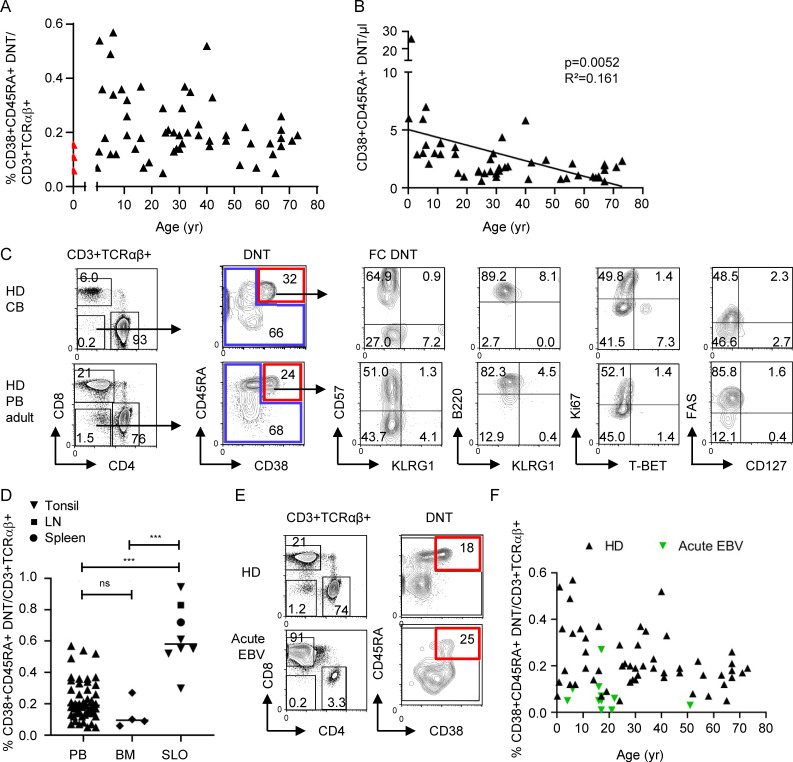
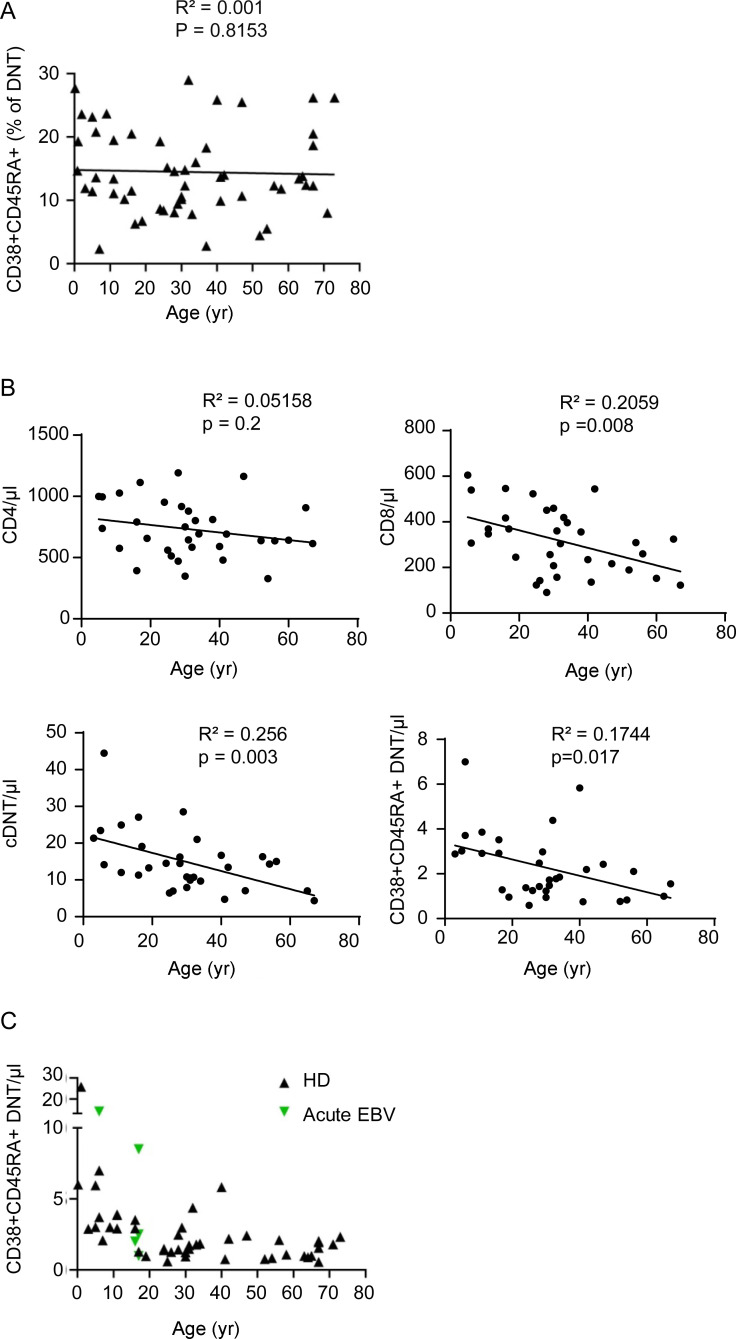
We further characterized FC T cells in healthy individuals. Frequencies of CD38+CD45RA+ FC DNTs were <1% of CD3+TCRαβ+ T cells and ranged between 2.3% and 29% (median, 13.4%) of total TCRαβ+ DNTs (Figs. 6 A and S4 A). Importantly, FC DNTs were already detectable in cord blood (Fig. 6 A). Absolute numbers in peripheral blood did not increase with age (Fig. 6 B), similarly to total CD8+ T cells and cDNTs in the same individuals (Fig. S4 B). Cord blood FC DNTs showed the characteristic B220+KLRG1−Ki67−/+EOMES+T-BET− features, while FAS expression was lower than in FC DNTs in peripheral blood in older individuals (Fig. 6 C). Moreover, FC DNT frequencies were significantly higher in secondary lymphoid organs (SLOs) than in blood and bone marrow (Fig. 6 D). During acute EBV infection, percentages and numbers of FC DNTs were not increased (Fig. 6, E and F; and Fig. S4 C). Thus, FC DNTs do not expand in the context of acute viral infection, are present before birth, do not accumulate with age, and are enriched in SLOs.
Figure 6.
Impact of age, localization, and acute infections on FC T cell pool size in HDs. (A and B) Percentages of CD38+CD45RA+ FC DNTs among CD3+TCRαβ+ T cells and absolute numbers of CD38+CD45RA+ FC DNTs/µl in peripheral blood of HDs (n = 54 and n = 47, respectively) are shown in relation to donor age in years. Values determined in HD cord blood are depicted in red. Solid lines represent linear regression analyses. R2 and P values are shown. (C) Example plots showing percentages and phenotypes of CD38+CD45RA+ FC DNTs in cord blood (CB; n = 3) compared with adult peripheral blood (PB; n = 10). (D) Percentages of CD38+CD45RA+ FC DNTs of CD3+TCRαβ+ T cells in HD PB (n = 54, age range the same as for A) compared with adult SLOs (n = 8) and adult bone marrow (BM; n = 4). Lines indicate medians. Statistical comparisons were performed using the Kruskal-Wallis test. ns, not significant; ***, P < 0.001. (E) Example plots showing percentages of CD38+CD45RA+ FC DNTs in one adult HD and one patient with acute EBV infection. (F) Summary plot depicting percentages of CD38+CD45RA+ FC DNTs of CD3+TCRαβ+ T cells in peripheral blood of HDs (n = 54) versus patients with acute EBV infection (n = 11) in relation to donor age.
Figure S4.
Frequencies of FC DNTs in peripheral blood of HDs. (A) Percentages of CD38+CD45RA+ cells among TCRαβ+ DNTs in peripheral blood of HDs are shown related to age in years. (B) Absolute values of CD4+, CD8+, cDNTs, and CD38+CD45RA+ DNTs in peripheral blood of HDs related to age. Solid lines represent linear regression analyses. R2 and P values are shown. (C) Absolute values of CD38+CD45RA DNTs/µl in peripheral blood of HDs (n = 43) versus patients with acute EBV infection (n = 5) related to age.
FC T cells in healthy individuals show increased susceptibility to FASL-induced apoptosis, and their survival is mTOR dependent
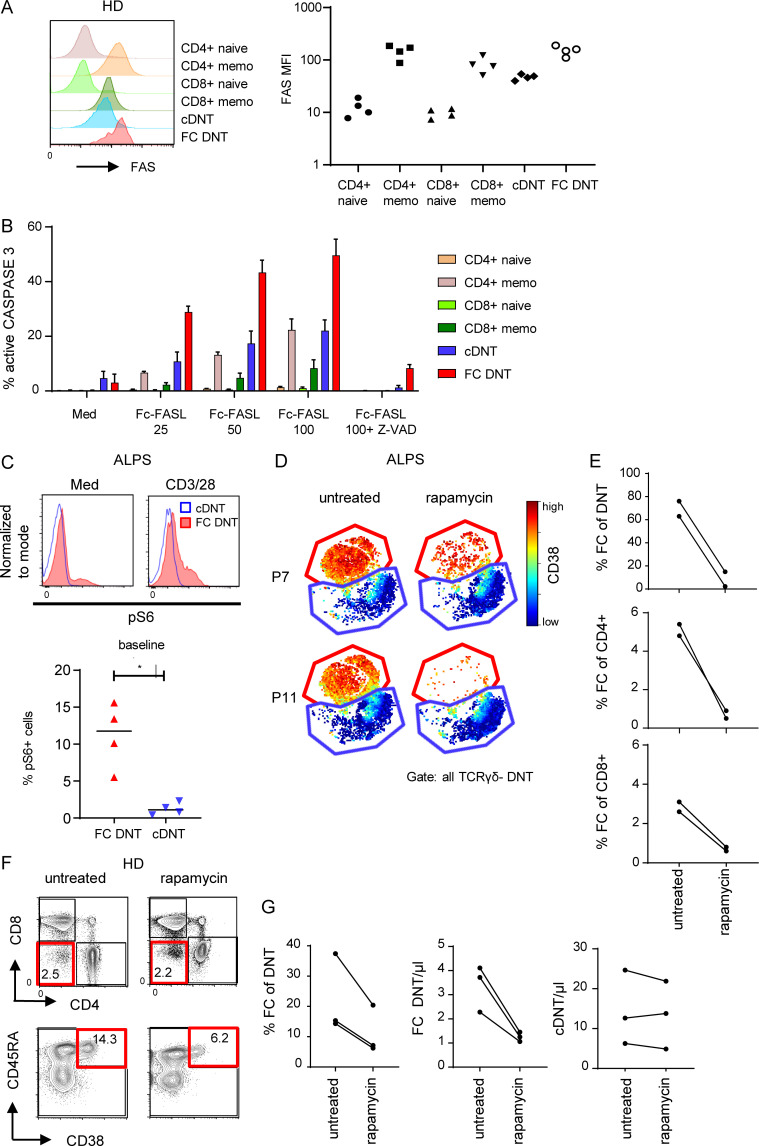
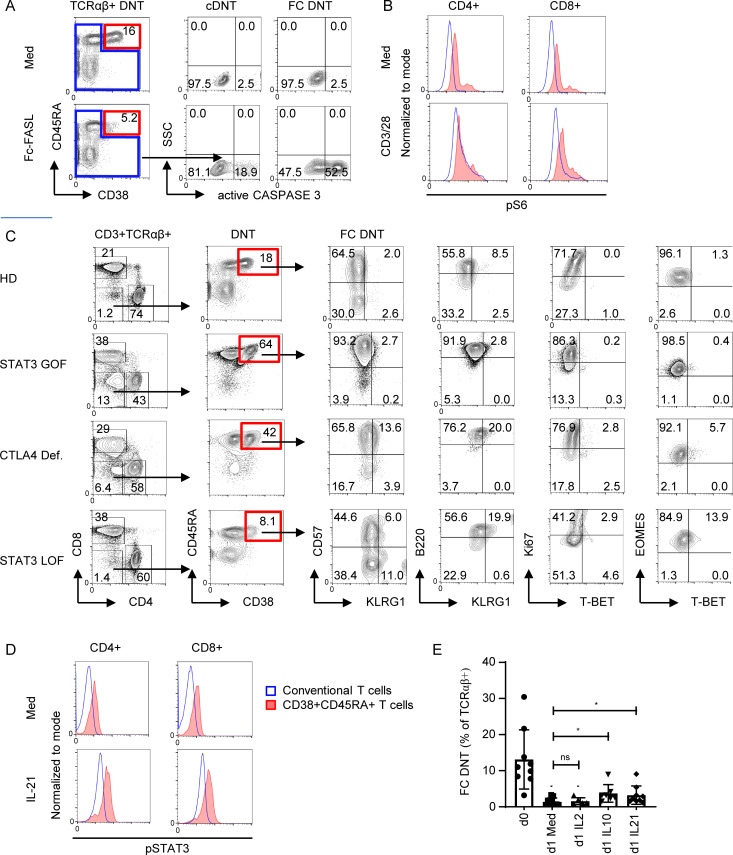
To address the molecular mechanisms involved in generation and maintenance of FC T cells, we first compared their sensitivity to FASL-induced apoptosis relative to other T cell populations. FC DNTs of HDs expressed FAS (Fig. 7 A) at levels similar to memory T cells but were more sensitive to FASL-induced apoptosis than the other T cell subsets, including cDNTs (Figs. 7 B and S5 A). This was not due to altered expression of the anti- and proapoptotic molecules BCL-2 and BIM (data not shown). We then analyzed whether the mTOR dependence previously described for ALPS DNTs (Völkl et al., 2016) is a characteristic and specific feature of FC T cells. Indeed, FC DNTs, but not cDNTs, showed increased baseline and anti-CD3/CD28–stimulated phosphorylation of S6 (Fig. 7 C), and this was confirmed in FC CD4+ and FC CD8+ T cells (Fig. S5 B). Moreover, CyTOF analysis revealed that rapamycin treatment leads to a selective decrease of FC T cells in ALPS patients (Fig. 7, D and E). Notably, the signature of the remaining FC DNTs was unchanged, indicating that mTOR does not drive their unconventional differentiation. Importantly, rapamycin given for other indications to immunologically healthy individuals also led to selective decrease of FC DNTs (Fig. 7, F and G). Thus, the FC T cell population is characterized by a selective hypersensitivity to FASL-induced apoptosis, and its maintenance is dependent on mTOR signaling.
Figure 7.
FC T cells are highly sensitive to FASL and depend on mTOR signaling. (A) Representative histogram showing FAS expression on CD4+ naive (CD45RA+CD57−), CD4+ memory (CD45RA−CD57+), CD8+ naive (CD45RA+CD57−), and CD8+ memory T cells (CD45RA−CD57+) and FC DNTs and cDNTs (CD38 and CD45RA used for gating). Summary graph showing FAS expression (median fluorescence intensity [MFI]) on annotated T cell subsets for four adult HDs. (B) Summary graph showing percentages of active caspase 3–positive T cells among annotated T cell populations after stimulation with increasing concentrations of Fc-FASL (ng/ml) in the presence or absence of the pan-caspase inhibitor Z-VAD determined in four HDs. Mean and SEM values are shown. (C) Representative plots showing pS6 in FC DNTs compared with cDNTs ex vivo and after anti-CD3/CD28 stimulation in one ALPS patient. Similar results were obtained in four ALPS patients. Summary plot depicting the percentages of pS6-positive cells among ex vivo FC DNTs and cDNTs of four ALPS patients. Lines indicate medians. Statistical comparison was performed using the Mann-Whitney U test. *, P < 0.05. (D) t-SNE plots representing TCRγδ− DNTs of two ALPS patients analyzed before and during treatment with rapamycin. (E) Summary graphs showing percentages of FC DNTs of total TCRαβ+ DNTs, FC CD4+ of total CD4+ T cells, and FC CD8+ of total CD8+ T cells in the two ALPS patients before and during rapamycin treatment. (F) Representative plots showing FC DNTs of one immunologically healthy individual before and during rapamycin treatment. (G) Summary plots showing percentages of FC DNTs of total TCRαβ+ DNTs and FC DNTs or cDNT absolute numbers of the three immunologically healthy individuals before and during rapamycin treatment.
Figure S5.
FC DNTs are regulated by FAS but also by mTOR, STAT3, and CTLA4. (A) Example plots showing proportions of active caspase 3–positive cells among FC DNTs and cDNTs after ex vivo treatment with recombinant Fc-FASL. Comparative results were seen in cells from four different HDs. (B) Representative plots of pS6 levels in FC CD4+ and FC CD8+ (CD38+CD45RA+; red) versus conventional CD4+ and CD8+ T cells (blue) of one ALPS-FAS patient with or without anti-CD3/CD28 stimulation. Comparable results were achieved using cells of three additional ALPS patients. (C) Example FACS plots showing percentages and phenotypes of CD38+CD45RA+ FC DNTs in patients with STAT3-activating mutation (STAT3 GOF), CTLA4 deficiency (CTLA4 Def.), and STAT3 loss of function mutation (STAT3 LOF). Additional patients (three with STAT3 GOF, three with CTLA-4 Def., and two with STAT3 LOF) were analyzed, with similar results found (data not shown). Numbers indicate percentages. (D) Representative plots of pSTAT3 levels in FC CD4+ and FC CD8+ of one ALPS-FAS patient with or without IL-21 stimulation. Analyses were performed in seven ALPS patients with similar results. (E) Summary plots showing the percentage of viable FC DNTs (CD38+CD45RA+) of total TCRαβ+ DNTs ex vivo and after 24-h incubation with or without the indicated cytokines. Levels of significance by mixed-effects analysis and Tukey’s multiple comparisons test. ns, not significant; *, P < 0.05.
CTLA4 and STAT3 contribute to the regulation of FC T cells
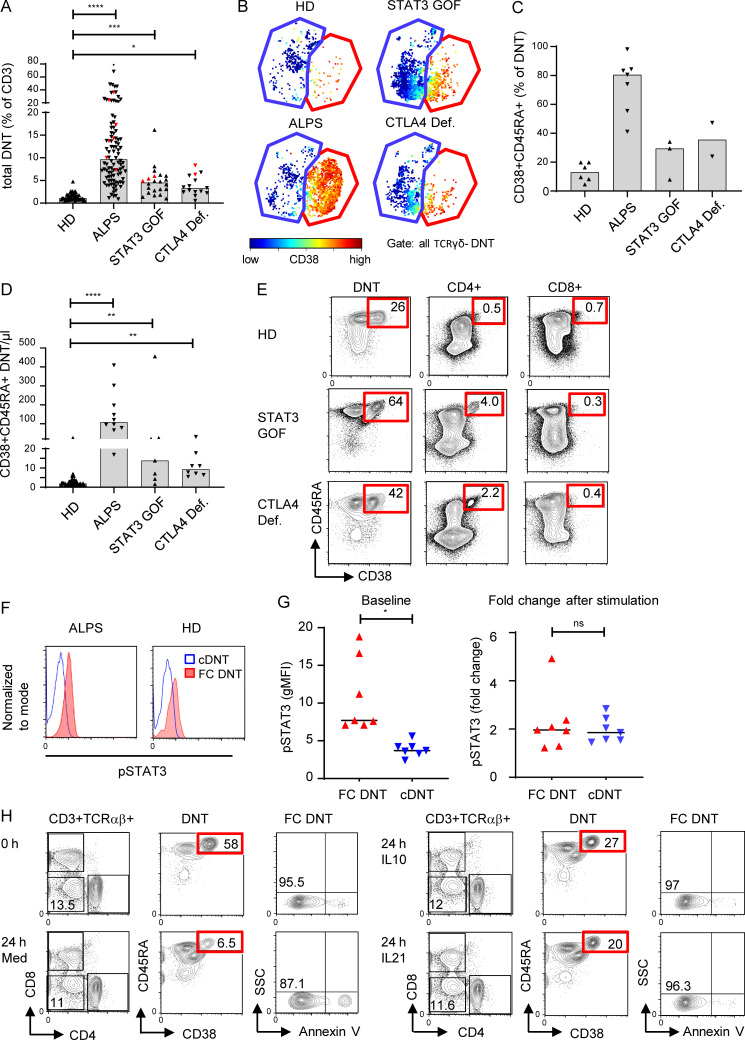
To further unravel the signals driving FC T cells, we followed the observation that TCRαβ+ DNTs can also be elevated in patients with activating STAT3 mutations or CTLA4 deficiency (Besnard et al., 2018; Milner et al., 2015; Nabhani et al., 2017; Fig. 8 A). We therefore analyzed T cells of such patients with DNTs >5% by CyTOF and found that FC DNTs with the characteristic phenotype were expanded (Fig. 8, B and C). Their absolute numbers were significantly raised compared with HDs in both diseases (Fig. 8 D). FC DNTs were not expanded in four patients with STAT3 loss-of-function mutations causing hyper-IgE syndrome (Fig. S5 C). Notably, FC CD4+ T cells were also increased in some patients with STAT3 gain of function (GOF) and CTLA4 deficiency (Fig. 8 E). While a role for CTLA4 in controlling DNT in lpr mice has been described (Takiguchi et al., 2000), the role of STAT3 signals is less clear. We found that ex vivo phosphorylated STAT3 (pSTAT3) levels were increased in FC T cells compared with cDNTs and total CD4+ or CD8+ T cells, indicating active STAT3 signaling in vivo (Figs. 8 F and S5 D). FC DNTs from HDs further increased STAT3 phosphorylation levels after stimulation with STAT3-activating cytokines in vitro, demonstrating full responsiveness (Fig. 8 G). Importantly, addition of the STAT3-activating cytokines IL-10 or IL-21 prolonged survival of FC DNTs, although it could not prevent the rapid ex vivo apoptosis known to occur in the majority of FC T cells (Figs. 8 H and S5 E). Thus, mechanistically, FC T cells are tightly controlled by FAS and CTLA4, while their survival is enhanced by mTOR and STAT3 signals.
Figure 8.
FC DNTs are regulated by CTLA4 and STAT3. (A) Percentages of total TCRαβ+ DNTs in whole blood of HDs and indicated disease groups (ALPS, n = 99; STAT3 GOF, n = 22; CTLA4 deficiency [Def.], n = 15). Values of patients included in the CyTOF analysis are highlighted with red symbols (ALPS, n = 7; STAT3 GOF, n = 3; CTLA4 Def., n = 2). Columns indicate median values. Statistical comparisons were performed using the Kruskal-Wallis test. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001. (B) Example t-SNE plots of all DNT (CD3+TCRγδ−) showing FC DNTs (red gate) and cDNTs (blue gate) and corresponding CD38 expression for indicated patient groups. (C) Percentages of FC DNTs among total DNTs as determined by CyTOF for the indicated patient groups. Columns indicate mean values. (D) Absolute numbers of FC DNTs according to CD38/CD45RA expression in whole blood are depicted for indicated patient groups (ALPS, n = 10; STAT3 GOF, n = 7; CTLA4 Def., n = 8). Columns indicate median values. Statistical comparisons were performed using the Kruskal-Wallis test. **, P < 0.01; ****, P < 0.0001. (E) Example plots showing percentages of CD38+CD45RA+ FC T cells among total TCRαβ+ DNTs and CD4+ and CD8+ T cells by flow cytometry in indicated disease groups. Similar results were obtained in 10 HDs, 3 patients with STAT3 GOF, and 3 patients with CTLA4 Def. (F) Representative histograms showing pSTAT3 in FC DNTs and cDNTs of one ALPS patient and one HD. Comparable results were obtained for six additional ALPS patients. (G) Summary plots showing baseline geometric mean fluorescence intensity (gMFI) levels for pSTAT3 in FC DNTs and cDNTs (left) and fold changes of pSTAT3 after stimulation with IL-21 (right) in cells of seven HDs. Statistical comparisons were performed using the Wilcoxon test. ns, not significant; *, P < 0.05. (H) Flow cytometry plots showing percentages of viable (annexin V−) FC DNTs of one ALPS patient at day 0 and after 24 h of stimulation with medium alone or with medium supplemented with IL-10 or IL-21. Data were generated in five to nine ALPS patients with similar results (Fig. S5 E). SSC, side scatter.
Discussion
Starting from a detailed characterization of DNT cells in ALPS patients, we describe a previously unrecognized physiological population of highly proliferative IL-10–producing T cells. This T cell population includes CD4+, CD8+, and DNT cells that share a unique molecular signature separating them from other well-defined T cell populations, including cDNTs and virus-activated T cells. They are generated before birth, are enriched in SLOs, do not expand in the context of acute viral infection, and are exceptionally sensitive to FAS-mediated apoptosis. Functionally, FC T cells represent highly proliferative, noncytotoxic T cells with elevated IL-10 expression. Mechanistically, their survival and expansion is controlled by FAS, CTLA4, mTOR, and STAT3 signaling. Our findings move these FC cells from the niche of aberrant coreceptor deficient T cells expanded in a rare disease to basic T cell immunology.
A first important finding of our study is the discrimination of two major DNT subsets, which we initially observed in ALPS patients and subsequently also detected in healthy individuals and in other patients with primary immunodeficiencies. This finding resolves the previously observed phenotypic differences between bulk TCRαβ+ DNTs of HDs and of patients with ALPS or other primary immunodeficiencies (Bleesing et al., 2002; Rensing-Ehl et al., 2014; and unpublished data): These differences reflect variable proportions of the two DNT subsets rather than different phenotypes of the same DNT population or different disease-specific DNT subsets. Notably, for diagnostic purposes, the two DNT populations can be distinguished by only two markers, CD38 and CD45RA. ALPS-FAS is unlikely if the proportion of CD38+CD45RA+ cells among DNTs is <25%. However, higher percentages are not specific for ALPS-FAS and can also occur, for example, in patients with activating STAT3 mutations or CTLA4 deficiency.
FC DNTs clustered separately from other T cell populations independent of inclusion or exclusion of CD4/CD8 in the t-SNE analysis, indicating their phenotypic difference from other T cell populations. In contrast, the more heterogeneous cDNTs were distributed among CD8+ T cells and TCRγδ+ cells and had a similar RNA expression pattern suggesting a possible CD8+ lineage relationship. In support of these data, cDNTs expressed IFNγ and cytolytic molecules similar to CD8+ T cells. Furthermore, three subclusters could be generated for cDNTs that could be aligned with differentiation patterns also described for conventional CD8+ T cells: naive (cluster 4: CD45RA+CD38−CD28+CD57−), effector memory (cluster 5: CD45RA−CD38−CD28intermediateCD57intermediate) and terminally differentiated T cells reexpressing CD45RA (cluster 3: CD45RA+CD38lowCD28−CD57+; Appay et al., 2008). In contrast, the two subclusters generated for FC DNTs shared all unconventional features, including combined expression of CD45RA+CD38+. The highest variability was seen in CD57 expression, which reflects the degree of glycosylation and/or number of cell divisions (Brenchley et al., 2003) and thus a more or less advanced differentiation state of the same population (Fig. 2 D). Overall, discrimination between FC DNTs and cDNTs was based not only on transcriptome and phenotype but also on transcription factor and effector molecule expression, STAT3 and mTOR signaling, and susceptibility to rapamycin.
The next unexpected finding was that FC DNTs are present in healthy individuals. The concept of signature identity between the prominent FC DNTs in ALPS patients and the small fraction of CD38+CD45RA+ DNTs in healthy individuals was initially based on CyTOF and flow cytometric analysis revealing the unconventional CD38+CD45RA+B220+KLRG1−CD127− differentiation pattern and unique transcription factor profile (MYB+OBF1+EOMES+T-BET). However, the transcriptional differences upon subsequent high-resolution RNA sequencing analysis were greater than expected and raised the question whether FC DNTs in patients with ALPS and HDs are different cell types or whether they represent variable differentiation states of the same cell type. We postulate cellular identity based on the following arguments: The shared dysregulated genes include the vast majority of genes characteristic of ALPS FC DNTs relative to other resting or in vivo activated T cells (acute EBV infection). Dysregulation of these genes not only is based on transcriptome data but also has largely been validated by quantitative PCR and/or protein expression. They include unique differentiation markers or unconventional marker combinations that have not been described in any other known T cell differentiation subset, a unique transcription factor profile, constitutive mTOR and STAT3 activation, proliferative behavior, characteristic effector molecule expression, and high sensitivity to FAS-mediated apoptosis induction. These features likely represent the “core signature” of FC T cells. Differences between the two populations mainly affected genes involved in cell cycle progression, glycolysis, and mTOR signaling. They likely reflect prolonged survival, activation, and uncontrolled proliferation of ALPS FC DNTs compared with their HD counterparts in the context of defective FAS signaling. Thus, although FC T cell fate determination occurs in a FAS-proficient environment with acquisition of key features already before birth, the differences between ALPS and HD FC DNTs indicate that lack of FAS-mediated apoptosis (and possibly also of FAS-mediated nonapoptotic signaling) prolong hyperactivation and uncontrolled proliferation, favoring further changes in transcription and protein expression.
Another key finding of our study is that the FC T cell signature is shared by subsets of CD4+ and CD8+ T cells in ALPS patients and HDs and therefore represents a cellularly and molecularly defined differentiation state that is not linked to CD4 or CD8 identity. High-resolution analysis revealed that the signature of FC SPTs and FC DNTs in ALPS patients is virtually identical. This may indicate that transcriptional downregulation of CD4/CD8 coreceptors is not an early event in FC T cell ontogeny, but a late differentiation step for some if not all FC T cells. It has been controversially debated for decades of murine and human research whether ALPS/lpr DNTs differentiate from chronically activated conventional T cells when sufficient FAS signals are lacking or whether they represent a rare physiological T cell subset that is efficiently eliminated in FAS-competent individuals and therefore has so far escaped detection (Bristeau-Leprince et al., 2008; Fortner et al., 2011; Pestano et al., 1999; Russell et al., 1993; Trimble et al., 2002; Mohamood et al., 2008; Hughes et al., 2008). Our data prove that these proliferating CD38+CD45RA+IL10+ FC T cells represent a physiological subset of the human T cell compartment that evolves independent of the presence or absence of intact FAS signaling. Moreover, the clear transcriptional difference to late differentiated CD57+CD8+ T cells as well as their separation upon clustering analysis renders the concept of transition from terminally differentiated conventional T cells highly unlikely.
FC T cells were already detectable in cord blood of HDs, and numbers slightly declined with age. Clinically, these findings are consistent with onset of lymphoproliferation in utero in patients with biallelic FAS mutations and reduction of lymphoproliferation in ALPS patients beyond adolescence (Price et al., 2014). They imply that FC T cells are generated and achieve their distinct transcriptional program prenatally, are maintained under homeostatic control by FAS, and decrease in parallel with age-dependent thymic involution. FC DNTs do not expand in the context of viral infection, indicating that they are poorly reactive to conventional stimulation or do not include antiviral TCR specificities.
The signature of FC T cells comprised expression of a unique combination of transcription factors. MYB, DNA-binding protein inhibitor ID3, and EOMES, all elevated in FC T cells, promote stemness and genome stability and further support high proliferative potential (Gautam et al., 2019; Ji et al., 2011; Intlekofer et al., 2005; Kaech and Cui, 2012). Of note, MYB also restrains effector differentiation via repression of the transcription factor ZEB2, which was indeed significantly down-regulated (log2 fold change, −4.4) in ALPS DNTs compared with LD CD8+ T cells and EBV-activated CD8+ T cells (not shown). FC T cells also lack T-BET, another driver of effector differentiation. The absence of T-BET in FC T cells was unexpected because T-BET upregulation has been linked to T cell activation and mTOR signaling (Intlekofer et al., 2005; Joshi et al., 2007; Rao et al., 2010). Indeed, all in vivo activated CD8+ T cell subsets, both effector and exhausted, highly up-regulated T-BET and its downstream target KLRG1 (Fig. 4 and data not shown). Interestingly, there was no heterogeneity among FC T cells regarding their EOMEShigh/T-BET− pattern, indicating early imprinting of this profile. Increased S6 phosphorylation and mTOR dependence, previously demonstrated for total ALPS DNTs (Teachey et al., 2009; Völkl et al., 2016), are selective features of FC T cells also in HDs. Consistently, cDNTs are not affected by rapamycin treatment of ALPS patients. Moreover, FC DNTs were also specifically reduced in otherwise healthy individuals treated with rapamycin for vascular malformations. Interestingly, in both ALPS patients and immunologically healthy subjects, the signature of the FC DNTs detectable under rapamycin was unchanged based on our CyTOF marker panel, indicating that rapamycin eliminates mTOR-dependent FC T cells but does not revert key features of their unconventional cellular program. This implies that mTOR promotes expansion of FC T cells but not FC fate determination.
Finally, our study provides insights into the molecular regulation of FC T cell homeostasis. FAS plays the key role in regulating their pool size. It is striking that among all FAS-expressing T cells, FC T cells are most sensitive to FAS-mediated apoptosis. This can be seen in vitro but is even more evident in vivo, where FC T cells constitute the great majority of accumulating cells in ALPS patients. Susceptibility to FAS-mediated apoptosis is regulated at multiple levels, including lipid raft localization, death-inducing signaling complex (DISC) composition, cellular FLICE (FADD-like IL-1β–converting enzyme)-inhibitory protein (c-FLIP) levels, expression of antiapoptotic molecules, activation and cell cycle status, metabolism, and signaling context (Buchbinder et al., 2018; Gajate and Mollinedo, 2015; Gulbins et al., 1996; Larsen et al., 2017; Wajant, 2014). FC T cells are highly activated and proliferating, both factors known to render T cells more susceptible to FAS-induced apoptosis, while BCL-2 and BIM expression levels were not different between HD FC DNTs, cDNTs, and memory CD4+/CD8+ T cells (not shown). The decisive factors determining enhanced FAS apoptosis sensitivity beyond features associated with their highly activated status remain to be defined but could involve the many uniquely dysregulated signal-modulating molecules. We show that STAT3 is another important regulator of FC T cell homeostasis. Baseline STAT3 phosphorylation in FC T cells of HDs is increased ex vivo. Moreover, IL-10 and IL-21 stimulation can prolong survival of FC T cells in short-term in vitro cultures. Consistently, we observed increased FC T cells in patients with activating STAT3 mutations. Notably, decreased sensitivity to FAS-mediated apoptosis has been reported in single patients with STAT3-activating mutations (Milner et al., 2015; Nabhani et al., 2017), suggesting that apart from enhancing survival, STAT3 signaling may contribute to FC T cell expansion by impacting FAS signaling. Finally, the increase of FC T cells in patients with CTLA4 deficiency indicates that CD28 also plays a role in FC T cell expansion. This is supported by the significant reduction of absolute DNT numbers after treatment of neonatal lpr mice with abatacept (Takiguchi et al., 2000).
Why are FC T cells generated, and what is the function of these cells? It has been proposed that Fas-regulated cells represent “useless” mis-selected CD8+ T cells that lack peripheral inhibitory peptide MHC interactions (Pestano et al., 1999) or potentially autoreactive T cells homeostatically proliferating in response to limited self-peptide–MHC contacts (Fortner et al., 2011; Trimble et al., 2002). However, it is not obvious why such “useless” cells should be so highly proliferative and secrete copious amounts of cytokines. Moreover, we found that they lack CD25 and IL-7 receptor α chain expression and do not respond to the homeostatic cytokines IL-7 and IL-15 in vitro (data not shown). Alternatively, they may represent T cells with suppressive function destined to populate nonlymphoid organs (Martina et al., 2016; Mohamood et al., 2008). Proliferation and IL-10 production are key features of FC T cells, while they do not express IFNγ and cytolytic molecules. Unfortunately, human FC T cells very rapidly die ex vivo, rendering their functional assessment difficult. The better definition of their molecular signature and the demonstration of their presence in healthy individuals in this study will spur interest in further exploration of the ontogeny and biological function of this unusual T cell population.
Conceptually, the view of FC T cells emerging from our data shares interesting parallels with “rogue” B cells: murine studies showed that Fas is dispensable for negative selection of conventional self-reactive B cells but selectively eliminates unconventional “rogue” B cells that undergo somatic hypermutation, survive independently of antigen, and develop into (auto)antibody-producing plasma cells (Butt et al., 2015). These “rogue” B cells represent a small, proliferating B cell population in wild-type mice tightly controlled by Fas. By analogy, Fas does not play a major role in the regulation of conventional virus-reactive T cells in vivo (Ehl et al., 1996; Zimmermann et al., 1996), while it controls a small proliferating T cell population. Hence, FC T cells may represent “rogue” T cells, potentially sharing key features with their B cell counterparts. A better understanding of these unusual cells and their differentiation program is relevant because their uncontrolled proliferation is associated with lymphoproliferative disease and a high incidence of autoimmunity, as illustrated in ALPS patients.
Materials and methods
Patients and HDs
42 patients with ALPS-FAS/sFAS, GOF STAT3 mutations, CTLA4 deficiency, or loss-of-function STAT3 mutations were included. Table S1 summarizes genetic and clinical characteristics. All patients were not receiving immunosuppressive treatment at the time of analysis, except for P23 (under low-dose rapamycin) and P28 (under low-dose cyclosporine and tocilizumab). Two ALPS patients (P7 and P11) were analyzed before and under rapamycin treatment (rapamycin levels in blood 3.2 and 3.7 ng/ml, respectively). 11 individuals (mean age, 18 [4–51] yr) with acute primary EBV infection and 54 HDs (i.e., without personal or family history of immunological disorders and normal blood counts; mean age, 32 [0–73] yr) were also studied. Three immunologically healthy individuals who started rapamycin treatment for vascular malformations were also included in the study. Rapamycin levels in blood ranged from 3.8 to 9.2 ng/ml. All participants gave informed consent. The study was conducted according to the Declaration of Helsinki and approved by ethics committees of the University of Freiburg (protocol numbers 409/16, 282/11, 610/15, 147/15, 174/13, 121/11, 251/13) and the University of Erlangen (protocol number 219_14 B).
Flow cytometry and cell sorting
Flow cytometry was performed with peripheral blood, buffy coats, cord blood, and lymphoid organ samples (Table S2). Antibodies used are summarized in Table S3. CD38 and CD45RA expression was measured in all HDs, and comparable expression patterns of B220, CD57, and KLRG1 and of CD27, CD28, HLA-DR, Ki67, EOMES, and T-BET (shown in Fig. 3 A) were confirmed in 20 and 10 HDs, respectively. For intranuclear staining, the Human Regulatory T Cell Staining Kit (eBioscience) was used. To measure cytokine production, 4 × 106 PBMCs were stimulated with PMA (Sigma-Aldrich; 50 ng/ml) and ionomycin (Sigma-Aldrich; 1 µg/ml) for 4 h and then analyzed using the Cytofix/Cytoperm Plus Kit and Golgi Plug (BD Biosciences). Similarly, IL-10 production was determined after 5 h of PMA/ionomycin stimulation as described elsewhere (Haug et al., 2019). S6 S240 phosphorylation was analyzed using Phosflow reagents as described previously (Völkl et al., 2016). pSTAT3 was measured using the PerFix Expose Kit (Beckman Coulter) and analyzed with or without cytokine stimulation for 15 min (100 ng/ml IL-21; Miltenyi Biotec). Data were acquired with a Gallios or Navios flow cytometer (Beckman Coulter). Data were analyzed using FlowJo 7.2.5 or 10.4 software (FlowJo LLC). Cell populations were either magnetically enriched using the Double-negative T cell Isolation Kit (Miltenyi Biotec) or sorted with the MoFlo Astrios (Beckman Coulter) or FACS Aria Fusion (Becton Dickinson) cell sorter.
Ex vivo apoptosis assay
3 × 106 PBMCs per well were seeded into 12-well plates and stimulated with increasing concentrations of recombinant Fc-FASL (Holler et al., 2003) for 2 h. Some cells were pretreated with the pan-caspase inhibitor Z-VAD-FMK 50 µM (Sigma-Aldrich) for 30 min before Fc-FASL treatment. Early apoptotic cells were determined by flow cytometry after intracellular staining of active caspase 3 (BD Biosciences).
Survival assay
2 × 105 PBMCs per well were seeded into 96-well plates and stimulated either with medium alone or with medium plus IL-2 (100 U/ml; R&D Systems), IL-21 (100 ng/ml; Miltenyi Biotec), or IL-10 (100 ng/ml; R&D Systems) for 24 h. Survival was measured by flow cytometry, and annexin V staining was performed using the annexin V binding buffer (BD PharMingen).
CyTOF
Sample preparation was performed as described previously (Fuchs et al., 2019) with antibodies listed in Table S4. A Helios mass cytometry instrument (Fluidigm) was used for sample acquisition. Data analysis was performed with Cytobank software with t-SNE for data visualization (Amir et al., 2013). UMAP version 2.1 (Becht et al., 2019) and FlowSOM (Van Gassen et al., 2015) were used as plugins for FlowJo version 10.4. Detailed information for figures depicting CyTOF data are provided in Table S5 (vi-SNE) and Table S6 (UMAP).
RNA sequencing analysis
DNT and CD8+ T cell populations of ALPS-FAS patients (P1 and P2: peripheral blood; P3: spleen) and blood of three HDs (HDs 1–3) were FACS sorted on the basis of CD28 and CD57 expression. In a second RNA sequencing experiment, DNT and CD8+ T cell subsets of three ALPS-FAS patients (P3: spleen; P6 and P22: peripheral blood) and three HDs (HDs 4–6: peripheral blood buffy coats) were sorted on the basis of CD28, CD57, CD38, and CD45RA expression. Because RNA amounts of HD FC DNTs were very low, we pooled RNA from four different HDs for the HD5 sample (HD5*). HD6 FC DNTs had to be excluded from the analysis because read numbers were too low. RNA sequencing steps included RNA isolation (Qiagen RNeasy Micro Kit), RNA quality analysis (2100 Bioanalyzer; Agilent Technologies), creation of barcoded RNA sequencing libraries (from 100 ng total RNA with Ovation Human RNA-Seq system; NuGEN), and depletion of ribosomal RNAs (insert-dependent adaptor cleavage method; NuGEN) as described previously (Eisenhut et al., 2016; Müller et al., 2017). To control sources of variability, a common set of external spike-in RNA controls (by the external RNA controls consortium) was used (Ambion). Libraries were subjected to single-end sequencing (101 bp) on a HighSeq 2500 platform (Illumina). Reads mapping to, for example, ribosomal RNAs, transfer RNAs, small nuclear RNAs, and interspersed repeats were first filtered out using bwa-mem version 0.7.8-r455 (Li, 2013), keeping only unmapped reads. Subsequently, reads were mapped against the hg19 reference genome with STAR aligner version 2.4.0j (Dobin et al., 2013) and STAR genome directory created by supplying an Ensembl gtf annotation file (version 2013-09) for hg19. Absolute read counts per gene were produced using Subread’s featureCounts program version 1.4.6-p2 (Liao et al., 2014) and the Ensembl gtf annotation file. Subsequent analyses were performed using R 3.2.1 (R Core Team, 2015). Genes with low expression (total raw counts across the 15 samples ≤5) were filtered out, and differential expression analysis was performed with the DESeq2 package version 1.8.1 (Love et al., 2014). Significantly regulated genes were selected based on their adjusted P value (Benjamini and Hochberg) <0.05.
Publicly available RNA sequencing data (Schmiedel et al., 2018; https://dice-database.org/landing) from sorted human regulatory FOXP3+ T cell subsets (naive T reg cells: CD3+CD4+CD25hiCD127lowCD45RA+; memory T reg cells: CD3+CD4+CD25hiCD127lowCD45RA−) were used to analyze expression of genes found dysregulated in ALPS DNTs in human T reg cells versus T naive (CD3+CD4+CD45RA+CCR7+).
Gene set enrichment analysis was performed using GAUGE R package version 2.36 (Luo et al., 2009) with MSigDB gene sets version 7.0 (Subramanian et al., 2005). An adjusted P value <0.05 was considered significant.
Quantitative RT-PCR
RNA was isolated with the RNeasy Mini Kit (Qiagen). For cDNA synthesis, the qScript cDNA Supermix (Quantabio) or the PrimeScript RT Reagent Kit (Takara Bio) was used. RT-PCR was performed using the Mastercycler Realplex instrument (Eppendorf) and FastStart Universal SYBR Green Master (ROX; Roche). Gene expression was normalized to hypoxanthine phosphoribosyltransferase (HPRT) and calculated with the 2−ΔCt method.
Statistical analysis
Statistical analyses were performed with GraphPad Prism software. t Tests (Mann-Whitney U tests), the Wilcoxon test or ANOVA, the Kruskal-Wallis test for multiple comparisons, and mixed-effects analysis with Tukey's multiple comparisons test were applied to compare T cell marker expression or frequencies.
Data-sharing statement
RNA sequencing data are available under Gene Expression Omnibus accession no. GSE154929.
Online supplemental material
Fig. S1 shows additional transcriptional features of ALPS DNTs and a comparison to T reg cells. Fig. S2 shows UMAP projections of the main T cell populations in ALPS patients versus HDs and a comparison of transcript levels in ALPS FC DNTs versus HD FC DNTs. Fig. 3 shows transcriptional and phenotypic features of FC CD4+ and FC CD8+ T cells as well as their percentages in ALPS patients. Fig. S4 reports percentages and absolute numbers of different T cell populations in HDs related to age. Fig. S5 shows representative plots of active caspase 3 staining, pS6 in FC CD4/CD8+ T cells, FC DNT phenotype in patients with different primary immunodeficiencies, and the impact of STAT3-activating cytokines on STAT3 phosphorylation and survival of FC T cells. Table S1 depicts clinical, immunological, and genetic features of included patients. Table S2 reports the sources of SLO and bone marrow samples. Table S3 lists flow cytometry antibodies. Table S4 lists antibodies used for CyTOF. Table S5 and Table S6 show CyTOF analysis information for t-SNE and UMAP visualizations, respectively.
Supplementary Material
depicts clinical, immunological, and genetic features of included patients.
reports the sources of SLOs and bone marrow samples.
lists the flow cytometry antibodies.
lists the CyTOF antibodies.
provides CyTOF analysis information for t-SNE.
lists CyTOF analysis information for UMAP.
Acknowledgments
We are grateful to the patients and their families who made this study possible. We thank the Center for Chronic Immunodeficiency Advanced Diagnostics Unit, the Center for Chronic Immunodeficiency Biobank as a partner of the FREEZE Biobank, and the Lighthouse Core Facility for excellent support. We thank Francesca Oprandi for figure processing.
This work was funded by the Wilhelm Sander Stiftung (1 027 087 401) and the German Federal Ministry of Education and Research (grant BMBF 01 EO 0803 to the Center of Chronic Immunodeficiency and grant BMBF 01GM1111B to the PID-Net Initiative) and was supported within the framework of the e:Med research and funding concept CoNfirm (FKZ 01ZX1708F to M. Boerries). S. Völkl and A. Mackensen were supported by the German Research Foundation (VO 1835/3-1 and MA1351/12-1). M.E. Maccari and F.G. Kapp were supported by an EXCEL-Fellowship of the Faculty of Medicine, University of Freiburg, funded by the Else–Kröner–Fresenius–Stiftung. M. Heeg was supported by the Berta-Ottenstein-Program for Clinician Scientists, Faculty of Medicine, University of Freiburg.
Author contributions: M.E. Maccari, S. Fuchs, P. Kury, S. Völkl, A. Rensing-Ehl, and S. Ehl designed experiments. M.E. Maccari, S. Fuchs, P. Kury, S. Völkl, B. Bengsch, S. Jägl, C.N. Castro, U. Warthorst, C. König, and A. Rensing-Ehl performed experiments and analyzed data. M.R. Lorenz, M. Führer, and K. Schwarz performed genetic investigations. S. Fuchs performed CyTOF. S. Fuchs, P. Kury, and T. Kalina analyzed CyTOF data. A.P. Frei and T. Kalina supervised CyTOF analyses. A. Ekici, A. Erxleben, R. Backofen, G. Andrieux, and M. Boerries performed and/or supervised RNA sequencing and bioinformatic analyses. M.E. Maccari, M. Heeg, M. Groß, C. Speckmann, J. Rohr, J. Thalhammer, F.G. Kapp, C.M. Niemeyer, C. Klemann, C. Schütz, C. König, M.G. Seidel, G. Dückers, S. Schönberger, C. Speckmann, R. Kobbe, D. Holzinger, P. Smisek, S. Owens, G. Horneff, R. Kolb, N. Naumann-Bartsch, M. Miano, J. Staniek, M. Rizzi, K. Warnatz, B. Grimbacher, H. Eibel, S. Völkl, A. Mackensen, A. Rensing-Ehl, and S. Ehl provided clinical information and patient and HD samples. M. Rizzi and P. Schneider provided experimental tools and expert advice. I. Fuchs supervised diagnostic work-up of patients. A. Rensing-Ehl and S. Ehl supervised the work. M.E. Maccari, S. Ehl, and A. Rensing-Ehl wrote the manuscript. All authors commented on the manuscript.
References
- Amir, A.D., Davis K.L., Tadmor M.D., Simonds E.F., Levine J.H., Bendall S.C., Shenfeld D.K., Krishnaswamy S., Nolan G.P., and Pe’er D.. 2013. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 31:545–552. 10.1038/nbt.2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appay, V., van Lier R.A., Sallusto F., and Roederer M.. 2008. Phenotype and function of human T lymphocyte subsets: consensus and issues. Cytometry A. 73:975–983. 10.1002/cyto.a.20643 [DOI] [PubMed] [Google Scholar]
- Becht, E., McInnes L., Healy J., Dutertre C.A., Kwok I.W.H., Ng L.G., Ginhoux F., and Newell E.W.. 2019. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37:38–44. 10.1038/nbt.4314 [DOI] [PubMed] [Google Scholar]
- Besnard, C., Levy E., Aladjidi N., Stolzenberg M.C., Magerus-Chatinet A., Alibeu O., Nitschke P., Blanche S., Hermine O., Jeziorski E., et al. Members of the French reference center for pediatric autoimmune cytopenias (CEREVANCE) . 2018. Pediatric-onset Evans syndrome: Heterogeneous presentation and high frequency of monogenic disorders including LRBA and CTLA4 mutations. Clin. Immunol. 188:52–57. 10.1016/j.clim.2017.12.009 [DOI] [PubMed] [Google Scholar]
- Bleesing, J.J., Brown M.R., Dale J.K., Straus S.E., Lenardo M.J., Puck J.M., Atkinson T.P., and Fleisher T.A.. 2001. TcR-alpha/beta(+) CD4(-)CD8(-) T cells in humans with the autoimmune lymphoproliferative syndrome express a novel CD45 isoform that is analogous to murine B220 and represents a marker of altered O-glycan biosynthesis. Clin. Immunol. 100:314–324. 10.1006/clim.2001.5069 [DOI] [PubMed] [Google Scholar]
- Bleesing, J.J., Brown M.R., Novicio C., Guarraia D., Dale J.K., Straus S.E., and Fleisher T.A.. 2002. A composite picture of TcR alpha/beta(+) CD4(-)CD8(-) T Cells (alpha/beta-DNTCs) in humans with autoimmune lymphoproliferative syndrome. Clin. Immunol. 104:21–30. 10.1006/clim.2002.5225 [DOI] [PubMed] [Google Scholar]
- Brenchley, J.M., Karandikar N.J., Betts M.R., Ambrozak D.R., Hill B.J., Crotty L.E., Casazza J.P., Kuruppu J., Migueles S.A., Connors M., et al. . 2003. Expression of CD57 defines replicative senescence and antigen-induced apoptotic death of CD8+ T cells. Blood. 101:2711–2720. 10.1182/blood-2002-07-2103 [DOI] [PubMed] [Google Scholar]
- Bristeau-Leprince, A., Mateo V., Lim A., Magerus-Chatinet A., Solary E., Fischer A., Rieux-Laucat F., and Gougeon M.L.. 2008. Human TCR alpha/beta+ CD4-CD8- double-negative T cells in patients with autoimmune lymphoproliferative syndrome express restricted Vbeta TCR diversity and are clonally related to CD8+ T cells. J. Immunol. 181:440–448. 10.4049/jimmunol.181.1.440 [DOI] [PubMed] [Google Scholar]
- Buchbinder, J.H., Pischel D., Sundmacher K., Flassig R.J., and Lavrik I.N.. 2018. Quantitative single cell analysis uncovers the life/death decision in CD95 network. PLOS Comput. Biol. 14:e1006368 10.1371/journal.pcbi.1006368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt, D., Chan T.D., Bourne K., Hermes J.R., Nguyen A., Statham A., O’Reilly L.A., Strasser A., Price S., Schofield P., et al. . 2015. FAS inactivation releases unconventional germinal center B cells that escape antigen control and drive IgE and autoantibody production. Immunity. 42:890–902. 10.1016/j.immuni.2015.04.010 [DOI] [PubMed] [Google Scholar]
- Dobin, A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehl, S., Hoffmann-Rohrer U., Nagata S., Hengartner H., and Zinkernagel R.. 1996. Different susceptibility of cytotoxic T cells to CD95 (Fas/Apo-1) ligand-mediated cell death after activation in vitro versus in vivo. J. Immunol. 156:2357–2360. [PubMed] [Google Scholar]
- Eisenhut, F., Heim L., Trump S., Mittler S., Sopel N., Andreev K., Ferrazzi F., Ekici A.B., Rieker R., Springel R., et al. . 2016. FAM13A is associated with non-small cell lung cancer (NSCLC) progression and controls tumor cell proliferation and survival. OncoImmunology. 6:e1256526 10.1080/2162402X.2016.1256526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, K., Voelkl S., Heymann J., Przybylski G.K., Mondal K., Laumer M., Kunz-Schughart L., Schmidt C.A., Andreesen R., and Mackensen A.. 2005. Isolation and characterization of human antigen-specific TCR alpha beta+ CD4(-)CD8- double-negative regulatory T cells. Blood. 105:2828–2835. 10.1182/blood-2004-07-2583 [DOI] [PubMed] [Google Scholar]
- Fisher, G.H., Rosenberg F.J., Straus S.E., Dale J.K., Middleton L.A., Lin A.Y., Strober W., Lenardo M.J., and Puck J.M.. 1995. Dominant interfering Fas gene mutations impair apoptosis in a human autoimmune lymphoproliferative syndrome. Cell. 81:935–946. 10.1016/0092-8674(95)90013-6 [DOI] [PubMed] [Google Scholar]
- Fortner, K.A., Lees R.K., MacDonald H.R., and Budd R.C.. 2011. Fas (CD95/APO-1) limits the expansion of T lymphocytes in an environment of limited T-cell antigen receptor/MHC contacts. Int. Immunol. 23:75–88. 10.1093/intimm/dxq466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs, S., Sawas N., Staedler N., Schubert D.A., D’Andrea A., Zeiser R., Piali L., Hruz P., and Frei A.P.. 2019. High-dimensional single-cell proteomics analysis identifies immune checkpoint signatures and therapeutic targets in ulcerative colitis. Eur. J. Immunol. 49:462–475. 10.1002/eji.201847862 [DOI] [PubMed] [Google Scholar]
- Gajate, C., and Mollinedo F.. 2015. Lipid rafts and raft-mediated supramolecular entities in the regulation of CD95 death receptor apoptotic signaling. Apoptosis. 20:584–606. 10.1007/s10495-015-1104-6 [DOI] [PubMed] [Google Scholar]
- Gautam, S., Fioravanti J., Zhu W., Le Gall J.B., Brohawn P., Lacey N.E., Hu J., Hocker J.D., Hawk N.V., Kapoor V., et al. . 2019. The transcription factor c-Myb regulates CD8+ T cell stemness and antitumor immunity. Nat. Immunol. 20:337–349. 10.1038/s41590-018-0311-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulbins, E., Coggeshall K.M., Brenner B., Schlottmann K., Linderkamp O., and Lang F.. 1996. Fas-induced apoptosis is mediated by activation of a Ras and Rac protein-regulated signaling pathway. J. Biol. Chem. 271:26389–26394. 10.1074/jbc.271.42.26389 [DOI] [PubMed] [Google Scholar]
- Haug, T., Aigner M., Peuser M.M., Strobl C.D., Hildner K., Mougiakakos D., Bruns H., Mackensen A., and Völkl S.. 2019. Human double-negative regulatory T-cells induce a metabolic and functional switch in effector T-cells by suppressing mTOR activity. Front. Immunol. 10:883 10.3389/fimmu.2019.00883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holler, N., Tardivel A., Kovacsovics-Bankowski M., Hertig S., Gaide O., Martinon F., Tinel A., Deperthes D., Calderara S., Schulthess T., et al. . 2003. Two adjacent trimeric Fas ligands are required for Fas signaling and formation of a death-inducing signaling complex. Mol. Cell. Biol. 23:1428–1440. 10.1128/MCB.23.4.1428-1440.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzelova, E., Vonarbourg C., Stolzenberg M.C., Arkwright P.D., Selz F., Prieur A.M., Blanche S., Bartunkova J., Vilmer E., Fischer A., et al. . 2004. Autoimmune lymphoproliferative syndrome with somatic Fas mutations. N. Engl. J. Med. 351:1409–1418. 10.1056/NEJMoa040036 [DOI] [PubMed] [Google Scholar]
- Hughes, P.D., Belz G.T., Fortner K.A., Budd R.C., Strasser A., and Bouillet P.. 2008. Apoptosis regulators Fas and Bim cooperate in shutdown of chronic immune responses and prevention of autoimmunity. Immunity. 28:197–205. 10.1016/j.immuni.2007.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai, Y., Yamashita Y., and Osawa T.. 1988. Enhancement of the activities of glycosyltransferases involved in the biosynthesis of mucin-type sugar chains in autoimmune MRL lpr/lpr mouse T cells. Mol. Immunol. 25:419–428. 10.1016/0161-5890(88)90161-7 [DOI] [PubMed] [Google Scholar]
- Intlekofer, A.M., Takemoto N., Wherry E.J., Longworth S.A., Northrup J.T., Palanivel V.R., Mullen A.C., Gasink C.R., Kaech S.M., Miller J.D., et al. . 2005. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat. Immunol. 6:1236–1244. 10.1038/ni1268 [DOI] [PubMed] [Google Scholar]
- Ji, Y., Pos Z., Rao M., Klebanoff C.A., Yu Z., Sukumar M., Reger R.N., Palmer D.C., Borman Z.A., Muranski P., et al. . 2011. Repression of the DNA-binding inhibitor Id3 by Blimp-1 limits the formation of memory CD8+ T cells. Nat. Immunol. 12:1230–1237. 10.1038/ni.2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi, N.S., Cui W., Chandele A., Lee H.K., Urso D.R., Hagman J., Gapin L., and Kaech S.M.. 2007. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 27:281–295. 10.1016/j.immuni.2007.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech, S.M., and Cui W.. 2012. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol. 12:749–761. 10.1038/nri3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebanoff, C.A., Scott C.D., Leonardi A.J., Yamamoto T.N., Cruz A.C., Ouyang C., Ramaswamy M., Roychoudhuri R., Ji Y., Eil R.L., et al. . 2016. Memory T cell-driven differentiation of naive cells impairs adoptive immunotherapy. J. Clin. Invest. 126:318–334. 10.1172/JCI81217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen, S.E., Bilenkin A., Tarasenko T.N., Arjunaraja S., Stinson J.R., McGuire P.J., and Snow A.L.. 2017. Sensitivity to Restimulation-induced cell death is linked to glycolytic metabolism in human T cells. J. Immunol. 198:147–155. 10.4049/jimmunol.1601218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gallo, M., Poissonnier A., Blanco P., and Legembre P.. 2017. CD95/Fas, non-apoptotic signaling pathways, and kinases. Front. Immunol. 8:1216 10.3389/fimmu.2017.01216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997 [q-bio.GN].
- Liao, Y., Smyth G.K., and Shi W.. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30:923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Love, M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, W., Friedman M.S., Shedden K., Hankenson K.D., and Woolf P.J.. 2009. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics. 10:161 10.1186/1471-2105-10-161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina, M.N., Noel S., Saxena A., Bandapalle S., Majithia R., Jie C., Arend L.J., Allaf M.E., Rabb H., and Hamad A.R.A.. 2016. Double-negative αβ T cells are early responders to AKI and are found in human kidney. J. Am. Soc. Nephrol. 27:1113–1123. 10.1681/ASN.2014121214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner, J.D., Vogel T.P., Forbes L., Ma C.A., Stray-Pedersen A., Niemela J.E., Lyons J.J., Engelhardt K.R., Zhang Y., Topcagic N., et al. . 2015. Early-onset lymphoproliferation and autoimmunity caused by germline STAT3 gain-of-function mutations. Blood. 125:591–599. 10.1182/blood-2014-09-602763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamood, A.S., Bargatze D., Xiao Z., Jie C., Yagita H., Ruben D., Watson J., Chakravarti S., Schneck J.P., and Hamad A.R.. 2008. Fas-mediated apoptosis regulates the composition of peripheral alphabeta T cell repertoire by constitutively purging out double negative T cells. PLoS One. 3:e3465 10.1371/journal.pone.0003465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountz, J.D., and Steinberg A.D.. 1989. Studies of c-myb gene regulation in MRL-lpr/lpr mice. Identification of a 5′ c-myb nuclear protein binding site and high levels of binding factors in nuclear extracts of lpr/lpr lymph node cells. J. Immunol. 142:328–335. [PubMed] [Google Scholar]
- Mountz, J.D., Steinberg A.D., Klinman D.M., Smith H.R., and Mushinski J.F.. 1984. Autoimmunity and increased c-myb transcription. Science. 226:1087–1089. 10.1126/science.6494925 [DOI] [PubMed] [Google Scholar]
- Müller, C.P., Kalinichenko L.S., Tiesel J., Witt M., Stöckl T., Sprenger E., Fuchser J., Beckmann J., Praetner M., Huber S.E., et al. . 2017. Paradoxical antidepressant effects of alcohol are related to acid sphingomyelinase and its control of sphingolipid homeostasis. Acta Neuropathol. 133:463–483. 10.1007/s00401-016-1658-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabhani, S., Schipp C., Miskin H., Levin C., Postovsky S., Dujovny T., Koren A., Harlev D., Bis A.M., Auer F., et al. . 2017. STAT3 gain-of-function mutations associated with autoimmune lymphoproliferative syndrome like disease deregulate lymphocyte apoptosis and can be targeted by BH3 mimetic compounds. Clin. Immunol. 181:32–42. 10.1016/j.clim.2017.05.021 [DOI] [PubMed] [Google Scholar]
- Ohga, S., Nomura A., Takahata Y., Ihara K., Takada H., Wakiguchi H., Kudo Y., and Hara T.. 2002. Dominant expression of interleukin 10 but not interferon gamma in CD4(-)CD8(-)alphabetaT cells of autoimmune lymphoproliferative syndrome. Br. J. Haematol. 119:535–538. 10.1046/j.1365-2141.2002.03848.x [DOI] [PubMed] [Google Scholar]
- Pestano, G.A., Zhou Y., Trimble L.A., Daley J., Weber G.F., and Cantor H.. 1999. Inactivation of misselected CD8 T cells by CD8 gene methylation and cell death. Science. 284:1187–1191. 10.1126/science.284.5417.1187 [DOI] [PubMed] [Google Scholar]
- Peter, M.E., Budd R.C., Desbarats J., Hedrick S.M., Hueber A.O., Newell M.K., Owen L.B., Pope R.M., Tschopp J., Wajant H., et al. . 2007. The CD95 receptor: apoptosis revisited. Cell. 129:447–450. 10.1016/j.cell.2007.04.031 [DOI] [PubMed] [Google Scholar]
- Price, S., Shaw P.A., Seitz A., Joshi G., Davis J., Niemela J.E., Perkins K., Hornung R.L., Folio L., Rosenberg P.S., et al. . 2014. Natural history of autoimmune lymphoproliferative syndrome associated with FAS gene mutations. Blood. 123:1989–1999. 10.1182/blood-2013-10-535393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, R.R., Li Q., Odunsi K., and Shrikant P.A.. 2010. The mTOR kinase determines effector versus memory CD8+ T cell fate by regulating the expression of transcription factors T-bet and Eomesodermin. Immunity. 32:67–78. 10.1016/j.immuni.2009.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team 2015. R: a language and environment for statistical computing. http://www.R-project.org/. (Accessed December 2019).
- Rensing-Ehl, A., Völkl S., Speckmann C., Lorenz M.R., Ritter J., Janda A., Abinun M., Pircher H., Bengsch B., Thimme R., et al. . 2014. Abnormally differentiated CD4+ or CD8+ T cells with phenotypic and genetic features of double negative T cells in human Fas deficiency. Blood. 124:851–860. 10.1182/blood-2014-03-564286 [DOI] [PubMed] [Google Scholar]
- Rieux-Laucat, F., Le Deist F., Hivroz C., Roberts I.A., Debatin K.M., Fischer A., and de Villartay J.P.. 1995. Mutations in Fas associated with human lymphoproliferative syndrome and autoimmunity. Science. 268:1347–1349. 10.1126/science.7539157 [DOI] [PubMed] [Google Scholar]
- Russell, J.H., Rush B., Weaver C., and Wang R.. 1993. Mature T cells of autoimmune lpr/lpr mice have a defect in antigen-stimulated suicide. Proc. Natl. Acad. Sci. USA. 90:4409–4413. 10.1073/pnas.90.10.4409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiedel, B.J., Singh D., Madrigal A., Valdovino-Gonzalez A.G., White B.M., Zapardiel-Gonzalo J., Ha B., Altay G., Greenbaum J.A., McVicker G., et al. . 2018. Impact of genetic polymorphisms on human immune cell gene expression. Cell. 175:1701–1715.e16. 10.1016/j.cell.2018.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takiguchi, M., Murakami M., Nakagawa I., Rashid M.M., Tosa N., Chikuma S., Hashimoto A., and Uede T.. 2000. Involvement of CD28/CTLA4-B7 costimulatory pathway in the development of lymphadenopathy and splenomegaly in MRL/lpr mice. J. Vet. Med. Sci. 62:29–36. 10.1292/jvms.62.29 [DOI] [PubMed] [Google Scholar]
- Teachey, D.T., Greiner R., Seif A., Attiyeh E., Bleesing J., Choi J., Manno C., Rappaport E., Schwabe D., Sheen C., et al. . 2009. Treatment with sirolimus results in complete responses in patients with autoimmune lymphoproliferative syndrome. Br. J. Haematol. 145:101–106. 10.1111/j.1365-2141.2009.07595.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimble, L.A., Prince K.A., Pestano G.A., Daley J., and Cantor H.. 2002. Fas-dependent elimination of nonselected CD8 cells and lpr disease. J. Immunol. 168:4960–4967. 10.4049/jimmunol.168.10.4960 [DOI] [PubMed] [Google Scholar]
- Van Gassen, S., Callebaut B., Van Helden M.J., Lambrecht B.N., Demeester P., Dhaene T., and Saeys Y.. 2015. FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A. 87:636–645. 10.1002/cyto.a.22625 [DOI] [PubMed] [Google Scholar]
- Völkl, S., Rensing-Ehl A., Allgäuer A., Schreiner E., Lorenz M.R., Rohr J., Klemann C., Fuchs I., Schuster V., von Bueren A.O., et al. . 2016. Hyperactive mTOR pathway promotes lymphoproliferation and abnormal differentiation in autoimmune lymphoproliferative syndrome. Blood. 128:227–238. 10.1182/blood-2015-11-685024 [DOI] [PubMed] [Google Scholar]
- Wajant, H. 2014. Principles and mechanisms of CD95 activation. Biol. Chem. 395:1401–1416. 10.1515/hsz-2014-0212 [DOI] [PubMed] [Google Scholar]
- Zimmermann, C., Rawiel M., Blaser C., Kaufmann M., and Pircher H.. 1996. Homeostatic regulation of CD8+ T cells after antigen challenge in the absence of Fas (CD95). Eur. J. Immunol. 26:2903–2910. 10.1002/eji.1830261215 [DOI] [PubMed] [Google Scholar]
- Zuo, J., Ge H., Zhu G., Matthias P., and Sun J.. 2007. OBF-1 is essential for the generation of antibody-secreting cells and the development of autoimmunity in MRL-lpr mice. J. Autoimmun. 29:87–96. 10.1016/j.jaut.2007.05.001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
depicts clinical, immunological, and genetic features of included patients.
reports the sources of SLOs and bone marrow samples.
lists the flow cytometry antibodies.
lists the CyTOF antibodies.
provides CyTOF analysis information for t-SNE.
lists CyTOF analysis information for UMAP.