Abstract
The R521K polymorphism of epidermal growth factor receptor has attenuated affinity in ligand binding and proto‐oncogene induction, which may affect the efficacy of cetuximab. We analyzed the effect of this polymorphism on the outcome of 112 patients with KRAS wild‐type metastatic colorectal carcinoma treated with first‐line cetuximab plus FOLFOX‐4. The associations of this polymorphism with vascular endothelial growth factor (VEGF) expression and clinicopathologic characteristics were also examined. The results showed that the frequencies of the G/G, G/A, and A/A genotypes were 32.1% (n = 36), 42.9% (n = 48), and 25.0% (n = 28), respectively. A marked decrease in VEGF expression levels (66.7% vs 28.9%, P < 0.01) was observed in patients with 521A allele variants (Arg/Lys or Lys/Lys), which were associated with a decreased tumor size (55.6% vs 31.6%, P = 0.02), good histological differentiation (63.9% vs 85.5%, P = 0.01), decreased lymphovascular invasion (69.4% vs 39.5%, P < 0.01), and a higher response rate to cetuximab plus FOLFOX treatment (55.6% vs 78.9%, P = 0.01). In addition, this polymorphism was associated with a longer progression‐free period (P = 0.001) and overall survival (P = 0.001). By multivariate analysis, this polymorphism was also identified as an independent prognostic factor. These data suggest that the R521K polymorphism of epidermal growth factor receptor, by reducing its activation and a consequential downregulation of its target genes, including VEGF, could be a key determinant of an increased response to cetuximab‐based chemotherapy and a longer survival for KRAS wild‐type colorectal carcinoma patients. (Cancer Sci 2012; 103: 791–796)
Cetuximab, a chimeric mAb, is an antibody against the extracellular domain of epidermal growth factor receptor (EGFR).1 It binds to EGFR with a high affinity and is able to compete with epidermal growth factor (EGF) binding, thereby inhibiting subsequent receptor activation and signalling.1 Cetuximab is approved for the treatment of patients with metastatic colorectal carcinoma (CRC) and squamous carcinoma of the head and neck. A favorable effect of cetuximab combined with chemotherapy in advanced non‐small‐cell lung cancer (NSCLC) has also been reported.2 The benefits of cetuximab‐based therapies are restricted to a particular subgroup of patients. The EGFR expression, as evaluated by immunohistochemistry, does not correlate with the response to cetuximab,3 but an increase in EGFR copy number identified by FISH, may predict a better response.4 Loss of phosphatase and tensin homolog (PTEN) protein expression is also associated with a lower response rate.5
Mutations of KRAS, a gene encoding a G protein that plays a key role in the downstream signaling of EGFR, lead to resistance to cetuximab.6 Mutations of KRAS occur in approximately 40% of CRC, and the mutation status of KRAS is considered a good predictive marker for cetuximab‐based treatment.6 Mutations of the v‐raf murine sarcoma viral oncogene homolog B1 (BRAF) and PIK3CA genes are also associated with a lower response rate to cetuximab, and the prediction of response rates could be improved by additional genotyping of these genes.7
In KRAS wild‐type populations, there remains a subgroup of patients who do not respond to cetuximab; therefore, additional markers for predicting the response are indicated. Genetic variations in EGFR or its ligand, EGF, may predict clinical outcome in patients treated with cetuximab‐based regimens.8, 9 For example, the A61G polymorphism of EGF predicts the effect of cetuximab plus irinotecan.8 In addition, the intron 1 CA dinucleotide repeat polymorphism of EGFR predicts survival of KRAS wild‐type patients treated with cetuximab‐based chemotherapy.9
A polymorphic variant in EGFR arising from a single nucleotide substitution (142285 G>A), leading to an arginine (R)/lysine (K) substitution in codon 521 in the extracellular domain of EGFR, has been identified.10 This polymorphism, previously described as R497K according to an older nomenclature, was found to be associated with a lower pelvic recurrence in rectal cancer patients treated with chemoradiation,11 longer survival in stage II/III CRC patients who received curative surgery, and a better response to oxaliplatin‐based chemotherapy.12 Compared with the wild‐type 521R allele, the 521K allele variant has attenuated affinity in ligand binding, tyrosine kinase activation, and induction of the proto‐oncogenes myc, fos, and jun.13 In breast cancer patients, the 521K allele variant was found to be associated with decreased lymph node metastasis and good histological differentiation.14 Interestingly, this polymorphism was associated with progression‐free survival in CRC patients treated with single‐agent cetuximab.15 In a retrospective study of 32 CRC patients with different KRAS status treated with cetuximab plus irinotecan, this polymorphism was found to be associated with higher response rate and longer survival; however, the enrolled patients were quite heterogeneous.4
Based on these earlier findings, we propose that the R521K polymorphism of EGFR might be associated with a better outcome in CRC patients receiving cetuximab‐based treatments. A study was carried out in 112 KRAS wild‐type CRC patients treated with first‐line cetuximab plus FOLFOX‐4, and the potential of using this polymorphism as a predictive/prognostic marker was evaluated.
Materials and Methods
Patient characteristics
We examined 118 consecutive patients with KRAS wild‐type metastatic CRC who had received cetuximab plus FOLFOX‐4 as a first‐line treatment, from January 2006 to December 2009. Among them, 112 patients were enrolled and analyzed. The remaining patients were excluded; these patients lacked measurable lesions (n = 3), did not have the primary tumor removed to examine the accurate T and N stages (n = 2), or were unwilling to participate (n = 1). Patients were treated with cetuximab (500 mg/m2, 2‐h infusion, day 1; Merck Serono, Darmstadt, Germany), followed by FOXFOX‐4. The FOLFOX‐4 regimen consisted of oxaliplatin (85 mg/m2, 1‐h infusion, day 1; Sanofi‐Aventis, Paris, France) and folinic acid (200 mg/m2, 2‐h infusion, days 1 and 2) before bolus 5‐fluorouracil (5‐FU; 400 mg/m2, days 1 and 2; Pharmachemie, Ga Haarlem, Holland), and infusional 5‐FU (600 mg/m2, 22‐h infusion immediately after bolus 5‐FU, days 1 and 2) given every 2 weeks.
The response to treatment was evaluated on the basis of standard RECIST criteria. In the case of intolerable toxicity or failure to respond to front‐line cetuximab plus FOLFOX‐4, the treatment was discontinued, and irinotecan‐based or fluoropyrimidine‐only regimens were begun according to the physicians' decision. During treatment, chest X‐ray and CT scan of the abdomen were carried out every 2 months. None of the patients was treated with antagonists that would interfere with the vascular endothelial growth factor (VEGF) pathway. All patients were followed up until disease progression, death, or loss to follow‐up at a similar intensity regardless of EGFR polymorphism status. Patients with different EGFR genotypes were followed up with a median duration of 20 months (range, 5–32 months). An institutional review board approved this study and informed consent was received from all patients before blood testing for genotyping.
Examination of KRAS mutations
Exon 1 of KRAS, the most frequent site of activating mutations in codons 12 and 13, was sequenced after PCR amplification according to a method previously described.16 DNA was extracted from patients' tumor tissue, including frozen (n = 46) or formalin‐fixed paraffin‐embedded samples (n = 66), using standard phenol–chloroform procedures. Adjacent sections were stained with H&E to confirm the presence of at least 50% carcinoma tissue at this location. Briefly, 0.1 μg genomic DNA, forward primer 5′‐ACT GAA TAT AAA CTT GTG GTA GTT GGA CCT‐3′ and reverse primer 5′‐TCA AAG AAT GGT CCT GGA CC‐3′, were used for PCR amplification. The PCR cycle conditions consisted of an initial denaturation step at 94°C for 5 min, followed by 35 cycles of 96°C for 60 s, 55°C for 60 s, 73°C for 30 s, and a final elongation at 72°C for 10 min. A negative (no DNA) control was run with each PCR analysis. After amplification, the PCR products were sequenced directly.
Examination of the R521K polymorphism of EGFR
Genomic DNA was extracted from patients' WBCs obtained from 0.5 mL whole blood using standard phenol–chloroform procedures. The R521K polymorphism of EGFR was examined by the PCR‐RFLP method as previously described.17 Briefly, 0.1 μg genomic DNA, forward primer 5′‐TGC TGT GAC CCA CTC TGT CT‐3′ and reverse primer 5′‐CAA CGC AAG GGG ATT AAA GA‐3′ were used for PCR amplification. The PCR cycle conditions consisted of an initial denaturation step at 94°C for 5 min, followed by 35 cycles of 94°C for 30 s, 63°C for 30 s, 72°C for 30 s, and a final elongation at 72°C for 10 min. The PCR products, after being digested by the StyI restriction enzyme (New England Biolabs, Beverly, MA, USA) at 37°C for 16 h, were separated on 3% ethidium bromide‐stained agarose gels. The RFLP data were compared with PCR‐direct sequencing results to avoid genotyping errors.
Examination of genetic polymorphisms involved in nucleotide excision repair (NER) pathway and expression of the thymidylate synthase gene
Because polymorphisms of genes that are involved in the NER pathway, including excision repair cross‐complementing group 1 (ERCC1) and xeroderma pigmentosum group D (XPD), contribute to resistance to FOLFOX‐4 treatment,18, 19 the influence of polymorphisms involved in the NER pathway warrants further study. Genomic DNA, extracted from patients' WBCs, was subjected to PCR amplification. ERCC1 codon 118 C→T and XPD K751Q polymorphisms were examined by the PCR‐RFLP method as previously described.18, 19 The PCR products, after being digested with BsrD1 (for ERCC1) or MboII (for XPD) restriction enzymes (New England Biolabs), were separated on Nusieve ethidium bromide‐stained agarose gels (Lonza, Basel, Switzerland) to determine different genotypes.
As 5‐FU was used in combination with oxaliplatin to treat these patients, and germ‐line polymorphisms of a 28‐bp tandemly repeated sequence in the 5′‐enhancer region of the thymidylate synthase gene (TSER) remarkably affect the response and survival of CRC patients receiving 5‐FU,20 the influence of this polymorphism on patients with different TSER genotypes warranted further analysis. The polymorphism of TSER was examined by PCR method as previously described.20 The amplified DNA fragments were analyzed by electrophoresis on a 4% agarose gel to determine the number of 28‐bp tandemly repeated sequences in TSER.
Immunohistochemistry
Because EGFR signaling regulates the synthesis of several pro‐angiogenic growth factors, including VEGF,21 we propose that the R521K polymorphism of EGFR, by attenuating its ligand binding and subsequent activation of downstream effectors, may be associated with reduced expression of VEGF. Paraffin‐embedded tumor tissue sections were stained with an anti‐VEGF antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA), using a streptavidin–biotin immunoperoxidase kit (BioGenex, San Ramon, CA, USA), according to the manufacturer's instructions. Two independent pathologists examined these slides microscopically, and the staining result of VEGF was divided into two groups according to the percentage of carcinoma cells showing specific IHC signals: less than 10%, and more than 10% positive cells.
Statistical analysis and survival curve plotting
Patients were divided into “G/G (wild‐type)” and “G/A or A/A (variant‐type)” groups according to the different EGFR codon 521 genotypes. Cause‐specific survival curves were plotted using the Kaplan–Meier product limit method, and the statistical differences in survival between the subgroups were compared using the log–rank test. The correlations of VEGF expression levels, tumor size, histological differentiation, lymphovascular invasion, genetic polymorphisms involved in the NER pathway, and response to cetuximab plus FOLFOX treatment, were analyzed separately according to the codon 521 status of EGFR. The statistical differences in these correlations were determined using the chi squared‐test. To assess the independent prognostic value of this polymorphism, we used Cox's proportional hazards regression analysis (multivariate), which included EGFR codon 521 status and other clinicopathologic parameters. All statistical analyses were carried out using spss for Windows (version 10.0; SPSS, Chicago, IL, USA).
Results
R521K polymorphism of EGFR correlates with reduced VEGF expression levels
An example of different KRAS mutation patterns analyzed by the PCR‐direct sequencing method is shown in Figure 1. Different allele patterns of the R521K polymorphism of EGFR analyzed by the PCR‐RFLP method are shown in Figure 2. The frequencies of EGFR codon 521 G/G (wild‐type), G/A, and A/A genotypes were 32.1% (n = 36), 42.9% (n = 48), and 25.0% (n = 28), respectively. As shown in Figure 3, a marked decrease in VEGF expression was observed in patients with the R521K polymorphism, as the percentage of patients with a higher VEGF expression level (more than 10% positive cells) in those with or without the R521K polymorphism was 28.9% and 66.7%, respectively (P < 0.01; Fig. 3).
Figure 1.
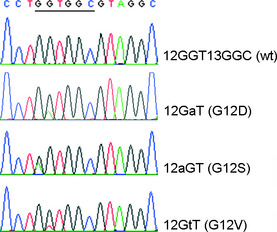
Representative patterns of different KRAS exon 1 genotypes. The PCR products containing codons 12 and 13 of KRAS were amplified with PCR. After amplification, the PCR products were sequenced directly. wt, wild type.
Figure 2.
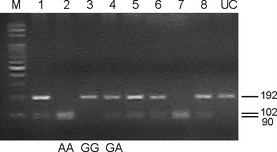
Representative PCR‐RFLP patterns of different EGFR 521 genotypes. Genomic DNA obtained from patients' WBCs was subjected to PCR amplification. After being digested by Sty I, the PCR products were separated by agarose gel electrophoresis. Lanes 1, 4, 5, 6, and 8 represent G/A; lanes 2 and 7 represent A/A; lane 3 represents G/G. M, marker; UC, PCR product that has not been digested.
Figure 3.
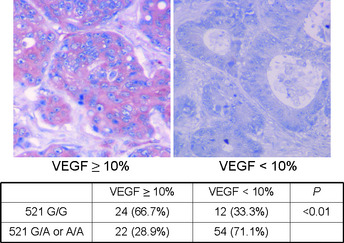
Representative immunohistochemical staining patterns of vascular endothelial growth factor (VEGF). The staining results for VEGF were divided into two groups, <10% and more than 10% positive cells, according to the percentage of carcinoma cells showing specific immune‐reactivity.
R521K polymorphism of EGFR associated with decreased tumor size, good histological differentiation, and lower rate of lymphovascular invasion
As the R521K polymorphism of EGFR dramatically decreases the activation of one of its downstream effectors, VEGF, alterations in clinicopathologic features and the prognosis of patients carrying this polymorphism were proposed. Therefore, the correlation between this polymorphism and the clinicopathologic characteristics of these patients were examined. As shown in Table 1, decreased tumor size (P = 0.02), good histological differentiation (P = 0.01), and a lower rate of lymphovascular invasion (P < 0.01), were clearly identified in patients with the R521K polymorphism. However, there were no between‐group differences in age, gender, performance status, or location of the primary tumor of patients with different EGFR codon 521 statuses (Table 1).
Table 1.
Clinicopathologic features according to epidermal growth factor receptor R521K polymorphism status in KRAS wild‐type colorectal carcinoma patients (n = 112)
| Characteristics | G/G (wild‐type) (%) | G/A or A/A (%) | P |
|---|---|---|---|
| All patients | 36 (100) | 76 (100) | |
| Age, years | |||
| <50 | 14 (38.9) | 26 (34.2) | 0.63 |
| ≥50 | 22 (61.1) | 50 (65.8) | |
| Gender | |||
| Male | 25 (69.4) | 55 (72.4) | 0.75 |
| Female | 11 (30.6) | 21 (27.6) | |
| Performance status | |||
| 0 | 26 (72.2) | 53 (69.7) | 0.79 |
| 1, 2 | 10 (27.8) | 23 (30.3) | |
| Primary tumor | |||
| Colon | 24 (62.1) | 57 (69.0) | 0.36 |
| Rectum | 12 (37.9) | 19 (31.0) | |
| VEGF expression | |||
| Higher (≥10% positive cells) | 24 (66.7) | 22 (28.9) | <0.01 |
| Lower (<10% positive cells) | 12 (33.3) | 54 (71.1) | |
| At least one tumor ≥6 cm | |||
| Present | 20 (55.6) | 24 (31.6) | 0.02 |
| Absent | 16 (44.4) | 52 (68.4) | |
| Histologic differentiation | |||
| Good/moderate | 23 (63.9) | 65 (85.5) | 0.01 |
| Poor | 13 (36.1) | 11 (14.5) | |
| Lymphovascular invasion | |||
| Present | 25 (69.4) | 30 (39.5) | <0.01 |
| Absent | 11 (30.6) | 46 (60.5) | |
| ERCC1 codon 118 genotype | |||
| C/C (wild‐type) | 17 (47.2) | 35 (46.1) | 0.91 |
| C/T or T/T | 19 (52.8) | 41 (53.9) | |
| XPD codon 751 status | |||
| Lys/Lys (wild‐type) | 31 (86.1) | 63 (82.9) | 0.67 |
| Lys/Gln | 5 (13.9) | 13 (17.1) | |
| TSER 28‐bp polymorphism | |||
| 2R/2R or 2R/3R | 12 (33.3) | 27 (35.5) | 0.82 |
| 3R/3R | 24 (66.7) | 49 (64.5) | |
bp, base pair; ERCC1, excision repair cross‐complementing group 1; TSER, 5′‐enhancer region of the thymidylate synthase gene; VEGF, vascular endothelial growth factor; XPD, xeroderma pigmentosum group D.
R521K polymorphism of EGFR correlates with a higher response rate to cetuximab plus FOLFOX and a favorable prognosis in KRAS wild‐type CRC patients
Preliminary studies have shown that the R497K (or R521K) polymorphism of EGFR is predictive of FOLFOX‐412 and cetuximab/irinotecan combination treatment benefit,4 so we speculated whether KRAS wild‐type CRC patients with this polymorphism might be more sensitive to cetuximab‐based treatment, which could translate into a favorable prognosis. As shown in Table 2, patients with G/A or A/A genotypes have a significantly higher response rate to treatment (78.9% vs 55.6%, P = 0.01), longer progression‐free period (8 vs 16 months, P < 0.01; Fig. 4A), and overall survival (16 vs 24 months, P < 0.01; Fig. 4B). With adjusted analysis, this polymorphism was identified as an independent prognostic factor (P = 0.02; Table 3). Second‐line chemotherapy also affects the survival of patients with metastatic CRC. In the current study, a subset of the patients (n = 96) was treated with second‐line chemotherapy, and improved survival was clearly shown (P = 0.02; Table 3).
Table 2.
Response rate to cetuximab plus FOLFOX‐4 in KRAS wild‐type colorectal carcinoma patients with different epidermal growth factor receptor codon 521 statuses (n = 112)
| Response | G/G (wild‐type) (%) | G/A or A/A (%) | P |
|---|---|---|---|
| All patients enrolled | 36 (100) | 76 (100) | |
| OR (CR + PR) | 20 (55.6) | 60 (78.9) | 0.01 |
| CR | 2 (5.6) | 3 (3.9) | |
| PR | 18 (50.0) | 57 (75.0) | |
| SD | 9 (25.0) | 11 (14.5) | |
| PD | 7 (19.4) | 5 (6.6) |
Comparison of overall response rate between patients with different EGFR R521K polymorphisms. CR, complete remission; OR, overall response; PD, progressive disease; PR, partial remission; SD, stable disease.
Figure 4.
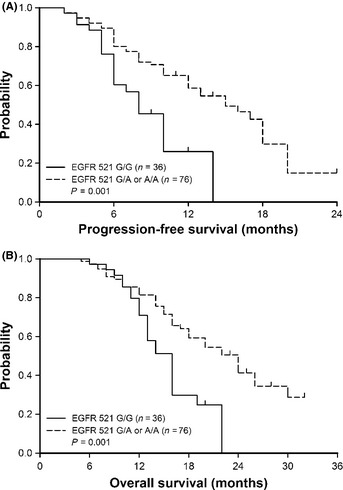
Epidermal growth factor receptor (EGFR) codon 521 G/A or A/A genotypes in KRAS wild‐type metastatic colorectal carcinoma patients are associated with longer progression‐free as well as overall survival. (A) Progression‐free survival curves of 112 metastatic colorectal carcinoma patients with different EGFR codon 521 genotypes were plotted by the Kaplan–Meier method (P = 0.001; log–rank test). (B) A similar method was used to plot overall survival curves (P = 0.001; log–rank test).
Table 3.
Analysis of factors that may affect the survival of patients with KRAS wild‐type colorectal carcinoma (n = 112)
| Characteristics | P (univariate) | P (multivariate) | HR |
|---|---|---|---|
| Age (years) | |||
| <50 vs ≥50 | 0.48 | NA | NA |
| Gender | |||
| Male vs female | 0.36 | NA | NA |
| Performance status | |||
| 0 vs 1, 2 | 0.21 | NA | NA |
| Primary tumor | |||
| Colon vs rectum | 0.45 | NA | NA |
| VEGF expression level | |||
| ≥10% vs <10% positive cells | 0.02 | 0.06 | 1.68 |
| At least one tumor ≥6 cm | |||
| Presence vs absence | 0.15 | NA | NA |
| Histologic differentiation | |||
| Good–moderate vs poor | 0.13 | NA | NA |
| Lymphovascular invasion | |||
| Presence vs absence | 0.06 | NA | NA |
| Metastasis at diagnosis | |||
| Yes vs No | 0.03 | <0.01 | 3.82 |
| Serum CEA level (ng/mL) | |||
| ≤6 vs >6 | 0.34 | NA | NA |
| Second‐line chemotherapy | |||
| No vs yes | 0.03 | 0.02 | 2.32 |
| EGFR R521K polymorphism | |||
| G/G (wild‐type) vs G/A or A/A | 0.03 | 0.02 | 2.41 |
| ERCC1 codon 118 genotype | |||
| C/T or T/T vs C/C | 0.01 | <0.01 | 4.51 |
| XPD codon 751 status | |||
| Lys/Gln vs Lys/Lys | 0.02 | <0.01 | 3.35 |
| TSER 28‐bp polymorphism | |||
| 2R/2R+2R/3R vs 3R/3R | 0.18 | NA | NA |
bp, base pair; CEA, carcinoembryonic antigen; EGFR, epidermal growth factor receptor; ERCC1, excision repair cross‐complementing group 1; HR, Cox hazard ratio; NA, not analyzed; TSER, 5′‐enhancer region of the thymidylate synthase gene; VEGF, vascular endothelial growth factor; XPD, xeroderma pigmentosum group D.
Although polymorphisms of the ERCC1 and XPD genes had a significant impact on survival of patients treated with cetuximab plus FOLFOX in this study (P < 0.01; Table 3), there were no between‐group differences in these genetic alterations among patients with different EGFR codon 521 genotypes (Table 1). Therefore, the influences of these polymorphisms in different EGFR genotypes could be neglected.
Discussion
Somatic mutations of EGFR are associated with increased sensitivity to EGFR‐tyrosine kinase inhibitors in NSCLC,22 but such mutations are rare or absent in CRC,23 and the addition of gefitinib to chemotherapy does not improve therapeutic efficacy in CRC patients.24 KRAS mutations lead to resistance to cetuximab;6 but in KRAS wild‐type populations, there remains a subgroup of patients that does not respond to cetuximab. Polymorphisms in EGFR have attracted a lot of attention because they affect not only their function in ligand binding,13 but also clinicopathologic features14 and response to current therapeutic agents,11, 12 including cetuximab.4, 15 In the current study, we focused on the extracellular domain of EGFR because this region has been shown to be highly polymorphic and to have a significant impact on the function in ligand binding and subsequent signaling.13 The R521K variant has also been described as being associated with cancer severity in EGFR‐expressing tumors, such as gliomas and lung cancer.25
In a retrospective study of 32 patients with metastatic CRC treated with cetuximab plus irinotecan, the R521K polymorphism of EGFR was found to be associated with higher response rate.4 However, the enrolled patients were quite heterogeneous. In 22 (68.8%) patients, cetuximab/irinotecan was given after two or more regimens failed, and a variety of previous chemotherapy was used in different individuals. One of them was even chemotherapy‐naive. In addition, the enrolled patients had different KRAS statuses, including 14 (43.8%) patients with KRAS mutations and another 18 with wild‐type KRAS, making difficult to ascertain the predictive/prognostic value of this EGFR polymorphism in this study.
The R521K (or R497K according to an older nomenclature) polymorphism of EGFR was previously found to be a prognostic marker for CRC patients because it was associated with longer survival in stage II/III CRC patients treated with curative surgery.12 In the current study, we further identified that this polymorphism could serve as a predictive marker for cetuximab‐based treatment in KRAS wild‐type CRC. We found that the EGFR R521K polymorphism correlates with a higher response rate (Table 2) and a favorable prognosis (Fig. 4). An attenuated EGFR signaling induced by the R521K polymorphism that could be more sensitive to targeted receptor inhibition was proposed. The resulting amino acid substitution, arginine to lysine, has been shown to reduce ligand binding and ligand‐induced EGFR signaling.13 Therefore, this genetic variant may alter the binding of its specific ligands, leading to an altered phenotype of EGFR signaling. Of particular interest, the expression of EGFR ligands epiregulin and amphiregulin, which may predict cetuximab benefit, has also been clearly shown.26 In fact, the resulting amino acid substitution is located at the boundary between EGFR domain III, which represents the direct interaction site with cetuximab, and domain IV.27 Therefore, the 521K variant could affect cetuximab binding and/or effects.
In the current study, G/A or A/A genotypes in the CRC patients were associated with smaller tumor size, better histologic differentiation, and lower rate of lymphovascular invasion (Table 1). These data are compatible with a previous report that the R521K polymorphism is correlated with a lower tumor grade and fewer lymph node metastases, and might be a prognostic factor in breast cancer patients.14 The association between the EGFR R521K polymorphism and lymph node metastasis deserves further consideration as a clinical indicator during pre‐operative evaluation. In fact, various studies have considered genetic factors involving cell mobility, vascular invasion and angiogenesis for predicting lymph node metastasis.28
We found that the R521K polymorphism of EGFR correlates with a higher response rate to cetuximab‐based treatment and a favorable prognosis in KRAS wild‐type CRC patients (Table 2, Fig. 4). However, patients with this polymorphism also have lower VEGF expression levels (Fig. 3); therefore, the confounding bias between EGFR polymorphism and VEGF status should be considered. Epidermal growth factor receptor signaling regulates the synthesis of several pro‐angiogenic growth factors, including VEGF.21 Previous studies have shown that the expression of VEGF is related to the extent of tumor vascularization and prognosis, and is predictive of resistance to chemotherapy.29 Interestingly, in a subset of human tumors, VEGF may promote the malignant progression of tumor cells by directly acting on its receptors through an endothelial cell‐independent pathway.30, 31 The expression of VEGF receptor 1 (VEGFR1) and VEGFR2 has been shown in CRC cells, and the activation of these receptors by VEGF leads to activation of the MAPK pathway and phenotypic changes in CRC cells.30, 31 Clinically, the addition of bevacizumab, a mAb directed against VEGF, to chemotherapy improves response rates and survival for patients with metastatic CRC.32
Discrepancies in the association between genotype of EGFR codon 521 and its impact on clinical outcome of anti‐EGFR therapy exist in different types of malignancy. The R521K polymorphism was associated with a longer progression‐free survival in CRC patients treated with cetuximab.15 In patients with NSCLC treated with gefitinib, although sensitivity to treatment strongly depends on the EGFR mutation status,33 the R521K polymorphism of EGFR was not associated with response rate.17 Whether this is due to different drugs used for treatment, or simply due to different tumor types, deserves further study. In addition, because FOLFOX‐4 was used in combination with cetuximab in the current study, it remains possible that the improvement in survival seen in patients with G/A and A/A genotypes is not a reflection of increased sensitivity to cetuximab, but rather that this group inherently has a higher sensitivity to FOLFOX‐4, or a longer survival owing to decreased EGFR signaling.12
Compared with Caucasian populations,4 a remarkably higher prevalence (67.9%) of the EGFR 521K allele variants was noted in our patients. Ethnic differences have a profound influence on the response and toxicity to chemotherapy in malignant diseases. For example, the UGT1A1*28 polymorphism is rare in Asian populations, which leads to a decreased risk of developing severe neutropenia after being treated with irinotecan.34 Due to a higher prevalence of EGFR mutations, gefitinib is very effective in Asian patients with NSCLC.33 In the current study, the frequency of EGFR codon 521 A allele variants was 67.9%, including 42.9% G/A and 25.0% A/A genotypes, which was significantly higher than that in Caucasian (37.5%) and Tunisian (39.2%) populations.4, 14 This result implied that Asian populations might have an attenuated EGFR ligand binding affinity and a better outcome to cetuximab‐based treatment, which deserves further study.
The activation of EGFR initiates intracellular proliferation signaling results in proliferation and survival through the Ras/Raf/MEK/ERK or PI3K/PTEN/AKT pathways, respectively.7 Therefore, alterations of the downstream effectors of the EGFR pathway may have influence in its signaling and the efficacy of EGFR‐targeted therapies. Loss of PTEN protein expression,5 and mutation of BRAF have been associated with lower efficacy of therapies directed against EGFR‐activated pathways.35 However, mutations in BRAF were identified in less than 5% of CRC patients,35 and this mutation is not routinely examined in clinical practice. Mutation of the PIK3CA gene, resulting in a mutant PI3‐kinase and constitutively activated Akt signaling,36 is also associated with a lower response rate to cetuximab.7 The prediction of response rates to cetuximab could be improved by additional genotyping of these genes, and prospective studies in clinical trial cohorts will be required to confirm the utility of these markers.
In summary, we showed that the R521K polymorphism of EGFR is associated with decreased VEGF expression in CRC cells. By reducing the activation of EGFR and consequential downregulation of its target genes, this polymorphism is likely to be one determinant of increased response to cetuximab‐based chemotherapy and longer survival for KRAS wild‐type CRC patients.
Disclosure Statement
The authors have no conflicts of interest.
Acknowledgments
The Taiwan Clinical Oncology Research Foundation supported this study. We thank Mr. Ping‐Liang Hung for excellent technical assistance.
References
- 1. Ng M, Cunningham D. Cetuximab (Erbitux)‐an emerging targeted therapy for epidermal growth receptor‐expressing tumors. Int J Clin Pract 2004; 58: 970–6. [DOI] [PubMed] [Google Scholar]
- 2. Pirker R, Pereira JR, Szczesna A et al Cetuximab plus chemotherapy in patients with advanced non‐small‐cell lung cancer (FLEX): an open‐label randomised phase III trial. Lancet 2009; 373: 1497–8. [DOI] [PubMed] [Google Scholar]
- 3. Chung KY, Shia J, Kemeny NE et al Cetuximab shows activity in colorectal cancer patients with tumors that do not express the epidermal growth factor receptor by immunohistochemistry. J Clin Oncol 2005; 23: 1803–10. [DOI] [PubMed] [Google Scholar]
- 4. Gonçalves A, Esteyries S, Taylor‐Smedra B et al A polymorphism of EGFR extracellular domain is associated with progression free‐survival in metastatic colorectal cancer patients receiving cetuximab‐based treatment. BMC Cancer 2008; 8: 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Frattini M, Saletti P, Romagnani E et al PTEN loss of expression predicts cetuximab efficacy in metastatic colorectal cancer patients. Br J Cancer 2007; 97: 1139–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lièvre A, Bachet JB, Le Corre D et al KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res 2006; 66: 3992–5. [DOI] [PubMed] [Google Scholar]
- 7. De Roock W, Claes B, Bernasconi D et al Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy‐refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol 2010; 11: 753–62. [DOI] [PubMed] [Google Scholar]
- 8. Garm Spindler KL, Pallisgaard N, Rasmussen AA et al The importance of KRAS mutations and EGF61 A>G polymorphism to the effect of cetuximab and irinotecan in metastatic colorectal cancer. Ann Oncol 2009; 20: 879–84. [DOI] [PubMed] [Google Scholar]
- 9. Pander J, Gelderblom H, Antonini NF et al Correlation of FCGR3A and EGFR germline polymorphisms with the efficacy of cetuximab in KRAS wild‐type metastatic colorectal cancer. Eur J Cancer 2010; 46: 1829–34. [DOI] [PubMed] [Google Scholar]
- 10. Zhang W, Stoehlmacher J, Park DJ et al Gene polymorphisms of epidermal growth factor receptor and its downstream effector, interleukin‐8, predict oxaliplatin efficacy in patients with advanced colorectal cancer. Clin Colorectal Cancer 2005; 5: 124–31. [DOI] [PubMed] [Google Scholar]
- 11. Zhang W, Park DJ, Lu B et al Epidermal growth factor receptor gene polymorphisms predict pelvic recurrence in patients with rectal cancer treated with chemoradiation. Clin Cancer Res 2005; 11: 600–5. [PubMed] [Google Scholar]
- 12. Wang WS, Chen PM, Chiou TJ et al Epidermal growth factor receptor R497K polymorphism is a favorable prognostic factor for patients with colorectal carcinoma. Clin Cancer Res 2007; 13: 3597–604. [DOI] [PubMed] [Google Scholar]
- 13. Moriai T, Kobrin MS, Hope C et al A variant epidermal growth factor receptor exhibits altered type alpha transforming growth factor binding and transmembrane signaling. Proc Natl Acad Sci USA 1994; 91: 10217–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kallel I, Rebai M, Khabir A et al Genetic Polymorphisms in the EGFR (R521K) and Estrogen Receptor (T594T) Genes, EGFR and ErbB‐2 Protein Expression, and Breast Cancer Risk in Tunisia. J Biomed Biotechnol 2009; 2009: 753683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lurje G, Nagashima F, Zhang W et al Polymorphisms in cyclooxygenase‐2 and epidermal growth factor receptor are associated with progression‐free survival independent of K‐ras in metastatic colorectal cancer patients treated with single‐agent cetuximab. Clin Cancer Res 2008; 14: 7884–95. [DOI] [PubMed] [Google Scholar]
- 16. Levi S, Urbano‐Ispizua A, Gill R et al Multiple K‐ras codon 12 mutations in cholangiocarcinomas demonstrated with a sensitive polymerase chain reaction technique. Cancer Res 1991; 51: 3497–502. [PubMed] [Google Scholar]
- 17. Ma F, Sun T, Shi Y et al Polymorphisms of EGFR predict clinical outcome in advanced non‐small‐cell lung cancer patients treated with Gefitinib. Lung Cancer 2009; 66: 114–9. [DOI] [PubMed] [Google Scholar]
- 18. Chang PM, Tzeng CH, Chen PM et al ERCC1 codon 118 C→T polymorphism associated with ERCC1 expression and outcome of FOLFOX‐4 treatment in Asian patients with metastatic colorectal carcinoma. Cancer Sci 2009; 100: 278–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lai JI, Tzeng CH, Chen PM et al Very low prevalence of XPD K751Q polymorphism and its association with XPD expression and outcomes of FOLFOX‐4 treatment in Asian patients with colorectal carcinoma. Cancer Sci 2009; 100: 1261–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Horie N, Aiba H, Oguro K et al Functional analysis and DNA polymorphism of the tandemly repeated sequences in the 5′‐terminal regulatory region of the human gene for thymidylate synthase. Cell Struct Funct 1995; 20: 191–7. [DOI] [PubMed] [Google Scholar]
- 21. De Luca A, Carotenuto A, Rachiglio A et al The role of the EGFR signaling in tumor microenvironment. J Cell Physiol 2008; 214: 559–67. [DOI] [PubMed] [Google Scholar]
- 22. Lynch TJ, Bell DW, Sordella R et al Activating mutations in the epidermal growth factor receptor underlying responsiveness of non‐small‐cell lung cancer to gefitinib. N Engl J Med 2004; 350: 2129–39. [DOI] [PubMed] [Google Scholar]
- 23. Barber TD, Vogelstein B, Kinzler KW et al Somatic mutations of EGFR in colorectal cancers and glioblastomas. N Engl J Med 2004; 351: 2883. [DOI] [PubMed] [Google Scholar]
- 24. Santoro A, Comandone A, Rimassa L et al A phase II randomized multicenter trial of gefitinib plus FOLFIRI and FOLFIRI alone in patients with metastatic colorectal cancer. Ann Oncol 2008; 19: 1888–93. [DOI] [PubMed] [Google Scholar]
- 25. Lassman AB, Rossi MR, Razier JR et al Molecular study of malignant gliomas treated with epidermal growth factor receptor inhibitors: tissue analysis from North American Brain Tumor Consortium trials 01‐03 and 00‐01. Clin Cancer Res 2005; 11: 7841–50. [DOI] [PubMed] [Google Scholar]
- 26. Khambata‐Ford S, Garrett CR, Meropol NJ et al Expression of epiregulin and amphiregulin and K‐ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol 2007; 25: 3230–7. [DOI] [PubMed] [Google Scholar]
- 27. Li S, Schmitz KR, Jeffrey PD et al Structural basis for inhibition of the epidermal growth factor receptor by cetuximab. Cancer Cell 2005; 7: 301–11. [DOI] [PubMed] [Google Scholar]
- 28. Carter CL, Allen C, Henson DE. Relation of tumor size, lymph node status, and survival in 24,740 breast cancer cases. Cancer 1989; 63: 181–7. [DOI] [PubMed] [Google Scholar]
- 29. Toi M, Matsumoto T, Bando H. Vascular endothelial growth factor: its prognostic, predictive, and therapeutic implications. Lancet Oncol 2001; 2: 667–73. [DOI] [PubMed] [Google Scholar]
- 30. Fan F, Wey JS, McCarty MF et al Expression and function of vascular endothelial growth factor receptor 1 on human colorectal cancer cells. Oncogene 2005; 24: 2647–53. [DOI] [PubMed] [Google Scholar]
- 31. Giatromanolaki A, Koukourakis MI, Sivridis E et al Activated VEGFR2/KDR pathway in tumor cells and tumor associated vessels of colorectal cancer. Eur J Clin Invest 2007; 37: 878–86. [DOI] [PubMed] [Google Scholar]
- 32. Hurwitz H, Fehrenbacher L, Novotny W et al Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med 2004; 350: 2335–42. [DOI] [PubMed] [Google Scholar]
- 33. Chang A, Parikh P, Thongprasert S et al Gefitinib (IRESSA) in patients of Asian origin with refractory advanced non‐small cell lung cancer: subset analysis from the ISEL study. J Thorac Oncol 2006; 1: 847–55. [PubMed] [Google Scholar]
- 34. Liu CY, Chen PM, Chiou TJ et al UGT1A1*28 polymorphism predicts irinotecan‐induced severe toxicities without affecting treatment outcome and survival in patients with metastatic colorectal carcinoma. Cancer 2008; 112: 1932–40. [DOI] [PubMed] [Google Scholar]
- 35. Laurent‐Puig P, Cayre A, Manceau G et al Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild‐type KRAS metastatic colon cancer. J Clin Oncol 2009; 27: 5924–30. [DOI] [PubMed] [Google Scholar]
- 36. Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3‐kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci USA 2005; 102: 802–7. [DOI] [PMC free article] [PubMed] [Google Scholar]


