Abstract
The objective of this study was to examine the association between the immunohistochemical Ki67 labeling index (IHC Ki67), Ki67 mRNA expression level, and first‐generation gene signatures in a cohort of breast cancer patients. We assessed associations between IHC Ki67 and first‐generation gene signatures in a panel of 39 tumor samples, using an oligonucleotide microarray. Gene expression analyses included Ki67 alone (MKi67), 21‐gene signature, mitosis kinome score signature, and genomic grade index. Correlation coefficients were calculated by Spearman's rank correlation test. In all cases, IHC Ki67, MKi67, and three genetic markers were highly correlated (ρ, 0.71–0.97). Estrogen receptor (ER)‐positive cases showed strong correlations between IHC Ki67 and other signatures (ρ, 0.79–0.83). The ER‐negative cases showed slightly lower correlations (ρ, 0.58–0.73). In ER‐positive cases, the low IHC Ki67 group showed significantly longer relapse‐free survival than the high IHC Ki67 group (P = 0.007). This difference was confirmed by multivariate analysis. Our data indicate that IHC Ki67 shows similar predictive power for proliferation in ER‐positive cancers as genomic markers. Further study of IHC Ki67 is needed to define prognostic factors and predictive factors for chemotherapy using central laboratory assessment. (Cancer Sci, doi: 10.1111/j.1349‐7006.2012.02319.x, 2012)
Immunohistochemical assessment of Ki67 labeling index (IHC Ki67) has been described as a prognostic and predictive marker for breast cancer. However, Ki67 is not included in routine clinical decision‐making due to a lack of clarity regarding how Ki67 measurement and thresholds should influence clinical decisions.1
According to the recent St. Gallen consensus conference, the Ki67 labeling index is chiefly important in the distinction between ‘luminal A’ and ‘luminal B (HER2‐negative)’ subtypes, and Ki67 was described as a predictive marker for chemotherapy.2 However, guidelines of the American Society of Clinical Oncology do not include Ki67 in the list of required routine biological markers.3 Recent studies have addressed the use of IHC in breast cancer to include assessment of the proliferation marker Ki67, leading to more refined definitions of good and poor prognosis in estrogen receptor (ER)‐positive cancers.4, 5, 6
Several genetic prognostic markers have been described for breast cancer, and at least two of them (the 21‐gene recurrence score and the 70‐gene prognostic signature) are commercially available and increasingly used in clinical practice.7, 8, 9, 10, 11, 12 These markers were developed from and validated in a mixed cohort of patients, including both ER‐positive and ER‐negative cancers as well as low and high histological grade tumors.
However, use of genetic markers is 50 times more expensive than IHC Ki67 and histological grading. Assessment using IHC Ki67 and histological grading is also predictive for prognosis and sensitivity to chemotherapy, and perhaps to endocrine therapy. These features are also associated with the first‐generation genomic prognostic assays, which invariably include many genes that capture clinical phenotype information.13, 14
One of our co‐authors previously showed that genetic markers and Ki67 mRNA expression (mRNA Ki67) had similar predictive power for both prognosis and sensitivity to chemotherapy.15 However, few clinical datasets provide information on correlations between genomic markers, mRNA Ki67, and IHC Ki67 in the same cohort of patients with survival data.
We hypothesized that IHC Ki67 had similar predictive power for prognosis as some first‐generation gene signatures and mRNA Ki67 alone. The objective of this study was to explore associations between IHC Ki67, mRNA Ki67, and various first‐generation gene signatures in a cohort of patients who had long‐term follow‐up for survival.
Patients and Methods
Sample analyses
A total of 39 samples, including 39 invasive ductal carcinomas, were selected for study. None of the patients received preoperative adjuvant hormone therapy or chemotherapy. This study was approved by an institutional ethics committee, and all patients provided written informed consent. The samples showed the following IHC phenotypes: 6.9% ER(+)/HER2(+); 8.6% ER(−)/HER2(−); 14.8% ER(−)/HER2(+); and 69.7% ER(+)/HER2(−). Ten samples from each category were serially collected, snap frozen in liquid nitrogen, and stored at −80°C for microarray analysis. However, in this analysis, we excluded one ER(−) and HER2(+) ductal carcinoma in situ, because this type of carcinoma has a different tumor biology. The remaining samples were fixed in 10% formalin for 48 h, embedded in paraffin, and subjected to IHC analysis as described below.
Immunohistochemistry and pathological evaluation
Estrogen receptor expression was stained using an automated staining system (Benchmark; Ventana Japan, Yokohama, Japan), and HER2 expression was detected using HercepTest (DakoCytomation, Carpinteria, CA, USA) in accordance with the manufacturer's instructions, and the use of positive and negative controls. Antibodies used included anti‐Ki‐67 (clone MIB‐1; DakoCytomation). Both ER and progesterone receptor (PgR) were evaluated as positive when positive cells accounted for more than 10% in an evaluated area. The Ki67 labeling index was evaluated as a percentage of positive cells among 500 cancer cells. All samples were evaluated pathologically according to World Health Organization classification standards16 and the Scarff–Bloom–Richardson grading system.17
Microarray analysis
Details of the microarray analysis were described previously.18 Total RNA was isolated by phenol–chloroform extraction (Sepazol‐I; Wako Pure Chemical Industries, Osaka, Japan) from 100–200 mg fresh frozen tissue. Extracted RNA was reverse transcribed to cDNA using T7‐oligo‐dT primer (Qiagen, Valencia, CA, USA) and converted to double‐stranded DNA, which was used for synthesis of biotin‐labeled cRNA using a MEGAscript Kit (Ambion, Austin, TX, USA). The cRNA was fragmented and hybridized to oligonucleotide microarray chips (GeneChips U95Av2; Affymetrix, Santa Clara, CA, USA), which contained 12 558 genes. Probe arrays were stained with streptavidin–phycoerythrin (Molecular Probes, Eugene, OR, USA) and scanned. Signal intensities were captured using Affymetrix GeneChip Expression Analysis Software (LIMS 5.0) according to the supplier's standard procedures, and analyzed with Microsoft Excel (Redmond, WA, USA).
Complete gene expression data are available in the Gene Expression Omnibus under accession number GSE6367. Expression data were normalized using the MAS5 algorithm and log2 transformed. We assessed associations between IHC Ki67 and first‐generation gene signatures, including MKI67 (419_at), 21‐gene signature,9 mitosis kinome score signature (MKS),19 and genomic grade index (GGI).11 If one gene has two or more probe sets, we retained only one probe set with the highest average gene expression. For the 21‐gene signature we used only Ki67, STK15, Survivin, CCNB1, and MYBL2 out of the 21 genes; these genes are all related to proliferation. In order to simplify genomic markers with distinct complex algorisms and compare multiple genomic markers with IHC Ki67 levels, we calculated average gene expressions for the 21‐gene signature, MKS, and GGI based on algorism normalized MAS5 log2 converted mRNA gene expression data.
Statistical methods
To assess the relationship between IHC Ki67 and survival outcomes, survival curves were calculated by the Kaplan–Meier method and compared with the log–rank test according to IHC Ki67. Cases were divided into two groups (high and low IHC Ki67) based on the median value of IHC Ki67. Relapse‐free survival (RFS) was defined as the time from the diagnosis to relapse. Overall survival (OS) was defined as the time from diagnosis to death. Survival was right censored at 10 years.
To assess the prognostic power of IHC Ki67 and other clinicopathological covariates, we applied a proportional hazards model by Cox's regression analysis to estimate hazard ratios and 95% confidence intervals for RFS. In multivariate analysis, to identify a model that best fit the dataset, we adopted the Akaike information criterion stepwise variable selection method. Statistical analyses were carried out using BRB‐ArrayTools version 4.1.0 (http://linus.nci.nih.gov/BRB-ArrayTools.html) and R software version 2.7.2 (http://www.r-project.org). Two‐sided P‐values < 0.05 were considered statistically significant.
Results
Patients and treatments
Samples from 39 patients were analyzed in this study. Patient characteristics are shown in Table 1. Median age at diagnosis was 51 years. Treatments included adjuvant chemotherapy (32 patients) and endocrine therapy (21 patients).
Table 1.
Characteristics of breast cancer patients who participated in this study (n = 39; median age at diagnosis, 51 years)
| Covariates | Number of samples |
|---|---|
| T stage | |
| 1 | 17 |
| 2 | 13 |
| 3 | 8 |
| 4 | 1 |
| N stage | |
| 0 | 20 |
| 1 | 16 |
| 2 | 1 |
| 3 | 2 |
| Unknown | 1 |
| Estrogen receptor status | |
| Positive | 20 |
| Negative | 19 |
| HER2 status | |
| Positive | 19 |
| Negative | 20 |
| Histological grade | |
| I | 11 |
| II | 10 |
| III | 17 |
| Adjuvant chemotherapy | |
| Yes | 32 |
| No | 7 |
| Adjuvant endocrine therapy | |
| Yes | 21 |
| No | 18 |
Associations between IHC Ki67, mRNA Ki67, and various first‐generation gene signatures
We compared the five markers and tumor grade using the Wilcoxon test (Fig. 1). Across the five markers, there were consistently significant differences between grades 3 and 1 and between grades 3 and 2, whereas there was no significant difference between grades 1 and 2. This result indicated that the power of IHC to capture grade information is similar to that of genomic markers.
Figure 1.
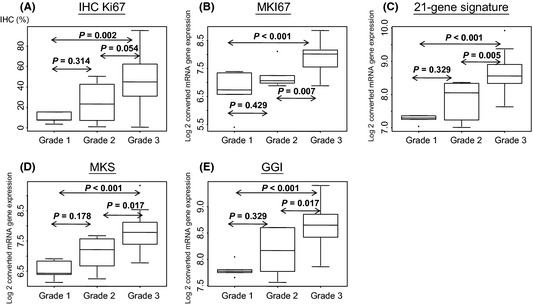
Boxplots with immunohistochemical (IHC) (using anti‐Ki‐670 clone, MIB‐1) and genomic markers by breast cancer grade (n = 39). P‐values were calculated by the Wilcoxon test. GGI, genomic grade index; MKi67, Ki67 alone; MKS, mitosis kinome score signature.
We also created a scatter plot matrix of the five markers and calculated correlation coefficients using Spearman's rank correlation test. When we plotted without any adjustment for ER status, IHC Ki67 and genomic markers were highly correlated (ρ, 0.71–0.78) (Fig. 2). All genomic markers were also highly correlated with each other (ρ, 0.87–0.97). The same analysis in ER‐positive cancers (n = 20) revealed similarly high correlations between IHC Ki67 and other genomic signatures (ρ, 0.77–0.83) (Fig. S1). In ER‐negative cancers (n = 19), correlation coefficients between IHC Ki67 and genomic markers were somewhat lower (ρ, 0.58–0.73) (Fig. S2). These results suggested that IHC Ki67 had predictive power for proliferation similar to that of genomic markers, and that MKI67‐mRNA gene expression level alone has similar predictive power to genomic markers with multiple genes. Moreover, IHC Ki67 information was more informative in ER‐positive cancers than in ER‐negative cancers, probably because all ER‐negative cases show similarly high proliferative rates.
Figure 2.

Scatter plot matrix for all breast tumor samples (n = 39). GGI, genomic grade index; IHC Ki67, immunohistochemical Ki67 labeling index; MKi67, Ki67 alone; MKS, mitosis kinome score signature.
Recurrence‐free and overall survival by IHC Ki67
We next analyzed the relationship between IHC Ki67 and survival. When we assessed RFS and OS without any adjustment based on ER status, there was no significant difference in RFS or OS between high Ki67 and low Ki67 (P = 0.445 and P = 0.622, respectively) (Fig. S3). The high Ki67 group showed a trend toward longer OS compared to low Ki67. However, assessment based on ER status revealed that in ER‐positive cases, high Ki67 was associated with significantly longer RFS (P = 0.007) (Fig. S4), whereas ER‐negative cases were unaffected by IHC Ki67 (P = 0.734) (Fig. S5). These results suggest that IHC Ki67 can serve as a prognostic marker in ER‐positive but not ER‐negative cases.
Table 2 shows the Cox proportional hazards model for 10‐year RFS. In univariate analysis, nodal status, histological grade, and type of surgery showed significant differences, and in multivariate analysis, progesterone receptor status, nodal status, type of surgery, and IHC Ki67 still had significant differences. This result further supports a role for IHC Ki67 as a prognostic marker in breast cancer patients.
Table 2.
Cox proportional hazards model for 10‐year recurrence‐free survival in breast cancer patients (n = 39)
| Clinicopathological markers | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| Hazard ratio | CI 95% | P‐value | Hazard ratio | CI 95% | P‐value | |
|
Estrogen receptor Positive (vs negative) |
0.948 | 0.34–2.62 | 0.917 | – | ||
|
Progesterone receptor Positive (vs negative) |
1.34 | 0.49–3.71 | 0.570 | 7.79 | 1.65–39.78 | 0.009 |
|
HER2 Negative (vs positive) |
1.57 | 0.56–4.42 | 0.392 | – | ||
|
Ta 3 + 4 (vs + 1 + 2) |
2.60 | 0.92–7.37 | 0.072 | 0.15 | 0.03–0.73 | 0.019 |
|
Na 1–3 (vs 0) |
6.06 | 1.70–21.67 | 0.006 | 26.42 | 4.53–154.12 | <0.001 |
|
Histological grade 3 (vs 1 + 2) |
7.36 | 0.97–56.08 | 0.054 | – | ||
|
Age High (vs low) |
0.68 | 0.25–1.88 | 0.459 | – | ||
|
Type of surgery Mastectomy (vs lumpectomy) |
3.32 | 1.05–10.47 | 0.040 | 7.95 | 1.77–35.77 | 0.007 |
|
Ki67b High (vs low) |
1.63 | 0.58–4.57 | 0.358 | 15.67 | 2.61–94.24 | 0.003 |
Clinical tumor size and nodal status according to TNM staging system.
Median value for Ki67 = 38.6%. Bold values indicate significance level.
CI, confidence interval; HER2, human epidermal growth factor receptor 2.
Discussion
Our data show that IHC Ki67 had similar predictive power for proliferation as genomic markers with multiple genes and using only one gene (Ki67). Furthermore, IHC Ki67 was more informative in ER‐positive cancers than in ER‐negative cancers.
Our data suggest that histological grade might still be a useful prognostic marker. Across the five markers, there were consistent significant differences between grades 3 and 1 and between grades 3 and 2. However, in stepwise multivariate analysis, grade was excluded and IHC Ki67 was retained, indicating that the two parameters have similar prognostic power. The other covariates, including PgR status, tumor size, nodal status, and type of surgery, may have distinct prognostic power independent of proliferative markers such as histological grade and IHC Ki67. Iwamoto et al.15 reported similar results indicating that clinical variables remained predictive of survival or chemotherapy response independent of genetic markers.
As a well‐established cell proliferation marker in breast cancer, IHC Ki67 is an excellent candidate biomarker for luminal B tumors in ER‐positive cases.2 Two meta‐analyses have shown a statistically significant association between high Ki67 expression and increased risk of breast cancer relapse and death.20, 21 However, lack of clarity regarding Ki67 measurement procedures and cut‐off points has hindered clinical application of IHC Ki67.1
Our data suggest that IHC Ki67 is correlated closely with first‐generation genomic markers. Other recent studies confirm the conclusion that IHC Ki67 has similar predictive power for proliferation as genomic markers. Cuzick and colleagues reported that four standard IHC assays (ER, PgR, HER2, and Ki67) carried out in a high quality laboratory can provide prognostic information similar to that provided by the 21‐gene signature, in endocrine‐treated ER‐positive breast cancer patients, using material from the ATAC trial.4 This approach has wide applicability and could extend the circumstances in which improved prognostic information is routinely available.4 Thus far, only IHC assays for ER, PgR, and HER2 have been widely adopted.
Williams and colleagues reported that assays using IHC markers of proliferation, such as Ki67, are easy and quick to carry out, relatively inexpensive, and correlate closely with the 21‐gene signature.22 However, further research using validated methods is necessary before widespread adoption23 in clinical laboratory settings. If validated, IHC Ki67 expression at $US30 per test represents an economical means of testing.23
In some cases, we obtained discordant results between IHC Ki67 and the 21‐gene score and other genomic markers. When multiple different genomic prognostic tests, IHC Ki67, and histological grade are used clinically and applied to the same case, discordant results can increase confusion in clinical decision making. When multiple predictors are applied to the same case, despite similar predictive performances, discordant risk predictions frequently occur. Currently, in such cases, we cannot identify the superior prognostic marker or predictor of sensitivity to chemotherapy.
Limitations of our study include small sample size and methods of collection of samples. We collected 10 samples from cases in four categories. Our study might include bias of collection. Also, genomic markers with multiple genes were assessed by average gene expression based on data normalized using the MAS5 algorithm, and log 2‐converted mRNA gene expression data, a departure from the original methods used in similar published reports.15
For determining prognosis and predicting sensitivity to chemotherapy, gene expression profiling (by, for example, the 21‐gene or 70‐gene signatures) remains the gold standard. Nevertheless, we suggest that IHC Ki67 could emerge as a cost‐effective alternative, defined and validated for worldwide clinical diagnostic use.
In conclusion, our data show that IHC Ki67 shows predictive power for proliferation similar to that of genomic markers in ER‐positive cancers. It is possible that IHC Ki67 could replace genetic markers for predicting prognosis and sensitivity to chemotherapy in the clinic. However, a lack of clarity regarding IHC Ki67 measurement and cut‐off points hinders clinical application. A prospective clinical trial of IHC Ki67 is needed to assess its prognostic value and ability to predict sensitivity to chemotherapy using central laboratory assessment.
Disclosure Statement
The authors have no conflicts of interest.
Supporting information
Fig. S1. Scatter plot matrix for estrogen receptor‐positive breast cancer cases (n = 20). GGI, genomic grade index; IHC Ki67, immunohistochemical Ki67 labeling index; MKi67, Ki67 alone; MKS, mitosis kinome score signature.
Fig. S2. Scatter plot matrix for estrogen receptor‐negative breast cancer cases (n = 19). GGI, genomic grade index; IHC Ki67, immunohistochemical Ki67 labeling index; MKi67, Ki67 alone; MKS, mitosis kinome score signature.
Fig. S3. Kaplan–Meier curves by immunohistochemical Ki67 labeling index for all breast cancer cases (n = 39).
Fig. S4. Kaplan–Meier curves by immunohistochemical Ki67 labeling index for estrogen receptor‐positive breast cancer cases (n = 20).
Fig. S5. Kaplan–Meier curves by immunohistochemical Ki67 labeling index for estrogen receptor‐negative breast cancer cases (n = 19).
Acknowledgments
We thank Mizuho Terada, Mayako Terao, Toru Morioka, and Risa Ohsitanai for compiling the clinical database, and Mitsuko Kobayashi for secretarial assistance. This research was supported in part by the MEXT‐ Supported Program for the Strategic Research Foundation at Private Universities, 2012–2016.
References
- 1. Dowsett M, Nielsen TO, A'Hern R et al Assessment of Ki67 in Breast Cancer: recommendations from the International Ki67 in Breast Cancer Working Group. JNCI: J Natl Cancer Inst 2011; 103: 1656–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Goldhirsch A, Wood WC, Coates AS et al Strategies for subtypes–dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol 2011; 22: 1736–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Harris L, Fritsche H, Mennel R et al American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol 2007; 25: 5287–312. [DOI] [PubMed] [Google Scholar]
- 4. Cuzick J, Dowsett M, Pineda S et al Prognostic value of a combined estrogen receptor, progesterone receptor, Ki‐67, and human epidermal growth factor receptor 2 immunohistochemical score and comparison with the genomic health recurrence score in early breast cancer. J Clin Oncol 2011; 29: 4273–8. [DOI] [PubMed] [Google Scholar]
- 5. Cheang MC, Chia SK, Voduc D et al Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst 2009; 101: 736–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hugh J, Hanson J, Cheang MCU et al Breast cancer subtypes and response to docetaxel in node‐positive breast cancer: use of an Immunohistochemical Definition in the BCIRG 001 Trial. J Clin Oncol 2009; 27: 1168–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sorlie T, Perou CM, Tibshirani R et al Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 2001; 98: 10869–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. van de Vijver MJ, He YD, van't Veer LJ et al A gene‐expression signature as a predictor of survival in breast cancer. N Engl J Med 2002; 347: 1999–2009. [DOI] [PubMed] [Google Scholar]
- 9. Paik S, Shak S, Tang G et al A multigene assay to predict recurrence of tamoxifen‐treated, node‐negative breast cancer. N Engl J Med 2004; 351: 2817–26. [DOI] [PubMed] [Google Scholar]
- 10. Wang Y, Klijn JG, Zhang Y et al Gene‐expression profiles to predict distant metastasis of lymph‐node‐negative primary breast cancer. Lancet 2005; 365: 671–9. [DOI] [PubMed] [Google Scholar]
- 11. Sotiriou C, Wirapati P, Loi S et al Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst 2006; 98: 262–72. [DOI] [PubMed] [Google Scholar]
- 12. Buyse M, Loi S, van't Veer L et al Validation and clinical utility of a 70‐gene prognostic signature for women with node‐negative breast cancer. J Natl Cancer Inst 2006; 98: 1183–92. [DOI] [PubMed] [Google Scholar]
- 13. Sotiriou C, Pusztai L. Gene‐expression signatures in breast cancer. N Engl J Med 2009; 360: 790–800. [DOI] [PubMed] [Google Scholar]
- 14. Wirapati P, Sotiriou C, Kunkel S et al Meta‐analysis of gene expression profiles in breast cancer: toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res 2008; 10: R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Iwamoto T, Lee JS, Bianchini G et al First generation prognostic gene signatures for breast cancer predict both survival and chemotherapy sensitivity and identify overlapping patient populations. Breast Cancer Res Treat 2011; 130: 155–64. [DOI] [PubMed] [Google Scholar]
- 16. The World Health Organization . Histological typing of breast tumors. Neoplasma 1983; 30: 113–23. [PubMed] [Google Scholar]
- 17. Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long‐term follow‐up. Histopathology 1991; 19: 403–10. [DOI] [PubMed] [Google Scholar]
- 18. Umemura S, Shirane M, Takekoshi S et al Overexpression of E2F‐5 correlates with a pathological basal phenotype and a worse clinical outcome. Br J Cancer 2009; 100: 764–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bianchini G, Iwamoto T, Qi Y et al Prognostic and therapeutic implications of distinct kinase expression patterns in different subtypes of breast cancer. Cancer Res 2010; 70: 8852–62. [DOI] [PubMed] [Google Scholar]
- 20. de Azambuja E, Cardoso F, de Castro G et al Ki‐67 as prognostic marker in early breast cancer: a meta‐analysis of published studies involving 12 155 patients. Br J Cancer 2007; 96: 1504–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stuart‐Harris R, Caldas C, Pinder SE, Pharoah P. Proliferation markers and survival in early breast cancer: a systematic review and meta‐analysis of 85 studies in 32,825 patients. Breast 2008; 17: 323–34. [DOI] [PubMed] [Google Scholar]
- 22. Williams DJ, Cohen C, Darrow M et al Proliferation (Ki‐67 and phosphohistone H3) and oncotype DX recurrence score in estrogen receptor‐positive breast cancer. Appl Immunohistochem Mol Morphol 2011; 19: 431–6. [DOI] [PubMed] [Google Scholar]
- 23. Yerushalmi R, Woods R, Ravdin PM et al Ki67 in breast cancer: prognostic and predictive potential. Lancet Oncol 2010; 11: 174–83. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Scatter plot matrix for estrogen receptor‐positive breast cancer cases (n = 20). GGI, genomic grade index; IHC Ki67, immunohistochemical Ki67 labeling index; MKi67, Ki67 alone; MKS, mitosis kinome score signature.
Fig. S2. Scatter plot matrix for estrogen receptor‐negative breast cancer cases (n = 19). GGI, genomic grade index; IHC Ki67, immunohistochemical Ki67 labeling index; MKi67, Ki67 alone; MKS, mitosis kinome score signature.
Fig. S3. Kaplan–Meier curves by immunohistochemical Ki67 labeling index for all breast cancer cases (n = 39).
Fig. S4. Kaplan–Meier curves by immunohistochemical Ki67 labeling index for estrogen receptor‐positive breast cancer cases (n = 20).
Fig. S5. Kaplan–Meier curves by immunohistochemical Ki67 labeling index for estrogen receptor‐negative breast cancer cases (n = 19).


