Abstract
One‐quarter of a century ago, identification of the human retinoblastoma gene (RB) loci proved Knudson's ‘two‐hit theory’ that tumor suppressor genes exist. Since then, numerous works delineated crucial roles for the RB protein (pRB)‐E2F transcription factor complex in G1‐S phase transition. In addition, discovering the relationship between pRB and tissue‐specific transcription factors enabled a better understanding of how cell cycle exit and terminal differentiation are coupled. Recent works provoked many exciting twists in views on pRB functions during cancer initiation and progression beyond its previously well‐appreciated roles. Various mitogenic and cytostatic cellular signals appeared to modulate pRB functions and thus affect a wide variety of effector molecules. In addition, genetic studies in mice as well as other creatures incessantly force us to revise our views on pRB functions. This review will focus particularly on the roles of pRB in regulating intracellular signaling, cell metabolism, chromatin function, stem cells and cancer stem cells. (Cancer Sci 2012; 103: 1182–1188)
RB mutation is found prevalently in retinoblastomas, osteosarcomas and small‐cell lung carcinomas, and such a spectrum of tumors is reproducible in Rb‐deficient mice.1 These findings suggest that in these tumors RB mutation occurs when tumors initiate. However, in the majority of cancers, including prostate, breast, bladder, esophageal, hepatic cancers, glioma or chronic myelogenous leukemia, inactivation of pRB functions caused by either mutation, gene deletion, promoter methylation, deregulated phosphorylation or decreased protein level usually occurs during cancer progression.2 Some evidence proposes that pRB is even ‘required’ for tumor initiation in these cancers due to its anti‐apoptotic function or cooperation with Ras‐transformation.3, 4 Despite having redundant functions for controlling G1‐S transition, other ‘pocket protein’ family members p107 and p130 are rarely mutated in cancers. These findings, as well as other discoveries (discussed later), suggest that pRB might possess many more multifaceted and unique functions than previously thought. Compared with its smaller protein abundance (in empirical terms), pRB appears to possess ‘too many’ functions, and some of them cause both positive and negative effects on the same biological event. This implies that a particular pRB function (or an effect of pRB inactivation) is selected depending on the cellular context or cancer stage.
Upstream Signals and Downstream Effectors of pRB
Various mitogenic signals (e.g. receptor tyrosine kinases/Ras, Akt, NF‐κB, Shh, Hippo, Wnt, Myc, Jak/STAT) merge more or less on the upregulation of D‐type cyclins, initiating pRB phosphorylation executed sequentially by cyclin D‐CDK4/6 and cyclin E‐CDK2. This prevents pRB from suppressing E2F function to transactivate the targeted genes. In addition, in response to DNA damage, ATM and Chk1/2 directly phosphorylate pRB.5 AMP‐activated kinase (AMPK) also directly phosphorylates pRB; this contributes to energy control favoring neural progenitor cell growth.6 Furthermore, pRb phosphorylation induced by AMPK leads to E2F1‐dependent cell death that occurs in inner ear cells when they sense mitochondrial defect (discussed later).7 Oxidative stress resets pRB phosphorylation via protein phosphatase 2A (PP2A).8 As is well documented, the CDK inhibitors (CDKI) suppress pRB phosphorylation from genetic upstream by attenuating the catalytic activity of cyclin‐CDK. However, the clinical outcome of CDKI inactivation is not always equivalent to that of pRB inactivation.9 pRB inactivation upregulates p16Ink4a by elevating Ras activity and its tumor suppressor role is taken over by p130.10 This, as well as previously described findings,9 indicate that the genetic interaction between CDKI and pRB is not linear. Furthermore, in addition to phosphorylation, many other types of post‐translational modification regulate pRB activity. For instance, pRB is acetylated by p300/CBP, PCAF and Tip60, deacetylated by Sirtuin1 (SIRT1), and methylated by Set7/9 and SMYD.2, 11 These modifications may alter the susceptibility of pRB to undergo CDK‐dependent phosphorylation or its binding affinity to other partners. Furthermore, pRB is sumoylated and is also catabolized following MDM2‐mediated ubiquitination or caspase 3 and 8‐mediated cleavage at the C‐terminus (Fig. 1).12, 13, 14 Mouse models demonstrated that E2F are crucial downstream mediators of the tumor suppressor function of pRB. The continuous advancement in our understanding of the E2F functions increases the number of possible functions that pRB might possess (discussed later). However, some mutant forms of pRB are defective in E2F binding and transcriptional repression, although the proteins partially retain their tumor suppressor activity.15 This fact led researchers to focus on different downstream effectors. pRB directly binds to Skp2, which allows APC‐Cdh1 to ubiquitinate Skp2. Thereby RB loss allows SCFSkp2 E3 ligase complex to bind to and then ubiquitinate phosphorylated p27Kip1. This nexus appears to be crucial in carcinogenesis since Rb +/−;Skp2 −/− mice are completely free of tumor.16 In addition, EID1 and KDM5A/Jarid1a/RBP2 are recognized as molecules likely to be involved in E2F‐independent functions of pRB (discussed later).17, 18, 19 The pRB family can interact with many enzymes that remodel histones to generate repressive chromatin (Fig. 1 and discussed later).20
Figure 1.
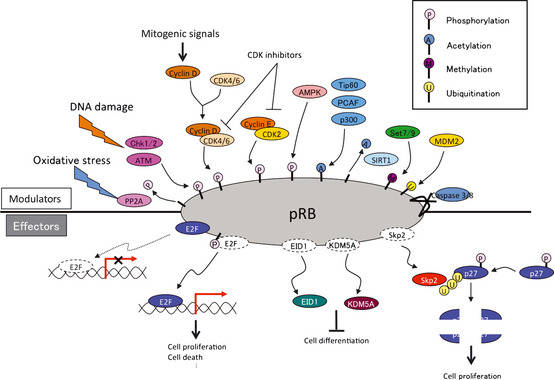
Modulators and effectors of RB protein (pRB) functions. DNA damage induces phosphorylation of pRB by ATM and Chk1/2, whereas oxidative stress induces dephosphorylation of pRB through PP2A. AMP‐activated kinase (AMPK) also phosphorylates pRB. pRB is acetylated by p300, PCAF, methylated by Set 7/9 and SMYD, and degradaded via MDM2‐mediated ubiquitination and caspase 3 and 8 activation. Several molecules, Skp2, KDM5A and EID1, are activated after dissociation from pRB in an E2F‐independent manner. Released Skp2 promotes ubiquitination and degradation of phospho‐p27kip1.
Function of pRB in Intracellular Signaling
Ras signal
Ras activity fluctuates in a cell cycle‐dependent manner.21 Epistasis studies in Caenorhabditis elegans proposed that class B synMuv genes including Rb (lin‐35) can control Ras (let‐60)‐mediated vulval development even from genetic upstream.22 Indeed, SV40 large T antigen‐mediated pRb inactivation or loss of Rb induced elevated Ras activity in mammalian cells.23, 24 The genetic interaction of Rb and Ras has been analyzed extensively in mouse embryos simultaneously lacking Rb and one of the ras isoforms.25 N‐ or K‐ras deletion significantly prolonged the life span of Rb‐null embryos. Their differentiation defects in different organs including muscle cells and erythrocytes were significantly rescued; however, aberrant cell proliferation and cell death persisted (results for erythrocytes are unpublished).26, 27 N‐ or K‐ras deletion in Rb‐deficient pituitary tumorigenesis attenuated tumor invasion with concomitant increase in differentiation degree without affecting tumor incidence.27, 28 These findings implicate that in addition to the previously appreciated pathway in which Ras is upstream of RB, Ras functions also downstream of RB in differentiation control and tumor progression (Fig. 2). Contrary to pituitary, N‐ras deletion converts Rb‐deficient calcitonin‐producing cell (C cell) adenoma or low grade adenocarcinoma to highly metastatic adenocarcinoma.28 The mechanism of this twist was explained in our later study10 as follows: Rb‐deficient C cell adenoma cells are sensitive to DNA damage response induced by mildly elevated N‐Ras activity (at most 10‐fold elevation in activity compared with that of Rb‐positive cells, whereas Ras with oncogenic mutation exhibits an approximately 60‐fold increase in activity). Subsequently, Rb‐deficient C cells undergo ‘paradoxical’ cellular senescence with the aid of p16Ink4a and p130, which protects them from further malignant progression.10 Consistent with this explanation, Rb‐heterozygous mice simultaneously lacking any of the Ink4a, Arf or Suv39h1 (senescence‐inducing genes) alleles directly developed highly malignant C cell tumors and at an earlier age (Fig. 3).10 This provided a further explanation why RB mutation is a relatively infrequent event during tumor initiation; RB loss‐induced carcinogenesis can be antagonized by cellular senescence in some types of cells. In a progenitor for human retinoblastoma, RB loss‐induced carcinogenesis might be antagonized by p53‐dependent apoptosis.29 However, retinoblastoma progression could be antagonized also by senescence since human RB‐null retinomas express elevated p16INK4a and p130 expression, and lose such marks during progression presumably due to chromosomal instability (CIN) (discussed later).30 To determine the mechanism that enables pRB to cease Ras activation status, pRb transcriptional targets were determined in Rb‐deficient N‐ras −/− mouse C cell tumor cells in which proliferation was not affected by the presence of pRb. This enabled the cell cycle‐independent function of pRB to be determined. The study10 detected many genes involved in protein farnesylation and geranylgeranylation (isoprenylation); these post‐translational modifications are essential for Ras to be matured and activated. In addition, the study discovered that these genes are dually innervated by E2F and sterol regulatory element‐binding protein (SREBP) transcription factors. The SREBP are also regulated by E2F (Fig. 4). Consistently, the same study demonstrated that the enhancement of pRb activity delays the trafficking of cytosolic N‐Ras to Golgi for which isoprenylation is essential.10 In addition to N‐Ras, many other small GTPases, CENP‐E and CENP‐F those with CAAX motifs are possibly regulated by pRB (Fig. 4). We also observed RhoA activity was actually suppressed by pRB.28
Figure 2.
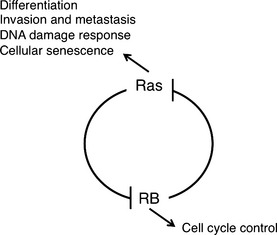
The genetic interaction between Ras and RB. The Ras signal induces RB protein (pRB) phosphorylation by upregulating cyclin D1; this constitutes one of primary mechanisms by which mitogenic signals control the cell cycle. Inversely, RB status can influence Ras activation status by regulating sterol regulatory element‐binding protein (SREBP) and isoprenylation‐related genes. Thereby, Ras mediates pRB function to control the indicated biological events.
Figure 3.
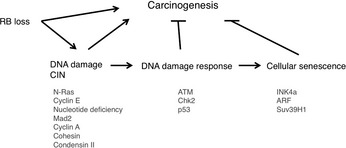
RB loss‐induced carcinogenesis is antagonized by cellular defense mechanisms such as DNA damage response or cellular senescence controlled by the indicated genes or events, depending on the cellular context.
Figure 4.
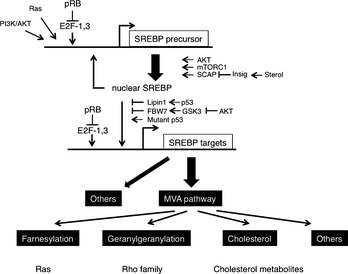
Various oncogenic signals merged on sterol regulatory element‐binding protein (SREBP) regulations (transcription, processing, nuclear localization, ubiquitination and transactivation). Because SREBP genes are dually regulated by E2F and SREBP, pRB gives rise to a different impact on genes involved in the mevalonate (MVA) pathway.
Other signals
A study by another group has demonstrated that AKTSer473 phosphorylation is specifically upregulated in cells lacking all RB family members.31 Although the exact mechanism is still unclear, our own preliminary study demonstrated that the kinetics in which acute pRb inactivation activates AKT seems to be different from that in which pRb inactivation increases GTP‐loaded Ras (Shunsuke Kitajima, unpublished data, 2011). In addition, elevated Ras activity reportedly prefers to induce phosphorylation at AKTSer308 rather than AKTSer473 via the PI3K pathway. Thus, mTORC2 function as well as Ras should be analyzed to discover more on this interaction. Of note, RB and TSC2 (downstream of PI3K/AKT signal) are in a synthetic lethal relationship.32 The genetic interaction between RB and PI3K/AKT/mTOR signal has just begun to be understood. Myc transcription factors are perhaps one of the most well‐recognized pRB transcriptional targets. N‐myc gene amplifications are found in retinoblastoma cases free from RB mutation, suggesting that Myc functions might be considerably overlapped with the signals induced by pRB inactivation.33 In addition, pRB might modulate intracellular signaling by regulating extracellular signaling molecules including VEGF, FGFR, bFGF, matrix metalloproteinases, interleukin‐8, hypoxia‐responsive gene products and Cox‐2; the mechanisms might involve E2F, Id2, Oct‐1, HIF‐1 or others.34, 35
Function of pRB in Cell Metabolism
Metabolic pathways
The aforementioned genetic interaction between RB and Ras mined a new genetic interaction between RB and SREBP; this further allocated a new role to pRB in lipid metabolism, since SREBP are master regulators of lipogenic and steroidogenic genes.10, 36 Indeed, in our initial study, pRb appeared to target many of the genes coding enzymes that participate in fatty acid and cholesterol biosynthesis. In their promoter, these genes possess either sterol regulatory elements (SRE) or E2F‐binding consensus sequences or both (Fig. 4).10 Recently, a new regulator of SREBP has emerged. Mutated p53 directly binds to SREBP‐2 and enhances its transactivation potential, thus contributing to the invasive morphology of breast cancer cells in 3D culture probably due to enhanced geranylgeranylation.37 SREBP transactivate most genes implicated in the mevalonate (MVA) pathway that governs farnesylation, geranylgeranylation and cholesterol synthesis (Fig. 4). This report, in addition to our own study, tightly linked two important tumor suppressors, pRB and p53, to the MVA pathway. Another regulator of SREBP is the PI3K/AKT signaling pathway. An activated AKT signal regulates the SCAP‐mediated processing of SREBP precursors, and also attenuates ubiquitination of mature (nuclear) SREBP by inhibiting GSK3 function to phosphorylate mature SREBP.38 A recent study demonstrated that Lipin1, which is a substrate for mTORC1 kinase activity, eliminates mature SREBP from the nucleus.39 Lipin1 could also link SREBP and p53.40 One of the SREBP targets, fatty acid synthase (FASN), has also been identified to be a pRB transcriptional target.10 This product uses NADPH provided by the shunt from the glycolytic pathway (pentose phosphate pathway) and fuels carbon sources into the MVA pathway. An elevated FASN level during tumor progression might well explain the ‘lipidogenic phenotype’ in cancer cells.41 This in conjugation with ‘aerobic glycolysis (Warburg's effect)’ constitutes two major metabolic perturbations featured in cancer cells. Current understanding is that these mechanisms synergize to efficiently produce and utilize NADPH for the synthesis of macromolecules including lipids and nucleotides. This occurs while avoiding ROS‐producing oxidative phosphorylation (OXPHOS) in mitochondria and preventing ATP production (Fig. 5).42 In addition, increased cellular cholesterol might suppress OXPHOS by altering the components of the mitochondria membrane.42 NADPH is also required for the synthesis of glutathione, an antioxidant. It is of note that wild‐type p53 controls glycolysis in a bipolar manner, for example, by upregulating hexokinase 2, which promotes glycolysis, and TIGAR, which suppresses glycolysis. In total, inactivation of the p53 function is thought to result in the shift of cell metabolism to glycolytic (Fig. 5). Not only sequential regulation of SREBP expression and maturation, but also an astonishing level of cooperation in regulating cancer cell metabolism is becoming evident between pRB and p53 (Fig. 5). AMPKα2 was identified to be in a PI3K‐sensitive gene group among E2F targets.43 This molecule functions as a subunit of a system that senses the cellular level of AMP and antagonizes many metabolic perturbations in cancer cells mostly driven by mTORC1, TSC2 or SREBP. Therefore, together with the phopshorylation of pRB by AMPK, these findings suggest a mutually suppressive genetic interaction between pRB and AMPK.6, 7 AMPK also functions downstream of another tumor suppressor, LKB1.44 Metformin, an AMPK agonist, was suggested to lower the cancer risk in individuals who were administered the drug.45 Although E2F‐AMPK genetic interaction was initially perceived in the context of apoptosis control, together with our knowledge on pRB genetic interaction with SREBP, Ras, AKT, Myc, p53, Oct‐1 and HIF‐1, this discovery will further our understanding of pRB functions in cancer cell metabolism.
Figure 5.
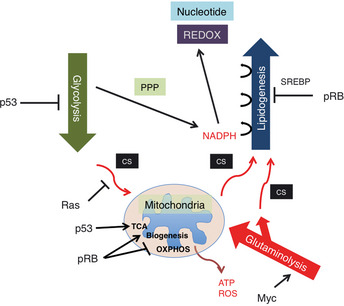
Various oncogenic signals contribute to metabolic perturbation in cancer cells. Simultaneous inactivation of RB protein (pRB) and p53 might optimize coupling of glycolytic and lipidogenic phenotypes, which is facilitated by NADPH production through the pentose phosphate pathway (PPP). Red arrows indicate the flow of carbon sources (CS).
Mitochondrial function
pRB controls mitochondrial biogenesis and function in dissimilar ways. For instance, pRB sustains mitochondrial biogenesis under particular stress conditions. Rb loss induces differentiation defects in cell cycle‐exiting erythrocytes and myotube‐forming muscle cells presumably by reducing the mitochondrial copy number.46, 47 The latter defect was associated with features of autophagy/mitophagy, and rescued by shifting cells to glycolytic status. This further prompted us to hypothesize that without glycolytic shift, pRB inactivation might induce a shortage of energy to support ATP‐consuming differentiation processes (i.e. hemoglobin or myogenic protein synthesis) or even tumor growth. The mechanism linking pRB inactivation to mitophagy might involve a genetic interaction between E2F and Bnip3.48 pRB also controls transcription of genes involved in mitochondrial functions. This also could overlap with the mechanism by which pRB controls mitochondrial biogenesis. pRB, probably through its functional interaction with KDM5A, is likely to upregulate a number of genes encoding mitochondrial proteins in human monocytic cells.49 Another study conducted in erythrocytes suggested that pRB affects mitochondrial biogenesis through regulating NRF1, NRF2a, PPARγ or PGC‐1β in a slightly more complicated manner.46 A recent twist in this field is that the pRB‐E2F‐1 complex appeared to suppress genes implicated in OXPHOS.50 Currently, bifurcated roles of pRB in controlling mitochondrial biogenesis and OXPHOS are unexplained, but this is an intriguing conundrum to solve (Fig. 5). In addition, pRB seems to participate in the regulation of cellular reactive oxygen species (ROS) levels, and undergoes feedback from ROS through CDKI, PP2A, SIRT1 or seladin.51 Various metabolic pathways controlled by pRB might counterbalance each other in order to keep homeostatic control of cellular metabolism.
Function of pRB in Chromatin Functions
CIN and DNA damage
Increased CIN is one of the representative events seen during progression of cancers. pRB inactivation undermines genome stability through various mechanisms that are E2F dependent (e.g. via Mad2, cyclin E‐driven hyper‐replication, nucleotide deficiency) or independent (e.g. via pRB complex with cohesin and condensin II).52, 53 As discussed above, CIN can contribute to retinoblastoma progression. Mad2‐induced CIN provides causality to not only tumorigenesis but also to tumor relapse.54 In addition to such mechanisms, pRB inactivation can induce DNA damage response and senescence due to mildly elevated Ras activity.10 Because of such diverged roles of pRB in genome stability, the nature of signals generated by the DNA damage response induced by pRB inactivation would be different from those induced by other stimuli, for example, oncogenic Ras or Raf. We are currently investigating how pRb status modulates the DNA damage response driven by ATM.
Epigenetic control
A recent study55 reported that epigenetic changes rather than CIN induced by RB loss contributes more strongly to retinoblastoma development. Among the genes that are epigenetically regulated by pRB, spleen tyrosine kinase (SYK) appears to play the most critical role.55 The exact role and mechanism of pRB in epigenetic control is still unclear. Many chromatin modifiers with the LxCxE motif, including DNMT1 (DNA methyltransferase), Suv39H1, Suv4‐20H1 (methyltransferase), HP1 (histone H3me3‐binding protein), Brm1, BRG1 (ATP‐dependent helicases) and HDAC (histone deacetylases), and many of the histone demethylases, including KDM5A, bind to pRB.20 In addition, new insights on the ‘metabolic reprogramming’ caused in part by pRB and p53 double inactivation has led us to investigate its indirect but noteworthy impact on the chemical modification of chromatin‐modifying enzymes by altering the cellular levels of metabolites such as nicotinamide adenine dinucleotide+ (NAD+), flavin adenine dinucleotide (FAD) or α‐ketoglutarate.
Function of pRB in Cell Fate Decision
The role of pRB in terminal differentiation has been well documented by its ability of binding to or functioning with lineage‐specific transcription factors including MyoD, C/EBPα, GRα, GATA‐1, PU1, CBFA‐1, Pdx1, Runx2 and NF‐Il6. In addition, pRB suppresses Id2, KDM5A and EID1, which disturb differentiation. pRB has a crucial role in cell fate decision. For instance, pRB status determines whether p53‐mutated osteosarcoma cells accept osteogenic or fat fate.56 This used to be explained by the functional interaction of pRB with RUNX2 and PPARγ. Nevertheless, the impact of ‘metabolic reprogramming’ on the differentiation program that stems from the synergistic p53 and pRB inactivation is also a possible explanation. Furthermore, the pRB‐p53 alliance appears to determine tumor subtypes through an unknown mechanism. The destruction of this mutually aiding partnership appears to shift breast and lung cancers to more primitive or epithelial‐mesenchymal transition (EMT)‐like types.57, 58
Function of pRB in Stem Cells and Cancer Stem Cells
Tissue stem cells
The intrinsic role of pRB in tissue stem cells has long been debated because studies on the extrinsic role of pRB in hematopoietic stem cells (HSC) for fetal–maternal nutrient exchange through placenta or bone marrow niche–hematopoietic stem cell adhesion had previously had a prevailing impact on the field.59, 60 However, recently the intrinsic role of pRB in HSC for oxidative stress response or mitochondrial biogenesis is also appreciated.46, 51 A surprising twist came from plant studies. Arabidopsis thaliana lacking a RB ortholog showed expansion in the root stem cell pool without losing its self‐renewal capacity; this again stimulated a debate on the role of pRB in stem cells.61, 62
Embryonic stem cells
Embryonic stem (ES) cell biology and induced pluripotent stem (iPS) cell technology indicated a requirement of carcinogenic signals for the induction of genome‐wide chromatin remodeling and ‘stemness’. Compared with adult somatic cells, ES or iPS cells exhibit an extraordinarily short G1 phase. The length of G1 could be one of the determinants of the fate of ES cells of whether to stay in a pluripotent state or to differentiate.63 A short G1 is at least in part due to pRB hyperphosphorylation and suppression of p53 transcription.64 Nanog seems to maintain pRB at the hyperphosphorylated status via CDK6 or CDC25A. 65 In contrast, loss of p53 promoted the efficacy of iPS cell induction.66 The p53‐mSin3a‐HDAC transcription suppressive complex present on the Nanog gene promoter might be a cell cycle‐independent mechanism of this.67 While an early published report denied the contribution of pRB depletion to iPS cell induction, one of the later studies has demonstrated caspase 3‐ and 8‐mediated pRB cleavage/inactivation facilitates iPS cell induction.14, 66 Since pRB is considered to be one of the acute targets of some of the pluripotent core factors, knockdown of RB might not show any more additive effect over acute gain of function of these factors.65 The ES cells lacking KDM5A fail prematurely to maintain Oct4 or Nanog expression under differentiation‐promoting conditions.68 KDM5A might also mediate the ability of pRB to control mitochondrial biogenesis or functions.49 Compared with somatic cells, ES and iPS cells contain fewer numbers of mitochondria; cells with far fewer numbers of organelle maintained high pluripotency but had lower teratoma‐forming activity (which might be relevant to cell proliferation). In contrast, cells with a comparatively high number of mitochondria lose pluripotency but gain higher proliferation activity.69 It would be intriguing to address whether the status of pRB affects mitochondria biogenesis and pluripotency in ES cells or in cancer stem cells.
Cancer stem cells
Previous studies have detected similarities between poorly differentiated cancer cells and ES cells.70 Core pluripotent genes, polycomb genes and Myc target genes might play pivotal roles in these shared features.71 The simultaneous disruption of pRb function and contact inhibition of cells allowed mouse fibroblasts to form a 3D aggregate expressing a variety of core pluripotent genes, and cells derived from such aggregate were also shown to form teratoma‐like tumors in immunodeficient mice.72 We developed a similar experimental model, and subsequently identified additional genetic changes that are in fact required for cancer stem cell‐like behaviors in Rb‐deficient cells (Shunsuke Kitajima, unpublished data, 2011). Detection of core pluripotent gene transcripts in cancers is often quoted to be circumstantial evidence of ‘cancer stemness’; however, its significance is totally unclear. Overexpression of Nanog transforms NIH3T3 fibroblasts and induces cancer stem‐like cells from astrocytes.73, 74 Sox2 (as an esophageal ‘oncogene’) and Nanog have been implicated in the anchorage‐independent growth of tumor cells.75, 76 Indeed, phenomenally, anchorage‐independent growth is hard to distinguish from sphere formation or cancer transplantability to immunodeficient mice. However, it is also possible that such circumstantial evidence is non‐specifically associated with a genome‐wide change in chromatin structure in transformed cells. Attenuation of Rb‐deficient pituitary cancers by deletion of the KDM5A loci implies Rb‐deficient carcinogenesis might depend on chromatin remodeling.68 Epigenetic mechanisms are known to affect the causality of human retinoblastoma progression.55 We observed pRb cooperates with a histone methyltransferase Suv39h1 in suppressing carcinogenesis.10 pRB inactivation induces cell cycle reentry or tumorigenicity in postmitotic fully differentiated cells while preserving or even reviving their differentiation potential.77, 78, 79, 80 Based on these observations, we might be able to establish new in vivo and in vitro models to address the exact role of pRB in stem cell‐like behaviors seen in cancer cells.
Conclusion
To achieve a greater understanding of the roles of pRB during tumor progression, future research should focus on the genome‐wide impact of pRB inactivation, which could be mediated by altered cell signaling, cellular metabolism and chromatin remodeling.
Disclosure Statement
The authors declare no potential conflict of interest.
Acknowledgments
The authors thank Susumu Kohno and Hayato Muranaka for critical reading, and Naoko Nagatani for formatting this manuscript. This work was supported by the Funding Program for Next Generation World‐Leading Researchers (NEXT), Grant‐in‐Aid for Scientific Research (MEXT), Astellas Foundation for Research on Metabolic Disorders, Takeda Science Foundation, the Naito Foundation, the Daiichi‐Sankyo Foundation for Life Science, the NOVARTIS Foundation (Japan) for the Promotion of Science, and the Hokkoku Foundation for Cancer Research. C.T. particularly thanks Dr Mark E. Ewen for his long‐standing encouragement.
References
- 1. Wikenheiser‐Brokamp KA. Retinoblastoma family proteins: insights gained through genetic manipulation of mice. Cell Mol Life Sci 2006; 63: 767–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Burkhart DL, Sage J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer 2008; 8: 671–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Viatour P, Sage J. Newly identified aspects of tumor suppression by RB. Dis Model Mech 2011; 4: 581–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Williams JP, Stewart T, Li B et al The retinoblastoma protein is required for Ras‐induced oncogenic transformation. Mol Cell Biol 2006; 26: 1170–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Inoue Y, Kitagawa M, Taya Y. Phosphorylation of pRB at Ser612 by Chk1/2 leads to a complex between pRB and E2F‐1 after DNA damage. EMBO J 2007; 26: 2083–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dasgupta B, Milbrandt J. AMP‐activated protein kinase phosphorylates retinoblastoma protein to control mammalian brain development. Dev Cell 2009; 16: 256–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Raimundo N, Song L, Shutt TE et al Mitochondrial Stress Engages E2F1 Apoptotic Signaling to Cause Deafness. Cell 2012; 148: 716–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cicchillitti L, Fasanaro P, Biglioli P et al Oxidative stress induces protein phosphatase 2A‐dependent dephosphorylation of the pocket proteins pRb, p107, and p130. J Biol Chem 2003; 278: 19509–17. [DOI] [PubMed] [Google Scholar]
- 9. Wikenheiser‐Brokamp KA. Retinoblastoma regulatory pathway in lung cancer. Curr Mol Med 2006; 6: 783–93. [DOI] [PubMed] [Google Scholar]
- 10. Shamma A, Takegami Y, Miki T et al Rb regulates DNA damage response and cellular senescence through E2F‐dependent suppression of N‐ras isoprenylation. Cancer Cell 2009; 15: 255–69. [DOI] [PubMed] [Google Scholar]
- 11. Wong S, Weber JD. Deacetylation of the retinoblastoma tumour suppressor protein by SIRT1. Biochem J 2007; 407: 451–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ledl A, Schmidt D, Muller S. Viral oncoproteins E1A and E7 and cellular LxCxE proteins repress SUMO modification of the retinoblastoma tumor suppressor. Oncogene 2005; 24: 3810–8. [DOI] [PubMed] [Google Scholar]
- 13. Uchida C, Miwa S, Kitagawa K et al Enhanced Mdm2 activity inhibits pRB function via ubiquitin‐dependent degradation. EMBO J 2005; 24: 160–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li F, He Z, Shen J et al Apoptotic caspases regulate induction of iPSCs from human fibroblasts. Cell Stem Cell 2010; 7: 508–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sellers WR, Novitch BG, Miyake S et al Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation, and suppress tumor cell growth. Genes Dev 1998; 12: 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang H, Bauzon F, Ji P et al Skp2 is required for survival of aberrantly proliferating Rb1‐deficient cells and for tumorigenesis in Rb1+/‐ mice. Nat Genet 2010; 42: 83–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Miyake S, Sellers WR, Safran M et al Cells degrade a novel inhibitor of differentiation with E1A‐like properties upon exiting the cell cycle. Mol Cell Biol 2000; 20: 8889–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. MacLellan WR, Xiao G, Abdellatif M et al A novel Rb‐ and p300‐binding protein inhibits transactivation by MyoD. Mol Cell Biol 2000; 20: 8903–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Benevolenskaya EV, Murray HL, Branton P et al Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Mol Cell 2005; 18: 623–35. [DOI] [PubMed] [Google Scholar]
- 20. Dick FA. Structure‐function analysis of the retinoblastoma tumor suppressor protein ‐ is the whole a sum of its parts? Cell Div 2007; 2: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Taylor SJ, Shalloway D. Cell cycle‐dependent activation of Ras. Curr Biol 1996; 6: 1621–7. [DOI] [PubMed] [Google Scholar]
- 22. Lu X, Horvitz HR. lin‐35 and lin‐53, two genes that antagonize a C. elegans Ras pathway, encode proteins similar to Rb and its binding protein RbAp48. Cell 1998; 95: 981–91. [DOI] [PubMed] [Google Scholar]
- 23. Raptis L, Brownell HL, Corbley MJ et al Cellular ras gene activity is required for full neoplastic transformation by the large tumor antigen of SV40. Cell Growth Differ 1997; 8: 891–901. [PubMed] [Google Scholar]
- 24. Lee KY, Ladha MH, McMahon C et al The retinoblastoma protein is linked to the activation of Ras. Mol Cell Biol 1999; 19: 7724–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Takahashi C, Ewen ME. Genetic interaction between Rb and N‐ras: differentiation control and metastasis. Cancer Res 2006; 66: 9345–8. [DOI] [PubMed] [Google Scholar]
- 26. Takahashi C, Bronson RT, Socolovsky M et al Rb and N‐ras function together to control differentiation in the mouse. Mol Cell Biol 2003; 23: 5256–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Takahashi C, Contreras B, Bronson RT et al Genetic interaction between Rb and K‐ras in the control of differentiation and tumor suppression. Mol Cell Biol 2004; 24: 10406–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Takahashi C, Contreras B, Iwanaga T et al Nras loss induces metastatic conversion of Rb1‐deficient neuroendocrine thyroid tumor. Nat Genet 2006; 38: 118–23. [DOI] [PubMed] [Google Scholar]
- 29. Laurie NA, Donovan SL, Shih CS et al Inactivation of the p53 pathway in retinoblastoma. Nature 2006; 444: 61–6. [DOI] [PubMed] [Google Scholar]
- 30. Dimaras H, Khetan V, Halliday W et al Loss of RB1 induces non‐proliferative retinoma: increasing genomic instability correlates with progression to retinoblastoma. Hum Mol Genet 2008; 17: 1363–72. [DOI] [PubMed] [Google Scholar]
- 31. El‐Naggar S, Liu Y, Dean DC. Mutation of the Rb1 pathway leads to overexpression of mTor, constitutive phosphorylation of Akt on serine 473, resistance to anoikis, and a block in c‐Raf activation. Mol Cell Biol 2009; 29: 5710–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li B, Gordon GM, Du CH et al Specific killing of Rb mutant cancer cells by inactivating TSC2. Cancer Cell 2010; 17: 469–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bremner R, Zacksenhaus E. Cyclins, Cdks, E2f, Skp2, and more at the first international RB Tumor Suppressor Meeting. Cancer Res 2010; 70: 6114–8. [DOI] [PubMed] [Google Scholar]
- 34. Gabellini C, Del Bufalo D, Zupi G. Involvement of RB gene family in tumor angiogenesis. Oncogene 2006; 25: 5326–32. [DOI] [PubMed] [Google Scholar]
- 35. Gauthier ML, Berman HK, Miller C et al Abrogated response to cellular stress identifies DCIS associated with subsequent tumor events and defines basal‐like breast tumors. Cancer Cell 2007; 12: 479–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jeon TI, Osborne TF. SREBPs: metabolic integrators in physiology and metabolism. Trends Endocrinol Metab 2012; 23: 65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Freed‐Pastor WA, Mizuno H, Zhao X et al Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell 2012; 148: 244–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Krycer JR, Sharpe LJ, Luu W et al The Akt‐SREBP nexus: cell signaling meets lipid metabolism. Trends Endocrinol Metab 2010; 21: 268–76. [DOI] [PubMed] [Google Scholar]
- 39. Peterson TR, Sengupta SS, Harris TE et al mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 2011; 146: 408–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Assaily W, Rubinger DA, Wheaton K et al ROS‐mediated p53 induction of Lpin1 regulates fatty acid oxidation in response to nutritional stress. Mol Cell 2011; 44: 491–501. [DOI] [PubMed] [Google Scholar]
- 41. Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer 2007; 7: 763–77. [DOI] [PubMed] [Google Scholar]
- 42. Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer 2011; 11: 85–95. [DOI] [PubMed] [Google Scholar]
- 43. Hallstrom TC, Mori S, Nevins JR. An E2F1‐dependent gene expression program that determines the balance between proliferation and cell death. Cancer Cell 2008; 13: 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shackelford DB, Shaw RJ. The LKB1‐AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer 2009; 9: 563–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Libby G, Donnelly LA, Donnan PT et al New users of metformin are at low risk of incident cancer: a cohort study among people with type 2 diabetes. Diabetes Care 2009; 32: 1620–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sankaran VG, Orkin SH, Walkley CR. Rb intrinsically promotes erythropoiesis by coupling cell cycle exit with mitochondrial biogenesis. Genes Dev 2008; 22: 463–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ciavarra G, Zacksenhaus E. Rescue of myogenic defects in Rb‐deficient cells by inhibition of autophagy or by hypoxia‐induced glycolytic shift. J Cell Biol 2010; 191: 291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tracy K, Dibling BC, Spike BT et al BNIP3 is an RB/E2F target gene required for hypoxia‐induced autophagy. Mol Cell Biol 2007; 27: 6229–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lopez‐Bigas N, Kisiel TA, Dewaal DC et al Genome‐wide analysis of the H3K4 histone demethylase RBP2 reveals a transcriptional program controlling differentiation. Mol Cell 2008; 31: 520–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Blanchet E, Annicotte JS, Lagarrigue S et al E2F transcription factor‐1 regulates oxidative metabolism. Nat Cell Biol 2011; 13: 1146–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Macleod KF. The role of the RB tumour suppressor pathway in oxidative stress responses in the haematopoietic system. Nat Rev Cancer 2008; 8: 769–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Manning AL, Dyson NJ. pRB, a tumor suppressor with a stabilizing presence. Trends Cell Biol 2011; 21: 433–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bester AC, Roniger M, Oren YS et al Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell 2011; 145: 435–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sotillo R, Schvartzman JM, Socci ND et al Mad2‐induced chromosome instability leads to lung tumour relapse after oncogene withdrawal. Nature 2010; 464: 436–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang J, Benavente CA, McEvoy J et al A novel retinoblastoma therapy from genomic and epigenetic analyses. Nature 2012; 481: 329–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Calo E, Quintero‐Estades JA, Danielian PS et al Rb regulates fate choice and lineage commitment in vivo . Nature 2010; 466: 1110–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jiang Z, Deng T, Jones R et al Rb deletion in mouse mammary progenitors induces luminal‐B or basal‐like/EMT tumor subtypes depending on p53 status. J Clin Invest 2010; 120: 3296–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Calbo J, van Montfort E, Proost N et al A functional role for tumor cell heterogeneity in a mouse model of small cell lung cancer. Cancer Cell 2011; 19: 244–56. [DOI] [PubMed] [Google Scholar]
- 59. Wu L, de Bruin A, Saavedra HI et al Extra‐embryonic function of Rb is essential for embryonic development and viability. Nature 2003; 421: 942–7. [DOI] [PubMed] [Google Scholar]
- 60. Walkley CR, Shea JM, Sims NA et al Rb regulates interactions between hematopoietic stem cells and their bone marrow microenvironment. Cell 2007; 129: 1081–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ebel C, Mariconti L, Gruissem W. Plant retinoblastoma homologues control nuclear proliferation in the female gametophyte. Nature 2004; 429: 776–80. [DOI] [PubMed] [Google Scholar]
- 62. Wildwater M, Campilho A, Perez‐Perez JM et al The RETINOBLASTOMA‐RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell 2005; 123: 1337–49. [DOI] [PubMed] [Google Scholar]
- 63. Orford KW, Scadden DT. Deconstructing stem cell self‐renewal: genetic insights into cell‐cycle regulation. Nat Rev Genet 2008; 9: 115–28. [DOI] [PubMed] [Google Scholar]
- 64. Miura T, Luo Y, Khrebtukova I et al Monitoring early differentiation events in human embryonic stem cells by massively parallel signature sequencing and expressed sequence tag scan. Stem Cells Dev 2004; 13: 694–715. [DOI] [PubMed] [Google Scholar]
- 65. Conklin JF, Sage J. Keeping an eye on retinoblastoma control of human embryonic stem cells. J Cell Biochem 2009; 108: 1023–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hong H, Takahashi K, Ichisaka T et al Suppression of induced pluripotent stem cell generation by the p53‐p21 pathway. Nature 2009; 460: 1132–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lin T, Chao C, Saito S et al p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat Cell Biol 2005; 7: 165–71. [DOI] [PubMed] [Google Scholar]
- 68. Lin W, Cao J, Liu J et al Loss of the retinoblastoma binding protein 2 (RBP2) histone demethylase suppresses tumorigenesis in mice lacking Rb1 or Men1. Proc Natl Acad Sci USA 2011; 108: 13379–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Schieke SM, Ma M, Cao L et al Mitochondrial metabolism modulates differentiation and teratoma formation capacity in mouse embryonic stem cells. J Biol Chem 2008; 283: 28506–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ben‐Porath I, Thomson MW, Carey VJ et al An embryonic stem cell‐like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet 2008; 40: 499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kim J, Woo AJ, Chu J et al A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell 2010; 143: 313–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Liu Y, Clem B, Zuba‐Surma EK et al Mouse fibroblasts lacking RB1 function form spheres and undergo reprogramming to a cancer stem cell phenotype. Cell Stem Cell 2009; 4: 336–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Piestun D, Kochupurakkal BS, Jacob‐Hirsch J et al Nanog transforms NIH3T3 cells and targets cell‐type restricted genes. Biochem Biophys Res Commun 2006; 343: 279–85. [DOI] [PubMed] [Google Scholar]
- 74. Moon JH, Kwon S, Jun EK et al Nanog‐induced dedifferentiation of p53‐deficient mouse astrocytes into brain cancer stem‐like cells. Biochem Biophys Res Commun 2011; 412: 175–81. [DOI] [PubMed] [Google Scholar]
- 75. Bass AJ, Watanabe H, Mermel CH et al SOX2 is an amplified lineage‐survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet 2009; 41: 1238–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Jeter CR, Badeaux M, Choy G et al Functional evidence that the self‐renewal gene NANOG regulates human tumor development. Stem Cells 2009; 27: 993–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Sage C, Huang M, Karimi K et al Proliferation of functional hair cells in vivo in the absence of the retinoblastoma protein. Science 2005; 307: 1114–8. [DOI] [PubMed] [Google Scholar]
- 78. Mantela J, Jiang Z, Ylikoski J et al The retinoblastoma gene pathway regulates the postmitotic state of hair cells of the mouse inner ear. Development 2005; 132: 2377–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Pajcini KV, Corbel SY, Sage J et al Transient inactivation of Rb and ARF yields regenerative cells from postmitotic mammalian muscle. Cell Stem Cell 2010; 7: 198–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ajioka I, Martins RA, Bayazitov IT et al Differentiated horizontal interneurons clonally expand to form metastatic retinoblastoma in mice. Cell 2007; 131: 378–90. [DOI] [PMC free article] [PubMed] [Google Scholar]


