Abstract
p62/SQSTM1 is a selective substrate of autophagy, and aberrant accumulation of p62 has been observed in various pathological conditions. To understand the roles p62 plays in non‐small‐cell lung cancer (NSCLC), we carried out immunohistochemical analyses of p62 expression in a cohort of patients with annotated clinicopathological data. As analyses of murine and human hepatocellular carcinomas suggested a correlation between p62 and Nrf2 accumulations, we also examined NRF2 expression in the same cohort. The expression of NRF2 and p62 was examined by immunohistochemical methods in 109 NSCLC cases, which included patients with adenocarcinoma (n = 72), squamous cell carcinoma (n = 31), and large cell carcinoma (n = 6). Accumulation of NRF2 and p62 was detected in 34% and 37% of NSCLC patients, respectively. The accumulations of p62 and NRF2 did not correlate with each other, but both were associated with worse lung cancer‐specific survival (P = 0.0003 for NRF2; P = 0.0130 for p62). NRF2 status had an impact on NSCLC prognosis irrespective of histology types, but p62 status did so particularly in adenocarcinoma (P = 0.037). Multivariate analysis indicated that positive immunoreactivities of NRF2 and p62 were both independent factors predicting worse lung cancer‐specific survival (P < 0.0001 for NRF2 and P = 0.04 for p62). This study revealed that both NRF2 and p62 are independent prognostic factors for NSCLC. The prognostic impact of p62 status was pronounced in adenocarcinoma patients, suggesting that molecular mechanisms underlying cancer evolution differ between adenocarcinoma and squamous cell carcinoma. (Cancer Sci 2012; 103: 760–766)
Non‐small‐cell lung cancer (NSCLC), which comprises mainly adenocarcinoma and squamous cell carcinoma, is one of the most common human cancers.1 Despite rigorous endeavors to develop anticancer therapies, the prognosis of lung cancer patients still remains poor. To predict recurrence after surgery and to judge indications of additional therapies appropriately, clinical and biological markers have long been sought.
The Keap1‐Nrf2 system plays a central role in protecting cells from electrophilic and oxidative stresses.2 Under unstressed conditions, Keap1 ubiquitinates Nrf2, and Nrf2 is degraded by the proteasome. Upon exposure to the stimuli, Keap1 is inactivated, and stabilized Nrf2 induces the transcription of many cytoprotective genes. Somatic mutations in the NRF2 or KEAP1 gene that cause constitutive stabilization of NRF2 have been found in many human cancers.3, 4, 5, 6, 7, 8, 9 Reduced expression of KEAP1 due to KEAP1 methylation has been suggested as another mechanism for NRF2 stabilization.10, 11 NRF2 stabilization and subsequent accumulation contribute to the poor prognosis of NSCLC patients.6, 7 This is because NRF2 confers resistance to anticancer therapies and an aggressive proliferative tendency on cancer cells.10, 12
Recently, still another cause for the constitutive stabilization of NRF2 in cancer cells was reported. One of the selective substrates for autophagy,13, 14 p62/SQTSM1, was found to stabilize Nrf2. Through disrupting the association between Keap1 and Nrf2, p62 inhibited the ubiquitination of Nrf2 and induced the accumulation of Nrf2.15 When autophagy was disrupted genetically in mouse liver, the robust accumulation of p62, simultaneous stabilization of Nrf2, and elevated expression of Nrf2 target genes were observed.16, 17 Interestingly, some cases of human hepatocellular carcinoma show NRF2 stabilization and p62 accumulation,17 suggesting that p62 accumulation leads to Nrf2 stabilization.
As a multifunctional protein, p62 serves as a scaffold for intracellular signaling.18, 19, 20 Targeted disruption of the p62 gene in mice has revealed the critical contribution of p62 to the cell signaling pathway controlling bone remodeling, obesity, and smooth muscle proliferation.21, 22, 23 p62 also serves as a carrier for sequestering ubiquitinated proteins into autophagosomes and is degraded through autophagy concomitant with the ubiquitinated proteins.13, 14 The analysis of autophagy‐deficient mice revealed that the autophagy plays a tumor‐suppressor role through eliminating p62.24 Abnormal p62 accumulation has been detected in gastrointestinal, prostate, liver, and breast cancer cases,25, 26, 27, 28 suggesting the presence of functional relationship between p62 accumulation and cancer progression. However, it remains to be delineated whether p62 is accumulated in lung cancer cases and what contribution p62 has to the prognosis of patients.
To elucidate the contribution of p62 to the development of lung cancers and their prognoses, we examined the expression of p62 and NRF2 in NSCLC patients and analyzed the association between the immunoreactivities and clinicopathological parameters. NRF2 status was strongly associated with poor clinical outcome irrespective of histology types, whereas p62 status was associated with poor prognosis in adenocarcinoma patients. Our results indicate that both NRF2 and p62 serve as feasible biomarkers for the prognostic prediction of lung cancer patients.
Materials and Methods
Patient and tissue specimens.
For this study, 109 specimens of NSCLC were obtained from patients who underwent surgical resection from 1993 to 1995 in the Department of Surgery at the Sendai Kousei Hospital (Sendai, Japan). The mean age of the patients was 65.6 years (range, 23–82 years). Patients examined in this study did not receive irradiation or chemotherapy before surgery. The mean follow‐up time was 1626 days (range, 17–3366 days). All specimens were fixed in 10% formalin and embedded in paraffin wax. The data collected from the patients were age, gender, smoking index, TNM stage, tumor size, lymph node metastasis, histology type, histological differentiation, and status of Ki‐67, p53, NRF2, p62, and NQO1. The Ethics Committees at Tohoku University School of Medicine (Sendai, Japan), Sendai Kousei Hospital (Sendai, Japan), and Miyagi Cancer Center Research Institute (Natori, Japan) approved the research protocols for this study.
Cell culture.
Two human NSCLC cell lines, A549 and H1650, were cultured in DMEM (Wako Pure Chemicals, Osaka, Japan) with 10% FBS. The H1650 cell line was a kind gift from Dr. Shibata at the National Cancer Center Research Institute in Tokyo, Japan. Immunohistochemistry for p62, NRF2, and NQO1 in these cell lines was carried out in cell blocks, which were fixed in formalin and embedded in paraffin.
Immunoblot analysis.
Whole cell extracts were prepared from A549 and H1650 cells. Cells were harvested after a 4‐h incubation with 100 μM diethylmaleate (DEM) or the vehicle DMSO in the cell culture medium. The protein samples were separated using SDS‐PAGE. NRF2, p62, and NQO1 were detected using anti‐NRF2 antibody,29 anti‐p62 antibody (GP62; Progen, Heidelberg, Germany) and anti‐NQO1 antibody (ab2346; Abcam, Cambridge, UK). Tubulin was detected as a loading control using anti‐tubulin (DH1A; Sigma, St. Louis, MO, USA) antibody.
Immunohistochemistry.
Paraffin sections were prepared and reacted with antibodies against NRF2 (sc‐13032X; Santa Cruz Biotechnology, Santa Cruz, CA, USA), p62 (GP62), NQO1 (ab2346), p53 (DO‐7; Novocastra, Newcastle upon Tyne, UK), and Ki‐67 (MIB1; Dako, Glostrup, Denmark). For the staining with anti‐p62 antibody, antigen retrieval was carried out by heating the slides in a microwave (500 W) for 10 min in citric acid buffer (2 mmol/L citric acid and 9 mmol/L trisodium citrate dehydrate [pH 6.0]). For the staining with anti‐NRF2 and Ki‐67 antibodies, antigen retrieval was carried out by heating the slides in an autoclave at 120°C for 5 min in citric acid buffer (2 mmol/L citric acid and 9 mmol/L trisodium citrate dehydrate [pH 6.0]). For the staining with anti‐NQO1 and p53 antibodies, antigen retrieval was carried out by heating the slides in a microwave (500 W) for 20 min in citric acid buffer (2 mmol/L citric acid and 9 mmol/L trisodium citrate dehydrate [pH 6.0]). The streptavidin–biotin amplification method was used for detecting the reacted antibodies. The antigen–antibody complex was visualized with diaminobenzidine solution (1 mmol/L diaminobenzidine, 50 mmol/L Tris‐HCl [pH 7.6] and 0.006% H2O2) and counterstained with hematoxylin. As a negative control, normal mouse or rabbit IgG was used instead of the primary antibody. Immunoreactivity of more than 10% of carcinoma cells was regarded as positive for NRF2, p62, NQO1, and p53.30 Ki‐67 immunoreactivity was scored in more than 1000 carcinoma cells for each case, and the percentage of immunoreactivity (i.e., labeling index) was determined.
Statistical analyses.
Statistical analyses were carried out using an unpaired two‐group t‐test or a cross‐table using the χ2‐test. The Kaplan–Meier analysis was carried out for lung cancer‐specific survival, which was defined from original cancer death. The statistical significance was evaluated using the log–rank test. Univariate and multivariate analyses were evaluated using a proportional hazard model (Cox). P‐values of less than 0.05 were considered significant in this study. The statistical analyses were carried out using StatView 5.0J software (SAS Institute, Cary, NC, USA).
Results
Immunohistochemical detection of p62 and NRF2.
We first optimized the immunostaining condition of p62 and NRF2 by using the human NSCLC cell lines A549 and H1650. NRF2 is constitutively stabilized in A549 cells due to a somatic mutation in the KEAP1 gene and the functional deficiency of KEAP1,11, 31 whereas the KEAP1‐NRF2 system is intact and NRF2 is present in a low level in H1650 cells. Immunoblot analysis showed abundant accumulation of NRF2 in A549 cells with or without the challenge of the electrophilic inducer DEM, whereas an increase in NRF2 was observed in H1650 cells when DEM was added to the medium (Fig. 1A). Intense signals were observed using an anti‐p62 antibody in A549 cells but not in H1650 cells (Fig. 1A).
Figure 1.

Immunohistochemical detection of NRF2, p62, and NQO1 in non‐small‐cell lung cancer cell lines and tissues. (A) Immunoblot analysis of A549 and H1650 cells. Whole cell extracts were prepared from cells with or without stimulation by diethylmaleate (DEM). (B–D) Immunostaining of A549 cells with antibodies against NRF2, p62, and NQO1. (E–G) Immunostaining of lung cancer tissue from a patient with squamous cell carcinoma. (H–M) Immunostaining of lung cancer tissues from patients with adenocarcinoma. Representative cases are shown for positive staining (H–J) and negative staining (K–M). (H,I) Double‐positive case; (K,L) double‐negative case. Scale bar = 50 μm (B–D). Scale bar = 100 μm (E–M).
Accordingly, we used A549 cells as a positive control for immunostaining with anti‐p62 and NRF2 antibodies. As one of the typical Nrf2 target genes, NQO1 expression was monitored. A block of A549 cells was embedded in paraffin, and sections were prepared. The staining conditions were optimized for each antibody so that sufficiently intense signals were obtained. Strong NRF2 signals were observed in the nucleus (Fig. 1B), and p62 and NQO1 signals were observed in the cytoplasm (Fig. 1C,D).
Using the same conditions, cancer samples surgically resected from NSCLC cases were stained with antibodies against NRF2, p62, and NQO1. In some of the cases of squamous cell carcinoma and adenocarcinoma, strong NRF2 signals were identified in the nucleus (Fig. 1E,H), and p62 and NQO1 signals were detected in the cytoplasm (Fig. 1F,G,I,J), showing very good agreement with the cell line studies. Consistent with the previous studies in mouse models,32 intense signals were not detected for either NRF2, p62, or NQO1 in the normal tissues surrounding the tumor tissues. In other cases, no staining was observed in tumors or in normal tissues (Fig. 1K–M). Therefore, these results indicate that the expression of NRF2, p62, and NQO1 can be appropriately evaluated by immunohistochemical examination.
When we stained cancer samples that were surgically resected from 42 NSCLC cases with anti‐NRF2 and anti‐p62 antibodies, we found that the samples were classified into four groups: double‐negative; NRF2‐single‐positive; p62‐single‐positive; and double‐positive (Fig. S1A). Based on our previous study,15 we considered that p62 stabilized NRF2 in the double‐positive cases. The NRF2‐single‐positive cases were expected to include the cases with somatic mutations in the KEAP1 or NRF2 gene and those with hypermethylation of the KEAP1 gene promoter. The KEAP1 expression in the tumor portion was not repressed in the NRF2‐single‐positive cases (Data S1, Fig. S1B), so the contribution of KEAP1 methylation, if any, to NRF2 stabilization was not substantial. The mutation analysis of KEAP1 and NRF2 genes (Table S1, Data S1) did not detect any non‐synonymous mutations in seven NRF2‐single‐positive cases whose tumor DNA was available (data not shown). Other unknown causes stabilized NRF2 in the NRF2‐single‐positive cases of this cohort.
Association of NRF2 stabilization with the clinicopathological features and clinical outcomes of NSCLC patients.
To examine the association between NRF2 immunoreactivity and clinicopathological features, we assembled another cohort of cases that contained information on survival after surgery. Cancer samples surgically resected from 109 NSCLC cases were stained with anti‐NRF2, ‐p62, and ‐NQO1 antibodies using the conditions identified above. Of the 109 cases, 37, 40, and 41 cases were positive for NRF2, p62, and NQO1, respectively (Tables 1, 2). We evaluated the association of NRF2 immunoreactivity with the clinicopathological parameters of the NSCLC cases (Table 1, left). We found that samples from male patients showed positive NRF2 staining more often than those from female patients (P = 0.04). This NRF2 stabilization was significantly associated with the positive immunoreactivity of NQO1 (P < 0.0001), which seems reasonable considering that the NQO1 gene is directly activated by NRF2. There were no significant correlations between NRF2 stabilization and other parameters examined in this study, including patient age, smoking index, TNM stage, tumor size, lymph node metastasis, histology type, histological differentiation, Ki‐67 staining, and p53 status.
Table 1.
Association between NRF2 immunoreactivity and clinicopathological parameters in 109 patients with non‐small‐cell lung cancer
| Value | NRF2 immunoreactivity | P‐value | P‐value | ||
|---|---|---|---|---|---|
| Positive (n = 37) | Negative (n = 72) | Adeno (n = 72)a | SCC (n = 31)a | ||
| Patient ageb | 67 ± 9 | 65 ± 11 | 0.1700 | 0.340 | 0.2300 |
| Gender | |||||
| Male | 31 | 47 | 0.0400 | 0.020 | 0.9900 |
| Female | 6 | 25 | |||
| Smoking index (pack‐years)b | 36 ± 36 | 32 ± 31 | 0.6200 | 0.840 | 0.8400 |
| TNM stage | |||||
| I | 21 | 48 | 0.3100 | 0.290 | 0.7700 |
| II and III | 16 | 24 | |||
| Tumor size (cm)b | 3.3 ± 1.3 | 3.4 ± 1.7 | 0.7200 | 0.930 | 0.7700 |
| Lymph node metastasis | |||||
| Positive | 14 | 21 | 0.4000 | 0.100 | 0.7800 |
| Negative | 23 | 51 | |||
| Histology type | |||||
| Adeno | 25 | 47 | 0.6500 | – | – |
| SCC | 11 | 20 | |||
| Large | 1 | 5 | |||
| Histological differentiation | |||||
| Well and moderately | 27 | 49 | 0.6000 | 0.740 | 0.9800 |
| Poorly | 10 | 23 | |||
| Ki‐67 LI (%)b | 26 ± 22 | 22 ± 19 | 0.3300 | 0.200 | 0.2200 |
| p53 | |||||
| Positive | 28 | 45 | 0.1600 | 0.490 | 0.3600 |
| Negative | 9 | 27 | |||
| NQO1 | |||||
| Positive | 27 | 14 | <0.0001 | 0.003 | <0.0001 |
| Negative | 10 | 58 | |||
P‐values less than 0.05 were considered significant and are shown in bold.
Adeno, adenocarcinoma; Large, large cell carcinoma; LI, labeling index; SCC, squamous cell carcinoma.
The association between p62 immunoreactivity and clinicopathological parameters was statistically evaluated according to each histological type.
Data are presented as the mean ± SD and were evaluated by an unpaired two‐group t‐test. All other values represent the number of cases and were evaluated by a cross‐table using the χ2‐test.
Table 2.
Association between p62 immunoreactivity and clinicopathological parameters in 109 patients with non‐small‐cell lung cancer
| Value | p62 immunoreactivity | P‐value | P‐value | ||
|---|---|---|---|---|---|
| Positive (n = 40) | Negative (n = 69) | Adeno (n = 72)b | SCC (n = 31)b | ||
| Patient agea | 66 ± 10 | 69 ± 10 | 0.630 | 0.73 | 0.89 |
| Gender | |||||
| Male | 29 | 49 | 0.870 | 0.34 | 0.99 |
| Female | 11 | 20 | |||
| Smoking index (pack‐years)‡ | 32 ± 32 | 34 ± 33 | 0.730 | 0.15 | 0.18 |
| TNM stage | |||||
| I | 21 | 48 | 0.070 | 0.10 | 0.83 |
| II and III | 19 | 21 | |||
| Tumor size (cm)a | 3.5 ± 1.6 | 3.3 ± 1.6 | 0.500 | 0.78 | 0.83 |
| Lymph node metastasis | |||||
| Positive | 18 | 17 | 0.030 | 0.03 | 0.44 |
| Negative | 22 | 52 | |||
| Histology type | |||||
| Adeno | 22 | 50 | 0.120 | – | – |
| SCC | 16 | 15 | |||
| Large | 2 | 4 | |||
| Histological differentiation | |||||
| Well and moderately | 26 | 50 | 0.420 | 0.15 | 0.20 |
| Poorly | 14 | 19 | |||
| Ki‐67 LI (%)a | 31 ± 22 | 20 ± 18 | 0.004 | 0.20 | 0.22 |
| p53 | |||||
| Positive | 26 | 47 | 0.740 | 0.15 | 0.17 |
| Negative | 14 | 22 | |||
| NRF2 | |||||
| Positive | 16 | 21 | 0.310 | 0.85 | 0.32 |
| Negative | 24 | 48 | |||
| NQO1 | |||||
| Positive | 17 | 24 | 0.640 | 0.79 | 0.16 |
| Negative | 23 | 45 | |||
P‐values less than 0.05 were considered significant and are shown in bold. Adeno, adenocarcinoma; Large, large cell carcinoma; LI, labeling index; SCC, squamous cell carcinoma.
The association between p62 immunoreactivity and clinicopathological parameters was statistically evaluated according to each histological type.
Data are presented as the mean ± SD and were evaluated by an unpaired two‐group t‐test. All other values represent the number of cases and were evaluated by a cross‐table using the χ2‐test.
When the association analysis was carried out within the two subgroups of histology types, adenocarcinoma and squamous cell carcinoma, the association between positive NRF2 staining and gender was more significant (P = 0.02 for adenocarcinoma patients and P = 0.99 for squamous cell carcinoma patients) (Table 1, right). In contrast, the association between positive NRF2 staining and NQO1 expression was significant irrespective of the histology type (P = 0.003 for adenocarcinoma and P < 0.0001 for squamous cell carcinoma), indicating that once stabilized, NRF2 functions as a transcription activator in cancer cells of both histology types.
We found that the NRF2 status showed a very clear association with the clinical outcome. The lung cancer‐specific survival of NRF2‐negative cases was much better than that of NRF2‐positive cases (P = 0.0003, Fig. 2A). This result is consistent with other reports on lung cancer cases.6, 7 Similar results were observed when we examined the lung cancer‐specific survival individually in subgroups of different histology types (P = 0.017 for adenocarcinoma patients [Fig. 2B] and P = 0.0026 for squamous cell carcinoma patients [Fig. 2C]). Taken together, these results indicate that NRF2 contributes to the malignant phenotype of NSCLC.
Figure 2.

Lung cancer‐specific survival of 109 patients with non‐small‐cell lung cancer according to NRF2 status (A–C) and p62 status (D–F). (A,D) Lung cancer‐specific survival of all patients (n = 109). (B,E) Lung cancer‐specific survival of patients with adenocarcinoma (n = 72). (C,F) Lung cancer‐specific survival of patients with squamous cell carcinoma (n = 31).
Association of p62 accumulation with the clinicopathological features and clinical outcomes of NSCLC patients.
We next examined the association between p62 immunoreactivity and the clinicopathological parameters of the NSCLC cases (Table 2, left) and found that p62 accumulation was associated with the presence of lymph node metastasis (P = 0.03) and a higher rate of Ki‐67 staining (P = 0.004). There were no significant correlations between p62 accumulation and other clinicopathological parameters examined in this study. The immunoreactivity of p62 was not associated with the positive staining of NRF2. Consistent with this observation, there was no correlation between p62 immunoreactivity and NQO1 immunoreactivity.
Similar association analysis within the two histology subgroups, adenocarcinoma and squamous cell carcinoma, showed that p62 status and Ki‐67 staining were not significantly correlated within either subgroup (Table 2, right). In contrast, the presence of lymph node metastasis was positively associated with p62 accumulation in adenocarcinoma patients (P = 0.03) but not in squamous cell carcinoma patients (P = 0.44). This result suggests that p62 accumulation exerts distinct effects on the cancer cell behavior in adenocarcinomas and squamous cell carcinomas.
Indeed, p62 accumulation showed a distinct association with the clinical outcomes of adenocarcinoma and squamous cell carcinoma. When we first examined all NSCLC patients in this study, we found that the lung cancer‐specific survival was significantly worse in p62‐positive cases (P = 0.013, Fig. 2D). When the adenocarcinoma and squamous cell carcinoma subgroups were examined separately, p62 accumulation was significantly associated with poor prognosis in adenocarcinoma patients (P = 0.037, Fig. 2E) but was not significantly associated with clinical outcomes in squamous cell carcinoma patients (P = 0.39, Fig. 2F). Thus, these results suggest that the accumulation of p62 may contribute to the malignant phenotype of adenocarcinoma.
Evaluation of the immunoreactivities of p62 and NRF2 as prognostic factors.
The results thus far have revealed that positive staining for NRF2 and p62 are both tightly associated with worse clinical outcomes for NSCLC patients. We observed similar results in another cohort of patients who underwent surgery during a different time period at a different institute (Fig. S2). In this cohort, the patients’ survival was evaluated using two different parameters, overall survival and relapse‐free survival, and both analyses gave similar tendencies. We further evaluated the relationship of NRF2 and p62 status to other factors in terms of the prognostic determination. In a univariate analysis, TNM stage (P < 0.0001), NRF2 status (P = 0.001), tumor size (P = 0.001), p62 status (P = 0.02), and p53 status (P = 0.03) were shown as significant prognostic factors for lung cancer‐specific survival (Table 3). The grade of histological differentiation and NQO1 immunoreactivity showed weak associations with worse clinical outcomes (P = 0.08 and P = 0.10, respectively), which did not reach statistical significance. No significant association was detected between Ki‐67 status (P = 0.27), histology type (P = 0.82), or smoking index (P = 0.91) and the clinical outcome of the patients.
Table 3.
Univariate and multivariate analyses for clinicopathological outcome in 109 patients with non‐small‐cell lung cancer
| Variable | Lung cancer‐specific survival | ||
|---|---|---|---|
| Univariate | Multivariate | ||
| P‐value | P‐value | Relative risk (95% CI) | |
| TNM stage (II and III vs I) | <0.0001 a | 0.0002 | 4.1 (1.9–8.9) |
| NRF2 (+ versus −) | 0.0010 a | <0.0001 | 5.0 (2.4–10.6) |
| Tumor size (9.0–0.8 cm) | 0.0010 a b | 0.0100 | 1.0 (1.0–1.1) |
| p62 (+ versus −) | 0.0200 a | 0.0400 | 2.0 (1.1–3.8) |
| p53 (+ versus −) | 0.0300 a | 0.2900 | N.A |
| Histological differentiation (poorly versus well and moderately) | 0.0800 | N.A | N.A |
| NQO1 (+ versus −) | 0.1000 | N.A | N.A |
| Gender (male versus female) | 0.1300 | N.A | N.A |
| Ki67 LI (≥10 vs <10) | 0.2700 | N.A | N.A |
| Histology typec (SCC vs Adeno) | 0.8200 | N.A | N.A |
| Smoking index (≥30 vs <30) | 0.9100 | N.A | N.A |
P‐values less than 0.05 were considered significant and are shown in bold. Adeno, adenocarcinoma; CI, confidence interval; LI, labeling index; SCC, squamous cell carcinoma.
Data were considered significant in the univariate analyses and were examined in the multivariate analyses.
Data were evaluated as continuous variables.
Cases of large cell carcinoma were excluded (n = 103). N.A, not analyzed.
To further characterize the association between positive staining of NRF2 and p62 and clinical outcome, we carried out a multivariate analysis. The analysis revealed that TNM stage (P = 0.0002), NRF2 status (P < 0.0001), tumor size (P = 0.01), and p62 status (P = 0.04) were all independent prognostic factors with a relative risk over 1.0. Although the statistical significance for p62 was marginal, pathways involving p62 and NRF2 were considered independently responsible for the malignant behavior of cancer cells of NSCLC patients.
We analyzed the association of the compound status of p62 and NRF2 with clinical outcome to examine whether p62 accumulation displayed an additive effect with NRF2 on the phenotype of cancers. We found that the simultaneous accumulation of p62 and NRF2 resulted in significantly worse lung cancer‐specific survival than the single accumulation of p62 (P = 0.027, Fig. 3), suggesting that the NRF2‐dependent pathway exerted further exacerbation of the prognosis in the presence of p62 accumulation. By contrast, the contribution of p62 to the prognosis became smaller in the presence of NRF2 stabilization. The p62‐single‐positive cases tended to display the worse prognosis than the double‐negative cases (P = 0.084), whereas the prognostic difference between the double‐positive cases and NRF2‐single‐positive cases was not significant (P = 0.20). Thus, the effect of NRF2 stabilization appeared to override that of p62 accumulation on cancer progression.
Figure 3.
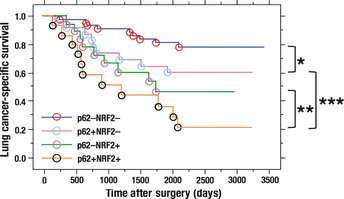
Lung cancer‐specific survival of 109 patients with non‐small‐cell lung cancer according to the combination of p62 and NRF2 status. *P = 0.084; **P = 0.20; ***P = 0.027.
Discussion
Multiple causes have been reported for the constitutive stabilization of NRF2, including the somatic mutation of the KEAP1 or NRF2 gene9 and the hypermethylation of the KEAP1 promoter.10, 11 The NRF2‐single‐positive cases that we examined did not show any reduction in KEAP1 expression nor did they harbor non‐synonymous mutations of KEAP1 and NRF2 genes. KEAP1 expression was rather higher in tumor tissues than in non‐tumor tissues, which appeared to be consistent with a result that KEAP1 is activated by NRF2 (NF, KT, HM, and MY, unpublished observation, 2011). A recent study reported that transcriptional upregulation of NRF2 by oncogene‐dependent signaling contributes to the increase of NRF2 protein level.33 Another report showed that an oncometabolite inactivates KEAP1 resulting in the stabilization of NRF2.34, 35 These mechanisms or other unknown causes stabilized NRF2 in the NRF2‐single‐positive cases.
In the double‐positive cases, accumulated p62 was considered to stabilize NRF2 because a disruptor of the Keap1–Nrf2 interaction, possibly p62 or p21, is another cause of NRF2 stabilization.15, 36 Because p62 is a specific substrate of autophagy, defective autophagy causes aberrant accumulation of p62.37 Recent studies suggested that autophagy plays a suppressive role in tumorigenesis through elimination of p62 in the liver.16, 17, 24 We independently found, in a study using genetically engineered mice and protein structure analysis, that p62 disrupts the Keap1–Nrf2 interaction and results in the stabilization of NRF2.15 This observation suggested that Nrf2 is one of the downstream effectors of the pathway involving p62 for the promotion of tumorigenesis. Supporting this notion, the pathway controlled by NRF2 was activated in p62‐positive cases of hepatocellular carcinomas.17 In the NSCLC cases analyzed in this study, however, accumulated p62 did not necessarily result in the stabilization of NRF2, implying that additional factors accompanying the increase of p62 might differentially influence NRF2 stabilization.
It is interesting to note that the association with lymph node metastasis in adenocarcinoma patients is unique to p62 accumulation. With very good agreement, analyses of breast cancers and digestive system cancers showed that the frequency of having distant metastasis was higher in p62‐positive cases.27, 38 Together with our current result, these data suggest that the pathway involving p62 may act to enhance the metastatic potential of cancer cells, which is likely to aggravate the clinical outcomes of adenocarcinoma.
It has been described that the increase of p62 leads to the activation of NFκB.39, 40 The interaction between p62 and TRAF6 seems to regulate phosphorylation and ubiquitination of the IKK complex and stimulate IKK activity, which results in the phosphorylation and degradation of IκB and the subsequent release and activation of NFκB. A recent report on melanoma showed that NFκB appeared to regulate metastasis‐associated genes,41 supporting the notion that p62‐positive cancer cells may possess higher metastatic activity through potentiating NFκB‐dependent transcription.
Together with the adenocarcinoma‐specific contribution of p62 to cancer development and progression, this study clearly indicates that the molecular mechanisms underlying cancer behavior are different between adenocarcinoma and squamous cell carcinoma. This study revealed that p62 could be used as a prognostic biomarker in lung adenocarcinoma patients. Elucidation of the molecular mechanisms through which p62 exacerbates the clinical outcome will uncover a new target molecule for anticancer therapy for adenocarcinoma patients.
Disclosure Statement
The authors have no conflict of interest.
Supporting information
Fig. S1. Status of NRF2/p62 and KEAP1 expression in 42 non‐small‐cell lung cancer samples.
Fig. S2. Survival curves of 70 patients with non‐small‐cell lung cancer.
Table S1. Primers used in the mutation analysis of KEAP1 and NRF2 genes. Data S1. Supplementary materials and methods.
Acknowledgments
We thank the Biomedical Research Core of Tohoku University Graduate School of Medicine for technical support. We appreciate the skillful technical assistance of Mr. Katsuhiko Ono (Department of Anatomical Pathology, Tohoku University Graduate School of Medicine) and Ms. Yayoi Takahashi (Department of Pathology, Tohoku University Hospital). We also appreciate the assistance of Ms. Eriko Naganuma and Ms. Sayoi Inomata (Department of Medical Biochemistry, Tohoku University Graduate School of Medicine). This work was supported by Grants‐in‐Aid for Creative Scientific Research (M.Y.), Scientific Research on Priority Areas (H.M. and M.Y.) and Scientific Research (T.S., H.M., and M.Y.) from the Ministry of Education, Science, Sports and Culture of Japan, the Tohoku University Global COE for Conquest of Signal Transduction Diseases with Network Medicine (M.Y.), the Naito Memorial Foundation (M.Y.), the Cell Science Research Foundation (H.M.), and a Research Grant from the Princess Takamatsu Cancer Research Fund 09‐24118 (H.M.).
References
- 1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011; 61: 69–90. [DOI] [PubMed] [Google Scholar]
- 2. Uruno A, Motohashi H. The Keap1‐Nrf2 system as an in vivo sensor for electrophiles. Nitric Oxide 2011; 25: 153–60. [DOI] [PubMed] [Google Scholar]
- 3. Singh A, Misra V, Thimmulappa RK et al Dysfunctional KEAP1‐NRF2 interaction in non‐small‐cell lung cancer. PloS Med 2006; 3: 1865–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ohta T, Iijima K, Miyamoto M et al Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res 2008; 68: 1303–9. [DOI] [PubMed] [Google Scholar]
- 5. Shibata T, Kokubu A, Gotoh M et al Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology 2008; 135: 1358–68. [DOI] [PubMed] [Google Scholar]
- 6. Shibata T, Ohta T, Tong KI et al Cancer related mutations in NRF2 impair its recognition by Keap1‐Cul3 E3 ligase and promote malignancy. Proc Natl Acad Sci USA 2008; 105: 13568–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Solis LM, Behrens C, Dong W et al Nrf2 and Keap1 abnormalities in non‐small cell lung carcinoma and association with clinicopathologic features. Clin Cancer Res 2010; 16: 3743–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim YR, Oh JE, Kim MS et al Oncogenic NRF2 mutations in squamous cell carcinomas of oesophagus and skin. J Pathol 2010; 220: 446–51. [DOI] [PubMed] [Google Scholar]
- 9. Taguchi K, Motohashi M, Yamamoto M. Molecular mechanisms of the Keap1‐Nrf2 pathway in stress response and cancer evolution. Genes Cells 2011; 16: 123–40. [DOI] [PubMed] [Google Scholar]
- 10. Zhang P, Singh A, Yegnasubramanian S et al Loss of Kelch‐like ECH‐associated protein 1 function in prostate cancer cells causes chemoresistance and radioresistance and promotes tumor growth. Mol Cancer Ther 2010; 9: 336–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang R, An J, Ji F, Jiao H, Sun H, Zhou D. Hypermethylation of the Keap1 gene in human lung cancer cell lines and lung cancer tissues. Biochem Biophys Res Commun 2008; 373: 151–4. [DOI] [PubMed] [Google Scholar]
- 12. Singh A, Boldin‐Adamsky S, Thimmulappa RK et al RNAi‐mediated silencing of nuclear factor erythroid‐2‐related factor 2 gene expression in non‐small cell lung cancer inhibits tumor growth and increases efficacy of chemotherapy. Cancer Res 2008; 68: 7975–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pankiv S, Clausen TH, Lamark T et al p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007; 282: 24131–45. [DOI] [PubMed] [Google Scholar]
- 14. Ichimura Y, Kumanomidou T, Sou YS et al Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem 2008; 283: 22847–57. [DOI] [PubMed] [Google Scholar]
- 15. Komatsu M, Kurokawa H, Waguri S et al The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol 2010; 12: 213–23. [DOI] [PubMed] [Google Scholar]
- 16. Takamura A, Komatsu M, Hara T et al Autophagy‐deficient mice develop multiple liver tumors. Genes Dev 2011; 25: 795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Inami Y, Waguri S, Sakamoto A et al Persostent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J Cell Biol 2011; 193: 275–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hiruma Y, Honjo T, Jelinek DF et al Increased signaling through p62 in the marrow microenvironment increases myeloma cell growth and osteoclast formation. Blood 2009; 113: 4894–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kurihara N, Hiruma Y, Zhou H et al Mutation of the sequestosome 1 (p62) gene increases osteoclastogenesis but does not induce Paget disease. J Clin Invest 2007; 117: 133–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jin Z, Li Y, Pitti R et al Cullin3‐based polyubiquitination and p62‐dependent aggregation of caspase‐8 mediate extrinsic apoptosis signaling. Cell 2009; 137: 721–35. [DOI] [PubMed] [Google Scholar]
- 21. Durán A, Serrano M, Leitges M et al The atypical PKC‐interacting protein p62 is an important mediator of RANK‐activated osteoclastogenesis. Dev Cell 2004; 6: 303–9. [DOI] [PubMed] [Google Scholar]
- 22. Rodriguez A, Durán A, Selloum M et al Mature‐onset obesity and insulin resistance in mice deficient in the signaling adapter p62. Cell Metab 2006; 3: 211–22. [DOI] [PubMed] [Google Scholar]
- 23. Sugimoto R, Warabi E, Katayanagi S et al Enhanced neointimal hyperplasia and carotid artery remodelling in sequestosome 1 deficient mice. J Cell Mol Med 2010; 14: 1546–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mathew R, Karp CM, Beaudoin B et al Autophagy suppresses tumorigenesis through elimination of p62. Cell 2009; 137: 1062–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stumptner C, Heid H, Fuchsbichler A et al Analysis of intracytoplasmic hyaline bodies in a hepatocellular carcinoma. Demonstration of p62 as major constituent. Am J Pathol 1999; 154: 1701–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thompson HG, Harris JW, Wold BJ, Lin F, Brody JP. p62 overexpression in breast tumors and regulation by prostate‐derived Ets factor in breast cancer cells. Oncogene 2003; 22: 2322–33. [DOI] [PubMed] [Google Scholar]
- 27. Su Y, Qian H, Zhang J, Wang S, Shi P, Peng X. The diversity expression of p62 in digestive system cancers. Clin Immunol 2005; 116: 118–23. [DOI] [PubMed] [Google Scholar]
- 28. Kitamura H, Torigoe T, Asanuma H et al Cytosolic overexpression of p62 sequestosome 1 in neoplastic prostate tissue. Histopathology 2006; 48: 157–61. [DOI] [PubMed] [Google Scholar]
- 29. Maruyama A, Tsukamoto S, Nishikawa K et al Nrf2 regulates the alternative first exons of CD36 in macrophages through specific antioxidant response elements. Arch Biochem Biophys 2008; 477: 139–45. [DOI] [PubMed] [Google Scholar]
- 30. Oka K, Suzuki T, Onodera Y et al Nudix‐type motif 2 in human breast carcinoma: a potent prognostic factor associated with cell proliferation. Int J Cancer 2011; 128: 1770–82. [DOI] [PubMed] [Google Scholar]
- 31. Taguchi K, Shimada M, Fujii S et al Redox cycling of 9,10‐phenanthraquinone to cause oxidative stress is terminated through its monoglucuronide conjugation in human pulmonary epithelial A549 cells. Free Radic Biol Med 2008; 44: 1645–55. [DOI] [PubMed] [Google Scholar]
- 32. Inoue D, Kubo H, Taguchi K et al Inducible disruption of autophagy in the lung causes airway hyper‐responsiveness. Biochem Biophys Res Commun 2011; 405: 13–8. [DOI] [PubMed] [Google Scholar]
- 33. DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K et al Oncogene‐induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 2011; 475: 106–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Adam J, Hatipoglu E, O'Flaherty L, Ternette N, Sahgal N, Lockstone H et al Renal cyst formation in Fh1‐deficient mice is independent of the Hif/Phd pathway: roles for fumarate in KEAP1 succination and Nrf2 signaling. Cancer Cell 2011; 20: 524–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kinch L, Grishin NV, Brugarolas J. Succination of Keap1 and activation of Nrf2‐dependent antioxidant pathways in FH‐deficient papillary renal cell carcinoma type 2. Cancer Cell 2011; 20: 418–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen WM, Sun Z, Wang XJ et al Direct interaction between Nrf2 and p21(Cip1/WAF) upregulates the Nrf2‐mediated antioxidant response. Mol Cell 2009; 34: 663–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Komatsu M, Waguri S, Koie M et al Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy‐deficient mice. Cell 2007; 131: 1149–63. [DOI] [PubMed] [Google Scholar]
- 38. Rolland P, Madjd Z, Durrant L, Ellis IO, Layfield R, Spendlove I. The ubiquitin‐binding protein p62 is expressed in breast cancers showing features of aggressive disease. Endocr Relat Cancer 2007; 14: 73–80. [DOI] [PubMed] [Google Scholar]
- 39. Moscat J, Diaz‐Meco MT, Wooten MW. Signal integration and diversification through the p62 scaffold protein. Trends Biochem Sci 2007; 32: 95–100. [DOI] [PubMed] [Google Scholar]
- 40. Nakamura K, Kimple AJ, Siderovski DP, Johnson GL. PB1 domain interaction of p62/sequestosome 1 and MEKK3 regulates NF‐kappaB activation. J Biol Chem 2010; 285: 2077–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Meir T, Dror R, Yu X et al Molecular characteristics of liver metastases from uveal melanoma. Invest Ophthalmol Vis Sci 2007; 48: 4890–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Status of NRF2/p62 and KEAP1 expression in 42 non‐small‐cell lung cancer samples.
Fig. S2. Survival curves of 70 patients with non‐small‐cell lung cancer.
Table S1. Primers used in the mutation analysis of KEAP1 and NRF2 genes. Data S1. Supplementary materials and methods.


