Watanabe et al. show that centrioles and their associated pericentriolar matrix can independently support spindle assembly. They delineate how the pericentriolar matrix acts without centrioles and describe cancer cell lines in which spindles can assemble in the absence of both centrioles and pericentriolar matrix.
Abstract
Centrosomes, composed of centrioles that recruit a pericentriolar material (PCM) matrix assembled from PCNT and CDK5RAP2, catalyze mitotic spindle assembly. Here, we inhibit centriole formation and/or remove PCNT–CDK5RAP2 in RPE1 cells to address their relative contributions to spindle formation. While CDK5RAP2 and PCNT are normally dispensable for spindle formation, they become essential when centrioles are absent. Acentriolar spindle assembly is accompanied by the formation of foci containing PCNT and CDK5RAP2 via a microtubule and Polo-like kinase 1–dependent process. Foci formation and spindle assembly require PCNT-CDK5RAP2–dependent matrix assembly and the ability of CDK5RAP2 to recruit γ-tubulin complexes. Thus, the PCM matrix can self-organize independently of centrioles to generate microtubules for spindle assembly; conversely, an alternative centriole-anchored mechanism supports spindle assembly when the PCM matrix is absent. Extension to three cancer cell lines revealed similar results in HeLa cells, whereas DLD1 and U2OS cells could assemble spindles in the absence of centrioles and PCNT-CDK5RAP2, suggesting cell type variation in spindle assembly mechanisms.
Introduction
Centrosomes are the primary microtubule organizing centers in metazoan cells. Centrosomes consist of a centriolar core that organizes a layered proteinaceous structure, called the pericentriolar material (PCM; Mennella et al., 2014). During mitotic entry, centrosomes increase in size to help meet the increased demand for microtubule generation for spindle assembly (Palazzo et al., 2000). This increase in size is due to expansion of an outer PCM matrix layer whose assembly requires the large coiled-coil proteins pericentrin (PCNT; pericentrin-like protein [PLP] in Drosophila) and CDK5RAP2/CEP215 (centrosomin [Cnn] in Drosophila, and SPD-5 in Caenorhabditis elegans; Fu and Glover, 2012; Lawo et al., 2012; Mennella et al., 2012, 2014; Woodruff et al., 2014). For convenience, we refer to this PCNT and CDK5RAP2–based matrix layer as the “PCM matrix,” noting that there are other PCM proteins, including CEP192 and NEDD1, that remain in a more centriole-proximal toroid when the outer PCM matrix layer is absent (Fu and Glover, 2012; Lawo et al., 2012; Mennella et al., 2012). The ability of centrosomes to nucleate microtubules is thought to be due, at least in part, to the ability of the PCM matrix to bind γ-tubulin–containing complexes (Moritz et al., 1995, 1998; Schnackenberg et al., 1998). One of the best-characterized γ-tubulin complex binding sites is a conserved sequence motif, termed CM1 (Samejima et al., 2008; Zhang and Megraw, 2007), in the CDK5RAP2 N-terminus (Choi et al., 2010; Fong et al., 2008). In Drosophila embryos, mutation of the Cnn CM1 motif reduced the amount of γ-tubulin at centrosomes (Zhang and Megraw, 2007); however, in human cells, reduction in centrosomal γ-tubulin upon mutation of the CM1 motif of CDK5RAP2 has not been noted (Kim and Rhee, 2014).
Within the centrosome, PCNT/PLP is thought to link the PCM matrix layer to the centriole and ensure its proper organization. In humans and Drosophila, PCNT/PLP associates with the outer centriole wall via its C-terminal pericentrin-AKAP450 centrosomal targeting (PACT) domain and, in interphase centrosomes, is oriented with its N-terminal domain facing outwards (Fu and Glover, 2012; Lawo et al., 2012; Mennella et al., 2012). CDK5RAP2/Cnn is thought to assemble on this PCNT foundation, as loss of PCNT leads to a significant reduction in centrosomal CDK5RAP2/Cnn (Fu and Glover, 2012; Lawo et al., 2012; Mennella et al., 2012). The mitotic expansion of the PCM matrix is controlled by Polo-like kinase (PLK) 1 (Cabral et al., 2019; Conduit et al., 2014; Dobbelaere et al., 2008; Haren et al., 2009; Lane and Nigg, 1996; Lee and Rhee, 2011; Woodruff et al., 2015). Structural work on the Drosophila proteins has suggested that PCM matrix expansion is driven by a phosphorylation-regulated self-interaction between Cnn molecules (Feng et al., 2017) in which the CM2 motif at the Cnn C-terminus interacts with an internal Cnn leucine zipper (Citron et al., 2018; Feng et al., 2017). The Cnn CM2 motif also interacts with an internal region in PLP, and it has been suggested that this interaction may help tether the Cnn-based matrix to the centriole (Citron et al., 2018). Although similar structural work has not yet been done on the human proteins, the dependence on PCNT for assembly of a CDK5RAP2/PCNT matrix layer (Fu and Glover, 2012; Lawo et al., 2012; Mennella et al., 2012), the dramatic expansion of the matrix layer when either PCNT or CDK5RAP2 is overexpressed (Lawo et al., 2012), and experiments showing that the CM2 domain of CDK5RAP2 is important for its ability to accumulate around centrioles and to interact with PCNT (Kim and Rhee, 2014; Wang et al., 2010) suggest that that the human PCM matrix assembles in a similar CM2-dependent manner.
Work across systems has suggested that cells lacking centrioles retain the ability to assemble a mitotic spindle but do so more slowly than control cells (Basto et al., 2006; Bazzi and Anderson, 2014; Khodjakov and Rieder, 2001; Lecland et al., 2013; Meitinger et al., 2016; Sir et al., 2013; Wong et al., 2015). Thus, the presence of centrioles significantly accelerates spindle assembly. Whether this acceleration is due to the ability of centrioles to organize PCM matrix or whether they have the capacity to accelerate spindle assembly independent of the matrix is not clear. Recent work has shown that spindle assembly in human cells that lack centrioles is accompanied by the coalescence of PCM proteins into foci at the spindle poles during mitosis (Meitinger et al., 2020; Yeow et al., 2020). Similar foci containing PCM proteins have also been observed in acentriolar Drosophila cells (Baumbach et al., 2015; Debec et al., 1995; Moutinho-Pereira et al., 2009). In Drosophila, foci formation requires Cnn, but the foci were not essential for spindle assembly (Baumbach et al., 2015; Moutinho-Pereira et al., 2009). Thus, key questions with respect to PCM foci during acentriolar mitosis in human cells are whether their formation represents bona fide centriole-independent assembly of PCM matrix, and whether the foci and their microtubule-nucleating capacity are important for spindle assembly.
In prior work, we developed centrinone, a selective inhibitor of the centriole duplication trigger kinase PLK4 (Wong et al., 2015). Treating cells with centrinone enables the generation of acentriolar cells. Here, we use centrinone in conjunction with PCNT and CDK5RAP2 inhibition to address the relative contributions of centrioles and the PCM matrix to spindle assembly in human cells. Using nontransformed RPE1 cells, we show that, whereas cells lacking either centrioles or PCM matrix proteins can assemble spindles, simultaneous loss of both PCM matrix and centrioles results in a severe spindle assembly defect. Both PCNT-CDK5RAP2 matrix assembly and γ-tubulin binding to the CDK5RAP2 CM1 domain are essential for acentriolar spindle assembly. We extended this analysis to three cancer cell types. In HeLa cells, the combined loss of centrioles and PCM matrix proteins also led to spindle assembly failure. Surprisingly, DLD1 and U2OS cells retained the ability to assemble a spindle in the absence of both centrioles and PCM matrix proteins, suggesting differential utilization of pathways for the generation of spindle microtubules that do not require either centrioles or PCM matrix proteins.
Results
The PCM matrix proteins PCNT and CDK5RAP2 are dispensable for spindle assembly
In human cells, the mitotic increase in centrosome size is due to expansion of a PCNT- and CDK5RAP2-dependent PCM layer (Lawo et al., 2012), which we refer to as the “PCM matrix.” To analyze the role of the PCM matrix in spindle assembly, we used CRISPR/Cas9 to individually knock out CDK5RAP2 and PCNT in nontransformed RPE1 cells (Fig. S1, A and B). Knockouts were generated in a USP28Δ background because USP28 is an essential component of the pathway that activates p53 and arrests cells in G1 in response to increased mitotic duration (Fong et al., 2016; Lambrus et al., 2016; Meitinger et al., 2016). For each knockout, a clonal cell line with no centrosomal localization of the target protein, consistent with genotyping data, was selected for further analysis (Fig. 1 A; and Fig. S1, A and B). For CDK5RAP2Δ, no band was detected; for PCNTΔ, a faint band was detected at high exposure (Fig. 1 A; and Fig. S1, A and B). As expected based on prior work, removal of either PCNT or CDK5RAP2 reduced but did not eliminate localization of CEP192 to the spindle poles and did not prevent spindle formation or chromosome segregation (Fig. 1, B and C; and Fig. S1 C; Fong et al., 2008; Gomez-Ferreria et al., 2007; Haren et al., 2009; Lawo et al., 2012; Wang et al., 2013). Mitotic duration, measured from nuclear envelope breakdown (NEBD) to chromosome decondensation, is a sensitive measure of defects in spindle assembly and chromosome–spindle interactions and was not increased by individual loss of PCNT or CDK5RAP2 (Fig. 1 C). Consistent with this, both mutant clones could be propagated long-term in culture.
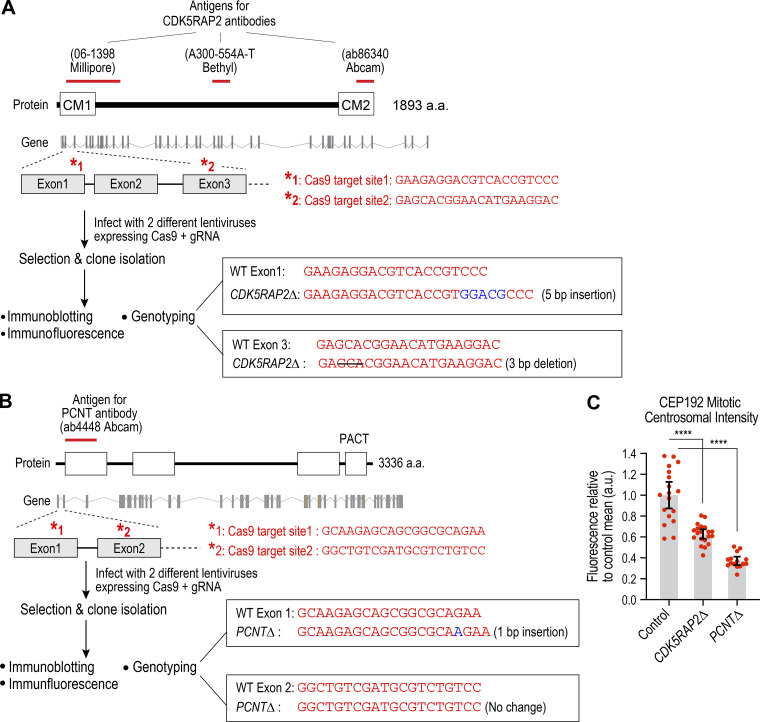
Figure S1.
PCNT and CDK5RAP2 knockout cell line generation scheme, genotyping, and analysis of centrosomal CEP192 intensity. (A and B) Schematics of the encoded protein, the genomic locus, and gRNAs employed to target the locus. The antigens for the antibodies used are depicted above the protein cartoon. Genotyping results for the knockout clones employed in the analysis are shown on the right. For CDK5RAP2, a 5-bp insertion in exon 1 and a 3-bp deletion in exon 3 were found in the clones. For PCNT, a single-bp insertion was observed in exon 1. Immunofluorescence and immunoblotting data are shown in Fig. 1 A. (C) Quantification of CEP192 centrosomal intensity in the parental Control (RPE1 USP28Δ) and knockout cell lines. Centrosomal intensity was measured in fixed mitotic cells; for example images, see Fig. 1 B. ****, P < 0.0001 from unpaired t tests. Error bars are 95% CI.
Figure 1.
PCNT and CDK5RAP2 are dispensable for spindle assembly and chromosome segregation in RPE1 cells. (A) Top: Images of fixed mitotic RPE1(USP28Δ) cells lacking PCNT or CDK5RAP2 after labeling with the indicated antibodies and a DNA dye. Scale bar, 10 µm. Bottom: Immunoblots analyzing CDK5RAP2 and PCNT in the parental line and knockout clones. α-tubulin is a loading control. (B) Effect of PCNT or CDK5RAP2 loss on localization of CEP192. Scale bar, 5 µm. (C) Live imaging of mitosis in cells lacking PCNT or CDK5RAP2. Cells were labeled with SiR-DNA for 2 h and then imaged at 5-min intervals. Quantification of mitotic duration is shown below for 20 cells per condition. Error bars represent SD. Scale bar, 10 µm. (D and E) CDK5RAP2 depletion removes residual centrosome-localized CDK5RAP2 in PCNTΔ cells. (D) Images of fixed cells for the indicated conditions; 2.5× magnified regions to the right show CDK5RAP2 signal in the centrosome region. Scale bar, 10 µm. (E) Centrosomal CDK5RAP2 intensity for the indicated conditions. Error bars represent SD. (F) Mitotic spindle morphology, 48 h after siRNA transfection, for the indicated conditions. Fixed cells were stained to label microtubules and DNA. Spindle morphology in prometaphase and metaphase stage cells was scored. MT, microtubules; ns, not significant; tub, tubulin. ****, P < 0.0001; ns, not significant (P > 0.05).
In agreement with prior work (Haren et al., 2009; Kim and Rhee, 2014; Lawo et al., 2012), PCNT loss reduced (to ∼15% of control levels) but did not abolish CDK5RAP2 recruitment to the vicinity of centrioles (Fig. 1, D and E). To address the possibility that PCNT and CDK5RAP2 might function redundantly, we used RNAi to deplete CDK5RAP2 in PCNTΔ cells (Fig. 1 D). In coinhibited cells, PCNT was absent, and no residual CDK5RAP2 was detected (Fig. 1 D). Coinhibited cells exhibited a mitotic phenotype essentially identical to loss of PCNT alone (Fig. 1, E and F). We conclude that the mild nature of the mitotic phenotypes in PCNTΔ and CDK5RAP2Δ cells does not result from redundancy between the two matrix proteins, but instead indicates that the mitotic PCM matrix comprised of PCNT and CDK5RAP2 is dispensable for spindle assembly.
A centriole-anchored mechanism generates microtubules for spindle assembly in the absence of PCM matrix
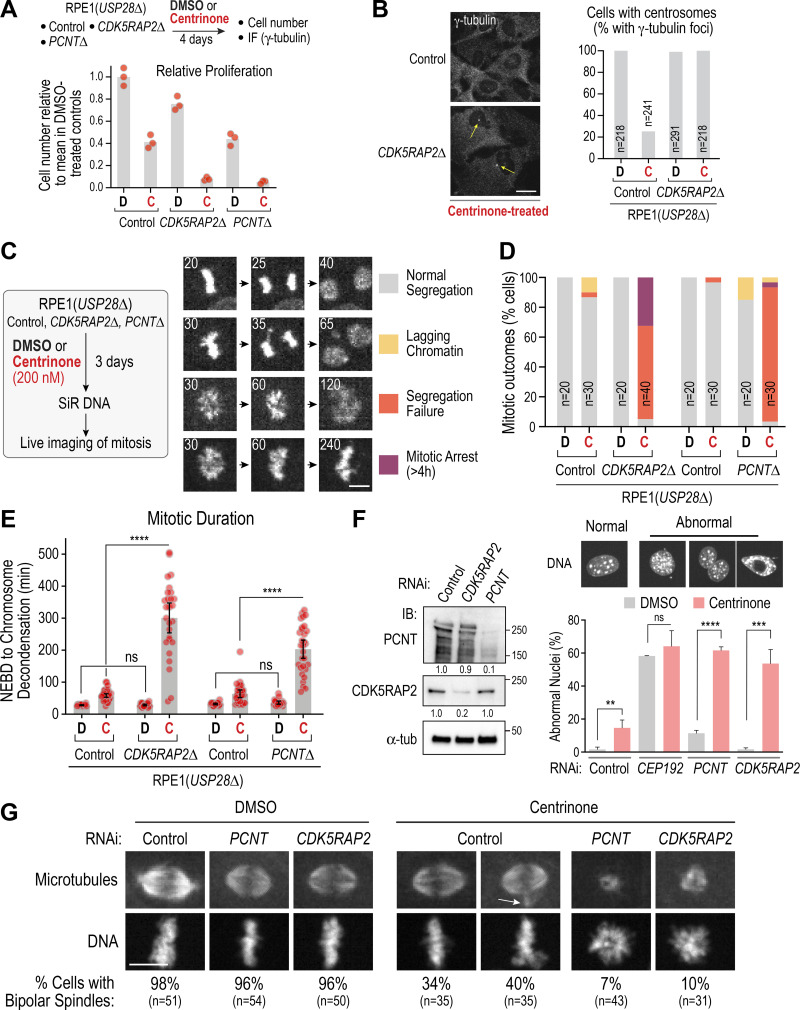
The above results indicate that in nontransformed RPE1 cells, loss of both conserved PCM matrix proteins does not prevent spindle formation, suggesting an additional mechanism for the generation of spindle microtubules. Since PCNTΔ and CDK5RAP2Δ cells retain centrioles that associate with spindle poles (Fig. 1 B), we tested whether centrioles anchor a PCM matrix–independent pathway for microtubule generation. For this purpose, we used centrinone to prevent new centriole formation. Measurement of cell number and normalization relative to DMSO-treated controls indicated that centrinone treatment reduced proliferation ∼10-fold for CDK5RAP2Δ cells, compared with ∼2.5-fold for parental RPE1 (USP28Δ) cells; similar results were obtained for PCNTΔ cells (Fig. 2 A). After 4 d in centrinone, ∼75% of control cells lacked centrioles, as indicated by absence of a focus of γ-tubulin staining (Fig. 2 B); by contrast, all remaining CDK5RAP2Δ cells retained centrioles (Fig. 2 B). These results suggest that centriole loss is synthetically lethal with the absence of PCNT or CDK5RAP2, so that in the absence of CDK5RAP2, only cells with residual centrioles survive centrinone treatment.
Figure 2.
PCNT-CDK5RAP2 and centrioles act in parallel to direct spindle assembly and chromosome segregation. (A and B) Analysis of relative proliferation (A) and centrosome number measured by counting γ-tubulin foci (B) following 4-d mock (DMSO [D]) or centrinone (C) treatment. Scale bar, 10 µm. (C) Live imaging of mitosis for the indicated conditions using the procedure on the left. Cells were first treated with DMSO (D) or centrinone (C) to generate centriole-less cells, labeled with SiR-DNA for 2 h, and then imaged at 5-min intervals. Mitotic outcomes were classified into four categories, representative examples of which are shown in the image series. Note that the “Normal Segregation” category images are from the Control sequence in Fig. 1 C. Numbers on the top left of image panels indicate time after NEBD in minutes. Colored squares to the right indicate the labeling scheme for each outcome. Scale bar, 5 µm. (D and E) Quantification of mitotic outcomes (D) and mitotic duration (E) for the indicated conditions. Error bars in E are the 95% CI. (F) RNAi analysis. Left: Immunoblot of indicated conditions. α-tubulin is a loading control; numbers below indicate relative total band intensities. Right: Quantification of nuclear morphology as a readout of mitotic outcomes. Mean and SD from three independent experiments are plotted. At least 200 nuclei were scored per condition per experiment. P values are from t tests. (G) Mitotic spindle morphology for the indicated conditions; for detailed protocol, see Fig. S2 A. Arrow marks extra microtubule focus frequently observed in centrinone-treated cells. Fixed cells were stained to label microtubules and DNA. Scale bar, 10 µm. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. IB, immunoblot; IF, immunofluorescence; ns, not significant; tub, tubulin.
To determine why centrioles are essential for cell proliferation in the absence of PCNT or CDK5RAP2, we imaged mitosis after treating cells with centrinone or DMSO for 3 d and classified the mitotic outcomes into four categories (Fig. 2 C). This analysis revealed penetrant chromosome segregation failure in centrinone-treated CDK5RAP2Δ or PCNTΔ cells (Fig. 2 D, Video 1, and Video 2). Mitotic duration was also significantly prolonged in centrinone-treated CDK5RAP2Δ and PCNTΔ cells, relative to centrinone treatment or the CDK5RAP2 or PCNT deletions treated with DMSO (Fig. 2 E). Thus, centriole loss in RPE1 cells lacking PCM matrix proteins leads to a high rate of mitotic failure.
Video 1.
Control and CDK5RAP2Δ RPE1 cells labeled with SiR-DNA and imaged at 20× magnification. 8 × 2–µm z-stacks were acquired every 5 min. Image frames shown are maximum intensity projections at each time point. Playback is sped up 900× relative to real-time. Elapsed time shown is in hours:minutes, and scale bar is 10 µm. KO, knockout.
Video 2.
Control and PCNTΔ RPE1 cells labeled with SiR-DNA and imaged at 20× magnification. 8 × 2–µm z-stacks were acquired every 5 min. Image frames shown are maximum intensity projections at each time point. Playback is sped up 900× relative to real-time. Elapsed time shown is in hours:minutes, and scale bar is 10 µm. KO, knockout.
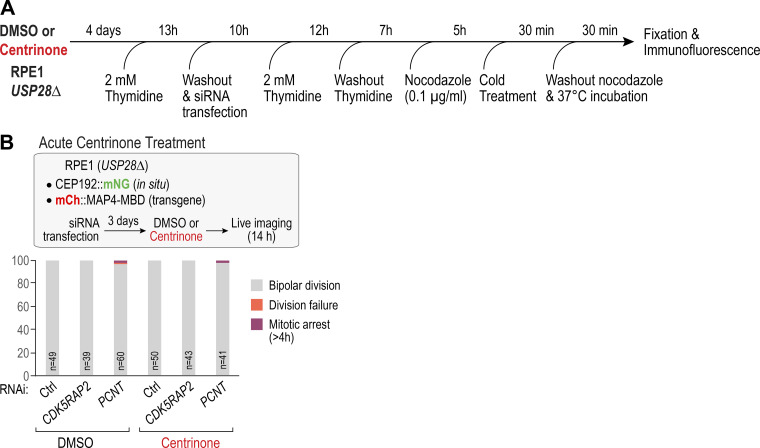
As centrioles could not be depleted in PCNTΔ or CDK5RAP2Δ cells, we inverted the order of the perturbations by using centrinone treatment to generate a population of cells lacking centrioles followed by RNAi to deplete PCNT or CDK5RAP2 (Fig. 2 F). An initial approach assessing mitotic outcomes by counting abnormal nuclei in fixed cells confirmed strong synergy between PCNT or CDK5RAP2 depletion and centrinone-mediated centriole removal (Fig. 2 F). In contrast to PCNT or CDK5RAP2, which were important only when centrioles were absent, CEP192 was important in both the presence and absence of centrioles (Fig. 2 F). Spindle assembly was assessed using a synchronization protocol in which cells were collected in mitosis in the presence of nocodazole and then fixed 30 min after nocodazole washout (Fig. 2 G and Fig. S2 A). PCNT or CDK5RAP2 depletion did not significantly perturb bipolar spindle formation under these conditions. Centrinone-treated cells exhibited reduced fidelity of bipolar spindle formation, primarily due to the frequent presence of an extra focus of microtubules (Fig. 2 G, white arrow). However, in centrinone-treated PCNT or CDK5RAP2-depleted cells, spindles failed to form, and only a small mass of disorganized microtubules was observed in the vicinity of the chromosomes (Fig. 2 G). This result suggests that defective spindle assembly underlies the mitotic failure observed in cells that lack both centrioles and PCM matrix proteins. To complement the fixed analysis, we used live imaging to monitor bipolar spindle formation, which indicated that both 1-centrosome and acentriolar cells failed to build bipolar spindles following CDK5RAP2 depletion (see Fig. 3 E and associated text below). Using acute centrinone treatment, we confirmed that the mitotic failure observed in centrinone-treated CDK5RAP2- or PCNT-depleted cells requires centriole loss and not just inhibition of PLK4 activity (Fig. S2 B).
Figure S2.
Analysis of spindle assembly following recovery from nocodazole treatment of synchronized cells and results of acute centrinone treatment. (A) Schematic of protocol used to synchronize cells, release into nocodazole, and recover from nocodazole before fixation and immunofluorescence. siRNA transfection was used to deplete CDK5RAP2 or PCNT in order to assess the impact on spindle assembly. (B) Analysis of acute centrinone treatment of CDK5RAP2- and PCNT-depleted cells. Centriole loss following centrinone treatment requires cell division; thus, in the first 24 h after addition of centrinone, PLK4 kinase activity is inhibited but centrioles are still present. Acute centrinone treatment did not result in a synthetic mitotic defect in combination with CDK5RAP2 or PCNT depletion. Ctrl, control.
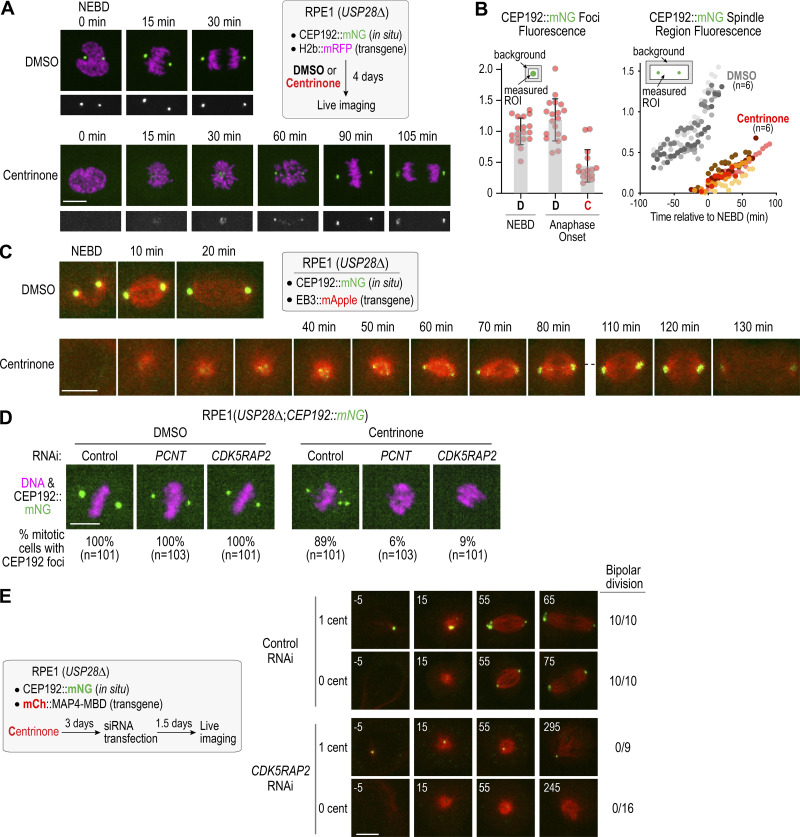
Figure 3.
PCM components form foci independently of centrioles after mitotic entry in a CDK5RAP2-PCNT dependent manner. (A) Live imaging of DMSO or centrinone-treated RPE1 cells with in situ mNG-tagged CEP192. Scale bar, 10 µm. (B) Fluorescence intensity quantification of CEP192 foci (left) and CEP192 in the spindle region (right). Error bars represent SD. (C) Live imaging of acentriolar foci formation following mitotic entry. Microtubules are visualized using a transgenic EB3::mApple fusion. Scale bar, 10 µm. (D) Analysis of CEP192 foci formation for the indicated conditions. Scale bar, 10 µm. (E) Live imaging of CEP192 foci formation and bipolar division in control and CDK5RAP2-depleted cells; microtubules are visualized using a transgenic fusion of mCherry with the microtubule-binding domain of MAP4. Numbers on top left of each panel indicate time in minutes after NEBD. Scale bar, 10 µm. C, centrinone; cent, centriole; D, DMSO; ROI, region of interest.
We conclude that that loss of PCM matrix proteins is well-tolerated when centrioles are present but highly detrimental for spindle assembly when centrioles are absent. Thus, PCM matrix proteins and centrioles can function independently of each other to promote spindle formation.
PCM matrix proteins are required for the formation of CEP192-containing foci during acentriolar mitosis
To understand how the PCM matrix contributes to acentriolar spindle assembly, we imaged RPE1(USP28Δ) cells with in situ mNeonGreen-tagged CEP192 (Gemble et al., 2019). After incubation with centrinone for 4 d (Fig. 3 A), a time point when ∼75% of the cell population lacks centrioles (Fig. 2 B; Wong et al., 2015), CEP192 foci were not observed during interphase. However, after NEBD, small CEP192 foci became visible and gradually coalesced into larger foci at the poles of acentriolar spindles (Fig. 3 A; Meitinger et al., 2020; Yeow et al., 2020); the polar foci persisted into anaphase but dispersed soon after (Video 3). CEP192 fluorescence in the polar foci at anaphase onset was about half of that at centrosomes in control cells at NEBD (Fig. 3 B). CEP192 fluorescence in the spindle region increased after NEBD, concurrent with coalescence of the mitotic foci (Fig. 3 B). Imaging CEP192::mNeonGreen (mNG) with a red fluorescent EB3 fusion that marks microtubules showed that multiple CEP192 foci were associated with microtubules and progressively concentrated at the spindle poles during self-organized acentriolar spindle assembly (Fig. 3 C and Video 4). CEP192 foci at the poles of acentriolar spindles also contain PCNT, CDK5RAP2, CEP152, PLK4, and γ-tubulin (Meitinger et al., 2020), none of which exhibited focal localization in interphase, indicating that acentriolar PCM foci form during mitosis. In contrast to centrinone-treated control RNAi cells, centrinone-treated cells depleted of PCNT or CDK5RAP2 did not form CEP192 foci or bipolar spindles (Fig. 3 D). We conclude that the CEP192-containing foci at the poles of acentriolar spindles require PCNT and CDK5RAP2 and resemble the mitotic PCM matrix that accumulates around centrioles during mitotic entry, except that foci form de novo after NEBD instead of before NEBD when templated by a centriole.
Video 3.
DMSO or centrinone-treated RPE1 cells expressing in situ mNeonGreen-tagged CEP192 and transgene-encoded mRFP-H2b. 5 × 2–µm z-stacks were acquired every 5 min at 60× magnification. Image frames shown are maximum intensity projections at each time point. Playback is sped up 900× relative to real-time. Elapsed time shown is in hours:minutes, and scale bar is 10 µm.
Video 4.
DMSO or centrinone-treated RPE1 cells expressing in situ mNeonGreen-tagged CEP192 and transgene-encoded mApple-fused EB3. 7 × 2–µm z-stacks were acquired every 5 min at 40× magnification. Image frames shown are maximum intensity projections at each time point. Playback is sped up 900× relative to real-time. Elapsed time shown is in hours:minutes, and scale bar is 10 µm.
Following inhibition of centriole formation with centrinone, an intermediate on the path to the acentriolar state is the formation of daughter cells with one instead of the normal two centrioles. To assess if the PCM matrix proteins are important for one-centriole mitosis, we treated RPE1(USP28Δ) cells expressing CEP192::mNG and an mCherry fusion with the MAP4 microtubule-binding domain (MBD) with centrinone. The number of CEP192 foci present before NEBD indicated if individual cells had one or zero centrioles (Fig. 3 E). In control RNAi cells treated with centrinone, all imaged one- and zero-centriole cells exhibited bipolar division, with CEP192 foci present at the acentriolar spindle poles (Fig. 3 E and Video 5). By contrast, following RNAi of CDK5RAP2, all one- and zero-centriole cells failed to assemble bipolar spindles, and CEP192 foci were only associated with the centriole-containing pole (Fig. 3 E and Video 5). Thus, CDK5RAP2 is required for bipolar spindle assembly in cells with either one or zero centrioles, and successful spindle assembly is associated with the CDK5RAP2- and PCNT-dependent formation of acentriolar PCM foci.
Video 5.
Centrinone-treated RPE1 cells expressing in situ mNeonGreen-tagged CEP192 and transgene-encoded mCherry-fused microtubule-binding domain of MAP4. One-centrosome and zero-centrosome cells are shown in control RNAi versus CDK5RAP2 RNAi. 8 × 2–µm z-stacks were acquired every 5 min at 40× magnification. Image frames shown are maximum intensity projections at each time point. Playback is sped up 900× relative to real-time. Elapsed time shown is in hours:minutes, and scale bar is 10 µm.
Formation of acentriolar PCM foci after mitotic entry requires microtubules and PLK1 activity
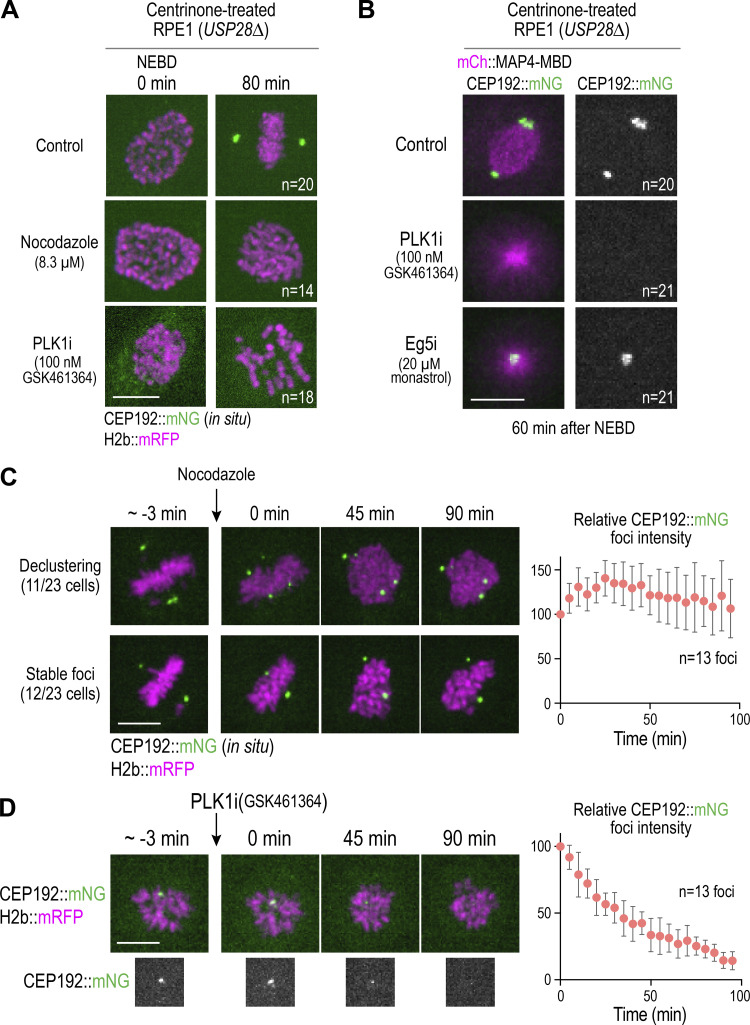
The PCM foci in acentriolar RPE1 cells form as microtubules assemble and are sorted into a self-organized bipolar spindle (Fig. 3 C), raising the question of whether microtubule assembly and reorganization are required for foci formation. To assess the involvement of microtubules, we treated acentriolar cells with nocodazole (Fig. 4 A). At a time after NEBD when live imaging of control acentriolar cells revealed robust CEP192 foci at the spindle poles, similar foci were not observed in nocodazole-treated acentriolar cells (Fig. 4 A and Video 6), indicating that formation of acentriolar PCM foci requires microtubule assembly. One possibility is that the PCM matrix proteins nucleate microtubules, and motor-dependent self-organization collects their minus ends to promote foci formation. Such a mechanism would be similar to the one previously suggested for the formation of acentriolar PCM foci in Drosophila somatic cells, which relies on minus-end–directed microtubule motors (Moutinho-Pereira et al., 2009).
Figure 4.
Requirements for assembly and maintenance of PCM foci in acentriolar cells. (A) Analysis of CEP192 foci formation after mitotic entry in control, nocodazole-treated, or PLK1-inhibited acentriolar cells generated by centrinone treatment. (B) CEP192 foci presence on monopolar spindles generated by PLK1 versus Eg5 inhibition. (C and D) Effect of depolymerizing microtubules (C) or inhibiting PLK1 (D) on maintenance of CEP192 foci formed after mitotic entry in acentriolar cells. Graph on right plots relative intensity of foci over time after inhibitor addition, normalized to the mean value at time 0. Error bars are 95% CI. Scale bars, 10 µm.
Video 6.
Centrinone-treated RPE1 cells expressing in situ mNeonGreen-tagged CEP192 and transgene-encoded mRFP-H2b either mock-treated with DMSO or treated with nocodazole or a PLK1 inhibitor. 7 × 2–µm z-stacks were acquired every 5 min at 60× magnification. Image frames shown are maximum intensity projections at each time point. Playback is sped up 900× relative to real-time. Elapsed time shown is in hours:minutes, and scale bar is 10 µm.
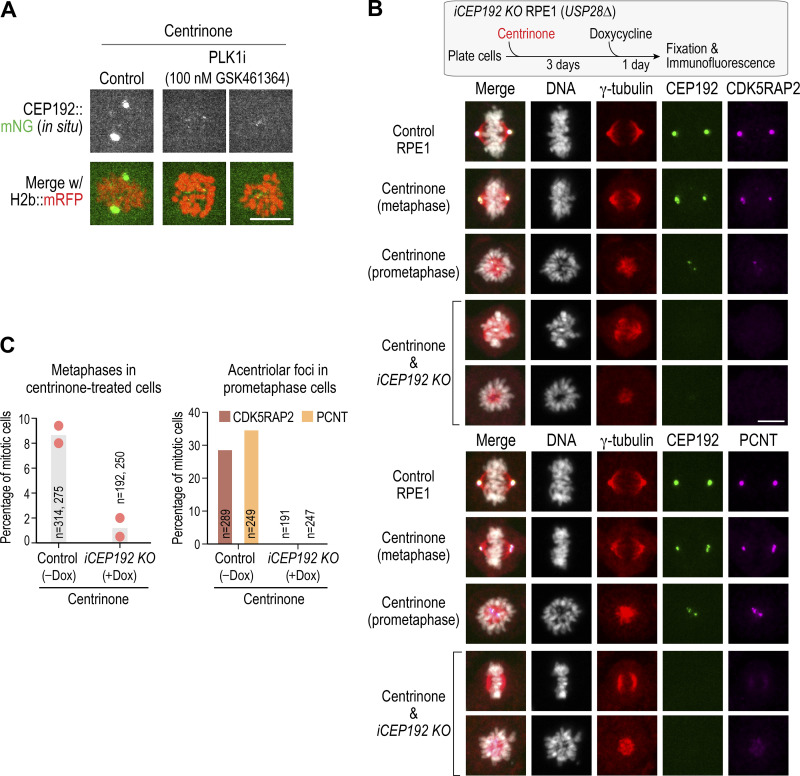
The assembly and maintenance of the mitotic PCM matrix around centrioles requires the activity of PLK1, which is thought to control the ability of matrix molecules to interact with each other (Cabral et al., 2019; Conduit et al., 2014; Dobbelaere et al., 2008; Haren et al., 2009; Lane and Nigg, 1996; Lee and Rhee, 2011; Woodruff et al., 2015). To determine whether the formation of acentriolar PCM foci also requires PLK1, we treated cells with the PLK1-specific inhibitor GSK461364 (Gilmartin et al., 2009). Robust CEP192 foci were observed in live imaging by 60 min after NEBD in DMSO-treated cells. Foci of similar size and intensity were never observed in GSK461364-treated cells (n = 21; Fig. 4, A and B; and Video 6); in four treated cells, minute foci were transiently observed (Fig. S3 A), but these failed to coalesce. As PLK1 inhibition prevents bipolar spindle formation and results in monopolar structures, we also analyzed monopolar mitotic arrays resulting from inhibition of the kinesin Eg5 (KIF11) in centrinone-treated cells. Robust CEP192 foci were present at the center of monopolar acentriolar arrays in all Eg5-inhibited cells (n = 21; Fig. 4 B). PLK1 targeting and function in PCM assembly around centrioles is thought to require CEP192 and its orthologues (Alvarez-Rodrigo et al., 2019; Decker et al., 2011; Meng et al., 2015). Consistent with this, inducibly knocking out CEP192, like PLK1 inhibition, also prevented the formation of acentriolar foci containing CDK5RAP2 and PCNT (Fig. S3, B and C). Thus, PLK1 kinase activity, potentially associated with CEP192, is required for the formation of acentriolar PCM foci during mitosis, analogous to the requirement for centrosome-targeted PLK1 in the expansion of the mitotic PCM matrix around centrioles.
Figure S3.
Examples of small CEP192 foci in PLK1-inhibited acentriolar mitotic cells and analysis of induced CEP192 knockout. (A) Example of small CEP192 foci observed in PLK1-inhibited acentriolar cells. Two examples are shown. Note that control cell image shown is 60 min after NEBD; the left PLK1-inhibited cell is 95 min after NEBD and the right PLK1-inhibited cell is 40 min after NEBD. Scale bar, 10 µm. (B) Top: Schematic of experimental approach. The inducible CEP192 knockout cell line was described previously (Meitinger et al., 2020). Cells were first treated with centrinone, and then CEP192 was inducibly deleted by adding doxycycline to express Cas9. Cells were then fixed and processed for immunofluorescence. Middle and bottom: Analysis of CDK5RAP2 (middle) and PCNT (bottom) localization in the indicated conditions. Note that the centrinone-only condition is the iCEP192 knockout cell line treated with centrinone but not with doxycycline. Scale bar, 10 µm. (C) Quantification of metaphase frequency (left) and acentriolar foci frequency (right) in the indicated conditions. Metaphase alignment is greatly reduced in frequency in centrinone-treated mitotic cells in which CEP192 has been knocked out; thus, the quantification of acentriolar foci only employed prometaphase stage cells from the control. Note that by metaphase, essentially all centrinone-treated control cells have robust acentriolar foci at their spindle poles (e.g., see Fig. S5 A). Dox, doxycycline; KO, knockout.
We next assessed if microtubules and PLK1 activity were required to maintain acentriolar CEP192 foci in mitotic cells. We imaged individual cells until they formed one or more clear CEP192 foci (∼1 h after NEBD), added nocodazole or the PLK1-specific inhibitor GSK461364, and continued imaging (Fig. 4, C and D). Nocodazole treatment resulted in CEP192 foci either declustering into smaller foci or remaining stable over an ∼100-min imaging period (Fig. 4 C). By contrast, PLK1 inhibition led to dissipation of acentriolar foci over time (Fig. 4 D). Thus, both microtubules and PLK1 activity are required to form acentriolar PCM foci during mitosis, but only PLK1 activity is continuously required to maintain them after their formation. By contrast, the requirement for microtubules is specific for acentriolar PCM foci, as prior work has suggested that microtubules are not required for the mitotic accumulation of PCM around centrioles (Khodjakov and Rieder, 1999).
The CM2 domain–containing C-terminal region of CDK5RAP2 is required to form PCM foci and for acentriolar spindle assembly
Acentriolar PCM foci appear to form via a mechanism similar to the mitotic expansion of the PCM matrix around centrioles. Human CDK5RAP2 and Drosophila Cnn have a conserved C-terminal CM2 motif that, based on structural work on Cnn, is thought to drive self-association by interacting with an internal leucine zipper–containing region in a PLK1 phosphorylation–dependent fashion (Citron et al., 2018; Feng et al., 2017; Wang et al., 2010). The CDK5RAP2 CM2 domain also mediates an interaction with PCNT. Disrupting the interaction between PCNT and CDK5RAP2 by mutating PCNT also disrupts mitotic PCM assembly (Kim and Rhee, 2014). Thus, the CDK5RAP2 CM2 domain likely promotes both self-interaction and an interaction with PCNT that underlie PCM matrix assembly.
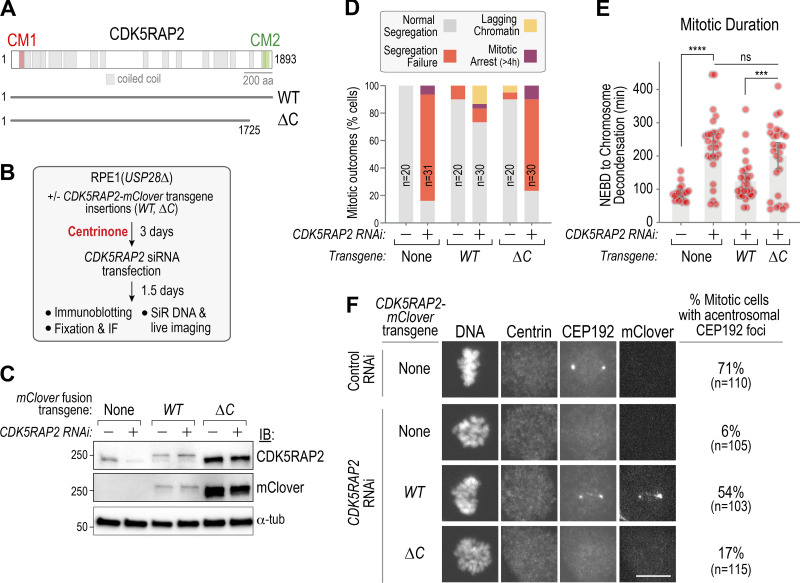
To determine if the C-terminal CM2 domain–containing region of CDK5RAP2 is required for the formation of acentriolar PCM foci, we treated RPE1(USP28Δ) cell lines stably expressing either WT or C-terminally truncated (ΔC) mClover-tagged CDK5RAP2 from RNAi-resistant transgenes (Fig. 5 A) with centrinone, followed by RNAi to deplete endogenous CDK5RAP2, and analyzed mitotic outcomes (Fig. 5 B). Immunoblotting confirmed RNAi efficacy and showed that WT CDK5RAP2 was expressed at near-endogenous levels, whereas ΔC CDK5RAP2 was expressed at a higher level (Fig. 5 C); unlike WT CDK5RAP2, ΔC CDK5RAP2 did not exhibit centrosome or Golgi localization (Fig. S4). While WT CDK5RAP2 restored CEP192 foci and significantly suppressed the mitotic defects observed following endogenous CDK5RAP2 depletion in acentriolar cells, ΔC CDK5RAP2 was unable to do so (Fig. 5, D–F).
Figure 5.
The C-terminal CM2-containing region of CDK5RAP2 is required for PCM foci formation and acentriolar mitosis. (A) Schematic of CDK5RAP2 highlighting predicted coiled-coils and the conserved CM1 and CM2 regions. The ΔC mutant deletes the C-terminal 168 amino acids that contain CM2. (B) Protocol for combining centrinone treatment and replacement of endogenous CDK5RAP2 with RNAi-resistant engineered variants. (C) Immunoblotting of the indicated conditions; mClover was detected using an anti-GFP antibody. α-Tubulin is a loading control. (D) Summary of mitotic outcomes in live imaging of the indicated conditions. (E) Mitotic duration for the indicated conditions. Error bars are 95% CI. P values are from t tests. (F) Immunofluorescence of acentrosomal CEP192 foci formation in fixed mitotic cells lacking centrioles. A centrin antibody was used to confirm centriole depletion. Scale bar, 10 µm. ***, P < 0.001; ****, P < 0.0001. IB, immunoblot; IF, immunofluorescence; ns, not significant; tub, tubulin.
Figure S4.
Localization of WT and C-terminally deleted CDK5RAP2 in control and centrinone-treated cells. CDK5RAP2 localizes to the centrosome and to the Golgi apparatus. The Golgi localization is retained in centrinone-treated cells that lack centrosomes. Neither centrosome nor Golgi localization is observed for ΔC CDK5RAP2. Scale bar, 10 µm.
We conclude that the CM2-containing C-terminal region of CDK5RAP2 is essential for the formation of acentriolar PCM foci and for spindle assembly in the absence of centrioles. This result, combined with PLK1 dependence, suggests that acentriolar PCM foci have a structural foundation similar to the PCM matrix that accumulates around centrioles during mitosis.
The γ-tubulin complex–binding CM1 motif of CDK5RAP2 is required for acentriolar spindle assembly
CDK5RAP2 family proteins have a short motif in the N-terminus, called CM1, that binds to and activates microtubule nucleation by γ-tubulin complexes (Fig. 6 A; Choi et al., 2010; Samejima et al., 2008; Zhang and Megraw, 2007). To test if γ-tubulin complex binding by CDK5RAP2 is important for acentriolar spindle formation, we expressed CDK5RAP2 with a F75A mutation (labeled CM1mut in figures; Fig. 6 A) that disrupts γ-tubulin complex binding by CDK5RAP2 (Choi et al., 2010). The F75A CM1 mutant, which was expressed at a modestly higher level than WT CDK5RAP2 (Fig. 6 B), failed to support bipolar spindle formation and exhibited penetrant mitotic phenotypes following endogenous CDK5RAP2 depletion in acentriolar cells (Fig. 6, C–E). Thus, CM1-dependent docking of γ-tubulin complexes onto CDK5RAP2 is essential for acentriolar spindle formation.
Figure 6.
The γ-tubulin complex–binding CM1 region of CDK5RAP2 is critical for acentriolar mitosis. (A) CDK5RAP2 schematic highlighting the N-terminal CM1 region; mutation of the conserved F75 residue to alanine blocks γ-tubulin binding (red asterisk). (B) Immunoblots of the indicated conditions. mClover was detected using an anti-GFP antibody; α-tubulin is a loading control; numbers below indicate relative total band intensities. (C) Mitotic outcomes observed by live imaging following centrinone treatment for the indicated conditions. (D) Mitotic duration in centrinone for the indicated conditions. Error bars are 95% CI. P value is from a t test. (E–G) Analysis of engineered CDK5RAP2 variants in the presence of endogenous CDK5RAP2. (E) Images of DMSO and centrinone-treated cells expressing WT or CM1 mutant CDK5RAP2; endogenous CDK5RAP2 is present in all conditions. (F) Immunoblots showing expression of the CM1 mutant, the C-terminal truncation, and the double mutant variants of CDK5RAP2. α-Tubulin is a loading control; numbers below indicate relative total band intensities. (G) Mitotic outcomes using live imaging following centrinone treatment for the indicated conditions. The key for the different outcomes is in C. ***, P < 0.001. IB, immunoblot; tub, tubulin.
While analyzing cells expressing CM1mut CDK5RAP2, we noticed that even without endogenous CDK5RAP2 depletion, centrinone treatment caused significant proliferation defects. This suggested that the F75A CM1 mutant, which is expressed at a higher level than endogenous CDK5RAP2 (Fig. 6 F), functioned as a dominant negative. Live imaging 3 d after centrinone treatment without endogenous CDK5RAP2 depletion revealed that whereas transgene-encoded WT CDK5RAP2 had no significant effect on mitotic outcomes, CM1mut CDK5RAP2 caused segregation failure at a frequency similar to CDK5RAP2 depletion (Fig. 6 G). Thus, the F75A CM1 mutant has a dominant-negative effect when centrioles are depleted, likely due to its higher expression relative to endogenous CDK5RAP2.
The dominant effect of the F75A mutant, defective in γ-tubulin complex interaction, may be due to the fact that this mutant retains a functional CM2 domain, which enables it to outcompete endogenous CDK5RAP2 for self-interaction and interaction with PCNT. To test this idea, we expressed a double F75A; ΔC mutant. The double mutant was expressed at a higher level than the F75A CM1 mutant alone (Fig. 6 F) but, in contrast to the single mutant, did not exhibit significant mitotic defects after 3-d centrinone treatment (Fig. 6 G). Thus, the dominant-negative effect of the F75A CM1 mutant following centrinone treatment requires the CM2-containing C-terminal region of CDK5RAP2. We conclude that acentriolar mitosis requires CM2-mediated assembly of a matrix with functional CM1 motifs that can dock γ-tubulin complexes.
Analysis of cancer cell lines suggests existence of PCNT-CDK5RAP2 and centriole-independent mechanism(s) for acentriolar spindle formation
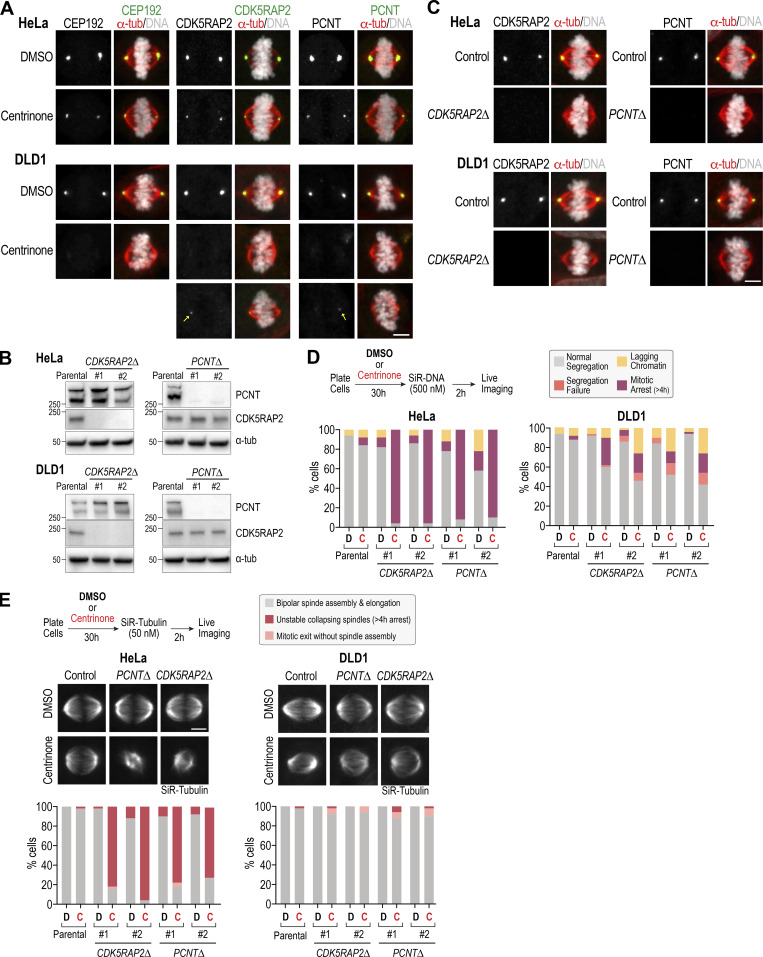
The finding that PCNT and CDK5RAP2 form acentriolar foci and become essential for spindle assembly in centrinone-treated RPE1 cells prompted us to analyze whether this was also true in other human cell lines derived from different tissue-origin cancers: HeLa (cervical carcinoma), DLD1 (colorectal carcinoma), and U2OS (osteosarcoma). As a first step, we analyzed PCM component localization in acentriolar mitotic spindles after centrinone treatment. In contrast to a recent report suggesting a lack of PCNT and CDK5RAP2 at the poles of acentriolar spindles in HeLa cells (Chinen et al., 2020), we found that HeLa cells, like RPE1 cells, reliably had foci containing CEP192, PCNT, and CDK5RAP2 at their spindle poles (Fig. 7 A and Fig. S5 A). While DLD1 cells assembled bipolar acentriolar spindles with aligned chromosomes, no CEP192 foci were detected at their spindle poles (Fig. 7 A and Fig. S5 A), and PCNT and CDK5RAP2 foci were only observed in a subset of cells (Fig. 7 A and Fig. S5 A). PCM foci were also observed at a reduced frequency in acentriolar spindles of U2OS cells (Fig. S5 B). This difference suggested that DLD1 and U2OS cells employ an acentriolar spindle assembly pathway that does not require PCM matrix foci.
Figure 7.
HeLa and DLD1 cells differ in their mechanism of acentriolar spindle assembly. (A) Immunofluorescence of control DMSO- versus centrinone-treated HeLa and DLD1 cells. Scale bar, 5 µm. (B) Immunoblots of knockout HeLa and DLD1 cells. Two independent clones are shown for each line. α-tubulin is a loading control. (C) Immunofluorescence of control and one of the knockout clones per target gene in HeLa and DLD1 cells. Scale bars, 5 µm. (D) Mitotic outcomes in live imaging of chromosome dynamics for the indicated conditions. Protocol is schematized on the top left. n = 50 cells per condition. (E) Microtubule imaging in mitotic cells for the indicated conditions. Protocol is schematized on the top left. n = 50 cells per condition. Scale bar, 5 µm. C, centrinone; D, DMSO; tub, tubulin.
Figure S5.
Quantification of acentriolar foci at spindle poles of HeLa and DLD1 cells and analysis of acentriolar mitosis in U2OS cells. (A) Left: Metaphase stage RPE1(USP28Δ) cells treated with DMSO or centrinone (for 4 d) and labeled for PCM components. Right: Quantification of acentriolar PCM foci at spindle poles in RPE1(USP28Δ), HeLa, and DLD1 cells. For images of HeLa and DLD1 cells, see Fig. 7 A. (B) Immunofluorescence analysis of control and centrinone-treated U2OS cells. Scale bar, 5 µm. (C) Immunoblot showing CDK5RAP2 depletion by RNAi in U2OS cells. (D) Functional analysis of mitotic outcomes and mitotic duration in CDK5RAP2-depleted acentriolar U2OS cells. Error bars are 95% CI. Unlike HeLa or RPE1 cells, a strong synergistic defect was not observed when comparing control and CDK5RAP2-depleted acentriolar U2OS cells. C, centrinone; D, DMSO; tub, tubulin.
To test this idea, we generated PCNT and CDK5RAP2 knockouts in HeLa and DLD1 cells; target loss was confirmed by immunoblotting and immunofluorescence (Fig. 7, B and C). Knockouts of PCNT and CDK5RAP2 were viable in HeLa and DLD1, indicating that, as in RPE1 cells, PCM matrix proteins are also not essential for viability when centrioles are present. Following centrinone treatment, the HeLa cell PCNT and CDK5RAP2 knockouts exhibited a severe spindle assembly defect (Fig. 7 D), with >90% of HeLa cells exhibiting misaligned chromosomes and long-term mitotic arrest (Fig. 7 D and Video 7). Imaging using Silicone Rhodamine (SiR)-tubulin to visualize microtubules (Lukinavičius et al., 2014) revealed that the spindles formed after centrinone treatment were small and unstable (Fig. 7 E and Video 8). We note that the centrinone treatment in this analysis was only for 30 h, and the majority of imaged cells were undergoing one-centrosome divisions. This was because, as with the RPE1 cells, longer centrinone treatment of the HeLa PCNT and CDK5RAP2 knockouts resulted in extensive lethality (data not shown).
Video 7.
Indicated HeLa cell lines labeled with SiR-DNA and imaged at 40× magnification. 5 × 2–µm z-stacks were acquired every 10 min. Image frames shown are maximum intensity projections at each time point. Playback is sped up 1,800× relative to real-time. Elapsed time shown is in hours:minutes, and scale bar is 10 µm.
Video 8.
Indicated HeLa cell lines labeled with SiR-tubulin and imaged at 40× magnification. 5 × 2–µm z-stacks were acquired every 10 min. Image frames shown are maximum intensity projections at each time point. Playback is sped up 1,800× relative to real-time. Elapsed time shown is in hours:minutes, and scale bar is 10 µm.
By contrast to HeLa and RPE1 cells, the DLD1 PCNT and CDK5RAP2 knockouts analyzed in parallel exhibited only mildly enhanced defects when treated with centrinone. In live imaging using SiR-DNA, 50–60% of the double-inhibited cells progressed through mitosis, aligning and separating their chromosomes without visible defects (Fig. 7 D and Video 9). Microtubule imaging revealed bipolar spindle formation (Fig. 7 E and Video 10); in fact, in the imaging analysis with SiR-tubulin, the 10–20% mitotic arrest observed in the analysis with SiR-DNA was not seen. This difference could be because SiR-DNA enhanced mitotic defects or because a mild microtubule-stabilizing effect of SiR-tubulin suppressed defects. While we did not generate knockouts in U2OS cells, RNAi-based depletion of CDK5RAP2 in conjunction with centrinone treatment suggested that they behave similarly to DLD1 cells (Fig. S5, C and D).
Video 9.
Indicated DLD1 cell lines labeled with SiR-DNA and imaged at 40× magnification. 5 × 2–µm z-stacks were acquired every 10 min. Image frames shown are maximum intensity projections at each time point. Playback is sped up 1,800× relative to real-time. Elapsed time shown is in hours:minutes, and scale bar is 10 µm.
Video 10.
Indicated DLD1 cell lines labeled with SiR-tubulin and imaged at 40× magnification. 5 × 2–µm z-stacks were acquired every 10 min. Image frames shown are maximum intensity projections at each time point. Playback is sped up 1,800× relative to real-time. Elapsed time shown is in hours:minutes, and scale bar is 10 µm.
Collectively, the analysis in RPE1, HeLa, DLD1, and U2OS cells suggests that some human cell types are able to assemble spindles capable of supporting chromosome alignment and segregation without centrioles or PCM matrix proteins. These findings illustrate the complexity of microtubule generation pathways that contribute to spindle assembly in human cells and highlight the utility of centrinone as a tool for analyzing microtubule generation mechanisms in diverse contexts.
Discussion
Centrosomes, composed of a centriolar core surrounded by a pericentriolar matrix, generate microtubules for spindle assembly. Classical experiments suggested a division of labor in which the centrioles recruit and organize a proteinaceous PCM matrix (Bobinnec et al., 1998; Mazia et al., 1960; Sluder and Rieder, 1985) that, in turn, docks γ-tubulin–containing complexes to nucleate microtubules (Frankel, 1976; Gould and Borisy, 1977; Keryer et al., 1984; Moritz et al., 1995, 1998; Osborn and Weber, 1976; Schnackenberg et al., 1998). Here, we test this organizational concept by deconstructing the centrosome in nontransformed RPE1 cells and three cancer cell lines: HeLa, DLD1, and U2OS. We characterize mitosis in cells with centrioles that lack the PCM matrix proteins PCNT and CDK5RAP2, in cells with PCNT and CDK5RAP2 that lack centrioles, and in cells lacking both. In two of the cell lines (RPE1 and HeLa), a similar phenomenon was observed in which cells lacking either centrioles or PCM matrix proteins were able to assemble spindles, but simultaneous loss of both PCM matrix and centrioles severely compromised spindle assembly. When centrioles were absent in these cell lines, acentriolar PCM foci formed that contributed to spindle assembly; conversely, when PCNT and CDK5RAP2 were absent, spindle assembly was supported by a centriole-based PCM matrix–independent pathway. Surprisingly, the two cancer cell lines DLD1 and U2OS retained the ability to assemble a spindle in the absence of both centrioles and PCM matrix proteins, suggesting that they can deploy microtubule generation pathways that do not require either centrioles or PCM matrix proteins.
Acentriolar PCM matrix foci can promote spindle assembly independently of centrioles
A key finding from our analysis is that formation of the PCM foci that support acentriolar spindle assembly resembles the mitotic accumulation of PCM matrix around centrioles. Like the mitotic PCM matrix, formation of acentriolar PCM foci depends on protein–protein interactions mediated by the CM2 domain of CDK5RAP2 (Citron et al., 2018; Feng et al., 2017; Kim and Rhee, 2014; Wang et al., 2010) and on PLK1 kinase activity (Cabral et al., 2019; Conduit et al., 2014; Dobbelaere et al., 2008; Haren et al., 2009; Lane and Nigg, 1996; Lee and Rhee, 2011; Woodruff et al., 2015). We additionally investigated the mechanism by which acentriolar PCM foci nucleate microtubules, finding that microtubule generation by acentriolar PCM foci depends on γ-tubulin complex binding to CDK5RAP2. Collectively, these results suggest that acentriolar spindle assembly requires the formation of bona fide PCM matrix with CM1-docked γ-tubulin complexes.
One difference between acentriolar and centriole-directed PCM matrix assembly is that the PCM matrix around centrioles begins to accumulate before NEBD and does not require microtubules, whereas acentriolar PCM foci assemble via a microtubule-dependent process after NEBD. One explanation for microtubule requirement may relate to the ability of PCNT to interact with dynein and be transported toward microtubule minus ends (Tynan et al., 2000; Young et al., 2000), a phenomenon recently shown to occur cotranslationally (Sepulveda et al., 2018). CDK5RAP2 also associates with the microtubule minus end–directed motor HSET (Chavali et al., 2016), which has both motor and nonmotor microtubule binding activities, and is implicated in clustering of microtubule-organizing centers (Kleylein-Sohn et al., 2012; Kwon et al., 2008). During acentriolar spindle assembly, minus end–directed transport of PCNT-CDK5RAP2 complexes may drive a feedback loop between microtubule generation and PCM material coalescence. We note that a similar minus end motor–based coalescence mechanism has been proposed for the formation of acentriolar PCM foci in Drosophila somatic cells (Moutinho-Pereira et al., 2009), suggesting that the ability of the PCM to assemble independently of centrioles via a microtubule-dependent process is likely to be conserved.
A centriole-anchored mechanism generates microtubules for spindle assembly in cells lacking PCM matrix proteins
RPE1 and HeLa cells undergo surprisingly normal mitosis in the absence of PCNT and CDK5RAP2, echoing prior work in chicken DT40 knockout and human cancer cell lines (Barr et al., 2010; Fong et al., 2008; Wang et al., 2013). Centrioles are present at the spindle poles of these cells, and removing them causes segregation failure, suggesting that the pathway driving spindle assembly in the absence of PCM matrix proteins is centriole-based. One potential candidate for acting in this pathway is CEP192, which functions as a complex scaffold at the centrosome, docking kinases important for centriole duplication and maturation and also binding to and promoting the centrosomal recruitment of γ-tubulin complexes (Gomez-Ferreria et al., 2007, 2012; Joukov and De Nicolo, 2018; Joukov et al., 2014; Meng et al., 2015; Zhu et al., 2008). Both candidate-focused and unbiased approaches will be needed to determine the molecular basis for the robust centriole-based microtubule generation pathway that is sufficient to support spindle assembly in the absence of the PCNT-CDK5RAP2 PCM matrix.
Centrinone reveals a diversity of acentriolar spindle assembly pathways
Our analysis in RPE1 and HeLa cells led us to expect that the synergy between PCNT-CDK5RAP2 inhibitions and centriole loss would be conserved in other human cell lines. Contrary to this expectation, our analysis of DLD1 and U2OS cells indicates that this is not the case. When acentriolar spindles in the four cell lines were compared, RPE1 and HeLa cell spindles reliably contained acentriolar PCM foci at their spindle poles, whereas DLD1 and U2OS cell spindles frequently did not. Notably, failure of spindle assembly when combining centriole loss and PCNT-CDK5RAP2 inhibitions was observed in the two cell lines that robustly formed acentriolar PCM foci. These data suggest that ability of cells to form acentriolar PCM foci correlates with the functional importance of PCM components in acentriolar spindle assembly. In addition, these results indicate the existence of additional mechanism(s) that can support acentriolar spindle assembly.
In recent work, the ubiquitin ligase TRIM37 has been suggested to control acentriolar spindle assembly following PLK4 inhibition–mediated centriole loss (Meitinger et al., 2020; Yeow et al., 2020). When TRIM37 is elevated, it targets CEP192 for degradation, which prevents the formation of PCM matrix foci and inhibits acentriolar spindle assembly (analysis of TRIM37 was conducted in RPE1, neuroblastoma CHP134, and breast cancer MCF7 cells). The correlation between loss of acentriolar PCM foci and spindle assembly failure in response to TRIM37 elevation suggests that in CHP134 and MCF7 cells, as in RPE1 and HeLa cells, acentriolar spindle assembly is reliant on the formation of PCM matrix–based foci. Consistent with this prior work, we show here that induced CEP192 knockout prevents the formation of acentriolar PCM foci by PCNT-CDK5RAP2. Addressing how CEP192 acts in both the centriole-based and acentriolar spindle assembly pathways is an important future goal emerging from this work.
Overall, our results highlight the fact that there can be significant differences in the mechanisms that generate microtubules for spindle assembly between human cell lines. Whether these differences arise due to cell transformation or are a consequence of their origins in distinct tissues will be an interesting question for future investigation. Understanding this diversity may suggest new avenues for specifically targeting the division of particular cancer cell types and could facilitate understanding tissue-specific phenotypes observed for mutations in human genes encoding centrosomal proteins (such as mutations in CDK5RAP2 that compromise the proliferation of neural progenitors in microcephaly; Jayaraman et al., 2018; Nano and Basto, 2017).
Materials and methods
Chemical inhibitors
The chemical inhibitors used in this study were the PLK4 inhibitor centrinone (LCR-263; 150–200 nM; synthesized by Sundia MediTech); the PLK1 inhibitor GSK461364 (PLK1i; 100 nM; Selleck Chemicals); the Kif11/Eg5 inhibitor monastrol (Eg5i; 20 µM; Tocris Bioscience), and nocodazole (8.3 µM; Sigma-Aldrich).
Antibodies
Antibodies against Cep192 (1–211 aa; used at 0.5 µg/ml for immunofluorescence), SAS6 (501–657 aa; used at 0.5 µg/ml for immunofluorescence), and PLK4 (814–970 aa; used at 0.5 µg/ml for immunofluorescence) were used as previously described (Meitinger et al., 2016). The following antibodies were purchased from commercial sources, with their working concentrations indicated in parentheses: anti-CDK5RAP2 (1:1,000 for immunofluorescence in Fig. 7, A and C; ab86340; Abcam) anti-CDK5RAP2 (1:1,000 for immunofluorescence in Fig. 1, A and D, and Western blotting in Figs. 2 F, 6 F and S5 C; 06–1398; Millipore), anti-CDK5RAP2 (1:500 for Western blotting in Fig. 1 A, Fig. 5 C, Fig. 6 B, and Fig. 7 B; A300-554A; Bethyl Laboratories, Inc.), anti-CEP152 (1:1,000; Abcam), anti-PCNT (1:2,000 for immunofluorescence, 1:500 for immunoblotting; ab4448; Abcam), GTU-88 (anti–γ-tubulin; 1:1,000; Sigma-Aldrich), DM1A (anti–α-tubulin; 1:5,000; Sigma-Aldrich), YOL1/34 (anti–α-tubulin; 1:500; Millipore), anti-GFP (1:500; 598; Medical and Biological Laboratories), and anti-Centrin1 (1:1,000; 20H5; Millipore). Antigens for anti-CDK5RAP2 and anti-PCNT antibodies are indicated in Fig. S1. Secondary antibodies were purchased from Jackson ImmunoResearch Laboratories and GE Healthcare.
Cell lines
All cell lines used in this study are listed in Table S1. hTERT-RPE1 (human telomerase reverse transcriptase-immortalized RPE-1) cells were grown in F12/DMEM, HeLa and Lenti-X 293T cells in DMEM, DLD1 cells in RPMI-1640, and U2OS cells in McCoy’s 5A. All growth media contained 10% FBS, 100 µg/ml streptomycin, and 100 U/ml penicillin. Cell lines were maintained in 37°C and 5% CO2. As the results of our localization analysis in HeLa cells differed from a prior report (Chinen et al., 2020), we submitted our HeLa cell line to the American Type Culture Collection for short tandem repeat profiling. The results indicated that our HeLa line is an exact match for American Type Culture Collection HeLa CCL-2.
The following transgenes, under control of the cytomegalovirus immediate early (CMV) or Ubiquitin C (UBC) promoters, were stably integrated into the genome using lentiviral constructs (Table S2): mCherry-MAP4 (CMV promoter; neomycin resistance gene); EB3-mApple (CMV promoter; neomycin resistance gene); CDK5RAP2-mClover (UBC promoter; neomycin resistance gene); CDK5RAP2-ΔC-mClover (UBC promoter; neomycin resistance gene); CDK5RAP2-F75A-mClover (UBC promoter; neomycin resistance gene), and CDK5RAP2- F75A-ΔC-mClover (UBC promoter; neomycin resistance gene). Cells were selected for 1 wk in 400 µg/ml G418.
For generating CRISPR/Cas9-mediated knockout lines, gRNAs targeting CDK5RAP2 (gRNA-1, GAAGAGGACGTCACCGTCCC; gRNA-2, GTCCTTCATGTTCCGTGCTC) or PCNT (gRNA-1, GCAAGAGCAGCGGCGCAGAA; gRNA-2, GGCTGTCGATGCGTCTGTCC) were cloned into the lentiviral vector lentiCRISPR v2 (a gift from F. Zhang, Broad Institute, Cambridge, MA; Addgene plasmid no. 52961; http://n2t.net/addgene:52961; RRID:Addgene_52961; Sanjana et al., 2014) and PX459v2 (a gift from F. Zhang; Addgene plasmid no. 48139; http://n2t.net/addgene:62988; RRID:Addgene_62988; Ran et al., 2013). To generate knockouts in DLD1, cells were transiently transfected with PX459v2 constructs (Table S2) using Lipofectamine 3000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. To generate knockouts in HeLa and RPE1, the Cas9 gene and two gRNAs were simultaneously delivered using lentiviral constructs (Table S2).
Viral particles were generated by transfecting the lentiviral packaging constructs into LentiX-293T cells using Lenti-X Packaging Single Shots (Clontech). 48 h after transfection, virus-containing medium was harvested and added to the growth medium of cells in combination with polybrene (EMD Millipore; 2.5 µg/ml for DLD1 and HeLa; 8 µg/ml for RPE1). Cells were selected with puromycin (4 µg/ml for DLD1; 10 µg/ml for RPE1; 3 µg/ml for HeLa) for 4–5 d and subsequently plated in low density (less than one cell per well) into 96-well plates. Single clones were screened by immunoblotting, immunofluorescence, and sequencing.
RPE1 CEP192-mNeonGreen were described previously (Meitinger et al., 2020). In brief, RPE1 cells were transiently transfected with pX459 expressing Cas9 as well as a gRNA (CGACTAATTGGTGAAGCTCT) that targets CEP192 close to the stop codon and subsequently infected with infectious recombinant adeno-associated virus (rAAV) particles containing the repair construct. The repair construct contained the left and right flanking region of the gRNA target site (960 bp and 672 bp; excluding the stop codon); the mNeonGreen coding sequence, for C-terminal fusion to CEP192, and the neomycin resistance gene aminoglycoside phosphotransferase from Tn5 were cloned between the left and right homology arms. The expression of the neomycin resistance gene is linked to endogenous CEP192-NeonGreen expression through a P2A sequence. Cells were selected in 400 µg/ml G418.
Cells were transfected with siRNAs (final 8 nM) using Lipofectamine RNAiMAX (Thermo Fisher Scientific) according to the manufacturer’s guidelines. The following siRNAs were used in this study (Sigma-Aldrich): MISSION siRNA Universal Negative Control #1; 5′-GCTAGTATGTCTGATACTTGG-3′ (Park et al., 2014) for CEP192; 5′-GCAGCUGAGCUGAAGGAGA[DT][DT]-3′ (Lee and Rhee, 2011) for PCNT; and 5′-UGGAAGAUCUCCUAACUAA[DT][DT]-3′ (Fong et al., 2008) for CDK5RAP2.
Proliferation assays
Experiments were performed as described previously (Meitinger et al., 2016). In brief, 100,000 cells/plate were seeded in 10-cm plates in triplicate per condition and grown in centrinone (200 nM) or DMSO. Cells were counted every 4 d and diluted to 100,000 cells/plate. Cell counting was performed using a TC20 automated cell counter (Bio-Rad).
Immunofluorescence
For immunofluorescence, 10,000 cells per well were seeded into 96-well plates 1 d before fixation. Cells were fixed in 100 µl ice-cold methanol for 5 min at −20°C, washed twice with 100 µl washing buffer (PBS containing 0.1% Triton X-100), and blocked with 100 µl blocking buffer (PBS containing 2–3% BSA, 0.1% Triton X-100, and 0.1% sodium azide) for 1 h at room temperature or overnight at 4°C. After blocking, cells were incubated with primary antibody in fresh blocking buffer (concentrations as indicated above) for 2 h at room temperature or overnight at 4°C. Cells were washed three times with washing buffer before 1 h incubation with the secondary antibody and DNA-staining Hoechst 33342 dye. Finally, cells were washed three times with washing buffer before inspection.
Imaging and quantification
Images were acquired on either a CellVoyager CV7000 or a CQ1 spinning disk confocal imaging system (Yokogawa Electric Corporation) equipped with a 20× (0.75 NA), 40× (0.95 NA), or 60× (water, 1.2 NA) U-PlanApo objective and a 2,560 × 2,160 pixel sCMOS camera (Andor). Images were taken either with no or 2 × 2 binning. Image acquisition and data analysis were performed using CellVoyager CV7000 software and ImageJ (Fiji), respectively. 5 × 2 µm z-sections were acquired with 25–50% laser power and 150 ms/z-section at 5-min intervals for 9–20 h, unless otherwise indicated.
For quantification of centrosomal or acentriolar foci signals, maximum intensity z-stack projections were analyzed using ImageJ. A rectangular box (area 1, A1) was drawn around the signal, and the mean intensity in the box was measured (mean intensity 1, I1). To correct for background signal, the box was expanded by 2 pixels on each side (area 2, A2), and the mean intensity was measured (mean intensity 2, I2). The signal and area differences between the two boxes were used to calculate the mean background signal per pixel {B = [(A2 × I2) − (A1 × I1)]/(A2 − A1)}. The centrosome and acentriolar foci intensities were calculated by subtracting the background signal (I1 − B).
For analysis of abnormal nuclei, cells with multiple nuclei, micronuclei, or abnormally shaped nuclei (with blebbing nuclei or with “holes” in the nucleus) were counted among total cells examined as shown in Fig. 2 F.
Live cell imaging
Schematics of experimental protocols are shown in the figures or described in the text. To monitor spindle formation and mitosis, the indicated cell lines were treated with DMSO or 200 nM centrinone for 3–4 d and/or transfected with siRNAs for 36 h before inspection. HeLa and DLD1 cell lines with and without CDK5RAP or PCNT knockouts were treated with 150 nM centrinone for 30 h and subsequently filmed every 10 min for 20 h. DNA and microtubules were labeled with SiR-DNA (50–500 nM) or SiR-tubulin (50 nM) 2 h before initiation of imaging.
For experiments in Fig. 4, cells were treated for 4 d in centrinone, transferred into 96-well imaging plates, and imaged every 5 min using CV7000. After 1–2 h, imaging was paused to add the indicated inhibitors (Nocodazole, PLKi, and Eg5i) and subsequently continued with the same settings for another 2 h to evaluate the effect on acentriolar CEP192 foci formation and maintenance.
For mitotic synchronization in combination with RNAi (Fig. 2 G, Fig. 3 D, and Fig. S2 A), cells were treated for 4 d with DMSO or 200 nM centrinone before a 13-h S-phase arrest in 2 mM thymidine. Subsequently, cells were washed twice with PBS before transfection of specific siRNA (control, CDK5RAP2, and PCNT). Cells were then transferred into 96-well plates and incubated for 9 h in DMSO or 200 nM centrinone. 2 mM thymidine was added for 12 h, and then the cells were released from the S-phase arrest for 7 h before addition of 100 ng/ml nocodazole for 5 h to enrich mitotic cells. The mitotic cells were incubated on ice in fresh medium to depolymerize microtubules, washed twice with PBS, and incubated for 30 min in fresh prewarmed medium at 37°C to allow mitotic spindle reformation.
Immunoblotting
For immunoblotting experiments shown in Fig. 2 F, Fig. 5 C, and Fig. 6, B and F, similar numbers of asynchronously growing cells were harvested from six-well dishes with 2 × Laemmli SDS sample buffer at 80–90% confluence, lysed by sonication, and directly applied to immunoblotting. To confirm CDK5RAP2 and PCNT knockouts (Fig. 1 A and Fig. 7 B), cells were cultured in 10-cm plates, harvested at 50–80% confluence, and lysed by sonication in RIPA buffer (Cell Signaling) with an additional protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific). Cell debris was removed by centrifugation. Cell extract concentrations were normalized based on a Protein Assay (Bio-Rad). For every sample, 30 µg protein/lane was run on Mini-PROTEAN gels (Bio-Rad) and transferred to polyvinylidene difluoride membranes using a TransBlot Turbo system (Bio-Rad). Blocking and antibody incubations were performed in Tris-buffered saline with 0.1% Tween-20 (TBS-T) + 5% nonfat dry milk. Detection was performed using HRP-conjugated secondary antibodies (GE Healthcare) with WesternBright Sirius (Advansta) or SuperSignal West Femto (Thermo Fisher Scientific) substrates. Membranes were imaged on a ChemiDoc MP system (Bio-Rad).
Statistical analysis
Statistical analysis was conducted using Prism v8 (GraphPad). P values were determined by t tests. In Fig. 1, C and E, Fig. 2, E and F, Fig. 5 E, Fig. 6 D, and Fig. S1 C, unpaired t tests assuming equal SD were performed. P > 0.05 (not significant [ns]); **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001. Data distribution was assumed to be normal, but this was not formally tested.
Online supplemental material
Fig. S1 shows the strategy for PCNT and CDK5RAP2 knockout generation, genotyping results, and analysis of CEP192 intensity. Fig. S2 shows the protocol employed for analysis of spindle assembly after nocodazole washout and results of acute centrinone treatment. Fig. S3 depicts further analysis of PLK1-inhibited acentriolar cells and analysis of an inducible CEP192 knockout. Fig. S4 shows localization of WT and ΔC CDK5RAP2. Fig. S5 shows quantification of acentriolar foci at spindle poles in different cell lines and RNAi-based analysis of U2OS cells. Video 1 and Video 2 show mitotic chromosome dynamics imaged using SiR-DNA in control and CDK5RAP2Δ (Video 1) or PCNTΔ (Video 2) RPE1 cells, treated with DMSO or centrinone. Video 3, Video 4, and Video 5 show in situ tagged CEP192::mNG in DMSO or centrinone-treated RPE1 cells, along with transgene-encoded mRFP::H2b (Video 3), mApple::EB3 (Video 4), or mCherry::MAP4 MBD (Video 5). Video 6 shows in situ tagged CEP192::mNG and transgene-encoded mRFP::H2b in centrinone-treated RPE1 cells, also treated with nocodazole or a PLK1 inhibitor. Video 7 and Video 8 show control and knockout HeLa lines treated with DMSO or centrinone and labeled with SiR-DNA (Video 7) or SiR-tubulin (Video 8). Video 9 and Video 10 show control and knockout DLD1 lines treated with DMSO or centrinone and labeled with SiR-DNA (Video 9) or SiR-tubulin (Video 10). Table S1 lists human cell lines used in the study, and Table S2 lists plasmids constructed for cell line engineering.
Supplementary Material
lists human cell lines used in the study.
lists plasmids constructed for cell line engineering.
Acknowledgments
We thank Gohta Goshima (Nagoya University, Nagoya, Japan) and Tomomi Kiyomitsu (Okinawa Institute of Science and Technology, Okinawa, Japan) for reagents, helpful discussions, and support; Dave Jenkins and Mehmet Kahraman (Small Molecule Discovery Program, Ludwig Institute, La Jolla, CA) for technical support with imaging experiments and small molecule inhibitors, respectively; and Midori Ohta for feedback on the manuscript.
This work was supported by grants from the National Institutes of Health (GM074207) to K. Oegema and from the Japan Society for the Promotion of Science (15H06270) to S. Watanabe. S. Watanabe was supported by a Uehara Memorial Foundation research fellowship and the Japan Society for the Promotion of Science Brain Circulation international exchange program (S2602). F. Meitinger received support from the German Science Foundation (ME 4713/1-1). A.K. Shiau, A. Desai, and K. Oegema received salary and other support from the Ludwig Institute for Cancer Research.
A.K. Shiau is an inventor on a patent application from the Ludwig Institute for Cancer Research, application number PCT/IB2016/000594: “Plk4 inhibitors.”
Author contributions: The study was conceptualized by S. Watanabe, F. Meitinger, K. Oegema, and A. Desai. Experiments and analysis were conducted by S. Watanabe and F. Meitinger, with input from K. Oegema, A.K. Shiau, and A. Desai. A.K. Shiau provided expertise in inhibitor treatments and facilitated live imaging analysis of mitosis. The manuscript draft was prepared by S. Watanabe, K. Oegema, and A. Desai and finalized with input from F. Meitinger and A.K. Shiau.
References
- Alvarez-Rodrigo, I., Steinacker T.L., Saurya S., Conduit P.T., Baumbach J., Novak Z.A., Aydogan M.G., Wainman A., and Raff J.W.. 2019. Evidence that a positive feedback loop drives centrosome maturation in fly embryos. eLife. 8:e50130. 10.7554/eLife.50130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr, A.R., Kilmartin J.V., and Gergely F.. 2010. CDK5RAP2 functions in centrosome to spindle pole attachment and DNA damage response. J. Cell Biol. 189:23–39. 10.1083/jcb.200912163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto, R., Lau J., Vinogradova T., Gardiol A., Woods C.G., Khodjakov A., and Raff J.W.. 2006. Flies without centrioles. Cell. 125:1375–1386. 10.1016/j.cell.2006.05.025 [DOI] [PubMed] [Google Scholar]
- Baumbach, J., Novak Z.A., Raff J.W., and Wainman A.. 2015. Dissecting the function and assembly of acentriolar microtubule organizing centers in Drosophila cells in vivo. PLoS Genet. 11:e1005261. 10.1371/journal.pgen.1005261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzi, H., and Anderson K.V.. 2014. Acentriolar mitosis activates a p53-dependent apoptosis pathway in the mouse embryo. Proc. Natl. Acad. Sci. USA. 111:E1491–E1500. 10.1073/pnas.1400568111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobinnec, Y., Khodjakov A., Mir L.M., Rieder C.L., Eddé B., and Bornens M.. 1998. Centriole disassembly in vivo and its effect on centrosome structure and function in vertebrate cells. J. Cell Biol. 143:1575–1589. 10.1083/jcb.143.6.1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral, G., Laos T., Dumont J., and Dammermann A.. 2019. Differential Requirements for Centrioles in Mitotic Centrosome Growth and Maintenance. Dev. Cell. 50:355–366. [DOI] [PubMed] [Google Scholar]
- Chavali, P.L., Chandrasekaran G., Barr A.R., Tátrai P., Taylor C., Papachristou E.K., Woods C.G., Chavali S., and Gergely F.. 2016. A CEP215-HSET complex links centrosomes with spindle poles and drives centrosome clustering in cancer. Nat. Commun. 7:11005. 10.1038/ncomms11005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinen, T., Yamamoto S., Takeda Y., Watanabe K., Kuroki K., Hashimoto K., Takao D., and Kitagawa D.. 2020. NuMA assemblies organize microtubule asters to establish spindle bipolarity in acentrosomal human cells. EMBO J. 39:e102378. 10.15252/embj.2019102378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, Y.K., Liu P., Sze S.K., Dai C., and Qi R.Z.. 2010. CDK5RAP2 stimulates microtubule nucleation by the gamma-tubulin ring complex. J. Cell Biol. 191:1089–1095. 10.1083/jcb.201007030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citron, Y.R., Fagerstrom C.J., Keszthelyi B., Huang B., Rusan N.M., Kelly M.J.S., and Agard D.A.. 2018. The centrosomin CM2 domain is a multi-functional binding domain with distinct cell cycle roles. PLoS One. 13:e0190530. 10.1371/journal.pone.0190530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit, P.T., Feng Z., Richens J.H., Baumbach J., Wainman A., Bakshi S.D., Dobbelaere J., Johnson S., Lea S.M., and Raff J.W.. 2014. The centrosome-specific phosphorylation of Cnn by Polo/Plk1 drives Cnn scaffold assembly and centrosome maturation. Dev. Cell. 28:659–669. 10.1016/j.devcel.2014.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debec, A., Détraves C., Montmory C., Géraud G., and Wright M.. 1995. Polar organization of gamma-tubulin in acentriolar mitotic spindles of Drosophila melanogaster cells. J. Cell Sci. 108:2645–2653. [DOI] [PubMed] [Google Scholar]
- Decker, M., Jaensch S., Pozniakovsky A., Zinke A., O’Connell K.F., Zachariae W., Myers E., and Hyman A.A.. 2011. Limiting amounts of centrosome material set centrosome size in C. elegans embryos. Curr. Biol. 21:1259–1267. 10.1016/j.cub.2011.06.002 [DOI] [PubMed] [Google Scholar]
- Dobbelaere, J., Josué F., Suijkerbuijk S., Baum B., Tapon N., and Raff J.. 2008. A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol. 6:e224. 10.1371/journal.pbio.0060224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, Z., Caballe A., Wainman A., Johnson S., Haensele A.F.M., Cottee M.A., Conduit P.T., Lea S.M., and Raff J.W.. 2017. Structural Basis for Mitotic Centrosome Assembly in Flies. Cell. 169:1078–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong, K.W., Choi Y.K., Rattner J.B., and Qi R.Z.. 2008. CDK5RAP2 is a pericentriolar protein that functions in centrosomal attachment of the gamma-tubulin ring complex. Mol. Biol. Cell. 19:115–125. 10.1091/mbc.e07-04-0371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong, C.S., Mazo G., Das T., Goodman J., Kim M., O’Rourke B.P., Izquierdo D., and Tsou M.F.. 2016. 53BP1 and USP28 mediate p53-dependent cell cycle arrest in response to centrosome loss and prolonged mitosis. eLife. 5:5e16270. 10.7554/eLife.16270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel, F.R. 1976. Organization and energy-dependent growth of microtubules in cells. Proc. Natl. Acad. Sci. USA. 73:2798–2802. 10.1073/pnas.73.8.2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, J., and Glover D.M.. 2012. Structured illumination of the interface between centriole and peri-centriolar material. Open Biol. 2:120104. 10.1098/rsob.120104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemble, S., Simon A., Pennetier C., Dumont M., Herve S., Meitinger F., Oegema K., Rodriguez R., Almouzni G., Fachinetti D., and Basto R.. 2019. Centromere Dysfunction Compromises Mitotic Spindle Pole Integrity. Curr. Biol. 29:3072–3080. [DOI] [PubMed] [Google Scholar]
- Gilmartin, A.G., Bleam M.R., Richter M.C., Erskine S.G., Kruger R.G., Madden L., Hassler D.F., Smith G.K., Gontarek R.R., Courtney M.P., et al. 2009. Distinct concentration-dependent effects of the polo-like kinase 1-specific inhibitor GSK461364A, including differential effect on apoptosis. Cancer Res. 69:6969–6977. 10.1158/0008-5472.CAN-09-0945 [DOI] [PubMed] [Google Scholar]
- Gomez-Ferreria, M.A., Rath U., Buster D.W., Chanda S.K., Caldwell J.S., Rines D.R., and Sharp D.J.. 2007. Human Cep192 is required for mitotic centrosome and spindle assembly. Curr. Biol. 17:1960–1966. 10.1016/j.cub.2007.10.019 [DOI] [PubMed] [Google Scholar]
- Gomez-Ferreria, M.A., Bashkurov M., Helbig A.O., Larsen B., Pawson T., Gingras A.C., and Pelletier L.. 2012. Novel NEDD1 phosphorylation sites regulate γ-tubulin binding and mitotic spindle assembly. J. Cell Sci. 125:3745–3751. 10.1242/jcs.105130 [DOI] [PubMed] [Google Scholar]
- Gould, R.R., and Borisy G.G.. 1977. The pericentriolar material in Chinese hamster ovary cells nucleates microtubule formation. J. Cell Biol. 73:601–615. 10.1083/jcb.73.3.601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haren, L., Stearns T., and Lüders J.. 2009. Plk1-dependent recruitment of gamma-tubulin complexes to mitotic centrosomes involves multiple PCM components. PLoS One. 4:e5976. 10.1371/journal.pone.0005976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman, D., Bae B.I., and Walsh C.A.. 2018. The Genetics of Primary Microcephaly. Annu. Rev. Genomics Hum. Genet. 19:177–200. 10.1146/annurev-genom-083117-021441 [DOI] [PubMed] [Google Scholar]
- Joukov, V., and De Nicolo A.. 2018. Aurora-PLK1 cascades as key signaling modules in the regulation of mitosis. Sci. Signal. 11:eaar4195. 10.1126/scisignal.aar4195 [DOI] [PubMed] [Google Scholar]
- Joukov, V., Walter J.C., and De Nicolo A.. 2014. The Cep192-organized aurora A-Plk1 cascade is essential for centrosome cycle and bipolar spindle assembly. Mol. Cell. 55:578–591. 10.1016/j.molcel.2014.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keryer, G., Ris H., and Borisy G.G.. 1984. Centriole distribution during tripolar mitosis in Chinese hamster ovary cells. J. Cell Biol. 98:2222–2229. 10.1083/jcb.98.6.2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov, A., and Rieder C.L.. 1999. The sudden recruitment of gamma-tubulin to the centrosome at the onset of mitosis and its dynamic exchange throughout the cell cycle, do not require microtubules. J. Cell Biol. 146:585–596. 10.1083/jcb.146.3.585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov, A., and Rieder C.L.. 2001. Centrosomes enhance the fidelity of cytokinesis in vertebrates and are required for cell cycle progression. J. Cell Biol. 153:237–242. 10.1083/jcb.153.1.237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S., and Rhee K.. 2014. Importance of the CEP215-pericentrin interaction for centrosome maturation during mitosis. PLoS One. 9:e87016. 10.1371/journal.pone.0087016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein-Sohn, J., Pöllinger B., Ohmer M., Hofmann F., Nigg E.A., Hemmings B.A., and Wartmann M.. 2012. Acentrosomal spindle organization renders cancer cells dependent on the kinesin HSET. J. Cell Sci. 125:5391–5402. 10.1242/jcs.107474 [DOI] [PubMed] [Google Scholar]
- Kwon, M., Godinho S.A., Chandhok N.S., Ganem N.J., Azioune A., Thery M., and Pellman D.. 2008. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 22:2189–2203. 10.1101/gad.1700908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrus, B.G., Daggubati V., Uetake Y., Scott P.M., Clutario K.M., Sluder G., and Holland A.J.. 2016. A USP28-53BP1-p53-p21 signaling axis arrests growth after centrosome loss or prolonged mitosis. J. Cell Biol. 214:143–153. 10.1083/jcb.201604054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane, H.A., and Nigg E.A.. 1996. Antibody microinjection reveals an essential role for human polo-like kinase 1 (Plk1) in the functional maturation of mitotic centrosomes. J. Cell Biol. 135:1701–1713. 10.1083/jcb.135.6.1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawo, S., Hasegan M., Gupta G.D., and Pelletier L.. 2012. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat. Cell Biol. 14:1148–1158. 10.1038/ncb2591 [DOI] [PubMed] [Google Scholar]
- Lecland, N., Debec A., Delmas A., Moutinho-Pereira S., Malmanche N., Bouissou A., Dupré C., Jourdan A., Raynaud-Messina B., Maiato H., and Guichet A.. 2013. Establishment and mitotic characterization of new Drosophila acentriolar cell lines from DSas-4 mutant. Biol. Open. 2:314–323. 10.1242/bio.20133327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K., and Rhee K.. 2011. PLK1 phosphorylation of pericentrin initiates centrosome maturation at the onset of mitosis. J. Cell Biol. 195:1093–1101. 10.1083/jcb.201106093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukinavičius, G., Reymond L., D’Este E., Masharina A., Göttfert F., Ta H., Güther A., Fournier M., Rizzo S., Waldmann H., et al. 2014. Fluorogenic probes for live-cell imaging of the cytoskeleton. Nat. Methods. 11:731–733. 10.1038/nmeth.2972 [DOI] [PubMed] [Google Scholar]
- Mazia, D., Harris P.J., and Bibring T.. 1960. The Multiplicity of the Mitotic Centers and the Time-Course of Their Duplication and Separation. J. Biophys. Biochem. Cytol. 7:1–20. 10.1083/jcb.7.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger, F., Anzola J.V., Kaulich M., Richardson A., Stender J.D., Benner C., Glass C.K., Dowdy S.F., Desai A., Shiau A.K., and Oegema K.. 2016. 53BP1 and USP28 mediate p53 activation and G1 arrest after centrosome loss or extended mitotic duration. J. Cell Biol. 214:155–166. 10.1083/jcb.201604081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger, F., Ohta M., Lee K.Y., Watanabe S., Davis R.L., Anzola J.V., Kabeche R., Jenkins D.A., Shiau A.K., Desai A., and Oegema K.. 2020. TRIM37 controls cancer-specific vulnerability to PLK4 inhibition. Nature. 585:440–446. 10.1038/s41586-020-2710-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, L., Park J.E., Kim T.S., Lee E.H., Park S.Y., Zhou M., Bang J.K., and Lee K.S.. 2015. Bimodal Interaction of Mammalian Polo-Like Kinase 1 and a Centrosomal Scaffold, Cep192, in the Regulation of Bipolar Spindle Formation. Mol. Cell. Biol. 35:2626–2640. 10.1128/MCB.00068-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennella, V., Keszthelyi B., McDonald K.L., Chhun B., Kan F., Rogers G.C., Huang B., and Agard D.A.. 2012. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat. Cell Biol. 14:1159–1168. 10.1038/ncb2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennella, V., Agard D.A., Huang B., and Pelletier L.. 2014. Amorphous no more: subdiffraction view of the pericentriolar material architecture. Trends Cell Biol. 24:188–197. 10.1016/j.tcb.2013.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz, M., Braunfeld M.B., Sedat J.W., Alberts B., and Agard D.A.. 1995. Microtubule nucleation by gamma-tubulin-containing rings in the centrosome. Nature. 378:638–640. 10.1038/378638a0 [DOI] [PubMed] [Google Scholar]
- Moritz, M., Zheng Y., Alberts B.M., and Oegema K.. 1998. Recruitment of the gamma-tubulin ring complex to Drosophila salt-stripped centrosome scaffolds. J. Cell Biol. 142:775–786. 10.1083/jcb.142.3.775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutinho-Pereira, S., Debec A., and Maiato H.. 2009. Microtubule cytoskeleton remodeling by acentriolar microtubule-organizing centers at the entry and exit from mitosis in Drosophila somatic cells. Mol. Biol. Cell. 20:2796–2808. 10.1091/mbc.e09-01-0011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nano, M., and Basto R.. 2017. Consequences of Centrosome Dysfunction During Brain Development. Adv. Exp. Med. Biol. 1002:19–45. 10.1007/978-3-319-57127-0_2 [DOI] [PubMed] [Google Scholar]
- Osborn, M., and Weber K.. 1976. Cytoplasmic microtubules in tissue culture cells appear to grow from an organizing structure towards the plasma membrane. Proc. Natl. Acad. Sci. USA. 73:867–871. 10.1073/pnas.73.3.867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo, R.E., Vogel J.M., Schnackenberg B.J., Hull D.R., and Wu X.. 2000. Centrosome maturation. Curr. Top. Dev. Biol. 49:449–470. 10.1016/S0070-2153(99)49021-0 [DOI] [PubMed] [Google Scholar]
- Park, S.Y., Park J.E., Kim T.S., Kim J.H., Kwak M.J., Ku B., Tian L., Murugan R.N., Ahn M., Komiya S., et al. 2014. Molecular basis for unidirectional scaffold switching of human Plk4 in centriole biogenesis. Nat. Struct. Mol. Biol. 21:696–703. 10.1038/nsmb.2846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran, F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., and Zhang F.. 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8:2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samejima, I., Miller V.J., Groocock L.M., and Sawin K.E.. 2008. Two distinct regions of Mto1 are required for normal microtubule nucleation and efficient association with the gamma-tubulin complex in vivo. J. Cell Sci. 121:3971–3980. 10.1242/jcs.038414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana, N.E., Shalem O., and Zhang F.. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 11:783–784. 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnackenberg, B.J., Khodjakov A., Rieder C.L., and Palazzo R.E.. 1998. The disassembly and reassembly of functional centrosomes in vitro. Proc. Natl. Acad. Sci. USA. 95:9295–9300. 10.1073/pnas.95.16.9295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulveda, G., Antkowiak M., Brust-Mascher I., Mahe K., Ou T., Castro N.M., Christensen L.N., Cheung L., Jiang X., Yoon D., et al. 2018. Co-translational protein targeting facilitates centrosomal recruitment of PCNT during centrosome maturation in vertebrates. eLife. 7:e34959. 10.7554/eLife.34959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sir, J.H., Pütz M., Daly O., Morrison C.G., Dunning M., Kilmartin J.V., and Gergely F.. 2013. Loss of centrioles causes chromosomal instability in vertebrate somatic cells. J. Cell Biol. 203:747–756. 10.1083/jcb.201309038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluder, G., and Rieder C.L.. 1985. Centriole number and the reproductive capacity of spindle poles. J. Cell Biol. 100:887–896. 10.1083/jcb.100.3.887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tynan, S.H., Purohit A., Doxsey S.J., and Vallee R.B.. 2000. Light intermediate chain 1 defines a functional subfraction of cytoplasmic dynein which binds to pericentrin. J. Biol. Chem. 275:32763–32768. 10.1074/jbc.M001536200 [DOI] [PubMed] [Google Scholar]
- Wang, Z., Wu T., Shi L., Zhang L., Zheng W., Qu J.Y., Niu R., and Qi R.Z.. 2010. Conserved motif of CDK5RAP2 mediates its localization to centrosomes and the Golgi complex. J. Biol. Chem. 285:22658–22665. 10.1074/jbc.M110.105965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Dantas T.J., Lalor P., Dockery P., and Morrison C.G.. 2013. Promoter hijack reveals pericentrin functions in mitosis and the DNA damage response. Cell Cycle. 12:635–646. 10.4161/cc.23516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, Y.L., Anzola J.V., Davis R.L., Yoon M., Motamedi A., Kroll A., Seo C.P., Hsia J.E., Kim S.K., Mitchell J.W., et al. 2015. Cell biology. Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science. 348:1155–1160. 10.1126/science.aaa5111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff, J.B., Wueseke O., and Hyman A.A.. 2014. Pericentriolar material structure and dynamics. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369:20130459. 10.1098/rstb.2013.0459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff, J.B., Wueseke O., Viscardi V., Mahamid J., Ochoa S.D., Bunkenborg J., Widlund P.O., Pozniakovsky A., Zanin E., Bahmanyar S., et al. 2015. Centrosomes. Regulated assembly of a supramolecular centrosome scaffold in vitro. Science. 348:808–812. 10.1126/science.aaa3923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeow, Z.Y., Lambrus B.G., Marlow R., Zhan K.H., Durin M.A., Evans L.T., Scott P.M., Phan T., Park E., Ruiz L.A., et al. 2020. Targeting TRIM37-driven centrosome dysfunction in 17q23-amplified breast cancer. Nature. 585:447–452. 10.1038/s41586-020-2690-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, A., Dictenberg J.B., Purohit A., Tuft R., and Doxsey S.J.. 2000. Cytoplasmic dynein-mediated assembly of pericentrin and gamma tubulin onto centrosomes. Mol. Biol. Cell. 11:2047–2056. 10.1091/mbc.11.6.2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., and Megraw T.L.. 2007. Proper recruitment of gamma-tubulin and D-TACC/Msps to embryonic Drosophila centrosomes requires Centrosomin Motif 1. Mol. Biol. Cell. 18:4037–4049. 10.1091/mbc.e07-05-0474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, F., Lawo S., Bird A., Pinchev D., Ralph A., Richter C., Müller-Reichert T., Kittler R., Hyman A.A., and Pelletier L.. 2008. The mammalian SPD-2 ortholog Cep192 regulates centrosome biogenesis. Curr. Biol. 18:136–141. 10.1016/j.cub.2007.12.055 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists human cell lines used in the study.
lists plasmids constructed for cell line engineering.