Abstract
Over the past several years, the success of genome‐wide association studies (GWAS) and pharmacogenomics has gradually begun to enable personalized medicine in some fields. In the field of liver diseases, host genetic factors are now very useful in clinical practice for predicting treatment outcome and adverse reactions for pegylated interferon plus ribavirin combination therapy against chronic hepatitis C virus (HCV) infection. Recently, three virus‐related hepatocellular carcinoma (HCC) GWAS were reported from Asia. One study examined hepatitis B virus‐related HCC in China, where hepatitis B is very prevalent, and the other two examined HCV‐related HCC in Japan. We identified a common variant in the DEPDC5 locus associated with HCV‐related HCC, and another group identified an association involving the MICA locus. In this review, we compare the results of these GWAS and earlier candidate gene studies. Further research is needed to determine the role of these single nucleotide polymorphisms on HCC risk, but identification of these markers could make it possible to assess the magnitude of the risk of cancer based on each patient's genetic background. Consideration of the genetic background of the patients will likely play a role in personalized medicine for HCC, and understanding the mechanism underlying the association could suggest novel promising therapeutic targets in the future. (Cancer Sci 2012; 103: 846–850)
Over the last several years, the success of GWAS and the International HapMap Project, a large‐scale database of SNPs, has identified genetic risk factors for more than 150 diseases, as well as genetic differences in drug response.1, 2, 3, 4 The success of these studies as well as pharmacogenomics has gradually begun to enable personalized medicine in some fields.5, 6, 7, 8 The goal of personalized medicine is to optimize the medical care and outcomes for each patient based on clinical, genetic, and environmental information.9 In the field of liver diseases, host genetic factors are now very useful in clinical practice for predicting treatment outcome and adverse reactions of PEG‐IFN‐α plus ribavirin combination therapy against chronic HCV infection,10, 11, 12, 13, 14, 15, 16, 17 which causes chronic hepatitis and HCC.
Epidemiology and Risk Factors of HCC
Hepatocellular carcinoma is the third leading cancer‐related cause of death and the seventh most common form of cancer worldwide.18 There are 750 000 new cases of HCC and nearly 700 000 deaths each year, making it a lethal form of cancer.18 A variety of risk factors for HCC have been reported, including hepatitis viruses, vinyl chloride, tobacco, aflatoxin B1, alcohol consumption, non‐alcoholic fatty liver disease, diabetes mellitus, obesity, diet, coffee, oral contraceptives, and hemochromatosis.19 Incidence of HCC varies around the world, largely reflecting the distribution of HBV and HCV. As HBV infection is highly prevalent in many Asian countries and in Africa, HBV is the most common etiology of HCC in these regions, whereas in many developed countries, including Japan, HCV infection is the most common risk factor for HCC.18, 19, 20, 21 Chronic hepatitis caused by HCV often leads to fibrosis and cirrhosis (stage F4 fibrosis), which markedly increase the risk of developing HCC.22 However, the incidence and progression of HCC varies by region, and only a fraction of HCV‐infected patients develop HCC. To date, many studies have examined patients with HCV and identified several predictive factors for HCC, including liver fibrosis, age, male gender, alcohol consumption, diabetes mellitus, obesity, ethnicity, and co‐infection with HBV.18, 23, 24, 25 In contrast to chronic HBV carriers, the influence of viral load and viral genotype on HCC is still controversial in chronic HCV carriers.26 In addition to these factors, multiple host genetic factors are thought to contribute to HCV‐related HCC development. Single nucleotide polymorphisms are the most common form of genomic variation, involving change at a single nucleotide in either coding or non‐coding DNA. The contribution of SNPs in the development of HCC has been investigated by various means. For decades, numerous studies have been undertaken using a candidate gene approach, in which candidate genes are selected prior to analysis on the basis of known functions thought to be relevant to disease risk, for example, inflammatory genes and oncogenes, and the corresponding genomic region is intensively screened for disease‐associated SNPs. For example, the association between HCV‐related HCC and SNPs in the region of the IL‐1beta, MDM2, and UGT1A7 genes have been reported from Japan and other countries.27, 28, 29, 30, 31, 32 It has been reported that these gene polymorphisms are also associated with HBV‐related HCC.33, 34, 35, 36 In addition, the influence of HFE and MnSOD gene polymorphisms on HCV‐related HCC has been reported from many countries, although not from East Asian countries.37, 38, 39 Gene polymorphisms associated with activity of hepatitis and liver fibrosis progression, which contribute to the development of HCC, have also been reported in HCV patients.40, 41 In spite of this effort, most studies had insufficient sample sizes, and the associations with HCV‐related HCC were not robust. Therefore, better predictive genetic markers are still needed.
Genome‐Wide Association Studies of HCV Treatment Response
Recently, methods for searching SNPs associated with diseases or drug responses have been changing dramatically. In contrast to the older candidate gene approach, the GWAS approach investigates not only the region around candidate genes with a known or predicted role in disease but across the entire genome using an SNP array, which simultaneously genotypes hundreds of thousands to millions of marker SNPs (also called tag SNPs). An SNP is often in strong linkage disequilibrium with multiple other SNPs in the same region, making it possible for tagging SNPs to serve as proxy markers for nearby SNPs that are not genotyped, and marker SNPs on genotyping platforms are selected to provide maximum coverage of the genome.42 Over the past few years, this new high‐throughput genotyping technology has revealed thousands of SNPs that are significantly associated with disease and drug responses, and this approach has been particularly promising in the field of liver diseases.
Anti‐HCV therapy is prescribed in many countries to prevent the progression of liver fibrosis and development of HCC.22, 43, 44 The current standard of care is PEG‐IFN plus ribavirin combination therapy, but this costly and poorly tolerated treatment leads to SVR in only 50% of patients with HCV genotype 1, which is the most prevalent genotype in many developed countries such as the USA, UK, France, Italy, Spain and Japan.45 To attempt to improve treatment efficacy, several viral and host factors responsible for SVR have been identified and studied extensively. Both HCV genotype and viral load are strong predictors of SVR.46 In HCV genotype 1b, amino acid substitutions at positions 70 and 91 of the HCV core protein and the presence of multiple substitutions in the interferon sensitivity determining region of the NS5A protein were also reported to affect treatment outcome, especially among Japanese patients.17, 47, 48 Host factors responsible for SVR include age, gender,15 degree of hepatic fibrosis,49 obesity, hepatic steatosis,50 low‐density lipoprotein cholesterol, gamma‐glutamyl transpeptidase,48 and insulin resistance.51 In addition, although the individual effects of genetic polymorphisms are typically small and of limited use for prediction, we recently identified an SNP in MAPKAPK3 that affects response to interferon therapy using a candidate gene approach.52 Using the GWAS approach, a series of studies independently revealed that a common polymorphism within the non‐coding region of the IL28 locus is strongly associated with both outcome of PEG‐IFN plus ribavirin therapy for chronic HCV infection10, 11, 12 as well as spontaneous clearance of the virus.53 Similarly, a polymorphism within the ITPA locus was found to strongly predict incidence of ribavirin‐induced anemia during therapy.13, 14 It is likely that future treatment regimens will involve screening for these and other SNPs in an effort to select the most promising treatment candidates, as well as to identify patients at risk for serious side‐effects. Direct‐acting antiviral agents, such as the protease inhibitors telaprevir and boceprevir, have recently become available, and in the near future triple therapy consisting of PEG‐IFN, ribavirin, and a protease inhibitor will likely become the standard of care.54, 55 In a recent clinical trial, we found that both IL28 and ITPA polymorphisms are also useful predictive factors for outcome and occurrence of side‐effects in triple therapy.56, 57
Genome‐Wide Association Studies of HCV‐Related HCC
The GWAS approach has also been used to identify HCV patients at greatest risk for developing HCC. The primary goal of antiviral therapy is to prevent development of HCC and advanced liver disease and improve prognosis of patients. Particularly among HCV and HBV patients who are unable to clear the virus, screening of additional SNPs associated with susceptibility to HCC may help improve prognosis and better target surveillance to high‐risk patients. As for HBV, which is the major cause of HCC in many Asian countries other than Japan, we identified variants in the HLA‐DP locus associated with persistent HBV infection in Japanese and Thai study groups using a GWAS approach,58 and this result was also confirmed in a Han Chinese patient group.59 Subsequently, in the first GWAS for HCC, Zhang et al.60 recently identified an SNP within the KIF1B locus associated with progression to HCC among chronic HBV carriers. However, it is known that the epidemiology is quite different between HBV‐related and HCV‐related HCC, and different virological effects of HBV and HCV have been reported.61, 62, 63 Hepatitis B infection alters pro‐apoptotic and DNA repair pathways, whereas HCV infection primarily affects anti‐apoptotic and inflammatory pathways.63 Two GWAS studies were reported very recently from Japan identifying genetic factors specific to HCV‐related HCC.64, 65 Kumar et al. identified the MICA locus associated with HCV‐related HCC, and we identified the DEPDC5 locus (Table 1).
Table 1.
Recently reported genome‐wide association studies of hepatocellular carcinoma (HCC)
| Etiology | Ethnicity | Characteristics | SNP | Chr. (locus) | Sample size | RAF | OR | 95% CI | P‐value | References | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case/control | Case | Control | Case | Control | ||||||||
| HBV | Chinese | Chronic HBV carriers with HCC/without HCC | rs17401966 | 1 (KIF1B) | 348 | 359 | 0.833 | 0.731 | 0.53 | 0.41–0.70 | 5.8 × 10−6 | 60 |
| HCV | Japanese | Chronic HCV carriers with HCC/non‐HCC controls | rs2596542 | 6 (MICA) | 721 | 2890 | 0.388 | 0.331 | 1.34 | 1.16–1.53 | 4.5 × 10−6 | 64 |
| HCV | Japanese | Chronic HCV carriers (age ≥ 55 years) with HCC/without HCC | rs1012068 | 22 (DEPDC5) | 212 | 765 | 0.189 | 0.095 | 2.20 | 1.64–2.97 | 8.0 × 10−8 | 65 |
Chr., chromosome; CI, confidence interval; HBV, hepatitis B virus; HCV, hepatitis C virus; OR, odds ratio; RAF, risk allele frequency; SNP, single nucleotide polymorphism.
Study design
A flowchart of our study is shown in Figure 1. To identify genetic markers associated with the risk of HCV‐related HCC development in the Japanese population, we carried out a two‐phase case–control study consisting of a GWAS and a replication study using a total of 3312 Japanese patients over the age of 55 with chronic HCV infection. An important point is that the controls used in this study were not healthy controls, but chronic HCV carriers who have the potential of developing HCC in the future. This choice of control helps to avoid confounding risk factors for developing HCV‐related HCC with risk factors for chronic HCV. Another important point is that we enrolled subjects over the age of 55 years (Fig. 2) because age at initial diagnosis of HCV‐related HCC has been increasing in Japan since the identification of HCV in 1989, and most patients are diagnosed at age 55 or older.23, 66, 67, 68 These two points represent major differences between the two Japanese GWAS studies of HCV‐related HCC, and we speculate that these differences partially explain their inconsistent results, even though both studies focus on Japanese patients (Table 1).
Figure 1.
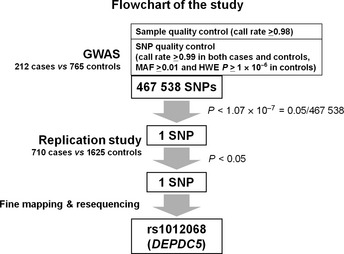
Flowchart of our two‐phase case–control study. For the genome‐wide association study (GWAS) stage, we used the Illumina HumanHap610‐Quad BeadChip. After we excluded two samples with call rate < 0.98, 467 538 single nucleotide polymorphisms (SNPs) passed the SNP quality control filters (call rate ≥ 0.99 in cases and controls, minor allele frequency [MAF] ≥0.01 and Hardy–Weinberg equilibrium [HWE] P‐value ≥ 1.0 × 10−6 in controls). Only one SNP, rs1012068, within the DEPDC5 gene reached statistical significance. We used multiplex‐PCR‐based Invader assays for the replication study and fine mapping. Finally, SNP rs1012068 had the strongest independent association with hepatitis C virus‐related hepatocellular carcinoma.65
Figure 2.
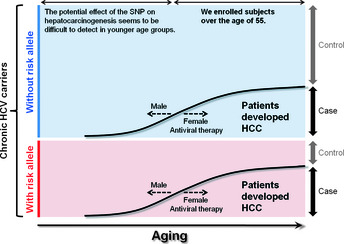
Scheme of our study design considering the age range for developing hepatocellular carcinoma (HCC). All subjects were Japanese patients with chronic hepatitis C virus (HCV) infection, therefore, the controls used in this study were not healthy controls but chronic HCV carriers.65 We enrolled subjects over the age of 55 years because most HCC patients are diagnosed at age 55 or older.23, 66, 67, 68 The potential effect of the SNP on hepatocarcinogenesis seems to be more difficult to detect in younger age groups, although males generally develop HCC at a younger age than females,23, 66, 67 and antiviral therapy may prevent development of HCC.22, 43, 44 SNP, single nucleotide polymorphism.
Results
We initially carried out a GWAS using the Illumina HumanHap610‐Quad BeadChip (Illumina, San Diego, CA, USA). After applying strict quality control filters, 467 538 autosomal SNPs remained and were analyzed using an additive model for genotype–phenotype association in 212 chronic HCV carriers with HCC (cases) and 765 chronic HCV carriers without HCC (controls). Principal component analysis revealed no population substructure in our study group, and the Cochran–Armitage trend test indicated a low probability of false‐positive associations resulting from population stratification. Only one intronic SNP, rs1012068, within the DEPDC5 locus on chromosome 22, showed a statistically significant association with HCC (P = 8.05 × 10−8) after Bonferroni correction for multiple testing (calculated as P < 0.05/467 538 = 1.07 × 10−7) with OR 2.20. To validate these results, we carried out a replication study using 710 cases and 1625 controls and confirmed the association between the SNP and HCC (P = 2.41 × 10−8, OR = 1.63). After adjusting for age, gender, and platelet count, which is known to correlate with the stage of liver fibrosis in HCV patients,22 the significance level of rs1012068 increased. However, there are many confounding factors in the analysis of HCC, so we cannot rule out the possibility that other confounding factors influenced the results. To investigate causative SNPs, we carried out fine mapping of the DEPDC5 locus including neighboring genes, and resequenced all 42 exons of the DEPDC5 gene, but found no SNP with a stronger association than rs1012068. In contrast to MICA, which has previously been proposed to have a functional association with HCC,69 DEPDC5 has not been reported in association with HCC, and its function remains unknown.70 Further functional analysis is needed to clarify which SNP is the true causative variant and to define the role of DEPDC5 on the susceptibility of HCV‐related HCC.
Limitations and future plans
An important limitation of our GWAS is the relatively small number of cases and the consequent lack of statistical power to detect other associations that are less robust, including rare variants and SNPs with weak effects. It remains to be determined whether other SNPs influence susceptibility to HCV‐related HCC in the Japanese population. For a process as complex as HCV‐related hepatocarcinogenesis, interactions among two or more SNPs as well as interactions with environmental factors should also be studied. In addition to SNPs, other types of genetic association, such as copy number variation, should be examined in the future. The question also remains whether the susceptibility loci within MICA and DEPDC5 are associated with HCV‐related HCC in other ethnic groups. Additional studies on other ethnic populations as well as stratification based on viral subgenotypes will provide more comprehensive information on the genetic etiology and heterogeneity of HCV‐related HCC.
Towards Personalized Medicine
In current clinical practice in Japan, patients with chronic hepatitis C are recommended for surveillance for progression of liver fibrosis and early detection of cancer.71, 72 The susceptibility SNPs are relatively weak markers, but in combination with other clinical predictors, SNP genotyping could constitute a useful addition to assess the magnitude of the risk of HCC (Fig. 3). Intervention using PEG‐IFN, ribavirin, and novel agents such as telaprevir54, 55, 56, 57 for reducing the risk for HCC22, 43, 44 is planned in the future, and some SNPs might provide information useful in deciding whether or not intervention should be carried out. Once HCC has developed, the most promising treatment is determined based on clinical practice guidelines that are mainly based on tumor stage as well as liver function.71, 72, 73, 74 For treating advanced HCC, various anticancer agents and new molecular‐targeted agents such as sorafenib have been advanced, but treatment outcome is still insufficient, and severe adverse drug reactions have occurred in some cases.75, 76, 77 Host genetic factors affecting drug responses have not yet been thoroughly studied, and recent research on HCC genomes have identified several previously uncharacterized mutation patterns.78, 79 Host as well as cancer genomes should be studied further, and both may bring about benefits to HCC treatment in the future.
Figure 3.
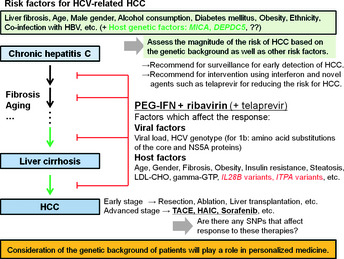
Suggested outline of management of hepatitis C virus (HCV)‐related hepatocellular carcinoma (HCC) incorporating genetic markers. Consideration of the genetic background of HCV patients will likely play a role in personalized medicine for HCV‐related HCC. HAIC, hepatic artery infusion chemotherapy; HBV, hepatitis B virus; gamma‐GTP, gamma‐glutamyl transpeptidase; LDL‐CHO, low‐density lipoprotein cholesterol; PEG‐IFN, pegylated interferon; SNP, single nucleotide polymorphism; TACE, transarterial chemoembolization.
Conclusion
In conclusion, consideration of the genetic background of HCV patients will likely play a role in personalized medicine for HCV‐related HCC, and understanding the mechanism underlying the association may suggest novel therapeutic targets.
Disclosure Statement
The authors have no conflicts of interest.
Abbreviations
- GWAS
genome‐wide association study
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- OR
odds ratio
- PEG‐IFN
pegylated interferon
- SNP
single nucleotide polymorphism
- SVR
sustained virological response
Acknowledgments
We thank the patients who agreed to participate in our study. We also thank the team members at Toranomon Hospital (Tokyo, Japan), Sapporo Kosei General Hospital (Hokkaido, Japan), Hiroshima University Hospital (Hiroshima, Japan), and the Hiroshima Liver Study Group for clinical sample collection. We thank T. Yokogi, Y. Hayashida, and K. Izumoto for technical assistance; J. Sakamiya for clerical assistance; and other members of the RIKEN Center for Genomic Medicine and Hiroshima University for assistance with various aspects of this study.
References
- 1. Ozaki K, Ohnishi Y, Iida A et al Functional SNPs in the lymphotoxin‐alpha gene that are associated with susceptibility to myocardial infarction. Nat Genet 2002; 32: 650–4. [DOI] [PubMed] [Google Scholar]
- 2. Tsunoda T, Lathrop GM, Sekine A et al Variation of gene‐based SNPs and linkage disequilibrium patterns in the human genome. Hum Mol Genet 2004; 13: 1623–32. [DOI] [PubMed] [Google Scholar]
- 3. Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest 2008; 118: 1590–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. International HapMap Consortium . A haplotype map of the human genome. Nature 2005; 437: 1299–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. International Warfarin Pharmacogenetics Consortium . Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med 2009; 360: 753–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hung SI, Chung WH, Jee SH et al Genetic susceptibility to carbamazepine‐induced cutaneous adverse drug reactions. Pharmacogenet Genomics 2006; 16: 297–306. [DOI] [PubMed] [Google Scholar]
- 7. Kiyotani K, Mushiroda T, Sasa M et al Impact of CYP2D6*10 on recurrence‐free survival in breast cancer patients receiving adjuvant tamoxifen therapy. Cancer Sci 2008; 99: 995–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chantarangsu S, Mushiroda T, Mahasirimongkol S et al Genome‐wide association study identifies variations in 6p21.3 associated with nevirapine‐induced rash. Clin Infect Dis 2011; 53: 341–8. [DOI] [PubMed] [Google Scholar]
- 9. Ginsburg GS, Willard HF. Genomic and personalized medicine: foundations and applications. Transl Res 2009; 154: 277–87. [DOI] [PubMed] [Google Scholar]
- 10. Ge D, Fellay J, Thompson AJ et al Genetic variation in IL28B predicts hepatitis C treatment‐induced viral clearance. Nature 2009; 461: 399–401. [DOI] [PubMed] [Google Scholar]
- 11. Suppiah V, Moldovan M, Ahlenstiel G et al IL28B is associated with response to chronic hepatitis C interferon‐alpha and ribavirin therapy. Nat Genet 2009; 41: 1100–4. [DOI] [PubMed] [Google Scholar]
- 12. Tanaka Y, Nishida N, Sugiyama M et al Genome‐wide association of IL28B with response to pegylated interferon‐alpha and ribavirin therapy for chronic hepatitis C. Nat Genet 2009; 41: 1105–9. [DOI] [PubMed] [Google Scholar]
- 13. Fellay J, Thompson AJ, Ge D et al ITPA gene variants protect against anaemia in patients treated for chronic hepatitis C. Nature 2010; 464: 405–8. [DOI] [PubMed] [Google Scholar]
- 14. Ochi H, Maekawa T, Abe H et al ITPA polymorphism affects ribavirin‐induced anemia and outcomes of therapy. Gastroenterology 2010; 139: 1190–7. [DOI] [PubMed] [Google Scholar]
- 15. Chayama K, Hayes CN, Yoshioka K et al Accumulation of refractory factors for pegylated interferon plus ribavirin therapy in older female patients with chronic hepatitis C. Hepatol Res 2010; 40: 1155–67. [DOI] [PubMed] [Google Scholar]
- 16. Afdhal NH, McHutchison JG, Zeuzem S et al Hepatitis C pharmacogenetics. Hepatology 2011; 53: 336–45. [DOI] [PubMed] [Google Scholar]
- 17. Hayes CN, Kobayashi M, Akuta N et al HCV substitutions and IL28B polymorphisms on outcome of peg‐interferon plus ribavirin combination therapy. Gut 2011; 60: 261–7. [DOI] [PubMed] [Google Scholar]
- 18. Yang JD, Roberts LR. Hepatocellular carcinoma: a global view. Nat Rev Gastroenterol Hepatol 2010; 7: 448–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Aravalli RN, Steer CJ, Cressman EN. Molecular mechanisms of hepatocellular carcinoma. Hepatology 2008; 48: 2047–63. [DOI] [PubMed] [Google Scholar]
- 20. Barrera JM, Bruguera M, Ercilla MG et al Persistent hepatitis C viremia after acute self‐limiting posttransfusion hepatitis C. Hepatology 1995; 21: 639–44. [PubMed] [Google Scholar]
- 21. Ikeda K, Saitoh S, Suzuki Y et al Disease progression and hepatocellular carcinogenesis in patients with chronic viral hepatitis. J Hepatol 1998; 28: 930–8. [DOI] [PubMed] [Google Scholar]
- 22. Yoshida H, Shiratori Y, Moriyama M et al Interferon therapy reduces the risk for hepatocellular carcinoma. Ann Intern Med 1999; 131: 174–81. [DOI] [PubMed] [Google Scholar]
- 23. Kiyosawa K, Umemura T, Ichijo T et al Hepatocellular carcinoma: recent trends in Japan. Gastroenterology 2004; 127: S17–26. [DOI] [PubMed] [Google Scholar]
- 24. Ohishi W, Fujiwara S, Cologne JB et al Risk factors for hepatocellular carcinoma in a Japanese population. Cancer Epidemiol Biomarkers Prev 2008; 17: 846–54. [DOI] [PubMed] [Google Scholar]
- 25. Yuen MF, Hou JL, Chutaputti A. Hepatocellular carcinoma in the Asia pacific region. J Gastroenterol Hepatol 2009; 24: 346–53. [DOI] [PubMed] [Google Scholar]
- 26. Yang HI, Yeh SH, Chen PJ et al Associations between hepatitis B virus genotype and mutants and the risk of hepatocellular carcinoma. J Natl Cancer Inst 2008; 100: 1134–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang Y, Kato N, Hoshida Y et al Interleukin‐1beta gene polymorphisms associated with hepatocellular carcinoma in hepatitis C virus infection. Hepatology 2003; 37: 65–71. [DOI] [PubMed] [Google Scholar]
- 28. Tanaka Y, Furuta T, Suzuki S et al Impact of interleukin‐1beta genetic polymorphisms on the development of hepatitis C virus‐related hepatocellular carcinoma in Japan. J Infect Dis 2003; 187: 1822–5. [DOI] [PubMed] [Google Scholar]
- 29. Sakamoto T, Higaki Y, Hara M et al Interaction between interleukin‐1beta ‐31T/C gene polymorphism and drinking and smoking habits on the risk of hepatocellular carcinoma among Japanese. Cancer Lett 2008; 271: 98–104. [DOI] [PubMed] [Google Scholar]
- 30. Okamoto K, Ishida C, Ikebuchi Y et al The genotypes of IL‐1 beta and MMP‐3 are associated with the prognosis of HCV‐related hepatocellular carcinoma. Intern Med 2010; 49: 887–95. [DOI] [PubMed] [Google Scholar]
- 31. Dharel N, Kato N, Muroyama R et al MDM2 promoter SNP309 is associated with the risk of hepatocellular carcinoma in patients with chronic hepatitis C. Clin Cancer Res 2006; 12: 4867–71. [DOI] [PubMed] [Google Scholar]
- 32. Wang Y, Kato N, Hoshida Y et al UDP‐glucuronosyltransferase 1A7 genetic polymorphisms are associated with hepatocellular carcinoma in Japanese patients with hepatitis C virus infection. Clin Cancer Res 2004; 10: 2441–6. [DOI] [PubMed] [Google Scholar]
- 33. Chen CC, Yang SY, Liu CJ et al Association of cytokine and DNA repair gene polymorphisms with hepatitis B‐related hepatocellular carcinoma. Int J Epidemiol 2005; 34: 1310–8. [DOI] [PubMed] [Google Scholar]
- 34. Migita K, Maeda Y, Abiru S et al Polymorphisms of interleukin‐1beta in Japanese patients with hepatitis B virus infection. J Hepatol 2007; 46: 381–6. [DOI] [PubMed] [Google Scholar]
- 35. Yoon YJ, Chang HY, Ahn SH et al MDM2 and p53 polymorphisms are associated with the development of hepatocellular carcinoma in patients with chronic hepatitis B virus infection. Carcinogenesis 2008; 29: 1192–6. [DOI] [PubMed] [Google Scholar]
- 36. Kong SY, Ki CS, Yoo BC, Kim JW. UGT1A7 haplotype is associated with an increased risk of hepatocellular carcinoma in hepatitis B carriers. Cancer Sci 2008; 99: 340–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ezzikouri S, El Feydi AE, El Kihal L et al Prevalence of common HFE and SERPINA1 mutations in patients with hepatocellular carcinoma in a Moroccan population. Arch Med Res 2008; 39: 236–41. [DOI] [PubMed] [Google Scholar]
- 38. Ezzikouri S, El Feydi AE, Chafik A et al Genetic polymorphism in the manganese superoxide dismutase gene is associated with an increased risk for hepatocellular carcinoma in HCV‐infected Moroccan patients. Mutat Res 2008; 649: 1–6. [DOI] [PubMed] [Google Scholar]
- 39. Nahon P, Sutton A, Pessayre D et al Manganese superoxide dismutase dimorphism and iron overload, hepatocellular carcinoma, and death in hepatitis C virus‐infected patients. Clin Gastroenterol Hepatol 2007; 5: 630–5. [DOI] [PubMed] [Google Scholar]
- 40. McIlroy D, Théodorou I, Ratziu V et al FAS promoter polymorphisms correlate with activity grade in hepatitis C patients. Eur J Gastroenterol Hepatol 2005; 17: 1081–8. [DOI] [PubMed] [Google Scholar]
- 41. Li CZ, Kato N, Chang JH et al Polymorphism of OAS‐1 determines liver fibrosis progression in hepatitis C by reduced ability to inhibit viral replication. Liver Int 2009; 29: 1413–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med 2010; 363: 166–76. [DOI] [PubMed] [Google Scholar]
- 43. Ikeda K, Saitoh S, Arase Y et al Effect of interferon therapy on hepatocellular carcinogenesis in patients with chronic hepatitis type C. Hepatology 1999; 29: 1124–30. [DOI] [PubMed] [Google Scholar]
- 44. Nishiguchi S, Kuroki T, Nakatani S et al Randomized trial of effects of interferon‐alpha on incidence of hepatocellular carcinoma in chronic active hepatitis C with cirrhosis. Lancet 1995; 346: 1051–5. [DOI] [PubMed] [Google Scholar]
- 45. Manns MP, McHutchison JG, Gordon SC et al Peginterferon alfa‐2b plus ribavirin compared with interferon alfa‐2b plus ribavirin for initial treatment of chronic hepatitis C. Lancet 2001; 358: 958–65. [DOI] [PubMed] [Google Scholar]
- 46. Ghany MG, Strader DB, Thomas DL, Seeff LB. Diagnosis, management, and treatment of Hepatitis C. Hepatology 2009; 49: 1335–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Enomoto N, Sakuma I, Asahina Y et al Comparison of full‐length sequences of interferon‐sensitive and resistant hepatitis C virus 1b. J Clin Invest 1995; 96: 224–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Akuta N, Suzuki F, Kawamura Y et al Predictive factors of early and sustained responses to peginterferon plus ribavirin combination therapy in Japanese patients infected with hepatitis C virus genotype 1b. J Hepatol 2007; 46: 403–10. [DOI] [PubMed] [Google Scholar]
- 49. Everson GT, Hoefs JC, Seeff LB et al Impact of disease severity on outcome of antiviral therapy for chronic hepatitis C. Hepatology 2006; 44: 1675–84. [DOI] [PubMed] [Google Scholar]
- 50. Dienstag JL, McHutchison JG. American Gastroenterological Association technical review on the management of hepatitis C. Gastroenterology 2006; 130: 231–64. [DOI] [PubMed] [Google Scholar]
- 51. Romero‐Gómez M, Del Mar Viloria M, Andrade RJ et al Insulin resistance impairs sustained response rate to peginterferon plus ribavirin in chronic hepatitis C patients. Gastroenterology 2005; 128: 636–41. [DOI] [PubMed] [Google Scholar]
- 52. Tsukada H, Ochi H, Maekawa T et al A polymorphism in MAPKAPK3 affects response to interferon therapy for chronic hepatitis C. Gastroenterology 2009; 136: 1796–805. [DOI] [PubMed] [Google Scholar]
- 53. Thomas DL, Thio CL, Martin MP et al Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature 2009; 461: 798–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kumada H, Toyota J, Okanoue T, Chayama K, Tsubouchi H, Hayashi N. Telaprevir with peginterferon and ribavirin for treatment‐naive patients chronically infected with HCV of genotype 1 in Japan. J Hepatol 2012; 56: 78–84. [DOI] [PubMed] [Google Scholar]
- 55. Chayama K, Takahashi S, Toyota J et al Dual therapy with the NS5A inhibitor BMS‐790052 and the NS3 protease inhibitor BMS‐650032 in HCV genotype 1b‐infected null responders. Hepatology 2012; 55: 742–8. [DOI] [PubMed] [Google Scholar]
- 56. Suzuki F, Suzuki Y, Akuta N et al Influence of ITPA polymorphisms on decreases of hemoglobin during treatment with pegylated interferon, ribavirin, and telaprevir. Hepatology 2011; 53: 415–21. [DOI] [PubMed] [Google Scholar]
- 57. Chayama K, Hayes CN, Abe H et al IL28B but not ITPA polymorphism is predictive of response to pegylated interferon, ribavirin, and telaprevir triple therapy in patients with genotype 1 hepatitis C. J Infect Dis 2011; 204: 84–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kamatani Y, Wattanapokayakit S, Ochi H et al A genome‐wide association study identifies variants in the HLA‐DP locus associated with chronic hepatitis B in Asians. Nat Genet 2009; 41: 591–5. [DOI] [PubMed] [Google Scholar]
- 59. Guo X, Zhang Y, Li J et al Strong influence of human leukocyte antigen (HLA)‐DP gene variants on development of persistent chronic hepatitis B virus carriers in the Han Chinese population. Hepatology 2011; 53: 422–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang H, Zhai Y, Hu Z et al Genome‐wide association study identifies 1p36.22 as a new susceptibility locus for hepatocellular carcinoma in chronic hepatitis B virus carriers. Nat Genet 2010; 42: 755–8. [DOI] [PubMed] [Google Scholar]
- 61. Tsuge M, Takahashi S, Hiraga N et al Effects of hepatitis B virus infection on the interferon response in immunodeficient human hepatocyte chimeric mice. J Infect Dis 2011; 204: 224–8. [DOI] [PubMed] [Google Scholar]
- 62. Tsuge M, Fujimoto Y, Hiraga N et al Hepatitis C virus infection suppresses the interferon response in the liver of the human hepatocyte chimeric mouse. PLoS ONE 2011; 6: e23856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ura S, Honda M, Yamashita T et al Differential microRNA expression between hepatitis B and hepatitis C leading disease progression to hepatocellular carcinoma. Hepatology 2009; 49: 1098–112. [DOI] [PubMed] [Google Scholar]
- 64. Kumar V, Kato N, Urabe Y et al Genome‐wide association study identifies a susceptibility locus for HCV‐induced hepatocellular carcinoma. Nat Genet 2011; 43: 455–8. [DOI] [PubMed] [Google Scholar]
- 65. Miki D, Ochi H, Hayes CN et al Variation in the DEPDC5 locus is associated with progression to hepatocellular carcinoma in chronic hepatitis C virus carriers. Nat Genet 2011; 43: 797–800. [DOI] [PubMed] [Google Scholar]
- 66. Ohishi W, Kitamoto M, Aikata H et al Impact of aging on the development of hepatocellular carcinoma in patients with hepatitis C virus infection in Japan. Scand J Gastroenterol 2003; 38: 894–900. [DOI] [PubMed] [Google Scholar]
- 67. Miki D, Aikata H, Uka K et al Clinicopathological features of elderly patients with hepatitis C virus‐related hepatocellular carcinoma. J Gastroenterol 2008; 43: 550–7. [DOI] [PubMed] [Google Scholar]
- 68. Taura N, Hamasaki K, Nakao K et al Aging of patients with hepatitis C virus‐associated hepatocellular carcinoma. Oncol Rep 2006; 16: 837–43. [PubMed] [Google Scholar]
- 69. Jinushi M, Takehara T, Tatsumi T et al Expression and role of MICA and MICB in human hepatocellular carcinomas and their regulation by retinoic acid. Int J Cancer 2003; 104: 354–61. [DOI] [PubMed] [Google Scholar]
- 70. Kharrat A, Millevoi S, Baraldi E, Ponting CP, Bork P, Pastore A. Conformational stability studies of the pleckstrin DEP domain. Biochim Biophys Acta 1998; 1385: 157–64. [DOI] [PubMed] [Google Scholar]
- 71. Arii S, Sata M, Sakamoto M et al Hepatol Res. Management of hepatocellular carcinoma. Hepatol Res 2010; 40: 667–85. [DOI] [PubMed] [Google Scholar]
- 72. Kudo M, Izumi N, Kokudo N et al Management of hepatocellular carcinoma in Japan. Dig Dis 2011; 29: 339–64. [DOI] [PubMed] [Google Scholar]
- 73. Bruix J, Sherman M, Llovet JM et al Clinical management of hepatocellular carcinoma. J Hepatol 2001; 35: 421–30. [DOI] [PubMed] [Google Scholar]
- 74. Benson AB 3rd, Abrams TA, Ben‐Josef E et al NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw 2009; 7: 350–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sakon M, Nagano H, Dono K et al Combined intraarterial 5‐fluorouracil and subcutaneous interferon‐alpha therapy for advanced hepatocellular carcinoma with tumor thrombi in the major portal branches. Cancer 2002; 94: 435–42. [DOI] [PubMed] [Google Scholar]
- 76. Kawaoka T, Aikata H, Katamura Y et al Hypersensitivity reactions to transcatheter chemoembolization with cisplatin and Lipiodol suspension for unresectable hepatocellular carcinoma. J Vasc Interv Radiol 2010; 21: 1219–25. [DOI] [PubMed] [Google Scholar]
- 77. Villanueva A, Llovet JM. Targeted therapies for hepatocellular carcinoma. Gastroenterology 2011; 140: 1410–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Totoki Y, Tatsuno K, Yamamoto S et al High‐resolution characterization of a hepatocellular carcinoma genome. Nat Genet 2011; 43: 464–9. [DOI] [PubMed] [Google Scholar]
- 79. Li M, Zhao H, Zhang X et al Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat Genet 2011; 43: 828–9. [DOI] [PMC free article] [PubMed] [Google Scholar]


