Abstract
Quantification of plasma cell-free Epstein Barr virus DNA (cf EBV DNA) has been suggested as a promising liquid biopsy assay for screening and early detection of nasopharyngeal carcinoma (NPC). However, the diagnostic value of this assay is currently not known in the population of Vietnam, one of the countries which contributed the most to the NPC cases. Herein, we have reported a highly sensitive quantitative polymerase chain reaction (qPCR)-based assay targeting cf EBV DNA for the detection of NPC. A standard curve with linear regression, R 2 = 0.9961 (range: 25-150 000 copies/mL) and a detection limit of 25 copies/mL were obtained using an EBV standard panel provided by the Chinese University of Hong Kong. The clinical performance of this assay was assessed using plasma samples obtained from 261 Vietnamese individuals. The optimized qPCR assay detected cf EBV DNA in plasma with a sensitivity of 97.4% and a specificity of 98.2%. The absolute quantitative results of pretreatment cf EBV DNA and patient overall clinical stages were statistically correlated (P < .05). In summary, the remarkably high sensitivity and specificity of our optimized qPCR assay strongly supports the wide use of cf EBV DNA quantification as a routine noninvasive method in early diagnosis and management of patients with NPC.
Keywords: nasopharyngeal carcinoma, cell-free Epstein-Barr virus DNA, quantitative PCR, early detection of cancer, Vietnam, liquid biopsy
Introduction
Nasopharyngeal carcinoma (NPC) is one of the malignancies that closely associate with Epstein-Barr virus (EBV).1-3 Interestingly, NPC shares almost the same geographical and ethnical distribution with the EBV infection.4 In particular, NPC is more prevalent in China and South-East Asian countries, with an incidence rate of up to 35 cases per 100 000 persons in some endemic areas.5 According to GLOBOCAN statistics in 2018, Vietnam was one of the countries which contributed the most to the NPC cases with 6212 new cases coressponding to an Age-Standardized Incidence Rate (ASR) = 5.7/100 000) and 4232 deaths (ASR = 3.9/100,000). In addition to EBV infection, risk factors for NPC may further include a family history of NPC, prolonged consumption of salted fish in early life, drinking, and smoking.6
Due to the fact that most patients with NPC present with nonspecific symptoms that are easily mistaken for innocuous problems such as headache, blocked ears, or a runny nose, NPC is often diagnosed at late stages, leading to a 5-year survival rate of merely 41%.7 By contrast, the survival rate could be increased up to 95% if the cancer is detected at earlier stages.7 Traditional diagnostic methods, such as biopsy and nasoendoscopy, are either invasive or ineffective for the early detection of NPC tumors in many circumstances.8-10 Thus, a noninvasive method for early diagnosis of NPC with high sensitivity is of critical need to improve patient survival.
The quantification of plasma cell-free EBV DNA (cf EBV DNA) has the potential to be a noninvasive method for early detection of NPC in endemic areas.11-13 Historically, the DNA, RNA, and proteins of EBV were detected in cancer cells and tissues from primary sites to various metastatic sites of most patients with NPC.14-22 More recently, cf EBV DNA was successfully detected in the plasma of patients with NPC by real-time quantitative polymerase chain reaction (qPCR) assays, the sensitivities of which vary from 31% to 97.1% in different study cohorts.6,13,23-26 However, the diagnostic performance of the plasma cf EBV DNA quantification in detecting NPC among Vietnamese population is not known. Although efforts have been made on the detection of EBV DNA in nasopharyngeal biopsies or brush samples, there is a lack of systemic research on establishing an optimal assay for the quantification of plasma cf EBV DNA.27,28
Herein, we aimed to propose a highly sensitive qPCR assay that measures cf EBV DNA in peripheral blood for the detection of NPC among Vietnamese patients. The sensitivity and specificity of the optimized qPCR assay were evaluated using plasma samples from 261 Vietnamese individuals.
Methods
Participants and Study Design
This cohort study involved 261 participants categorizing into 2 groups: NPC patient group (n = 152) and non-NPC individuals (n = 109) as the control group. The patient group included patients with NPC from 2 referral hospitals for cancer treatment in Hanoi, Vietnam (108 Military Central Hospital and Hanoi National Cancer Hospital K3). Inclusion criteria were: (1) biopsy-proven NPC and (2) receiving primary treatment. Participants who were diagnosed with squamous cell carcinoma, non-NPC head and neck cancers, or stage IVC NPC were excluded from our study. A total of 109 healthy volunteers were recruited to serve as controls for patient group with eligibility criteria such as (1) no risk factors for NPC (family history of NPC, smoking, alcohol drinking, frequent consumption of preserved or salted food) and (2) no occupational exposure to dusk or hazardous chemicals. Healthy volunteers have been followed up for at least 12 months after diagnosis. All participants provided written consent prior to the workup and were undergone endoscopic examination with or without biopsy for clinical staging.
Data Collection
The demographic and clinical characteristics of the patients were collected from medical records of the participants. Patients’ data included age, gender, location, and tumor characteristics (TNM staging, overall stages). Follow-up study on the control group was conducted by tele-interviews on a monthly basis.
Clinical Specimen Preparation
Peripheral blood (5 mL) was taken from each participant, placed in an EDTA-treated tube, and centrifuged at 3214g for 10 minutes (Centrifuge 5810R; Eppendorf). The plasma was then carefully transferred into 1.5-mL microtubes. The obtained plasma samples were stored at −80 °C until further processing.
The EBV Standard Preparation
The EBV standard panel used in this study was kindly donated by Prof. Allen Chan at The Chinese University of Hong Kong. Briefly, plasmid DNA bearing EBV genome is extracted from the diploid Namalwa cell line that contains 2 integrated EBV genomes per cell. A conversion factor of 6.6 pg of DNA per diploid cell was used to correlate the equivalent amounts of genomic and viral single-copy genes as previously described.11,29,30
The EBV DNA Extraction From Plasma
Plasma DNA was extracted using QIAamp Circulating Nucleic Acid Kit (Qiagen). Prior to DNA extraction, the plasma samples were thawed and centrifuged at 20 000g for 5 minutes. About 1 mL of each plasma sample per column (supplied in the QIAamp kit) was used for DNA extraction; 50 µL of distilled water was used to elute the DNA from the extraction column.
Real-Time Quantitative PCR for EBV DNA
Primer and probe design
Primers for BamHI-W target were designed and validated by Oligo Primer Analysis Software (Molecular Biology Insights). The sequences of the forward and reverse primers were 5′-CCAGACGAGTCCGTAGAAGG-3′ and 5′-AGCCTAATCCCACCCAGACT-3′, respectively, as previously described.31 BLAST sequence analysis was employed to ensure no cross-reactivity occurred with other viruses or genomic DNA. A dual fluorescence-labeled oligomer, 5′-(FAM) AGAGGAGGTGGTAAGCGGTT (BHQ1)-3′, synthesized by Integrated DNA Technologies was used as probe.
The procedure of qPCR assay
The EBV DNA in plasma was analyzed by a real-time PCR instrument system (Rotor-Gene Q), and the levels of cf EBV DNA were expressed as the number of EBV genome copies per milliliter of plasma; 8.6 µL of extracted DNA template was used in each 20 µL-qPCR reaction. Each analysis consisting of patients’ cf DNA, standard calibrators, multiple no-template controls as negative controls was done in duplicate. A standard curve, running in parallel and in duplicate with each analysis, was established by plotting threshold cycle (Ct) values against relative standard EBV DNA copy numbers. Data were collected and analyzed using Rotor-Gene Q software. Amplification signal observed in any replicate was treated as a positive result regardless of the signal level. For participants with unexpected positive test results (those who are among healthy controls), another blood sample was obtained approximately 4 weeks later for reanalysis. All DNA samples were also subjected to real-time quantitative PCR for the human β-actin gene which served as an internal control to ensure the quality of PCR amplification and to normalize real-time PCR signal. Reproducibility tests consisting of quadruplicate of standard dilutions (101-105 copies/5 μL) were performed prior to testing.
Optimization of PCR conditions
Variables affecting real-time quantitative PCR assay performance including primer concentration, Taqman probe concentration, and dimethyl sulfoxide (DMSO) concentration were optimized. Each change in variables’ concentration was tested in triplicate, and a negative control was included in each reaction to ensure no contamination is present in any of the PCR reagents. Agarose electrophoresis was used as a reference method to validate the specific amplified PCR product.
Primer concentrations
SYBR Green dye-based assays were employed to optimize the primer concentration. In these assays, Taqman probe was not used while all other reaction conditions were kept unchanged. Five different concentrations of BamHI-W primer pair ranging from 0.1 to 0.5 µM were investigated.
Taqman probe concentration
For the optimization of Taqman probe conditions, 3 different probe concentrations (0.05, 0.1, and 0.2 µM) were tested.
DMSO concentration
DMSO is known to increase PCR amplification yield and specificity of GC-rich DNA. To examine whether such additive might improve our PCR assay amplification, different concentrations of DMSO (0%, 2.5%, 5%, and 7.5%) were tested. The reaction with 0% DMSO was used as control.
Statistical Analysis
A plasma specimen is considered negative for EBV if PCR signal for internal control (β-actin gene) is valid (properly amplified) and the signal for EBV DNA is not detected. For the purposes of data analysis, samples with undetectable EBV DNA were considered to have a viral load of 0. The association between NPC clinical stages and plasma EBV-DNA levels was assessed by the Pearson χ2 test. All statistical tests were 2-sided, and a P value of less than .05 was considered to indicate statistical significance. Analyses were performed with the use of SPSS software (version 21.0; IBM Corporation).
Results and Discussion
Demographic and Clinical Characteristics of Participants
The demographic and clinical characteristics of 152 individuals diagnosed with NPC are listed in Table 1. The median age of the patient cohort at diagnosis was 50 years (range, 18-77 years); 7 (4.6%) patients were aged ≤30, 31 (20.39%) were aged 30 to 39, 35 (23.02%) were aged 40 to 49, 38 (25%) were aged 50 to 59, and 41 (26.99%) were aged ≥60. Male patients of NPC outnumbered female patients by 98 (64.5%) to 54 (35.5%). Of 152 patients with NPC, 46 had stage I and II and 106 had stage III and IV NPC. Meaning 69.74% of patients with NPC in this study were lately diagnosed (stage III and IV), while only 30.26% of them were early diagnosed (stage I and II). In general, the numbers showed that patients with NPC are mostly at the age of 60 and older (26.99%) and that males are more likely to be diagnosed with NPC than females.
Table 1.
Demographic Characteristics of 152 Patients.
| Characteristics | Value |
|---|---|
| Ethnicity, n (%) | |
| Northern Vietnam | 125 (82) |
| Central Vietnam | 27 (18) |
| Gender, n (%) | |
| Male | 98 (65) |
| Female | 54 (35) |
| Age, years | |
| Median | 50 |
| Range | 18-77 |
| Age distribution, years (%) | |
| <30 | 7 (5) |
| 30-39 | 31 (20) |
| 40-49 | 35 (23) |
| 50-59 | 38 (25) |
| ≥60 | 41 (27) |
| T stage, n (%) | |
| 1 | 48 (32) |
| 2 | 40 (26) |
| 3 | 27 (18) |
| 4 | 37 (24) |
| N stage, n (%) | |
| 0 | 25 (16) |
| 1 | 54 (36) |
| 2 | 42 (28) |
| 3 | 25 (16) |
| Undefined | 6 (4) |
| Overall stage, n (%) | |
| I and II | 46 (30) |
| III and IV | 106 (70) |
Consensus in Choosing cf EBV DNA as an NPC Biomarker
In qPCR-based diagnosis method for NPC detection, the target gene for amplification is the BamHI-W region, which is a major internal repeat sequence (primarily <181 bp) in EBV genome.32,33 Among the naturally occurring EBV isolates, the number of BamHI-W repeats varies from 5 to 11 with a mean number of 6 repeats.33 However, the use of BamHI-W in qPCR assay for NPC diagnosis may be subjected to debate due to variations caused by different repeat numbers of this sequence in quantification standard and tested samples.34 Recent research studies continue to provide evidence for the effectiveness of this target gene.35-38 Furthermore, good quantitative correlations between qPCR results for BamHI-W and single repeat EBV target genes (such as EBNA1, LMP2, or POL1)35,37,39,40 additionally consolidate the validity of this NPC biomarker. As a result, the BamHI-W has been utilized as the target gene for the design of our qPCR assay.
Optimization of PCR Assay
Details of optimal component concentrations and the final protocol for the developed qPCR assay are presented in Supplementary Tables 1 and 2. Briefly, 0.2 µM of primer, 0.05 µM of Taqman probe, and 2.5% of DMSO were used in our assay.
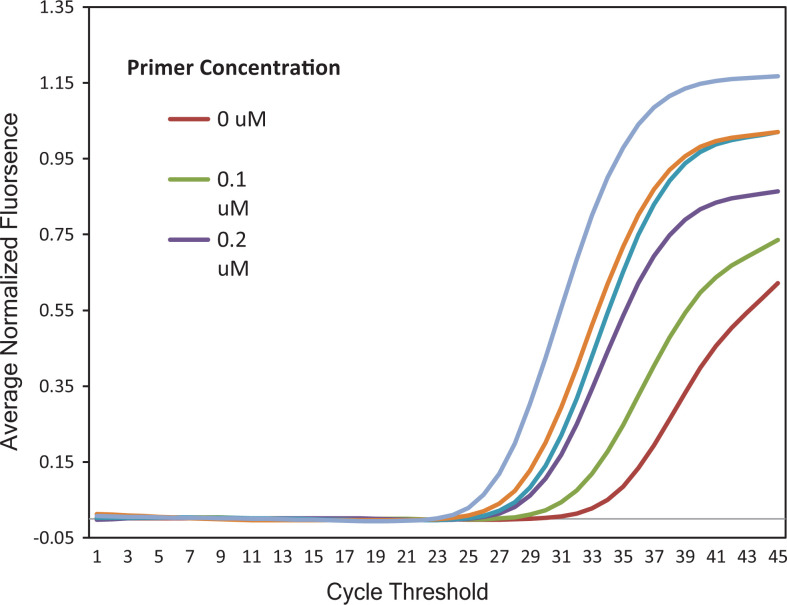
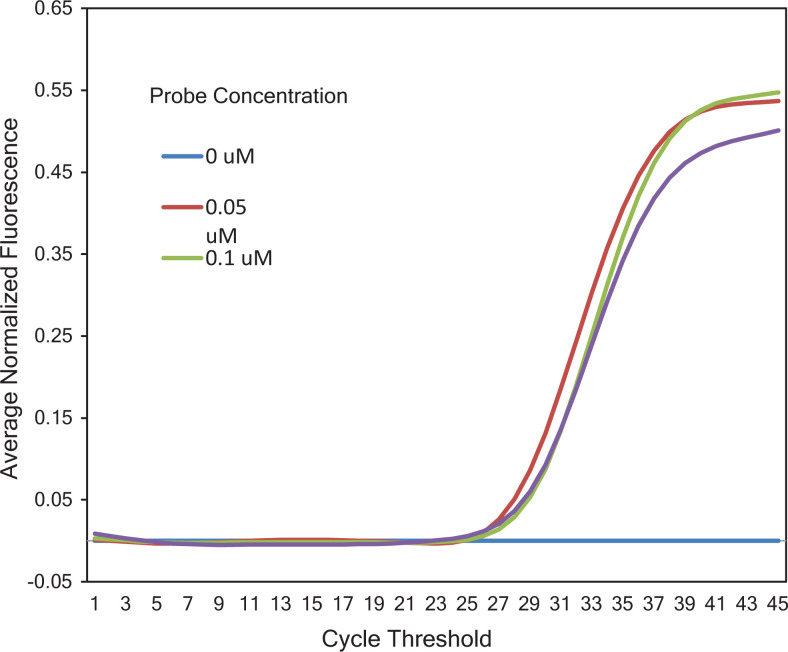
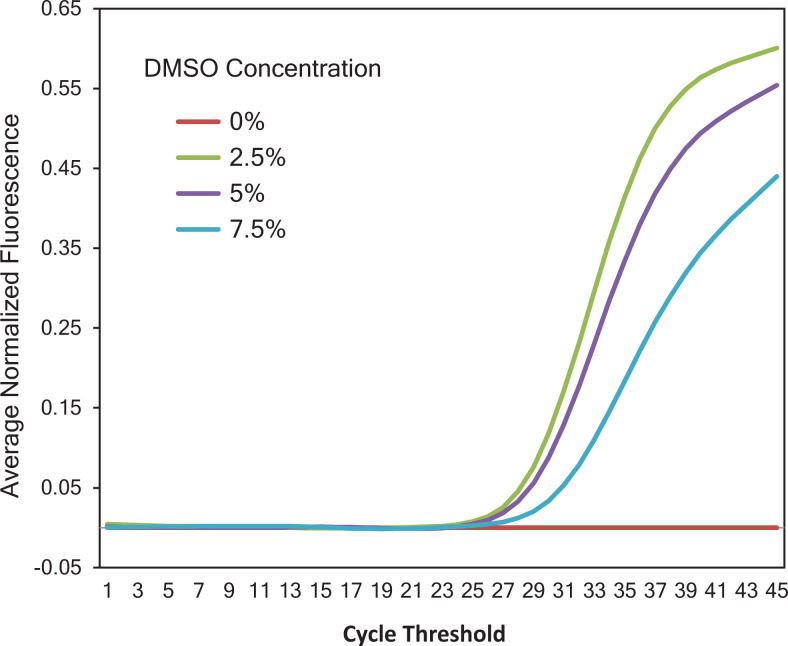
At primer concentration of 0.1 µM, no amplification signal associated with target amplicon was observed, while reactions with primer concentrations ranging from 0.2 to 0.5 µM yielded equivalent Ct values ranging from 30.89 to 31.90 (Figure 1). As a result, a final concentration of 0.2 µM was chosen as the optimal primer concentration. In reactions with different Taqman probe concentrations (0.05, 0.1, and 0.2 µM), the earliest amplification signal was observed in a reaction with probe concentration of 0.05 µM. Consequently, 0.05 µM was determined as the optimal concentration for Taqman probe in our qPCR assay (Figure 2). Figure 3 showed that on the same tested sample, reaction with 2.5% of DMSO showed the earliest amplification signal. Hence, 2.5% of DMSO is optimal for our qPCR assay. The qPCR reactions were performed with the following conditions: 15 minutes of hot start at 95 °C, followed by 45 cycles of 94 °C (15 seconds) and 63 °C (30 seconds).
Figure 1.
Optimization of the primer concentrations for quantitative polymerase chain reaction (qPCR) assay. 0.1Q, 0.2Q, 0.3Q, 0.4Q, and 0.5Q indicated primer concentrations of 0.1, 0.2, 0.3, 0.4, and 0.5 µM, respectively, in qPCR reactions with the same amount of DNA template. am Q was the negative control with primer concentration of 0.2 µM. As a result, the optimal primer concentration for the qPCR assay was determined to be 0.2 µM.
Figure 2.
Optimization of the Taqman probe concentration. Three different titers of Taqman probe at −0.2, 0.1, and 0.05 µM were used with the same amount of template DNA. (35 0.2_1-) was the negative control with a primer concentration of 0.2 µM and probe concentration of 0.2 µM.
Figure 3.
Optimization of the dimethyl sulfoxide (DMSO) concentration. Different concentrations of DMSO: 0% (SS_Q), 2.5% (2.5%_Q), 5% (5%_Q), 7.5% (7.5%_Q), and 10% (10%_Q) were tested against intact conditions of primer, probe, and DNA template concentrations. The sample with 2.5% of DMSO (2.5%_Q) gave the earliest amplification signal among those samples and therefore was chosen to be the optimal DMSO concentration for our polymerase chain reaction (PCR) assay.
Precision of the Assay
Namalwa DNA standards were serially diluted to make 5 concentrations ranging from 101 to 105 copies/5 µL (103-107 copies/mL), and quintuplicate measurements at each concentration level were subjected to qPCR assay on 4 separate days (25 total replicates per assay/day). Coefficient of variation and SD for cycle threshold (Ct) values obtained from 5 titers of EBV DNA were calculated (Table 2). Cycle threshold in qPCR is defined as the number of cycles required for the fluorescent signal to cross the threshold (background level).
Table 2.
Assay Precision Evaluated by Ct Values Obtained From Real-Time Quantitative PCR Analysis of Standard EBV DNA.
| Level | Average observed EBV DNA titer (log10 copies/mL) | Within-run precision | Between-run precision | Total precision | |||
|---|---|---|---|---|---|---|---|
| SD | CV (%) | SD | CV (%) | SD | CV (%) | ||
| 1 | 6.079 | 0.081 | 0% | 0.277 | 1% | 0.231 | 1% |
| 2 | 5.079 | 0.081 | 0% | 0.192 | 1% | 0.166 | 1% |
| 3 | 4.380 | 0.195 | 1% | 0.287 | 1% | 0.263 | 1% |
| 4 | 4.079 | 0.170 | 1% | 0.658 | 2% | 0.594 | 2% |
| 5 | 3.778 | 0.918 | 3% | 0.488 | 1% | 0.766 | 2% |
Abbreviation: CV, coefficient of variation; EBV, Epstein Barr virus; PCR, polymerase chain reaction.
Standard Curve Establishment and Limit of Detection
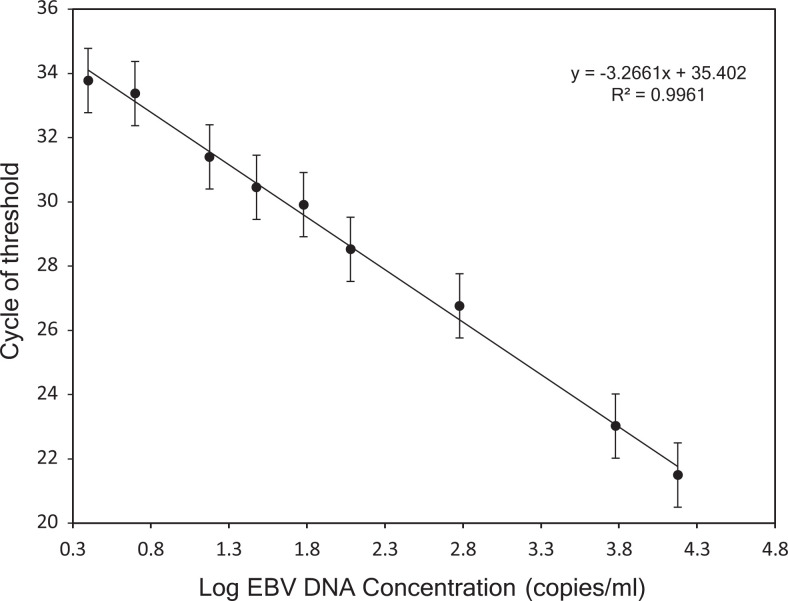
To determine the dynamic range of real-time quantitative PCR, EBV DNA standards were serially diluted to 150 000; 60 000; 6000; 1200; 600; 300; 150; 50; 25 copies/mL and tested. Triplicate at each dilution was tested on 6 separate analyses (18 total replicates per dilution). A standard curve was constructed by plotting the logarithm of the EBV DNA copy number against the measured Ct values (R 2 = 0.9961). A linear range for quantitative PCR detection of cf EBV DNA (range, 50-150, 000 copies/mL) was obtained with a detection limit of 25 copies/mL (Figure 4). All standard DNA dilutions were detected in 18 out of 18 replicates except for the lowest one (25 copies/mL) that was detected in 17 (95%) of 18 replicates. No amplification signal was observed for negative control, indicating high specificity of our assay.
Figure 4.
Linearity of the optimized quantitative polymerase chain reaction (qPCR) assay for plasma cell-free Epstein Barr virus DNA (cf EBV DNA) quantification based on international EBV standard panel. The log of known concentrations of standard (150 000, 60 000, 6000, 1200, 600, 300, 150, 50, 25 copies/mL) on the x-axis is plotted against the corresponding Ct values on the y-axis. A linear range for quantitative PCR detection of cf EBV DNA (50-150 000 copies/mL) was obtained with a detection limit of 25 copies/mL.
Results on Clinical Samples
Among 152 patients with NPC who were confirmed for NPC by endoscopic biopsy, 148 out of 152 patients (sensitivity of 97.4%) were detected by our real-time quantitative PCR assay. Meanwhile, 2 out of 109 healthy volunteers show positive results (specificity of 98.2%). These 2 patients had <40 copies/mL of EBV DNA in plasma, and the plasma samples, which were recollected after 2 weeks from these patients, were negative with EBV DNA.
Plasma samples that bear ≥300 copies/mL of EBV DNA accounted for the highest number of positives at 74.34%, followed by 23.03% for <300 copies/mL EBV DNA samples and 2.63% for 0 copies/mL (undetectable EBV DNA). The mean value of EBV-DNA load calculated from all NPC patient specimens was 12 327 copies/mL (range, 0-383,000 copies/mL; Table 3).
Table 3.
Distribution of NPC Patients (N = 152) by Mean of EBV DNA Load.
| EBV-DNA titer (copies/mL) | EBV-DNA- negative patients, n = 4 (2.63%) | EBV-DNA- positive patients, n = 148 (97.3%) | |
|---|---|---|---|
| >0 | <300 | n.a | 35 (23.03%) |
| ≥300 | n.a | 113 (74.34%) | |
| Min | 0 | 13 | |
| Max | 0 | 383,000 | |
| Mean | 0 | 12,327 | |
| SEM | 0 | 3087 | |
Abbreviations: EBV, Epstein Barr virus; n.a, not applicable; NPC, nasopharyngeal carcinoma; SEM, standard error of the mean.
a Three levels of EBV DNA (0, <300, and ≥300 copies/mL) were used to categorize diagnosed participants.
Relationship Between Pretreatment EBV DNA Levels and Overall Clinical Stages
Statistical regressions from NPC studies revealed that the pretreatment circulating cf EBV DNA levels accurately reflect clinical stages of patients with NPC.12,31,41 Meanwhile, the posttreatment EBV DNA levels are statistically associated with the possibility of recurrence,23,42 the chance of survival,25,43 and the presence of residual disease in patients with NPC.44,45 Taken together, cf EBV DNA may comprehensively indicate a patient’s tumor burden and hence could be confidently used as a biomarker in the diagnosis and treatment of NPC.46
Among 46 patients with confirmed stage I and II NPC, 44 (96%) had detectable EBV DNA in plasma with a mean copy number of 6920 copies/mL (range, 13-73 000). Of those 44 patient specimens, 13 had <300 copies/mL and 31 had ≥300 copies/mL of EBV DNA. The sensitivity of 96% in detecting patients with stage I and II NPC suggested the potential use of this noninvasive assay for early detection of NPC among Vietnamese population.
In 106 patients with confirmed stage III and IV NPC, EBV DNA was detected in 104 patients (98.11%) with a mean viral load of 15 116 copies/mL (range, 16.5-383 000). It is noted that a significantly higher proportion of these patients had ≥300 copies/mL of EBV DNA in their plasma than those who had <300 copies/mL of EBV DNA (77.4% vs 20.8%).
A correlation between the mean level of pretreatment EBV DNA and patient clinical overall stage (P = .016 and < .05) suggested that the quantification of circulating EBV DNA reliably reflects the tumor burden in patients with NPC (Table 4). This result, in consistence with previously published studies,31,39,42,47-49 again validated the effectiveness of circulating EBV DNA as a biomarker for NPC in early diagnosis and disease assessment.
Table 4.
Distributions of NPC Patients by Overall Clinical Stages and Relationships with their EBV DNA Levels.
| Variable | No. of patients (N = 152) | Undetectable EBV DNA, n (%) | EBV DNA (copies/mL) | P valuea | ||
|---|---|---|---|---|---|---|
| <300 copies/mL, n (%) | ≥300 copies/mL, n (%) | Mean ± SE (range) | ||||
| Stage I and II | 46 | 2 (4.3%) | 13 (28.3%) | 31 (67.4%) | 6920 ± 2314 (0-73 000 copies/mL) | .016 |
| Stage III and IV | 106 | 2 (1.9%) | 22 (20.8%) | 82 (77.4%) | 15 116 ± 4299 (0-383 000 copies/mL) | |
Abbreviations: EBV, Epstein Barr virus; NPC, nasopharyngeal carcinoma; SE, standard error.
a Comparison was performed using Pearson χ2 test.
Conclusion
In summary, we reported a validated highly sensitive qPCR assay for the detection of cf EBV DNA in peripheral blood of patients. The assay, with a detection limit of 25 copies/mL, provides a remarkably high sensitivity of 97.4% and high specificity of 98.2% in the detection of NPC. In addition to outstanding diagnostic performance, the advantages of convenience and noninvasiveness support this liquid biopsy assay, in combination with traditional methods, to be a new approach for future NPC screening. While further investigations with large sample size are highly warranted, it could be expected that a further endoscopy focusing on the high-risk population (cf EBV DNA positive) could improve the confirmation ability of NPC diagnosis, thus possibly eliminating the needs for repeated screening, minimizing invasive procedures, and eventually resulting in improved management of NPC and patient survival rates.
Supplemental Material
Supplemental_Material_Text for Validation of a Highly Sensitive qPCR Assay for the Detection of Plasma Cell-Free Epstein-Barr Virus DNA in Nasopharyngeal Carcinoma Diagnosis by Vu Nguyen Quynh Anh, Nguyen Van Ba, Do Tram Anh, Nguyen Dinh Ung, Nguyen Hoang Hiep, Vu Thi Ly, Dinh Thi Thu Hang, Bui Tien Sy, Hoang Dao Chinh, Le Minh Ky, Vu Truong Phong, Nguyen Kim Luu, Nguyen Thanh Trung, Ho Anh Son, Hoang Van Luong, Nghiem Duc Thuan, Ngo Thanh Tung and Ho Huu Tho in Cancer Control
Acknowledgments
The authors would like to express sincere thanks to the participants in this study. The authors also thank the medical and nursing staff of the participating hospitals for their assistance in sampling. Thanks to Prof. Allen Chan (The Chinese University of Hong Kong) for the donation of EBV standard samples. The authors thank Pham Van Quyen for excellent technical assistance and Dr Pham The Tai, Dr Hoang Xuan Su, Duong Thuy Linh, Do Lan Huong, Prof. Benjamin Pinsky, and Prof. Quynh Thu Le for their helpful discussion.
Authors’ Note: Vu Nguyen Quynh Anh, Nguyen Van Ba, and Do Tram Anh are co-first authors. This study was conducted under approval of Human Research Ethics Committee at Vietnam Military Medical University (Approval No. 2446/QĐ-HVQY 01 August 2017). All patients provided written informed consent prior to enrollment in the study.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by Hanoi Department of Science and Technology, Vietnam, under grant number 01C-08/06-2016-3.
ORCID iD: Nguyen Hoang Hiep, PhD  https://orcid.org/0000-0002-6116-6106
https://orcid.org/0000-0002-6116-6106
Ho Huu Tho, MD, PhD  https://orcid.org/0000-0002-8363-1427
https://orcid.org/0000-0002-8363-1427
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Maruo S. Epstein-Barr virus and cancers. Biotherapy. 2005;19(4):295–302. doi:10.1158/1078-0432.CCR-0670-3 [Google Scholar]
- 2. Hitt MM, Allday MJ, Hara T, et al. EBV gene expression in an NPC-related tumour. EMBO J. 1989;8(9):2639–2651. doi:10.1002/j.1460-2075.1989.tb08404.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Busson P, Keryer C, Ooka T, Corbex M. EBV-associated nasopharyngeal carcinomas: from epidemiology to virus-targeting strategies. Trends Microbiol. 2004;12(8):356–360. doi:10.1016/j.tim.2004.06.005 [DOI] [PubMed] [Google Scholar]
- 4. Ma BBY, Hui EP, Chan ATC. Investigational drugs for nasopharyngeal carcinoma. Expert Opin Investig Drugs. 2017;26(6):677–685. doi:10.1080/13543784.2017.1324568 [DOI] [PubMed] [Google Scholar]
- 5. Torre LA, Bray F, Siegel RL, Ferlay J, Tieulent JL, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi:10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 6. Chan KCA, Woo JKS, King A, et al. Analysis of plasma Epstein–Barr virus DNA to screen for nasopharyngeal cancer. N Engl J Med. 2017;377(6):513–522. doi:10.1056/NEJMoa1701717 [DOI] [PubMed] [Google Scholar]
- 7. Chai SJ, Pua KC, Saleh A, et al. Clinical significance of plasma Epstein-Barr virus DNA loads in a large cohort of Malaysian patients with nasopharyngeal carcinoma. J Clin Virol. 2012;55(1):34–39. doi:10.1016/j.jcv.2012.05.017 [DOI] [PubMed] [Google Scholar]
- 8. Zhang J, Shu C, Song Y, Li Q, Huang J, Ma X. Epstein-Barr virus DNA level as a novel prognostic factor in nasopharyngeal carcinoma: a meta-analysis. Medicine (Baltimore). 2016;95(40): e5130 doi:10.1097/MD.0000000000005130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chan KCA, Hung ECW, Woo JKS, et al. Early detection of nasopharyngeal carcinoma by plasma Epstein-Barr virus DNA analysis in a surveillance program. Cancer. 2013;119(10):1838–1844. doi:10.1002/cncr.28001 [DOI] [PubMed] [Google Scholar]
- 10. Ji MF, Huang QH, Yu X, et al. Evaluation of plasma Epstein-Barr virus DNA load to distinguish nasopharyngeal carcinoma patients from healthy high-risk populations in Southern China. Cancer. 2014;120(9):1353–1360. doi:10.1002/cncr.28564 [DOI] [PubMed] [Google Scholar]
- 11. Lo YMD, Chan LYS, Lo KW, et al. Quantitative analysis of cell-free Epstein-Barr virus DNA in plasma of patients with nasopharyngeal carcinoma. Cancer Res. 1999;59(6):1188–1191. [PubMed] [Google Scholar]
- 12. Lin JC, Wang WY, Chen KY, et al. Quantification of plasma Epstein–Barr virus DNA in patients with advanced nasopharyngeal carcinoma. N Engl J Med. 2004;350(24):2461–2470. doi:10.1056/NEJMoa032260 [DOI] [PubMed] [Google Scholar]
- 13. Mutirangura A, Pornthanakasem W, Theamboonlers A, et al. Epstein-Barr viral DNA in serum of patients with nasopharyngeal carcinoma. Clin Cancer Res. 1998;4(3):665–669. [PubMed] [Google Scholar]
- 14. Chang YS, Tyan YS, Liu ST, Tsai MS, Pao CC. Detection of Epstein-Barr virus DNA sequences in nasopharyngeal carcinoma cells by enzymatic DNA amplification. J Clin Microbiol. 1990;28(11):2398–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wu TC, Mann RB, Epstein JI, et al. Abundant expression of EBER1 small nuclear RNA in nasopharyngeal carcinoma. A morphologically distinctive target for detection of Epstein-Barr virus in formalin-fixed paraffin-embedded carcinoma specimens. Am J Pathol. 1991;138(6):1461–1469. Accessed October 18, 2019 http://www.ncbi.nlm.nih.gov/pubmed/1647139. [PMC free article] [PubMed] [Google Scholar]
- 16. Chen CL, Wen WN, Chen JY, Hsu MM, Hsu HC. Detection of Epstein-Barr virus genome in nasopharyngeal carcinoma by in situ DNA hybridization. Intervirology. 1993;36(2):91–98. doi:10.1159/000150327 [DOI] [PubMed] [Google Scholar]
- 17. Pathmanathan R, Prasad U, Chandrika G, Sadler R, Flynn K, Traub NR. Undifferentiated, nonkeratinizing, and squamous cell carcinoma of the nasopharynx. Variants of Epstein-Barr virus-infected neoplasia. Am J Pathol. 1995;146(6):1355–1367. Accessed October 18, 2019 http://www.ncbi.nlm.nih.gov/pubmed/7778675 [PMC free article] [PubMed] [Google Scholar]
- 18. Lee WY, Hsiao JR, Jin YT, Tsai ST. Epstein-Barr virus detection in neck metastases by in-situ hybridization in fine-needle aspiration cytologic studies: an aid for differentiating the primary site. Head Neck. 2000;22(4):336–340. doi:10.1002/1097-0347(200007)22:4<336:: aid-hed4>3.0.co;2-t [DOI] [PubMed] [Google Scholar]
- 19. Chao TY, Chow KC, Chang JY, et al. Expression of Epstein-Barr virus-encoded RNAs as a marker for metastatic undifferentiated nasopharyngeal carcinoma. Cancer.1996;78(1):24–29.doi:10.1002/(SICI)1097-0142(19960701)78:1<24:: AID-CNCR5>3.0.CO;2-H [DOI] [PubMed] [Google Scholar]
- 20. Macdonald MR, Freeman JL, Hui MF, et al. Role of Epstein-Barr virus in fine-needle aspirates of metastatic neck nodes in the diagnosis of nasopharyngeal carcinoma. Head Neck. 1995;17(6):487–493. doi:10.1002/hed.2880170606 [DOI] [PubMed] [Google Scholar]
- 21. Tsai ST, Jin YT, Su IJ. Expression of EBER1 in primary and metastatic nasopharyngeal carcinoma tissues using in situ hybridization. A correlation with WHO histologic subtypes. Cancer. 1996;77(2):231–236. doi:10.1002/(SICI)1097-0142(19960115)77:2<231:: AID-CNCR2>3.0.CO;2-P [DOI] [PubMed] [Google Scholar]
- 22. Niedobitek G. Epstein-Barr virus infection in the pathogenesis of nasopharyngeal carcinoma. J Clin Pathol Mol Pathol. 2000;53(5):248–254. doi:10.1136/mp.53.5.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lo YMD, Chan LYS, Chan ATC, et al. Quantitative and temporal correlation between circulating cell-free Epstein-Barr virus DNA and tumor recurrence in nasopharyngeal carcinoma. Cancer Res. 1999;59(21):5452–5455. [PubMed] [Google Scholar]
- 24. Hsiao JR, Jin YT, Tsai ST. Detection of cell free Epstein-Barr virus DNA in sera from patients with nasopharyngeal carcinoma. Cancer. 2002;94(3):723–729. doi:10.1002/cncr.10251 [DOI] [PubMed] [Google Scholar]
- 25. Lo YMD, Chan ATC, Chan LYS, et al. Molecular prognostication of nasopharyngeal carcinoma by quantitative analysis of circulating Epstein-Barr virus DNA. Cancer Res. 2000;60(24):6878–6881. [PubMed] [Google Scholar]
- 26. Shotelersuk K, Khorprasert C, Sakdikul S, Pornthanakasem W, Voravud N, Mutirangura A. Epstein-Barr virus DNA in serum/plasma as a tumor marker for nasopharyngeal cancer. Clin Cancer Res. 2000;6(3):1046–1051. [PubMed] [Google Scholar]
- 27. Nguyen HAT, Lao DT, Ngo DK, et al. Epstein-BARR virus detection in Vietnamese nasopharyngeal cancer patients based on BALF5 gene In: International Conference on the Development of Biomedical Engineering in Vietnam. Springer Verlag; 2018:255–258. doi:10.1007/978-981-10-4361-1_42 [Google Scholar]
- 28. Lao TD, Nguyen DH, Nguyen TM, Le TAH. Molecular screening for Epstein-Barr virus (EBV): detection of genomic EBNA-1, EBNA-2, LMP-1, LMP-2 among Vietnamese patients with nasopharyngeal brush samples. Asian Pacific J Cancer Prev. 2017;18(6):1675–1679. doi:10.22034/APJCP.2017.18.6.1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lawrence JB, Villnave CA, Singer RH. Sensitive, high-resolution chromatin and chromosome mapping in situ: presence and orientation of two closely integrated copies of EBV in a lymphoma line. Cell. 1988;52(1):51–61. doi:10.1016/0092-8674(88)90530-2 [DOI] [PubMed] [Google Scholar]
- 30. Whitaker AM. The chromosomes of the Namalwa cell line. J Biol Stand. 1985;13(2):173–175. Accessed October 29, 2019 http://www.ncbi.nlm.nih.gov/pubmed/3997900 [DOI] [PubMed] [Google Scholar]
- 31. Lo YMD, Leung SF, Chan LYS, et al. Plasma cell-free Epstein-Barr virus DNA quantitation in patients with nasopharyngeal carcinoma: correlation with clinical staging. Ann N Y Acad Sci. 2006;906(1):99–101. doi:10.1111/j.1749-6632.2000.tb06597.x [DOI] [PubMed] [Google Scholar]
- 32. Jones MD, Griffin BE. Clustered repeat sequences in the genome of Epstein Barr virus. Nucleic Acids Res. 1983;11(12):3919–3937. doi:10.1093/nar/11.12.3919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Allan GJ, Rowe DT. Size and stability of the Epstein-Barr virus major internal repeat (IR-1) in Burkitt’s lymphoma and lymphoblastoid cell lines. Virology. 1989;173(2):489–498. doi:10.1016/0042-6822(89)90561-8 [DOI] [PubMed] [Google Scholar]
- 34. Ryan JL, Fan H, Glaser SL, Schichman SA, Raab-Traub N, Gulley ML. Epstein-Barr virus quantitation by real-time PCR targeting multiple gene segments: a novel approach to screen for the virus in paraffin-embedded tissue and plasma. J Mol Diagn. 2004;6(4):378–385. doi:10.1016/S1525-1578(10)60535 -1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Abeynayake J, Johnson R, Libiran P, et al. Commutability of the Epstein-Barr virus WHO international standard across two quantitative PCR methods. J Clin Microbiol. 2014;52(10):3802–3804. doi:10.1128/JCM.01676-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mukohyama J, Iwakiri D, Zen Y, et al. Evaluation of the risk of lymphomagenesis in xenografts by the PCR-based detection of EBV BamHI W region in patient cancer specimens. Oncotarget. 2016;7(31):50150–50160. doi:10.18632/oncotarget.10322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sanosyan A, De Maudave AF, Bollore K, et al. The impact of targeting repetitive BamHI-W sequences on the sensitivity and precision of EBV DNA quantification. PLoS One. 2017;12(8):1–12. doi:10.1371/journal.pone.0183856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim KY, Le QT, Yom SS, et al. Current state of PCR-based Epstein-Barr virus DNA testing for nasopharyngeal cancer. J Natl Cancer Inst. 2017;109(4):1–7. doi:10.1093/jnci/djx007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vo JH, Nei WL, Hu M, et al. Comparison of circulating tumour cells and circulating cell-free Epstein-Barr virus DNA in patients with nasopharyngeal carcinoma undergoing radiotherapy. Sci Rep. 2016;6(1):1–9. doi:10.1038/s41598-016-0006-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Le QT, Jones CD, Yau TK, et al. A comparison study of different PCR assays in measuring circulating plasma Epstein-Barr virus DNA levels in patients with nasopharyngeal carcinoma. Clin Cancer Res. 2005;11(16):5700–5707. doi:10.1158/1078-0432.CCR-05-0648 [DOI] [PubMed] [Google Scholar]
- 41. Alfieri S, Iacovelli NA, Marceglia S, et al. Circulating pre-treatment Epstein-Barr virus DNA as prognostic factor in locally-advanced nasopharyngeal cancer in a nonendemic area. Oncotarget. 2017;8(29):47780–47789. doi:10.18632/oncotarget.17822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ferrari D, Codecà C, Bertuzzi C, et al. Role of plasma EBV DNA levels in predicting recurrence of nasopharyngeal carcinoma in a western population. BMC Cancer. 2012;12:208 doi:10.1186/1471-2407-12-208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li WF, Zhang Y, Huang XB, et al. Prognostic value of plasma Epstein–Barr virus DNA level during posttreatment follow-up in the patients with nasopharyngeal carcinoma having undergone intensity-modulated radiotherapy. Chin J Cancer. 2017;36(1):1–9. doi:10.1186/s40880-017-0256-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chan ATC. Plasma Epstein-Barr virus DNA and residual disease after radiotherapy for undifferentiated nasopharyngeal carcinoma. CancerSpectrum Knowl Environ. 2002;94(21):1614–1619. doi:10.1093/jnci/94.21.1614 [DOI] [PubMed] [Google Scholar]
- 45. Lin JC, Wang WY, Chen KY, et al. Quantification of plasma Epstein-Barr virus DNA in patients with advanced nasopharyngeal carcinoma. N Engl J Med. 2004;350(24):2461–2470. doi:10.1056/NEJMoa032260 [DOI] [PubMed] [Google Scholar]
- 46. Kanakry J, Ambinder R. The biology and clinical utility of EBV monitoring in blood. Curr Top Microbiol Immunol. 2015;391:475–499. doi:10.1007/978-3-319-22834-1_17 [DOI] [PubMed] [Google Scholar]
- 47. Zhang L, Tang LQ, Chen QY, et al. Plasma Epstein-Barr viral DNA complements TNM classification of nasopharyngeal carcinoma in the era of intensity-modulated radiotherapy. Oncotarget. 2016;7(5):6221–6230. doi:10.18632/oncotarget.6754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhao F-P, Liu X, Chen X-M, et al. Levels of plasma Epstein-Barr virus DNA prior and subsequent to treatment predicts the prognosis of nasopharyngeal carcinoma. Oncol Lett. 2015;10(5):2888–2894. doi:10.3892/ol.2015.3628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lee VHF, Kwong DLW, Leung TW, et al. Prognostication of serial post-intensity-modulated radiation therapy undetectable plasma EBV DNA for nasopharyngeal carcinoma. Oncotarget. 2017;8(3):5292–5308. doi:10.18632/oncotarget.14137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental_Material_Text for Validation of a Highly Sensitive qPCR Assay for the Detection of Plasma Cell-Free Epstein-Barr Virus DNA in Nasopharyngeal Carcinoma Diagnosis by Vu Nguyen Quynh Anh, Nguyen Van Ba, Do Tram Anh, Nguyen Dinh Ung, Nguyen Hoang Hiep, Vu Thi Ly, Dinh Thi Thu Hang, Bui Tien Sy, Hoang Dao Chinh, Le Minh Ky, Vu Truong Phong, Nguyen Kim Luu, Nguyen Thanh Trung, Ho Anh Son, Hoang Van Luong, Nghiem Duc Thuan, Ngo Thanh Tung and Ho Huu Tho in Cancer Control