Abstract
The prognostic assessment of patients with hepatocellular carcinoma (HCC) after resection is an important clinical issue. The present study investigated those genes associated with high serum alpha‐fetoprotein (AFP), and their clinical significance, including prognosis and recurrence after hepatectomy. Based on gene expression analysis of 110 training HCC cases, 20 genes whose mRNA expression levels were significantly upregulated and 50 genes that were downregulated correlated with high serum AFP‐associated HCC patients. Gene expression profiles of Villin1 (Vil1) were obtained in high serum AFP‐associated HCC tumor tissues. In the present analysis, only VIL1 was significantly correlated with the recurrence of HCC. The results were validated independently using Taqman gene expression assays and immunostaining analysis. Results showed that the upregulation of VIL1 mRNA was also correlated with high serum PIVKAII, vascular invasion (P < 0.05), poor differentiation, an advanced cancer stage (P < 0.01) and recurrence‐free survival (P = 0.017). The upregulation of VIL1 mRNA was observed more frequently in the early recurrence patients as compared to the late recurrence patients. Cox regression univariate and multivariate analyses indicated that high serum AFP levels (overall survival, HR 1.675, P = 0.002; FRS, HR 1.359, P = 0.039) and Vil1 protein expression (overall survival, HR 0.253, P = 0.009; FRS, HR 0.401, P = 0.041) were independent, unfavorable prognostic factors for overall and recurrence‐free survival of patients. We demonstrated that the VIL1 gene is a potential candidate molecular marker for high serum AFP‐associated HCC and a predictive candidate for the postoperative recurrence and poorer prognosis of HCC. (Cancer Sci, doi: 10.1111/j.1349‐7006.2012.02315.x, 2012)
Hepatocellular carcinoma (HCC) is the sixth most common cancer in the world, and the 5‐year survival rate of individuals with HCC is lower than 10%.1, 2 The prognosis of HCC patients remains dismal, and novel diagnostic modalities as well as improvements in therapeutic strategies currently in use are crucial to improve the clinical outcome for HCC patients. Surgical resection is not the only curative treatment for HCC. Liver transplant is another option for selected patients; however, the rapid recurrence of these tumors is observed frequently, even after apparently curative resections.3, 4 In ten years, various molecules have been proposed as predictive markers for the recurrence of HCC, but none have proved useful clinically, because different patterns of recurrence have not been considered.5, 6 In Tanaka et al., 7 we report that aberrant Aurora B expression in primary HCC might be a predictive factor of HCC recurrence exceeding the Milan criteria after a curative hepatectomy. The risk factors of recurrence after a curative resection for macroscopic vascular invasion negative hepatocellular carcinoma are closely related to serum alpha‐fetoprotein (AFP) levels, multiple tumor formation and non‐anatomic hepatic resection. Network expression analysis revealed that distinct signaling pathways of epithelial–mesenchymal transitions are associated with recurrence after an anatomically systematized hepatectomy.8
Alpha‐fetoprotein is one of several embryo‐specific proteins and a useful diagnostic tool for HCC.9, 10 AFP mRNA is also reported to be a predictive marker for tumor recurrence, and the efficacy of predicting HCC recurrence after a curative resection using AFP mRNA detection has been confirmed in many published studies.11, 12, 13, 14 The function of AFP is unknown, but it has been suggested that it has immunoregulatory properties, and that it might influence cell proliferation and growth, and directly promote proliferation in cultured cells. AFP might also function as an apoptotic factor.15, 16, 17, 18, 19 However, the molecular background of HCC associated with increased AFP levels, the mechanisms responsible for the recurrence of HCC are presently unclear.
Gene expression analysis by cDNA microarray offers a systematic approach to acquire comprehensive information regarding gene transcription profiles. It is now possible to perform a large‐scale expression survey to identify candidate target genes and to develop a novel molecular‐targeted therapy for HCC. The present study sought a biomarker associated with and predictive of recurrences after a curative resection for high serum AFP‐associated HCC, by examining the genome‐wide expression profile of HCC generated by cDNA microarray experiments. Using a prediction system obtained from studies based on training analysis, we identified one candidate gene. This gene was validated independently in patients and its significance was also analyzed clinicopathologically.
Materials and Methods
Patients and tissue samples
In this study, we enrolled 200 HCC at the Tokyo Medical and Dental University Hospital patients who underwent hepatectomy for the first time between 2001 and 2009. Written informed consent was obtained from these patients, and the institutional review board approved the study. A total of 110 patients were randomly assigned to a microarray training set, and the remaining 90 to a validation set. Tissues were collected during surgery, snap frozen in liquid nitrogen and then stored at −80°C. Part of the tissue sample was fixed in 10% formaldehyde and embedded in paraffin for histopathological analysis. The histological diagnosis was made when two pathologists specializing in liver disease reached a consensus. Patient follow up involved assays of serum levels of AFP and protein induced by vitamin K absence or antagonists II every month, and ultrasonography, computed tomography and magnetic resonance imaging every 3 months.
Preparation of tissues and cell lines for gene expression
Tissue specimens were obtained from surgically resected materials. The human HCC cell lines Hep3B and SK‐Hep1 were obtained from the American Type Culture Collection (Manassas, VA, USA). Huh1, Huh6, Huh7, HLE, HLF, HepG2 JHH‐2 and JHH‐4 were obtained from the Human Science Research Resources Bank (Osaka, Japan). The cell culture conditions were as reported previously.20 For gene expression analysis, total RNA was extracted from the tissues and cell lines using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA; Hilden, Germany) and treated with RNase‐free DNase I, according to the manufacturer's instructions. The integrity of the RNA obtained was assessed using the Agilent 2100 BioAnalyzer (Agilent Technologies, Palo Alto, CA, USA). All samples had an RNA integrity number > 5.0. Using 2 μg of total tissue or cell line RNA, cRNA were prepared using one cycle target labeling and control reagents with Affymetrix (Affymetrix, Santa Clara, CA, USA). Hybridization and signal detection of the HG‐U133 plus 2.0 arrays (Affymetrix) were performed following the manufacturer's instructions.
Gene expression analysis
A total of 110 microarray datasets were normalized using the robust multi‐array average method with R statistical software version 2.12.1, together with the BioConductor package. The estimated gene expression levels were obtained in log2‐transformed values, and 62 control probe sets were removed for further analysis. The fold change (FC) values were calculated using the ratio of the geometric means of the gene expression levels between AFP ≥ 100 ng/mL and AFP < 100 ng/mL groups. The 8 cell lines of microarray data were calculated using the AFP gene expression levels between AFP high expression and AFP low expression groups. A Wilcoxon rank sum test was performed to estimate the significance levels of the differences in gene expression between the two groups. For each statistical test, the P‐values obtained from the multiple hypothetical testing were adjusted by a false discovery rate (FDR), and the probe sets with a FDR < 0.01. The Cox proportional hazards regression model was used to estimate the relationship between the gene expression pattern and the tumor recurrence‐free survival rate for each probe set. The probe sets that satisfied P < 0.01 according to the likelihood ratio test and those with a more than 1.5‐fold change in mean expression value between the recurrence group and the non‐recurrence groups were selected.
TaqMan MGB probe real‐time PCR
Two micrograms of tissue or cell line total RNA was reverse‐transcribed to cDNA using a High‐Capacity cDNA Reverse Transcription Kit (Applied Biosytems, Foster City, CA, USA) using the random primer contained in the kit, following the manufacturer's directions. TaqMan gene expression assays for VIL1 (Hs00200229_m1) were performed using a TaqMan Fast Universal PCR master mix 2× on the AB 7500 Fast Real Time PCR System (Applied Biosystems). For quantitative analysis of specific mRNA expression, CT values were calculated using 7500 SDS software. The data were analyzed using the △△CT method (Applied Biosystems User Bulletin #2, 1997), and the samples were normalized to the 18s‐rRNA transcript.
Immunohistochemical analysis
To further validate the expression of the candidate gene by the microarray, immunohistochemical studies were performed on 70 HCC samples. The primary antibody, anti‐mouse monoclonal against Villin (ab739, Abcam), was used at a 1:50 dilution in PBS containing 1% BSA. The tissue sections were stained with an automated immunostainer (BenchMark XT; Ventana Medical Systems, Tucson, AZ, USA) using heat‐induced epitope retrieval and a standard diaminobenzidine detection kit (Ventana). Staining was evaluated by two independent observers and interpreted to be positive when > 10% of the tumor cells showed the positive signal.
Statistical analysis
Statistical comparisons of the clinicopathological characteristics for significance were performed using the chi‐square ‐test or Fisher's exact test. Differences in mRNA levels between the groups and the association between clinicopathological factors were analyzed using the paired or unpaired t‐test. The data were expressed as means ± SD. The overall survival curve and recurrence‐free survival rate was calculated using the Kaplan–Meier method, and the rates were reported with 95% confidence intervals. Differences were tested for significance using the log‐rank test. A P < 0.05 was deemed to be statistically significant. To investigate the factors that predicted recurrence and survival, univariate and multivariate analyses were performed using Cox proportional hazard models.
Results
Clinicopathological factors associated with hepatocellular carcinoma recurrence
We examined the association between clinicopathological factors and the tumor recurrence rate in 200 patients with HCC. The median follow‐up period was 1006 days (range, 2–3085). A total of 130 patients were recurrent during the follow‐up period or at the end point of this study, and the median recurrence period was 336 days (range, 1–2715). Univariate analysis of HCC recurrence revealed aspartate amino transferase, prothrombin time, albumin, AFP, maximal tumor size, portal vein invasion, hepatic vein invasion, vascular invasion, advanced stage of cancer and poor tumor differentiation as risk factors. In the multivariate analysis, aspartate amino transferase (P = 0.004), AFP ≥ 100 ng/mL (P = 0.023) and macrovascular invasion (P = 0.021) were identified as independent factors for recurrence. The other clinicopathological factors were not statistically significant (Table 1).
Table 1.
Clinicopathological features and risk factor of recurrence in 200 primary hepatocellular carcinomas (HCC)
| Clinicopathologic factors | Primary HCC | Univariate | Multivariate | ||
|---|---|---|---|---|---|
| (n = 200) | χ 2 | P–value | χ 2 | P–value | |
| Age (years, mean ± SD) | 64.8 ± 10.8 | 0.537 | 0.464 | ||
| Gender (male/female) | 154/45 | 0.178 | 0.673 | ||
| Viral infection (HBV/HCV/NBNC) | 43/99/57 | 3.386 | 0.184 | ||
| Child‐Pugh classification (A/B/C) | 146/51/3 | 3.322 | 0.073 | ||
| AST (IU/L, mean ± SD) | 54.12 ± 33.8 | 11.30 | 0.001 | 8.524 | 0.004 |
| ALT (IU/L, mean ± SD) | 49.92 ± 38.9 | 1.191 | 0.275 | ||
| Plt (×109/L, mean ± SD) | 15.57 ± 8.7 | 1.680 | 0.195 | ||
| ICG‐R15 (%, mean ± SD) | 19.02 ± 11.4 | 1.047 | 0.306 | ||
| PT% (mean ± SD) | 84.89 ± 14.3 | 8.620 | 0.003 | 1.564 | 0.211 |
| T.bil (mg/dL, mean ± SD) | 0.94 ± 0.76 | 0.449 | 0.503 | ||
| Alb (g/dL, mean ± SD) | 3.86 ± 0.54 | 5.038 | 0.025 | 0.826 | 0.364 |
| AFP (ng/mL) ≥ 100 vs < 100 | 65/135 | 9.933 | 0.002 | 5.154 | 0.023 |
| PIVKA‐II (mAU/mL) ≥ 100 vs < 100 | 102/98 | 2.629 | 0.105 | ||
| Tumor max size (mean ± SD) | 5.06 ± 3.7 | 4.154 | 0.042 | 2.892 | 0.089 |
| Solitary or multiple | 136/64 | 0.498 | 0.481 | ||
| Capsular formation (pfc) (−/+) | 45/155 | 0.512 | 0.474 | ||
| Capsular invasion (pfc‐inf) (−/+) | 81/119 | 0.014 | 0.907 | ||
| Portal vein invasion (pvp) (−/+) | 102/98 | 7.312 | 0.007 | 0.034 | 0.854 |
| Macro vascular invasion (vp0–1/vp > 2) | 159/41 | 16.709 | <0.001 | 5.338 | 0.021 |
| Hepatic vein invasion (pvv) (−/+) | 171/29 | 6.260 | 0.012 | 1.624 | 0.225 |
| Stages (I/II/III/IV) | 14/87/66/52 | 8.048 | 0.005 | 0.648 | 0.421 |
| Background of liver (CH/LC/NL) | 87/102/11 | 1.689 | 0.430 | ||
| Degree of differention (well/mod/poor) | 49/115/36 | 7.982 | 0.019 | 0.542 | 0.762 |
AFP, α‐fetoprotein; Alb, albumin; ALT, alanine aminotransferase; AST, aspartate amino transferase; CH, chronic hepatitis; ICG‐R15, indocyanine green retention rate at 15 min; LC, cirrhosis; NL, normal liver; PIVKAII, protein induced by vitamin K absence or antagonists II; PLT, platelet; PT%, prothrombin time; T.bil, total bilirubin; +, positive; −, negative.
Selection of genes for predictive of recurrence of high serum alpha‐fetoprotein‐associated hepatocellular carcinoma
To identify the candidate genes associated with high serum AFP and the prediction of recurrence in HCC, we applied a combination of criteria for the selection of candidate genes. First, the gene expression changes in 100 probe sets associated with serum AFP levels were evaluated by microarray on 110 (AFP ≥ 100ng/mL [n = 37] and AFP < 100ng/mL [n = 73]) specimens of HCC tissue (Table S1). Among the 54 613 probes, an FDR < 0.01, Fc > 1.5 and Wilcoxon P < 0.0001 were detected in 29 probes (20 known genes), whose expression levels were significantly upregulated, and in 71 probes (50 known genes) downregulated in the high serum AFP cases as compared with the low serum AFP cases. Figure 1A shows the hierarchical clustering of the 100 probes. As shown in Supplementary Table S1, it is of interest to note that the genes associated with actin binding protein were upregulated in high serum AFP‐associated HCC tissues. These were: Villin1, transcriptional repressor, cadherin‐associated protein (cell adhesion molecule); catenin (CTNND2), preferentially expressed antigen in melanoma (PRAME), hypermethylated in cancer 2 protein (HIC2) (alternative name: HIC1‐related gene on chromosome 22 protein 2), Oct4‐regulated enzyme; tet oncogene 1 (TET1), and the novel oncofetal antigen glypican 3 (GPC3) were upregulated in high serum AFP associated HCC tissues. Next, the gene expression changes in the top 59 probe sets associated with AFP high transcription levels were evaluated using gene expression profiles of 8 of the hepatoma cell lines (Wilcoxon P < 0.01; FC > 10) (Table S2). Figure 1B shows the heatmap of the genes upregulated in the cell lines with high AFP transcription levels. As shown in Supplementary Table S2, Villin1, GPC3 and EPCAM were also upregulated in the cell lines with high AFP transcription levels. The genes that were upregulated in both tissues and cell lines were identified as candidate genes associated with high AFP levels. Table 2 shows the upregulated genes that were associated with high serum AFP and high transcription levels in both primary HCC tissues and hepatoma cell lines (P < 0.05, FC > 1.5). To identify candidate genes for the prediction of recurrence in high serum AFP‐associated HCC, a Cox proportional hazards regression model was used to estimate the relationship between a gene expression pattern and the tumor recurrence‐free survival for each probe set (Table S1). Among these 9 upregulated genes, only VIL1 was significantly correlated with the recurrence of high serum AFP‐associated HCC (P < 0.01, FC > 1.5). Depending on the expression level of the genes, the tissue samples were divided into two groups: H or L. The H group showed higher mRNA levels than average, and the L group revealed lower than average. Figure 1C shows all of the microarray‐examined patients, with HCC classified into high and low VIL1 mRNA expression groups (H versus L groups). There was a statistically significant difference between the H and L groups in terms of recurrence‐free survival (n = 53/57; P = 0.0026). Figure 1D shows that there were significant correlations between AFP mRNA expression and VIL1 mRNA expression in the microarray‐examined hepatoma cell lines (R = 0.809; P = 0.015).
Figure 1.
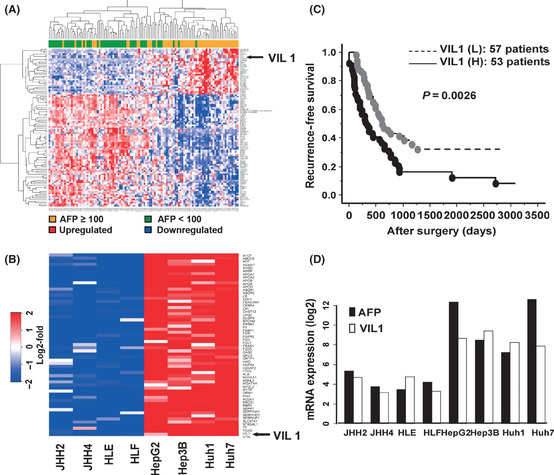
Identification of genes responsible for high serum alpha‐fetoprotein (AFP)‐associated hepatocellular carcinoma (HCC). (A) Hierarchical clustering of gene expression profiles of selected 100 probe sets obtained from 110 HCC patients: the expression profile of high serum AFP‐associated genes that were identified by false discovery rate < 0.01, Wilcoxon signed rank test (P < 0.0001) and more than 1.5‐fold change between serum AFP ≥ 100 ng/mL (denoted as orange in vertical side bar) and serum AFP < 100ng/m/L (denoted as green). (B) Heatmap of genes upregulated in cell lines with AFP high transcription levels. For genes with multiple probe sets, the probe set with the lowest P‐value was chosen for representative expression values of each sample. (C) Villin1 (Vil1) mRNA expression and recurrence‐free survival of HCC patients. All of the microarray‐examined patients with HCC classified into VIL1 mRNA expression high (H) and low (L) groups. (D) Correlation analysis of AFP mRNA and VIL1 mRNA expression in hepatoma cell lines using cDNA microarrays.
Table 2.
Upregulated genes associated with high serum and high transcription α‐fetoprotein (AFP) levels in both primary hepatocellular carcinoma (HCC) tissues and hepatoma cell lines
| Symbol | Title | HCC tissues | Hepatoma cell lines | ||||
|---|---|---|---|---|---|---|---|
| P‐value | FC | FDR | P‐value | FC | FDR | ||
| AFP | Alpha‐fetoprotein | 7.85E‐11 | 8.98 | 0.000 | 0.001863 | 62.28 | 0.2450 |
| VIL1 | Villin 1 | 5.25E‐06 | 2.98 | 0.004 | 1.28E‐06 | 36.72 | 0.0032 |
| C5orf13 | Chromosome 5 open reading frame 13 | 9.51E‐07 | 2.17 | 0.002 | 0.031979 | 2.59 | 0.7213 |
| GPC3 | Glypican 3 | 3.99E‐04 | 1.60 | 0.028 | 8.30E‐06 | 62.70 | 0.0113 |
| HIC2 | Hypermethylated in cancer 2 | 2.40E‐06 | 2.03 | 0.002 | 0.006328 | 4.64 | 0.4138 |
| TET1 | Tet oncogene 1 | 3.97E‐05 | 1.74 | 0.010 | 0.020374 | 2.64 | 0.6362 |
| SALL2 | Sal‐like 2 (Drosophila) | 4.59E‐06 | 1.82 | 0.003 | 0.016742 | 3.20 | 0.5978 |
| ARID3A | AT rich interactive domain 3A (BRIGHT‐like) | 1.60E‐07 | 1.86 | 0.001 | 0.005683 | 6.01 | 0.3963 |
| ZNF813 | zinc finger protein 813 | 1.26E‐07 | 1.89 | 0.001 | 0.04018 | 1.90 | 0.7770 |
FC, fold change; FDR, false discovery rate.
Validation study on TaqMan gene expression assays
Among the 9 upregulated genes, VIL1 was ranked as one of the top for increased AFP‐associated gene expression in both tissues and cell lines (P < 0.001, FC > 2.0 and FDR > 0.01). Therefore, we focused on the VIL1 gene; this result was validated independently in 90 patients by TaqMan gene expression assay. We also tested the microarray results of the training set of 20 patients. There were significant correlations between the cDNA microarray and TaqMan gene expression assays results for VIL1 (r = 0.945, P = 9.10E‐10−5), as shown in Figure 2A. A total of 180 tissue samples of TaqMan gene expression data were normalized against the expression of the control gene, and the estimated gene expression levels were log2‐transformed. After quantitative PCR, the results from agarose gel electrophoresis confirmed that specific products and no nonspecific products were obtained upon amplification of VIL1 (Fig. S1A). The mRNA expression levels of VIL1 were statistically significant different between HCC and the corresponding non‐tumor liver (7.20 ± 2.81 vs 5.74 ± 1.60, P < 0.01, Fig. 2B); VIL1 was more strongly expressed in patients with serum AFP ≥ 100 ng/mL than in patients with serum AFP < 100 ng/mL (8.19 ± 2.80 vs 6.75 ± 2.72, P = 0.024, Fig. 2C). The expression levels of VIL1 mRNA were also evaluated in 10 hepatoma cell lines. The RT‐PCR results showed that VIL1 mRNA was strongly expressed in the HCC cell lines, and there was the same expression pattern in the VIL1 microarray and Taqman gene expression assays (Fig. S1B).
Figure 2.
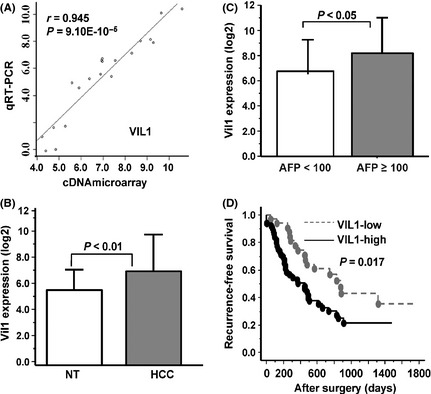
Validation analyses of Villin1 (Vil1) mRNA expression in hepatocellular carcinoma patients. (A) Correlation of test samples for VIL1 mRNA expression by cDNA microarray and TaqMan gene expression assay. (B) expression of VIL1 was compared between cancer and adjacent non‐cancerous tissues (NT), (C) between serum alpha‐fetoprotein (AFP) ≥ 100 ng/mL and serum AFP < 100 ng/mL patients. (D) VIL1 mRNA expression level (high versus low) and recurrence‐free survival curves of validation cases.
Clinicopathological and survival analysis of Villin1 in hepatocellular carcinoma tissues
The clinicopathological significance of VIL1 mRNA expression was analyzed in 90 HCC patients. Depending on the expression level of VIL1 mRNA, the clinicopathologic factors were divided into two groups. As shown in Table 3, there was a statistically significant difference in VIL1 expression levels between each category of clinicopathologic group, such as patients with high serum PIVKAII levels (P = 0.046), poor differentiation (P = 0.004), portal vein invasion (P = 0.011) and at advanced cancer stage (P = 0.005). There were no statistically significant differences in age, gender, size of tumor, number of tumors and other factors between the two groups. We also explored the expression of VIL1 mRNA levels and the recurrence‐free survival of patients. The median follow‐up period was 871 days (range, 4–1827). A total of 53 patients were recurrent during the follow‐up period or at the end point of this study and the median recurrence period was 279 days (range, 1–1319). The H group VIL1 expression was associated with better recurrence‐free survival rates (P = 0.017) (Fig. 2D). Tumor recurrence present within 12 months after hepatectomy was evaluated as early recurrence and others as late recurrence. Early recurrence after resection was observed in 29 out of 53 cases (54%). The median recurrence time was 157 days (range, 1–365). Early recurrence was statistically correlated with patient age, virus infection, prothrombin time (%), hepatic vein invasion and VIL1 mRNA expression. The multivariate analysis on early recurrence was carried out with these five factors, tumor hepatic vein invasion (P = 0.047) and VIL1 mRNA expression as independent predictors of early recurrence after HCC resection (HR, 1.164; P = 0.028) (Table 4).
Table 3.
Villin 1 (VIL1) mRNA expression and clinicopathologic findings in hepatocellular carcinoma (HCC) patients
| Clinicopathologic factor | Feature | HCC (n = 90) | VIL1 expression | P‐value |
|---|---|---|---|---|
| Age | <60 | 20 | 7.50 ± 2.89 | 0.585 |
| ≥60 | 70 | 7.11 ± 2.80 | ||
| Gender | Male | 70 | 7.06 ± 2.89 | 0.372 |
| Female | 20 | 7.70 ± 2.49 | ||
| PIVKAII (mAU/mL) | <100 | 41 | 6.56 ± 3.11 | 0.046* |
| ≥100 | 49 | 7.74 ± 2.42 | ||
| Child–Pugh classification | A | 76 | 7.18 ± 2.68 | 0.890 |
| B | 14 | 7.32 ± 3.51 | ||
| Number of tumor | Solitary | 58 | 6.87 ± 2.61 | 0.139 |
| Multiple | 32 | 7.79 ± 3.09 | ||
| Tumor size (cm) | <5 | 50 | 6.83 ± 2.96 | 0.159 |
| ≥5 | 40 | 7.67 ± 2.56 | ||
| Degree of differentiation | Well | 23 | 5.99 ± 3.34 | 0.004* |
| Poor | 13 | 8.65 ± 1.77 | ||
| Moderate | 54 | 7.36 ± 2.59 | 0.042 | |
| Poor | 13 | 8.65 ± 1.77 | ||
| Portal vein invasion | Absent | 43 | 6.42 ± 2.75 | 0.011* |
| Present | 47 | 7.91 ± 2.69 | ||
| Hepatic vein invasion | Absent | 74 | 7.23 ± 2.83 | 0.840 |
| Present | 16 | 7.07 ± 2.76 | ||
| Vascular invasion | Absent | 38 | 6.44 ± 2.76 | 0.029* |
| Present | 52 | 7.75 ± 2.73 | ||
| Infiltration to capsule | Absent | 39 | 7.12 ± 2.96 | 0.812 |
| Present | 51 | 7.26 ± 2.71 | ||
| Tumor stages | I + II | 37 | 6.22 ± 2.81 | 0.005* |
| III + IV | 53 | 7.88 ± 2.62 |
AFP, α‐fetoprotein; PIVKAII, protein induced by vitamin K absence or antagonists II. *Statistical significance, unpaired t‐test.
Table 4.
Univariate and multivariate analyses of early recurrence
| Clinicopathologic factors | Early recurrence (n = 29) | Late recurrence (n = 24) | ||||||
|---|---|---|---|---|---|---|---|---|
| Univariate (HR, 95% CI) | P‐value | Multivariate (HR, 95% CI) | P‐value | Univariate (HR 95% CI) | P‐value | Multivariate (HR, 95% CI) | P‐value | |
| Age | 0.954 (0.919–0.990) | 0.012 | 0.965 (0.900–1.034) | 0.307 | 0.966(0.925–1.010) | 0.125 | ||
|
Gender (female versus male) |
1.051 (0.421–2.627) | 0.915 | 1.408 (0.498–3.985) | 0.519 | ||||
| Viral infection | 0.025 | 0.938 | 0.218 | |||||
| NBNC | NA | NA | NA | |||||
| HBV | 5.339 (1.522–18.728) | 0.922 (0.103–8.251) | 2.629 (0.784–8.815) | |||||
| HCV | 1.276 (0.521–3.126) | 0.815 (0.260–2.558) | 2.212 (0.803–6.095) | |||||
| AST | 0.999 (0.989–1.009) | 0.854 | 1.004 (0.989–1.019) | 0.622 | ||||
| ALT | 0.993 (0.981–1.004) | 0.210 | 1.004 (0.997–1.011) | 0.245 | ||||
| Plt | 0.983 (0.929–1.039) | 0.540 | 0.947 (0.897–1.004) | 0.068 | ||||
| ICG‐R15 | 1.014 (0.983–1.045) | 0.388 | 1.005 (0.961–1.051) | 0.835 | ||||
| PT% | 0.962 (0.932–0.992) | 0.015 | 0.971 (0.933–1.011) | 0.151 | 0.998 (0.980–1.016) | 0.798 | ||
| T.bil | 2.069 (0.858–4.985) | 0.105 | 3.082 (1.112–8.543) | 0.030 | 4.580 (1.202–17.46) | 0.026 | ||
| Alb | 1.409 (0.491–4.046) | 0.524 | 1.543 (0.475–5.018) | 0.471 | ||||
|
AFP, ng/mL ≥100 vs < 100) |
0.580 (0.271–1.240) | 0.160 | 0.467 (0.148–1.475) | 0.194 | ||||
|
PIVKA‐II, mAU/mL (≥100 vs < 100) |
0.778 (0.361–1.677) | 0.522 | 0.875 (0.363–2.108) | 0.765 | ||||
|
Tumor max size (≥5.0 vs < 5.0) |
0.921 (0.427–1.985) | 0.833 | 3.588 (1.164–11.06) | 0.026 | 3.932 (1.205–12.83) | 0.023 | ||
|
Number of tumors (multiple versus solitary) |
1.153 (0.502–2.650) | 0.738 | 1.143 (0.480–2.718) | 0.763 | ||||
| Capsular invasion | 0.802 (0.361–1.784) | 0.589 | 0.849 (0.368–1.957) | 0.700 | ||||
| Portal vein invasion | 0.535 (0.247–1.158) | 0.112 | 1.965 (0.808–4.781) | 0.137 | ||||
| Hepatic vein invasion | 0.323 (0.124–0.846) | 0.022 | 0.357 (0.129–0.987) | 0.047 | 1.223 (0.359–4.174) | 0.748 | ||
| Vascular invasion | 0.486 (0.215–1.095) | 0.082 | 1.965 (0.808–4.781) | 0.137 | ||||
| Stages (I + II versus III + IV) | 0.526 (0.233–1.187) | 0.122 | 1.549 (0.635–3.778) | 0.336 | ||||
| VIL1 mRNA expression | 1.145 (1.008–1.301) | 0.037 | 1.164 (1.016–1.333) | 0.028 | 0.882 (0.767–1.014) | 0.079 | ||
AFP, α‐fetoprotein; Alb, albumin; ALT, alanine aminotransferase; AST, aspartate amino transferase; ICG‐R15, indocyanine green retention rate at 15 min; PIVKAII, protein induced by vitamin K absence or antagonists II; PLT, platelet; PT%, prothrombin time; T.bil, total bilirubin.
Overexpression of Villin1 in hepatocellular carcinoma tissues associated with poor prognosis
Immunohistochemical analysis was performed to evaluate the clinical significance of VIL1 expression by using tissue samples from 70 patients with HCC. Cytoplasmic and nuclear membrane immunostaining of VIL1 was detected in the cancer tissues, but not in any of the non‐cancerous tissues. Consequently, the specific expression of VIL1 was recognized in 25 out of 70 cases (35.7%). Figure 3A,B shows representative cases of positive or negative immunostaining for VIL1. Survival analysis showed that the positive immunostaining of VIL1 was significantly correlated with poor overall survival (P = 0.004, Fig. 3C) and the recurrence‐free survival of HCC (P = 0.025, Fig. 3D). According to Cox regression multivariate analyses, overall survival rates revealed that not only the AFP (HR, 1.359; P = 0.039), but also the VIL1 (HR, 0.401; P = 0.041) and portal vein invasion (HR, 0.454; P = 0.043) were identified as independent predictors of recurrence of HCC (Table 5).
Figure 3.
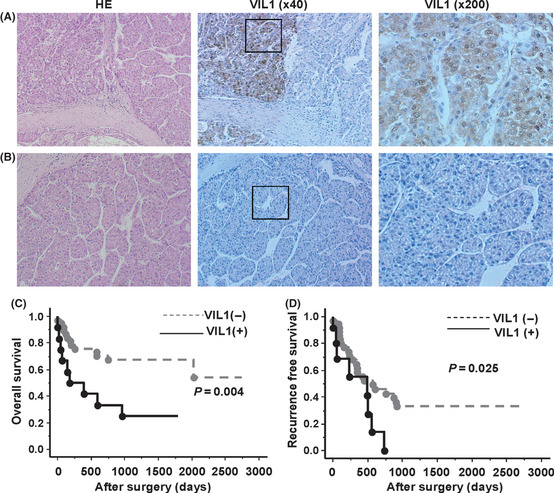
VIL1 protein expression in hepatocellular carcinoma (HCC) tissues and survival curves of patients. (A,B) Immunohistochemistry staining in HCC patients; the representative cases with Villin1 (Vil1) positive or negative staining are shown. (C,D) Expression of VIL1 protein and overall and recurrence‐free survival of patients.
Table 5.
Cox regression univariate and multivariate analyses of overall survival and recurrence free survival in the 70 validation cases
| Clinicopathologic factors | Overall survival | P‐value | Recurrence free survival | P‐value | ||||
|---|---|---|---|---|---|---|---|---|
| Univariate HR (95% CI) | P‐value | Multivariate HR (95% CI) | Univariate HR (95% CI) | P‐value | Multivariate HR (95% CI) | |||
| Clinicopathologic factors | ||||||||
| Age | 1.012 (0.967–1.060) | 0.602 | 1.013 (0.971–1.057) | 0.538 | ||||
|
Gender (female versus male) |
1.390 (0.539–3.586) | 0.496 | 1.415 (0.624–3.209) | 0.407 | ||||
|
Viral infection (absent versus present) |
0.262 (0.035–1.952) | 0.191 | 1.135 (0.433–2.976) | 0.797 | ||||
| AST | 1.007 (0.997–1.017) | 0.198 | 1.010 (1.000–1.019) | 0.047 | 1.007 (0.997–1.016) | 0.185 | ||
| ALT | 1.002 (0.992–1.012) | 0.704 | 1.004 (0.994–1.014) | 0.463 | ||||
| Plt | 1.019 (0.975–1.064) | 0.406 | 0.983 (0.935–1.034) | 0.505 | ||||
| ICG‐R15 | 1.006 (0.973–1.041) | 0.718 | 1.026 (0.998–1.056) | 0.071 | ||||
| PT% | 0.986 (0.967–1.006) | 0.166 | 0.996 (0.977–1.015) | 0.676 | ||||
| T.bil | 1.947 (1.176–3.221) | 0.0095 | 2.214 (1.260–3.887) | 0.006 | 1.345 (0.824–2.196) | 0.236 | ||
| Alb | 0.585 (0.207–1.655) | 0.312 | 0.740 (0.304–1.802) | 0.507 | ||||
| AFP (ng/mL) | 1.579 (1.200–2.079) | 0.001 | 1.675 (1.205–2.331) | 0.002 | 1.456 (1.108–1.913) | 0.007 | 1.359 (1.015–1.819) | 0.039 |
| PIVKA‐II (mAU/mL) | 1.512 (1.087–2.104) | 0.014 | 1.196 (0.873–1.640) | 0.265 | ||||
|
Tumor max size (≥5.0 vs < 5.0) |
0.265 (0.106–0.661) | 0.004 | 0.625 (0.302–1.293) | 0.205 | ||||
| Tumor number (multiple) | 2.051 (0.874–4.813) | 0.098 | 0.923 (0.395–2.158) | 0.853 | ||||
| Capsular invasion | 1.025 (0.436–2.409) | 0.955 | 1.263 (0.610–2.612) | 0.529 | ||||
| Portal vein invasion | 0.407 (0.157–1.056) | 0.065 | 0.425 (0.201–0.901) | 0.026 | 0.454 (0.211–0.976) | 0.043 | ||
| Hepatic vein invasion | 1.042 (0.307–3.542) | 0.947 | 1.336 (0.466–3.833) | 0.590 | ||||
| Vascular invasion | 0.357 (0.130–0.978) | 0.045 | 0.514 (0.243–1.085) | 0.081 | ||||
| Stages (I + II versus III + IV) | 0.237 (0.078–0.715) | 0.011 | 0.153 (0.030–0.770) | 0.023 | 0.646 (0.310–1.347) | 0.244 | ||
| Molecule factor | ||||||||
|
VIL1 protein expression |
0.303 (0.127–0.722) | 0.007 | 0.253 (0.090–0.712) | 0.009 | 0.395 (0.170–0.918) | 0.031 | 0.401 (0.167–0.964) | 0.041 |
AFP, α‐fetoprotein; Alb, albumin; ALT, Alanine aminotransferase; AST, aspartate amino transferase; HR, hepatocellular carcinoma resection; ICG‐R15, indocyanine green retention rate at 15 min; PIVKAII, protein induced by vitamin K absence or antagonists II; PLT, platelet; PT%, prothrombin time; T.bil, total bilirubin.
Discussion
In the present study on predicting recurrence, we focused on high serum AFP‐associated HCC. According to our results, postoperative recurrence was associated with three factors: AST, high serum AFP and severe vascular invasion, which might regulate intrahepatic metastasis (Table 1). Using a microarray technique prediction system, we identified 9 genes whose expression was concordant with that of AFP in both HCC tumor tissues and cell lines. The coordinated expression of these genes might contribute to the malignant phenotypes of AFP‐producing HCC. Based on training analyses, the genes associated with high serum AFP levels, especially VIL1, were significantly upregulated, and high VIL1 expression was significantly correlated with the recurrence‐free survival of patients. This result was validated independently in patients. A strong correlation between the qRT‐PCR expression data and the microarray values was observed. VIL1 was overexpressed more in the tumors than in the adjacent non‐tumor specimens. Overexpression was also observed in the high serum AFP cases and not in the low serum AFP HCC cases. Here, we also studied the significance of VIL1 mRNA expression from the clinical viewpoint of the progression of hepatocellular carcinomas, and showed that the expression of VIL1 was also associated with the levels of serum PIVKAII, tumor poor differentiation, vascular invasion and an advanced cancer stage in HCC patients. In our validation studies, the clinical significance of VIL1 on recurrence‐free survival was observed for both mRNA and protein expression (Tables 4 and 5). More importantly, the overexpression of VIL1 mRNA was identified as an independent predictor for early recurrence in HCC.
In this study, we also investigated VIL1 expressed in cultured human HCC cell lines. In 3 out of 10 cell lines, the VIL1 mRNA levels were lower than others, such as HLE and HLF, which are derived from epithelial and fibroblastic colonies from the same undifferentiated hepatoma, and do not produce AFP and albumin. Skhep1 is an immortal human cell line derived from the ascitic fluid of a patient with an adenocarcinoma of the liver. The expression pattern of the cells was similar to the AFP expression reported by Saito et al. 21 and to our study data on the gene expression analysis of HCC cell lines by microarray (Fig. 1D). This suggests that the biological behavior of the VIL1 gene could be closely associated with AFP.
Our study demonstrated for the first time the novel biomarker of Villin1 in HCC. VIL1 is a gastrointestinal‐related cytoskeletal protein that is associated with the microfilament bundles of brush border microvilli. A major structural component of the brush border cytoskeleton, VIL1 binds actin in a calcium‐dependent manner.22, 23, 24 Under normal physiological conditions, villin1 is expressed in epithelial cells of the intestinal mucosa, gall bladder, renal proximal tubules and ductuli efferentes of the testis.25, 26 Wang et al. report VIL1 to be an epithelial cell‐specific anti‐apoptotic protein, and to have an important function in regulating actin dynamics, cell morphology, epithelial‐to‐mesenchymal transitions, cell migration and cell survival.27, 28 Furthermore, the overexpression of villin has been reported in many diseases, such as gastrointestinal neuroendocrine tumors and29 colon cancer;30, 31 and in lung cancers, villin1 is reported to be a useful marker to distinguish large cell neuroendocrine carcinomas from squamous cell carcinomas from the sera of patients with lung cancer.32 As a diagnostic marker for cervical and endometrial adenocarcinomas, VIL1 is positively associated with the epithelial cell surface marker EpCAM, and the mesenchymal stem cell marker CD44.33, 34 Recently, we reported that EpCAM is overexpressed in HCC, and is closely related to morphological progression in HCC. In particular, EpCAM might play a critical role in the aggressiveness of the confluent multinodular type of HCC.35 The possibility that aberrant VIL1 expression may be related to the recurrence of HCC and to accelerated hepatocarcinogenesis may be mediated via epithelial‐to‐mesenchymal malignant transitions, the positive regulation of hepato‐epithelial cell migration, and the epidermal growth factor receptor signaling pathway. Further study is required to verify these possibilities.
In conclusion, in the present study, we used a genome‐wide gene expression analysis approach to identify several genes closely associated with high serum AFP‐associated HCC that can be used as useful biomarkers. We found that VIL1 was one of the novel biomarker predictive for postoperative recurrence and a poorer prognosis of patients with high serum AFP‐associated HCC.
Disclosure statement
The authors have no conflict of interest to declare.
Supporting information
Fig. S1. Expression of VIL1 mRNA in HCC tissues and cell lines.
Table S1. The list of 100 probe sets identified as differently expressed genes associated with high serum alpha‐fetoprotein (AFP) levels in HCC tissues.
Table S2. The list of 59 probe sets identified as differently expressed genes associated with high alpha‐fetoprotein (AFP) transcription levels in hepatoma cell lines.
Acknowledgments
This work was supported by Special Coordination Funds for Promoting Science and Technology (Japan Science and Technology Agency) and a Grant‐in‐Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan. The authors thank Hiromi Ohnari and Ayumi Shioya for clerical and technical assistance.
References
- 1. Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer 2006; 6: 674–87. [DOI] [PubMed] [Google Scholar]
- 2. But DYK, Lai CL, Yuen MF. Natural history of hepatitis‐related hepatocellular carcinoma. World J Gastroenterol 2008; 14: 1652–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arii S, Yamaoka Y, Futagawa S et al Results of surgical and nonsurgical treatment for small‐sized hepatocellular carcinomas: a retrospective and nationwide survey in Japan. The Liver Cancer Study Group Japan. Hepatol 2000; 32: 1224–9. [DOI] [PubMed] [Google Scholar]
- 4. Tanaka S, Noguchi N, Ochiai T et al Outcomes and recurrence of initially resection of hepatocellular carcinoma meeting Milan Criteria: raionale for partial hepatectomy as first strategy. J Am Coll Surg 2007; 204: 1–6. [DOI] [PubMed] [Google Scholar]
- 5. Iizuka N, Oka M, Yamada‐Okabe H et al Oligonucleotide microarray for prediction of early intrahepatic recurrence of hepatocellular carcinoma after curative resection. Lancet 2003; 361: 923–9. [DOI] [PubMed] [Google Scholar]
- 6. Ho MC, Lin JJ, Chen CN et al A gene expression profile for vascular invasion can predict the recurrence after resection of hepatocellular carcinoma: a microarray approach. Ann Surg Oncol 2006; 13: 1474–84. [DOI] [PubMed] [Google Scholar]
- 7. Tanaka S, Arii S, Yasen M et al Aurora kinase B is a predictive factor for aggressive recurrence of hepatocellular carcinoma after curative hepatectomy. Br J Surg 2008; 95: 611–9. [DOI] [PubMed] [Google Scholar]
- 8. Tanaka S, Mogushi K, Yasen M et al Surgical contribution to recurrence‐free survival in patients with macrovascular–invasion‐negative hepatocellular carcinoma. J Am Coll Surg 2009; 208: 368–74. [DOI] [PubMed] [Google Scholar]
- 9. Arrigoni A, Andriulli A, Gindro T, Piantino P, Capussotti L, Rizzetto M. Pattern analysis of serum alpha‐fetoprotein in the early diagnosis of hepatocellular carcinoma in liver cirrhosis. Int J Biol Markers 1988; 3: 172–6. [DOI] [PubMed] [Google Scholar]
- 10. Tangkijvanich P, Anukulkarnkusol N, Suwangool P et al Clinical characteristics and prognosis of hepatocellular carcinoma: analysis based on serum alpha‐fetoprotein levels. J Clin Gastroenterol 2000; 31: 302–8. [DOI] [PubMed] [Google Scholar]
- 11. Morimoto O, Nagano H, Miyamoto A et al Association between recurrence of hepatocellular carcinoma and alpha‐fetoprotein messenger RNA levels in peripheral blood. Surg Today 2005; 35: 1033–41. [DOI] [PubMed] [Google Scholar]
- 12. Kamiyama T, Takahashi M, Nakagawa T et al AFP mRNA detected in bone marrow by real‐time quantitative RT‐PCR analysis predicts survival and recurrence after curative hepatectomy for hepatocellular carcinoma. Ann Surg 2006; 244: 451–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Okuda N, Nakao A, Takeda S et al Clinical significance of alpha‐fetoprotein mRNA during perioperative period in HCC. Hepatogastroenterology 1999; 46: 381–6. [PubMed] [Google Scholar]
- 14. Jeng KS, Sheen IS, Tsai YC. Circulating messenger RNA of alpha‐fetoprotein: a possible risk factor of recurrence after resection of hepatocellular carcinoma. Arch Surg 2004; 139: 1055–60. [DOI] [PubMed] [Google Scholar]
- 15. Tomasi TB Jr. Structure and function of alpha‐fetoprotein. Annu Rev Med 1977; 28: 453–65. [DOI] [PubMed] [Google Scholar]
- 16. Wang XW, Xie H. Alpha‐fetoprotein enhances the proliferation of human hepatoma cells in vitro. Life Sci 1999; 64: 17–23. [DOI] [PubMed] [Google Scholar]
- 17. Dudich E, Semenkova L, Gorbatova E et al Growth‐regulative activity of human alpha‐fetoprotein for different types of tumor and normal cells. Tumour Biol 1998; 19: 30–40. [DOI] [PubMed] [Google Scholar]
- 18. Semenkova LN, Dudich EI, Dudich IV. Induction of apoptosis in human hepatoma cells by alpha‐fetoprotein. Tumour Biol 1997; 18: 261–73. [DOI] [PubMed] [Google Scholar]
- 19. Dudich E, Semenkova L, Dudich I et al Alpha‐fetoprotein causes apoptosis in tumor cells via a pathway independent of CD95, TNFR1 and TNFR2 through activation of caspase‐3‐like proteases. Eur J Biochem 1999; 266: 750–61. [DOI] [PubMed] [Google Scholar]
- 20. Yasen M, Mizushima H, Mogushi K et al Expression of Aurora B and alternative forms in hepatocellular carcinoma and adjacent tissue. Cancer Sci 2009; 100: 472–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shigeru S, Hidenori O, Hitoshi I, Setsuo H, Tadashi K. Molecular background of a‐fetoprotein in liver cancer cells as revealed by global RNA expression analysis. Cancer Sci 2008; 99: 2402–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bretscher A, Weber K. Villin is a major protein of the microvillus cytoskeleton which binds both G and F actin in a calcium‐dependent manner. Cell 1980; 20: 839–47. [DOI] [PubMed] [Google Scholar]
- 23. Nambu Y, Iannettoni MD, Orringer MB, Beer DG. Unique expression patterns and alterations in the intestinal protein villin in primary and metastatic pulmonary adenocarcinomas. Mol Carcinog 1998; 23: 234–42. [PubMed] [Google Scholar]
- 24. Khurana P, Henty JL, Huang S, Staiger AM, Blanchoin L, Staiger CJ. Arabidopsis VILLIN1 and VILLIN3 have overlapping and distinct activities in actin bundle formation and turnover. Plant Cell 2010; 22: 2727–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moll R, Robine S, Dudouet B, Louvard D. Villin: a cytoskeletal protein and a differentiation marker expressed in some human adenocarcinomas. Virchows Arch B Cell Pathol Incl Mol Pathol 1987; 54: 155–69. [DOI] [PubMed] [Google Scholar]
- 26. Bacchi CE, Gown AM. Distribution and pattern of expression of villin, a gastrointestinal‐associated cytoskeletal protein, in human carcinomas: a study employing paraffin‐embedded tissue. Lab Invest 1991; 64: 418–24. [PubMed] [Google Scholar]
- 27. Wang Y, Srinivasan K, Siddiqui MR, George SP, Tomar A, Khurana S. A novel role for villin in intestinal epithelial cell survival and homeostasis. J Biol Chem 2008; 283: 9454–64. [DOI] [PubMed] [Google Scholar]
- 28. Khurana S, George SP. Regulation of cell structure and function by actin‐binding proteins: Villin's perspective. FEBS Lett 2008; 582: 2128–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang PJ, Harris KR, Alobeid B, Brooks JJ. Immunoexpression of villin in neuroendocrine tumors and its diagnostic implications. Arch Pathol Lab Med 1999; 123: 812–6. [DOI] [PubMed] [Google Scholar]
- 30. Rimm DL, Holland TE, Morrow JS, Anderson JM. Autoantibodies specific for villin found in patients with colon cancer and other colitides. Dig Dis Sci 1995; 40: 389–95. [DOI] [PubMed] [Google Scholar]
- 31. Xi L, Gooding W, McCarty K, Godfrey TE, Hughes SJ. Identification of mRNA markers for molecular staging of lymph nodes in colorectal cancer. Clin Chem 2006; 52: 520–3. [DOI] [PubMed] [Google Scholar]
- 32. Nagashio R, Sato Y, Jiang SX et al Detection of tumor‐specific autoantibodies in sera of patients with lung cancer. Lung Cancer 2008; 62: 364–73. [DOI] [PubMed] [Google Scholar]
- 33. Nakamura E, Iwakawa M, Furuta R et al Villin1, a novel diagnostic marker for cervical adenocarcinoma. Cancer Biol Ther 2009; 8: 1146–53. [DOI] [PubMed] [Google Scholar]
- 34. Nakamura E, Satoh T, Iwakawa M et al Villin1, a diagnostic marker for endometrial adenocarcinoma with high grade nuclear atypia. Cancer Biol Ther 2011; 12: 181–90. [DOI] [PubMed] [Google Scholar]
- 35. Murakata A, Tanaka S, Mogushi K et al Gene expression signature of the gross morphology in hepatocellular carcinoma. Ann Surg 2011; 253: 94–100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Expression of VIL1 mRNA in HCC tissues and cell lines.
Table S1. The list of 100 probe sets identified as differently expressed genes associated with high serum alpha‐fetoprotein (AFP) levels in HCC tissues.
Table S2. The list of 59 probe sets identified as differently expressed genes associated with high alpha‐fetoprotein (AFP) transcription levels in hepatoma cell lines.


