Abstract
The respiratory complex I transduces redox energy into an electrochemical proton gradient in aerobic respiratory chains, powering energy-requiring processes in the cell. However, despite recently resolved molecular structures, the mechanism of this gigantic enzyme remains poorly understood. By combining large-scale quantum and classical simulations with site-directed mutagenesis and biophysical experiments, we show here how the conformational state of buried ion-pairs and water molecules control the protonation dynamics in the membrane domain of complex I and establish evolutionary conserved long-range coupling elements. We suggest that an electrostatic wave propagates in forward and reverse directions across the 200 Å long membrane domain during enzyme turnover, without significant dissipation of energy. Our findings demonstrate molecular principles that enable efficient long-range proton–electron coupling (PCET) and how perturbation of this PCET machinery may lead to development of mitochondrial disease.
Introduction
With up to 45 subunits and a molecular mass of 1.0 MDa, the respiratory complex I (NADH:ubiquinone oxidoreductase) is one of the largest and most complicated membrane-bound proteins.1−4 This gigantic enzyme functions as the initial electron acceptor in respiratory chains of mitochondria and aerobic bacteria by transferring electrons between nicotinamide adenine dinucleotide (NADH, Em = −320 mV) and ubiquinone (Q, Em = +90 mV), and the 0.8 V redox span is employed to pump four protons across the membrane.5 The established proton motive force (pmf) is further used for active transport and synthesis of adenosine triphosphate (ATP) by the rotary motion of FoF1-ATP synthase.6,7
In recent years, several structures of complex I from different organisms have been resolved,8−11 showing that the electron transfer (eT) and proton pumping machineries are separated into unique domains of the enzyme. The eT module in the hydrophilic domain extends up to 100 Å above the membrane plane, whereas proton pumping is catalyzed by the 200 Å long membrane domain (Figure 1).8,9 The complete charge transfer (CT) process thus spans nearly 300 Å, but despite its large spatial extent, the process is fully reversible and strongly coupled. Consistent with the principle of microscopic reversibility, perturbations at the far end of the pumping module result in decreased Q oxidoreduction activity.12−17 Although molecular principles of this remarkable long-range proton-coupled electron transfer (PCET) process have recently started to emerge,1−4,8−11,18−21 the overall mechanism still remains one of the most complex unsolved questions in biochemistry (cf. ref (22) and references therein for definition and discussion of both short- and long-range PCET).
Figure 1.
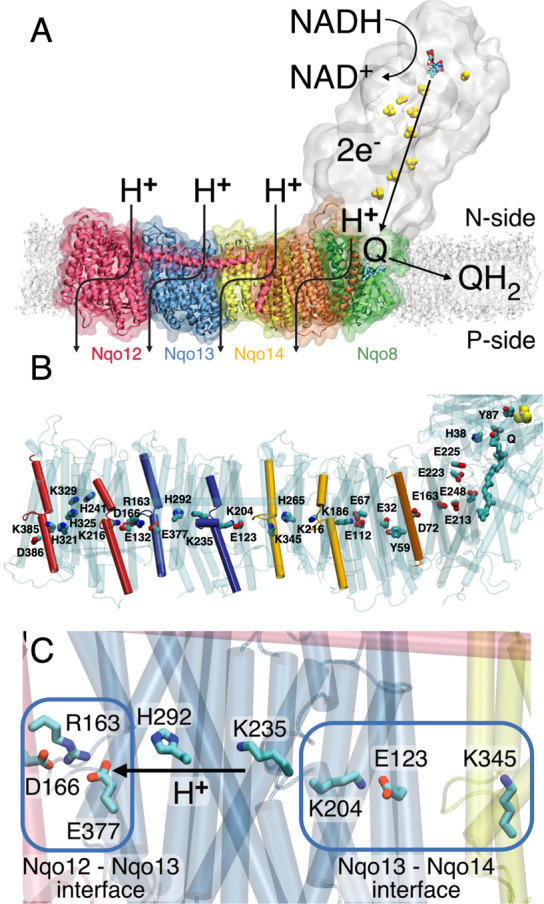
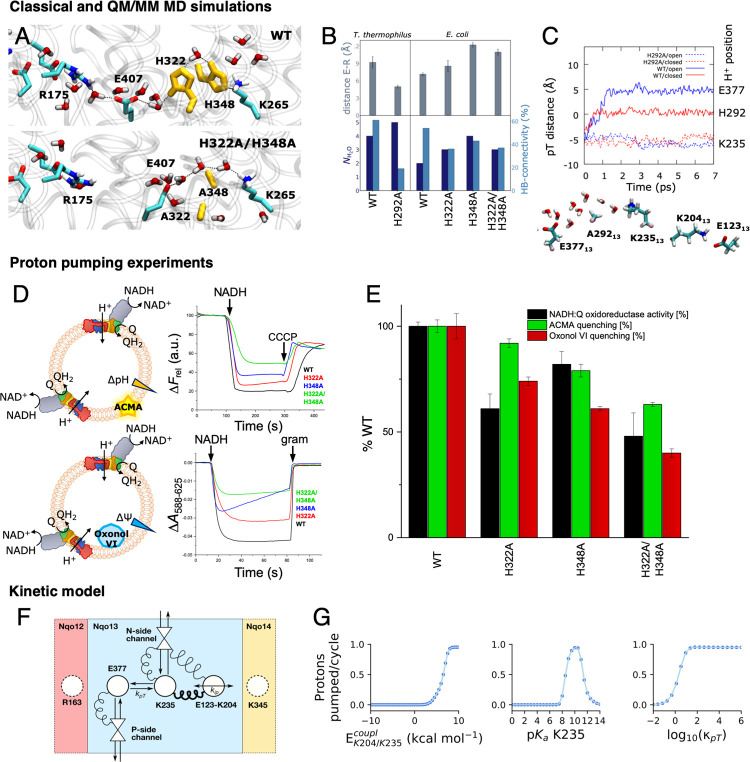
(A) Complex I catalyzes electron transfer from NADH to Q in its hydrophilic domain (in white) and pumps protons across the Nqo12 (red), Nqo13 (blue), and Nqo14 (yellow), as well as in the Nqo8 (green) and Nqo7/10/11 (in orange) subunits of the membrane domain. (B) An axis of conserved charged and polar residues spans the membrane domain from the Q channel to the terminal Nqo12 subunit. (C) Nqo13 is connected to its neighboring subunits by charged interfaces (blue squares) that enable formation of ion-pairs between the subunits. The lateral pT from Lys23513 via His29213 to Glu37713 is marked with an arrow.
Structural data in combination with computational and biophysical experiments provide a starting point to derive a molecular understanding of the functional principles in complex I.1−4,8−10,23−30 Recent studies suggest that proton pumping is triggered by electrostatic and conformational transitions resulting from the reduction of quinone,23,28 bound around 20 Å above the membrane plane in a protein cavity near the N2 iron–sulfur (FeS) center (Figure 1A).30 The thermodynamic driving force (ΔG = 0.8 V) for the pumping process is likely to originate from motion and coupled conformational transitions of the Q-species from a low potential site near the FeS chain (−300 mV) to a second, transient Q-binding site in the membrane domain with a redox potential most likely close to that of Q in membranes (+90 mV),1,30−32 which still remains to be experimentally validated.
The membrane domain comprises the antiporter-like subunits Nqo12/13/14 (T. thermophilus nomenclature) that contain a 200 Å long array of buried charged residues located at the center of the membrane and provide key elements in the pumping machinery (Figure 1B). Molecular simulations1,25,27,29,33 suggest that proton channels are established around broken helix motifs TM7a/b and TM12a/b within five-helical transmembrane (TM) bundle segments of the antiporter-like subunits, with several residues along these pathways indirectly supported by site-directed mutagenesis experiments.12−17 The proton transfer (pT) involves hydration changes within TM4–8 and TM9–13 in Nqo12/13/14, and indications of a fourth proton channel have been found near Nqo8/Nqo10 that also involves the TM2–6 bundle segment of Nqo8.8,10,27 The proton channels start from the negatively charged side (N-side) of the membrane and connect via a horizontal axis along the membrane plane to output sites at the positively charged side (P-side) of the membrane. Recent data25,27 suggest that the pumping process is mediated by a central lysine residue (Lys21614/Lys23513/Lys32912) that pushes the protons laterally toward a terminal lysine (Lys34514/Lys38512) in Nqo12/14 or a glutamate (Glu37713) residue in Nqo13, with pT reactions that are controlled by the conformational state of the conserved Lys/Glu (Asp) ion-pair at the interface of each subunit (Figure 1).1,25,27 Long-range proton pumping was suggested to propagate by an electrostatic wave in forward and reverse pulses across the membrane domain.1 It was also suggested that complex I could drive pumping by direct conformational changes, e.g., along the 100 Å horizontal helix connecting Nqo12 to Nqo148,26 or β-sheet motifs between subunits.4,8,26 Site-directed mutagenesis experiments34−36 do not currently support such “steam engine/piston”-models, whereas local conformational/electrostatic coupling between the Q site and Nqo8 has indirectly been supported by structural and biochemical studies.23,28
Our previous work has shown that complex I employs proton pathways in symmetry-related locations25,27,29 to transfer protons across the membrane, with both hydration changes27,29 and ion-pair dynamics25,27 forming important elements for the long-range charge transfer process. Here we elucidate how the ion-pair dynamics determines both the wetting/dewetting transitions and proton transfer energetics, and vice versa, using both classical and hybrid quantum/classical (QM/MM) free energy simulations. We show how the hydration state of the antiporter-like subunit Nqo13 strongly modulates the protonation and ion-pair dynamics and further validate the findings by site-directed mutagenesis experiments, pumping measurements, and kinetic models.
Results
Ion-Pair Conformation Is Determined by the Hydration State of the Proton Channels
To probe the coupling between the hydration state and the proton transfer reactions, we performed classical molecular dynamics (MD) simulations on complex I from Thermus thermophilus (PDB ID: 4HEA(8)) embedded in a lipid membrane environment. Consistent with previous simulations,25,27 the membrane domain spontaneously hydrates on 0.2 μs time scales, and water molecules establish quasi-one-dimensional wires that connect the N-side bulk with buried charged and polar residues at the center of the antiporter-like subunits (Figure S1A). In Nqo13, the water molecules enter TM7a from the N-side solvent connecting to Lys23513, from which the proton wire continues further via His29213 to Glu37713 of TM12a and to the P-side of the membrane (Figure S1A), with similar hydration patterns observed also in Nqo12 and Nqo14.25,27 The simulations support that the channel hydration strongly depends on the protonation state of the middle Lys23513 (Figure S1B), consistent with previous data.25,27
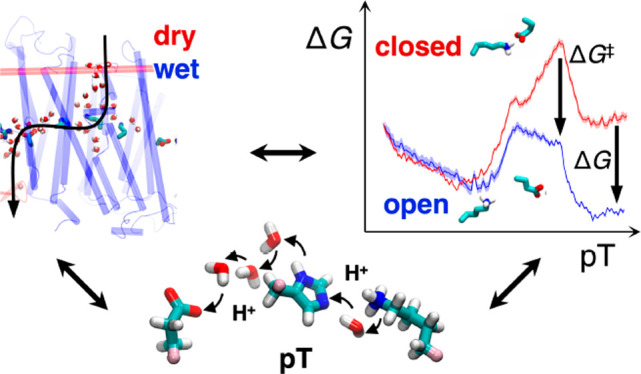
On the basis of selected snapshots of the hydration trajectory, we next computed free energy profiles for opening the central Lys/Glu ion-pair in Nqo13 (Lys20413/Glu12313) in a dry state (N < 5 water molecules), at intermediate channel hydration levels (N ≈ 10 water molecules), and in a fully hydrated “wet state” of the channel (N > 15 water molecules) (Figure 2). The wet/dry transition is likely to represent key intermediate states in the pumping process of complex I, as supported by the steady state equilibrium reached during the MD and free energy simulations (Figures S1, S10). Rigorous grand-canonical hydration free energy calculations37 are outside the scope of our large complex I simulations, with close to 1 million atoms. In the open ion-pair conformation, formed by shifting Glu12313 to establish a new ion-pair with Lys34514 (Figure 2B), Lys20413 is stabilized by π–cation interactions with Tyr23413/Trp21313 (Figure S2B), which pushes Lys23513 closer to His29213 by ∼1 Å (Figure S2D) and enabled by the flexibility of the broken helix TM7a/b. However, despite key conformational changes at the local side-chain level, the average (maximum) root-mean square deviation on the protein backbone is <1.5 Å (<2 Å) (Figure S2C), suggesting that subtle conformational changes are sufficient to support the switching dynamics.
Figure 2.
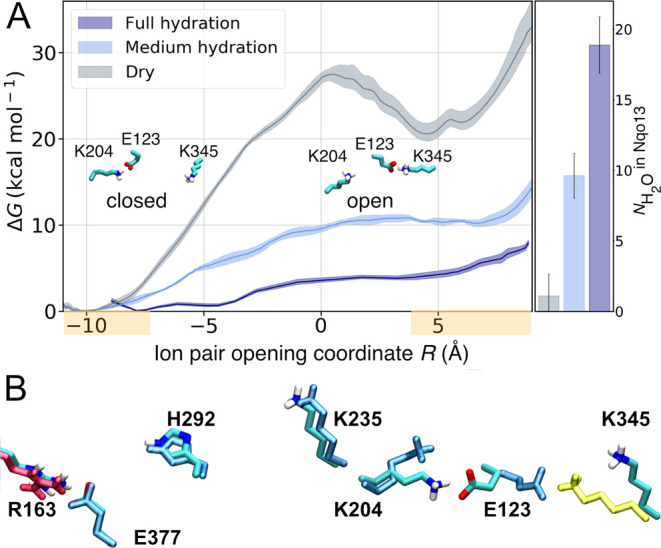
(A) Free energy profiles for opening the Lys20413/Glu12313 ion-pair at the Nqo13/Nqo14 interface in the fully hydrated state (dark blue), at medium hydration levels (light blue), and in the dry state (gray) of Nqo13. The Glu/Lys residues were modeled in their charged state (see Table S10). The ion-pair opening (shifting) reaction coordinate, R, is defined as d(E12313-K20413) – d(E12313-K34514). Closed ion-pair R ∼ (−9 ± 2) Å and open ion pair R∼ (+5 ± 2) Å are marked in orange on the R-axis. Ion-pair opening is favored by the increased hydration state of the proton channel. (B) In the closed conformation, the ion-pair interaction is established between Lys20413 and Glu12313, whereas in the open conformation, Glu12313 forms a contact with Lys34514 (in yellow).
In the dry state, our replica-exchange umbrella sampling (REUS) calculations result in a high dissociation free energy barrier of Lys20413/Glu12313 ion-pair of 27 kcal mol–1 with an open state free energy of >20 kcal mol–1 above the closed ion-pair configuration (Figure 2) that is both kinetically and thermodynamically expected to be blocked during complex I turnover on the millisecond time scale.1−4 Interestingly, the free energy barrier drops to ∼10 kcal mol–1 at intermediate hydration levels, whereas in the fully hydrated state, the ion-pair opening barrier is only ∼4 kcal mol–1 (Figure 2). This suggests that the opening dynamics becomes kinetically accessible in the nano- to-microsecond time scales upon channel wetting but is kinetically prevented when the proton channels are dry.
To further validate the computed free energies, we performed unbiased MD simulations of the fully hydrated state, mimicking states prior to and after the pT reaction. When Lys23513 is protonated, the electrostatic repulsion between the middle lysine and Lys20413 rapidly closes the Lys20413/Glu12313 ion-pair. However, in stark contrast, when the proton is moved from the middle lysine to Glu37713, the ion-pair remains open for at least 100 ns (Figure S3A,B), further supporting the tight coupling between the protonation and ion-pair dynamics.
Ion-Pair Dynamics Triggers Proton Transfer
Having established that the channel hydration controls the ion-pair dynamics, we next probed how the pT reaction itself is affected by the conformational state of the ion-pair. As classical simulations cannot capture bond-formation/breaking processes, we employed density functional theory (DFT)-based QM/MM molecular dynamics simulations and free energy calculations to elucidate reaction barriers and thermodynamic driving forces for the pT reaction (see Materials and Methods). In the QM/MM calculations, we opened up a reactive region of nearby polar protein residues and water molecules connecting Lys23513 via His29213 to Glu37713 while electrostatically coupling the system to the surrounding protein (Figure 3A). When the ion-pair is in the closed conformation, we obtain an overall endergonic free energy profile for the pT to Glu37713 with a barrier of ∼10 kcal mol–1, whereas dissociation of the ion-pair leads to an exergonic pT reaction of approximately −2 kcal mol–1, and the reaction barrier drops to ∼4 kcal mol–1, making the overall reaction accessible to nanosecond time scales (Figure 3C). This suggests that the formation of the open ion-pair (Glu12313/Lys20413) conformation rather than pT between Lys23513 and Glu37713 itself becomes rate-limiting for the overall process. The dynamics observed in our unbiased QM/MM MD simulations is consistent with the calculated free energy profiles, with the proton remaining bound to the initial Lys23513/His29213 bridge in the closed ion-pair conformation, whereas spontaneous pT to Glu37713 is observed in selected snapshots of the open ion-pair state (Figure 3D). The free energy barrier for the dry-state pT reaction is >20 kcal mol–1 in both closed and open ion-pair conformations (Figure S3D), suggesting that the pT reaction is water-gated.
Figure 3.
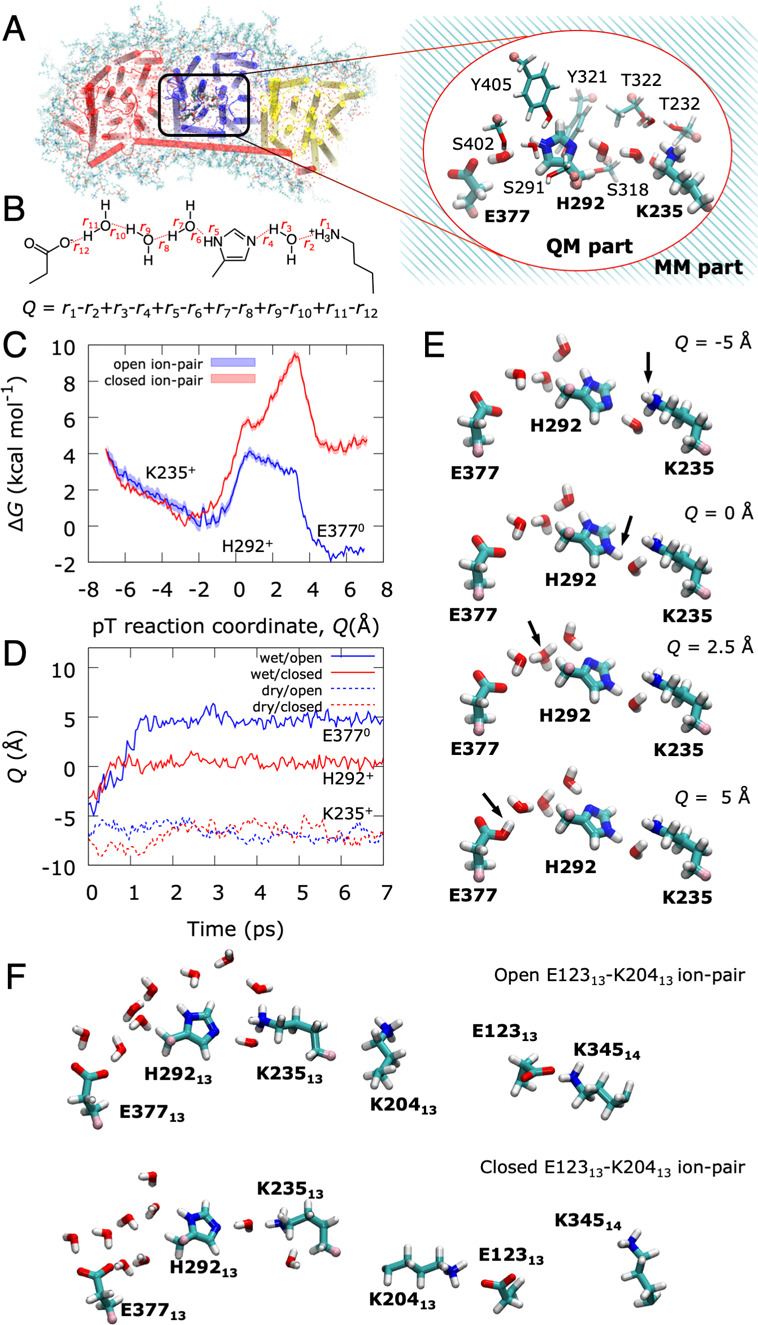
Free energy and dynamics of lateral proton transfer in Nqo13. Prior to the pT reaction, all Glu/Lys residues were modeled in their charged states (see Table S10). (A) QM/MM models for the pT reaction in the Nqo13 subunit, showing the atoms in the QM region (inset). (B) The proton transfer (pT) reaction coordinate (Q) is represented by a linear combination of bond breaking and bond forming distances. (C) QM/MM free energy profiles for the lateral pT from Lys23513 via His29213 to Glu37713 in the fully hydrated state with closed (in red) and open (in blue) ion-pair conformations, respectively. (D) Unbiased QM/MM MD simulations for the pT process. In the medium-hydrated state (red and blue dashed lines), the pT stalls at Lys23513 in both open and closed ion-pair conformations. In the fully hydrated state with a closed ion-pair, the proton equilibrates between Lys23513 and His29213 within ∼1 ps (solid red line) but does not progress onward during the remaining simulation. In stark contrast, the ion-pair opening induces full pT to Glu37713 within ∼2 ps (solid blue line). (E) Snapshots of the QM/MM free energy profiles showing the transferred proton on Lys23513 (top), His29213 (second panel), at an intermediate Zundel ion (third panel), and Glu37713 (bottom). See Movie S1 for animation of the pT dynamics. (F) Snapshots of the open ion-pair conformation (top, with intact Glu12313/Lys34514 ion-pair) and the closed ion-pair conformation (bottom, intact Lys20413/Glu12313 ion-pair).
Perturbing the Proton Pumping Machinery
To perturb the lateral pT process in Nqo13, we mutated the bridging His29213 that provides an important intermediate site in the pT process into an alanine residue, as well as the homologous His322M and another bridging histidine, His348M of the NuoM subunit in E. coli complex I26 (Ser31813, in the T. thermophilus, Figure S9; see Figure S11 and Table S11 for comparison of structure and sequence of the two organisms). The mutagenesis studies were feasible only for E. coli complex I. The proton pumping subunits share a ∼30% sequence similarity but have a high structural similarity and are thus expected to employ similar pumping mechanisms (see Table S12 and Figure S9). His348M stabilizes the proton wire in MD simulations of the E. coli complex I (Figure S4C). In classical MD simulations of the H292A13 and H322AM/H348AM variants, the introduced substitution results in a long proton wire with N > 5 water molecules relative to 2–3 bridging water molecules between the lysine and glutamate in the wild type (WT) system (Figure 4A, Figure S4A–F). The water wire is partially disrupted by the histidine substitutions, leading to an overall lowered hydrogen-bonded connectivity relative to wild type model (Figure 4B, Figure S4). The substitution also results in conformational changes that could electrostatically modulate the proton affinity of the proton acceptor Glu37713 (Figure 4A, Figure S4), in particular by shorter distances to Arg16312 of Nqo12 (Arg175L in E. coli, Figure 4A,B), which could electrostatically tune the pKa of the Glu37713.
Figure 4.
Perturbing the proton transfer dynamics in complex I. (A) Snapshots of pT wires in the WT (top) and H322A/H348A variant (bottom) of E. coli complex I after 100 ns MD simulations (see Figure S4). (B) Substitution of the bridging histidine residues leads to perturbation in the Glu37713/Arg16312 (Glu407M/Arg175L) and Lys23513/Glu37713 (Lys265M/Glu407M) distances in classical MD simulations of T. thermophilus (T) and E. coli (E) complex I. Additional water molecules close the gap between the lysine and glutamate residues but with a lowered occupancy relative to the WT. (C) pT dynamics from QM/MM MD simulations of the WT and H292A variant (top) and snapshot of the H292A variant, showing the position of the pT wire and the Lys20413-Glu12313 ion-pair in closed conformation (bottom). (D) ACMA (top) and oxonol-VI (bottom) assays from proton pumping experiments in E. coli complex I. The ACMA and oxonol-VI assays monitor the ΔpH and ΔΨ gradient across the proteoliposome membrane, respectively (see also Figure S6D). (E) Summary of NADH:Q oxidoreduction activity, ACMA, and oxonol quenching experiments of WT and alanine variants. All measurements were performed in triplicates, with errors given as standard deviations. All activities were measured from the maximum level of the optical changes without extrapolation to zero time point using the plateau levels as a base. The NADH:Q oxidoreductase activity was determined from monitoring NADH decrease at 340 nm. See Supporting Information Methods and Figure S6 for further details. (F) Schematic representation of the proton pumping kinetic model of complex I with N- and P-side channels, Glu37713, Lys23513, and the ion-pair Glu12313/Lys20413 (see Materials and Methods). (G) Effect of the parameters of the kinetic model on the proton pumping across the membrane (see Materials and Methods). Left: coupling energy between ion-pair and Lys23513. Center: the pKa of Lys23513. Right: the K235/E377 intrinsic pT rate. See Supporting Information Methods and Figures S7, S13 for a detailed description of the kinetic model.
In QM/MM simulations of the H292A13 variant, we do not observe spontaneous deprotonation from Lys23513, resembling the pT behavior in the dry state (Figure 4C), with drastically increased pT barrier in DFT models (Figure S5). The perturbed barriers are also supported in DFT models constructed based on the E. coli complex I simulations (Figure S12). The simulations on the E. coli complex I support that both histidine residues provide important gating residues for the pT process (Figure 4C–F, Figure S12).
To experimentally assess the effect of a perturbed pT reaction, we introduced both single (H322AM, H348AM) and the double substitutions (H322AM/H348AM) into NuoM of E. coli complex I. Figure 4D and Figure 4E show pumping experiments, where we monitored the proton translocation of complex I reconstituted into proteoliposomes. To this end, we used both an ACMA fluorescence assay that is quenched as a response to pT across the membrane (ΔpH), as well as by monitoring absorbance changes using the membrane-potential (ΔΨ) sensitive dye, oxonol-VI, providing an independent measure of the pumping process. The absorbance signal is linear to the ΔΨ, whereas the fluorescence signal has a more complex dependence on the established ΔpH.38 The ΔpH/ΔΨ generation for both the single and double mutant variants was measured and compared to that of wild type complex I. All variants were fully assembled, stable and active, and reconstituted with similar orientations into liposomes (Figure S6, Tables S1 and S2). On the basis of the ACMA quenching, the histidine substitutions lead to a significant decrease in the proton pumping activity by approximately 40% for the double mutant (Figure 4D,E, Table 1, Table S2), whereas the individual substitutions have a smaller effect of 10–30%. The MD simulations suggest that the protons might take alternative pathways in the single variants to partially rescue the perturbed wiring (Figure 4C–F). A similar but slightly stronger effect on the ΔΨ generation was inferred by monitoring absorbance changes with the oxonol assay, where the double mutant is reduced to 40% of the WT, and the single mutants show a residual absorbance of 61% and 74%, respectively (Figure 4D,E, Table S2). Part of the difference might arise from proton leaks in the vesicles, indicative of the initial burst phase followed by constant decrease in the signal (Figure 4D), in addition to the nonlinearity of the fluorescence signal with the proton concentration. However, the proteoliposome background leak levels, assembly, and orientation of proteoliposomes containing the WT and mutant variants are similar. Our data thus show that the characterized variants, with mutations around 100 Å from the electron transfer module, result in lower electron transfer rates and reduced proton pumping (Figure S6, Supporting Information Methods). The histidine substitutions result in a significant decrease of the oxidoreduction activity up to 52% for the double variant, whereas a 20–40% reduction in activity is observed with the single substitutions (Figure 4E, Table 1). The experiments thus support that the simulated proton transfer reactions in Nqo13 (NuoM) could affect complex I activity.
Table 1. Proton Pumping and NADH:Q Oxidoreductase Activity of Wild Type and NuoM Variants of Complex Ia.
| pumping |
activity | ||
|---|---|---|---|
| construct | ACMA (rel WT [%]) | oxonol-VI (rel WT [%]) | NADH:Q oxidoreductase (rel WT [%]) |
| WT | 100 ± 3 | 100 ± 6 | 100 ± 2 |
| H322A | 92 ± 2 | 74 ± 2 | 61 ± 7 |
| H348A | 79 ± 3 | 61 ± 1 | 82 ± 6 |
| H322A/H348A | 63 ± 1 | 40 ± 7 | 48 ± 11 |
The values are shown relative to the wild type complex I. See Tables S1 and S2 for absolute values.
Proton Pumping Kinetics
To better understand the role of the histidine residues in proton pumping, we constructed a kinetic model of the pumping process using a kinetic master equation approach, with relative barriers derived from our atomistic simulations (Figure 4F,G, Figure S7, Tables S3 and S4; see Materials and Methods). To this end, we modeled the Glu/Lys ion-pair and the pumping elements in Nqo13 with proton acceptor/donor sites, mimicking the middle lysine residue (Lys23513), connected to a proton acceptor site (Glu37713). We coupled the ion-pair opening and the pT transfer reaction with relative rates similar to the wet/dry states transition and induced opening of the ion-pair by applying a nonequilibrium external force (see Materials and Methods). The kinetic model predicts that one proton is taken up from the N-side and pumped across to the P-side (Figure 4F,G) when the coupling between the ion-pair and the middle lysine residue reaches a threshold of >5 kcal mol–1. To mathematically model the effect of the histidine substitutions, we next slowed down the pT rate along the channel (kpT). Interestingly, when the pT rate in the channel becomes competing with pT uptake/release rates to the bulk, we observed a strong lowering of the proton pumping stoichiometry, consistent with our experimental observations. The pumping is also sensitive to the pKa of the middle lysine, with efficient proton translocation when the pKa resides in the 10 ± 2 range, within the optimized parameter set. The kinetic model thus further supports that the ion-pair opening modulates both kinetically and thermodynamically the protonation signal to effectively propagate across the membrane domain to achieve pumping.
Discussion
Our combined findings suggest that proton pumping along the 200 Å long membrane domain in complex I is established by conformational switching of the conserved ion-pairs that enables lateral pT within each proton pumping module. The pT reactions are gated by water molecules, whereas the channel wetting is in turn determined by the protonation state of the middle lysine residues.12,25,27 We could show that the ion-pair dissociation is kinetically accessible only when the proton channel is in a hydrated state but kinetically blocked in the dry state.
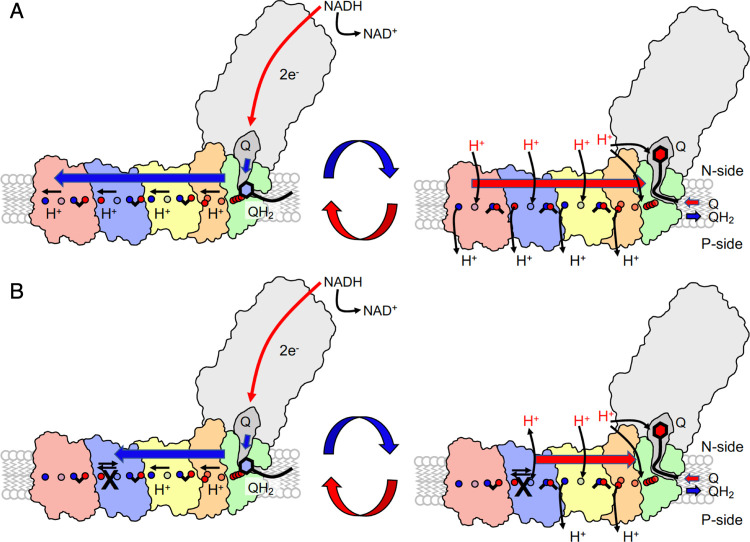
Our findings provide support for a recently proposed long-range pumping mechanism1 in which pumping is established by forward and backward propagation of a protonation signal across the membrane domain (Figure 5A). In the proposed mechanism, the thermodynamic driving force for the pumping originates from quinone reduction that could trigger proton uptake and conformational changes at or near the Nqo8 subunit.23,28 After quinone motion to a putative second binding site within the membrane domain,30 the free energy is transduced to the pumping module by pushing the proton toward the terminal end of the Nqo8/10/11 subunits, near the first ion-pair in Nqo14. This positive partial charge accumulates near the Nqo14 edge and triggers opening of the Glu-Lys ion-pair in Nqo14, pushing the proton laterally toward the Nqo13 interface. The charge imbalance further induces sequential ion-pair opening, hydration changes, and pT in Nqo13 and similarly in Nqo12. Upon ejection of a proton at the terminal end of Nqo12 to the P-side, a new proton is taken up from the N-side followed by ion-pair closure within the same subunit. This destabilizes the proton at the terminal edge of Nqo13, which is then ejected to the P-side. Reprotonation from the N-side and ion-pair closure at Nqo13 ejects the pumped proton at Nqo14 and a similar event could lead to ejection of the proton at the Nqo8/10/11 interface. Exchange of the quinol with a new quinone and reprotonation of the initial quinone reduction site lead to a new reaction cycle.
Figure 5.
Putative proton pumping mechanism in complex I. (A) Left: reduction of quinone (Q) to quinol (QH2) triggers stepwise proton transfer and ion-pair opening steps that propagate as a forward electrostatic pulse (blue arrow) to the terminal Nqo12 edge (in red) of the membrane domain. Right: The pumped protons are released by proton uptake from the N-side (H+ in red) and ion-pair closure during the backward signal (red arrow) to the membrane-bound Q binding site. Quinol/quinone exchange and reprotonation of the Q-site reset the pumping machinery for the next catalytic cycle. (B) Introduced mutations in Nqo13 (black cross) perturb the lateral pT from Nqo13 onward. The electrostatic backward pulse is reflected prematurely back from Nqo13 to the Q-channel, establishing pumping by Nqo14 (in yellow) and Nqo8/7/10/11 (green/orange) but at perturbed timing that affects the Q oxidoreduction activity.
By partially blocking the pT elements in Nqo13, we created a complex I variant that is unable to effectively propagate the electrostatic pulse from the Nqo13 subunit onward, with terminal protons most likely ejected from the Nqo13/14 interface (Figure 5B). We expect that the Nqo14 and the Nqo8/10/11 modules can, nevertheless, still pump protons at near wild type activities in the mutant variants, thus resulting in roughly halved pT activity observed in our pumping experiments. The perturbed quinone oxidoreductase activity provides possible experimental support for the proposed electrostatic backwave1 upon which the protons are ejected to the P-side of the membrane. The reduced activity could arise from slips in forward/reverse signals that partially propagate in two directions or in a too rapid backwave-signal to the putative second quinone binding site30 that results in the uncoupling effects, although detailed experiments are still required to fully quantify the effects. Nevertheless, we argue that both the experimental validation and kinetic models provide important boundaries for the computational prediction and allow testing implication beyond the microscopic description of the proton pumping process. Interestingly, perturbed timing in protonation reactions has been shown to uncouple eT and pT steps also in cytochrome oxidase (CcO), another redox-driven proton pump in respiratory chains.39 We have demonstrated here that water molecules, although not yet experimentally resolved in complex I, provide key elements in gating the proton translocation process by conformational switching and modulation of the kinetic proton transfer barriers. Similar electrostatically driven functional elements based on hydration dynamics40 could be mechanistically important in charge transfer reactions of cytochrome oxidase,41 light-driven ion-pumps,42 photosystem II,43 or other PCET-catalyzing enyzmes.44 Our findings illustrate how complex I enables an action-at-a-distance coupling of elementary electron and proton transfer processes by combining electrostatic and conformational switching with specific wetting-transitions of its proton channels. The findings provide a blueprint for understanding conserved mechanisms in the complex I superfamily and how mutations within this charge-transfer machinery may result in human disease.
Materials and Methods
Molecular Dynamics Simulations
The X-ray structure of complex I from T. thermophilus (PDB ID: 4HEA(8)) was embedded in a POPC/POPE-membrane, solvated with TIP3P water, 150 mM NaCl (see Table S7), with protonation states based on Poisson–Boltzmann continuum electrostatic calculations and previous studies.28 The system was modeled using the CHARMM36m force field,45 with cofactor parameters derived from DFT calculations.27,28 The FeS centers were modeled in the oxidized states except for N2, which was kept reduced in all simulations. The simulation setups comprised about 900 000 atoms. MD simulations were performed in the NPT ensemble at 310 K and 1 bar pressure, using a 2 fs integration time step and modeling long-range electrostatics using the particle mesh Ewald (PME) method.46 After initial minimization and equilibration, the systems were simulated for 200 ns to establish hydration of the water channels in the membrane domain. Simulations were also carried out with His29213 substituted by an alanine and for the WT and NuoM-variants of the membrane domain of E. coli complex I (PDB ID: 3RKO(26)), constructed using the same conditions as for T. thermophilus complex I. All classical MD simulations were performed using NAMD2.47 See Table S7 for a summary of all performed classical simulations.
Classical Free Energy Calculations
Free energy profiles for the Lys/Glu ion-pair dissociation in Nqo13 were computed using replica exchange umbrella sampling (REUS). To this end, the center-of-mass difference between Lys20413/Glu 12313 (r1) and Glu12313/Lys34514 (r2) headgroups was used as reaction coordinate, Rc = r2 – r1. To generate initial structures for the 20 REUS windows, 10 ns steered molecular dynamics (SMD) were performed. The SMDs were started from a 100 ns relaxed MD simulation of complex I of the dry state or after 200 ns MD simulations of the hydrated state. A harmonic bias with a force constant of 10 kcal mol–1 Å–1 was applied on the Rc. The center of the harmonic potential was scanned from −9.5 Å to +9.5 Å during the SMD simulations, from which 20 windows with equidistant values of the Rc from −10 Å (closed) to +8 Å (open) were equilibrated for 1 ns each. A harmonic force constant of 2.5 kcal mol–1 Å–1 was used for the bias, and exchange between neighboring windows was attempted every 10 ps. Each REUS window was simulated for 36 ns (720 ns in total for each free energy profile). Free energy profiles were computed using the dynamic histogram analysis method (DHAM)48 with a bin width of 0.01 Å. Previously calculated free energies27 were also remapped on the Rc based on REUS simulations performed using the complementary r1 reaction coordinate. Free energy barriers were converted into rates using transition state theory with a standard pre-exponential factor of 0.16 ps–1, a reflection coefficient κ = 1 of and at T = 310 K. See Figure S8 for convergence of free energy simulations, Table S7 for summary of all performed classical simulations, and Table S10 for summary of key protonation states.
QM/MM Dynamics and Free Energies
Hybrid DFT-based QM/MM MD simulations on the pT in Nqo13 were performed based on structures selected from the classical MD simulations in different hydration states, with open and closed ion-pair conformations. The system was trimmed to ∼50 000 atoms, including the Nqo12, Nqo13, and Nqo14 subunits, and lipid/water/ions in their vicinity (Figure 3A). The QM region comprised 125–137 atoms, including Lys235, His292, Glu377, Ser291, Ser318, Thr232, Thr322, Tyr321, Tyr405, Ser402 of Nqo13 and up to 10 water molecules obtained from the MD simulations. Link-atoms were introduced between Cα and Cβ atoms of the protein residues. The QM region was described at the B3LYP-D3/def2-SVP level,49−52 and the MM region with the CHARMM36m force field.45 The classically relaxed structures were optimized at the QM/MM level using the adopted-basis Newton–Raphson algorithm and allowing all QM and MM atoms around 10 Å of the QM region to relax. QM/MM potential energy surfaces of the pT reaction were computed by scanning the reaction coordinate Q, defined as a linear combination of all breaking and forming bond distances, between −7.0 Å and +7.0 Å, with a harmonic restrain of 3000 kcal mol–1 Å–2 on Q. QM/MM free energy profiles were calculated at the B3LYP-D3/def2-SVP level49−52 using umbrella sampling (US)/weighted histogram analysis method (WHAM), with 58 windows spanning Q and a harmonic restraint of 100 kcal mol–1 Å–2. Each window was simulated for 1.6 ps at T = 310 K using a 1 fs integration time step, with total sampling of ∼93 ps for each free energy profile. The pT reaction was also studied using unbiased QM/MM MD simulations starting from selected snapshots of wet and dry states and for the H292A mutant. All QM/MM calculations were performed with the CHARMM/TURBOMOLE interface.53 See Figure S8 for convergence of free energy simulations and Table S8 for summary of all performed QM/MM simulations.
DFT Energy Profiles
DFT models were built based on MD snapshots from the fully hydrated trajectories of the WT and the H292A variant. The models included residues Lys235, His292/Ala292, Glu377, Ser291, Ser318, Thr232, Thr322, Tyr321, Tyr405, Ser402, Ala288, Met293, Leu314, Val399 of Nqo13 subunit and N = 10 (WT)/13 (H292A) water molecules, comprising 179–181 atoms. Cα–Cβ positions were cut and saturated with hydrogen atoms, with Cβ positions fixed during geometry optimizations to simulate the protein framework. Geometry optimizations were performed at the B3LYP-D3/def2-SVP level, using an implicit polarizable medium with ε = 4 to model the protein environment,49−52,54,55 with single point energies computed at the B3LYP-D3/def2-TZVP/ε = 4 level. Entropic effects (TΔS at T = 310 K) and zero-point energies (ZPE) were evaluated at the B3LYP-D3/def2-SVP/ε = 4 level. Reaction pathways for the pT process were optimized using a chain-of-states method.55,56 All QM calculations were performed with TURBOMOLE version 7.3.57
Kinetic Model and Master Equation
A five-site kinetic master equation model58−60 was created to probe the proton pumping kinetics in Nqo13 by modeling proton channels from the N- and P-sides of the membrane, connected to middle (Lys23513) and terminal (Glu37713) proton acceptor sites, respectively, and a switchable ion-pair that was opened upon application of external force (Figure 4F). The energy of each microstate was expressed as a sum of site energies (Enintr) and coupling energies (Enm),
| 1 |
where occupied/empty (protonated/deprotonated, open/closed) states correspond to χ = 1/0, resulting in a total of 25 = 32 microstates. After parameter optimization (see below), the intrinsic energy of all sites was scanned across a pKa range of 0–14, with Eintr = −RT (pH–pKa) model for protonation sites, and in the range of −10 kcal mol–1 to +10 kcal mol–1 for channel and ion-pair sites. The coupling strengths were scanned in the range of −10 kcal mol–1 to +10 kcal mol–1. Transition rates, kij, between microstates i and j, were modeled as
| 2 |
with optimized parameters and intrinsic rate coefficients κij given in Tables S3 and S4. The population dynamics was described with the master equation,
| 3 |
that was numerically integrated using an ordinary differential equation (ODE) solver as implemented in the SciPy library.61 After an initial equilibrium distribution obtained at steady state (dpi/dt = 0), a biasing force was applied to open the ion-pair for t time steps,
| 4 |
where ΔEbiasmax is 400 mV and tperiod is set to 1 ms based on the turnover of complex I. The pumping flux from the N-side to the P-side was measured as the net number of protons exiting (entering) the system in one cycle during the application of the external force. See Supporting Information Methods for more detailed description of the kinetic model and dependence on the model parameters.
Construction of Complex I Mutants
A derivative of the E. coli strain BW2511362 with deleted ndh gene and the nuo operon replaced by a resistance cartridge (nptII) was created by genomic replacement63 and used to overproduce complex I and the variants. E. coli DH5αΔnuo64 and plasmid pBADnuoHis65 were used for generation of the mutants (see Table S5). All mutations were confirmed by DNA sequencing (GATC Eurofins, Konstanz, Germany). Cells were grown at 37 °C in baffled flasks using autoinduction medium66 (Table S6). Cells were disrupted by three passages through an EmulsiFlex (Avestin), and cytoplasmic membranes were isolated by differential centrifugation as described before.62 Cytoplasmic membranes were suspended in buffer A*pH6.8 (Table S6).
Purification and Reconstitution of Complex I
All steps were carried out at 4 °C. 2% (w/v) LMNG (Anatrace) was added to the membrane suspension, followed by incubation for 1 h with gentle stirring at RT and 20 min centrifugation at 160 000g. The supernatant was adjusted to 20 mM imidazole and applied on a ProBond Ni2+-IDA column (35 mL, Invitrogen) equilibrated in binding buffer (Table S6). Bound proteins were eluted with 308 mM imidazole (60% elution buffer; Table S6). Fractions with NADH/ferricyanide oxidoreductase activity were concentrated by ultrafiltration (100 kDa MWCO Amicon Ultra-15 centrifugal filter; Millipore) and further applied onto a Superose 6 size-exclusion chromatography column (300 mL, GE Healthcare) equilibrated in buffer A*MNG (Table S6). The fractions with the highest NADH/ferricyanide oxidoreductase activity were concentrated, and the protein was either directly used for biophysical measurements or stored at −80 °C.
Reconstitution of Complex I into Liposomes
E. coli polar lipids (25 mg mL–1 in CHCl3; Avanti) were evaporated and dissolved in a 5-fold volume lipid buffer (Table S6). The suspension was frozen in liquid nitrogen and thawed at 29 °C seven times to obtain unilamellar vesicles, followed by extrusion (0.1 μM polycarbonate membrane, Mini Extruder; Avanti).71 For reconstitution, 0.5–1 mg of complex I was mixed with reconstitution buffer (Table S6) and incubated for 5 min on ice. An amount of 250 μL of liposomes was mixed with 8 μL of sodium cholate (20%, w/v) and mixed with complex I in the reconstitution buffer and incubated for 15 min at RT. The solution was applied onto a size-exclusion column (PD-10 desalting column, 8.3 mL, Sephadex G-25, GE Healthcare) equilibrated in lipid buffer in order to remove the excess detergent. The eluate was centrifuged (4 °C, 2 bar air pressure, 30 min, 150 000g, Rotor A-100, Beckman Airfuge), and sedimented proteoliposomes were resuspended in 500 μL of lipid buffer. The NADH/ferricyanide oxidoreductase activity of the proteoliposomes was used to adjust an equal complex I content in the pumping assays.
Measurement of ΔpH and ΔΨ
The proton gradient (ΔpH) generated by WT and variants was probed by monitoring the fluorescence quenching of 9-amino-6-chloro-2-methoxyacridine (ACMA, Sigma), with an assay containing 100 μM decylubiquinone (Sigma), 0.2 μM ACMA, and 50 μL of proteoliposomes (with 50–100 μg of complex I) in the ACMA buffer (Table S6). Measurements were performed at 30 °C by starting the reaction with 130 μM NADH. The fluorescence was detected with an LS 55 fluorescence spectrometer (PerkinElmer) at excitation/emission wavelengths of 430 nm/480 nm. Addition of 10 μM carbonyl cyanide 3-chlorophenylhydrazone (CCCP) was used to dissipate ΔpH. The generation of ΔΨ was determined by monitoring absorption change at 588–625 nm of oxonol-VI (Sigma), with an assay containing 0.5 μM oxonol-VI, 50 μM Q0, and proteoliposomes in the oxonol buffer (Table S6).67 The process was studied at 30 °C by starting the reaction with 100 μM NADH. The absorbance changes were measured with a diode-array spectrometer (Hellma; TIDAS II, J&M Aalen). Addition of 2 μg mL–1 gramicidin was used to dissipate the ΔΨ. Pumping activities were characterized from the maximum level of the optical changes without extrapolation to zero time point using the plateau levels as a base. Exact proton pumping stoichiometry was not characterized (cf. e.g., refs (68−70)). All measurements were performed in triplicates for all three constructs and errors are reported as standard deviations. See Supporting Information Methods for further experimental details.
Activity Measurements
The NADH:decylubiquinone oxidoreductase activity was measured by the decrease of the NADH concentration at 340 nm.65 The assay contained 60 μM decylubiquinone, 2 μg of complex I, and a 10-fold molar excess (5 μg) of E. coli cytochrome bo3 in buffer A*MNG. The reaction was started by an addition of 150 μM NADH. All activity assays were performed at 30 °C. See Supporting Information Methods for further details.
Acknowledgments
This work received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 Research and Innovation Program (Grant Agreement 715311, to V.R.I.K.), and the Deutsche Forschungsgemeinschaft within the SPP 1927 (Grant FR 1140/11-1, to T.F.). V.R.I.K. is also supported by the Knut and Alice Wallenberg (KAW) Foundation. Computing time was provided by the Leibniz Rechenzentrum (LRZ) (SuperMUC Project pr27xu) and PRACE (Project pr1ejk).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.0c02789.
Supplementary methods; figures showing hydration, proton transfer and conformational dynamics, DFT reaction barriers, kinetic model, convergence of free energy calculations, and multiple sequence alignments; experimental details of biophysical experiments, oligonucleotide sequences of primers used for site-directed mutagenesis, buffers used for expression, purification, reconstitution, and biophysical experiments; and summary of QM/MM and MD simulations (PDF)
Movie showing pT reaction in QM/MM models (MP4)
Author Contributions
∥ M.E.M. and P.S. contributed equally.
The authors declare no competing financial interest.
Notes
Data supporting the findings of this manuscript are available from the corresponding authors upon reasonable request.
Supplementary Material
References
- Kaila V. R. I. Long-Range Proton-Coupled Electron Transfer in Biological Energy Conversion: Towards Mechanistic Understanding of Respiratory Complex I. J. R. Soc., Interface 2018, 15, 20170916. 10.1098/rsif.2017.0916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J. Mitochondrial Complex I. Annu. Rev. Biochem. 2013, 82, 551–575. 10.1146/annurev-biochem-070511-103700. [DOI] [PubMed] [Google Scholar]
- Brandt U. Energy Converting NADH: Quinone Oxidoreductase (Complex I). Annu. Rev. Biochem. 2006, 75, 69–92. 10.1146/annurev.biochem.75.103004.142539. [DOI] [PubMed] [Google Scholar]
- Sazanov L. A. A giant molecular proton pump: Structure and mechanism of respiratory complex I. Nat. Rev. Mol. Cell Biol. 2015, 16, 375–388. 10.1038/nrm3997. [DOI] [PubMed] [Google Scholar]
- Wikström M. Two protons are pumped from the mitochondrial matrix per electron transferred between NADH and ubiquinone. FEBS Lett. 1984, 169, 300–304. 10.1016/0014-5793(84)80338-5. [DOI] [PubMed] [Google Scholar]
- Mitchell P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature 1961, 191, 144–148. 10.1038/191144a0. [DOI] [PubMed] [Google Scholar]
- Yoshida M.; Muneyuki E.; Hisabori T. ATP synthase - A marvellous rotary engine of the cell. Nat. Rev. Mol. Cell Biol. 2001, 2, 669–677. 10.1038/35089509. [DOI] [PubMed] [Google Scholar]
- Baradaran R.; Berrisford J. M.; Minhas G. S.; Sazanov L. A. Crystal structure of the entire respiratory complex I. Nature 2013, 494, 443–448. 10.1038/nature11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agip A. N. A.; Blaza J. N.; Bridges H. R.; Viscomi C.; Rawson S.; Muench S. P.; Hirst J. Cryo-EM structures of complex I from mouse heart mitochondria in two biochemically defined states. Nat. Struct. Mol. Biol. 2018, 25, 548–556. 10.1038/s41594-018-0073-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickermann V.; Wirth C.; Nasiri H.; Siegmund K.; Schwalbe H.; Hunte C.; Brandt U. Mechanistic insight from the crystal structure of mitochondrial complex I. Science 2015, 347, 44–49. 10.1126/science.1259859. [DOI] [PubMed] [Google Scholar]
- Schuller J. M.; Saura P.; Thiemann J.; Schuller S. K.; Gamiz-Hernandez A. P.; Kurisu G.; Nowaczyk M. M.; Kaila V. R. I. Redox-coupled proton pumping drives carbon concentration in the photosynthetic complex I. Nat. Commun. 2020, 11, 494. 10.1038/s41467-020-14347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euro L.; Belevich G.; Verkhovsky M. I.; Wikström M.; Verkhovskaya M. Conserved lysine residues of the membrane subunit NuoM are involved in energy conversion by the proton-pumping NADH:ubiquinone oxidoreductase (Complex I). Biochim. Biophys. Acta, Bioenerg. 2008, 1777, 1166–1172. 10.1016/j.bbabio.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Nakamaru-Ogiso E.; Kao M. C.; Chen H.; Sinha S. C.; Yagi T.; Ohnishi T. The membrane subunit NuoL(ND5) is involved in the indirect proton pumping mechanism of Escherichia coli complex I. J. Biol. Chem. 2010, 285, 39070–39078. 10.1074/jbc.M110.157826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarneh B.; Vik S. B. Mutagenesis of subunit N of the Escherichia coli Complex I. Identification of the initiation codon and the sensitivity of mutants to decylubiquinone. Biochemistry 2003, 42, 4800–4808. 10.1021/bi0340346. [DOI] [PubMed] [Google Scholar]
- Michel J.; DeLeon-Rangel J.; Zhu S.; van Ree K.; Vik S. B. Mutagenesis of the L, M, and N subunits of complex I from Escherichia coli indicates a common role in function. PLoS One 2011, 6, e17420. 10.1371/journal.pone.0017420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha P. K.; Torres-Bacete J.; Nakamaru-Ogiso E.; Castro-Guerrero N.; Matsuno-Yagi A.; Yagi T. Critical roles of subunit NuoH (ND1) in the assembly of peripheral subunits with the membrane domain of Escherichia coli NDH-1. J. Biol. Chem. 2009, 284, 9814–9823. 10.1074/jbc.M809468200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Bacete J.; Nakamaru-Ogiso E.; Matsuno-Yagi A.; Yagi T. Characterization of the NuoM (ND4) subunit in Escherichia coli NDH-1: Conserved charged residues essential for energy-coupled activities. J. Biol. Chem. 2007, 282, 36914–36922. 10.1074/jbc.M707855200. [DOI] [PubMed] [Google Scholar]
- Sazanov L. A. The mechanism of coupling between electron transfer and proton translocation in respiratory complex I. J. Bioenerg. Biomembr. 2014, 46, 247–253. 10.1007/s10863-014-9554-z. [DOI] [PubMed] [Google Scholar]
- Wikström M.; Sharma V.; Kaila V. R. I.; Hosler J. P.; Hummer G. New Perspectives on Proton Pumping in Cellular Respiration. Chem. Rev. 2015, 115, 2196–2221. 10.1021/cr500448t. [DOI] [PubMed] [Google Scholar]
- Le Breton N.; Wright J. J.; Jones A. J. Y.; Salvadori E.; Bridges H. R.; Hirst J.; Roessler M. M. Spectroscopy to define the proton-coupled electron trasnfer reaction at Fe-S cluster N2 in respiratory complex I. J. Am. Chem. Soc. 2017, 139, 16319–16326. 10.1021/jacs.7b09261. [DOI] [PubMed] [Google Scholar]
- Saura P.; Kaila V. R. I. Energetics and dynamics of proton-coupled electron transfer in the NADH/FMN site of respiratory complex I. J. Am. Chem. Soc. 2019, 141, 5710–5719. 10.1021/jacs.8b11059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammes-Schiffer S. Proton-coupled electron transfer: classification scheme and guide to theoretical methods. Energy Environ. Sci. 2012, 5, 7696–7703. 10.1039/c2ee03361e. [DOI] [Google Scholar]
- Cabrera-Orefice A.; Yoga E. G.; Wirth C.; Siegmund K.; Zwicker K.; Guerrero-Castillo S.; Zickermann V.; Hunte C.; Brandt U. Locking loop movement in the ubiquinone pocket of complex I disengages the proton pumps. Nat. Commun. 2018, 9, 4500. 10.1038/s41467-018-06955-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor J. G.; Jones A. J. Y.; Di Luca A.; Kaila V. R. I.; Hirst J. Correlating kinetic and structural data on ubiquinone binding and reduction by respiratory complex I. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, 12737–12742. 10.1073/pnas.1714074114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Luca A.; Mühlbauer M. E.; Saura P.; Kaila V. R. I. How inter-subunit contacts in the membrane domain of complex I affect proton transfer energetics. Biochim. Biophys. Acta, Bioenerg. 2018, 1859, 734–741. 10.1016/j.bbabio.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremov R. G.; Sazanov L. A. Structure of the membrane domain of respiratory complex I. Nature 2011, 476, 414–420. 10.1038/nature10330. [DOI] [PubMed] [Google Scholar]
- Di Luca A.; Gamiz-Hernandez A. P.; Kaila V. R. I. Symmetry-related proton transfer pathways in respiratory complex I. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, E6314–E6321. 10.1073/pnas.1706278114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V.; Belevich G.; Gamiz-Hernandez A. P.; Róg T.; Vattulainen I.; Verkhovskaya M. L.; Wikström M.; Hummer G.; Kaila V. R. I. Redox-induced activation of the proton pump in the respiratory complex I. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 11571–11576. 10.1073/pnas.1503761112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaila V. R. I.; Wikström M.; Hummer G. Electrostatics, hydration, and proton transfer dynamics in the membrane domain of respiratory complex I. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, 6988–6993. 10.1073/pnas.1319156111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnau J.; Sharma V.; Gamiz-Hernandez A. P.; Di Luca A.; Haapanen O.; Vattulainen I.; Wikström M.; Hummer G.; Kaila V. R. I. Redox-Coupled Quinone Dynamics in the Respiratory Complex I. Proc. Natl. Acad. Sci. U. S. A. 2018, 115, E8413–E8420. 10.1073/pnas.1805468115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunte C.; Zickermann V.; Brandt U. Functional modules and structural basis of conformational coupling in mitochondrial complex I. Science 2010, 329, 448–451. 10.1126/science.1191046. [DOI] [PubMed] [Google Scholar]
- Gamiz-Hernandez A. P.; Jussupow A.; Johansson M. P.; Kaila V. R. I. Terminal Electron-Proton Transfer Dynamics in the Quinone Reduction of Respiratory Complex I. J. Am. Chem. Soc. 2017, 139, 16282–16288. 10.1021/jacs.7b08486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haapanen O.; Sharma V. Role of water and protein dynamics in proton pumping by respiratory complex I. Sci. Rep. 2017, 7, 7747. 10.1038/s41598-017-07930-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belevich G.; Knuuti J.; Verkhovsky M. I.; Wikström M.; Verkhovskaya M. Probing the mechanistic role of the long α-helix in subunit L of respiratory Complex I from Escherichia coli by site-directed mutagenesis. Mol. Microbiol. 2011, 82, 1086–1095. 10.1111/j.1365-2958.2011.07883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S.; Vik S. B. Constraining the lateral helix of respiratory Complex I by cross-linking does not impair enzyme activity or proton translocation. J. Biol. Chem. 2015, 290, 20761–20773. 10.1074/jbc.M115.660381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steimle S.; Schnick C.; Burger E. M.; Nuber F.; Krämer D.; Dawitz H.; Brander S.; Matlosz B.; Schäfer J.; Maurer K.; Glessner U.; Friedrich T. Cysteine scanning reveals minor local rearrangements of the horizontal helix of respiratory complex I. Mol. Microbiol. 2015, 98, 151–161. 10.1111/mmi.13112. [DOI] [PubMed] [Google Scholar]
- Rasaiah J. C.; Garde S.; Hummer G. Water in Nonpolar Confinement: From Nanotubes to Proteins and Beyond. Annu. Rev. Phys. Chem. 2008, 59, 713–740. 10.1146/annurev.physchem.59.032607.093815. [DOI] [PubMed] [Google Scholar]
- D’Alessandro M.; Turina P.; Melandri B. A. Quantitative evaluation of the intrinsic uncoupling modulated by ADP and Pi in the reconstituted ATP synthase of Escherichia coli. Biochim. Biophys. Acta, Bioenerg. 2011, 1807, 130–143. 10.1016/j.bbabio.2010.08.011. [DOI] [PubMed] [Google Scholar]
- Pawate A. S.; Morgan J.; Namslauer A.; Mills D.; Brzezinski P.; Ferguson-Miller S.; Gennis R. B. A mutation in subunit I of cytochrome oxidase from Rhodobacter sphaeroides results in an increase in steady-state activity but completely eliminates proton pumping. Biochemistry 2002, 41, 13417–13423. 10.1021/bi026582+. [DOI] [PubMed] [Google Scholar]
- Saura P.; Frey D. M.; Gamiz-Hernandez A. P.; Kaila V. R. I. Electric field modulated redox-driven protonation and hydration energetics in energy converting enzymes. Chem. Commun. 2019, 55, 6078–6081. 10.1039/C9CC01135H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supekar S.; Kaila V. R. I. Dewetting transitions coupled to K-channel activation in cytochrome c oxidase. Chem. Sci. 2018, 9, 6703–6710. 10.1039/C8SC01587B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomivuori C.-M.; Gamiz-Hernandez A. P.; Sundholm D.; Kaila V. R. I. Energetics and dynamics of a light-driven sodium-pumping rhodopsin. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, 7043–7048. 10.1073/pnas.1703625114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugur I.; Rutherford A. W.; Kaila V. R. I. Redox-coupled substrate water reorganization in the active site of Photosystem II - The role of calcium in substrate water delivery. Biochim. Biophys. Acta, Bioenerg. 2016, 1857, 740–748. 10.1016/j.bbabio.2016.01.015. [DOI] [PubMed] [Google Scholar]
- Kaila V. R. I.; Hummer G. Energetics of Direct and Water-Mediated Proton-Coupled Electron Transfer. J. Am. Chem. Soc. 2011, 133, 19040–19043. 10.1021/ja2082262. [DOI] [PubMed] [Google Scholar]
- Huang J.; Rauscher S.; Nawrocki G.; Ran T.; Feig M.; De Groot B. L.; Grubmüller H.; MacKerell A. D. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods 2017, 14, 71–73. 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essmann U.; Perera L.; Berkowitz M. L.; Darden T.; Lee H.; Pedersen L. G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. 10.1063/1.470117. [DOI] [Google Scholar]
- Phillips J. C.; Braun R.; Wang W.; Gumbart J.; Tajkhorshid E.; Villa E.; Chipot C.; Skeel R. D.; Kalé L.; Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzl L. S.; Kells A.; Rosta E.; Hummer G. Dynamic Histogram Analysis To Determine Free Energies and Rates from Biased Simulations. J. Chem. Theory Comput. 2017, 13, 6328–6342. 10.1021/acs.jctc.7b00373. [DOI] [PubMed] [Google Scholar]
- Lee C.; Yang W.; Parr R. G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B: Condens. Matter Mater. Phys. 1988, 37, 785–789. 10.1103/PhysRevB.37.785. [DOI] [PubMed] [Google Scholar]
- Becke A. D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648–5652. 10.1063/1.464913. [DOI] [Google Scholar]
- Schäfer A.; Horn H.; Ahlrichs R. Fully optimized contracted Gaussian basis sets for atoms Li to Kr. J. Chem. Phys. 1992, 97, 2571–2577. 10.1063/1.463096. [DOI] [Google Scholar]
- Grimme S.; Antony J.; Ehrlich S.; Krieg H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J. Chem. Phys. 2010, 132, 154104–1–19. 10.1063/1.3382344. [DOI] [PubMed] [Google Scholar]
- Riahi S.; Rowley C. N. The CHARMM-TURBOMOLE interface for efficient and accurate QM/MM molecular dynamics, free energies, and excited state properties. J. Comput. Chem. 2014, 35, 2076–2086. 10.1002/jcc.23716. [DOI] [PubMed] [Google Scholar]
- Klamt A.; Schüürmann G. COSMO: A new approach to dielectric screening in solvents with explicit expressions for the screening energy and its gradient. J. Chem. Soc., Perkin Trans. 2 1993, 2, 799–805. 10.1039/P29930000799. [DOI] [Google Scholar]
- Plessow P. Reaction path optimization without NEB springs or interpolation algorithms. J. Chem. Theory Comput. 2013, 9, 1305–1310. 10.1021/ct300951j. [DOI] [PubMed] [Google Scholar]
- E W.; Ren W.; Vanden-Eijnden E. String method for the study of rare events. Phys. Rev. B: Condens. Matter Mater. Phys. 2002, 66, 052301. 10.1103/PhysRevB.66.052301. [DOI] [Google Scholar]
- Ahlrichs R.; Bär M.; Häser M.; Horn H.; Kölmel C. Electronic structure calculations on workstation computers: The program system turbomole. Chem. Phys. Lett. 1989, 162, 165–169. 10.1016/0009-2614(89)85118-8. [DOI] [Google Scholar]
- Hill T. L.Free Energy Transduction in Biology; Academic: New York, 1977. [Google Scholar]
- Kim Y. C.; Wikström M.; Hummer G. Kinetic models of redox-coupled proton pumping. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 2169–2174. 10.1073/pnas.0611114104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. C.; Hummer G. Proton-pumping mechanism of cytochrome c oxidase: A kinetic master-equation approach. Biochim. Biophys. Acta, Bioenerg. 2012, 1817, 526–536. 10.1016/j.bbabio.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtanen P.; Gommers R.; Oliphant T. E.; Haberland M.; Reddy T.; Cournapeau D.; Burovski E.; Peterson P.; Weckesser W.; Bright J.; van der Walt M.; Brett M.; Wilson J.; Millman K. J.; Mayorov N.; Nelson A. R. J.; Jones E.; Kern R.; Larson E.; Carey C. J.; Polat I.; Feng Y.; Moore E. W.; VanderPlas J.; Laxalde D.; Perktold J.; Cimrman R.; Henriksen I.; Quintero E. A.; Harris C. R.; Archibald A. M.; Ribeiro A. H.; Pedregosa F.; van Mulbregt P. and SciPy 1.0 Contributors. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nat. Methods 2020, 17 (3), 261–272. 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko K. A.; Wanner B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 2000, 97, 6640–6645. 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burschel S.; Kreuzer Decovic D.; Nuber F.; Stiller M.; Hofmann M.; Zupok A.; Siemiatkowska B.; Gorka M.; Leimkühler S.; Friedrich T. Iron-sulfur cluster carrier proteins involved in the assembly of Escherichia coli NADH:ubiquinone oxidoreductase (complex I). Mol. Microbiol. 2019, 111, 31–45. 10.1111/mmi.14137. [DOI] [PubMed] [Google Scholar]
- Pohl T.; Uhlmann M.; Kaufenstein M.; Friedrich T. Lambda red-mediated mutagenesis and efficient large scale affinity purification of the Escherichia coli NADH:Ubiquinone oxidoreductase (complex I). Biochemistry 2007, 46, 10694–10702. 10.1021/bi701057t. [DOI] [PubMed] [Google Scholar]
- Pohl T.; Bauer T.; Dörner K.; Stolpe S.; Sell P.; Zocher G.; Friedrich T. Iron-sulfur cluster N7 of the NADH:Ubiquinone oxidoreductase (complex I) is essential for stability but not involved in electron transfer. Biochemistry 2007, 46, 6588–6596. 10.1021/bi700371c. [DOI] [PubMed] [Google Scholar]
- Studier W. F. Stable expression clones and auto-induction for protein production in E. coli. Methods Mol. Biol. 2014, 1091, 17–32. 10.1007/978-1-62703-691-7_2. [DOI] [PubMed] [Google Scholar]
- Verkhovskaya M.; Knuuti J.; Wikström M. Role of Ca2+ in structure and function of Complex I from Escherichia coli. Biochim. Biophys. Acta, Bioenerg. 2011, 1807, 36–41. 10.1016/j.bbabio.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Dröse S.; Krack S.; Sokolova L.; Zwicker K.; Barth H. D.; Morgner N.; Heide H.; Steger M.; Nübel E.; Zickermann V.; Kerscher S.; Brutschy B.; Radermacher M.; Brandt U. Functional Dissection of the Proton Pumping Modules of Mitochondrial Complex I. PLoS Biol. 2011, 9, e1001128–e1001128. 10.1371/journal.pbio.1001128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A. J. Y.; Blaza J. N.; Varghese F.; Hirst J. Respiratory Complex I in Bos Taurus and Paracoccus Denitrificans Pumps Four Protons across the Membrane for Every NADH Oxidized. J. Biol. Chem. 2017, 292, 4987–4995. 10.1074/jbc.M116.771899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin A.; Dröse S.; Brandt U. The Proton Pumping Stoichiometry of Purified Mitochondrial Complex I Reconstituted into Proteoliposomes. Biochim. Biophys. Acta, Bioenerg. 2006, 1757 (12), 1575–1581. 10.1016/j.bbabio.2006.10.001. [DOI] [PubMed] [Google Scholar]
- von Ballmoos C.; Biner O.; Nilsson T.; Brzezinski P. Mimicking respiratory phosphorylation using purified enzymes. Biochim. Biophys. Acta, Bioenerg. 2016, 1857, 321–331. 10.1016/j.bbabio.2015.12.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.