Abstract
Familial aggregation of hepatocellular carcinoma (HCC), the third leading cause of cancer death worldwide, has shown to be a common phenomenon. We investigated the association between the genetic background and HCC familial aggregation. Serum samples were collected from HCC family members and normal control family members for screening the differentially expressed protein peaks with the approach of surface‐enhanced laser desorption ionization time‐of‐flight mass spectrometry. Potential genetically associated protein peaks were selected and further identified by matrix assisted laser desorption ionization‐time of flight mass spectrometry. A panel of six protein peaks (m/z 6432.94, 8478.35, 9381.91, 17284.67, 17418.34, and 18111.04) were speculated to reflect the genetic susceptibility of HCC familial aggregation. Three of them (m/z 6432.94, 8478.35, and 9381.91) were selected to identify as the candidate proteins. Nine identified proteins, including mostly apolipoprotein family (ApoA1, ApoA2, ApoC3, ApoE) and serum amyloid A protein (SAA), were found overexpressed in the multiple HCC cases family members. The comparative proteomic profiles have suggested that genetic factors ought to be taken into account for familial aggregation of HCC.
Hepatocellular carcinoma (HCC) is one of the most commonly diagnosed cancers, and ranks the third cause of cancer related death in the world (9.2% of the total).1, 2 Among different geographical regions, large variation of morbidity and risk factors were observed, with the highest rate in Asian, and lowest rate in the US as well as Europe countries.3, 4 Hepatitis B virus (HBV) infection, hepatitis C virus (HCV) infection, and chronic exposure to aflatoxin play crucial roles in hepatocarcinogenesis among Asian people, while fatty liver disease and high consumption of alcohol are the dominant causes among low incidence regions.5, 6 The HCC incidence rate is remarkably high in China, accounting for approximately 55% of annual new cases in the world.7 Particularly in Guangxi province of China, an endemic area of HBV infection, the HCC mortality has risen as the first cause of malignant cancer‐relative death.8
It is well‐accepted that HBV infection constitutes the main etiological factor of HCC, while the process of hepatocarcinogenesis integrates multi‐stages and multi‐factors, including interactions of HBV, chemical carcinogen, and genetic susceptibility.9 In addiction, the literature conveys that the lifetime risk of developing HCC for HBV infected individuals has been estimated to be 40%,10 implying that a wide individual discrepancy exists in the HCC susceptibility, and that environmental factors alone are insufficient to fully address the familial aggregation of HCC.11, 12 We have observed that most HCC occurs obviously in clusters by geographical distribution or by family aggregation in Guangxi, China. Family aggregation of HCC has received researchers' attention for decades. Pedigree studies are frequently carried out to validate the underlying mechanisms of the HCC family aggregation. HBV infection accounts for the most likely cause involving the clustering phenomenon, and genetic factors should play a crucial role as well, since siblings of HCC subjects are significantly associated with liver cancer development even without HBV infection.13, 14, 15, 16
A great many HCC biomarkers have been reported by numerous independent relative studies in an effort to help effectively distinguish the onset of liver malignancy; however, most of them are conducted in HCC patients who have already progressed to advanced stage when the diagnosis is made.17, 18 To a certain extent, the clinical values of these biologically significant markers are not as beneficial for early HCC detection as expected, because serological proteins manifest closely related to pathophysiologic process.19 Researchers have emphasized the importance of detecting available and valuable markers for early prediction of preclinical high risk samples.18
The genetic effects upon the HCC family aggregation remain poorly investigated. We hypothesized that the potential genetic factors of the HCC family members might differ from those of the non‐HCC family members, which may promote the onset of HCC in an independent manner or coordinate with the carcinogenesis function of HBV. With the application of proteomic approaches, proteomic profiles could provide more comprehensive evidence than DNA arrays or mRNA, since the post‐translational modifications like phosphorylation, acetylation, and glycosylation are conferred at the same time.20
In the current study, we have for the first time developed the global differentially expressed protein profiles for individuals from the HCC families and non‐HCC families using comparative proteomic approches. For instance, surface‐enhanced laser desorption ionization time‐of‐flight mass spectrometry (SELDI‐TOF‐MS) was used to screen the differentially expressed low abundance proteins in serum, and we further identified the proteins using matrix assisted laser desorption ionization‐time of flight mass spectrometry (MALDI‐TOF‐MS). With the aim of elucidating the genetic mechanism of HCC development, this study attempted to remove the greatest HBV risk factor, which is recognized as a critical cause to familial effects of HCC.
Materials and Methods
Study population
Probands were 73 HCC patients pathologically diagnosed in the first affiliated hospital of Guangxi medical university in 2008–2009. All of the patients were native village residents in Guangxi. Briefly, four‐generation pedigree members with genetic connection to the HCC patients were recruited from 31 case families as studied subjects. Genetically unrelated family members such as spouses and in‐laws were excluded to avoid confounding factors.
The healthy control subjects were matched for age, gender, race, career, and alcohol use. They were recruited from families without cancer history in the same community coordinated with the case families.
Pedigree information and sample collection
Essential information was obtained through face‐to‐face interviews by trained investigators based on a well‐designed questionnaire including age, gender, race, occupation, family history, personal history, and relationship with the proband cases. Considering that the precise category may help to detect mild differences, we established four groups as follows: a multiple cases group for members from families with 2 or >2 HCC cases; a single case group for members from families with only one case; a HBV positive control group for individuals with HBV infection from HCC families; and a normal control group for members from non‐HCC families. Fifteen milliliters of peripheral blood of each participant was collected and stored at −80°C. The use of the blood for research purposes was approved by the patients and the hospital's ethical committee.
HBV, HCV serologic markers and liver function
Five HBV serologic markers (HBsAg, HBsAb, HBeAg, HBeAb, HBcAb) were determined by ELISA Kits (Xinchuang, Xiamen, China) following the manufacturer's instruction. Anti‐HCV was measured by enzyme immunoassay (Xinchuang). Liver function was assessed by automatic biochemical analyzer (TMS‐1024i; TOKYO BOEKI, Tokyo, Japan) with commercially available reagents.
SELDI‐TOF‐MS and data analysis
Serum samples were thawed on ice and we removed the high abundance proteins with acetonitrile at 4°C. Then the supernatant of each sample was centrifugated at 112g for 5 min, and mixed with U9 buffer (containing DTT) then vortexed vigorously for 30 min with ice‐incubation. Briefly, the proteins were denatured by weak cation exchange (WCX) (Beijing SED Science & Technology, Beijing, China). Meanwhile, the WCX2 chips were placed in a biochip bioprocessor. A total of 200 μL WCX2 buffer was added to each spot and vibrated twice, 5 min for each time. After the WCX2 buffer was discarded carefully, 100 μL of the denatured serum sample was added to the spot, and incubated for 1 h at 4°C. Unbound proteins were discarded. The washing steps with the WCX2 as described above were repeated twice. The proteinchip was washed with 200 μL HEPES. The chips were then taken out and dried in the air at room temperature. Finally, 0.5 μL sinapinic acid (SPA) was spotted onto the protein chip, then dried, and 0.5 μL SPA was added again. Following the air drying of the chips, the chip reading was performed with PBSII‐C type proteinchip reader and analyzed with Biomarker Wizard and Biomarker Patterns Software of Ciphergen Company (Fremont, CA, USA).
To minimize the quality bias (≤0.1%), the system was corrected with ALL‐IN‐ONE standard protein (Ciphergen). The key instrument parameters were adopted as follows: 50 kD for highest molecular weight, 2–20 kD for optimization range, 235 for laser intensity, nine for detection sensitivity, and 130 shots for every chip spot. Considering the baseline peak biases, the peaks <1 kD were filtered.
MALDI‐TOF‐MS and data analysis
Based on the differential protein peaks revealed by SELDI‐TOF‐MS, the potential genetically significant proteins were further identified using MALDI‐TOF‐MS, which incorporates features like enhanced sensitivity, accurate mass/charge measurement, larger affinity surface, and high through‐output.21, 22
Half of the samples were randomly selected from the multiple cases family group, the single case family group, and the non‐cancer family group, respectively. The samples of the HBV infective control group remained identical. These samples were submitted to MALDI‐TOF‐MS. The high abundance proteins were removed with acetonitrile at 4°C as mentioned above. Then the serum protein concentration was adjusted to 1.5 g/L. In brief, proteins were processed to separation procedure by SDS PAGE MES for about 3 h at a voltage of 100 V. Target gels were excised, decolorized, dehydrated, digested, and eventually scanned by MALDI‐TOF‐MS. The obtained peptide mass fingerprint spectrum and series were compared in NCBInr database using Mascot software (http://www.matrixscience.com/). Based on the definite molecular mass, we also searched SWISS‐PROT protein database to identify the candidate proteins. The function of the identified proteins was retrieved from NCBI protein databanks.
Statistical analysis
Ciphergen ProteinChip Software 3.2.0 was applied to normalize the profiling spectra of serum samples. Normalized protein expression intensities were comparisons between each studied group and the control group using t‐test. Following that the protein peaks were labeled by Biomarker Wizard software. The significant protein profiles were submitted to Biomarker Pattern Software (BPS) and evaluated using the classification model. Two tails P < 0.05 was considered statistically significant.
Results
Of the 73 probands, 60 were from multiple case families, and 13 were from single case families. The morbidity rate in multiple case families of males was significantly higher than that of females (19.75% vs 7.45%). The HCC incidences in the single case families were uniformly male. This study has recruited 355 case family members (208/147 for male/female), and 313 control family members (128/186 for male/female). All of the participants were submitted to the HBV and HCV screening. No anti‐HCV positive individual was found. To minimize the effect of HBV confounder, four groups were established as follows: (i) multiple cases family group; (ii) single case family group; (iii) HBV infective case family group; and (iv) normal control family group. Briefly, the multiple cases family group, single case family group, and normal control family group consisted of 100 HBsAg negative members from corresponding families, respectively. Importantly, 40 HBsAg positive members from case families constructed a HBV infective control group to restrict the environmental risk factors. The flow chart of recruitment was shown in Fig. 1. The essential information of the studied populations was listed in Table 1.
Figure 1.
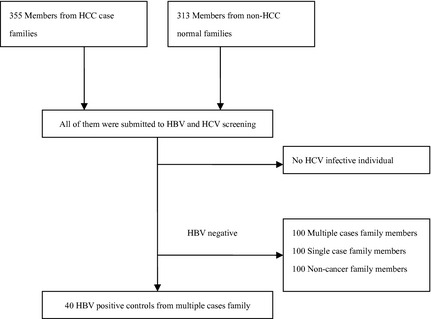
The flow chart of recruitment for studied individuals. HBV, hepatitis B virus; HCC, hepatocellular carcinoma.
Table 1.
Characteristic information of the participants
| Group | Age | HBsAg (+/−) | Anti‐HCV (+/−) | ALT (u/L) |
|---|---|---|---|---|
| Multiple cases family | 30.06 ± 19.19 | 51/192 | 0/243 | 21.81 ± 12.88 |
| Single case family | 34.33 ± 16.24 | 37/75 | 0/112 | 29.11 ± 18.10 |
| Non‐HCC family | 36.48 ± 18.91 | 48/265 | 0/313 | 24.37 ± 13.77 |
These data were obtained according to the original materials of each participant, before the further precise category. ALT, alanine aminotransferase; HBsAg, hepatitis B surface antigen; HCC, hepatocellular carcinoma.
SELDI WCX2 binding chips were applied to produce different serum protein profiles in terms of number and resolution of protein peaks. After intensity normalization by using Biomarker Wizard (version 3.2), proteomic data demonstrated up to 85 protein peaks between m/z 1 KD and m/z 45 KD among the four groups. Taking ion suppression effects into consideration, the discriminated peaks <2 KD were eliminated. Sixty discriminated peaks located between m/z 2 KD and m/z 20 KD, while 13 located between m/z 20 KD and m/z 45 KD.
Based on comparison with control family group, there were 21 significant differentially expressed protein peaks in the multiple case family group with 16 upregulated expression and five downregulated expression, 42 in the single case family group with 21 upregulated expression and 21 downregulated expression, and 50 in the HBV positive control group with 23 upregulated expression and 27 downregulated expression (only data of multiple cases group were presented in Table 2). The protein fingerprinting of four groups and electrophoretograms are presented in Fig. 2.
Table 2.
Differentially expressed proteins identified in multiple cases group†
| Molecular weight (m/z) | Expression tendency‡ | M ± SD§ | P‐value |
|---|---|---|---|
| 4356.84 | ↑ | 3.00 ± 2.61 | 0.000 |
| 7680.39 | ↑ | 0.45 ± 0.23 | 0.000 |
| 8581.35 | ↑ | 23.06 ± 8.46 | 0.000 |
| 8709.52 | ↑ | 30.11 ± 9.92 | 0.000 |
| 8399.43 | ↑ | 0.27 ± 0.21 | 0.001 |
| 3944.13 | ↓ | 1.49 ± 1.22 | 0.001 |
| 17418.34 | ↑ | 1.17 ± 0.95 | 0.002 |
| 13777.56 | ↑ | 1.47 ± 1.17 | 0.002 |
| 15357.71 | ↑ | 0.46 ± 0.43 | 0.003 |
| 3281.51 | ↑ | 0.14 ± 0.23 | 0.004 |
| 17284.67 | ↑ | 1.01 ± 0.76 | 0.012 |
| 15150.87 | ↑ | 0.34 ± 0.31 | 0.014 |
| 18111.04 | ↑ | 0.16 ± 0.10 | 0.016 |
| 8478.35 | ↑ | 4.02 ± 3.98 | 0.018 |
| 15211.97 | ↑ | 0.34 ± 0.25 | 0.022 |
| 6432.94 | ↑ | 10.46 ± 7.13 | 0.024 |
| 4098.68 | ↓ | 7.80 ± 4.91 | 0.027 |
| 7771.15 | ↓ | 1.15 ± 2.43 | 0.028 |
| 14072.17 | ↑ | 0.73 ± 0.53 | 0.029 |
| 4973.53 | ↓ | 1.09 ± 1.43 | 0.047 |
| 9381.91 | ↑ | 3.29 ± 2.01 | 0.050 |
†The result was gained from comparison with normal control group. ‡“↑” Represented upregulated expression, “↓” represented downregulated expression. §Expression intensity is represented by mean ± standard deviation.
Figure 2.
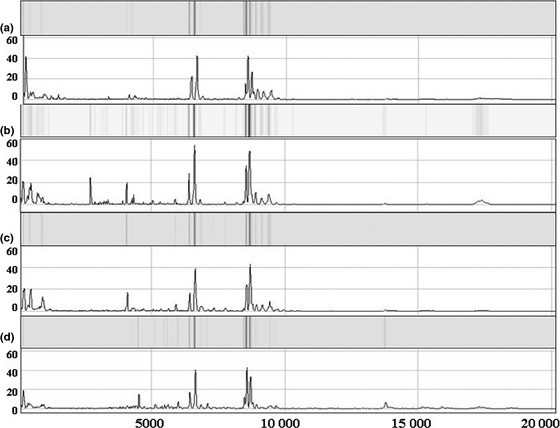
Protein fingerprinting and corresponding electrophoretograms of four groups (a represents single case group with chip303‐C; b represents multiple cases family with chip10‐D; c represents normal control group with chip322‐F; d represents HBV positive control group with chip 203‐D). The m/z spectrum ranges from 5000 to 20 000 Da.
Identification of the differentially expressed proteins associated with HCC
Given the reliability and stability of the results, the three protein peaks (m/z value 3281.51, 7771.15, and 4973.53) with standard deviation (SD) > mean value were missed from the further comparison. Five protein peaks (m/z value 3944.13, 8399.43, 15150.87, 15211.97, 15357.71), which were observed (SD) > mean in the single case family group or HBV control group, were also excluded from the study. Among the remaining 13 potential HCC associated discriminated peaks, six were simultaneously found in the single case group and the HBV control group (m/z value 4098.68, 4356.84, 8581.35, 8709.52, 9381.91, and 13777.56), five of which (m/z 4098.68, 4356.84, 8581.35, 8709.52, and 13777.56) showed identical tendency of up or downregulation. It implied that these five protein peaks may relate to HBV infection. In addition, m/z 9381.91 was only observed to be upregulated in the multiple cases group, but downregulated in the single case and HBV control groups, which suggested that m/z 9381.91 may be associated with HCC family aggregation. No significant difference was found for seven peaks (m/z 6432.94, 7680.39, 8478.35, 14072.17, 17284.67, 17418.34, and 18111.04) between the multiple cases group and single case group, when m/z 7680.39 was found upregulation in the HBV control group as well. Although m/z 6432.94 and 14072.17 were demonstrated to be differentially expressed in the HBV control group, the expressed level appeared to be in independent manners with upregulation in the multiple case family group, and downregulation in the HBV control group. Four differentially expressed protein peaks m/z 8478.35, 17284.67, 17418.34 and 18111.04 were observed exclusively upregulated in the multiple cases group.
A total of six protein peaks were speculated to relate to HBV infection. Alternatively, another six protein peaks were predominantly discriminated in the multiple case family group, and addressed as HCC genetically associated proteins (m/z value 6432.94, 8478.35, 9381.91, 17284.67, 17418.34, and 18111.04). The latter six were selected to construct the diagnostic model and yielded a sensitivity of 82% and specificity of 77% (Fig. 2 and Table 3). The corresponding receiver operating characteristics curve with an integral of 0.858 was presented (Fig. 3).
Table 3.
Candidate protein peaks associated with genetic factor for hepatocellular carcinoma (HCC) familial aggregation
| Molecular weight (m/z) | Group | Expression tendency† | M ± SD‡ | P‐value |
|---|---|---|---|---|
| 6432.94 | Normal control family | – | 8.30 ± 4.54 | – |
| Multiple cases family | ↑ | 10.46 ± 7.13 | 0.024 | |
| Single case family | – | 8.71 ± 4.27 | 0.54 | |
| HBV positive control | ↓ | 6.75 ± 2.95 | 0.027 | |
| 8478.35 | Normal control family | – | 2.87 ± 2.67 | – |
| Multiple cases family | ↑ | 4.02 ± 3.98 | 0.018 | |
| Single case family | – | 3.58 ± 3.14 | 0.122 | |
| HBV positive control | – | 3.24 ± 2.91 | 0.222 | |
| 9381.91 | Normal control family | – | 2.73 ± 1.56 | – |
| Multiple cases family | ↑ | 3.29 ± 2.01 | 0.050 | |
| Single case family | ↓ | 2.31 ± 1.80 | 0.005 | |
| HBV positive control | ↓ | 1.57 ± 0.75 | 0.000 | |
| 17284.67 | Normal control family | – | 0.74 ± 0.57 | – |
| Multiple cases family | ↑ | 1.01 ± 0.76 | 0.012 | |
| Single case family | – | 0.73 ± 0.51 | 0.893 | |
| HBV positive control | – | 0.77 ± 0.57 | 0.825 | |
| 17418.34 | Normal control family | – | 0.75 ± 0.50 | – |
| Multiple cases family | ↑ | 1.17 ± 0.95 | 0.002 | |
| Single case family | – | 0.88 ± 0.63 | 0.270 | |
| HBV positive control | – | 0.87 ± 0.57 | 0.268 | |
| 18111.04 | Normal control family | – | 0.13 ± 0.10 | – |
| Multiple cases family | ↑ | 0.16 ± 0.10 | 0.016 | |
| Single case family | – | 0.11 ± 0.09 | 0.207 | |
| HBV positive control | – | 0.11 ± 0.07 | 0.285 |
†“↑”represents upregulated expression, “↓” represents downregulated expression, “–” represents not detected. ‡Expression intensity is represented by mean ± standard deviation.
Figure 3.
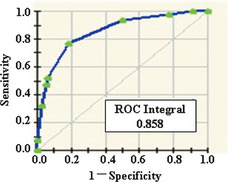
Corresponding receiver operating characteristics curve with an integral of 0.858.
To confirm the presence of proteins in gels when separated by SDS‐PAGE,23 three peaks with detectable high molecular masses (m/z 6432.94, 8478.35, and 9381.91) were selected for further identification using MALDI.
The bands at 6500, 8500, 9500 KD were further detected by mass spectrometry and identified as the candidate proteins listed in Table 4. Among them, cDNAFLJ53910 was most likely to be keratin, which was considered to be contaminated. LOC100133107 hypothetical protein was unable to be validated. Anti‐Mpl scFv possibly assumed to be proteolytic fragment from larger protein. The other nine identified proteins included mostly apolipoprotein families, while serum amyloid A protein was widely confirmed cancer biomarker in various malignancies.
Table 4.
Candidate proteins associated with hepatocellular carcinoma (HCC) familial aggregation
| Bands molecular weight† | SWISS‐PROT Serial number | Candidate protein | Theoretical MW (Da) | Possible function‡ |
|---|---|---|---|---|
| 6.5 KD | IPI00021854 | Apolipoprotein A‐II (APOA2) | 11167.9 | Signal transduction |
| IPI00657670 | Apolipoprotein C‐III variant 1 | 12807.5 | Lipid transport | |
| IPI00828105 | Anti‐Mpl scFv(fragment) | 25246.3 | – | |
| 8.5 KD | IPI00657670 | Apolipoprotein C‐III variant 1 | 12807.5 | Lipid transport |
| IPI00853525 | Apolipoprotein A1 (APOA1) | 27891.3 | Signal transduction | |
| IPI00828105 | Anti‐Mpl scFv (fragment) | 25246.3 | – | |
| IPI00871389 | LOC100133107 hypothetical protein | 22994.1 | Calcium ion binding | |
| 9.5 KD | IPI00657670 | Apolipoprotein C‐III variant 1 | 12807.5 | Lipid transport |
| IPI00021842 | APOE Apolipoprotein E | 36131.8 | Signal transduction | |
| IPI00552578 | Serum amyloid A protein (SAA) | 13523.5 | Signal transduction | |
| IPI00021854 | Apolipoprotein A‐II (APOA2) | 11167.9 | Signal transduction | |
| IPI00871389 | LOC100133107n hypothetical protein | 22994.1 | Calcium ion binding |
†The molecular weight of the excised band. ‡Possible function is retrieved from NCBI protein databanks.
Discussion
The HCC family aggregative phenomenon is not arbitrary, but raises the attention of researchers. HBV infection is responsible for the leading cause of HCC familial aggregation, while genetic factors ought to be taken into consideration. This work was firstly conducted for the HCC family members with proteomic techniques in an attempt to provide insights into the underlying genetic background of the HCC family members. We obtained significant differentially expressed protein peaks with 21 for the multiple cases family group, 42 for the single case family group, and 50 for the HBV positive control group. To some degree, the numbers could reflect the differences of protein backgrounds among the three studied groups.
In this study, the HBV positive rates of the HCC family members (incorporating the multiple case family and the single case family) appeared remarkably higher than the normal control family members (24.8% vs 15.3%, P = 0.002). This result has confirmed the prior statement that HBV infection plays a dominant role in the familial aggregation of HCC.9, 24
To better access the genetic factor involving the HCC susceptibility in the case family population, two proteomic approaches were combined for the analyses here. Different from those published proteomic studies that mainly concentrated on the comparison of the protein data for HCC patients and healthy controls,18, 25, 26 our study was performed for the healthy family members of the HCC case family and the control family by detecting the molecular differences.
Six differentially expressed protein peaks with m/z value at 6432.94, 8478.35, 9381.91, 17284.67, 17418.34, and 18111.04 were found to be potentially associated with familial clustering of HCC. Given the feasibility and possibility of detecting the HCC associated proteins in serum samples, three peaks (m/z 6432.94, 8478.35, and 9381.91) were put forward to protein identification in combination with MALDI. Eleven proteins were consequently validated, for two of which there were previously no data in terms of hypothetical proteins.
It was noteworthy that most identified proteins belonged to the apolipoprotein family. Both Apo A‐1 and Apo A‐2 have been reported to be differentially expressed in HCC patients, and treated as potential biomarkers.27, 28 ApoE was illustrated as overexpression in the multiple cases family member group, and this protein has already shown upregulated expression in various cancers such as liver,29, 30 brain,31 and ovary.32 Although the association between ApoE and carcinogenesis remains unclear, we suspected ApoE may function through modulating immune system like suppressing T cell proliferation, and regulating inflammation as well as oxidation.33 Most previous studies of ApoC3 focus on its association with hypertriglyceridemia and diabetes, while Qiu et al. has demonstrated its abnormal expression level in portal vein tumor thrombi in HCC.19, 34 All three differentially expressed protein peaks could isolate the ApoC3, which suggested the importance of further investigation about its relationship with cancer.
Another noteworthy protein in the low abundance fraction was serum amyloid A (SAA), which is known as an acute phase protein responsible for kinds of inflammatory stimuli and injuries. SAA expression level was already found elevated in HCC patients.35 Moreover, cancer studies of lung cancer,36 renal cell cancer,37 or ovarian carcinoma38 have implied the potential biomarker role of SAA. Obviously, it is not appropriate to use SAA as an exclusive biomarker for any specific cancer type, whereas it could coordinate with other markers for cancer surveillance.39
Although it is unclear whether the presence of these altered proteins plays either a causal or epiphenomenal role in the hepatocarcinogenesis, it has been suggested that there is a different genetic background of the HCC family members from those of the normal control families at the time when all of them are free from HBV infection. We propose that this novel evidence implies that the HCC family members were predisposed to severe liver injury if infected by HBV. Meanwhile this hypothesis was confirmed by our ongoing community‐based screening study.
Concerning the limitations of this study, the serum protein profile could be changed constantly in a manner coordinated with disease states; therefore, the results obtained from transverse analysis are dismal to address the development and progression of a disease. We suggest that a longitudinal regular survey of these identified proteins in an extended cohort is of great importance. Besides, the HBV positive and the normal control sample sizes were relatively small for the reason that control populations should be well‐matched with studied populations. Additional controls are needed. A study strength is that the studied and control individuals were native village residents recruited from a similar geographic distribution, which can minimize lifestyle confounders.
Our data suggest that the genetic factors ought to play a role in familial aggregation of HCC. Large case–control studies focused on the genetic effects upon the HCC familial aggregation are required. The effects and early diagnostic value of the identified proteins in our study remain a field to be explored in future.
Disclosure statement
The authors have no conflict of interest.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No. 30760219 and No. 30960170).
(Cancer Sci, doi: 10.1111/j.1349‐7006.2012.02368.x, 2012)
References
- 1. Ferlay J, Shin HR, Bray F et al Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010; 127: 2893–917. [DOI] [PubMed] [Google Scholar]
- 2. Gomaa AI, Khan SA, Toledano MB et al Hepatocellular carcinoma: epidemiology, risk factors and pathogenesis. World J Gastroenterol 2008; 14: 4300–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Venook AP, Papandreou C, Furuse J, de Guevara LL. The incidence and epidemiology of hepatocellular carcinoma: a global and regional perspective. Oncologist 2010; 15 (Suppl 4): 5–13. [DOI] [PubMed] [Google Scholar]
- 4. Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin 2005; 55: 74–108. [DOI] [PubMed] [Google Scholar]
- 5. Kim BK, Han KH, Ahn SH. Prevention of hepatocellular carcinoma in patients with chronic hepatitis B virus infection. Oncology 2011; 81 (Suppl 1): 41–9. [DOI] [PubMed] [Google Scholar]
- 6. McGlynn KA, London WT. The global epidemiology of hepatocellular carcinoma: present and future. Clin Liver Dis 2011; 15: 223–43, vii–x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schutte K, Bornschein J, Malfertheiner P. Hepatocellular carcinoma‐epidemiological trends and risk factors. Dig Dis 2009; 27: 80–92. [DOI] [PubMed] [Google Scholar]
- 8. Zhang CY, Huang TR, Yu JH et al Epidemiological analysis of primary liver cancer in the early 21st century in Guangxi province of China. Chin J Cancer 2010; 29: 545–50. [DOI] [PubMed] [Google Scholar]
- 9. Yu MW, Chang HC, Chen PJ et al Increased risk for hepatitis B‐related liver cirrhosis in relatives of patients with hepatocellular carcinoma in northern Taiwan. Int J Epidemiol 2002; 31: 1008–15. [DOI] [PubMed] [Google Scholar]
- 10. Beasley RP. Hepatitis B virus. The major etiology of hepatocellular carcinoma. Cancer 1988; 61: 1942–56. [DOI] [PubMed] [Google Scholar]
- 11. Dragani TA. Risk of HCC: genetic heterogeneity and complex genetics. J Hepatol 2010; 52: 252–7. [DOI] [PubMed] [Google Scholar]
- 12. Khoury MJ, Beaty TH, Liang KY. Can familial aggregation of disease be explained by familial aggregation of environmental risk factors? Am J Epidemiol 1988; 127: 674–83. [DOI] [PubMed] [Google Scholar]
- 13. Gong HM. [Family aggregation of hepatocarcinoma–preliminary analysis.] Zhonghua Zhong Liu Za Zhi 1985; 7: 408–10. (In Chinese.) [PubMed] [Google Scholar]
- 14. Yu MW, Chang HC, Liaw YF et al Familial risk of hepatocellular carcinoma among chronic hepatitis B carriers and their relatives. J Natl Cancer Inst 2000; 92: 1159–64. [DOI] [PubMed] [Google Scholar]
- 15. Hassan MM, Spitz MR, Thomas MB et al The association of family history of liver cancer with hepatocellular carcinoma: a case–control study in the United States. J Hepatol 2009; 50: 334–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gao Y, Jiang Q, Zhou X et al HBV infection and familial aggregation of liver cancer: an analysis of case–control family study. Cancer Causes Control 2004; 15: 845–50. [DOI] [PubMed] [Google Scholar]
- 17. Qiao B, Wang J, Xie J et al Detection and identification of peroxiredoxin 3 as a biomarker in hepatocellular carcinoma by a proteomic approach. Int J Mol Med 2012; 29: 832–40. [DOI] [PubMed] [Google Scholar]
- 18. Wu FX, Wang Q, Zhang ZM et al Identifying serological biomarkers of hepatocellular carcinoma using surface‐enhanced laser desorption/ionization‐time‐of‐flight mass spectroscopy. Cancer Lett 2009; 279: 163–70. [DOI] [PubMed] [Google Scholar]
- 19. Qiu JG, Fan J, Liu YK et al Screening and detection of portal vein tumor thrombi‐associated serum low molecular weight protein biomarkers in human hepatocellular carcinoma. J Cancer Res Clin Oncol 2008; 134: 299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pan J, Sun LC, Tao YF et al ATP synthase ecto‐alpha‐subunit: a novel therapeutic target for breast cancer. J Transl Med 2011; 9: 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gatlin‐Bunai CL, Cazares LH, Cooke WE et al Optimization of MALDI‐TOF MS detection for enhanced sensitivity of affinity‐captured proteins spanning a 100 kDa mass range. J Proteome Res 2007; 6: 4517–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Callesen AK, Christensen R, Madsen JS et al Reproducibility of serum protein profiling by systematic assessment using solid‐phase extraction and matrix‐assisted laser desorption/ionization mass spectrometry. Rapid Commun Mass Spectrom 2008; 22: 291–300. [DOI] [PubMed] [Google Scholar]
- 23. Tirumalai RS, Chan KC, Prieto DA et al Characterization of the low molecular weight human serum proteome. Mol Cell Proteomics 2003; 2: 1096–103. [DOI] [PubMed] [Google Scholar]
- 24. Tai DI, Changchien CS, Hung CS, Chen CJ. Replication of hepatitis B virus in first‐degree relatives of patients with hepatocellular carcinoma. Am J Trop Med Hyg 1999; 61: 716–9. [DOI] [PubMed] [Google Scholar]
- 25. Zinkin NT, Grall F, Bhaskar K et al Serum proteomics and biomarkers in hepatocellular carcinoma and chronic liver disease. Clin Cancer Res 2008; 14: 470–7. [DOI] [PubMed] [Google Scholar]
- 26. Junrong T, Huancheng Z, Feng H et al Proteomic identification of CIB1 as a potential diagnostic factor in hepatocellular carcinoma. J Biosci 2011; 36: 659–68. [DOI] [PubMed] [Google Scholar]
- 27. Pleguezuelo M, Lopez‐Sanchez LM, Rodriguez‐Ariza A et al Proteomic analysis for developing new biomarkers of hepatocellular carcinoma. World J Hepatol 2010; 2: 127–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jiang ZH, Zhang ZY, He M et al [The screening and identification of Apolipoprotein A‐II from serum differential proteins in hepatocellular carcinoma patients.] Zhonghua Gan Zang Bing Za Zhi 2010; 18: 445–9. (In Chinese.) [DOI] [PubMed] [Google Scholar]
- 29. Blanc JF, Lalanne C, Plomion C et al Proteomic analysis of differentially expressed proteins in hepatocellular carcinoma developed in patients with chronic viral hepatitis C. Proteomics 2005; 5: 3778–89. [DOI] [PubMed] [Google Scholar]
- 30. Yokoyama Y, Kuramitsu Y, Takashima M et al Protein level of apolipoprotein E increased in human hepatocellular carcinoma. Int J Oncol 2006; 28: 625–31. [PubMed] [Google Scholar]
- 31. Lichtor T. Gene expression of apolipoprotein E in human brain tumors. Neurosci Lett 1992; 138: 287–90. [DOI] [PubMed] [Google Scholar]
- 32. Hough CD, Sherman‐Baust CA, Pizer ES et al Large‐scale serial analysis of gene expression reveals genes differentially expressed in ovarian cancer. Cancer Res 2000; 60: 6281–7. [PubMed] [Google Scholar]
- 33. Zhang HL, Wu J, Zhu J. The immune‐modulatory role of apolipoprotein E with emphasis on multiple sclerosis and experimental autoimmune encephalomyelitis. Clin Dev Immunol 2010; 2010: doi: 10.1155/2010/186813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kozlitina J, Boerwinkle E, Cohen JC, Hobbs HH. Dissociation between APOC3 variants, hepatic triglyceride content and insulin resistance. Hepatology 2011; 53: 467–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. He QY, Zhu R, Lei T et al Toward the proteomic identification of biomarkers for the prediction of HBV related hepatocellular carcinoma. J Cell Biochem 2008; 103: 740–52. [DOI] [PubMed] [Google Scholar]
- 36. Sung HJ, Ahn JM, Yoon YH et al Identification and validation of SAA as a potential lung cancer biomarker and its involvement in metastatic pathogenesis of lung cancer. J Proteome Res 2011; 10: 1383–95. [DOI] [PubMed] [Google Scholar]
- 37. Vermaat JS, Gerritse FL, van der Veldt AA et al Validation of Serum Amyloid alpha as an independent biomarker for progression‐free and overall survival in metastatic renal cell cancer patients. Eur Urol 2012; doi: 10.1016/j.eururo.2012.01.020. [DOI] [PubMed] [Google Scholar]
- 38. Urieli‐Shoval S, Finci‐Yeheskel Z, Dishon S et al Expression of serum amyloid a in human ovarian epithelial tumors: implication for a role in ovarian tumorigenesis. J Histochem Cytochem 2010; 58: 1015–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li J, Xie Z, Shi L et al Purification, identification and profiling of serum amyloid A proteins from sera of advanced‐stage cancer patients. J Chromatogr B Analyt Technol Biomed Life Sci 2012; 890: 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]


